microRNA information: hsa-miR-132-3p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-132-3p | miRbase |
| Accession: | MIMAT0000426 | miRbase |
| Precursor name: | hsa-mir-132 | miRbase |
| Precursor accession: | MI0000449 | miRbase |
| Symbol: | MIR132 | HGNC |
| RefSeq ID: | NR_029674 | GenBank |
| Sequence: | UAACAGUCUACAGCCAUGGUCG |
Reported expression in cancers: hsa-miR-132-3p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-132-3p | breast cancer | downregulation | "This study aims to investigate the role and the un ......" | 25450365 | |
| hsa-miR-132-3p | breast cancer | downregulation | "Aberrant Expression of Breast Development Related ......" | 27382390 | qPCR |
| hsa-miR-132-3p | cervical and endocervical cancer | downregulation | "In this study we found that miR-212 and miR-132 fr ......" | 25988335 | |
| hsa-miR-132-3p | colorectal cancer | downregulation | "To investigate the biological role and underlying ......" | 24914372 | qPCR |
| hsa-miR-132-3p | colorectal cancer | downregulation | "Downregulation of microRNA 132 by DNA hypermethyla ......" | 26675712 | qPCR |
| hsa-miR-132-3p | colorectal cancer | downregulation | "Down Regulation of microRNA 132 is Associated with ......" | 26868958 | qPCR |
| hsa-miR-132-3p | liver cancer | downregulation | "Epigenetic repression of miR 132 expression by the ......" | 23376496 | qPCR |
| hsa-miR-132-3p | liver cancer | downregulation | "To observe the biological role and underlying mech ......" | 25676267 | qPCR |
| hsa-miR-132-3p | liver cancer | downregulation | "Downregulation of microRNA 132 indicates progressi ......" | 27698698 | qPCR |
| hsa-miR-132-3p | ovarian cancer | downregulation | "The aim of this study was to find specific profile ......" | 23542579 | Microarray; qPCR |
| hsa-miR-132-3p | pancreatic cancer | downregulation | "Downregulation of miR 132 by promoter methylation ......" | 21665894 | Microarray |
| hsa-miR-132-3p | prostate cancer | downregulation | "DNA methylation silences miR 132 in prostate cance ......" | 22310291 |
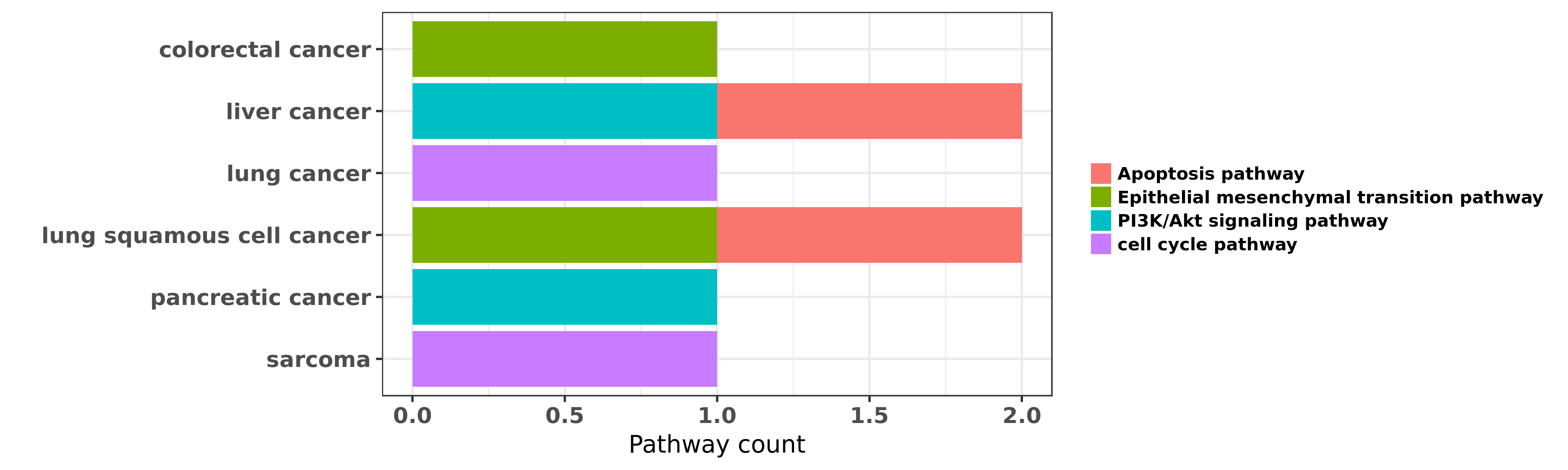
Reported cancer pathway affected by hsa-miR-132-3p
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-132-3p | colorectal cancer | Epithelial mesenchymal transition pathway | "miR 132 inhibits colorectal cancer invasion and me ......" | 24914372 | Transwell assay; Western blot; Luciferase |
| hsa-miR-132-3p | liver cancer | PI3K/Akt signaling pathway | "Epigenetic repression of miR 132 expression by the ......" | 23376496 | Colony formation |
| hsa-miR-132-3p | liver cancer | Apoptosis pathway | "Effects of MicroRNA 132 transfection on the prolif ......" | 25676267 | Flow cytometry; Western blot |
| hsa-miR-132-3p | liver cancer | Apoptosis pathway | "MiR 132 inhibits cell proliferation invasion and m ......" | 26252738 | Colony formation; Luciferase; Western blot |
| hsa-miR-132-3p | liver cancer | Apoptosis pathway | "An Encapsulation of Gene Signatures for Hepatocell ......" | 27467251 | |
| hsa-miR-132-3p | liver cancer | Apoptosis pathway | "Effect of co transfection of miR 520c 3p and miR 1 ......" | 27633306 | Western blot; Flow cytometry |
| hsa-miR-132-3p | lung cancer | cell cycle pathway | "Furthermore miR-212/132 overexpression induced cel ......" | 25435090 | |
| hsa-miR-132-3p | lung squamous cell cancer | Apoptosis pathway | "Hsa miR 132 regulates apoptosis in non small cell ......" | 24158730 | |
| hsa-miR-132-3p | lung squamous cell cancer | Apoptosis pathway | "Decreased microRNA 132 and its function in human n ......" | 25607827 | Cell migration assay; MTT assay |
| hsa-miR-132-3p | lung squamous cell cancer | Epithelial mesenchymal transition pathway | "MicroRNA 132 inhibits migration invasion and epith ......" | 27735039 | Wound Healing Assay; Western blot |
| hsa-miR-132-3p | pancreatic cancer | PI3K/Akt signaling pathway | "Downregulation of miR 132 by promoter methylation ......" | 21665894 | Colony formation |
| hsa-miR-132-3p | sarcoma | cell cycle pathway | "miR 132 targeting cyclin E1 suppresses cell prolif ......" | 24449507 |
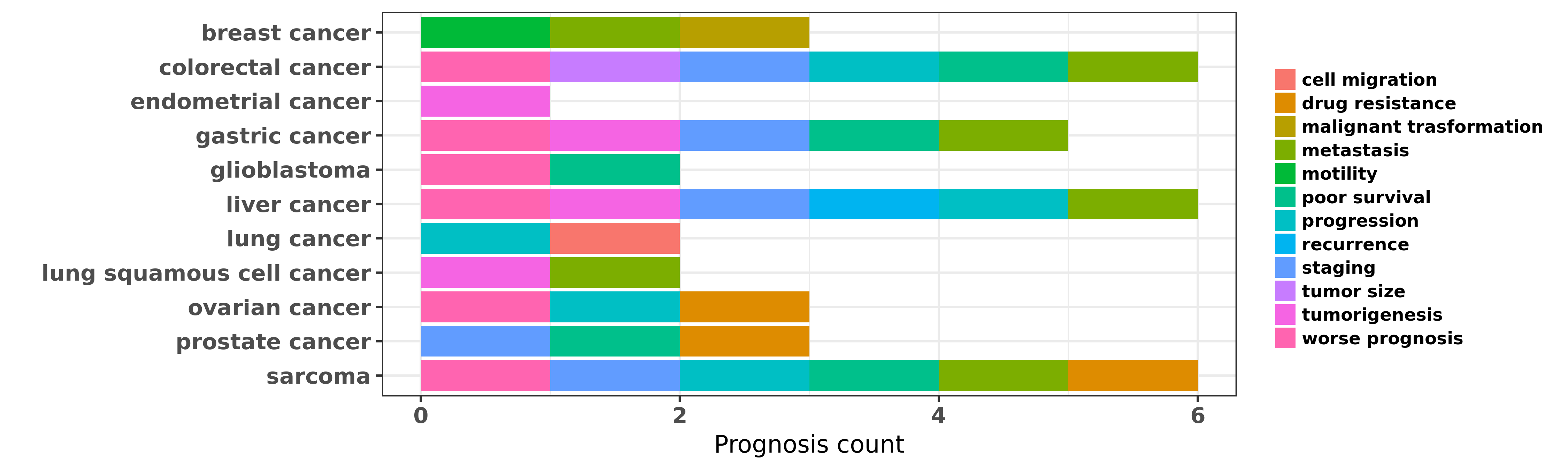
Reported cancer prognosis affected by hsa-miR-132-3p
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-132-3p | breast cancer | malignant trasformation | "In support of these findings in vitro functional s ......" | 24104550 | |
| hsa-miR-132-3p | breast cancer | motility | "Here we have defined novel epithelial as well as s ......" | 25122196 | |
| hsa-miR-132-3p | breast cancer | metastasis | "MiR 132 prohibits proliferation invasion migration ......" | 25450365 | |
| hsa-miR-132-3p | colorectal cancer | metastasis; progression; staging; tumor size; poor survival | "miR 132 inhibits colorectal cancer invasion and me ......" | 24914372 | Transwell assay; Western blot; Luciferase |
| hsa-miR-132-3p | colorectal cancer | worse prognosis | "Downregulation of microRNA 132 by DNA hypermethyla ......" | 26675712 | |
| hsa-miR-132-3p | colorectal cancer | worse prognosis; metastasis; poor survival | "Down Regulation of microRNA 132 is Associated with ......" | 26868958 | Luciferase |
| hsa-miR-132-3p | colorectal cancer | poor survival | "The expression levels of miR-206 miR-219 miR-192 m ......" | 27666868 | |
| hsa-miR-132-3p | endometrial cancer | tumorigenesis | "Hypermethylation of miR-345 and miR-132 associated ......" | 25767621 | |
| hsa-miR-132-3p | gastric cancer | worse prognosis; staging; metastasis; poor survival | "The expression and clinical significance of miR 13 ......" | 24621117 | |
| hsa-miR-132-3p | gastric cancer | tumorigenesis | "miR 132 upregulation promotes gastric cancer cell ......" | 26298723 | Luciferase |
| hsa-miR-132-3p | glioblastoma | poor survival; worse prognosis | "Correlation of MicroRNA 132 Up regulation with an ......" | 24466377 | |
| hsa-miR-132-3p | liver cancer | tumorigenesis | "Epigenetic repression of miR 132 expression by the ......" | 23376496 | Colony formation |
| hsa-miR-132-3p | liver cancer | worse prognosis | "An Encapsulation of Gene Signatures for Hepatocell ......" | 27467251 | |
| hsa-miR-132-3p | liver cancer | progression; recurrence; staging; metastasis | "Downregulation of microRNA 132 indicates progressi ......" | 27698698 | |
| hsa-miR-132-3p | lung cancer | cell migration; progression | "miR 132 inhibits lung cancer cell migration and in ......" | 26543603 | Transwell assay; Wound Healing Assay; Western blot; Luciferase |
| hsa-miR-132-3p | lung squamous cell cancer | tumorigenesis | "Hsa miR 132 regulates apoptosis in non small cell ......" | 24158730 | |
| hsa-miR-132-3p | lung squamous cell cancer | metastasis | "MicroRNA 132 inhibits migration invasion and epith ......" | 27735039 | Wound Healing Assay; Western blot |
| hsa-miR-132-3p | ovarian cancer | progression | "miR 132 targeting E2F5 suppresses cell proliferati ......" | 27186275 | Colony formation |
| hsa-miR-132-3p | ovarian cancer | worse prognosis; drug resistance | "MicroRNAs such as miR-26a and miR-132 have been in ......" | 27673409 | |
| hsa-miR-132-3p | prostate cancer | drug resistance; staging; poor survival | "DNA methylation silences miR 132 in prostate cance ......" | 22310291 | |
| hsa-miR-132-3p | sarcoma | worse prognosis; poor survival; staging; metastasis; drug resistance; progression | "Loss of microRNA 132 predicts poor prognosis in pa ......" | 23801049 | |
| hsa-miR-132-3p | sarcoma | metastasis | "MicroRNA 132 inhibits cell growth and metastasis i ......" | 26352673 | Western blot; Luciferase |
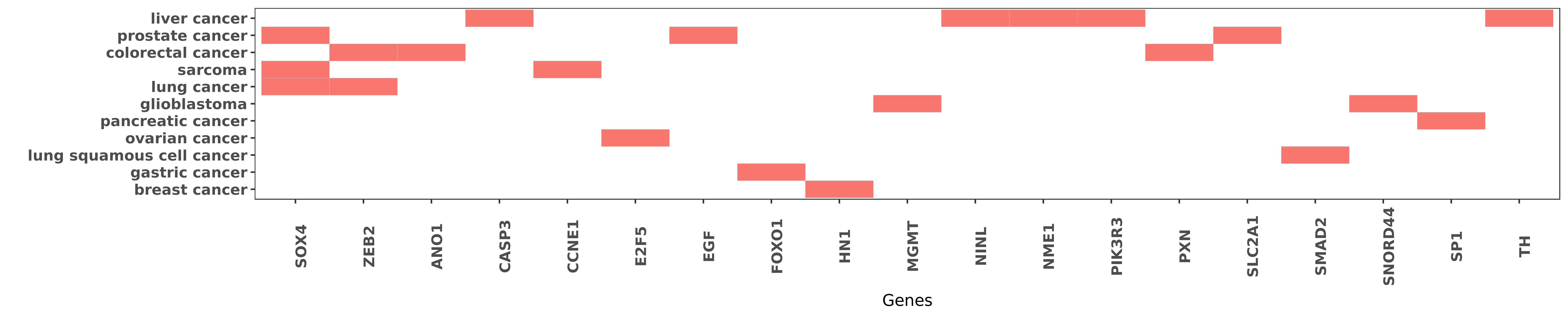
Reported gene related to hsa-miR-132-3p
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-132-3p | lung cancer | SOX4 | "miR 132 inhibits lung cancer cell migration and in ......" | 26543603 |
| hsa-miR-132-3p | prostate cancer | SOX4 | "Immunohistochemistry IHC was used to detect the ex ......" | 27527117 |
| hsa-miR-132-3p | sarcoma | SOX4 | "MicroRNA 132 inhibits cell growth and metastasis i ......" | 26352673 |
| hsa-miR-132-3p | colorectal cancer | ZEB2 | "miR 132 inhibits colorectal cancer invasion and me ......" | 24914372 |
| hsa-miR-132-3p | lung cancer | ZEB2 | "MiR 132 suppresses the migration and invasion of l ......" | 24626466 |
| hsa-miR-132-3p | colorectal cancer | ANO1 | "The luciferase reporter assay revealed that anocta ......" | 26868958 |
| hsa-miR-132-3p | liver cancer | CASP3 | "After miR-132 transfectionthe expression of Caspas ......" | 25676267 |
| hsa-miR-132-3p | sarcoma | CCNE1 | "Of significance contrary to CCNE1 expression level ......" | 24449507 |
| hsa-miR-132-3p | ovarian cancer | E2F5 | "miR 132 targeting E2F5 suppresses cell proliferati ......" | 27186275 |
| hsa-miR-132-3p | prostate cancer | EGF | "Two pro-survival proteins-heparin-binding epiderma ......" | 22310291 |
| hsa-miR-132-3p | gastric cancer | FOXO1 | "miR 132 upregulation promotes gastric cancer cell ......" | 26298723 |
| hsa-miR-132-3p | breast cancer | HN1 | "MiR 132 prohibits proliferation invasion migration ......" | 25450365 |
| hsa-miR-132-3p | glioblastoma | MGMT | "miR-132 was a stronger prognostic indicator than E ......" | 24466377 |
| hsa-miR-132-3p | liver cancer | NINL | "Various assays were performed to explore the role ......" | 27467251 |
| hsa-miR-132-3p | liver cancer | NME1 | "Spearman correlation analysis demonstrated that mi ......" | 27698698 |
| hsa-miR-132-3p | liver cancer | PIK3R3 | "MiR 132 inhibits cell proliferation invasion and m ......" | 26252738 |
| hsa-miR-132-3p | colorectal cancer | PXN | "Bioinformatics analysis indicated that the mechani ......" | 26675712 |
| hsa-miR-132-3p | prostate cancer | SLC2A1 | "miR 132 mediates a metabolic shift in prostate can ......" | 27398313 |
| hsa-miR-132-3p | lung squamous cell cancer | SMAD2 | "Moreover expression of EMT-related markers and Sma ......" | 27735039 |
| hsa-miR-132-3p | glioblastoma | SNORD44 | "miR-132 levels relative to RNU44 were assessed by ......" | 24466377 |
| hsa-miR-132-3p | pancreatic cancer | SP1 | "miR-132 was transcribed by RNA polymerase II and S ......" | 21665894 |
| hsa-miR-132-3p | liver cancer | TH | "Th e associations between miR-132 expression level ......" | 27698698 |
Expression profile in cancer corhorts:

Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-132-3p | KBTBD3 | 13 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; OV; PRAD; SARC; STAD | miRNAWalker2 validate | TCGA BLCA -0.238; TCGA BRCA -0.175; TCGA COAD -0.257; TCGA ESCA -0.223; TCGA HNSC -0.138; TCGA KIRC -0.209; TCGA LIHC -0.149; TCGA LUAD -0.188; TCGA LUSC -0.293; TCGA OV -0.125; TCGA PRAD -0.082; TCGA SARC -0.136; TCGA STAD -0.23 |
hsa-miR-132-3p | NDUFB10 | 11 cancers: BLCA; ESCA; KIRC; LGG; LIHC; LUAD; PAAD; PRAD; SARC; THCA; STAD | miRNAWalker2 validate | TCGA BLCA -0.149; TCGA ESCA -0.22; TCGA KIRC -0.108; TCGA LGG -0.062; TCGA LIHC -0.123; TCGA LUAD -0.1; TCGA PAAD -0.19; TCGA PRAD -0.148; TCGA SARC -0.086; TCGA THCA -0.119; TCGA STAD -0.147 |
hsa-miR-132-3p | S100A1 | 9 cancers: BLCA; COAD; LIHC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.322; TCGA COAD -0.284; TCGA LIHC -0.256; TCGA LUAD -0.471; TCGA LUSC -0.479; TCGA PAAD -0.304; TCGA PRAD -0.251; TCGA STAD -0.453; TCGA UCEC -0.584 |
hsa-miR-132-3p | TSPAN6 | 10 cancers: BLCA; COAD; KIRC; LGG; LIHC; LUAD; OV; PAAD; PRAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.187; TCGA COAD -0.725; TCGA KIRC -0.16; TCGA LGG -0.178; TCGA LIHC -0.271; TCGA LUAD -0.161; TCGA OV -0.172; TCGA PAAD -0.445; TCGA PRAD -0.092; TCGA UCEC -0.078 |
hsa-miR-132-3p | ZNF652 | 9 cancers: BLCA; BRCA; ESCA; LGG; LIHC; LUAD; LUSC; THCA; UCEC | miRNAWalker2 validate; MirTarget | TCGA BLCA -0.265; TCGA BRCA -0.205; TCGA ESCA -0.36; TCGA LGG -0.095; TCGA LIHC -0.175; TCGA LUAD -0.147; TCGA LUSC -0.121; TCGA THCA -0.103; TCGA UCEC -0.066 |
hsa-miR-132-3p | CAMTA1 | 9 cancers: BLCA; CESC; COAD; HNSC; LIHC; LUAD; LUSC; PRAD; STAD | MirTarget | TCGA BLCA -0.066; TCGA CESC -0.144; TCGA COAD -0.146; TCGA HNSC -0.185; TCGA LIHC -0.174; TCGA LUAD -0.13; TCGA LUSC -0.126; TCGA PRAD -0.087; TCGA STAD -0.24 |
hsa-miR-132-3p | ZNF133 | 11 cancers: BLCA; CESC; COAD; HNSC; KIRP; LGG; LUAD; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.199; TCGA CESC -0.112; TCGA COAD -0.213; TCGA HNSC -0.222; TCGA KIRP -0.159; TCGA LGG -0.122; TCGA LUAD -0.116; TCGA PRAD -0.139; TCGA THCA -0.053; TCGA STAD -0.142; TCGA UCEC -0.14 |
hsa-miR-132-3p | SAP30L | 9 cancers: BLCA; BRCA; COAD; ESCA; KIRC; KIRP; LIHC; LUAD; LUSC | MirTarget; miRNATAP | TCGA BLCA -0.062; TCGA BRCA -0.094; TCGA COAD -0.098; TCGA ESCA -0.184; TCGA KIRC -0.12; TCGA KIRP -0.158; TCGA LIHC -0.184; TCGA LUAD -0.103; TCGA LUSC -0.213 |
hsa-miR-132-3p | NFE2L2 | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LGG; LIHC; PAAD; SARC; STAD | MirTarget | TCGA BLCA -0.099; TCGA BRCA -0.072; TCGA COAD -0.108; TCGA ESCA -0.304; TCGA HNSC -0.145; TCGA LGG -0.082; TCGA LIHC -0.212; TCGA PAAD -0.113; TCGA SARC -0.117; TCGA STAD -0.112 |
hsa-miR-132-3p | PIGY | 10 cancers: BLCA; COAD; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; PRAD; STAD | MirTarget | TCGA BLCA -0.068; TCGA COAD -0.213; TCGA HNSC -0.113; TCGA KIRP -0.094; TCGA LGG -0.052; TCGA LIHC -0.248; TCGA LUAD -0.07; TCGA LUSC -0.154; TCGA PRAD -0.1; TCGA STAD -0.126 |
hsa-miR-132-3p | EPM2AIP1 | 10 cancers: BLCA; BRCA; COAD; KIRC; LIHC; LUAD; LUSC; OV; SARC; UCEC | MirTarget | TCGA BLCA -0.203; TCGA BRCA -0.086; TCGA COAD -0.697; TCGA KIRC -0.155; TCGA LIHC -0.316; TCGA LUAD -0.174; TCGA LUSC -0.16; TCGA OV -0.114; TCGA SARC -0.148; TCGA UCEC -0.369 |
hsa-miR-132-3p | BLCAP | 10 cancers: BLCA; BRCA; COAD; HNSC; KIRC; KIRP; LIHC; OV; PRAD; UCEC | MirTarget | TCGA BLCA -0.259; TCGA BRCA -0.089; TCGA COAD -0.233; TCGA HNSC -0.137; TCGA KIRC -0.128; TCGA KIRP -0.187; TCGA LIHC -0.317; TCGA OV -0.063; TCGA PRAD -0.075; TCGA UCEC -0.078 |
hsa-miR-132-3p | SRCIN1 | 9 cancers: BLCA; BRCA; COAD; LIHC; LUAD; LUSC; OV; SARC; UCEC | mirMAP | TCGA BLCA -0.634; TCGA BRCA -0.219; TCGA COAD -0.537; TCGA LIHC -0.206; TCGA LUAD -0.575; TCGA LUSC -0.287; TCGA OV -0.297; TCGA SARC -0.355; TCGA UCEC -0.433 |
hsa-miR-132-3p | ZNF629 | 10 cancers: BLCA; BRCA; COAD; ESCA; KIRC; LGG; LUAD; OV; PRAD; THCA | mirMAP | TCGA BLCA -0.156; TCGA BRCA -0.084; TCGA COAD -0.125; TCGA ESCA -0.178; TCGA KIRC -0.107; TCGA LGG -0.087; TCGA LUAD -0.113; TCGA OV -0.116; TCGA PRAD -0.054; TCGA THCA -0.071 |
hsa-miR-132-3p | C1orf21 | 9 cancers: BLCA; BRCA; ESCA; LIHC; LUAD; LUSC; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.187; TCGA BRCA -0.229; TCGA ESCA -0.514; TCGA LIHC -0.269; TCGA LUAD -0.308; TCGA LUSC -0.245; TCGA SARC -0.223; TCGA THCA -0.227; TCGA STAD -0.424 |
hsa-miR-132-3p | RC3H1 | 9 cancers: BLCA; BRCA; CESC; COAD; LGG; LIHC; LUAD; SARC; STAD | mirMAP | TCGA BLCA -0.093; TCGA BRCA -0.104; TCGA CESC -0.081; TCGA COAD -0.11; TCGA LGG -0.055; TCGA LIHC -0.157; TCGA LUAD -0.102; TCGA SARC -0.063; TCGA STAD -0.088 |
hsa-miR-132-3p | SHANK2 | 11 cancers: BLCA; BRCA; COAD; KIRC; LIHC; LUAD; OV; PAAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.316; TCGA BRCA -0.237; TCGA COAD -0.32; TCGA KIRC -0.425; TCGA LIHC -0.339; TCGA LUAD -0.266; TCGA OV -0.242; TCGA PAAD -0.208; TCGA THCA -0.189; TCGA STAD -0.346; TCGA UCEC -0.293 |
hsa-miR-132-3p | KSR2 | 9 cancers: BLCA; BRCA; KIRC; LUAD; LUSC; OV; SARC; THCA; UCEC | mirMAP; miRNATAP | TCGA BLCA -0.54; TCGA BRCA -0.193; TCGA KIRC -0.684; TCGA LUAD -0.27; TCGA LUSC -0.435; TCGA OV -0.232; TCGA SARC -0.354; TCGA THCA -0.126; TCGA UCEC -0.259 |
hsa-miR-132-3p | PBX1 | 9 cancers: BLCA; BRCA; COAD; ESCA; KIRC; LUSC; OV; SARC; STAD | mirMAP | TCGA BLCA -0.393; TCGA BRCA -0.283; TCGA COAD -0.528; TCGA ESCA -0.594; TCGA KIRC -0.492; TCGA LUSC -0.311; TCGA OV -0.136; TCGA SARC -0.348; TCGA STAD -0.423 |
hsa-miR-132-3p | SAMD12 | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; UCEC | miRNATAP | TCGA BLCA -0.696; TCGA BRCA -0.299; TCGA COAD -0.271; TCGA ESCA -0.737; TCGA HNSC -0.354; TCGA KIRC -0.296; TCGA LUAD -0.274; TCGA LUSC -0.383; TCGA PAAD -0.336; TCGA UCEC -0.51 |
hsa-miR-132-3p | EIF4A2 | 12 cancers: BLCA; CESC; COAD; HNSC; KIRC; KIRP; LIHC; LUSC; OV; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.168; TCGA CESC -0.136; TCGA COAD -0.177; TCGA HNSC -0.147; TCGA KIRC -0.082; TCGA KIRP -0.175; TCGA LIHC -0.167; TCGA LUSC -0.183; TCGA OV -0.101; TCGA SARC -0.139; TCGA STAD -0.184; TCGA UCEC -0.073 |
hsa-miR-132-3p | PUM2 | 9 cancers: BLCA; BRCA; COAD; KIRC; LIHC; LUSC; OV; SARC; UCEC | miRNATAP | TCGA BLCA -0.124; TCGA BRCA -0.079; TCGA COAD -0.153; TCGA KIRC -0.087; TCGA LIHC -0.152; TCGA LUSC -0.095; TCGA OV -0.088; TCGA SARC -0.074; TCGA UCEC -0.05 |
hsa-miR-132-3p | DCAF8 | 11 cancers: BLCA; BRCA; ESCA; KIRC; KIRP; LIHC; LUAD; LUSC; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.108; TCGA BRCA -0.08; TCGA ESCA -0.178; TCGA KIRC -0.124; TCGA KIRP -0.109; TCGA LIHC -0.289; TCGA LUAD -0.135; TCGA LUSC -0.141; TCGA SARC -0.096; TCGA STAD -0.174; TCGA UCEC -0.096 |
hsa-miR-132-3p | PHKG2 | 9 cancers: BLCA; BRCA; CESC; HNSC; LUAD; LUSC; OV; PAAD; PRAD | miRNATAP | TCGA BLCA -0.079; TCGA BRCA -0.075; TCGA CESC -0.112; TCGA HNSC -0.183; TCGA LUAD -0.206; TCGA LUSC -0.129; TCGA OV -0.07; TCGA PAAD -0.186; TCGA PRAD -0.095 |
hsa-miR-132-3p | ATP6V0E1 | 10 cancers: BRCA; CESC; COAD; HNSC; KIRP; LGG; LUAD; PRAD; THCA; STAD | miRNAWalker2 validate | TCGA BRCA -0.1; TCGA CESC -0.103; TCGA COAD -0.138; TCGA HNSC -0.093; TCGA KIRP -0.139; TCGA LGG -0.152; TCGA LUAD -0.118; TCGA PRAD -0.066; TCGA THCA -0.116; TCGA STAD -0.136 |
hsa-miR-132-3p | TUSC2 | 9 cancers: BRCA; COAD; HNSC; KIRC; LUSC; OV; PRAD; SARC; STAD | miRNAWalker2 validate | TCGA BRCA -0.083; TCGA COAD -0.114; TCGA HNSC -0.104; TCGA KIRC -0.106; TCGA LUSC -0.119; TCGA OV -0.061; TCGA PRAD -0.083; TCGA SARC -0.078; TCGA STAD -0.083 |
hsa-miR-132-3p | NVL | 9 cancers: BRCA; COAD; KIRP; LGG; LUAD; LUSC; PAAD; PRAD; SARC | MirTarget | TCGA BRCA -0.176; TCGA COAD -0.146; TCGA KIRP -0.102; TCGA LGG -0.136; TCGA LUAD -0.142; TCGA LUSC -0.076; TCGA PAAD -0.08; TCGA PRAD -0.107; TCGA SARC -0.058 |
hsa-miR-132-3p | SERP1 | 11 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRP; LGG; LIHC; LUSC; PAAD; PRAD | MirTarget | TCGA BRCA -0.103; TCGA CESC -0.178; TCGA COAD -0.091; TCGA ESCA -0.177; TCGA HNSC -0.16; TCGA KIRP -0.116; TCGA LGG -0.078; TCGA LIHC -0.196; TCGA LUSC -0.222; TCGA PAAD -0.181; TCGA PRAD -0.114 |
hsa-miR-132-3p | EBPL | 10 cancers: CESC; COAD; HNSC; LGG; LIHC; LUSC; OV; PAAD; PRAD; THCA | miRNAWalker2 validate | TCGA CESC -0.183; TCGA COAD -0.308; TCGA HNSC -0.237; TCGA LGG -0.143; TCGA LIHC -0.455; TCGA LUSC -0.147; TCGA OV -0.16; TCGA PAAD -0.213; TCGA PRAD -0.14; TCGA THCA -0.141 |
hsa-miR-132-3p | TFDP2 | 9 cancers: COAD; HNSC; KIRC; LGG; LIHC; LUSC; PAAD; PRAD; STAD | mirMAP | TCGA COAD -0.228; TCGA HNSC -0.204; TCGA KIRC -0.237; TCGA LGG -0.064; TCGA LIHC -0.095; TCGA LUSC -0.236; TCGA PAAD -0.134; TCGA PRAD -0.094; TCGA STAD -0.177 |
Enriched cancer pathways of putative targets