microRNA information: hsa-miR-1-3p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-1-3p | miRbase |
| Accession: | MIMAT0000416 | miRbase |
| Precursor name: | hsa-mir-1-1 | miRbase |
| Precursor accession: | MI0000651 | miRbase |
| Symbol: | MIR1-1 | HGNC |
| RefSeq ID: | NR_029780 | GenBank |
| Sequence: | UGGAAUGUAAAGAAGUAUGUAU |
Reported expression in cancers: hsa-miR-1-3p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-1-3p | bladder cancer | downregulation | "We investigated the miRNA expression signature of ......" | 21304530 | |
| hsa-miR-1-3p | bladder cancer | deregulation | "Expression levels of miRNAs were assessed by quant ......" | 23169479 | qPCR |
| hsa-miR-1-3p | bladder cancer | upregulation | "Hsa miR 1 downregulates long non coding RNA urothe ......" | 25015192 | |
| hsa-miR-1-3p | bladder cancer | deregulation | "In GBC miRNAs with tumor suppressor activity miR-1 ......" | 26855538 | |
| hsa-miR-1-3p | bladder cancer | deregulation | "Thus the current study focused on a comprehensive ......" | 27350368 | Microarray |
| hsa-miR-1-3p | breast cancer | downregulation | "In the present study we reported that hsa-miR-1 wa ......" | 26275461 | |
| hsa-miR-1-3p | colon cancer | downregulation | "Among the miRNAs demonstrating the largest fold up ......" | 21694772 | |
| hsa-miR-1-3p | colorectal cancer | downregulation | "Tumor suppressor miR 1 restrains epithelial mesenc ......" | 25196260 | |
| hsa-miR-1-3p | colorectal cancer | downregulation | "The potential value of miR 1 and miR 374b as bioma ......" | 26045793 | qPCR |
| hsa-miR-1-3p | colorectal cancer | downregulation | "The expression levels of miR-1 and -133b were rela ......" | 26980745 | |
| hsa-miR-1-3p | esophageal cancer | downregulation | "In the present study in order to determine the inv ......" | 25672418 | Microarray; qPCR |
| hsa-miR-1-3p | gastric cancer | upregulation | "miRNA expression was determined using a custom-des ......" | 22112324 | Microarray |
| hsa-miR-1-3p | gastric cancer | upregulation | "We previously identified five miRNAs miR-1 miR-20a ......" | 24122958 | Reverse transcription PCR; qPCR |
| hsa-miR-1-3p | head and neck cancer | downregulation | "Our studies of microRNA miRNA expression signature ......" | 27169691 | |
| hsa-miR-1-3p | kidney renal cell cancer | downregulation | "MiR 1 downregulation correlates with poor survival ......" | 26036633 | |
| hsa-miR-1-3p | liver cancer | downregulation | "The relationship between microRNA-1 miR-1 expressi ......" | 26554254 | qPCR |
| hsa-miR-1-3p | lung cancer | downregulation | "Here we report that micro-RNA-1 miR-1 abundant in ......" | 18818206 | |
| hsa-miR-1-3p | prostate cancer | downregulation | "Our recent analyses of miRNA expression signatures ......" | 22068816 | |
| hsa-miR-1-3p | prostate cancer | downregulation | "We previously reported that miR-1 is among the mos ......" | 22210864 | |
| hsa-miR-1-3p | prostate cancer | downregulation | "miR 1 and miR 133b are differentially expressed in ......" | 24967583 | Microarray; qPCR |
| hsa-miR-1-3p | sarcoma | upregulation | "MicroRNAs miRNAs are endogenous short approximatel ......" | 19710019 | |
| hsa-miR-1-3p | thyroid cancer | downregulation | "In this study we evaluated miRNA expression as a m ......" | 21537871 | Microarray |
Reported cancer pathway affected by hsa-miR-1-3p
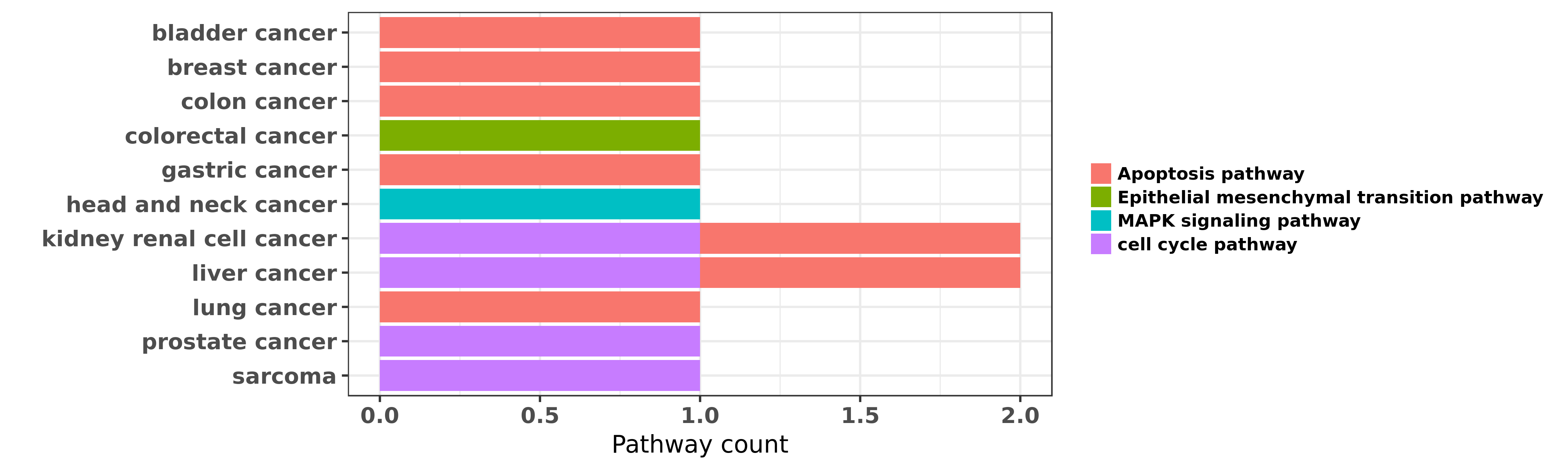
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-1-3p | bladder cancer | Apoptosis pathway | "The tumour suppressive function of miR 1 and miR 1 ......" | 21304530 | |
| hsa-miR-1-3p | bladder cancer | Apoptosis pathway | "We have previously found that restoration of tumor ......" | 22178073 | Luciferase |
| hsa-miR-1-3p | bladder cancer | Apoptosis pathway | "Hsa miR 1 downregulates long non coding RNA urothe ......" | 25015192 | |
| hsa-miR-1-3p | breast cancer | Apoptosis pathway | "Hsa miR 1 suppresses breast cancer development by ......" | 26275461 | |
| hsa-miR-1-3p | colon cancer | Apoptosis pathway | "The expression profiles of miRNAs were examined in ......" | 26459459 | MTT assay |
| hsa-miR-1-3p | colorectal cancer | Epithelial mesenchymal transition pathway | "Tumor suppressor miR 1 restrains epithelial mesenc ......" | 25196260 | |
| hsa-miR-1-3p | gastric cancer | Apoptosis pathway | "In order to identify miRNA signatures for gastric ......" | 22112324 | |
| hsa-miR-1-3p | head and neck cancer | MAPK signaling pathway | "Dual receptor EGFR and c MET inhibition by tumor s ......" | 27169691 | |
| hsa-miR-1-3p | kidney renal cell cancer | cell cycle pathway; Apoptosis pathway | "The functional significance of miR 1 and miR 133a ......" | 21745735 | Luciferase |
| hsa-miR-1-3p | kidney renal cell cancer | cell cycle pathway | "MiR 1 downregulation correlates with poor survival ......" | 26036633 | |
| hsa-miR-1-3p | liver cancer | cell cycle pathway; Apoptosis pathway | "miRNAs are small non-coding RNAs which target comp ......" | 24496037 | |
| hsa-miR-1-3p | liver cancer | Apoptosis pathway | "To investigate the effect of microRNA-1 miR-1 on t ......" | 26019452 | Flow cytometry; Transwell assay; MTT assay |
| hsa-miR-1-3p | lung cancer | Apoptosis pathway | "Down regulation of micro RNA 1 miR 1 in lung cance ......" | 18818206 | |
| hsa-miR-1-3p | prostate cancer | cell cycle pathway | "We previously reported that miR-1 is among the mos ......" | 22210864 | |
| hsa-miR-1-3p | sarcoma | cell cycle pathway | "Downregulation of microRNAs miR 1 206 and 29 stabi ......" | 22330340 | |
| hsa-miR-1-3p | sarcoma | cell cycle pathway | "miRNA expression profile in human osteosarcoma: ro ......" | 23229283 | |
| hsa-miR-1-3p | sarcoma | cell cycle pathway | "MicroRNA-1 miR-1 has been shown to function as a c ......" | 24969180 | Luciferase |
Reported cancer prognosis affected by hsa-miR-1-3p
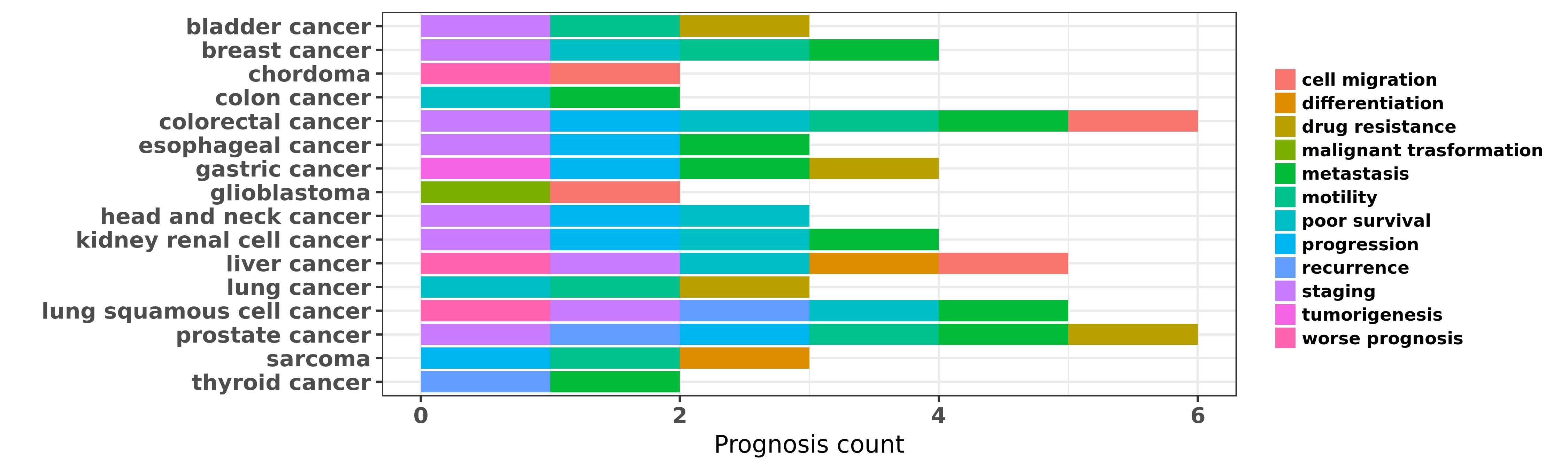
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-1-3p | bladder cancer | staging | "Expression levels of miRNAs were assessed by quant ......" | 23169479 | |
| hsa-miR-1-3p | bladder cancer | drug resistance | "To solve the problem we applied miRNA response ele ......" | 23442927 | Luciferase |
| hsa-miR-1-3p | bladder cancer | motility | "miR 1 and miR 145 act as tumor suppressor microRNA ......" | 24966896 | Colony formation |
| hsa-miR-1-3p | bladder cancer | motility | "Hsa miR 1 downregulates long non coding RNA urothe ......" | 25015192 | |
| hsa-miR-1-3p | breast cancer | motility; metastasis; poor survival | "Hsa miR 1 suppresses breast cancer development by ......" | 26275461 | |
| hsa-miR-1-3p | breast cancer | staging; metastasis; poor survival | "Abnormal expression of miR 1 in breast carcinoma a ......" | 26331797 | |
| hsa-miR-1-3p | chordoma | worse prognosis | "Prognostic significance of miRNA 1 miR 1 expressio ......" | 24501096 | Cell proliferation assay; Cell Proliferation Assay |
| hsa-miR-1-3p | chordoma | cell migration | "MicroRNA 1 miR 1 inhibits chordoma cell migration ......" | 24760686 | MTT assay; Western blot |
| hsa-miR-1-3p | colon cancer | metastasis | "MiR 1 downregulation cooperates with MACC1 in prom ......" | 22179665 | |
| hsa-miR-1-3p | colon cancer | poor survival | "The expression profiles of miRNAs were examined in ......" | 26459459 | MTT assay |
| hsa-miR-1-3p | colorectal cancer | staging | "Among the miRNAs we found to be dysregulated many ......" | 21734013 | |
| hsa-miR-1-3p | colorectal cancer | progression; motility | "miRNA profiling in colorectal cancer highlights mi ......" | 22343615 | |
| hsa-miR-1-3p | colorectal cancer | cell migration | "The miR 1 NOTCH3 Asef pathway is important for col ......" | 24244701 | |
| hsa-miR-1-3p | colorectal cancer | metastasis; progression | "Tumor suppressor miR 1 restrains epithelial mesenc ......" | 25196260 | |
| hsa-miR-1-3p | colorectal cancer | staging | "The potential value of miR 1 and miR 374b as bioma ......" | 26045793 | |
| hsa-miR-1-3p | colorectal cancer | poor survival | "Two miRNAs infrequently expressed in the populatio ......" | 27517623 | |
| hsa-miR-1-3p | esophageal cancer | progression; staging | "In the present study in order to determine the inv ......" | 25672418 | |
| hsa-miR-1-3p | esophageal cancer | metastasis; progression | "The tumor suppressive function of miR 1 by targeti ......" | 26414725 | Luciferase; Western blot |
| hsa-miR-1-3p | gastric cancer | drug resistance | "In order to identify miRNA signatures for gastric ......" | 22112324 | |
| hsa-miR-1-3p | gastric cancer | progression; tumorigenesis | "MicroRNA-206 miR-206 as a homolog of miR-1 plays i ......" | 23751352 | |
| hsa-miR-1-3p | gastric cancer | metastasis; progression; tumorigenesis | "Insights into roles of the miR 1 133 and 206 famil ......" | 27349337 | |
| hsa-miR-1-3p | glioblastoma | malignant trasformation | "Extracellular vesicles modulate the glioblastoma m ......" | 24310399 | |
| hsa-miR-1-3p | glioblastoma | cell migration | "MicroRNA-1 miR-1 expression is deregulated in GBM ......" | 25374033 | Flow cytometry; Cell migration assay |
| hsa-miR-1-3p | head and neck cancer | poor survival | "In this study miRNA expression profiles of head an ......" | 19179615 | |
| hsa-miR-1-3p | head and neck cancer | progression | "Several miRNAs miRLet-7A miR-1 miR-206 miR-153 miR ......" | 26239836 | |
| hsa-miR-1-3p | head and neck cancer | staging | "Dual receptor EGFR and c MET inhibition by tumor s ......" | 27169691 | |
| hsa-miR-1-3p | kidney renal cell cancer | metastasis; poor survival; staging; progression | "MiR 1 downregulation correlates with poor survival ......" | 26036633 | |
| hsa-miR-1-3p | liver cancer | staging; poor survival | "Here we investigated the prognostic potential of m ......" | 23810247 | |
| hsa-miR-1-3p | liver cancer | cell migration | "miRNAs are small non-coding RNAs which target comp ......" | 24496037 | |
| hsa-miR-1-3p | liver cancer | worse prognosis; staging; differentiation; poor survival | "The relationship between microRNA-1 miR-1 expressi ......" | 26554254 | |
| hsa-miR-1-3p | lung cancer | poor survival; motility; drug resistance | "Down regulation of micro RNA 1 miR 1 in lung cance ......" | 18818206 | |
| hsa-miR-1-3p | lung cancer | drug resistance | "mir 1 mediated paracrine effect of cancer associat ......" | 27035564 | |
| hsa-miR-1-3p | lung squamous cell cancer | staging; poor survival | "In the discovery stage Solexa sequencing followed ......" | 20194856 | |
| hsa-miR-1-3p | lung squamous cell cancer | worse prognosis; metastasis; recurrence | "Correlation between the expression levels of miR 1 ......" | 24328409 | Western blot |
| hsa-miR-1-3p | prostate cancer | metastasis; recurrence; progression; motility | "We previously reported that miR-1 is among the mos ......" | 22210864 | |
| hsa-miR-1-3p | prostate cancer | progression | "miR 1 and miR 133b are differentially expressed in ......" | 24967583 | |
| hsa-miR-1-3p | prostate cancer | drug resistance; staging | "Heat shock protein HSPB1 attenuates microRNA miR 1 ......" | 24982356 | Western blot |
| hsa-miR-1-3p | prostate cancer | metastasis | "MicroRNA 1 miR-1 levels are decreased in clinical ......" | 25802280 | |
| hsa-miR-1-3p | prostate cancer | metastasis | "EGF Receptor Promotes Prostate Cancer Bone Metasta ......" | 26071255 | |
| hsa-miR-1-3p | sarcoma | differentiation | "Distinct roles for miR 1 and miR 133a in the proli ......" | 20466878 | |
| hsa-miR-1-3p | sarcoma | differentiation | "In order to investigate the involvement of miRNAs ......" | 23133552 | |
| hsa-miR-1-3p | sarcoma | motility | "miRNA expression profile in human osteosarcoma: ro ......" | 23229283 | |
| hsa-miR-1-3p | sarcoma | progression; differentiation | "Therefore in this review we focus on the regulatio ......" | 24831881 | |
| hsa-miR-1-3p | sarcoma | progression | "MicroRNA-1 miR-1 has been shown to function as a c ......" | 24969180 | Luciferase |
| hsa-miR-1-3p | thyroid cancer | metastasis; recurrence | "In this study we evaluated miRNA expression as a m ......" | 21537871 |
Reported gene related to hsa-miR-1-3p
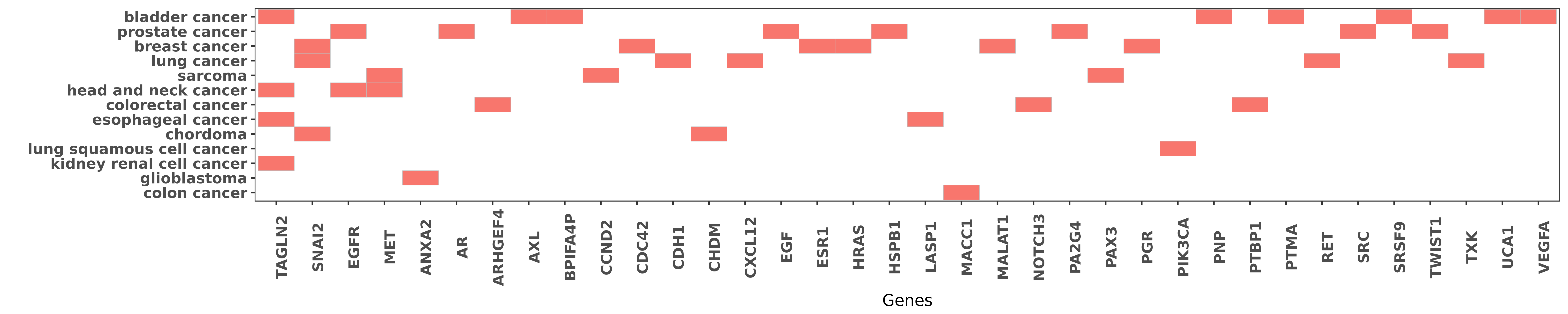
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-1-3p | bladder cancer | TAGLN2 | "The tumour suppressive function of miR 1 and miR 1 ......" | 21304530 |
| hsa-miR-1-3p | esophageal cancer | TAGLN2 | "The tumor suppressive function of miR 1 by targeti ......" | 26414725 |
| hsa-miR-1-3p | head and neck cancer | TAGLN2 | "miR 1 as a tumor suppressive microRNA targeting TA ......" | 21378409 |
| hsa-miR-1-3p | kidney renal cell cancer | TAGLN2 | "The expression level of TAGLN2 mRNA was significan ......" | 21745735 |
| hsa-miR-1-3p | prostate cancer | AR | "We show that AR binds to the miR-1-2 regulatory re ......" | 25802280 |
| hsa-miR-1-3p | prostate cancer | AR | "In this study an assumed regulation of Ebp1 by the ......" | 24798454 |
| hsa-miR-1-3p | prostate cancer | AR | "Shortened isoforms of the androgen receptor are re ......" | 24222130 |
| hsa-miR-1-3p | breast cancer | MALAT1 | "Hsa miR 1 suppresses breast cancer development by ......" | 26275461 |
| hsa-miR-1-3p | breast cancer | MALAT1 | "MALAT1 induced migration and invasion of human bre ......" | 26926567 |
| hsa-miR-1-3p | breast cancer | MALAT1 | "Reciprocal regulation of Hsa miR 1 and long noncod ......" | 26676637 |
| hsa-miR-1-3p | breast cancer | SNAI2 | "Slug was identified as a direct target of miR-1 ......" | 26676637 |
| hsa-miR-1-3p | chordoma | SNAI2 | "MicroRNA 1 miR 1 inhibits chordoma cell migration ......" | 24760686 |
| hsa-miR-1-3p | lung cancer | SNAI2 | "These structural and tumorigenic properties induce ......" | 23142026 |
| hsa-miR-1-3p | chordoma | CHDM | "MicroRNA 1 miR 1 inhibits chordoma cell migration ......" | 24760686 |
| hsa-miR-1-3p | chordoma | CHDM | "Prognostic significance of miRNA 1 miR 1 expressio ......" | 24501096 |
| hsa-miR-1-3p | head and neck cancer | EGFR | "Dual receptor EGFR and c MET inhibition by tumor s ......" | 27169691 |
| hsa-miR-1-3p | prostate cancer | EGFR | "In this study we demonstrated a role for EGFR tran ......" | 26071255 |
| hsa-miR-1-3p | prostate cancer | HSPB1 | "Shortened isoforms of the androgen receptor are re ......" | 24222130 |
| hsa-miR-1-3p | prostate cancer | HSPB1 | "Heat shock protein HSPB1 attenuates microRNA miR 1 ......" | 24982356 |
| hsa-miR-1-3p | head and neck cancer | MET | "Dual receptor EGFR and c MET inhibition by tumor s ......" | 27169691 |
| hsa-miR-1-3p | sarcoma | MET | "Moreover both EGCG and miR-1 mimic inhibited c-MET ......" | 26499783 |
| hsa-miR-1-3p | glioblastoma | ANXA2 | "The mRNA encoding Annexin A2 ANXA2 one of the most ......" | 24310399 |
| hsa-miR-1-3p | colorectal cancer | ARHGEF4 | "The miR 1 NOTCH3 Asef pathway is important for col ......" | 24244701 |
| hsa-miR-1-3p | bladder cancer | AXL | "The ectopic expression of miR-1 and miR-145 in NOZ ......" | 24966896 |
| hsa-miR-1-3p | bladder cancer | BPIFA4P | "On the base of the microRNA miRNA expression signa ......" | 21304530 |
| hsa-miR-1-3p | sarcoma | CCND2 | "Downregulation of microRNAs miR 1 206 and 29 stabi ......" | 22330340 |
| hsa-miR-1-3p | breast cancer | CDC42 | "MALAT1 induced migration and invasion of human bre ......" | 26926567 |
| hsa-miR-1-3p | lung cancer | CDH1 | "These structural and tumorigenic properties induce ......" | 23142026 |
| hsa-miR-1-3p | lung cancer | CXCL12 | "Moreover we also confirmed that the expression lev ......" | 27035564 |
| hsa-miR-1-3p | prostate cancer | EGF | "EGF Receptor Promotes Prostate Cancer Bone Metasta ......" | 26071255 |
| hsa-miR-1-3p | breast cancer | ESR1 | "miR-1 ISH status was significantly associated with ......" | 26331797 |
| hsa-miR-1-3p | breast cancer | HRAS | "Hsa miR 1 suppresses breast cancer development by ......" | 26275461 |
| hsa-miR-1-3p | esophageal cancer | LASP1 | "The tumor suppressive function of miR 1 by targeti ......" | 26414725 |
| hsa-miR-1-3p | colon cancer | MACC1 | "MiR 1 downregulation cooperates with MACC1 in prom ......" | 22179665 |
| hsa-miR-1-3p | colorectal cancer | NOTCH3 | "The miR 1 NOTCH3 Asef pathway is important for col ......" | 24244701 |
| hsa-miR-1-3p | prostate cancer | PA2G4 | "In this study an assumed regulation of Ebp1 by the ......" | 24798454 |
| hsa-miR-1-3p | sarcoma | PAX3 | "Downregulation of microRNAs miR 1 206 and 29 stabi ......" | 22330340 |
| hsa-miR-1-3p | breast cancer | PGR | "miR-1 ISH status was significantly associated with ......" | 26331797 |
| hsa-miR-1-3p | lung squamous cell cancer | PIK3CA | "Correlation between the expression levels of miR 1 ......" | 24328409 |
| hsa-miR-1-3p | bladder cancer | PNP | "PTMA and PNP were identified as new target genes r ......" | 22378464 |
| hsa-miR-1-3p | colorectal cancer | PTBP1 | "Our results suggested that PTBP1 and PTBP1-associa ......" | 26980745 |
| hsa-miR-1-3p | bladder cancer | PTMA | "PTMA and PNP were identified as new target genes r ......" | 22378464 |
| hsa-miR-1-3p | lung cancer | RET | "Exogenous miR-1 significantly reduced expression o ......" | 18818206 |
| hsa-miR-1-3p | prostate cancer | SRC | "SRC has been implicated as a critical factor in bo ......" | 25802280 |
| hsa-miR-1-3p | bladder cancer | SRSF9 | "In this study we focused on splicing factor serine ......" | 22178073 |
| hsa-miR-1-3p | prostate cancer | TWIST1 | "EGF Receptor Promotes Prostate Cancer Bone Metasta ......" | 26071255 |
| hsa-miR-1-3p | lung cancer | TXK | "Exogenous miR-1 significantly reduced expression o ......" | 18818206 |
| hsa-miR-1-3p | bladder cancer | UCA1 | "Here we report that downregulated hsa-miR-1 and up ......" | 25015192 |
| hsa-miR-1-3p | bladder cancer | VEGFA | "The ectopic expression of miR-1 and miR-145 in NOZ ......" | 24966896 |
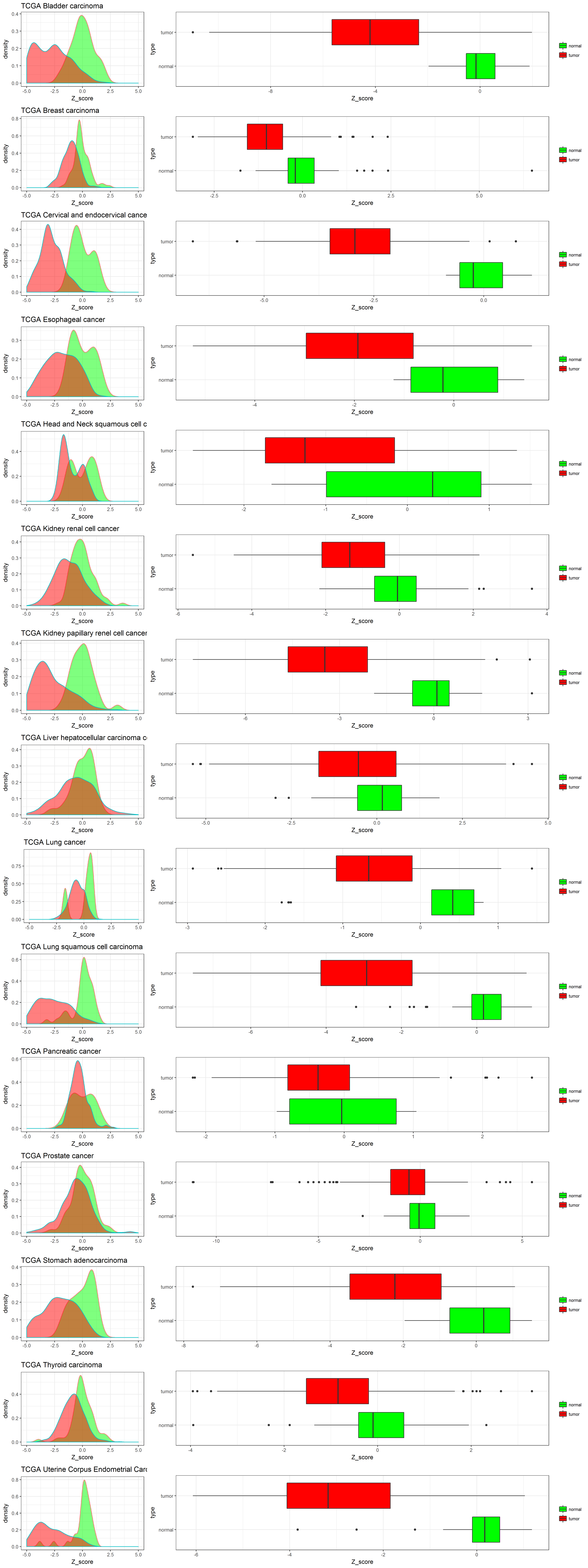
Expression profile in cancer corhorts:
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-1-3p | PNP | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; LGG; LUAD; SARC; STAD; UCEC | miRTarBase | TCGA BLCA -0.053; TCGA BRCA -0.103; TCGA CESC -0.082; TCGA COAD -0.126; TCGA ESCA -0.085; TCGA LGG -0.096; TCGA LUAD -0.099; TCGA SARC -0.082; TCGA STAD -0.104; TCGA UCEC -0.096 |
hsa-miR-1-3p | HSPD1 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; LIHC; LUAD; LUSC; PRAD; STAD; UCEC | miRTarBase; MirTarget | TCGA BLCA -0.074; TCGA BRCA -0.096; TCGA CESC -0.112; TCGA COAD -0.066; TCGA ESCA -0.129; TCGA LIHC -0.086; TCGA LUAD -0.182; TCGA LUSC -0.201; TCGA PRAD -0.214; TCGA STAD -0.216; TCGA UCEC -0.085 |
hsa-miR-1-3p | HSPA4 | 9 cancers: BLCA; BRCA; CESC; ESCA; LUAD; LUSC; PRAD; STAD; UCEC | miRTarBase | TCGA BLCA -0.057; TCGA BRCA -0.071; TCGA CESC -0.059; TCGA ESCA -0.058; TCGA LUAD -0.057; TCGA LUSC -0.092; TCGA PRAD -0.108; TCGA STAD -0.075; TCGA UCEC -0.084 |
hsa-miR-1-3p | AP1S1 | 11 cancers: BLCA; BRCA; ESCA; KIRP; LIHC; LUSC; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.09; TCGA BRCA -0.181; TCGA ESCA -0.135; TCGA KIRP -0.072; TCGA LIHC -0.095; TCGA LUSC -0.142; TCGA PRAD -0.072; TCGA SARC -0.079; TCGA THCA -0.09; TCGA STAD -0.125; TCGA UCEC -0.066 |
hsa-miR-1-3p | VEGFA | 13 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; PAAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.203; TCGA BRCA -0.08; TCGA COAD -0.116; TCGA ESCA -0.177; TCGA HNSC -0.072; TCGA KIRC -0.377; TCGA LIHC -0.078; TCGA LUAD -0.173; TCGA LUSC -0.104; TCGA PAAD -0.191; TCGA SARC -0.061; TCGA STAD -0.16; TCGA UCEC -0.063 |
hsa-miR-1-3p | TWF1 | 9 cancers: BLCA; COAD; ESCA; LGG; LUAD; LUSC; PRAD; THCA; STAD | MirTarget | TCGA BLCA -0.057; TCGA COAD -0.079; TCGA ESCA -0.068; TCGA LGG -0.072; TCGA LUAD -0.109; TCGA LUSC -0.146; TCGA PRAD -0.087; TCGA THCA -0.06; TCGA STAD -0.103 |
hsa-miR-1-3p | NETO2 | 9 cancers: BLCA; BRCA; ESCA; KIRC; LUAD; LUSC; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.159; TCGA BRCA -0.134; TCGA ESCA -0.131; TCGA KIRC -0.308; TCGA LUAD -0.191; TCGA LUSC -0.18; TCGA PRAD -0.341; TCGA STAD -0.184; TCGA UCEC -0.123 |
hsa-miR-1-3p | RAD54B | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.152; TCGA BRCA -0.213; TCGA CESC -0.084; TCGA COAD -0.106; TCGA ESCA -0.153; TCGA HNSC -0.079; TCGA KIRC -0.134; TCGA KIRP -0.066; TCGA LIHC -0.113; TCGA LUAD -0.258; TCGA LUSC -0.302; TCGA PRAD -0.264; TCGA STAD -0.23; TCGA UCEC -0.111 |
hsa-miR-1-3p | FAM91A1 | 9 cancers: BLCA; BRCA; ESCA; LUAD; LUSC; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.055; TCGA BRCA -0.075; TCGA ESCA -0.097; TCGA LUAD -0.1; TCGA LUSC -0.109; TCGA PRAD -0.192; TCGA SARC -0.069; TCGA STAD -0.176; TCGA UCEC -0.053 |
hsa-miR-1-3p | NXT2 | 10 cancers: BLCA; ESCA; LGG; LIHC; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.063; TCGA ESCA -0.065; TCGA LGG -0.051; TCGA LIHC -0.057; TCGA LUSC -0.1; TCGA PAAD -0.08; TCGA PRAD -0.08; TCGA SARC -0.091; TCGA STAD -0.175; TCGA UCEC -0.078 |
hsa-miR-1-3p | HOOK1 | 10 cancers: BLCA; BRCA; COAD; KIRP; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.177; TCGA BRCA -0.224; TCGA COAD -0.055; TCGA KIRP -0.096; TCGA LUAD -0.115; TCGA LUSC -0.065; TCGA PAAD -0.103; TCGA PRAD -0.174; TCGA STAD -0.251; TCGA UCEC -0.14 |
hsa-miR-1-3p | WWC1 | 10 cancers: BLCA; BRCA; CESC; COAD; KIRP; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.153; TCGA BRCA -0.155; TCGA CESC -0.273; TCGA COAD -0.055; TCGA KIRP -0.071; TCGA PAAD -0.13; TCGA PRAD -0.103; TCGA SARC -0.138; TCGA STAD -0.166; TCGA UCEC -0.12 |
hsa-miR-1-3p | E2F5 | 11 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LGG; LIHC; LUAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.088; TCGA BRCA -0.198; TCGA CESC -0.151; TCGA ESCA -0.134; TCGA HNSC -0.06; TCGA LGG -0.14; TCGA LIHC -0.106; TCGA LUAD -0.133; TCGA PRAD -0.323; TCGA STAD -0.146; TCGA UCEC -0.087 |
hsa-miR-1-3p | SOX9 | 10 cancers: BLCA; BRCA; ESCA; KIRC; KIRP; LGG; LUSC; PAAD; SARC; STAD | MirTarget | TCGA BLCA -0.105; TCGA BRCA -0.08; TCGA ESCA -0.18; TCGA KIRC -0.106; TCGA KIRP -0.262; TCGA LGG -0.145; TCGA LUSC -0.205; TCGA PAAD -0.149; TCGA SARC -0.16; TCGA STAD -0.242 |
hsa-miR-1-3p | HSP90B1 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.057; TCGA BRCA -0.085; TCGA CESC -0.077; TCGA COAD -0.091; TCGA ESCA -0.084; TCGA LUAD -0.138; TCGA LUSC -0.139; TCGA PAAD -0.071; TCGA PRAD -0.079; TCGA SARC -0.06; TCGA STAD -0.12; TCGA UCEC -0.085 |
hsa-miR-1-3p | PHF6 | 9 cancers: BLCA; BRCA; ESCA; LGG; LUAD; LUSC; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.092; TCGA BRCA -0.052; TCGA ESCA -0.055; TCGA LGG -0.055; TCGA LUAD -0.085; TCGA LUSC -0.119; TCGA PRAD -0.22; TCGA STAD -0.146; TCGA UCEC -0.057 |
hsa-miR-1-3p | LRRC59 | 11 cancers: BRCA; CESC; COAD; ESCA; LGG; LIHC; LUAD; LUSC; PRAD; SARC; STAD | MirTarget | TCGA BRCA -0.157; TCGA CESC -0.13; TCGA COAD -0.081; TCGA ESCA -0.116; TCGA LGG -0.052; TCGA LIHC -0.057; TCGA LUAD -0.116; TCGA LUSC -0.102; TCGA PRAD -0.104; TCGA SARC -0.096; TCGA STAD -0.121 |
Enriched cancer pathways of putative targets