microRNA information: hsa-miR-133a-3p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-133a-3p | miRbase |
| Accession: | MIMAT0000427 | miRbase |
| Precursor name: | hsa-mir-133a-1 | miRbase |
| Precursor accession: | MI0000450 | miRbase |
| Symbol: | MIR133A1 | HGNC |
| RefSeq ID: | NR_029675 | GenBank |
| Sequence: | UUUGGUCCCCUUCAACCAGCUG |
Reported expression in cancers: hsa-miR-133a-3p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-133a-3p | bladder cancer | downregulation | "We identified a subset of 7 miRNAs miR-145 miR-30a ......" | 19378336 | qPCR |
| hsa-miR-133a-3p | bladder cancer | downregulation | "We have recently identified down-regulated microRN ......" | 20160723 | |
| hsa-miR-133a-3p | bladder cancer | downregulation | "We investigated the miRNA expression signature of ......" | 21304530 | |
| hsa-miR-133a-3p | bladder cancer | downregulation | "We previously demonstrated that miR-133a is a tumo ......" | 21396852 | |
| hsa-miR-133a-3p | breast cancer | downregulation | "In this study the expression profile of microRNA-1 ......" | 23786162 | |
| hsa-miR-133a-3p | cervical and endocervical cancer | downregulation | "microRNA-133a miR-133a has been reported to play a ......" | 26134491 | Reverse transcription PCR; qPCR |
| hsa-miR-133a-3p | colorectal cancer | downregulation | "Tumor suppressor functions of miR 133a in colorect ......" | 23723074 | Microarray |
| hsa-miR-133a-3p | colorectal cancer | downregulation | "In recent studies of microRNA expression miR-133a ......" | 23968734 | |
| hsa-miR-133a-3p | colorectal cancer | downregulation | "The clinicopathological significance of miR 133a i ......" | 25104873 | qPCR |
| hsa-miR-133a-3p | colorectal cancer | downregulation | "Decreased expression of miR 133a correlates with p ......" | 25170220 | qPCR |
| hsa-miR-133a-3p | colorectal cancer | downregulation | "MicroRNAs miRs are a type of small non-coding RNA ......" | 25621061 | |
| hsa-miR-133a-3p | colorectal cancer | downregulation | "Expression of 16 miRNAs miRNA-9 21 30d 31 106a 127 ......" | 25773836 | qPCR |
| hsa-miR-133a-3p | colorectal cancer | downregulation | "Screening miRNAs for early diagnosis of colorectal ......" | 27022275 | RNA-Seq |
| hsa-miR-133a-3p | esophageal cancer | downregulation | "A comparison of miRNA signatures from ESCC and our ......" | 21351259 | |
| hsa-miR-133a-3p | esophageal cancer | downregulation | "Re-analysis of a public microarray database reveal ......" | 22641236 | Microarray |
| hsa-miR-133a-3p | esophageal cancer | downregulation | "microRNA-133a miR-133a and miR-133b located on chr ......" | 25280517 | qPCR |
| hsa-miR-133a-3p | gastric cancer | downregulation | "Here we found that miR-145 miR-133a and miR-133b w ......" | 24613927 | |
| hsa-miR-133a-3p | gastric cancer | downregulation | "We also found that miR-133 was down-regulated in 1 ......" | 25152372 | |
| hsa-miR-133a-3p | gastric cancer | downregulation | "Although miR-133a has been shown to function as a ......" | 25620172 | |
| hsa-miR-133a-3p | gastric cancer | downregulation | "To investigate the function and mechanism of miR-1 ......" | 25780292 | qPCR |
| hsa-miR-133a-3p | gastric cancer | downregulation | "Here a heatmap analysis of the miRNomes was perfor ......" | 26276722 | |
| hsa-miR-133a-3p | gastric cancer | downregulation | "Circulatory miR-133a is a marker shared by several ......" | 27268657 | |
| hsa-miR-133a-3p | head and neck cancer | upregulation | "Down-regulation of the miRNA miR-133a in many type ......" | 21109942 | |
| hsa-miR-133a-3p | kidney renal cell cancer | upregulation | "The most relevant differentially expressed microRN ......" | 24475095 | qPCR |
| hsa-miR-133a-3p | liver cancer | downregulation | "A growing body of evidence showed that microRNA-13 ......" | 26156803 | |
| hsa-miR-133a-3p | lung squamous cell cancer | downregulation | "MiR 133a is downregulated in non small cell lung c ......" | 25903369 | Reverse transcription PCR; qPCR |
| hsa-miR-133a-3p | lung squamous cell cancer | downregulation | "Down regulation of miR 133a as a poor prognosticat ......" | 27282282 | qPCR |
| hsa-miR-133a-3p | ovarian cancer | downregulation | "It has been demonstrated that microRNA miR-133a is ......" | 24944666 | |
| hsa-miR-133a-3p | pancreatic cancer | downregulation | "However the roles of miR-133a and its related mole ......" | 25198665 | qPCR |
| hsa-miR-133a-3p | prostate cancer | downregulation | "Our recent analyses of miRNA expression signatures ......" | 22068816 | |
| hsa-miR-133a-3p | sarcoma | downregulation | "The expression levels of miR-133a and miR-539 were ......" | 26388701 | qPCR |
Reported cancer pathway affected by hsa-miR-133a-3p

| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-133a-3p | bladder cancer | Apoptosis pathway | "The tumour suppressive function of miR 1 and miR 1 ......" | 21304530 | |
| hsa-miR-133a-3p | bladder cancer | Apoptosis pathway | "MiR 133a induces apoptosis through direct regulati ......" | 21396852 | Flow cytometry; Luciferase; Western blot |
| hsa-miR-133a-3p | breast cancer | cell cycle pathway | "In this study the expression profile of microRNA-1 ......" | 23786162 | Luciferase |
| hsa-miR-133a-3p | cervical and endocervical cancer | Apoptosis pathway | "miR 133a inhibits cervical cancer growth by target ......" | 26134491 | Colony formation; Western blot; Luciferase |
| hsa-miR-133a-3p | colorectal cancer | cell cycle pathway; Apoptosis pathway | "Tumor suppressor functions of miR 133a in colorect ......" | 23723074 | |
| hsa-miR-133a-3p | gastric cancer | cell cycle pathway | "MiR 145 miR 133a and miR 133b inhibit proliferatio ......" | 24613927 | |
| hsa-miR-133a-3p | gastric cancer | Apoptosis pathway | "Although miR-133a has been shown to function as a ......" | 25620172 | |
| hsa-miR-133a-3p | gastric cancer | Apoptosis pathway | "Tumor suppressor role of miR 133a in gastric cance ......" | 25780292 | Colony formation; Western blot; Luciferase |
| hsa-miR-133a-3p | gastric cancer | Apoptosis pathway | "In the present study downregulation of microRNA mi ......" | 25815687 | |
| hsa-miR-133a-3p | gastric cancer | Apoptosis pathway | "Impairment of growth of gastric carcinoma by miR 1 ......" | 26076812 | MTT assay; Luciferase |
| hsa-miR-133a-3p | gastric cancer | cell cycle pathway | "Ursolic Acid Inhibits the Proliferation of Gastric ......" | 26629938 | |
| hsa-miR-133a-3p | glioblastoma | Apoptosis pathway | "Growth of glioblastoma is inhibited by miR 133 med ......" | 26138587 | Flow cytometry; MTT assay; Luciferase |
| hsa-miR-133a-3p | kidney renal cell cancer | cell cycle pathway; Apoptosis pathway | "The functional significance of miR 1 and miR 133a ......" | 21745735 | Luciferase |
| hsa-miR-133a-3p | liver cancer | Apoptosis pathway | "HepG2 cells transfected with versican 3'-UTR displ ......" | 23180826 | Colony formation |
| hsa-miR-133a-3p | liver cancer | cell cycle pathway; Apoptosis pathway | "A growing body of evidence showed that microRNA-13 ......" | 26156803 | Colony formation; Luciferase |
| hsa-miR-133a-3p | liver cancer | Apoptosis pathway | "Our study aimed to reveal the roles of microRNA mi ......" | 26173501 | Western blot; Luciferase |
| hsa-miR-133a-3p | lung cancer | Epithelial mesenchymal transition pathway | "MicroRNA miR was implicated in the tumorigenesis o ......" | 26858166 | |
| hsa-miR-133a-3p | ovarian cancer | cell cycle pathway | "miR 133a suppresses ovarian cancer cell proliferat ......" | 24127040 | Colony formation |
| hsa-miR-133a-3p | ovarian cancer | Apoptosis pathway | "It has been demonstrated that microRNA miR-133a is ......" | 24944666 | |
| hsa-miR-133a-3p | pancreatic cancer | Apoptosis pathway | "miR 133a functions as a tumor suppressor and direc ......" | 25198665 | Luciferase |
| hsa-miR-133a-3p | pancreatic cancer | Apoptosis pathway | "The effects of propofol on Panc-1 cell proliferati ......" | 26214431 | MTT assay |
| hsa-miR-133a-3p | sarcoma | Apoptosis pathway | "Although miR-133a has been shown to be important i ......" | 23756231 |
Reported cancer prognosis affected by hsa-miR-133a-3p
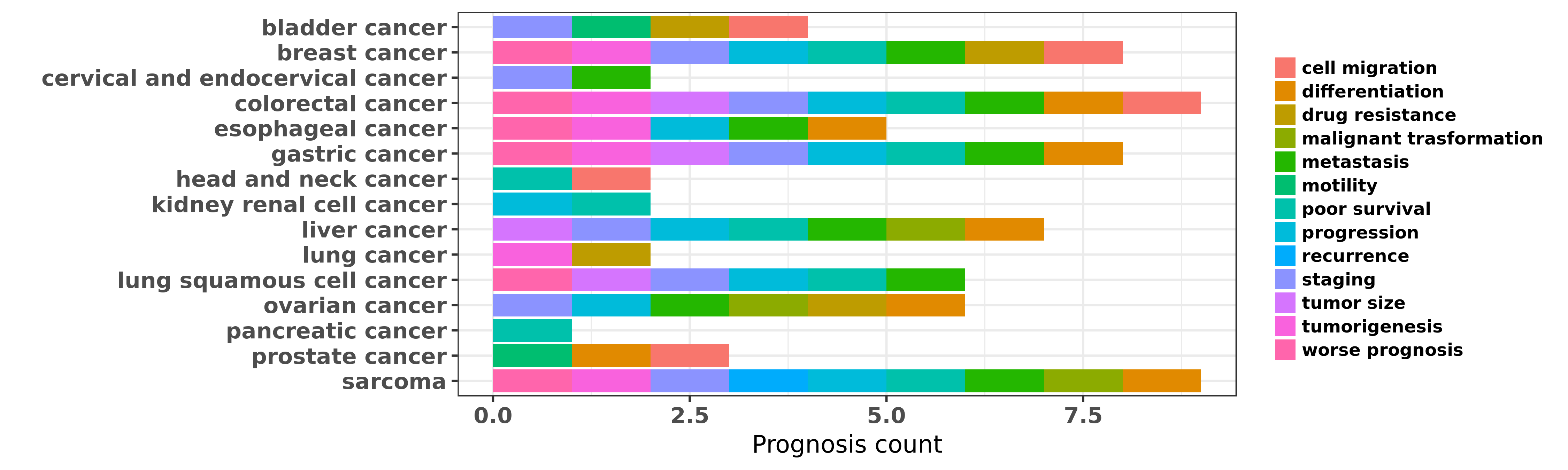
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-133a-3p | bladder cancer | staging | "Expression levels of miRNAs were assessed by quant ......" | 23169479 | |
| hsa-miR-133a-3p | bladder cancer | cell migration | "MicroRNA-133a miR-133a and microRNA-133b miR-133b ......" | 23206218 | Western blot; Luciferase; Cell migration assay |
| hsa-miR-133a-3p | bladder cancer | drug resistance | "To solve the problem we applied miRNA response ele ......" | 23442927 | Luciferase |
| hsa-miR-133a-3p | bladder cancer | motility | "A profile of miRNA expression was determined using ......" | 24966896 | |
| hsa-miR-133a-3p | breast cancer | poor survival; staging; metastasis; cell migration; progression; worse prognosis | "Loss of miR 133a expression associated with poor s ......" | 22292984 | Luciferase |
| hsa-miR-133a-3p | breast cancer | tumorigenesis | "In this study the expression profile of microRNA-1 ......" | 23786162 | Luciferase |
| hsa-miR-133a-3p | breast cancer | staging | "Global miRNA analysis was performed on serum from ......" | 24694649 | |
| hsa-miR-133a-3p | breast cancer | drug resistance | "MiR 133a Is Functionally Involved in Doxorubicin R ......" | 26107945 | |
| hsa-miR-133a-3p | breast cancer | poor survival | "We aimed to investigate the effect of UTMD of miR- ......" | 27465833 | MTT assay; Western blot |
| hsa-miR-133a-3p | cervical and endocervical cancer | staging; metastasis | "miR 133a inhibits cervical cancer growth by target ......" | 26134491 | Colony formation; Western blot; Luciferase |
| hsa-miR-133a-3p | colorectal cancer | progression | "Furthermore several of these miRNAs were associate ......" | 19843336 | |
| hsa-miR-133a-3p | colorectal cancer | metastasis; cell migration | "miR 133a represses tumour growth and metastasis in ......" | 23968734 | |
| hsa-miR-133a-3p | colorectal cancer | staging; metastasis; poor survival; worse prognosis | "The clinicopathological significance of miR 133a i ......" | 25104873 | |
| hsa-miR-133a-3p | colorectal cancer | worse prognosis; tumorigenesis; staging; metastasis; poor survival; differentiation; tumor size; progression | "Decreased expression of miR 133a correlates with p ......" | 25170220 | |
| hsa-miR-133a-3p | colorectal cancer | progression | "MicroRNAs miRs are a type of small non-coding RNA ......" | 25621061 | |
| hsa-miR-133a-3p | colorectal cancer | worse prognosis | "Screening miRNAs for early diagnosis of colorectal ......" | 27022275 | |
| hsa-miR-133a-3p | esophageal cancer | progression; tumorigenesis | "CD47 expression regulated by the miR 133a tumor su ......" | 22641236 | |
| hsa-miR-133a-3p | esophageal cancer | metastasis; differentiation | "Among these miRNAs that displayed unique miRNA exp ......" | 23516093 | |
| hsa-miR-133a-3p | esophageal cancer | worse prognosis | "FSCN1 and MMP14 were evaluated by immunohistochemi ......" | 24196787 | |
| hsa-miR-133a-3p | esophageal cancer | progression | "MiR 133a suppresses the migration and invasion of ......" | 26396670 | |
| hsa-miR-133a-3p | gastric cancer | progression | "MiR 145 miR 133a and miR 133b inhibit proliferatio ......" | 24613927 | |
| hsa-miR-133a-3p | gastric cancer | metastasis; tumor size; worse prognosis; poor survival | "miR 133 is a key negative regulator of CDC42 PAK p ......" | 25152372 | |
| hsa-miR-133a-3p | gastric cancer | metastasis | "Although miR-133a has been shown to function as a ......" | 25620172 | |
| hsa-miR-133a-3p | gastric cancer | tumorigenesis; staging; differentiation | "Tumor suppressor role of miR 133a in gastric cance ......" | 25780292 | Colony formation; Western blot; Luciferase |
| hsa-miR-133a-3p | gastric cancer | poor survival | "Impairment of growth of gastric carcinoma by miR 1 ......" | 26076812 | MTT assay; Luciferase |
| hsa-miR-133a-3p | gastric cancer | poor survival; staging; metastasis; differentiation | "Then we further investigated these GC specific miR ......" | 27173517 | |
| hsa-miR-133a-3p | head and neck cancer | poor survival | "In this study miRNA expression profiles of head an ......" | 19179615 | |
| hsa-miR-133a-3p | head and neck cancer | cell migration | "Caveolin 1 mediates tumor cell migration and invas ......" | 21109942 | Western blot; Luciferase |
| hsa-miR-133a-3p | head and neck cancer | cell migration | "Our expression signatures of human cancers includi ......" | 22378351 | Luciferase |
| hsa-miR-133a-3p | kidney renal cell cancer | progression; poor survival | "The most relevant differentially expressed microRN ......" | 24475095 | |
| hsa-miR-133a-3p | liver cancer | poor survival | "HepG2 cells transfected with versican 3'-UTR displ ......" | 23180826 | Colony formation |
| hsa-miR-133a-3p | liver cancer | staging; metastasis; differentiation; tumor size; poor survival | "A growing body of evidence showed that microRNA-13 ......" | 26156803 | Colony formation; Luciferase |
| hsa-miR-133a-3p | liver cancer | malignant trasformation; progression | "Our study aimed to reveal the roles of microRNA mi ......" | 26173501 | Western blot; Luciferase |
| hsa-miR-133a-3p | lung cancer | drug resistance | "To address this issue we inserted microRNA respons ......" | 25132116 | |
| hsa-miR-133a-3p | lung cancer | tumorigenesis | "MicroRNA miR was implicated in the tumorigenesis o ......" | 26858166 | |
| hsa-miR-133a-3p | lung squamous cell cancer | poor survival; progression; worse prognosis | "MiR-133a is known as a key regulator in skeletal a ......" | 24816813 | |
| hsa-miR-133a-3p | lung squamous cell cancer | poor survival; staging; metastasis; tumor size | "MiR 133a is downregulated in non small cell lung c ......" | 25903369 | |
| hsa-miR-133a-3p | lung squamous cell cancer | progression; staging; poor survival | "Down regulation of miR 133a as a poor prognosticat ......" | 27282282 | |
| hsa-miR-133a-3p | ovarian cancer | progression; malignant trasformation; staging; metastasis; differentiation | "It has been demonstrated that microRNA miR-133a is ......" | 24944666 | |
| hsa-miR-133a-3p | ovarian cancer | drug resistance | "Of these 10 miRNAs miR-193a-5p miR-375 miR-339-3p ......" | 26485143 | |
| hsa-miR-133a-3p | pancreatic cancer | poor survival | "miR 133a functions as a tumor suppressor and direc ......" | 25198665 | Luciferase |
| hsa-miR-133a-3p | prostate cancer | differentiation; motility; cell migration | "microRNA-133 miR-133 has long been recognized as a ......" | 22407299 | Western blot; Luciferase |
| hsa-miR-133a-3p | sarcoma | differentiation | "Distinct roles for miR 1 and miR 133a in the proli ......" | 20466878 | |
| hsa-miR-133a-3p | sarcoma | progression; tumorigenesis; worse prognosis | "Although miR-133a has been shown to be important i ......" | 23756231 | |
| hsa-miR-133a-3p | sarcoma | malignant trasformation; metastasis; poor survival; worse prognosis | "We identified key microRNAs miRNAs that were dereg ......" | 24715690 | |
| hsa-miR-133a-3p | sarcoma | worse prognosis; staging; metastasis; recurrence; poor survival; progression | "Down regulation of miR 133a and miR 539 are associ ......" | 26388701 | |
| hsa-miR-133a-3p | sarcoma | metastasis; progression | "A growing body of evidence showed that microRNA-13 ......" | 26845446 | Luciferase |
Reported gene related to hsa-miR-133a-3p
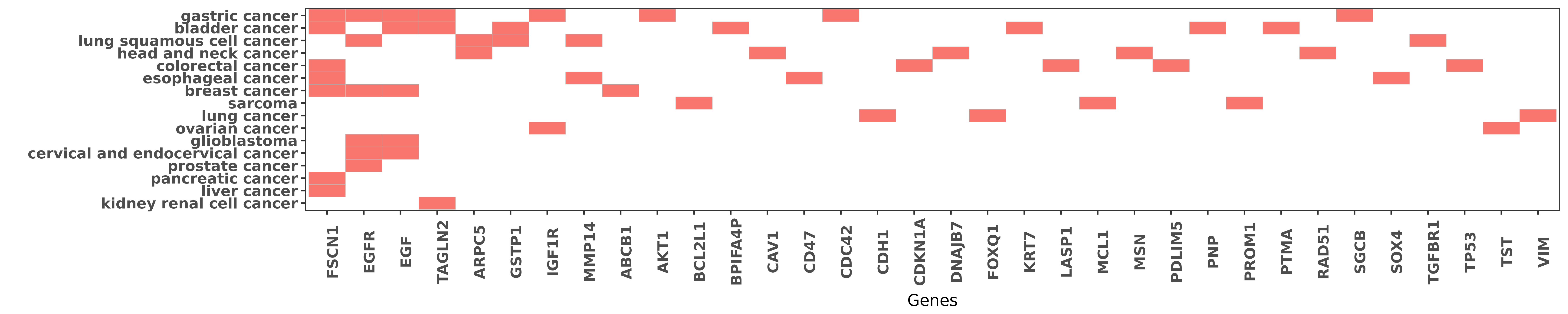
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-133a-3p | breast cancer | EGFR | "Epidermal growth factor receptor EGFR expression a ......" | 27465833 |
| hsa-miR-133a-3p | breast cancer | EGFR | "Bioinformatics prediction showed that epidermal gr ......" | 23786162 |
| hsa-miR-133a-3p | cervical and endocervical cancer | EGFR | "miR 133a inhibits cervical cancer growth by target ......" | 26134491 |
| hsa-miR-133a-3p | gastric cancer | EGFR | "Moreover the 3'-untranslated region 3'UTR of Her-2 ......" | 26076812 |
| hsa-miR-133a-3p | glioblastoma | EGFR | "Growth of glioblastoma is inhibited by miR 133 med ......" | 26138587 |
| hsa-miR-133a-3p | lung squamous cell cancer | EGFR | "MiR-133a expression was negatively correlated to l ......" | 25903369 |
| hsa-miR-133a-3p | lung squamous cell cancer | EGFR | "MiR-133a can inhibit cell invasiveness and cell gr ......" | 24816813 |
| hsa-miR-133a-3p | prostate cancer | EGFR | "We also provide the first evidence that miR-133 ma ......" | 22407299 |
| hsa-miR-133a-3p | bladder cancer | FSCN1 | "miR 145 and miR 133a function as tumour suppressor ......" | 20160723 |
| hsa-miR-133a-3p | breast cancer | FSCN1 | "Luciferase assay was performed to assess miR-133a ......" | 22292984 |
| hsa-miR-133a-3p | colorectal cancer | FSCN1 | "Furthermore the Fascin1 FSCN1 gene was identified ......" | 25621061 |
| hsa-miR-133a-3p | esophageal cancer | FSCN1 | "miR 145 miR 133a and miR 133b: Tumor suppressive m ......" | 21351259 |
| hsa-miR-133a-3p | esophageal cancer | FSCN1 | "FSCN1 and MMP14 were evaluated by immunohistochemi ......" | 24196787 |
| hsa-miR-133a-3p | gastric cancer | FSCN1 | "In the present study downregulation of microRNA mi ......" | 25815687 |
| hsa-miR-133a-3p | liver cancer | FSCN1 | "Our study aimed to reveal the roles of microRNA mi ......" | 26173501 |
| hsa-miR-133a-3p | pancreatic cancer | FSCN1 | "miR 133a functions as a tumor suppressor and direc ......" | 25198665 |
| hsa-miR-133a-3p | bladder cancer | EGF | "The first evidence was provided that miR-133a and ......" | 23206218 |
| hsa-miR-133a-3p | breast cancer | EGF | "Epidermal growth factor receptor EGFR expression a ......" | 27465833 |
| hsa-miR-133a-3p | breast cancer | EGF | "Bioinformatics prediction showed that epidermal gr ......" | 23786162 |
| hsa-miR-133a-3p | cervical and endocervical cancer | EGF | "The epidermal growth factor receptor EGFR was conf ......" | 26134491 |
| hsa-miR-133a-3p | gastric cancer | EGF | "Moreover the 3'-untranslated region 3'UTR of Her-2 ......" | 26076812 |
| hsa-miR-133a-3p | glioblastoma | EGF | "Bioinformatic analysis was performed showing that ......" | 26138587 |
| hsa-miR-133a-3p | bladder cancer | TAGLN2 | "The tumour suppressive function of miR 1 and miR 1 ......" | 21304530 |
| hsa-miR-133a-3p | gastric cancer | TAGLN2 | "It was found that the expression of miR-133a was d ......" | 25620172 |
| hsa-miR-133a-3p | kidney renal cell cancer | TAGLN2 | "The expression level of TAGLN2 mRNA was significan ......" | 21745735 |
| hsa-miR-133a-3p | head and neck cancer | ARPC5 | "The genome-wide gene expression analysis and bioin ......" | 22378351 |
| hsa-miR-133a-3p | lung squamous cell cancer | ARPC5 | "Furthermore transient transfection of miR-133a rep ......" | 22089643 |
| hsa-miR-133a-3p | head and neck cancer | DNAJB7 | "Restoration of miR-133a in HNSCC cell lines FaDu H ......" | 22266319 |
| hsa-miR-133a-3p | head and neck cancer | DNAJB7 | "In HSC3 cells restoration of miR-133a or silencing ......" | 22378351 |
| hsa-miR-133a-3p | bladder cancer | GSTP1 | "MiR 133a induces apoptosis through direct regulati ......" | 21396852 |
| hsa-miR-133a-3p | lung squamous cell cancer | GSTP1 | "Furthermore transient transfection of miR-133a rep ......" | 22089643 |
| hsa-miR-133a-3p | gastric cancer | IGF1R | "Tumor suppressor role of miR 133a in gastric cance ......" | 25780292 |
| hsa-miR-133a-3p | ovarian cancer | IGF1R | "Through in silico search we found that the 3'-untr ......" | 24127040 |
| hsa-miR-133a-3p | esophageal cancer | MMP14 | "FSCN1 and MMP14 were evaluated by immunohistochemi ......" | 24196787 |
| hsa-miR-133a-3p | lung squamous cell cancer | MMP14 | "The levels of miR-133a were negatively correlated ......" | 27282282 |
| hsa-miR-133a-3p | breast cancer | ABCB1 | "MiR 133a Is Functionally Involved in Doxorubicin R ......" | 26107945 |
| hsa-miR-133a-3p | gastric cancer | AKT1 | "Overexpression of miR-133a increased the G1 phase ......" | 26629938 |
| hsa-miR-133a-3p | sarcoma | BCL2L1 | "Furthermore bioinformatic prediction and experimen ......" | 23756231 |
| hsa-miR-133a-3p | bladder cancer | BPIFA4P | "On the base of the microRNA miRNA expression signa ......" | 21304530 |
| hsa-miR-133a-3p | head and neck cancer | CAV1 | "Caveolin 1 mediates tumor cell migration and invas ......" | 21109942 |
| hsa-miR-133a-3p | esophageal cancer | CD47 | "CD47 expression regulated by the miR 133a tumor su ......" | 22641236 |
| hsa-miR-133a-3p | gastric cancer | CDC42 | "miR 133 is a key negative regulator of CDC42 PAK p ......" | 25152372 |
| hsa-miR-133a-3p | lung cancer | CDH1 | "Further investigation revealed that this inhibitio ......" | 26858166 |
| hsa-miR-133a-3p | colorectal cancer | CDKN1A | "Cell-cycle analysis revealed that miR-133a induced ......" | 23723074 |
| hsa-miR-133a-3p | lung cancer | FOXQ1 | "We have identified miR-133 as a putative regulator ......" | 26858166 |
| hsa-miR-133a-3p | bladder cancer | KRT7 | "Gain-of-function analysis revealed that KRT7 mRNA ......" | 19378336 |
| hsa-miR-133a-3p | colorectal cancer | LASP1 | "In contrast to the phenotypes induced by miR-133a ......" | 23968734 |
| hsa-miR-133a-3p | sarcoma | MCL1 | "Furthermore bioinformatic prediction and experimen ......" | 23756231 |
| hsa-miR-133a-3p | head and neck cancer | MSN | "Genome-wide gene expression analysis of miR-133a t ......" | 22266319 |
| hsa-miR-133a-3p | colorectal cancer | PDLIM5 | "miR 133a represses tumour growth and metastasis in ......" | 23968734 |
| hsa-miR-133a-3p | bladder cancer | PNP | "PTMA and PNP were identified as new target genes r ......" | 22378464 |
| hsa-miR-133a-3p | sarcoma | PROM1 | "Furthermore in a clinical study high expression le ......" | 24715690 |
| hsa-miR-133a-3p | bladder cancer | PTMA | "PTMA and PNP were identified as new target genes r ......" | 22378464 |
| hsa-miR-133a-3p | head and neck cancer | RAD51 | "The genome-wide gene expression analysis and bioin ......" | 22378351 |
| hsa-miR-133a-3p | gastric cancer | SGCB | "Furthermore miR-133a inhibited tumor growth and xe ......" | 25780292 |
| hsa-miR-133a-3p | esophageal cancer | SOX4 | "MiR 133a suppresses the migration and invasion of ......" | 26396670 |
| hsa-miR-133a-3p | lung squamous cell cancer | TGFBR1 | "MiR-133a can inhibit cell invasiveness and cell gr ......" | 24816813 |
| hsa-miR-133a-3p | colorectal cancer | TP53 | "We further revealed that miR-133a markedly increas ......" | 23723074 |
| hsa-miR-133a-3p | ovarian cancer | TST | "Quantitative polymerase chain reaction was employe ......" | 24944666 |
| hsa-miR-133a-3p | lung cancer | VIM | "Further investigation revealed that this inhibitio ......" | 26858166 |
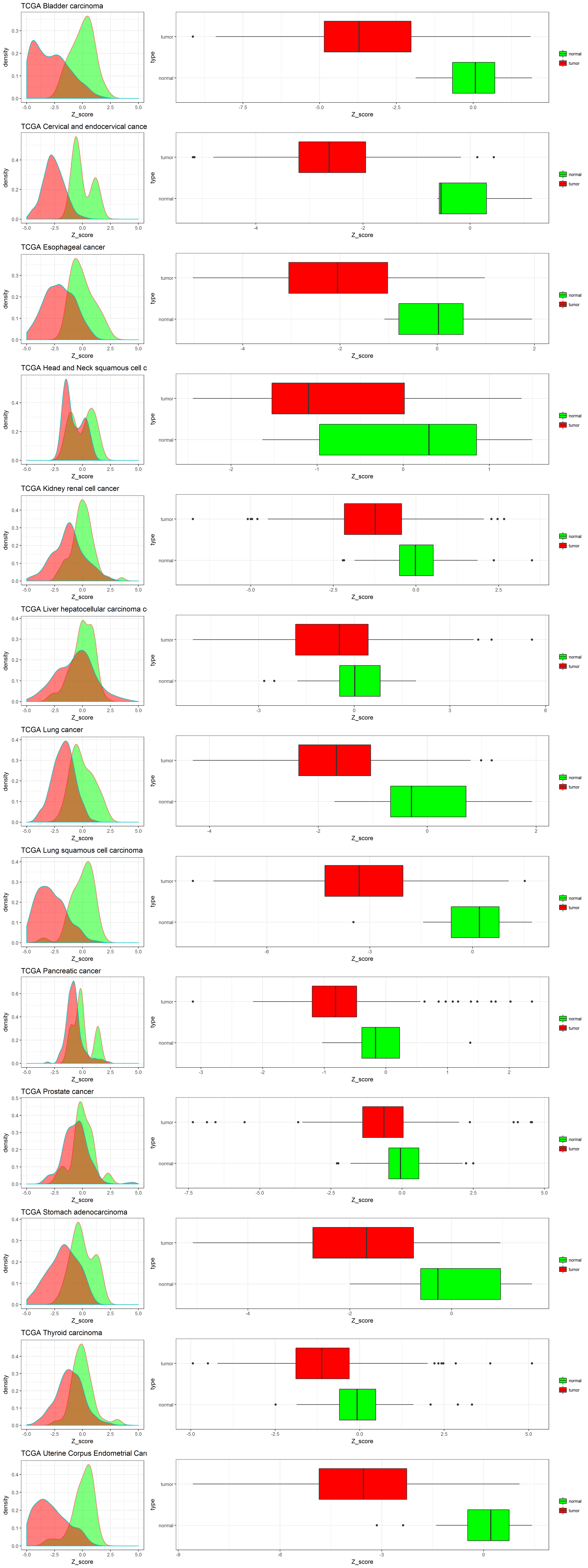
Expression profile in cancer corhorts:
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-133a-3p | ARL6IP1 | 9 cancers: BLCA; COAD; ESCA; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.108; TCGA COAD -0.089; TCGA ESCA -0.128; TCGA LUAD -0.119; TCGA LUSC -0.105; TCGA PAAD -0.103; TCGA PRAD -0.231; TCGA STAD -0.144; TCGA UCEC -0.068 |
hsa-miR-133a-3p | PRELID1 | 13 cancers: BLCA; COAD; ESCA; KIRC; LIHC; LUAD; LUSC; OV; PAAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.058; TCGA COAD -0.077; TCGA ESCA -0.105; TCGA KIRC -0.131; TCGA LIHC -0.085; TCGA LUAD -0.078; TCGA LUSC -0.066; TCGA OV -0.078; TCGA PAAD -0.061; TCGA SARC -0.052; TCGA THCA -0.057; TCGA STAD -0.051; TCGA UCEC -0.071 |
hsa-miR-133a-3p | VEGFA | 12 cancers: BLCA; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; PAAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.191; TCGA COAD -0.089; TCGA ESCA -0.163; TCGA HNSC -0.064; TCGA KIRC -0.335; TCGA LIHC -0.068; TCGA LUAD -0.105; TCGA LUSC -0.087; TCGA PAAD -0.179; TCGA SARC -0.051; TCGA STAD -0.175; TCGA UCEC -0.056 |
hsa-miR-133a-3p | RFC3 | 9 cancers: BLCA; COAD; ESCA; LGG; LIHC; LUAD; LUSC; PRAD; STAD | MirTarget | TCGA BLCA -0.125; TCGA COAD -0.12; TCGA ESCA -0.189; TCGA LGG -0.058; TCGA LIHC -0.09; TCGA LUAD -0.216; TCGA LUSC -0.187; TCGA PRAD -0.185; TCGA STAD -0.245 |
hsa-miR-133a-3p | RBMX | 10 cancers: BLCA; COAD; KIRC; LGG; LIHC; LUAD; LUSC; OV; PRAD; STAD | MirTarget | TCGA BLCA -0.054; TCGA COAD -0.066; TCGA KIRC -0.061; TCGA LGG -0.076; TCGA LIHC -0.053; TCGA LUAD -0.065; TCGA LUSC -0.07; TCGA OV -0.058; TCGA PRAD -0.108; TCGA STAD -0.069 |
hsa-miR-133a-3p | DOLPP1 | 9 cancers: BLCA; COAD; ESCA; LIHC; LUAD; LUSC; PRAD; SARC; STAD | MirTarget | TCGA BLCA -0.089; TCGA COAD -0.092; TCGA ESCA -0.113; TCGA LIHC -0.087; TCGA LUAD -0.092; TCGA LUSC -0.161; TCGA PRAD -0.102; TCGA SARC -0.095; TCGA STAD -0.08 |
hsa-miR-133a-3p | NFS1 | 9 cancers: BLCA; ESCA; LIHC; LUAD; LUSC; OV; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.054; TCGA ESCA -0.067; TCGA LIHC -0.058; TCGA LUAD -0.079; TCGA LUSC -0.15; TCGA OV -0.053; TCGA PRAD -0.087; TCGA STAD -0.056; TCGA UCEC -0.066 |
hsa-miR-133a-3p | EIF4A1 | 9 cancers: BLCA; COAD; ESCA; LGG; LUAD; LUSC; PRAD; SARC; STAD | MirTarget | TCGA BLCA -0.067; TCGA COAD -0.062; TCGA ESCA -0.081; TCGA LGG -0.092; TCGA LUAD -0.091; TCGA LUSC -0.09; TCGA PRAD -0.062; TCGA SARC -0.105; TCGA STAD -0.083 |
hsa-miR-133a-3p | ZSCAN16 | 11 cancers: BLCA; CESC; COAD; LGG; LIHC; LUAD; OV; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.096; TCGA CESC -0.074; TCGA COAD -0.071; TCGA LGG -0.108; TCGA LIHC -0.071; TCGA LUAD -0.09; TCGA OV -0.084; TCGA PRAD -0.155; TCGA THCA -0.072; TCGA STAD -0.058; TCGA UCEC -0.083 |
hsa-miR-133a-3p | UBE2Q1 | 10 cancers: BLCA; ESCA; KIRC; LIHC; LUAD; LUSC; PAAD; PRAD; THCA; STAD | MirTarget | TCGA BLCA -0.058; TCGA ESCA -0.058; TCGA KIRC -0.05; TCGA LIHC -0.079; TCGA LUAD -0.093; TCGA LUSC -0.09; TCGA PAAD -0.091; TCGA PRAD -0.101; TCGA THCA -0.056; TCGA STAD -0.054 |
hsa-miR-133a-3p | RCE1 | 9 cancers: BLCA; COAD; ESCA; LGG; LUAD; LUSC; PAAD; SARC; STAD | MirTarget | TCGA BLCA -0.07; TCGA COAD -0.06; TCGA ESCA -0.072; TCGA LGG -0.058; TCGA LUAD -0.075; TCGA LUSC -0.125; TCGA PAAD -0.054; TCGA SARC -0.05; TCGA STAD -0.107 |
hsa-miR-133a-3p | SF3B4 | 9 cancers: BLCA; ESCA; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; STAD | MirTarget | TCGA BLCA -0.067; TCGA ESCA -0.078; TCGA LGG -0.09; TCGA LIHC -0.06; TCGA LUAD -0.114; TCGA LUSC -0.155; TCGA PAAD -0.068; TCGA PRAD -0.073; TCGA STAD -0.061 |
hsa-miR-133a-3p | HAPLN1 | 9 cancers: BLCA; CESC; HNSC; KIRC; LUAD; LUSC; PAAD; THCA; UCEC | MirTarget | TCGA BLCA -0.377; TCGA CESC -0.2; TCGA HNSC -0.087; TCGA KIRC -0.41; TCGA LUAD -0.195; TCGA LUSC -0.314; TCGA PAAD -0.274; TCGA THCA -0.428; TCGA UCEC -0.261 |
hsa-miR-133a-3p | SEC61B | 9 cancers: COAD; ESCA; KIRC; LGG; LIHC; LUAD; LUSC; PRAD; SARC | miRNAWalker2 validate; MirTarget | TCGA COAD -0.088; TCGA ESCA -0.07; TCGA KIRC -0.074; TCGA LGG -0.087; TCGA LIHC -0.083; TCGA LUAD -0.089; TCGA LUSC -0.052; TCGA PRAD -0.058; TCGA SARC -0.073 |
hsa-miR-133a-3p | SEPHS2 | 9 cancers: COAD; ESCA; LIHC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | MirTarget | TCGA COAD -0.056; TCGA ESCA -0.082; TCGA LIHC -0.076; TCGA LUAD -0.119; TCGA LUSC -0.056; TCGA PRAD -0.071; TCGA SARC -0.052; TCGA STAD -0.077; TCGA UCEC -0.084 |
hsa-miR-133a-3p | TIMM17A | 9 cancers: COAD; ESCA; LIHC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA COAD -0.081; TCGA ESCA -0.072; TCGA LIHC -0.111; TCGA LUAD -0.111; TCGA LUSC -0.102; TCGA PAAD -0.055; TCGA PRAD -0.051; TCGA STAD -0.088; TCGA UCEC -0.072 |
hsa-miR-133a-3p | CLTA | 9 cancers: COAD; ESCA; LIHC; LUSC; OV; PAAD; SARC; THCA; STAD | MirTarget | TCGA COAD -0.065; TCGA ESCA -0.073; TCGA LIHC -0.073; TCGA LUSC -0.071; TCGA OV -0.081; TCGA PAAD -0.088; TCGA SARC -0.068; TCGA THCA -0.091; TCGA STAD -0.057 |
hsa-miR-133a-3p | SLC39A1 | 9 cancers: ESCA; KIRC; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; STAD | MirTarget | TCGA ESCA -0.052; TCGA KIRC -0.071; TCGA LGG -0.138; TCGA LIHC -0.061; TCGA LUAD -0.084; TCGA LUSC -0.055; TCGA PAAD -0.089; TCGA PRAD -0.058; TCGA STAD -0.073 |
Enriched cancer pathways of putative targets