microRNA information: hsa-let-7e-5p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-let-7e-5p | miRbase |
| Accession: | MIMAT0000066 | miRbase |
| Precursor name: | hsa-let-7e | miRbase |
| Precursor accession: | MI0000066 | miRbase |
| Symbol: | MIRLET7E | HGNC |
| RefSeq ID: | NR_029482 | GenBank |
| Sequence: | UGAGGUAGGAGGUUGUAUAGUU |
Reported expression in cancers: hsa-let-7e-5p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-let-7e-5p | breast cancer | deregulation | "Three of these miRNAs were up-regulated miR-141 mi ......" | 25451164 | |
| hsa-let-7e-5p | colorectal cancer | deregulation | "In our CD patients six miRNAs were upregulated fro ......" | 22241525 | |
| hsa-let-7e-5p | esophageal cancer | deregulation | "Differentially-expressed miRNAs were analyzed usin ......" | 23534712 | Reverse transcription PCR |
| hsa-let-7e-5p | lung squamous cell cancer | downregulation | "Differential expression of miR 125a 5p and let 7e ......" | 24945821 | qPCR |
| hsa-let-7e-5p | ovarian cancer | deregulation | "Among them 10 miRNA were down-regulated including ......" | 21122376 | |
| hsa-let-7e-5p | retinoblastoma | upregulation | "Identification of miRNAs associated with tumorigen ......" | 18818933 | Microarray; Northern blot; in situ hybridization |
| hsa-let-7e-5p | sarcoma | upregulation | "There were 21 significantly up-regulated miRNAs in ......" | 21213367 |
Reported cancer pathway affected by hsa-let-7e-5p
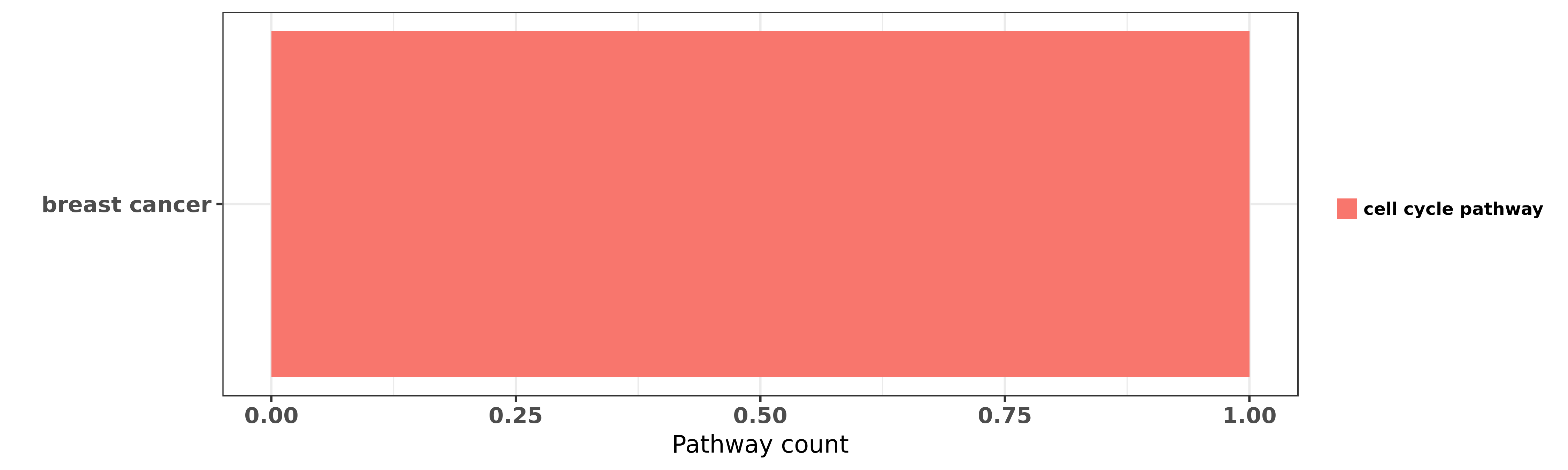
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-let-7e-5p | breast cancer | cell cycle pathway | "Jumonji/ARID1 B JARID1B protein promotes breast tu ......" | 21969366 | RNAi |
Reported cancer prognosis affected by hsa-let-7e-5p
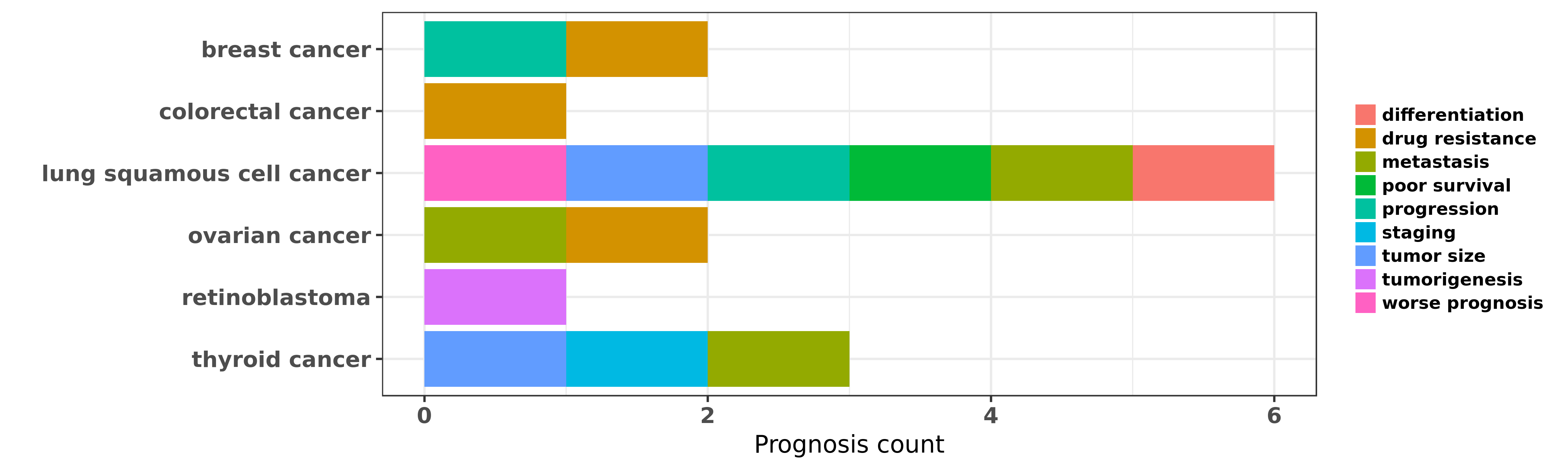
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-let-7e-5p | breast cancer | progression | "Jumonji/ARID1 B JARID1B protein promotes breast tu ......" | 21969366 | RNAi |
| hsa-let-7e-5p | breast cancer | drug resistance | "We created a doxorubicin-resistant MCF-7 MCF-/Adr ......" | 25451164 | MTT assay |
| hsa-let-7e-5p | colorectal cancer | drug resistance | "We investigated this issue by profiling the expres ......" | 20881268 | |
| hsa-let-7e-5p | lung squamous cell cancer | metastasis; differentiation; tumor size; poor survival; worse prognosis | "High-throughput microarray was used to measure miR ......" | 22618509 | |
| hsa-let-7e-5p | lung squamous cell cancer | progression; worse prognosis; differentiation; poor survival | "Differential expression of miR 125a 5p and let 7e ......" | 24945821 | |
| hsa-let-7e-5p | ovarian cancer | drug resistance | "High-throughput analysis of the miRNA profile in a ......" | 18823650 | |
| hsa-let-7e-5p | ovarian cancer | metastasis | "miRNA microarray was applied to compare the miRNA ......" | 21122376 | |
| hsa-let-7e-5p | retinoblastoma | tumorigenesis | "Identification of miRNAs associated with tumorigen ......" | 18818933 | |
| hsa-let-7e-5p | thyroid cancer | staging; metastasis; tumor size | "Genome-wide serum miRNA expression profiles were d ......" | 22472564 | |
| hsa-let-7e-5p | thyroid cancer | staging; metastasis; tumor size | "Expression levels of miRs-21 -34b -130b -135b -146 ......" | 26414548 |
Reported gene related to hsa-let-7e-5p
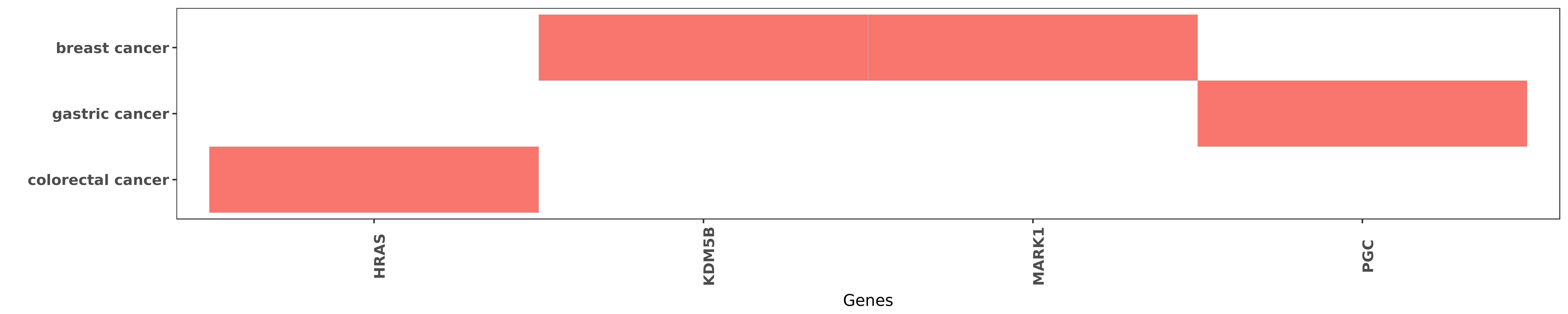
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-let-7e-5p | colorectal cancer | HRAS | "On the basis of these data we suggest that the dow ......" | 20881268 |
| hsa-let-7e-5p | breast cancer | KDM5B | "Jumonji/ARID1 B JARID1B protein promotes breast tu ......" | 21969366 |
| hsa-let-7e-5p | breast cancer | MARK1 | "Chromatin immunoprecipitation analysis demonstrate ......" | 21969366 |
| hsa-let-7e-5p | gastric cancer | PGC | "Also let-7e rs8111742 PGC rs6458238 and H pylori i ......" | 26988755 |
Expression profile in cancer corhorts:
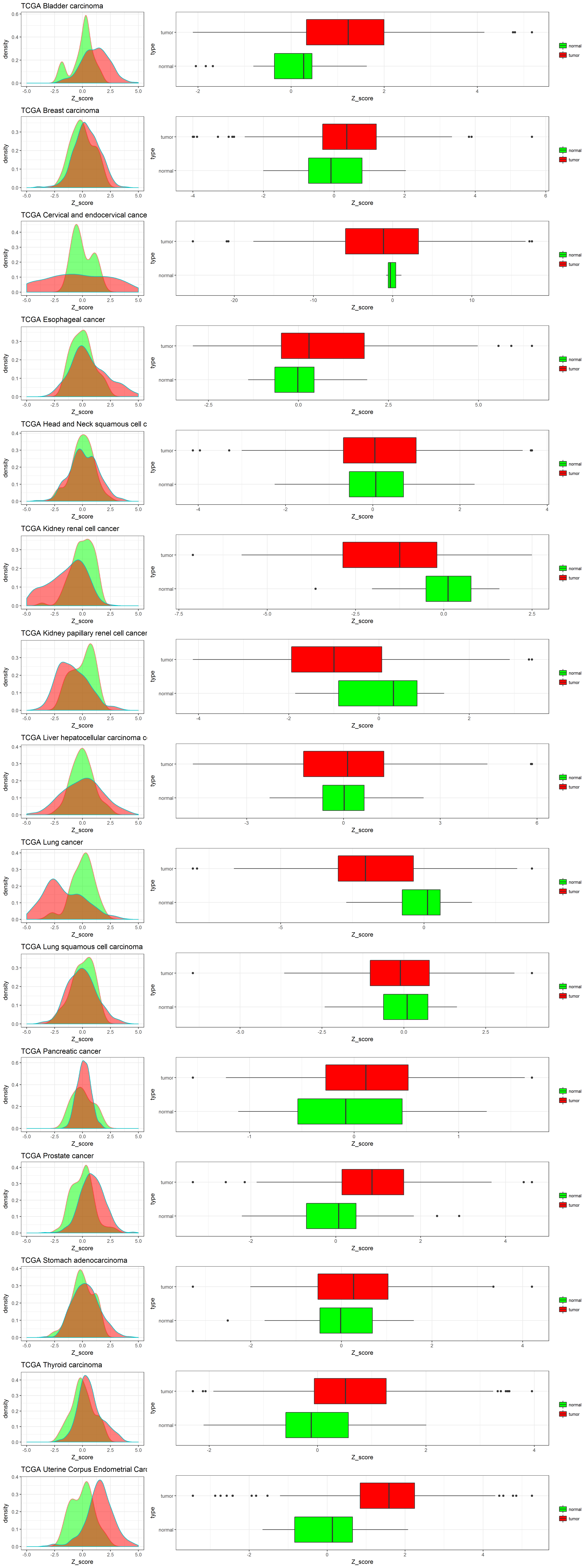
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-let-7e-5p | CNBP | 9 cancers: BLCA; CESC; HNSC; LGG; LIHC; LUSC; OV; SARC; STAD | miRNAWalker2 validate | TCGA BLCA -0.064; TCGA CESC -0.113; TCGA HNSC -0.159; TCGA LGG -0.061; TCGA LIHC -0.103; TCGA LUSC -0.13; TCGA OV -0.148; TCGA SARC -0.097; TCGA STAD -0.063 |
hsa-let-7e-5p | GLUL | 9 cancers: BLCA; HNSC; LIHC; LUSC; PAAD; PRAD; SARC; THCA; UCEC | miRNAWalker2 validate | TCGA BLCA -0.197; TCGA HNSC -0.141; TCGA LIHC -0.233; TCGA LUSC -0.33; TCGA PAAD -0.134; TCGA PRAD -0.213; TCGA SARC -0.204; TCGA THCA -0.075; TCGA UCEC -0.091 |
hsa-let-7e-5p | GTPBP8 | 10 cancers: BLCA; BRCA; CESC; HNSC; LGG; LUSC; OV; PAAD; SARC; STAD | miRNAWalker2 validate | TCGA BLCA -0.142; TCGA BRCA -0.099; TCGA CESC -0.103; TCGA HNSC -0.122; TCGA LGG -0.075; TCGA LUSC -0.215; TCGA OV -0.166; TCGA PAAD -0.054; TCGA SARC -0.064; TCGA STAD -0.151 |
hsa-let-7e-5p | MRPS2 | 9 cancers: BLCA; HNSC; LGG; LIHC; LUAD; LUSC; OV; THCA; STAD | miRNAWalker2 validate | TCGA BLCA -0.121; TCGA HNSC -0.097; TCGA LGG -0.124; TCGA LIHC -0.156; TCGA LUAD -0.095; TCGA LUSC -0.2; TCGA OV -0.169; TCGA THCA -0.051; TCGA STAD -0.214 |
hsa-let-7e-5p | SPN | 9 cancers: BLCA; BRCA; CESC; KIRC; KIRP; PRAD; SARC; THCA; UCEC | miRNAWalker2 validate | TCGA BLCA -0.578; TCGA BRCA -0.191; TCGA CESC -0.215; TCGA KIRC -0.596; TCGA KIRP -0.373; TCGA PRAD -0.191; TCGA SARC -0.334; TCGA THCA -0.306; TCGA UCEC -0.331 |
hsa-let-7e-5p | TDRD7 | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUSC; PRAD; SARC; STAD | miRNAWalker2 validate | TCGA BLCA -0.234; TCGA CESC -0.172; TCGA COAD -0.107; TCGA ESCA -0.132; TCGA HNSC -0.155; TCGA LUSC -0.261; TCGA PRAD -0.072; TCGA SARC -0.172; TCGA STAD -0.181 |
hsa-let-7e-5p | CRTAM | 11 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.561; TCGA BRCA -0.183; TCGA CESC -0.384; TCGA HNSC -0.182; TCGA KIRC -0.91; TCGA KIRP -0.279; TCGA PRAD -0.205; TCGA SARC -0.245; TCGA THCA -0.573; TCGA STAD -0.281; TCGA UCEC -0.296 |
hsa-let-7e-5p | LBR | 9 cancers: BLCA; BRCA; HNSC; KIRP; LGG; LUSC; SARC; THCA; UCEC | MirTarget | TCGA BLCA -0.108; TCGA BRCA -0.114; TCGA HNSC -0.069; TCGA KIRP -0.197; TCGA LGG -0.168; TCGA LUSC -0.175; TCGA SARC -0.329; TCGA THCA -0.166; TCGA UCEC -0.154 |
hsa-let-7e-5p | ADRB2 | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRP; LIHC; SARC; THCA; UCEC | MirTarget | TCGA BLCA -0.396; TCGA BRCA -0.362; TCGA CESC -0.454; TCGA ESCA -0.427; TCGA HNSC -0.199; TCGA KIRP -0.704; TCGA LIHC -0.556; TCGA SARC -0.235; TCGA THCA -0.216; TCGA UCEC -0.535 |
hsa-let-7e-5p | FASLG | 11 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LUAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.681; TCGA BRCA -0.337; TCGA CESC -0.312; TCGA HNSC -0.353; TCGA KIRC -0.957; TCGA KIRP -0.243; TCGA LUAD -0.243; TCGA SARC -0.382; TCGA THCA -0.51; TCGA STAD -0.406; TCGA UCEC -0.402 |
hsa-let-7e-5p | GRIN3A | 10 cancers: BLCA; BRCA; CESC; KIRC; KIRP; LGG; LIHC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.379; TCGA BRCA -0.191; TCGA CESC -0.251; TCGA KIRC -0.358; TCGA KIRP -0.156; TCGA LGG -0.484; TCGA LIHC -0.127; TCGA THCA -0.162; TCGA STAD -0.185; TCGA UCEC -0.336 |
hsa-let-7e-5p | VSNL1 | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRP; LIHC; LUSC; THCA; UCEC | miRNATAP | TCGA BLCA -0.863; TCGA BRCA -0.304; TCGA CESC -1.16; TCGA ESCA -0.44; TCGA HNSC -0.276; TCGA KIRP -1.039; TCGA LIHC -0.637; TCGA LUSC -0.319; TCGA THCA -0.341; TCGA UCEC -0.553 |
hsa-let-7e-5p | NUDT8 | 10 cancers: BRCA; CESC; ESCA; HNSC; LIHC; LUAD; LUSC; OV; THCA; STAD | miRNAWalker2 validate | TCGA BRCA -0.364; TCGA CESC -0.38; TCGA ESCA -0.193; TCGA HNSC -0.415; TCGA LIHC -0.335; TCGA LUAD -0.273; TCGA LUSC -0.506; TCGA OV -0.211; TCGA THCA -0.191; TCGA STAD -0.202 |
hsa-let-7e-5p | RPL10 | 11 cancers: BRCA; COAD; ESCA; HNSC; KIRC; LGG; LIHC; LUSC; OV; STAD; UCEC | miRNAWalker2 validate | TCGA BRCA -0.056; TCGA COAD -0.118; TCGA ESCA -0.1; TCGA HNSC -0.071; TCGA KIRC -0.246; TCGA LGG -0.232; TCGA LIHC -0.114; TCGA LUSC -0.109; TCGA OV -0.233; TCGA STAD -0.159; TCGA UCEC -0.105 |
hsa-let-7e-5p | ANKRD49 | 10 cancers: BRCA; COAD; HNSC; KIRC; LGG; LUAD; LUSC; OV; SARC; THCA | MirTarget; miRNATAP | TCGA BRCA -0.064; TCGA COAD -0.242; TCGA HNSC -0.115; TCGA KIRC -0.099; TCGA LGG -0.095; TCGA LUAD -0.065; TCGA LUSC -0.158; TCGA OV -0.094; TCGA SARC -0.117; TCGA THCA -0.063 |
hsa-let-7e-5p | DTX2 | 11 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PRAD; STAD | miRNATAP | TCGA BRCA -0.113; TCGA CESC -0.199; TCGA COAD -0.154; TCGA ESCA -0.164; TCGA HNSC -0.176; TCGA KIRC -0.418; TCGA KIRP -0.351; TCGA LUAD -0.179; TCGA LUSC -0.142; TCGA PRAD -0.1; TCGA STAD -0.152 |
hsa-let-7e-5p | ZNF57 | 10 cancers: BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUSC; OV; PAAD; STAD | miRNATAP | TCGA BRCA -0.116; TCGA CESC -0.307; TCGA COAD -0.274; TCGA ESCA -0.111; TCGA HNSC -0.258; TCGA LIHC -0.127; TCGA LUSC -0.267; TCGA OV -0.106; TCGA PAAD -0.13; TCGA STAD -0.221 |
hsa-let-7e-5p | USE1 | 10 cancers: CESC; ESCA; HNSC; KIRC; LGG; LIHC; LUAD; LUSC; OV; STAD | miRNAWalker2 validate | TCGA CESC -0.118; TCGA ESCA -0.156; TCGA HNSC -0.098; TCGA KIRC -0.207; TCGA LGG -0.088; TCGA LIHC -0.196; TCGA LUAD -0.116; TCGA LUSC -0.251; TCGA OV -0.144; TCGA STAD -0.148 |
hsa-let-7e-5p | DCAF15 | 9 cancers: CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; STAD | MirTarget; miRNATAP | TCGA CESC -0.164; TCGA COAD -0.08; TCGA ESCA -0.101; TCGA HNSC -0.108; TCGA KIRC -0.164; TCGA KIRP -0.063; TCGA LUAD -0.201; TCGA LUSC -0.11; TCGA STAD -0.082 |
hsa-let-7e-5p | MXD1 | 10 cancers: CESC; COAD; ESCA; HNSC; KIRC; LGG; LUSC; PRAD; STAD; UCEC | MirTarget; miRNATAP | TCGA CESC -0.588; TCGA COAD -0.178; TCGA ESCA -0.341; TCGA HNSC -0.233; TCGA KIRC -0.104; TCGA LGG -0.113; TCGA LUSC -0.141; TCGA PRAD -0.125; TCGA STAD -0.196; TCGA UCEC -0.164 |
hsa-let-7e-5p | TRABD | 9 cancers: CESC; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; THCA; STAD | miRNATAP | TCGA CESC -0.107; TCGA ESCA -0.114; TCGA HNSC -0.163; TCGA KIRC -0.296; TCGA KIRP -0.201; TCGA LGG -0.1; TCGA LUAD -0.293; TCGA THCA -0.184; TCGA STAD -0.226 |
hsa-let-7e-5p | EZH2 | 11 cancers: CESC; COAD; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; PAAD; THCA; STAD | miRNATAP | TCGA CESC -0.21; TCGA COAD -0.186; TCGA HNSC -0.378; TCGA KIRC -0.536; TCGA KIRP -0.304; TCGA LGG -0.288; TCGA LUAD -0.352; TCGA LUSC -0.339; TCGA PAAD -0.154; TCGA THCA -0.214; TCGA STAD -0.183 |
hsa-let-7e-5p | RPL27A | 9 cancers: COAD; HNSC; KIRC; LGG; LIHC; LUSC; OV; THCA; STAD | miRNAWalker2 validate | TCGA COAD -0.131; TCGA HNSC -0.188; TCGA KIRC -0.27; TCGA LGG -0.189; TCGA LIHC -0.09; TCGA LUSC -0.16; TCGA OV -0.237; TCGA THCA -0.061; TCGA STAD -0.179 |
hsa-let-7e-5p | RPLP2 | 9 cancers: COAD; HNSC; KIRC; LGG; LUAD; LUSC; OV; THCA; STAD | miRNAWalker2 validate | TCGA COAD -0.135; TCGA HNSC -0.179; TCGA KIRC -0.28; TCGA LGG -0.211; TCGA LUAD -0.077; TCGA LUSC -0.177; TCGA OV -0.28; TCGA THCA -0.119; TCGA STAD -0.229 |
hsa-let-7e-5p | TARBP2 | 11 cancers: COAD; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; OV; THCA; STAD | miRNATAP | TCGA COAD -0.11; TCGA HNSC -0.098; TCGA KIRC -0.126; TCGA KIRP -0.299; TCGA LGG -0.086; TCGA LIHC -0.077; TCGA LUAD -0.169; TCGA LUSC -0.088; TCGA OV -0.123; TCGA THCA -0.082; TCGA STAD -0.083 |
hsa-let-7e-5p | SLC35D2 | 9 cancers: COAD; HNSC; KIRC; LIHC; LUSC; PRAD; THCA; STAD; UCEC | miRNATAP | TCGA COAD -0.221; TCGA HNSC -0.133; TCGA KIRC -0.061; TCGA LIHC -0.186; TCGA LUSC -0.197; TCGA PRAD -0.07; TCGA THCA -0.129; TCGA STAD -0.254; TCGA UCEC -0.16 |
hsa-let-7e-5p | TIMM17B | 11 cancers: COAD; HNSC; KIRC; LGG; LIHC; LUAD; LUSC; OV; SARC; THCA; STAD | miRNATAP | TCGA COAD -0.15; TCGA HNSC -0.15; TCGA KIRC -0.083; TCGA LGG -0.104; TCGA LIHC -0.124; TCGA LUAD -0.195; TCGA LUSC -0.105; TCGA OV -0.188; TCGA SARC -0.084; TCGA THCA -0.127; TCGA STAD -0.149 |
Enriched cancer pathways of putative targets