microRNA information: hsa-let-7b-3p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-let-7b-3p | miRbase |
| Accession: | MIMAT0004482 | miRbase |
| Precursor name: | hsa-let-7b | miRbase |
| Precursor accession: | MI0000063 | miRbase |
| Symbol: | MIRLET7B | HGNC |
| RefSeq ID: | NR_029479 | GenBank |
| Sequence: | CUAUACAACCUACUGCCUUCCC |
Reported expression in cancers: hsa-let-7b-3p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-let-7b-3p | acute myeloid leukemia | downregulation | "Among them miR-128a and -128b are significantly ov ......" | 18056805 | qPCR |
| hsa-let-7b-3p | breast cancer | downregulation | "Previously we found that let-7 miRNAs were downreg ......" | 21826373 | |
| hsa-let-7b-3p | breast cancer | downregulation | "The heterochronic microRNA let 7 inhibits cell mot ......" | 23339187 | |
| hsa-let-7b-3p | breast cancer | downregulation | "Breast cancer specific TRAIL expression mediated b ......" | 25150312 | qPCR |
| hsa-let-7b-3p | colon cancer | downregulation | "Recently let-7 miRNAs were found to regulate human ......" | 16651716 | |
| hsa-let-7b-3p | colorectal cancer | upregulation | "Of the 99 mCRC patients we studied differential mi ......" | 23098991 | Microarray |
| hsa-let-7b-3p | colorectal cancer | upregulation | "High expression of let-7 g was associated with a g ......" | 23932154 | |
| hsa-let-7b-3p | head and neck cancer | deregulation | "Significance was determined using significance ana ......" | 18798260 | Microarray |
| hsa-let-7b-3p | liver cancer | downregulation | "In present study we found that the tumor suppresso ......" | 21814544 | |
| hsa-let-7b-3p | liver cancer | downregulation | "Downregulation of let-7 miRNA in many human cancer ......" | 22289550 | |
| hsa-let-7b-3p | liver cancer | downregulation | "Let-7 family microRNAs have been reported to be do ......" | 24724096 | |
| hsa-let-7b-3p | lung cancer | downregulation | "Reduced expression of the let 7 microRNAs in human ......" | 15172979 | |
| hsa-let-7b-3p | lung cancer | downregulation | "Suppression of non small cell lung tumor developme ......" | 18308936 | |
| hsa-let-7b-3p | lung cancer | downregulation | "Indeed some transcripts of the let-7 family that a ......" | 19275611 | |
| hsa-let-7b-3p | lung cancer | downregulation | "Down-regulation of let-7 microRNA miRNA plays an i ......" | 20033209 | |
| hsa-let-7b-3p | lung cancer | downregulation | "Most miRs including all members of the let-7 famil ......" | 20068076 | qPCR |
| hsa-let-7b-3p | lung cancer | downregulation | "let 7b and miR 126 are down regulated in tumor tis ......" | 23029111 | qPCR |
| hsa-let-7b-3p | melanoma | downregulation | "A microRNA expression screen was performed analyzi ......" | 18379589 | qPCR |
| hsa-let-7b-3p | ovarian cancer | downregulation | "Using microRNA microarrays we identify several miR ......" | 18560586 | Microarray |
| hsa-let-7b-3p | ovarian cancer | downregulation | "The aim of this study was to find specific profile ......" | 23542579 | Microarray; qPCR |
| hsa-let-7b-3p | pancreatic cancer | upregulation | "The rapamycin effects centered on decreased mTOR-r ......" | 25576058 | |
| hsa-let-7b-3p | prostate cancer | downregulation | "Previous work has shown reduced expression levels ......" | 20418948 | |
| hsa-let-7b-3p | prostate cancer | downregulation | "Multiple tumor-suppressive miRNAs were downregulat ......" | 22719071 | |
| hsa-let-7b-3p | sarcoma | downregulation | "We detected decreased expression levels of some le ......" | 21412931 |
Reported cancer pathway affected by hsa-let-7b-3p

| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-let-7b-3p | breast cancer | Wnt signaling pathway; Apoptosis pathway; Notch signaling pathway | "The miRNAs and their clusters such as the miR-200 ......" | 26712794 | |
| hsa-let-7b-3p | breast cancer | Apoptosis pathway | "The tumor-suppressive let-7 family of microRNAs mi ......" | 27574028 | Western blot; Luciferase |
| hsa-let-7b-3p | esophageal cancer | Epithelial mesenchymal transition pathway | "HMGA2 is down regulated by microRNA let 7 and asso ......" | 24612219 | |
| hsa-let-7b-3p | head and neck cancer | Epithelial mesenchymal transition pathway | "The tumour suppressor roles of the let-7 family th ......" | 23340306 | |
| hsa-let-7b-3p | liver cancer | Apoptosis pathway | "The let 7 family of microRNAs inhibits Bcl xL expr ......" | 20347499 | Western blot |
| hsa-let-7b-3p | lung cancer | cell cycle pathway | "Reduced miR 31 and let 7 maintain the balance betw ......" | 22301433 | |
| hsa-let-7b-3p | lung cancer | Epithelial mesenchymal transition pathway | "Specifically let-7 and miR-9 are deregulated in bo ......" | 23365639 | |
| hsa-let-7b-3p | lung cancer | Apoptosis pathway | "Specifically following HDAC inhibition MYC which n ......" | 26915294 | |
| hsa-let-7b-3p | lung squamous cell cancer | cell cycle pathway | "Flow cytometry analysis revealed these miRNAs indu ......" | 19688090 | Flow cytometry |
| hsa-let-7b-3p | melanoma | cell cycle pathway | "MicroRNA let 7b targets important cell cycle molec ......" | 18379589 | |
| hsa-let-7b-3p | melanoma | Epithelial mesenchymal transition pathway | "An exploratory miRNA analysis of 666 miRs by low d ......" | 20302635 | |
| hsa-let-7b-3p | melanoma | Apoptosis pathway | "Let 7b and microRNA 199a inhibit the proliferation ......" | 23162627 | |
| hsa-let-7b-3p | pancreatic cancer | cell cycle pathway | "The rapamycin effects centered on decreased mTOR-r ......" | 25576058 | |
| hsa-let-7b-3p | pancreatic cancer | cell cycle pathway | "LIN28B suppresses microRNA let 7b expression to pr ......" | 26609473 | Flow cytometry; RNAi |
| hsa-let-7b-3p | prostate cancer | cell cycle pathway | "Distinct microRNA expression profiles in prostate ......" | 22719071 | |
| hsa-let-7b-3p | retinoblastoma | cell cycle pathway | "In addition hsa-miR-25 hsa-miR-18a and hsa-miR-20a ......" | 26730174 | |
| hsa-let-7b-3p | sarcoma | Epithelial mesenchymal transition pathway | "Using miRNA microarrays capable of detecting a kno ......" | 24457375 | Cell proliferation assay; Cell Proliferation Assay |
Reported cancer prognosis affected by hsa-let-7b-3p

| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-let-7b-3p | acute myeloid leukemia | differentiation | "We studied miRNA expression of leukemic blasts of ......" | 20425795 | |
| hsa-let-7b-3p | bladder cancer | poor survival | "A let 7 microRNA binding site polymorphism in the ......" | 27620744 | |
| hsa-let-7b-3p | breast cancer | tumorigenesis; staging; malignant trasformation | "Let 7 family miRNAs regulate estrogen receptor alp ......" | 20535543 | Luciferase |
| hsa-let-7b-3p | breast cancer | metastasis | "To identify microRNAs miRNA involved in metastasis ......" | 21057539 | |
| hsa-let-7b-3p | breast cancer | drug resistance | "let 7 microRNAs induce tamoxifen sensitivity by do ......" | 21826373 | Luciferase |
| hsa-let-7b-3p | breast cancer | tumorigenesis | "LIN28: a regulator of tumor suppressing activity o ......" | 22081076 | |
| hsa-let-7b-3p | breast cancer | motility; metastasis; cell migration | "The heterochronic microRNA let 7 inhibits cell mot ......" | 23339187 | |
| hsa-let-7b-3p | breast cancer | malignant trasformation | "Here we identified 33 miRNAs with similar deregula ......" | 24104550 | |
| hsa-let-7b-3p | breast cancer | drug resistance | "Breast cancer specific TRAIL expression mediated b ......" | 25150312 | Luciferase |
| hsa-let-7b-3p | breast cancer | metastasis; motility | "Among these we identified miRNAs previously associ ......" | 25277099 | |
| hsa-let-7b-3p | breast cancer | recurrence | "Multiple Let-7 family members were negatively corr ......" | 26717565 | |
| hsa-let-7b-3p | breast cancer | drug resistance | "Specifically we will discuss key miRNAs involved i ......" | 27721895 | |
| hsa-let-7b-3p | colon cancer | staging | "Association study of the let 7 miRNA complementary ......" | 24727325 | |
| hsa-let-7b-3p | colon cancer | metastasis | "Nine dysregulated miRNA pairs fell into three miRN ......" | 25287248 | |
| hsa-let-7b-3p | colorectal cancer | poor survival | "Genetic modulation of the Let 7 microRNA binding t ......" | 20177422 | |
| hsa-let-7b-3p | colorectal cancer | drug resistance; poor survival | "A let 7 microRNA binding site polymorphism in 3' u ......" | 20603437 | |
| hsa-let-7b-3p | colorectal cancer | drug resistance | "We investigated this issue by profiling the expres ......" | 20881268 | |
| hsa-let-7b-3p | colorectal cancer | staging; poor survival | "A let 7 microRNA SNP in the KRAS 3'UTR is prognost ......" | 21994416 | |
| hsa-let-7b-3p | colorectal cancer | metastasis | "We investigated expression of the reported metasta ......" | 22120473 | |
| hsa-let-7b-3p | colorectal cancer | drug resistance | "The LCS6 polymorphism in the binding site of let 7 ......" | 23324806 | |
| hsa-let-7b-3p | colorectal cancer | worse prognosis | "High expression of let-7 g was associated with a g ......" | 23932154 | |
| hsa-let-7b-3p | colorectal cancer | metastasis | "We investigated the expression of metastasis-assoc ......" | 24653631 | |
| hsa-let-7b-3p | colorectal cancer | drug resistance | "We identified a miRNA profile that was analysed by ......" | 25197016 | |
| hsa-let-7b-3p | colorectal cancer | staging | "We performed miRNA profiling of 1547 human miRNAs ......" | 26045793 | |
| hsa-let-7b-3p | colorectal cancer | progression | "rs712 polymorphism within let 7 microRNA binding s ......" | 26543374 | |
| hsa-let-7b-3p | colorectal cancer | progression | "Of these 3 upregulated let-7b miR-1290 and miR-126 ......" | 27698796 | |
| hsa-let-7b-3p | colorectal cancer | poor survival; immune evasion; immune resistance; worse prognosis | "MicroRNA let 7 T cells and patient survival in col ......" | 27737877 | |
| hsa-let-7b-3p | esophageal cancer | metastasis; tumorigenesis | "Role of microRNA let 7 and effect to HMGA2 in esop ......" | 21598109 | Western blot |
| hsa-let-7b-3p | gastric cancer | metastasis; tumorigenesis | "Let 7 microRNA family is selectively secreted into ......" | 20949044 | Western blot |
| hsa-let-7b-3p | gastric cancer | metastasis; tumorigenesis | "MicroRNA let 7b suppresses human gastric cancer ma ......" | 25613480 | Luciferase; Western blot |
| hsa-let-7b-3p | head and neck cancer | metastasis; poor survival | "Let-7 has been shown to function as a tumour suppr ......" | 21292542 | |
| hsa-let-7b-3p | liver cancer | motility; drug resistance | "We evaluated the expression of microRNA in human H ......" | 21352471 | |
| hsa-let-7b-3p | liver cancer | tumorigenesis | "In the present study we analyzed mCRP expression i ......" | 24297460 | Western blot |
| hsa-let-7b-3p | lung cancer | poor survival | "Reduced expression of the let 7 microRNAs in human ......" | 15172979 | |
| hsa-let-7b-3p | lung cancer | differentiation | "Reduced miR 31 and let 7 maintain the balance betw ......" | 22301433 | |
| hsa-let-7b-3p | lung cancer | poor survival | "let 7b and miR 126 are down regulated in tumor tis ......" | 23029111 | |
| hsa-let-7b-3p | lung cancer | drug resistance | "For better understanding of the cellular response ......" | 25451482 | |
| hsa-let-7b-3p | lung cancer | poor survival | "Specifically following HDAC inhibition MYC which n ......" | 26915294 | |
| hsa-let-7b-3p | lung squamous cell cancer | progression | "Flow cytometry analysis revealed these miRNAs indu ......" | 19688090 | Flow cytometry |
| hsa-let-7b-3p | lung squamous cell cancer | metastasis; staging; poor survival | "Different members of let-7 family have been report ......" | 23981581 | |
| hsa-let-7b-3p | lung squamous cell cancer | poor survival | "Systemic nanodelivery of miR-34 and let-7 suppress ......" | 25174400 | |
| hsa-let-7b-3p | lung squamous cell cancer | drug resistance | "Combinatorial Action of MicroRNAs let 7 and miR 34 ......" | 25714397 | |
| hsa-let-7b-3p | melanoma | malignant trasformation; progression | "MicroRNA let 7b targets important cell cycle molec ......" | 18379589 | |
| hsa-let-7b-3p | melanoma | metastasis; differentiation | "An exploratory miRNA analysis of 666 miRs by low d ......" | 20302635 | |
| hsa-let-7b-3p | melanoma | progression | "In situ analysis of melanoma samples using progres ......" | 23728176 | |
| hsa-let-7b-3p | melanoma | differentiation | "Let-7 microRNAs miRNAs are highly conserved well-e ......" | 25669981 | |
| hsa-let-7b-3p | ovarian cancer | worse prognosis | "The microRNA expression profiles were examined usi ......" | 18451233 | |
| hsa-let-7b-3p | ovarian cancer | worse prognosis | "We highlight the role of the let-7 and miR-200 fam ......" | 20083225 | |
| hsa-let-7b-3p | ovarian cancer | drug resistance | "Luciferase assays and real-time PCRs showed that t ......" | 21109987 | Luciferase |
| hsa-let-7b-3p | ovarian cancer | drug resistance | "Let-7 is a family of small non-coding RNAs regulat ......" | 21571355 | |
| hsa-let-7b-3p | ovarian cancer | worse prognosis; staging | "Two families of miRNAs miR-200 and let-7 are frequ ......" | 23237306 | |
| hsa-let-7b-3p | ovarian cancer | differentiation | "These overwhelmingly observed oncogenic genes were ......" | 24129674 | |
| hsa-let-7b-3p | ovarian cancer | tumorigenesis | "To explore miRNAs involved in the tumorigenesis in ......" | 25273528 | |
| hsa-let-7b-3p | ovarian cancer | motility | "LncRNA HOST2 regulates cell biological behaviors i ......" | 25292198 | |
| hsa-let-7b-3p | pancreatic cancer | progression | "let 7 MicroRNA transfer in pancreatic cancer deriv ......" | 19323605 | |
| hsa-let-7b-3p | pancreatic cancer | poor survival | "The expression of miR-21 was significantly higher ......" | 21139804 | |
| hsa-let-7b-3p | pancreatic cancer | tumorigenesis | "miR-122 let-7 family and miR-101 are down-regulate ......" | 22303361 | |
| hsa-let-7b-3p | pancreatic cancer | metastasis | "We therefore established a screening strategy to f ......" | 23733161 | Western blot |
| hsa-let-7b-3p | pancreatic cancer | poor survival | "The rapamycin effects centered on decreased mTOR-r ......" | 25576058 | |
| hsa-let-7b-3p | pancreatic cancer | staging | "In addition from 10 to 50 weeks of age stage-speci ......" | 26516699 | |
| hsa-let-7b-3p | pancreatic cancer | drug resistance | "In particular modulations of let-7 miR-29a miR-17- ......" | 27539232 | |
| hsa-let-7b-3p | prostate cancer | progression; poor survival | "Distinct microRNA expression profile in prostate c ......" | 23798998 | |
| hsa-let-7b-3p | prostate cancer | drug resistance; metastasis | "Loss of let 7 microRNA upregulates IL 6 in bone ma ......" | 23977098 | Luciferase |
| hsa-let-7b-3p | prostate cancer | malignant trasformation | "Role of miRNA let 7 and its major targets in prost ......" | 25276782 | |
| hsa-let-7b-3p | prostate cancer | worse prognosis | "miRNA let 7b modulates macrophage polarization and ......" | 27157642 | |
| hsa-let-7b-3p | retinoblastoma | metastasis | "In addition hsa-miR-25 hsa-miR-18a and hsa-miR-20a ......" | 26730174 | |
| hsa-let-7b-3p | sarcoma | malignant trasformation | "Let 7 microRNA and HMGA2 levels of expression are ......" | 21412931 | |
| hsa-let-7b-3p | thyroid cancer | differentiation | "Effects of let 7 microRNA on Cell Growth and Diffe ......" | 19956384 |
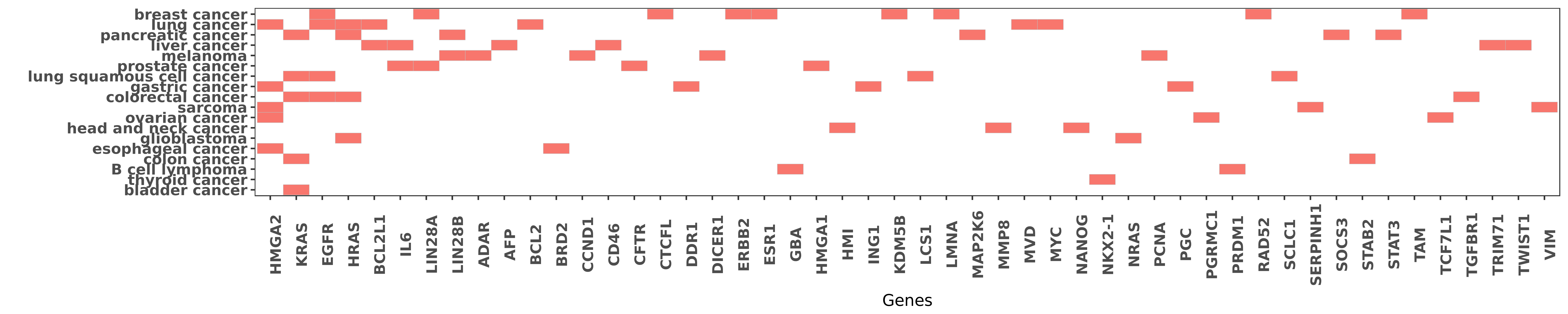
Reported gene related to hsa-let-7b-3p
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-let-7b-3p | bladder cancer | KRAS | "A let 7 microRNA binding site polymorphism in the ......" | 27620744 |
| hsa-let-7b-3p | colon cancer | KRAS | "Association study of the let 7 miRNA complementary ......" | 24727325 |
| hsa-let-7b-3p | colorectal cancer | KRAS | "A let 7 microRNA binding site polymorphism in 3' u ......" | 20603437 |
| hsa-let-7b-3p | colorectal cancer | KRAS | "Let 7 microRNA binding site polymorphism in the 3' ......" | 24890702 |
| hsa-let-7b-3p | colorectal cancer | KRAS | "The LCS6 polymorphism in the binding site of let 7 ......" | 23324806 |
| hsa-let-7b-3p | colorectal cancer | KRAS | "A let 7 microRNA SNP in the KRAS 3'UTR is prognost ......" | 21994416 |
| hsa-let-7b-3p | colorectal cancer | KRAS | "Let 7 miRNA binding site polymorphism in the KRAS ......" | 23167843 |
| hsa-let-7b-3p | colorectal cancer | KRAS | "Genetic modulation of the Let 7 microRNA binding t ......" | 20177422 |
| hsa-let-7b-3p | colorectal cancer | KRAS | "A let 7 microRNA binding site polymorphism in KRAS ......" | 25183481 |
| hsa-let-7b-3p | colorectal cancer | KRAS | "Our results showed that miR-143 but not let-7b inc ......" | 26581910 |
| hsa-let-7b-3p | colorectal cancer | KRAS | "Preclinical and experimental data in vivo indicate ......" | 22584434 |
| hsa-let-7b-3p | lung squamous cell cancer | KRAS | "A SNP in a let 7 microRNA complementary site in th ......" | 18922928 |
| hsa-let-7b-3p | pancreatic cancer | KRAS | "Interestingly the differential expression of miRNA ......" | 26516699 |
| hsa-let-7b-3p | esophageal cancer | HMGA2 | "Role of microRNA let 7 and effect to HMGA2 in esop ......" | 21598109 |
| hsa-let-7b-3p | esophageal cancer | HMGA2 | "HMGA2 is down regulated by microRNA let 7 and asso ......" | 24612219 |
| hsa-let-7b-3p | gastric cancer | HMGA2 | "Since let-7 miRNAs generally play a tumor-suppress ......" | 20949044 |
| hsa-let-7b-3p | gastric cancer | HMGA2 | "Recent studies report that HMGA2 is negatively reg ......" | 18413822 |
| hsa-let-7b-3p | lung cancer | HMGA2 | "MiR-98 a member in the let-7 family acts as a nega ......" | 23700794 |
| hsa-let-7b-3p | ovarian cancer | HMGA2 | "HMGA2 mRNA and let-7 family microRNA were detected ......" | 23073586 |
| hsa-let-7b-3p | sarcoma | HMGA2 | "Let 7 microRNA and HMGA2 levels of expression are ......" | 21412931 |
| hsa-let-7b-3p | colorectal cancer | HRAS | "On the basis of these data we suggest that the dow ......" | 20881268 |
| hsa-let-7b-3p | glioblastoma | HRAS | "Transfection of let-7 miRNA reduced expression of ......" | 20607356 |
| hsa-let-7b-3p | lung cancer | HRAS | "Using an established orthotopic mouse lung cancer ......" | 18344688 |
| hsa-let-7b-3p | lung cancer | HRAS | "k-Ras and c-Myc two key oncogenes in lung cancer h ......" | 20033209 |
| hsa-let-7b-3p | pancreatic cancer | HRAS | "Because let-7 microRNA targets the K-ras oncogene ......" | 19323605 |
| hsa-let-7b-3p | breast cancer | EGFR | "The tumours in the TMAs were assigned to subtypes ......" | 22294324 |
| hsa-let-7b-3p | colorectal cancer | EGFR | "The LCS6 polymorphism in the binding site of let 7 ......" | 23324806 |
| hsa-let-7b-3p | lung cancer | EGFR | "Our mathematical model explains the signaling link ......" | 23365639 |
| hsa-let-7b-3p | lung squamous cell cancer | EGFR | "This effect was observed over a wide range of miRN ......" | 25714397 |
| hsa-let-7b-3p | breast cancer | LIN28A | "Moreover further studies showed that p21 Rb and Le ......" | 22808086 |
| hsa-let-7b-3p | breast cancer | LIN28A | "LIN28 is a RNA-binding protein that has been well ......" | 22081076 |
| hsa-let-7b-3p | breast cancer | LIN28A | "The Wnt β catenin pathway represses let 7 microRN ......" | 23613467 |
| hsa-let-7b-3p | prostate cancer | LIN28A | "Furthermore let-7c expression is downregulated in ......" | 22479342 |
| hsa-let-7b-3p | liver cancer | BCL2L1 | "The let 7 family of microRNAs inhibits Bcl xL expr ......" | 20347499 |
| hsa-let-7b-3p | lung cancer | BCL2L1 | "As a result transcript levels of the tumor-suppres ......" | 26915294 |
| hsa-let-7b-3p | melanoma | CCND1 | "The effect of let-7b on protein expression was due ......" | 18379589 |
| hsa-let-7b-3p | melanoma | CCND1 | "Cyclin D1 expression was significantly higher in c ......" | 23162627 |
| hsa-let-7b-3p | breast cancer | ERBB2 | "The tumours in the TMAs were assigned to subtypes ......" | 22294324 |
| hsa-let-7b-3p | breast cancer | ERBB2 | "For instance an abundant expression of multiple le ......" | 25907662 |
| hsa-let-7b-3p | breast cancer | ESR1 | "Let 7 family miRNAs regulate estrogen receptor alp ......" | 20535543 |
| hsa-let-7b-3p | breast cancer | ESR1 | "let 7 microRNAs induce tamoxifen sensitivity by do ......" | 21826373 |
| hsa-let-7b-3p | liver cancer | IL6 | "Knocking down IL-6 and Twist in HSCs significantly ......" | 21352471 |
| hsa-let-7b-3p | prostate cancer | IL6 | "Loss of let 7 microRNA upregulates IL 6 in bone ma ......" | 23977098 |
| hsa-let-7b-3p | melanoma | LIN28B | "Mechanistically we discovered that Lin28B a well-c ......" | 26071398 |
| hsa-let-7b-3p | pancreatic cancer | LIN28B | "LIN28B suppresses microRNA let 7b expression to pr ......" | 26609473 |
| hsa-let-7b-3p | lung cancer | MYC | "Our mathematical model explains the signaling link ......" | 23365639 |
| hsa-let-7b-3p | lung cancer | MYC | "k-Ras and c-Myc two key oncogenes in lung cancer h ......" | 20033209 |
| hsa-let-7b-3p | melanoma | PCNA | "The effect of let-7b on protein expression was due ......" | 18379589 |
| hsa-let-7b-3p | melanoma | PCNA | "Cyclin D1 expression was significantly higher in c ......" | 23162627 |
| hsa-let-7b-3p | melanoma | ADAR | "Importantly we discovered that ADAR1 fundamentally ......" | 23728176 |
| hsa-let-7b-3p | liver cancer | AFP | "As a single marker α-fetoprotein AFP and miR-122 ......" | 26264553 |
| hsa-let-7b-3p | lung cancer | BCL2 | "As a result transcript levels of the tumor-suppres ......" | 26915294 |
| hsa-let-7b-3p | esophageal cancer | BRD2 | "Let-7 expressions were detected in esophageal canc ......" | 21598109 |
| hsa-let-7b-3p | liver cancer | CD46 | "The present study suggests that CD46 plays an impo ......" | 24297460 |
| hsa-let-7b-3p | prostate cancer | CFTR | "Validation of the data using an independent high-r ......" | 23798998 |
| hsa-let-7b-3p | breast cancer | CTCFL | "Circulating cell free cancer testis MAGE A RNA BOR ......" | 24983365 |
| hsa-let-7b-3p | gastric cancer | DDR1 | "Let-7 g improved the effects of X-rays on GC cells ......" | 25972194 |
| hsa-let-7b-3p | melanoma | DICER1 | "Importantly we discovered that ADAR1 fundamentally ......" | 23728176 |
| hsa-let-7b-3p | B cell lymphoma | GBA | "Let-7 in particular let-7b is overexpressed in DLB ......" | 20651244 |
| hsa-let-7b-3p | prostate cancer | HMGA1 | "Furthermore we identified HMGA1 a non-histone prot ......" | 23798998 |
| hsa-let-7b-3p | head and neck cancer | HMI | "Subsequently microarray analysis followed by Ingen ......" | 27588466 |
| hsa-let-7b-3p | gastric cancer | ING1 | "MicroRNA let 7b suppresses human gastric cancer ma ......" | 25613480 |
| hsa-let-7b-3p | breast cancer | KDM5B | "In MCF-7 cells with stable RNAi-mediated suppressi ......" | 21969366 |
| hsa-let-7b-3p | lung squamous cell cancer | LCS1 | "The purpose of this study was to identify single n ......" | 18922928 |
| hsa-let-7b-3p | breast cancer | LMNA | "Twenty-five differentially expressed miRNAs P < 0. ......" | 20535543 |
| hsa-let-7b-3p | pancreatic cancer | MAP2K6 | "Restoring let-7 levels in cancer-derived cell line ......" | 19323605 |
| hsa-let-7b-3p | head and neck cancer | MMP8 | "However the role of let-7 in head and neck cancer ......" | 21292542 |
| hsa-let-7b-3p | lung cancer | MVD | "We demonstrated significantly higher MVD and decre ......" | 23029111 |
| hsa-let-7b-3p | head and neck cancer | NANOG | "Most importantly overexpression of let-7 or knockd ......" | 21292542 |
| hsa-let-7b-3p | thyroid cancer | NKX2-1 | "In addition let-7 enhanced transcriptional express ......" | 19956384 |
| hsa-let-7b-3p | glioblastoma | NRAS | "Transfection of let-7 miRNA reduced expression of ......" | 20607356 |
| hsa-let-7b-3p | gastric cancer | PGC | "This study examined the expression patterns of ser ......" | 25549793 |
| hsa-let-7b-3p | ovarian cancer | PGRMC1 | "In conclusion we propose that PGRMC1 expression is ......" | 21109987 |
| hsa-let-7b-3p | B cell lymphoma | PRDM1 | "Data obtained from expression analysis luciferase ......" | 20651244 |
| hsa-let-7b-3p | breast cancer | RAD52 | "We found that individuals with AC OR 0.684 95%CI 0 ......" | 27726100 |
| hsa-let-7b-3p | lung squamous cell cancer | SCLC1 | "Among them five downregulated miRNAs let-7 miR-20 ......" | 23117485 |
| hsa-let-7b-3p | sarcoma | SERPINH1 | "Using a quantitative shotgun proteomics approach w ......" | 24457375 |
| hsa-let-7b-3p | pancreatic cancer | SOCS3 | "MicroRNA let 7 downregulates STAT3 phosphorylation ......" | 24491408 |
| hsa-let-7b-3p | colon cancer | STAB2 | "Nine dysregulated miRNA pairs fell into three miRN ......" | 25287248 |
| hsa-let-7b-3p | pancreatic cancer | STAT3 | "MicroRNA let 7 downregulates STAT3 phosphorylation ......" | 24491408 |
| hsa-let-7b-3p | breast cancer | TAM | "Transfection of let-7 mimics to Tam-resistant MCF7 ......" | 21826373 |
| hsa-let-7b-3p | ovarian cancer | TCF7L1 | "These overwhelmingly observed oncogenic genes were ......" | 24129674 |
| hsa-let-7b-3p | colorectal cancer | TGFBR1 | "rs868 is contained in a let-7 miRNA binding site i ......" | 27234654 |
| hsa-let-7b-3p | liver cancer | TRIM71 | "Lin-41 is a stem cell-specific E3 ligase and a kno ......" | 23097274 |
| hsa-let-7b-3p | liver cancer | TWIST1 | "Knocking down IL-6 and Twist in HSCs significantly ......" | 21352471 |
| hsa-let-7b-3p | sarcoma | VIM | "Using a quantitative shotgun proteomics approach w ......" | 24457375 |
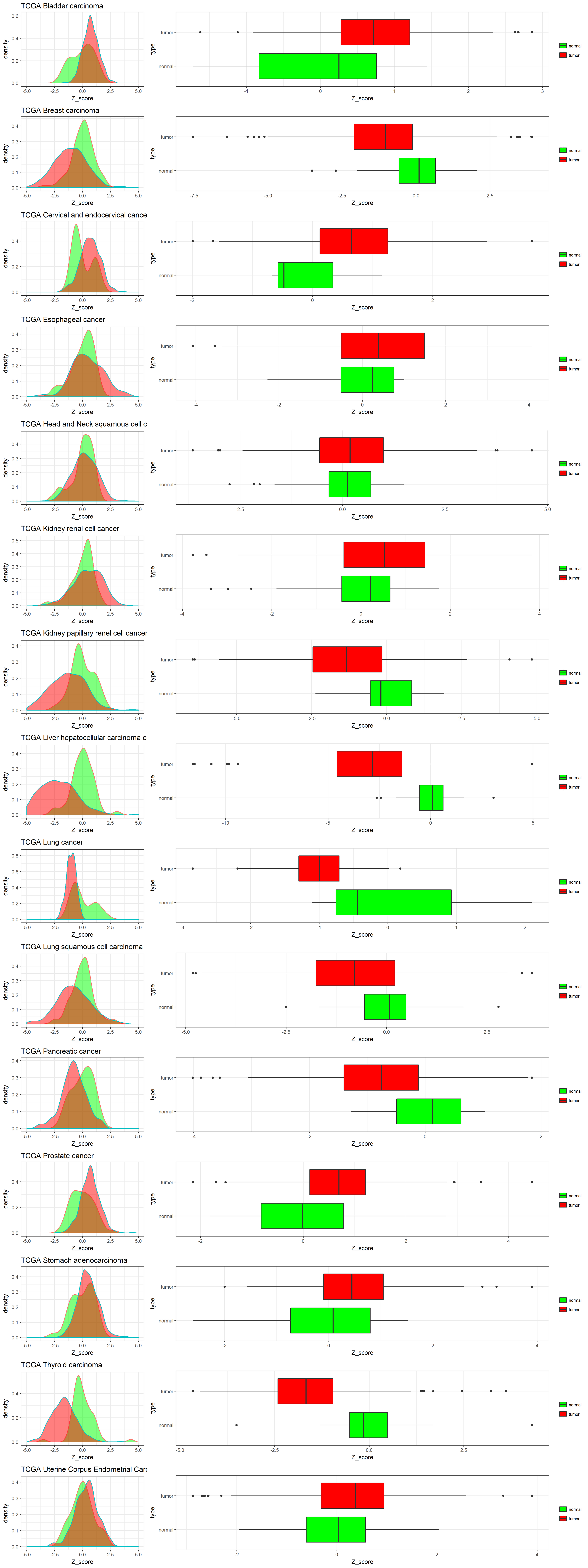
Expression profile in cancer corhorts:
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-let-7b-3p | CACNG4 | 11 cancers: BLCA; CESC; COAD; KIRC; LIHC; PAAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.477; TCGA CESC -0.641; TCGA COAD -0.828; TCGA KIRC -0.745; TCGA LIHC -0.531; TCGA PAAD -0.639; TCGA PRAD -0.521; TCGA SARC -0.48; TCGA THCA -1.215; TCGA STAD -0.872; TCGA UCEC -0.538 |
hsa-let-7b-3p | NLGN1 | 9 cancers: BLCA; COAD; KIRP; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.65; TCGA COAD -0.575; TCGA KIRP -0.339; TCGA LUSC -0.358; TCGA PAAD -0.49; TCGA PRAD -0.513; TCGA SARC -0.64; TCGA STAD -1.508; TCGA UCEC -0.447 |
hsa-let-7b-3p | MIF4GD | 9 cancers: BLCA; CESC; HNSC; KIRP; LGG; LUSC; PAAD; SARC; THCA | MirTarget | TCGA BLCA -0.091; TCGA CESC -0.101; TCGA HNSC -0.24; TCGA KIRP -0.148; TCGA LGG -0.054; TCGA LUSC -0.09; TCGA PAAD -0.217; TCGA SARC -0.148; TCGA THCA -0.239 |
hsa-let-7b-3p | LRRC34 | 11 cancers: BLCA; ESCA; HNSC; KIRC; KIRP; LUSC; OV; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.179; TCGA ESCA -0.579; TCGA HNSC -0.157; TCGA KIRC -0.21; TCGA KIRP -0.207; TCGA LUSC -0.331; TCGA OV -0.184; TCGA PAAD -0.265; TCGA PRAD -0.25; TCGA STAD -0.323; TCGA UCEC -0.172 |
hsa-let-7b-3p | ITM2C | 10 cancers: BLCA; BRCA; CESC; ESCA; LIHC; LUAD; OV; PAAD; PRAD; UCEC | MirTarget | TCGA BLCA -0.328; TCGA BRCA -0.183; TCGA CESC -0.23; TCGA ESCA -0.373; TCGA LIHC -0.218; TCGA LUAD -0.127; TCGA OV -0.231; TCGA PAAD -0.278; TCGA PRAD -0.287; TCGA UCEC -0.476 |
hsa-let-7b-3p | MAP4K3 | 9 cancers: BLCA; ESCA; KIRC; LGG; LUAD; LUSC; OV; STAD; UCEC | mirMAP | TCGA BLCA -0.112; TCGA ESCA -0.223; TCGA KIRC -0.178; TCGA LGG -0.05; TCGA LUAD -0.095; TCGA LUSC -0.132; TCGA OV -0.104; TCGA STAD -0.189; TCGA UCEC -0.096 |
hsa-let-7b-3p | CNR1 | 10 cancers: BLCA; CESC; COAD; ESCA; KIRP; LIHC; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.577; TCGA CESC -0.37; TCGA COAD -0.532; TCGA ESCA -1.301; TCGA KIRP -0.385; TCGA LIHC -0.631; TCGA PRAD -0.509; TCGA SARC -0.839; TCGA STAD -1.7; TCGA UCEC -0.288 |
hsa-let-7b-3p | MAP1B | 9 cancers: BLCA; CESC; COAD; ESCA; LUSC; PAAD; PRAD; SARC; STAD | mirMAP | TCGA BLCA -0.481; TCGA CESC -0.287; TCGA COAD -0.339; TCGA ESCA -0.347; TCGA LUSC -0.306; TCGA PAAD -0.502; TCGA PRAD -0.578; TCGA SARC -0.242; TCGA STAD -0.984 |
hsa-let-7b-3p | SCN3A | 9 cancers: BLCA; CESC; COAD; ESCA; KIRC; PAAD; PRAD; SARC; STAD | mirMAP | TCGA BLCA -0.454; TCGA CESC -0.464; TCGA COAD -0.414; TCGA ESCA -0.446; TCGA KIRC -0.437; TCGA PAAD -0.673; TCGA PRAD -0.201; TCGA SARC -0.729; TCGA STAD -0.784 |
hsa-let-7b-3p | UQCRB | 11 cancers: BLCA; BRCA; HNSC; KIRC; LIHC; LUAD; LUSC; PAAD; SARC; THCA; UCEC | mirMAP | TCGA BLCA -0.106; TCGA BRCA -0.103; TCGA HNSC -0.288; TCGA KIRC -0.135; TCGA LIHC -0.218; TCGA LUAD -0.132; TCGA LUSC -0.175; TCGA PAAD -0.178; TCGA SARC -0.169; TCGA THCA -0.079; TCGA UCEC -0.179 |
hsa-let-7b-3p | RIC3 | 9 cancers: BLCA; COAD; KIRC; LUSC; OV; PAAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.597; TCGA COAD -0.657; TCGA KIRC -0.588; TCGA LUSC -0.373; TCGA OV -0.308; TCGA PAAD -0.603; TCGA SARC -0.499; TCGA STAD -1.371; TCGA UCEC -0.356 |
hsa-let-7b-3p | SLC24A5 | 10 cancers: BLCA; BRCA; CESC; KIRC; KIRP; LGG; OV; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.516; TCGA BRCA -0.226; TCGA CESC -0.519; TCGA KIRC -0.279; TCGA KIRP -0.311; TCGA LGG -0.086; TCGA OV -0.269; TCGA SARC -0.594; TCGA THCA -0.567; TCGA STAD -1.447 |
hsa-let-7b-3p | ZNF84 | 11 cancers: BLCA; CESC; ESCA; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; OV; PAAD | mirMAP | TCGA BLCA -0.107; TCGA CESC -0.086; TCGA ESCA -0.188; TCGA HNSC -0.126; TCGA KIRP -0.163; TCGA LGG -0.053; TCGA LIHC -0.092; TCGA LUAD -0.129; TCGA LUSC -0.156; TCGA OV -0.128; TCGA PAAD -0.131 |
hsa-let-7b-3p | ZNF248 | 11 cancers: BLCA; CESC; ESCA; HNSC; LIHC; LUSC; OV; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.171; TCGA CESC -0.121; TCGA ESCA -0.231; TCGA HNSC -0.132; TCGA LIHC -0.095; TCGA LUSC -0.148; TCGA OV -0.114; TCGA PAAD -0.135; TCGA PRAD -0.079; TCGA STAD -0.465; TCGA UCEC -0.125 |
hsa-let-7b-3p | NFYB | 9 cancers: BLCA; ESCA; HNSC; KIRP; LUSC; PAAD; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.084; TCGA ESCA -0.136; TCGA HNSC -0.124; TCGA KIRP -0.086; TCGA LUSC -0.121; TCGA PAAD -0.161; TCGA PRAD -0.066; TCGA STAD -0.184; TCGA UCEC -0.105 |
hsa-let-7b-3p | PCSK2 | 9 cancers: BLCA; BRCA; CESC; COAD; HNSC; PRAD; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -1.1; TCGA BRCA -0.25; TCGA CESC -0.592; TCGA COAD -0.528; TCGA HNSC -0.458; TCGA PRAD -0.826; TCGA THCA -1.1; TCGA STAD -1.813; TCGA UCEC -0.237 |
hsa-let-7b-3p | KIF5C | 10 cancers: BLCA; CESC; COAD; ESCA; LUAD; LUSC; PAAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.503; TCGA CESC -0.268; TCGA COAD -0.285; TCGA ESCA -0.608; TCGA LUAD -0.172; TCGA LUSC -0.232; TCGA PAAD -0.654; TCGA SARC -0.397; TCGA STAD -0.75; TCGA UCEC -0.181 |
hsa-let-7b-3p | STXBP5L | 10 cancers: BLCA; CESC; ESCA; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.801; TCGA CESC -0.353; TCGA ESCA -0.671; TCGA LUAD -0.498; TCGA LUSC -0.619; TCGA PAAD -1.121; TCGA PRAD -0.598; TCGA SARC -1.045; TCGA STAD -1.108; TCGA UCEC -0.231 |
hsa-let-7b-3p | NOVA1 | 9 cancers: BLCA; COAD; ESCA; HNSC; LUSC; PAAD; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.897; TCGA COAD -0.546; TCGA ESCA -0.646; TCGA HNSC -0.307; TCGA LUSC -0.314; TCGA PAAD -0.975; TCGA PRAD -0.262; TCGA STAD -1.429; TCGA UCEC -0.554 |
hsa-let-7b-3p | NGDN | 9 cancers: BRCA; COAD; HNSC; LIHC; LUAD; LUSC; OV; PAAD; UCEC | MirTarget | TCGA BRCA -0.058; TCGA COAD -0.132; TCGA HNSC -0.107; TCGA LIHC -0.116; TCGA LUAD -0.135; TCGA LUSC -0.107; TCGA OV -0.114; TCGA PAAD -0.092; TCGA UCEC -0.18 |
hsa-let-7b-3p | TSFM | 12 cancers: BRCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; OV; PAAD; THCA; UCEC | MirTarget | TCGA BRCA -0.126; TCGA HNSC -0.148; TCGA KIRC -0.161; TCGA KIRP -0.082; TCGA LGG -0.103; TCGA LIHC -0.103; TCGA LUAD -0.199; TCGA LUSC -0.166; TCGA OV -0.162; TCGA PAAD -0.169; TCGA THCA -0.081; TCGA UCEC -0.106 |
hsa-let-7b-3p | PDCL3 | 11 cancers: BRCA; HNSC; KIRP; LIHC; LUAD; LUSC; OV; PAAD; SARC; THCA; UCEC | MirTarget | TCGA BRCA -0.097; TCGA HNSC -0.173; TCGA KIRP -0.09; TCGA LIHC -0.143; TCGA LUAD -0.16; TCGA LUSC -0.115; TCGA OV -0.083; TCGA PAAD -0.094; TCGA SARC -0.08; TCGA THCA -0.056; TCGA UCEC -0.119 |
hsa-let-7b-3p | POLR2K | 9 cancers: BRCA; COAD; HNSC; KIRC; LIHC; LUAD; LUSC; SARC; UCEC | MirTarget | TCGA BRCA -0.07; TCGA COAD -0.2; TCGA HNSC -0.104; TCGA KIRC -0.107; TCGA LIHC -0.218; TCGA LUAD -0.194; TCGA LUSC -0.115; TCGA SARC -0.158; TCGA UCEC -0.188 |
hsa-let-7b-3p | RNF8 | 9 cancers: BRCA; COAD; HNSC; LIHC; LUSC; OV; PAAD; PRAD; UCEC | mirMAP | TCGA BRCA -0.06; TCGA COAD -0.115; TCGA HNSC -0.109; TCGA LIHC -0.205; TCGA LUSC -0.089; TCGA OV -0.118; TCGA PAAD -0.097; TCGA PRAD -0.108; TCGA UCEC -0.066 |
hsa-let-7b-3p | ATP5E | 9 cancers: BRCA; COAD; HNSC; KIRC; LIHC; LUAD; PAAD; THCA; UCEC | mirMAP | TCGA BRCA -0.149; TCGA COAD -0.125; TCGA HNSC -0.29; TCGA KIRC -0.181; TCGA LIHC -0.237; TCGA LUAD -0.128; TCGA PAAD -0.237; TCGA THCA -0.131; TCGA UCEC -0.179 |
hsa-let-7b-3p | TBCA | 10 cancers: BRCA; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; SARC; THCA; UCEC | mirMAP | TCGA BRCA -0.086; TCGA HNSC -0.173; TCGA KIRP -0.094; TCGA LIHC -0.172; TCGA LUAD -0.144; TCGA LUSC -0.198; TCGA PAAD -0.243; TCGA SARC -0.115; TCGA THCA -0.115; TCGA UCEC -0.076 |
hsa-let-7b-3p | RBBP7 | 9 cancers: BRCA; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; SARC; STAD | miRNATAP | TCGA BRCA -0.054; TCGA HNSC -0.075; TCGA KIRP -0.095; TCGA LGG -0.065; TCGA LIHC -0.096; TCGA LUAD -0.128; TCGA LUSC -0.129; TCGA SARC -0.137; TCGA STAD -0.106 |
hsa-let-7b-3p | GDPD1 | 9 cancers: CESC; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; OV; PAAD | MirTarget | TCGA CESC -0.308; TCGA ESCA -0.658; TCGA HNSC -0.159; TCGA KIRP -0.138; TCGA LIHC -0.295; TCGA LUAD -0.196; TCGA LUSC -0.211; TCGA OV -0.313; TCGA PAAD -0.287 |
hsa-let-7b-3p | PLAG1 | 10 cancers: CESC; COAD; HNSC; KIRC; LIHC; OV; PRAD; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA CESC -0.554; TCGA COAD -0.534; TCGA HNSC -0.283; TCGA KIRC -0.436; TCGA LIHC -0.45; TCGA OV -0.332; TCGA PRAD -0.548; TCGA THCA -0.299; TCGA STAD -0.269; TCGA UCEC -0.517 |
hsa-let-7b-3p | MRS2 | 10 cancers: CESC; COAD; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; OV; UCEC | mirMAP | TCGA CESC -0.093; TCGA COAD -0.1; TCGA HNSC -0.087; TCGA KIRC -0.167; TCGA KIRP -0.072; TCGA LIHC -0.07; TCGA LUAD -0.086; TCGA LUSC -0.15; TCGA OV -0.123; TCGA UCEC -0.075 |
hsa-let-7b-3p | PCMTD2 | 10 cancers: CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; OV; STAD; UCEC | mirMAP | TCGA CESC -0.126; TCGA COAD -0.194; TCGA ESCA -0.167; TCGA HNSC -0.098; TCGA KIRC -0.099; TCGA LUAD -0.075; TCGA LUSC -0.19; TCGA OV -0.134; TCGA STAD -0.154; TCGA UCEC -0.141 |
hsa-let-7b-3p | PM20D2 | 10 cancers: COAD; KIRC; LGG; LIHC; LUAD; LUSC; PAAD; SARC; STAD; UCEC | MirTarget | TCGA COAD -0.429; TCGA KIRC -0.139; TCGA LGG -0.072; TCGA LIHC -0.257; TCGA LUAD -0.203; TCGA LUSC -0.299; TCGA PAAD -0.184; TCGA SARC -0.216; TCGA STAD -0.254; TCGA UCEC -0.17 |
hsa-let-7b-3p | SYT1 | 9 cancers: COAD; LIHC; LUAD; LUSC; OV; PRAD; THCA; STAD; UCEC | MirTarget; mirMAP; miRNATAP | TCGA COAD -0.476; TCGA LIHC -0.471; TCGA LUAD -0.455; TCGA LUSC -0.397; TCGA OV -0.209; TCGA PRAD -0.335; TCGA THCA -0.713; TCGA STAD -0.898; TCGA UCEC -0.304 |
hsa-let-7b-3p | TIGD7 | 11 cancers: COAD; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; OV; SARC; STAD; UCEC | MirTarget | TCGA COAD -0.256; TCGA HNSC -0.123; TCGA KIRP -0.124; TCGA LGG -0.065; TCGA LIHC -0.255; TCGA LUAD -0.102; TCGA LUSC -0.151; TCGA OV -0.124; TCGA SARC -0.135; TCGA STAD -0.356; TCGA UCEC -0.185 |
hsa-let-7b-3p | HSDL1 | 9 cancers: COAD; HNSC; KIRC; LGG; LIHC; LUAD; LUSC; OV; STAD | mirMAP | TCGA COAD -0.109; TCGA HNSC -0.053; TCGA KIRC -0.084; TCGA LGG -0.056; TCGA LIHC -0.178; TCGA LUAD -0.099; TCGA LUSC -0.117; TCGA OV -0.165; TCGA STAD -0.168 |
hsa-let-7b-3p | MYBL1 | 9 cancers: COAD; KIRP; LIHC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | mirMAP; miRNATAP | TCGA COAD -0.224; TCGA KIRP -0.342; TCGA LIHC -0.153; TCGA LUAD -0.381; TCGA LUSC -0.283; TCGA PRAD -0.188; TCGA SARC -0.362; TCGA STAD -0.462; TCGA UCEC -0.091 |
hsa-let-7b-3p | GDAP1 | 10 cancers: ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; STAD; UCEC | MirTarget | TCGA ESCA -0.244; TCGA HNSC -0.125; TCGA KIRC -0.123; TCGA KIRP -0.118; TCGA LIHC -0.206; TCGA LUAD -0.136; TCGA LUSC -0.26; TCGA PAAD -0.325; TCGA STAD -0.369; TCGA UCEC -0.294 |
hsa-let-7b-3p | LBR | 11 cancers: ESCA; KIRP; LGG; LIHC; LUAD; LUSC; OV; PRAD; SARC; STAD; UCEC | mirMAP | TCGA ESCA -0.292; TCGA KIRP -0.093; TCGA LGG -0.078; TCGA LIHC -0.171; TCGA LUAD -0.1; TCGA LUSC -0.12; TCGA OV -0.115; TCGA PRAD -0.092; TCGA SARC -0.241; TCGA STAD -0.264; TCGA UCEC -0.187 |
hsa-let-7b-3p | RAB6B | 9 cancers: ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; STAD; UCEC | miRNATAP | TCGA ESCA -0.374; TCGA HNSC -0.409; TCGA KIRP -0.318; TCGA LIHC -0.13; TCGA LUAD -0.203; TCGA LUSC -0.685; TCGA PAAD -0.416; TCGA STAD -0.574; TCGA UCEC -0.159 |
hsa-let-7b-3p | RIMKLB | 9 cancers: HNSC; KIRC; KIRP; LGG; LUSC; OV; PRAD; STAD; UCEC | MirTarget | TCGA HNSC -0.119; TCGA KIRC -0.151; TCGA KIRP -0.139; TCGA LGG -0.07; TCGA LUSC -0.142; TCGA OV -0.118; TCGA PRAD -0.192; TCGA STAD -0.63; TCGA UCEC -0.134 |
hsa-let-7b-3p | TMEM117 | 9 cancers: KIRC; KIRP; LGG; LUAD; LUSC; PAAD; PRAD; THCA; STAD | MirTarget | TCGA KIRC -0.173; TCGA KIRP -0.195; TCGA LGG -0.074; TCGA LUAD -0.117; TCGA LUSC -0.239; TCGA PAAD -0.149; TCGA PRAD -0.173; TCGA THCA -0.309; TCGA STAD -0.143 |
Enriched cancer pathways of putative targets