microRNA information: hsa-miR-127-3p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-127-3p | miRbase |
| Accession: | MIMAT0000446 | miRbase |
| Precursor name: | hsa-mir-127 | miRbase |
| Precursor accession: | MI0000472 | miRbase |
| Symbol: | MIR127 | HGNC |
| RefSeq ID: | NR_029696 | GenBank |
| Sequence: | UCGGAUCCGUCUGAGCUUGGCU |
Reported expression in cancers: hsa-miR-127-3p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-127-3p | breast cancer | downregulation | "Prognostic and biological significance of microRNA ......" | 25477702 | Reverse transcription PCR |
| hsa-miR-127-3p | esophageal cancer | downregulation | "Quantitative RT-PCR qRT-PCR was used to compare mi ......" | 27645894 | qPCR |
| hsa-miR-127-3p | gastric cancer | downregulation | "The Tumor Suppressor Roles of miR 433 and miR 127 ......" | 23880861 | |
| hsa-miR-127-3p | glioblastoma | downregulation | "In this study using next generation sequencing ana ......" | 24517116 | RNA-Seq |
| hsa-miR-127-3p | ovarian cancer | downregulation | "In the present study the tumor-suppressive role of ......" | 27571744 | qPCR |
| hsa-miR-127-3p | pancreatic cancer | downregulation | "MicroRNA 127 is aberrantly downregulated and acted ......" | 27571739 | |
| hsa-miR-127-3p | sarcoma | downregulation | "MiR-127-3p was found at reduced levels in OS tissu ......" | 26707641 |
Reported cancer pathway affected by hsa-miR-127-3p
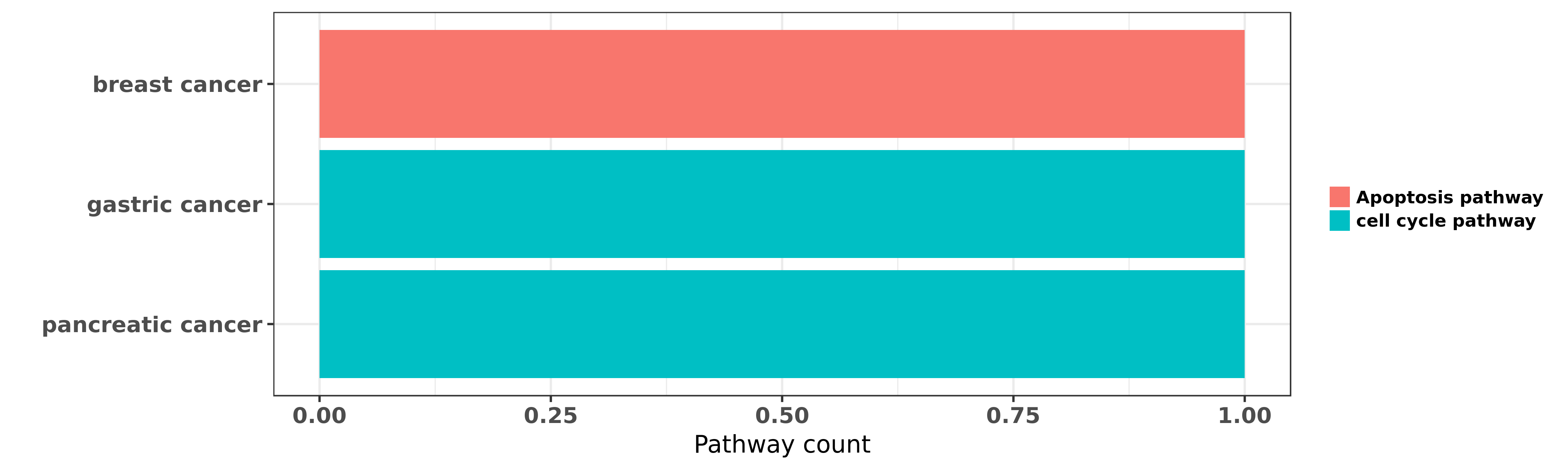
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-127-3p | breast cancer | Apoptosis pathway | "Prognostic and biological significance of microRNA ......" | 25477702 | |
| hsa-miR-127-3p | gastric cancer | cell cycle pathway | "The Tumor Suppressor Roles of miR 433 and miR 127 ......" | 23880861 | |
| hsa-miR-127-3p | pancreatic cancer | cell cycle pathway | "MicroRNA 127 is aberrantly downregulated and acted ......" | 27571739 | Luciferase |
Reported cancer prognosis affected by hsa-miR-127-3p
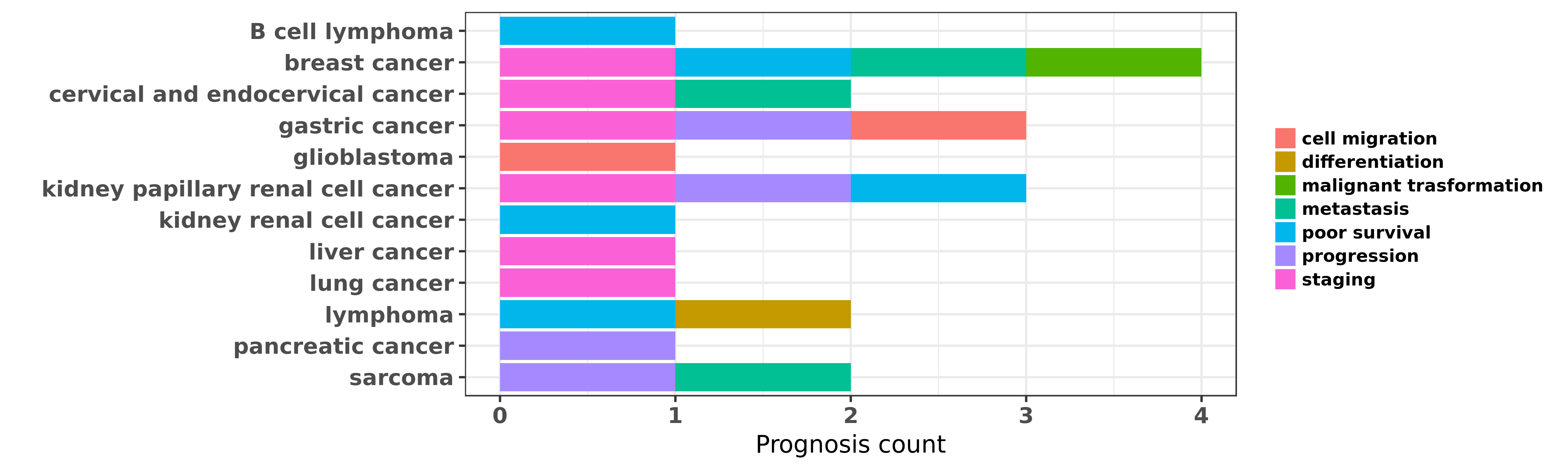
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-127-3p | B cell lymphoma | poor survival | "Finally eight miRNAs were found to correlate with ......" | 18537969 | |
| hsa-miR-127-3p | breast cancer | poor survival | "To investigate the global expression profile of mi ......" | 18812439 | |
| hsa-miR-127-3p | breast cancer | metastasis | "MicroRNA 127 is downregulated by Tudor SN protein ......" | 24155205 | |
| hsa-miR-127-3p | breast cancer | staging | "Circulating miR-127-3p miR-376a and miR-652 select ......" | 24194846 | |
| hsa-miR-127-3p | breast cancer | malignant trasformation; staging; metastasis; poor survival | "Prognostic and biological significance of microRNA ......" | 25477702 | |
| hsa-miR-127-3p | cervical and endocervical cancer | staging; metastasis | "In this study we profiled miRNA expression in 10 e ......" | 18451214 | |
| hsa-miR-127-3p | gastric cancer | staging; progression; cell migration | "The Tumor Suppressor Roles of miR 433 and miR 127 ......" | 23880861 | |
| hsa-miR-127-3p | glioblastoma | cell migration | "MicroRNA 127 3p promotes glioblastoma cell migrati ......" | 24604520 | |
| hsa-miR-127-3p | kidney papillary renal cell cancer | staging; poor survival; progression | "In the training stage the expression levels of 12 ......" | 25906110 | |
| hsa-miR-127-3p | kidney renal cell cancer | poor survival | "Global miRNA expression profiles were obtained by ......" | 22492545 | |
| hsa-miR-127-3p | liver cancer | staging | "MicroRNA 127 post transcriptionally downregulates ......" | 24854842 | Western blot; Luciferase |
| hsa-miR-127-3p | lung cancer | staging | "Individuals with the highest levels of methylated ......" | 24665010 | |
| hsa-miR-127-3p | lymphoma | differentiation | "Here we investigated the expression of specific mi ......" | 19530237 | |
| hsa-miR-127-3p | lymphoma | differentiation | "Epstein Barr nuclear antigen 1 induces expression ......" | 22941339 | |
| hsa-miR-127-3p | lymphoma | poor survival | "Fourteen miRs were differentially expressed betwee ......" | 23835716 | |
| hsa-miR-127-3p | pancreatic cancer | progression | "MicroRNA 127 is aberrantly downregulated and acted ......" | 27571739 | Luciferase |
| hsa-miR-127-3p | sarcoma | metastasis; progression | "MicroRNA 127 3p inhibits proliferation and invasio ......" | 26707641 |
Reported gene related to hsa-miR-127-3p
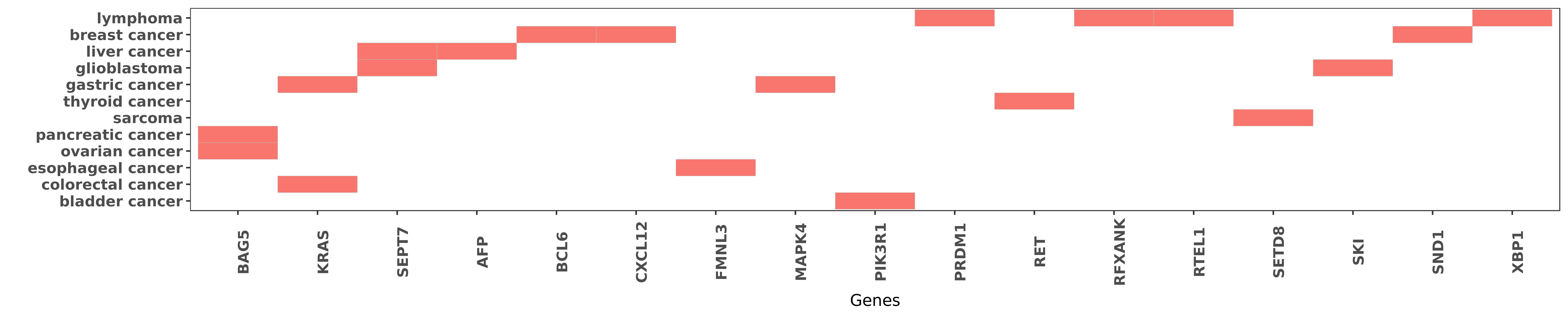
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-127-3p | ovarian cancer | BAG5 | "MicroRNA 127 3p acts as a tumor suppressor in epit ......" | 27571744 |
| hsa-miR-127-3p | pancreatic cancer | BAG5 | "Dual-luciferase reporter assay and qRT-PCR were pe ......" | 27571739 |
| hsa-miR-127-3p | colorectal cancer | KRAS | "Increased expression of miR-127-3p and miR-92a in ......" | 21922590 |
| hsa-miR-127-3p | gastric cancer | KRAS | "Furthermore the ectopic expression of miR-433 and ......" | 23880861 |
| hsa-miR-127-3p | glioblastoma | SEPT7 | "MicroRNA 127 3p promotes glioblastoma cell migrati ......" | 24604520 |
| hsa-miR-127-3p | liver cancer | SEPT7 | "MicroRNA 127 post transcriptionally downregulates ......" | 24854842 |
| hsa-miR-127-3p | liver cancer | AFP | "MiR-127 is downregulated in 69.7% of HCC tissues c ......" | 24854842 |
| hsa-miR-127-3p | breast cancer | BCL6 | "Functional analyses showed that upregulation of mi ......" | 25477702 |
| hsa-miR-127-3p | breast cancer | CXCL12 | "Analyses of miRNA expression profiles identified n ......" | 21343399 |
| hsa-miR-127-3p | esophageal cancer | FMNL3 | "MicroRNA 127 is a tumor suppressor in human esopha ......" | 27645894 |
| hsa-miR-127-3p | gastric cancer | MAPK4 | "Furthermore the ectopic expression of miR-433 and ......" | 23880861 |
| hsa-miR-127-3p | bladder cancer | PIK3R1 | "Integrated gene network analysis and text mining r ......" | 24004856 |
| hsa-miR-127-3p | lymphoma | PRDM1 | "In addition we found evidence that hsa-miR-127 is ......" | 19530237 |
| hsa-miR-127-3p | thyroid cancer | RET | "Significantly lower miR-127 levels were observed i ......" | 22747440 |
| hsa-miR-127-3p | lymphoma | RFXANK | "Here we investigated the expression of specific mi ......" | 19530237 |
| hsa-miR-127-3p | lymphoma | RTEL1 | "Fourteen miRs were differentially expressed betwee ......" | 23835716 |
| hsa-miR-127-3p | sarcoma | SETD8 | "MicroRNA 127 3p inhibits proliferation and invasio ......" | 26707641 |
| hsa-miR-127-3p | glioblastoma | SKI | "Next generation sequencing analysis of miRNAs: MiR ......" | 24517116 |
| hsa-miR-127-3p | breast cancer | SND1 | "MicroRNA 127 is downregulated by Tudor SN protein ......" | 24155205 |
| hsa-miR-127-3p | lymphoma | XBP1 | "In addition we found evidence that hsa-miR-127 is ......" | 19530237 |
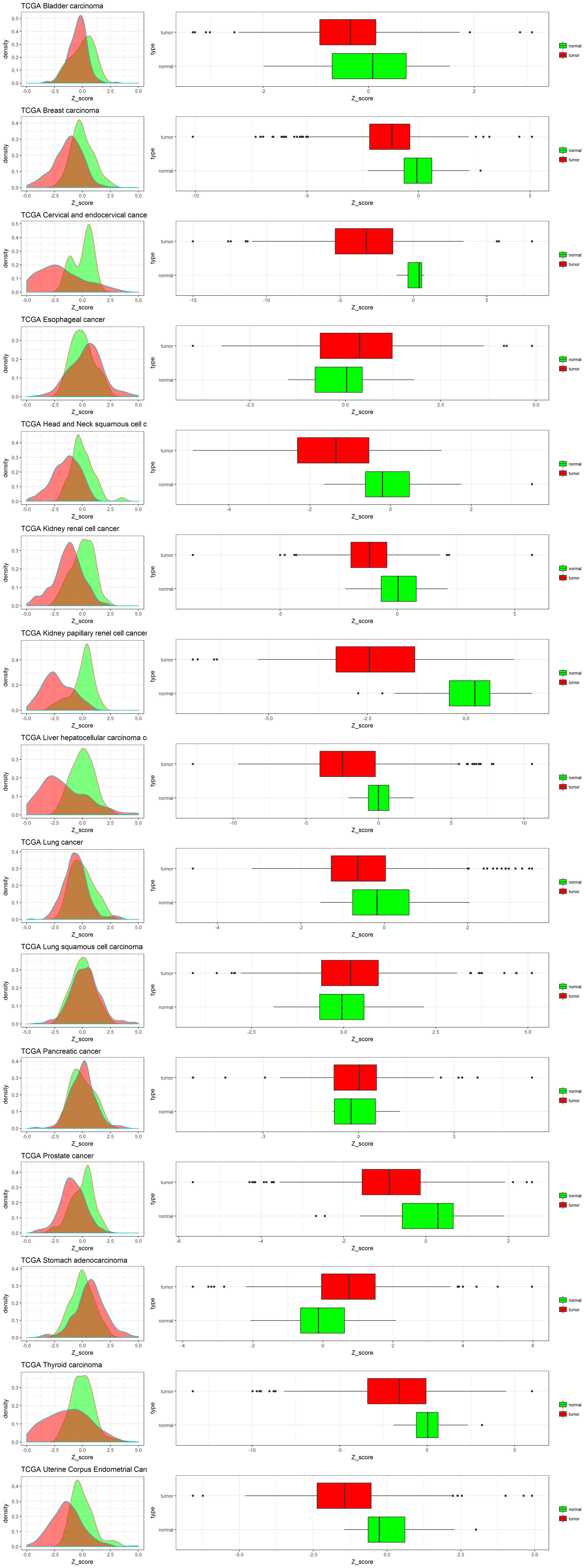
Expression profile in cancer corhorts:
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-127-3p | MTCP1 | 11 cancers: BLCA; BRCA; HNSC; KIRC; KIRP; LGG; LIHC; LUSC; OV; PAAD; SARC | PITA; miRanda | TCGA BLCA -0.162; TCGA BRCA -0.082; TCGA HNSC -0.094; TCGA KIRC -0.334; TCGA KIRP -0.238; TCGA LGG -0.06; TCGA LIHC -0.082; TCGA LUSC -0.103; TCGA OV -0.093; TCGA PAAD -0.111; TCGA SARC -0.145 |
hsa-miR-127-3p | BSG | 9 cancers: BLCA; BRCA; COAD; HNSC; KIRP; OV; PAAD; PRAD; THCA | miRanda | TCGA BLCA -0.098; TCGA BRCA -0.169; TCGA COAD -0.091; TCGA HNSC -0.068; TCGA KIRP -0.085; TCGA OV -0.061; TCGA PAAD -0.182; TCGA PRAD -0.12; TCGA THCA -0.07 |
hsa-miR-127-3p | TARBP2 | 10 cancers: BLCA; BRCA; HNSC; KIRP; LGG; LUAD; OV; PAAD; PRAD; UCEC | miRanda | TCGA BLCA -0.087; TCGA BRCA -0.114; TCGA HNSC -0.115; TCGA KIRP -0.086; TCGA LGG -0.073; TCGA LUAD -0.054; TCGA OV -0.094; TCGA PAAD -0.122; TCGA PRAD -0.285; TCGA UCEC -0.051 |
hsa-miR-127-3p | CUL9 | 10 cancers: BLCA; BRCA; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; STAD | miRanda | TCGA BLCA -0.126; TCGA BRCA -0.058; TCGA ESCA -0.141; TCGA HNSC -0.108; TCGA LIHC -0.082; TCGA LUAD -0.066; TCGA LUSC -0.125; TCGA PAAD -0.082; TCGA PRAD -0.135; TCGA STAD -0.104 |
hsa-miR-127-3p | POLR2H | 11 cancers: BLCA; BRCA; HNSC; KIRC; KIRP; LGG; LUAD; OV; PAAD; PRAD; UCEC | miRanda | TCGA BLCA -0.086; TCGA BRCA -0.173; TCGA HNSC -0.194; TCGA KIRC -0.087; TCGA KIRP -0.068; TCGA LGG -0.07; TCGA LUAD -0.084; TCGA OV -0.107; TCGA PAAD -0.21; TCGA PRAD -0.325; TCGA UCEC -0.086 |
hsa-miR-127-3p | MTHFD2L | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LGG; LUAD; LUSC; PRAD; SARC; STAD; UCEC | miRanda | TCGA BLCA -0.093; TCGA BRCA -0.105; TCGA COAD -0.154; TCGA ESCA -0.169; TCGA HNSC -0.08; TCGA LGG -0.074; TCGA LUAD -0.057; TCGA LUSC -0.078; TCGA PRAD -0.342; TCGA SARC -0.108; TCGA STAD -0.179; TCGA UCEC -0.053 |
hsa-miR-127-3p | ZNF7 | 12 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; LGG; LUAD; OV; PAAD; PRAD | miRanda | TCGA BLCA -0.064; TCGA BRCA -0.13; TCGA CESC -0.064; TCGA COAD -0.147; TCGA HNSC -0.069; TCGA KIRC -0.072; TCGA KIRP -0.053; TCGA LGG -0.069; TCGA LUAD -0.093; TCGA OV -0.086; TCGA PAAD -0.065; TCGA PRAD -0.207 |
hsa-miR-127-3p | ZNF232 | 9 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRP; LGG; PRAD; STAD | miRanda | TCGA BLCA -0.077; TCGA BRCA -0.099; TCGA CESC -0.091; TCGA COAD -0.219; TCGA HNSC -0.086; TCGA KIRP -0.059; TCGA LGG -0.089; TCGA PRAD -0.131; TCGA STAD -0.09 |
hsa-miR-127-3p | FAM98C | 13 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; STAD | miRanda | TCGA BLCA -0.1; TCGA BRCA -0.144; TCGA COAD -0.12; TCGA ESCA -0.121; TCGA HNSC -0.127; TCGA KIRC -0.097; TCGA KIRP -0.055; TCGA LIHC -0.072; TCGA LUAD -0.092; TCGA LUSC -0.125; TCGA PAAD -0.145; TCGA PRAD -0.26; TCGA STAD -0.12 |
hsa-miR-127-3p | HMG20B | 11 cancers: BLCA; BRCA; HNSC; KIRP; LGG; LIHC; OV; PAAD; PRAD; SARC; STAD | miRanda | TCGA BLCA -0.083; TCGA BRCA -0.156; TCGA HNSC -0.108; TCGA KIRP -0.095; TCGA LGG -0.134; TCGA LIHC -0.064; TCGA OV -0.088; TCGA PAAD -0.241; TCGA PRAD -0.308; TCGA SARC -0.061; TCGA STAD -0.068 |
hsa-miR-127-3p | BOLA1 | 14 cancers: BLCA; BRCA; COAD; KIRC; LGG; LIHC; LUAD; LUSC; OV; PAAD; PRAD; THCA; STAD; UCEC | miRanda | TCGA BLCA -0.091; TCGA BRCA -0.245; TCGA COAD -0.111; TCGA KIRC -0.067; TCGA LGG -0.081; TCGA LIHC -0.141; TCGA LUAD -0.074; TCGA LUSC -0.12; TCGA OV -0.116; TCGA PAAD -0.186; TCGA PRAD -0.187; TCGA THCA -0.062; TCGA STAD -0.107; TCGA UCEC -0.058 |
hsa-miR-127-3p | HMGCL | 10 cancers: BLCA; BRCA; COAD; ESCA; OV; PAAD; PRAD; THCA; STAD; UCEC | miRanda | TCGA BLCA -0.067; TCGA BRCA -0.066; TCGA COAD -0.092; TCGA ESCA -0.167; TCGA OV -0.168; TCGA PAAD -0.07; TCGA PRAD -0.146; TCGA THCA -0.058; TCGA STAD -0.097; TCGA UCEC -0.084 |
hsa-miR-127-3p | HINT2 | 10 cancers: BLCA; BRCA; CESC; LGG; LIHC; LUAD; LUSC; OV; PRAD; STAD | miRanda | TCGA BLCA -0.059; TCGA BRCA -0.119; TCGA CESC -0.091; TCGA LGG -0.073; TCGA LIHC -0.106; TCGA LUAD -0.065; TCGA LUSC -0.101; TCGA OV -0.165; TCGA PRAD -0.229; TCGA STAD -0.095 |
hsa-miR-127-3p | PTK6 | 10 cancers: BLCA; BRCA; KIRC; KIRP; LGG; LUAD; PAAD; PRAD; SARC; UCEC | miRanda | TCGA BLCA -0.229; TCGA BRCA -0.191; TCGA KIRC -0.191; TCGA KIRP -0.236; TCGA LGG -0.169; TCGA LUAD -0.172; TCGA PAAD -0.514; TCGA PRAD -0.298; TCGA SARC -0.05; TCGA UCEC -0.298 |
hsa-miR-127-3p | KIAA0907 | 17 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; OV; PAAD; PRAD; SARC; STAD; UCEC | miRanda | TCGA BLCA -0.206; TCGA BRCA -0.189; TCGA CESC -0.085; TCGA COAD -0.11; TCGA ESCA -0.172; TCGA HNSC -0.151; TCGA KIRC -0.172; TCGA KIRP -0.149; TCGA LGG -0.084; TCGA LUAD -0.186; TCGA LUSC -0.154; TCGA OV -0.107; TCGA PAAD -0.206; TCGA PRAD -0.326; TCGA SARC -0.053; TCGA STAD -0.104; TCGA UCEC -0.061 |
hsa-miR-127-3p | RAD52 | 10 cancers: BLCA; BRCA; COAD; KIRC; KIRP; LGG; LUAD; LUSC; PRAD; STAD | miRanda | TCGA BLCA -0.083; TCGA BRCA -0.076; TCGA COAD -0.122; TCGA KIRC -0.083; TCGA KIRP -0.184; TCGA LGG -0.108; TCGA LUAD -0.087; TCGA LUSC -0.079; TCGA PRAD -0.082; TCGA STAD -0.08 |
hsa-miR-127-3p | KLHL17 | 12 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; PAAD; PRAD; UCEC | miRanda | TCGA BLCA -0.09; TCGA BRCA -0.187; TCGA CESC -0.115; TCGA HNSC -0.225; TCGA KIRC -0.116; TCGA KIRP -0.187; TCGA LGG -0.108; TCGA LIHC -0.076; TCGA LUAD -0.131; TCGA PAAD -0.292; TCGA PRAD -0.465; TCGA UCEC -0.131 |
hsa-miR-127-3p | RAD9A | 13 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; OV; PAAD; PRAD; THCA | miRanda; miRNATAP | TCGA BLCA -0.124; TCGA BRCA -0.205; TCGA CESC -0.121; TCGA HNSC -0.224; TCGA KIRC -0.117; TCGA KIRP -0.204; TCGA LGG -0.095; TCGA LUAD -0.064; TCGA LUSC -0.069; TCGA OV -0.107; TCGA PAAD -0.206; TCGA PRAD -0.288; TCGA THCA -0.081 |
hsa-miR-127-3p | NEDD4L | 10 cancers: BLCA; BRCA; COAD; ESCA; LIHC; LUAD; LUSC; PRAD; STAD; UCEC | miRanda | TCGA BLCA -0.092; TCGA BRCA -0.081; TCGA COAD -0.158; TCGA ESCA -0.167; TCGA LIHC -0.103; TCGA LUAD -0.115; TCGA LUSC -0.172; TCGA PRAD -0.264; TCGA STAD -0.221; TCGA UCEC -0.074 |
hsa-miR-127-3p | TTC14 | 10 cancers: BLCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; PRAD; SARC; THCA | miRanda | TCGA BLCA -0.131; TCGA HNSC -0.056; TCGA KIRC -0.089; TCGA KIRP -0.217; TCGA LGG -0.068; TCGA LUAD -0.076; TCGA LUSC -0.15; TCGA PRAD -0.164; TCGA SARC -0.057; TCGA THCA -0.066 |
hsa-miR-127-3p | LUC7L | 11 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; PAAD; PRAD | miRNATAP | TCGA BLCA -0.108; TCGA BRCA -0.067; TCGA CESC -0.051; TCGA HNSC -0.157; TCGA KIRC -0.118; TCGA KIRP -0.288; TCGA LGG -0.114; TCGA LUAD -0.095; TCGA LUSC -0.075; TCGA PAAD -0.13; TCGA PRAD -0.371 |
hsa-miR-127-3p | CCDC40 | 9 cancers: BRCA; KIRC; KIRP; LGG; LIHC; PRAD; THCA; STAD; UCEC | miRanda | TCGA BRCA -0.094; TCGA KIRC -0.116; TCGA KIRP -0.215; TCGA LGG -0.091; TCGA LIHC -0.174; TCGA PRAD -0.319; TCGA THCA -0.068; TCGA STAD -0.141; TCGA UCEC -0.109 |
hsa-miR-127-3p | ELL3 | 10 cancers: BRCA; ESCA; HNSC; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | miRanda | TCGA BRCA -0.207; TCGA ESCA -0.132; TCGA HNSC -0.098; TCGA LUAD -0.14; TCGA LUSC -0.106; TCGA OV -0.089; TCGA PAAD -0.235; TCGA PRAD -0.25; TCGA STAD -0.149; TCGA UCEC -0.081 |
hsa-miR-127-3p | FBXO43 | 11 cancers: BRCA; COAD; HNSC; KIRC; KIRP; LGG; LIHC; OV; PAAD; PRAD; SARC | miRanda | TCGA BRCA -0.398; TCGA COAD -0.229; TCGA HNSC -0.205; TCGA KIRC -0.175; TCGA KIRP -0.215; TCGA LGG -0.14; TCGA LIHC -0.155; TCGA OV -0.156; TCGA PAAD -0.154; TCGA PRAD -0.402; TCGA SARC -0.118 |
hsa-miR-127-3p | CENPA | 9 cancers: BRCA; CESC; COAD; HNSC; KIRC; LGG; PAAD; PRAD; UCEC | miRanda | TCGA BRCA -0.437; TCGA CESC -0.12; TCGA COAD -0.144; TCGA HNSC -0.269; TCGA KIRC -0.164; TCGA LGG -0.111; TCGA PAAD -0.292; TCGA PRAD -0.505; TCGA UCEC -0.162 |
hsa-miR-127-3p | AP4B1 | 9 cancers: BRCA; ESCA; HNSC; LIHC; OV; PAAD; PRAD; STAD; UCEC | miRanda | TCGA BRCA -0.069; TCGA ESCA -0.105; TCGA HNSC -0.062; TCGA LIHC -0.059; TCGA OV -0.058; TCGA PAAD -0.121; TCGA PRAD -0.134; TCGA STAD -0.113; TCGA UCEC -0.06 |
hsa-miR-127-3p | TMEM27 | 9 cancers: COAD; HNSC; KIRP; LGG; OV; PRAD; SARC; THCA; STAD | miRanda | TCGA COAD -0.304; TCGA HNSC -0.145; TCGA KIRP -0.219; TCGA LGG -0.098; TCGA OV -0.152; TCGA PRAD -0.224; TCGA SARC -0.174; TCGA THCA -0.063; TCGA STAD -0.218 |
hsa-miR-127-3p | MYB | 9 cancers: ESCA; HNSC; KIRC; KIRP; LUSC; OV; PAAD; PRAD; UCEC | miRanda | TCGA ESCA -0.541; TCGA HNSC -0.183; TCGA KIRC -0.162; TCGA KIRP -0.15; TCGA LUSC -0.228; TCGA OV -0.162; TCGA PAAD -0.293; TCGA PRAD -0.192; TCGA UCEC -0.193 |
Enriched cancer pathways of putative targets