microRNA information: hsa-miR-129-5p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-129-5p | miRbase |
| Accession: | MIMAT0000242 | miRbase |
| Precursor name: | hsa-mir-129-1 | miRbase |
| Precursor accession: | MI0000252 | miRbase |
| Symbol: | MIR129-1 | HGNC |
| RefSeq ID: | NR_029596 | GenBank |
| Sequence: | CUUUUUGCGGUCUGGGCUUGC |
Reported expression in cancers: hsa-miR-129-5p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-129-5p | breast cancer | downregulation | "The down regulation of miR 129 in breast cancer an ......" | 22907300 | |
| hsa-miR-129-5p | breast cancer | downregulation | "Levels of miR-129-5p were low in breast cancer tis ......" | 26460733 | |
| hsa-miR-129-5p | colorectal cancer | downregulation | "miR 129 promotes apoptosis and enhances chemosensi ......" | 23744359 | |
| hsa-miR-129-5p | gastric cancer | downregulation | "Growth inhibitory effects of three miR 129 family ......" | 24055727 | |
| hsa-miR-129-5p | gastric cancer | downregulation | "Expression of microRNA-129-5p miR-129-5p has been ......" | 27468870 | qPCR |
| hsa-miR-129-5p | liver cancer | deregulation | "miR 129 suppresses tumor cell growth and invasion ......" | 26116538 | |
| hsa-miR-129-5p | prostate cancer | downregulation | "The present study was designed to explore the clin ......" | 26823749 | qPCR |
| hsa-miR-129-5p | retinoblastoma | upregulation | "Identification of miRNAs associated with tumorigen ......" | 18818933 | Microarray; Northern blot; in situ hybridization |
Reported cancer pathway affected by hsa-miR-129-5p
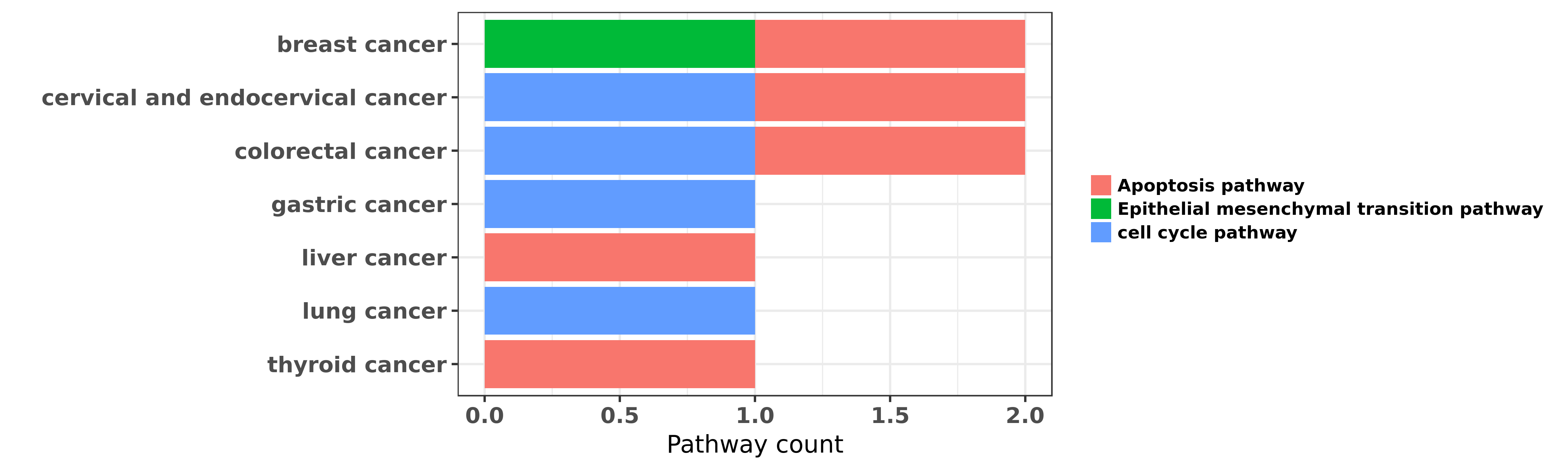
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-129-5p | breast cancer | Epithelial mesenchymal transition pathway | "Down regulation of miR 129 5p via the Twist1 Snail ......" | 26460733 | |
| hsa-miR-129-5p | breast cancer | Apoptosis pathway | "Downregulation of miR 129 2 by promoter hypermethy ......" | 26935022 | |
| hsa-miR-129-5p | cervical and endocervical cancer | cell cycle pathway; Apoptosis pathway | "In this study we found that miR-129-5p to be a can ......" | 24358111 | |
| hsa-miR-129-5p | colorectal cancer | Apoptosis pathway; cell cycle pathway | "miR 129 promotes apoptosis and enhances chemosensi ......" | 23744359 | |
| hsa-miR-129-5p | colorectal cancer | Apoptosis pathway | "MTT assay was used to detect the cell viability in ......" | 25218158 | MTT assay; Flow cytometry; Western blot |
| hsa-miR-129-5p | gastric cancer | cell cycle pathway | "Growth inhibitory effects of three miR 129 family ......" | 24055727 | |
| hsa-miR-129-5p | liver cancer | Apoptosis pathway | "It was found that miR-129-5p directly inhibited th ......" | 22536440 | |
| hsa-miR-129-5p | liver cancer | Apoptosis pathway | "miR 129 suppresses tumor cell growth and invasion ......" | 26116538 | RNAi |
| hsa-miR-129-5p | lung cancer | cell cycle pathway | "Epigenetic regulation of miR 129 2 and its effects ......" | 26081366 | |
| hsa-miR-129-5p | thyroid cancer | Apoptosis pathway | "MiR 129 5p is down regulated and involved in the g ......" | 24631532 |
Reported cancer prognosis affected by hsa-miR-129-5p
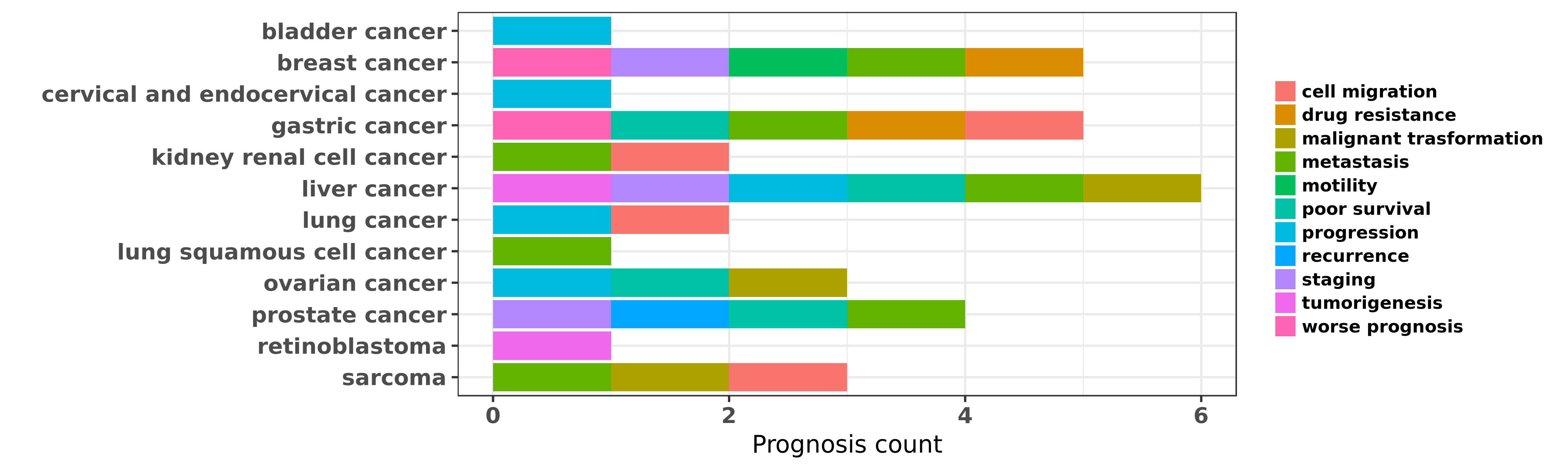
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-129-5p | bladder cancer | progression | "Genomic profiling of microRNAs in bladder cancer: ......" | 19487295 | Luciferase |
| hsa-miR-129-5p | breast cancer | motility; metastasis | "The down regulation of miR 129 in breast cancer an ......" | 22907300 | Cell migration assay; Wound Healing Assay |
| hsa-miR-129-5p | breast cancer | worse prognosis; staging | "Down regulation of miR 129 5p via the Twist1 Snail ......" | 26460733 | |
| hsa-miR-129-5p | breast cancer | drug resistance | "MiR 129 3p promotes docetaxel resistance of breast ......" | 26487539 | Luciferase |
| hsa-miR-129-5p | breast cancer | drug resistance | "mir 129 5p Attenuates Irradiation Induced Autophag ......" | 26720492 | |
| hsa-miR-129-5p | cervical and endocervical cancer | progression | "In this study we found that miR-129-5p to be a can ......" | 24358111 | |
| hsa-miR-129-5p | gastric cancer | worse prognosis | "lncRNA AC130710 targeting by miR 129 5p is upregul ......" | 24969565 | |
| hsa-miR-129-5p | gastric cancer | cell migration; metastasis | "miR 129 1 3p inhibits cell migration by targeting ......" | 25008064 | Transwell assay |
| hsa-miR-129-5p | gastric cancer | drug resistance | "Methylation of miR 129 5p CpG island modulates mul ......" | 25344911 | |
| hsa-miR-129-5p | gastric cancer | poor survival | "Then we further investigated these GC specific miR ......" | 27173517 | |
| hsa-miR-129-5p | gastric cancer | cell migration | "MiR 129 5p is down regulated and involved in migra ......" | 27468870 | Colony formation; Luciferase |
| hsa-miR-129-5p | kidney renal cell cancer | metastasis; cell migration | "miR 129 3p as a diagnostic and prognostic biomarke ......" | 24802708 | Western blot |
| hsa-miR-129-5p | liver cancer | progression | "It was found that miR-129-5p directly inhibited th ......" | 22536440 | |
| hsa-miR-129-5p | liver cancer | progression; metastasis; poor survival | "MiR-129-5p is deregulated in various human cancers ......" | 25912876 | |
| hsa-miR-129-5p | liver cancer | staging; malignant trasformation; progression; tumorigenesis | "miR 129 suppresses tumor cell growth and invasion ......" | 26116538 | RNAi |
| hsa-miR-129-5p | lung cancer | cell migration; progression | "Epigenetic regulation of miR 129 2 and its effects ......" | 26081366 | |
| hsa-miR-129-5p | lung squamous cell cancer | metastasis | "MiR 129 regulates MMP9 to control metastasis of no ......" | 25716201 | |
| hsa-miR-129-5p | ovarian cancer | malignant trasformation; progression; poor survival | "Here we report a microRNA miR-129-5p directly repr ......" | 25895125 | |
| hsa-miR-129-5p | prostate cancer | staging; metastasis; recurrence; poor survival | "Downregulation of miR 129 in peripheral blood mono ......" | 26823749 | |
| hsa-miR-129-5p | retinoblastoma | tumorigenesis | "Identification of miRNAs associated with tumorigen ......" | 18818933 | |
| hsa-miR-129-5p | sarcoma | malignant trasformation; metastasis; cell migration | "Demethylation mediated miR 129 5p up regulation in ......" | 25566966 | Western blot; Luciferase |
Reported gene related to hsa-miR-129-5p
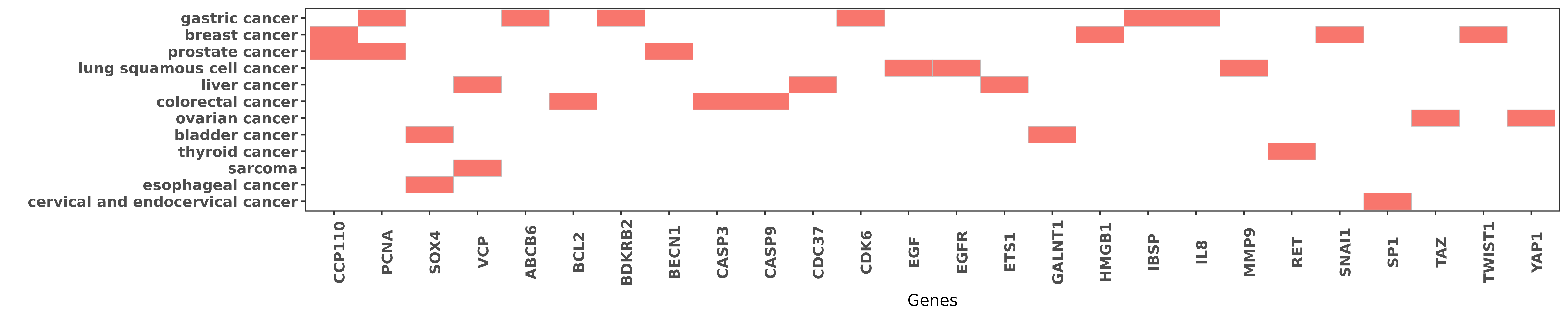
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-129-5p | colorectal cancer | BCL2 | "Piceatannol induced apoptosis by promoting express ......" | 25218158 |
| hsa-miR-129-5p | colorectal cancer | BCL2 | "In this study we discovered a novel mechanism medi ......" | 23744359 |
| hsa-miR-129-5p | breast cancer | CCP110 | "MiR 129 3p promotes docetaxel resistance of breast ......" | 26487539 |
| hsa-miR-129-5p | prostate cancer | CCP110 | "miR 129 3p controls centrosome number in metastati ......" | 26918338 |
| hsa-miR-129-5p | gastric cancer | PCNA | "Interestingly we showed that cyclin dependent kina ......" | 24055727 |
| hsa-miR-129-5p | prostate cancer | PCNA | "The results of in vitro CCK-8 assays as well as pr ......" | 26823749 |
| hsa-miR-129-5p | bladder cancer | SOX4 | "Using luciferase assays we documented a direct lin ......" | 19487295 |
| hsa-miR-129-5p | esophageal cancer | SOX4 | "miR 129 2 suppresses proliferation and migration o ......" | 23677061 |
| hsa-miR-129-5p | liver cancer | VCP | "It was found that miR-129-5p directly inhibited th ......" | 22536440 |
| hsa-miR-129-5p | sarcoma | VCP | "In the present study we found a significantly nega ......" | 25566966 |
| hsa-miR-129-5p | gastric cancer | ABCB6 | "Methylation of miR 129 5p CpG island modulates mul ......" | 25344911 |
| hsa-miR-129-5p | gastric cancer | BDKRB2 | "miR 129 1 3p inhibits cell migration by targeting ......" | 25008064 |
| hsa-miR-129-5p | prostate cancer | BECN1 | "Norcantharidin induces autophagy related prostate ......" | 26638170 |
| hsa-miR-129-5p | colorectal cancer | CASP3 | "The intrinsic apoptotic pathway triggered by miR-1 ......" | 23744359 |
| hsa-miR-129-5p | colorectal cancer | CASP9 | "The intrinsic apoptotic pathway triggered by miR-1 ......" | 23744359 |
| hsa-miR-129-5p | liver cancer | CDC37 | "Ectopic restoration of miR-129-5p expression in HC ......" | 25912876 |
| hsa-miR-129-5p | gastric cancer | CDK6 | "Interestingly we showed that cyclin dependent kina ......" | 24055727 |
| hsa-miR-129-5p | lung squamous cell cancer | EGF | "However miR-129 level was not affected by EGF stim ......" | 25716201 |
| hsa-miR-129-5p | lung squamous cell cancer | EGFR | "Here we reported significant decrease in miR-129 a ......" | 25716201 |
| hsa-miR-129-5p | liver cancer | ETS1 | "Ectopic restoration of miR-129-5p expression in HC ......" | 25912876 |
| hsa-miR-129-5p | bladder cancer | GALNT1 | "Using luciferase assays we documented a direct lin ......" | 19487295 |
| hsa-miR-129-5p | breast cancer | HMGB1 | "mir 129 5p Attenuates Irradiation Induced Autophag ......" | 26720492 |
| hsa-miR-129-5p | gastric cancer | IBSP | "Using microarray and BSP Bisulfate Sequencing PCR ......" | 25344911 |
| hsa-miR-129-5p | gastric cancer | IL8 | "MiR 129 5p is down regulated and involved in migra ......" | 27468870 |
| hsa-miR-129-5p | lung squamous cell cancer | MMP9 | "MiR 129 regulates MMP9 to control metastasis of no ......" | 25716201 |
| hsa-miR-129-5p | thyroid cancer | RET | "MiR 129 5p is down regulated and involved in the g ......" | 24631532 |
| hsa-miR-129-5p | breast cancer | SNAI1 | "Down regulation of miR 129 5p via the Twist1 Snail ......" | 26460733 |
| hsa-miR-129-5p | cervical and endocervical cancer | SP1 | "SP1 is a direct target of miR-129-5p in Hela cells ......" | 24358111 |
| hsa-miR-129-5p | ovarian cancer | TAZ | "Hence we demonstrate a novel mechanism for YAP and ......" | 25895125 |
| hsa-miR-129-5p | breast cancer | TWIST1 | "Down regulation of miR 129 5p via the Twist1 Snail ......" | 26460733 |
| hsa-miR-129-5p | ovarian cancer | YAP1 | "Hence we demonstrate a novel mechanism for YAP and ......" | 25895125 |
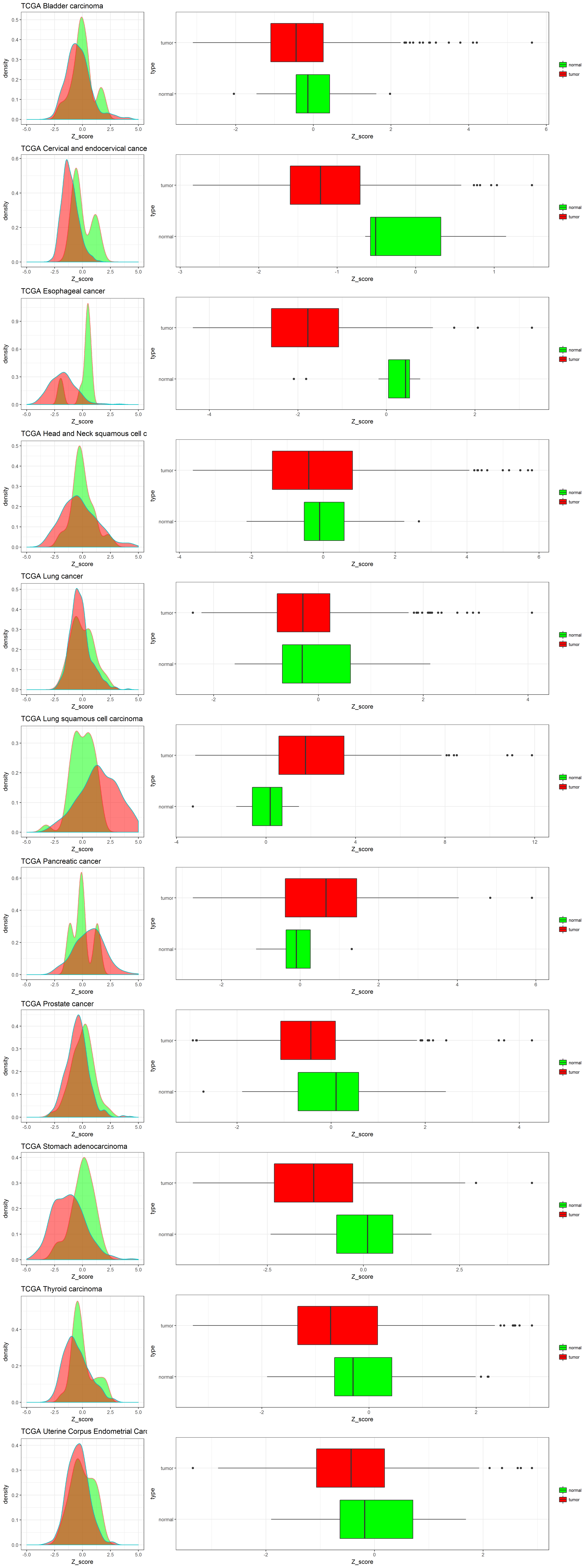
Expression profile in cancer corhorts:
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-129-5p | IFIH1 | 10 cancers: BLCA; CESC; ESCA; HNSC; LGG; LUAD; LUSC; OV; SARC; STAD | MirTarget | TCGA BLCA -0.09; TCGA CESC -0.141; TCGA ESCA -0.166; TCGA HNSC -0.164; TCGA LGG -0.069; TCGA LUAD -0.083; TCGA LUSC -0.104; TCGA OV -0.137; TCGA SARC -0.144; TCGA STAD -0.081 |
hsa-miR-129-5p | UPP1 | 11 cancers: BLCA; CESC; ESCA; HNSC; LGG; LUAD; OV; PAAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.074; TCGA CESC -0.113; TCGA ESCA -0.292; TCGA HNSC -0.158; TCGA LGG -0.084; TCGA LUAD -0.126; TCGA OV -0.144; TCGA PAAD -0.058; TCGA SARC -0.129; TCGA STAD -0.136; TCGA UCEC -0.081 |
hsa-miR-129-5p | PMEPA1 | 9 cancers: BLCA; ESCA; HNSC; LUAD; LUSC; OV; PAAD; STAD; UCEC | MirTarget; miRanda; miRNATAP | TCGA BLCA -0.161; TCGA ESCA -0.147; TCGA HNSC -0.076; TCGA LUAD -0.084; TCGA LUSC -0.194; TCGA OV -0.162; TCGA PAAD -0.116; TCGA STAD -0.119; TCGA UCEC -0.126 |
hsa-miR-129-5p | XAF1 | 9 cancers: BLCA; CESC; ESCA; HNSC; LUAD; LUSC; PAAD; SARC; STAD | MirTarget; miRanda | TCGA BLCA -0.115; TCGA CESC -0.204; TCGA ESCA -0.179; TCGA HNSC -0.213; TCGA LUAD -0.109; TCGA LUSC -0.148; TCGA PAAD -0.135; TCGA SARC -0.178; TCGA STAD -0.1 |
hsa-miR-129-5p | ISG20 | 9 cancers: BLCA; CESC; ESCA; HNSC; LGG; LUSC; OV; PAAD; SARC | MirTarget | TCGA BLCA -0.084; TCGA CESC -0.123; TCGA ESCA -0.157; TCGA HNSC -0.169; TCGA LGG -0.08; TCGA LUSC -0.115; TCGA OV -0.117; TCGA PAAD -0.156; TCGA SARC -0.169 |
hsa-miR-129-5p | SP100 | 9 cancers: BLCA; ESCA; HNSC; LGG; LUSC; OV; PAAD; PRAD; SARC | MirTarget; PITA; miRanda | TCGA BLCA -0.079; TCGA ESCA -0.072; TCGA HNSC -0.097; TCGA LGG -0.106; TCGA LUSC -0.116; TCGA OV -0.077; TCGA PAAD -0.104; TCGA PRAD -0.072; TCGA SARC -0.129 |
hsa-miR-129-5p | PLEK | 9 cancers: BLCA; CESC; HNSC; LGG; LUAD; LUSC; OV; PRAD; SARC | MirTarget | TCGA BLCA -0.176; TCGA CESC -0.188; TCGA HNSC -0.078; TCGA LGG -0.107; TCGA LUAD -0.063; TCGA LUSC -0.165; TCGA OV -0.155; TCGA PRAD -0.098; TCGA SARC -0.296 |
hsa-miR-129-5p | DOK3 | 9 cancers: BLCA; CESC; HNSC; LGG; LUAD; LUSC; OV; SARC; UCEC | MirTarget; miRanda | TCGA BLCA -0.113; TCGA CESC -0.118; TCGA HNSC -0.089; TCGA LGG -0.144; TCGA LUAD -0.094; TCGA LUSC -0.126; TCGA OV -0.08; TCGA SARC -0.164; TCGA UCEC -0.058 |
hsa-miR-129-5p | FCGR1B | 9 cancers: BLCA; CESC; HNSC; LGG; LUAD; LUSC; OV; SARC; STAD | MirTarget | TCGA BLCA -0.257; TCGA CESC -0.231; TCGA HNSC -0.148; TCGA LGG -0.061; TCGA LUAD -0.106; TCGA LUSC -0.167; TCGA OV -0.159; TCGA SARC -0.212; TCGA STAD -0.188 |
hsa-miR-129-5p | UBD | 11 cancers: BLCA; CESC; ESCA; HNSC; LGG; LUSC; OV; PAAD; PRAD; SARC; STAD | miRanda | TCGA BLCA -0.318; TCGA CESC -0.265; TCGA ESCA -0.265; TCGA HNSC -0.149; TCGA LGG -0.166; TCGA LUSC -0.196; TCGA OV -0.176; TCGA PAAD -0.296; TCGA PRAD -0.203; TCGA SARC -0.295; TCGA STAD -0.325 |
hsa-miR-129-5p | LILRB4 | 10 cancers: BLCA; CESC; HNSC; LGG; LUAD; LUSC; OV; PRAD; SARC; STAD | miRanda | TCGA BLCA -0.291; TCGA CESC -0.194; TCGA HNSC -0.097; TCGA LGG -0.149; TCGA LUAD -0.123; TCGA LUSC -0.104; TCGA OV -0.163; TCGA PRAD -0.064; TCGA SARC -0.317; TCGA STAD -0.12 |
hsa-miR-129-5p | CD86 | 10 cancers: BLCA; CESC; HNSC; LGG; LUAD; LUSC; OV; PRAD; SARC; STAD | miRanda | TCGA BLCA -0.159; TCGA CESC -0.186; TCGA HNSC -0.085; TCGA LGG -0.098; TCGA LUAD -0.086; TCGA LUSC -0.16; TCGA OV -0.128; TCGA PRAD -0.064; TCGA SARC -0.263; TCGA STAD -0.077 |
hsa-miR-129-5p | TMEM51 | 10 cancers: BLCA; CESC; ESCA; HNSC; LGG; LUAD; LUSC; OV; PAAD; SARC | miRanda | TCGA BLCA -0.06; TCGA CESC -0.098; TCGA ESCA -0.157; TCGA HNSC -0.14; TCGA LGG -0.054; TCGA LUAD -0.07; TCGA LUSC -0.129; TCGA OV -0.064; TCGA PAAD -0.077; TCGA SARC -0.09 |
hsa-miR-129-5p | LCP2 | 9 cancers: BLCA; CESC; ESCA; HNSC; LGG; LUAD; LUSC; OV; SARC | miRanda | TCGA BLCA -0.143; TCGA CESC -0.201; TCGA ESCA -0.098; TCGA HNSC -0.087; TCGA LGG -0.077; TCGA LUAD -0.062; TCGA LUSC -0.184; TCGA OV -0.157; TCGA SARC -0.224 |
hsa-miR-129-5p | CLEC7A | 10 cancers: BLCA; CESC; ESCA; HNSC; LGG; LUAD; LUSC; OV; SARC; STAD | miRanda | TCGA BLCA -0.149; TCGA CESC -0.285; TCGA ESCA -0.203; TCGA HNSC -0.193; TCGA LGG -0.079; TCGA LUAD -0.111; TCGA LUSC -0.245; TCGA OV -0.162; TCGA SARC -0.24; TCGA STAD -0.119 |
hsa-miR-129-5p | LAP3 | 9 cancers: BLCA; CESC; ESCA; HNSC; LGG; LUSC; OV; SARC; STAD | miRanda | TCGA BLCA -0.103; TCGA CESC -0.065; TCGA ESCA -0.102; TCGA HNSC -0.152; TCGA LGG -0.091; TCGA LUSC -0.101; TCGA OV -0.094; TCGA SARC -0.098; TCGA STAD -0.088 |
hsa-miR-129-5p | ICAM1 | 10 cancers: BLCA; CESC; ESCA; HNSC; LGG; LUSC; PAAD; SARC; STAD; UCEC | miRanda; miRNATAP | TCGA BLCA -0.163; TCGA CESC -0.1; TCGA ESCA -0.137; TCGA HNSC -0.093; TCGA LGG -0.123; TCGA LUSC -0.273; TCGA PAAD -0.098; TCGA SARC -0.134; TCGA STAD -0.123; TCGA UCEC -0.073 |
hsa-miR-129-5p | STAT1 | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUAD; OV; PAAD; SARC; STAD | miRanda | TCGA BLCA -0.131; TCGA CESC -0.133; TCGA COAD -0.093; TCGA ESCA -0.151; TCGA HNSC -0.142; TCGA LUAD -0.059; TCGA OV -0.077; TCGA PAAD -0.053; TCGA SARC -0.118; TCGA STAD -0.152 |
hsa-miR-129-5p | TNFSF10 | 11 cancers: BLCA; CESC; ESCA; HNSC; LUAD; LUSC; OV; PAAD; PRAD; SARC; STAD | miRanda | TCGA BLCA -0.172; TCGA CESC -0.208; TCGA ESCA -0.159; TCGA HNSC -0.135; TCGA LUAD -0.095; TCGA LUSC -0.09; TCGA OV -0.178; TCGA PAAD -0.087; TCGA PRAD -0.133; TCGA SARC -0.256; TCGA STAD -0.097 |
hsa-miR-129-5p | SAMD9L | 11 cancers: BLCA; CESC; ESCA; HNSC; LGG; LUAD; LUSC; OV; PAAD; SARC; STAD | miRanda | TCGA BLCA -0.108; TCGA CESC -0.194; TCGA ESCA -0.238; TCGA HNSC -0.151; TCGA LGG -0.1; TCGA LUAD -0.099; TCGA LUSC -0.181; TCGA OV -0.118; TCGA PAAD -0.111; TCGA SARC -0.143; TCGA STAD -0.09 |
hsa-miR-129-5p | RAC2 | 11 cancers: BLCA; CESC; ESCA; HNSC; LGG; LUAD; LUSC; OV; PAAD; PRAD; SARC | miRanda | TCGA BLCA -0.097; TCGA CESC -0.123; TCGA ESCA -0.109; TCGA HNSC -0.1; TCGA LGG -0.163; TCGA LUAD -0.053; TCGA LUSC -0.136; TCGA OV -0.107; TCGA PAAD -0.134; TCGA PRAD -0.089; TCGA SARC -0.138 |
hsa-miR-129-5p | SAMSN1 | 9 cancers: BLCA; CESC; HNSC; LGG; LUAD; LUSC; OV; PRAD; SARC | miRanda; miRNATAP | TCGA BLCA -0.174; TCGA CESC -0.193; TCGA HNSC -0.094; TCGA LGG -0.098; TCGA LUAD -0.073; TCGA LUSC -0.179; TCGA OV -0.136; TCGA PRAD -0.112; TCGA SARC -0.257 |
hsa-miR-129-5p | LILRB3 | 10 cancers: BLCA; CESC; HNSC; LGG; LUAD; LUSC; OV; PRAD; SARC; STAD | miRanda | TCGA BLCA -0.198; TCGA CESC -0.125; TCGA HNSC -0.108; TCGA LGG -0.237; TCGA LUAD -0.079; TCGA LUSC -0.204; TCGA OV -0.119; TCGA PRAD -0.099; TCGA SARC -0.243; TCGA STAD -0.079 |
hsa-miR-129-5p | SOD2 | 9 cancers: BLCA; CESC; HNSC; LGG; LUSC; OV; PRAD; SARC; STAD | miRanda; mirMAP | TCGA BLCA -0.096; TCGA CESC -0.133; TCGA HNSC -0.081; TCGA LGG -0.06; TCGA LUSC -0.106; TCGA OV -0.16; TCGA PRAD -0.066; TCGA SARC -0.156; TCGA STAD -0.075 |
hsa-miR-129-5p | NMI | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; LGG; LUSC; OV; PAAD; PRAD; SARC; STAD | miRanda | TCGA BLCA -0.102; TCGA CESC -0.109; TCGA COAD -0.115; TCGA ESCA -0.175; TCGA HNSC -0.112; TCGA LGG -0.139; TCGA LUSC -0.053; TCGA OV -0.057; TCGA PAAD -0.134; TCGA PRAD -0.091; TCGA SARC -0.101; TCGA STAD -0.117 |
hsa-miR-129-5p | CD53 | 9 cancers: BLCA; CESC; HNSC; LGG; LUAD; LUSC; OV; PRAD; SARC | miRanda | TCGA BLCA -0.197; TCGA CESC -0.199; TCGA HNSC -0.104; TCGA LGG -0.087; TCGA LUAD -0.072; TCGA LUSC -0.18; TCGA OV -0.16; TCGA PRAD -0.073; TCGA SARC -0.27 |
hsa-miR-129-5p | IL18 | 9 cancers: BLCA; ESCA; HNSC; LGG; LUSC; OV; PAAD; SARC; STAD | miRanda | TCGA BLCA -0.118; TCGA ESCA -0.235; TCGA HNSC -0.088; TCGA LGG -0.132; TCGA LUSC -0.11; TCGA OV -0.146; TCGA PAAD -0.194; TCGA SARC -0.278; TCGA STAD -0.164 |
hsa-miR-129-5p | TAPBP | 11 cancers: BLCA; CESC; COAD; ESCA; HNSC; LGG; LUSC; OV; PAAD; SARC; STAD | miRanda | TCGA BLCA -0.099; TCGA CESC -0.058; TCGA COAD -0.076; TCGA ESCA -0.072; TCGA HNSC -0.097; TCGA LGG -0.076; TCGA LUSC -0.084; TCGA OV -0.126; TCGA PAAD -0.101; TCGA SARC -0.114; TCGA STAD -0.051 |
hsa-miR-129-5p | IRF1 | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; LGG; LUAD; LUSC; OV; PAAD; SARC; STAD | miRanda; mirMAP | TCGA BLCA -0.124; TCGA CESC -0.184; TCGA COAD -0.121; TCGA ESCA -0.143; TCGA HNSC -0.131; TCGA LGG -0.162; TCGA LUAD -0.061; TCGA LUSC -0.144; TCGA OV -0.167; TCGA PAAD -0.083; TCGA SARC -0.162; TCGA STAD -0.102 |
hsa-miR-129-5p | HLA-F | 10 cancers: BLCA; CESC; ESCA; HNSC; LGG; LUAD; LUSC; OV; PAAD; SARC | miRanda | TCGA BLCA -0.14; TCGA CESC -0.148; TCGA ESCA -0.226; TCGA HNSC -0.193; TCGA LGG -0.11; TCGA LUAD -0.071; TCGA LUSC -0.16; TCGA OV -0.136; TCGA PAAD -0.124; TCGA SARC -0.211 |
hsa-miR-129-5p | LY86 | 9 cancers: BLCA; CESC; HNSC; LGG; LUAD; LUSC; OV; PRAD; SARC | miRanda | TCGA BLCA -0.191; TCGA CESC -0.181; TCGA HNSC -0.165; TCGA LGG -0.097; TCGA LUAD -0.079; TCGA LUSC -0.206; TCGA OV -0.107; TCGA PRAD -0.059; TCGA SARC -0.232 |
hsa-miR-129-5p | VDR | 9 cancers: BLCA; CESC; ESCA; HNSC; LUSC; OV; PAAD; STAD; UCEC | miRanda | TCGA BLCA -0.1; TCGA CESC -0.145; TCGA ESCA -0.276; TCGA HNSC -0.13; TCGA LUSC -0.148; TCGA OV -0.131; TCGA PAAD -0.087; TCGA STAD -0.094; TCGA UCEC -0.139 |
hsa-miR-129-5p | C2 | 9 cancers: BLCA; COAD; HNSC; LGG; LUSC; OV; PAAD; PRAD; SARC | miRanda | TCGA BLCA -0.241; TCGA COAD -0.156; TCGA HNSC -0.111; TCGA LGG -0.157; TCGA LUSC -0.204; TCGA OV -0.163; TCGA PAAD -0.155; TCGA PRAD -0.151; TCGA SARC -0.223 |
hsa-miR-129-5p | GPR183 | 9 cancers: BLCA; CESC; HNSC; LGG; LUAD; LUSC; OV; PRAD; SARC | miRanda | TCGA BLCA -0.135; TCGA CESC -0.115; TCGA HNSC -0.09; TCGA LGG -0.072; TCGA LUAD -0.084; TCGA LUSC -0.158; TCGA OV -0.133; TCGA PRAD -0.079; TCGA SARC -0.276 |
hsa-miR-129-5p | PTPN22 | 12 cancers: BLCA; CESC; ESCA; HNSC; LGG; LUAD; LUSC; OV; PAAD; PRAD; SARC; STAD | miRanda | TCGA BLCA -0.175; TCGA CESC -0.21; TCGA ESCA -0.12; TCGA HNSC -0.15; TCGA LGG -0.183; TCGA LUAD -0.1; TCGA LUSC -0.212; TCGA OV -0.198; TCGA PAAD -0.136; TCGA PRAD -0.097; TCGA SARC -0.171; TCGA STAD -0.077 |
hsa-miR-129-5p | CTSS | 10 cancers: BLCA; CESC; ESCA; HNSC; LGG; LUSC; OV; PAAD; SARC; STAD | miRanda | TCGA BLCA -0.13; TCGA CESC -0.098; TCGA ESCA -0.125; TCGA HNSC -0.06; TCGA LGG -0.171; TCGA LUSC -0.191; TCGA OV -0.147; TCGA PAAD -0.115; TCGA SARC -0.237; TCGA STAD -0.088 |
hsa-miR-129-5p | HLA-E | 9 cancers: BLCA; CESC; ESCA; HNSC; LGG; LUSC; OV; PAAD; SARC | miRanda | TCGA BLCA -0.099; TCGA CESC -0.076; TCGA ESCA -0.096; TCGA HNSC -0.143; TCGA LGG -0.065; TCGA LUSC -0.144; TCGA OV -0.107; TCGA PAAD -0.065; TCGA SARC -0.133 |
hsa-miR-129-5p | IRF8 | 9 cancers: BLCA; CESC; ESCA; HNSC; LGG; LUSC; OV; PAAD; SARC | miRanda | TCGA BLCA -0.18; TCGA CESC -0.132; TCGA ESCA -0.16; TCGA HNSC -0.136; TCGA LGG -0.059; TCGA LUSC -0.184; TCGA OV -0.101; TCGA PAAD -0.114; TCGA SARC -0.2 |
hsa-miR-129-5p | MLKL | 11 cancers: BLCA; CESC; ESCA; HNSC; LGG; LUSC; OV; PAAD; SARC; STAD; UCEC | miRanda | TCGA BLCA -0.077; TCGA CESC -0.124; TCGA ESCA -0.162; TCGA HNSC -0.112; TCGA LGG -0.11; TCGA LUSC -0.112; TCGA OV -0.151; TCGA PAAD -0.148; TCGA SARC -0.141; TCGA STAD -0.088; TCGA UCEC -0.093 |
hsa-miR-129-5p | MMP7 | 10 cancers: BLCA; ESCA; LGG; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | miRanda | TCGA BLCA -0.254; TCGA ESCA -0.296; TCGA LGG -0.252; TCGA LUAD -0.13; TCGA LUSC -0.17; TCGA OV -0.246; TCGA PAAD -0.281; TCGA PRAD -0.194; TCGA STAD -0.374; TCGA UCEC -0.194 |
hsa-miR-129-5p | CYTIP | 9 cancers: BLCA; CESC; HNSC; LGG; LUSC; OV; PAAD; PRAD; SARC | miRanda | TCGA BLCA -0.168; TCGA CESC -0.196; TCGA HNSC -0.101; TCGA LGG -0.125; TCGA LUSC -0.194; TCGA OV -0.135; TCGA PAAD -0.118; TCGA PRAD -0.082; TCGA SARC -0.245 |
hsa-miR-129-5p | TBXAS1 | 10 cancers: BLCA; CESC; HNSC; LGG; LUAD; LUSC; OV; PAAD; PRAD; SARC | miRanda | TCGA BLCA -0.175; TCGA CESC -0.117; TCGA HNSC -0.09; TCGA LGG -0.1; TCGA LUAD -0.062; TCGA LUSC -0.195; TCGA OV -0.09; TCGA PAAD -0.106; TCGA PRAD -0.054; TCGA SARC -0.207 |
hsa-miR-129-5p | RNF213 | 9 cancers: BLCA; CESC; ESCA; HNSC; LGG; OV; PAAD; SARC; STAD | miRanda; mirMAP | TCGA BLCA -0.08; TCGA CESC -0.089; TCGA ESCA -0.095; TCGA HNSC -0.057; TCGA LGG -0.069; TCGA OV -0.144; TCGA PAAD -0.067; TCGA SARC -0.073; TCGA STAD -0.081 |
hsa-miR-129-5p | UNC13D | 10 cancers: BLCA; CESC; ESCA; HNSC; LGG; LUAD; LUSC; OV; PAAD; SARC | miRanda | TCGA BLCA -0.182; TCGA CESC -0.13; TCGA ESCA -0.246; TCGA HNSC -0.11; TCGA LGG -0.146; TCGA LUAD -0.118; TCGA LUSC -0.212; TCGA OV -0.106; TCGA PAAD -0.208; TCGA SARC -0.132 |
hsa-miR-129-5p | HAVCR2 | 10 cancers: BLCA; CESC; ESCA; HNSC; LGG; LUAD; LUSC; OV; SARC; STAD | miRanda | TCGA BLCA -0.218; TCGA CESC -0.217; TCGA ESCA -0.115; TCGA HNSC -0.13; TCGA LGG -0.112; TCGA LUAD -0.1; TCGA LUSC -0.164; TCGA OV -0.147; TCGA SARC -0.24; TCGA STAD -0.133 |
hsa-miR-129-5p | TNFAIP8 | 10 cancers: BLCA; CESC; ESCA; HNSC; LGG; LUAD; LUSC; PAAD; PRAD; SARC | miRanda | TCGA BLCA -0.055; TCGA CESC -0.09; TCGA ESCA -0.077; TCGA HNSC -0.085; TCGA LGG -0.167; TCGA LUAD -0.082; TCGA LUSC -0.05; TCGA PAAD -0.14; TCGA PRAD -0.138; TCGA SARC -0.063 |
hsa-miR-129-5p | WAS | 9 cancers: BLCA; CESC; HNSC; LGG; LUAD; LUSC; OV; PRAD; SARC | miRanda | TCGA BLCA -0.161; TCGA CESC -0.202; TCGA HNSC -0.128; TCGA LGG -0.121; TCGA LUAD -0.067; TCGA LUSC -0.17; TCGA OV -0.098; TCGA PRAD -0.059; TCGA SARC -0.229 |
hsa-miR-129-5p | LAPTM5 | 10 cancers: BLCA; CESC; HNSC; LGG; LUAD; LUSC; OV; PAAD; PRAD; SARC | miRanda | TCGA BLCA -0.15; TCGA CESC -0.157; TCGA HNSC -0.114; TCGA LGG -0.128; TCGA LUAD -0.079; TCGA LUSC -0.175; TCGA OV -0.157; TCGA PAAD -0.074; TCGA PRAD -0.055; TCGA SARC -0.214 |
hsa-miR-129-5p | CASP10 | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; LGG; LUAD; LUSC; OV; PAAD; SARC; STAD | mirMAP | TCGA BLCA -0.085; TCGA CESC -0.096; TCGA COAD -0.069; TCGA ESCA -0.154; TCGA HNSC -0.082; TCGA LGG -0.065; TCGA LUAD -0.057; TCGA LUSC -0.152; TCGA OV -0.088; TCGA PAAD -0.133; TCGA SARC -0.159; TCGA STAD -0.075 |
hsa-miR-129-5p | ARPC1B | 9 cancers: BLCA; ESCA; HNSC; LGG; OV; PAAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.072; TCGA ESCA -0.191; TCGA HNSC -0.085; TCGA LGG -0.126; TCGA OV -0.089; TCGA PAAD -0.101; TCGA SARC -0.084; TCGA STAD -0.055; TCGA UCEC -0.059 |
hsa-miR-129-5p | CCL4L2 | 9 cancers: CESC; ESCA; HNSC; LUAD; LUSC; OV; PRAD; SARC; UCEC | MirTarget | TCGA CESC -0.23; TCGA ESCA -0.19; TCGA HNSC -0.1; TCGA LUAD -0.088; TCGA LUSC -0.141; TCGA OV -0.159; TCGA PRAD -0.107; TCGA SARC -0.219; TCGA UCEC -0.126 |
hsa-miR-129-5p | ARHGEF10L | 9 cancers: CESC; HNSC; LGG; LUAD; LUSC; OV; SARC; STAD; UCEC | MirTarget; PITA; miRanda; miRNATAP | TCGA CESC -0.071; TCGA HNSC -0.077; TCGA LGG -0.07; TCGA LUAD -0.064; TCGA LUSC -0.125; TCGA OV -0.088; TCGA SARC -0.172; TCGA STAD -0.059; TCGA UCEC -0.077 |
hsa-miR-129-5p | CTSC | 9 cancers: CESC; ESCA; HNSC; LGG; LUAD; PAAD; SARC; STAD; UCEC | MirTarget; miRanda; mirMAP | TCGA CESC -0.072; TCGA ESCA -0.116; TCGA HNSC -0.09; TCGA LGG -0.17; TCGA LUAD -0.082; TCGA PAAD -0.061; TCGA SARC -0.108; TCGA STAD -0.063; TCGA UCEC -0.058 |
hsa-miR-129-5p | PRRG2 | 9 cancers: CESC; ESCA; HNSC; LGG; LUAD; LUSC; OV; STAD; UCEC | miRanda | TCGA CESC -0.063; TCGA ESCA -0.089; TCGA HNSC -0.081; TCGA LGG -0.06; TCGA LUAD -0.069; TCGA LUSC -0.081; TCGA OV -0.113; TCGA STAD -0.077; TCGA UCEC -0.074 |
hsa-miR-129-5p | OAS2 | 11 cancers: CESC; ESCA; HNSC; LGG; LUAD; LUSC; OV; PAAD; PRAD; SARC; STAD | miRanda | TCGA CESC -0.205; TCGA ESCA -0.202; TCGA HNSC -0.159; TCGA LGG -0.08; TCGA LUAD -0.08; TCGA LUSC -0.106; TCGA OV -0.122; TCGA PAAD -0.166; TCGA PRAD -0.137; TCGA SARC -0.218; TCGA STAD -0.132 |
hsa-miR-129-5p | CASP1 | 9 cancers: CESC; ESCA; HNSC; LGG; LUAD; LUSC; OV; SARC; STAD | miRanda | TCGA CESC -0.163; TCGA ESCA -0.187; TCGA HNSC -0.131; TCGA LGG -0.156; TCGA LUAD -0.074; TCGA LUSC -0.121; TCGA OV -0.183; TCGA SARC -0.244; TCGA STAD -0.079 |
hsa-miR-129-5p | DPYD | 9 cancers: CESC; ESCA; HNSC; LGG; LUAD; LUSC; OV; PAAD; SARC | miRanda | TCGA CESC -0.19; TCGA ESCA -0.135; TCGA HNSC -0.136; TCGA LGG -0.145; TCGA LUAD -0.074; TCGA LUSC -0.237; TCGA OV -0.174; TCGA PAAD -0.066; TCGA SARC -0.169 |
hsa-miR-129-5p | IL15 | 10 cancers: CESC; COAD; ESCA; HNSC; LGG; LUAD; LUSC; OV; PAAD; SARC | miRanda | TCGA CESC -0.278; TCGA COAD -0.109; TCGA ESCA -0.13; TCGA HNSC -0.149; TCGA LGG -0.118; TCGA LUAD -0.07; TCGA LUSC -0.156; TCGA OV -0.24; TCGA PAAD -0.063; TCGA SARC -0.242 |
hsa-miR-129-5p | TRIM14 | 9 cancers: CESC; ESCA; HNSC; LGG; LUAD; OV; PAAD; SARC; STAD | miRanda | TCGA CESC -0.058; TCGA ESCA -0.129; TCGA HNSC -0.064; TCGA LGG -0.091; TCGA LUAD -0.063; TCGA OV -0.093; TCGA PAAD -0.067; TCGA SARC -0.124; TCGA STAD -0.071 |
hsa-miR-129-5p | MIR155HG | 9 cancers: CESC; ESCA; LGG; LUAD; PAAD; PRAD; SARC; STAD; UCEC | miRanda | TCGA CESC -0.124; TCGA ESCA -0.111; TCGA LGG -0.157; TCGA LUAD -0.091; TCGA PAAD -0.076; TCGA PRAD -0.08; TCGA SARC -0.185; TCGA STAD -0.125; TCGA UCEC -0.067 |
hsa-miR-129-5p | SLC43A3 | 9 cancers: ESCA; HNSC; LGG; LUAD; LUSC; OV; PAAD; SARC; STAD | MirTarget | TCGA ESCA -0.156; TCGA HNSC -0.07; TCGA LGG -0.121; TCGA LUAD -0.089; TCGA LUSC -0.206; TCGA OV -0.099; TCGA PAAD -0.066; TCGA SARC -0.08; TCGA STAD -0.077 |
hsa-miR-129-5p | DCBLD1 | 9 cancers: ESCA; HNSC; LUAD; LUSC; OV; PAAD; SARC; STAD; UCEC | miRanda | TCGA ESCA -0.214; TCGA HNSC -0.106; TCGA LUAD -0.053; TCGA LUSC -0.09; TCGA OV -0.065; TCGA PAAD -0.053; TCGA SARC -0.088; TCGA STAD -0.091; TCGA UCEC -0.108 |
Enriched cancer pathways of putative targets