microRNA information: hsa-miR-140-5p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-140-5p | miRbase |
| Accession: | MIMAT0000431 | miRbase |
| Precursor name: | hsa-mir-140 | miRbase |
| Precursor accession: | MI0000456 | miRbase |
| Symbol: | MIR140 | HGNC |
| RefSeq ID: | NR_029681 | GenBank |
| Sequence: | CAGUGGUUUUACCCUAUGGUAG |
Reported expression in cancers: hsa-miR-140-5p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-140-5p | breast cancer | downregulation | "Several reports have indicated that miR-140 a poss ......" | 23060440 | |
| hsa-miR-140-5p | breast cancer | downregulation | "Downregulation of miR 140 promotes cancer stem cel ......" | 23752191 | |
| hsa-miR-140-5p | breast cancer | downregulation | "Roles of microRNA 140 in stem cell associated earl ......" | 25426255 | |
| hsa-miR-140-5p | breast cancer | downregulation | "Tumor tissue had higher miR-140-5p expression than ......" | 25983620 | |
| hsa-miR-140-5p | cervical and endocervical cancer | deregulation | "Previous studies have revealed that MicroRNA-140-5 ......" | 27588393 | |
| hsa-miR-140-5p | colorectal cancer | downregulation | "We have systematically identified targets of hsa-m ......" | 25980495 | |
| hsa-miR-140-5p | colorectal cancer | downregulation | "The aim of this study was to investigate the clini ......" | 26402430 | qPCR |
| hsa-miR-140-5p | esophageal cancer | deregulation | "RT-PCR and Western blot were used to detect the ex ......" | 25322669 | |
| hsa-miR-140-5p | gastric cancer | upregulation | "Plasma samples from 3 independent groups comprise ......" | 26607322 | Microarray |
| hsa-miR-140-5p | lung cancer | deregulation | "Therefore we conduct to systematically identify mi ......" | 26870998 | Microarray |
| hsa-miR-140-5p | lung squamous cell cancer | downregulation | "miR 140 suppresses tumor growth and metastasis of ......" | 24039995 | |
| hsa-miR-140-5p | ovarian cancer | downregulation | "Dysregulation of miRNAs is a common feature in hum ......" | 26297547 |
Reported cancer pathway affected by hsa-miR-140-5p

| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-140-5p | bladder cancer | cell cycle pathway | "A functional variant in TP63 at 3q28 associated wi ......" | 26695686 | |
| hsa-miR-140-5p | colorectal cancer | cell cycle pathway | "Inhibition of colorectal cancer stem cell survival ......" | 25980495 | |
| hsa-miR-140-5p | gastric cancer | cell cycle pathway | "MicroRNA 140 Inhibits Cell Proliferation in Gastri ......" | 27353653 | Flow cytometry; Colony formation |
| hsa-miR-140-5p | liver cancer | Epithelial mesenchymal transition pathway | "Long non coding RNA Unigene56159 promotes epitheli ......" | 27597739 | |
| hsa-miR-140-5p | ovarian cancer | Apoptosis pathway | "miR 140 5p inhibits ovarian cancer growth partiall ......" | 26297547 |
Reported cancer prognosis affected by hsa-miR-140-5p
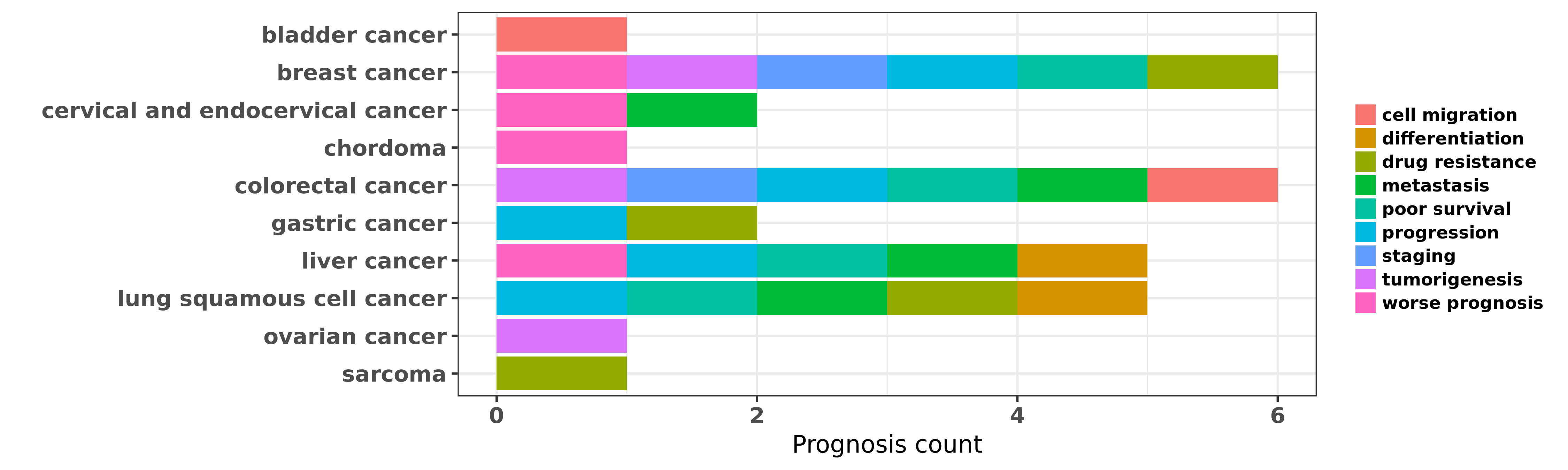
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-140-5p | bladder cancer | cell migration | "A functional variant in TP63 at 3q28 associated wi ......" | 26695686 | |
| hsa-miR-140-5p | breast cancer | tumorigenesis; drug resistance; poor survival | "Estrogen receptor α signaling regulates breast tu ......" | 23060440 | Luciferase |
| hsa-miR-140-5p | breast cancer | staging | "Downregulation of miR 140 promotes cancer stem cel ......" | 23752191 | |
| hsa-miR-140-5p | breast cancer | staging; progression; drug resistance | "Roles of microRNA 140 in stem cell associated earl ......" | 25426255 | |
| hsa-miR-140-5p | breast cancer | worse prognosis; drug resistance | "The aim of this study was to investigate the role ......" | 27746365 | |
| hsa-miR-140-5p | cervical and endocervical cancer | metastasis; worse prognosis | "MicroRNA 140 5p targets insulin like growth factor ......" | 27588393 | |
| hsa-miR-140-5p | chordoma | worse prognosis | "Identification of miR 140 3p as a marker associate ......" | 25197358 | |
| hsa-miR-140-5p | colorectal cancer | cell migration; metastasis | "microRNA 140 suppresses the migration and invasion ......" | 25567303 | Western blot; Wound Healing Assay |
| hsa-miR-140-5p | colorectal cancer | poor survival; metastasis; progression; staging | "Inhibition of colorectal cancer stem cell survival ......" | 25980495 | |
| hsa-miR-140-5p | colorectal cancer | progression; staging; poor survival; tumorigenesis | "MicroRNA 140 5p inhibits the progression of colore ......" | 26402430 | Transwell assay; Luciferase |
| hsa-miR-140-5p | gastric cancer | drug resistance; progression | "MicroRNA 140 Inhibits Cell Proliferation in Gastri ......" | 27353653 | Flow cytometry; Colony formation |
| hsa-miR-140-5p | liver cancer | metastasis; poor survival; differentiation; progression; worse prognosis | "MicroRNA 140 5p suppresses tumor growth and metast ......" | 23401231 | |
| hsa-miR-140-5p | lung squamous cell cancer | metastasis; progression | "miR 140 suppresses tumor growth and metastasis of ......" | 24039995 | |
| hsa-miR-140-5p | lung squamous cell cancer | differentiation; poor survival | "Monocyte to macrophage differentiation associated ......" | 24971538 | |
| hsa-miR-140-5p | lung squamous cell cancer | drug resistance | "Associating drug response with micro-RNA expressio ......" | 27247353 | |
| hsa-miR-140-5p | ovarian cancer | tumorigenesis | "miR 140 5p inhibits ovarian cancer growth partiall ......" | 26297547 | |
| hsa-miR-140-5p | sarcoma | drug resistance | "Mechanism of chemoresistance mediated by miR 140 i ......" | 19734943 | |
| hsa-miR-140-5p | sarcoma | drug resistance | "miR 140 5p attenuates chemotherapeutic drug induce ......" | 27582507 |
Reported gene related to hsa-miR-140-5p
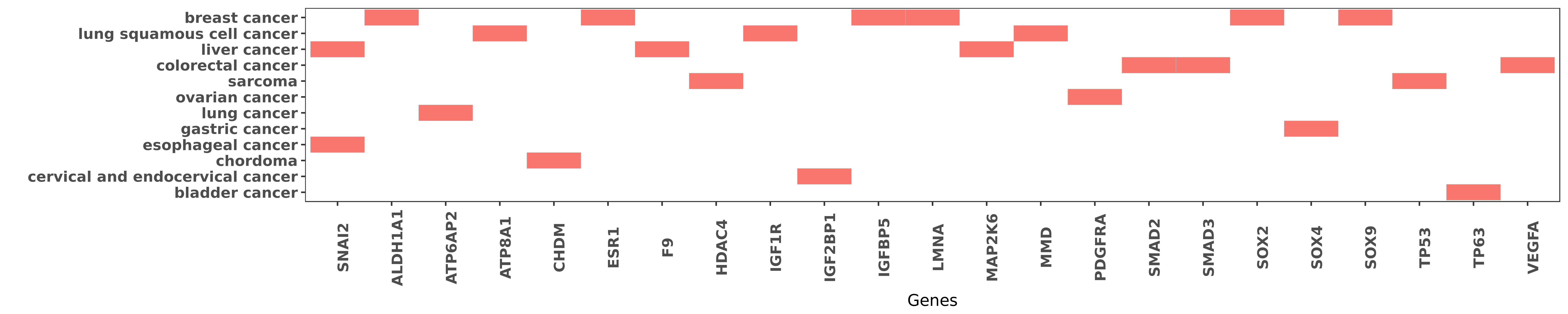
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-140-5p | esophageal cancer | SNAI2 | "Down regulation of miR 140 induces EMT and promote ......" | 25322669 |
| hsa-miR-140-5p | liver cancer | SNAI2 | "Mechanistically Unigene56159 could directly bind t ......" | 27597739 |
| hsa-miR-140-5p | breast cancer | SOX2 | "We found that the stem cell self-renewal regulator ......" | 23060440 |
| hsa-miR-140-5p | breast cancer | SOX2 | "miR-140 plays an important tumor suppressive role ......" | 25426255 |
| hsa-miR-140-5p | breast cancer | SOX9 | "Furthermore we found that SOX9 and ALDH1 the most ......" | 23752191 |
| hsa-miR-140-5p | breast cancer | SOX9 | "miR-140 plays an important tumor suppressive role ......" | 25426255 |
| hsa-miR-140-5p | breast cancer | ALDH1A1 | "Furthermore we found that SOX9 and ALDH1 the most ......" | 23752191 |
| hsa-miR-140-5p | lung cancer | ATP6AP2 | "MicroRNA 140 3p inhibits proliferation migration a ......" | 26722475 |
| hsa-miR-140-5p | lung squamous cell cancer | ATP8A1 | "MiR 140 3p suppressed cell growth and invasion by ......" | 26415732 |
| hsa-miR-140-5p | chordoma | CHDM | "Identification of miR 140 3p as a marker associate ......" | 25197358 |
| hsa-miR-140-5p | breast cancer | ESR1 | "We initiated studies that examined estrogen recept ......" | 23060440 |
| hsa-miR-140-5p | liver cancer | F9 | "MicroRNA 140 5p suppresses tumor growth and metast ......" | 23401231 |
| hsa-miR-140-5p | sarcoma | HDAC4 | "Histone deacetylase 4 HDAC4 was confirmed to be on ......" | 19734943 |
| hsa-miR-140-5p | lung squamous cell cancer | IGF1R | "Integrated analysis identified IGF1R as a direct a ......" | 24039995 |
| hsa-miR-140-5p | cervical and endocervical cancer | IGF2BP1 | "MicroRNA 140 5p targets insulin like growth factor ......" | 27588393 |
| hsa-miR-140-5p | breast cancer | IGFBP5 | "Experimental and bioinformatics studies have shown ......" | 25983620 |
| hsa-miR-140-5p | breast cancer | LMNA | "The purpose of this review is to examine the cance ......" | 25426255 |
| hsa-miR-140-5p | liver cancer | MAP2K6 | "Multipathway reporter arrays suggested that miR-14 ......" | 23401231 |
| hsa-miR-140-5p | lung squamous cell cancer | MMD | "Monocyte to macrophage differentiation associated ......" | 24971538 |
| hsa-miR-140-5p | ovarian cancer | PDGFRA | "miR 140 5p inhibits ovarian cancer growth partiall ......" | 26297547 |
| hsa-miR-140-5p | colorectal cancer | SMAD2 | "In this study we show that hsa-miR-140-5p directly ......" | 25980495 |
| hsa-miR-140-5p | colorectal cancer | SMAD3 | "microRNA 140 suppresses the migration and invasion ......" | 25567303 |
| hsa-miR-140-5p | gastric cancer | SOX4 | "MicroRNA 140 Inhibits Cell Proliferation in Gastri ......" | 27353653 |
| hsa-miR-140-5p | sarcoma | TP53 | "miR-140 induced p53 and p21 expression accompanied ......" | 19734943 |
| hsa-miR-140-5p | bladder cancer | TP63 | "A functional variant in TP63 at 3q28 associated wi ......" | 26695686 |
| hsa-miR-140-5p | colorectal cancer | VEGFA | "MicroRNA 140 5p inhibits the progression of colore ......" | 26402430 |
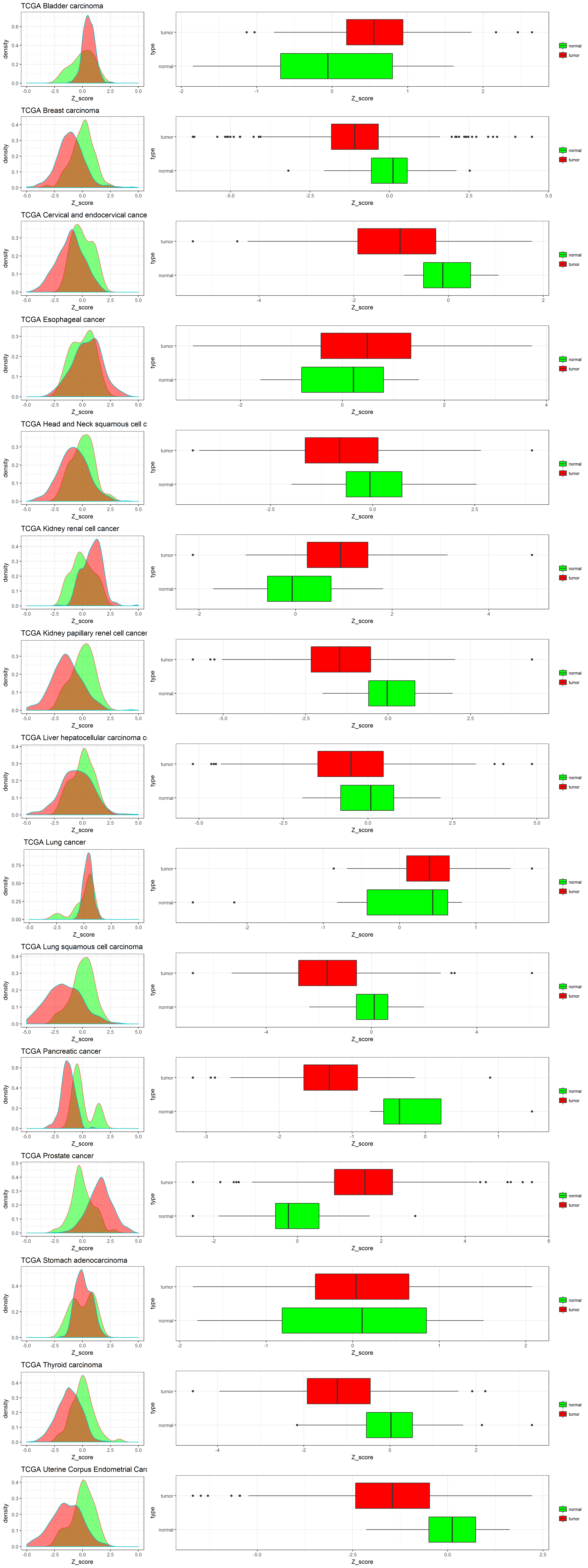
Expression profile in cancer corhorts:
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-140-5p | BCL9 | 10 cancers: BLCA; BRCA; KIRC; LUAD; LUSC; PAAD; SARC; THCA; STAD; UCEC | MirTarget; PITA; miRanda; miRNATAP | TCGA BLCA -0.162; TCGA BRCA -0.21; TCGA KIRC -0.317; TCGA LUAD -0.149; TCGA LUSC -0.235; TCGA PAAD -0.232; TCGA SARC -0.223; TCGA THCA -0.254; TCGA STAD -0.228; TCGA UCEC -0.134 |
hsa-miR-140-5p | MED13 | 11 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; THCA; STAD | MirTarget; miRanda; miRNATAP | TCGA BLCA -0.086; TCGA CESC -0.08; TCGA COAD -0.1; TCGA ESCA -0.137; TCGA HNSC -0.116; TCGA KIRC -0.106; TCGA KIRP -0.105; TCGA LUAD -0.207; TCGA LUSC -0.13; TCGA THCA -0.377; TCGA STAD -0.093 |
hsa-miR-140-5p | STRN3 | 10 cancers: BLCA; CESC; ESCA; HNSC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | MirTarget; PITA; miRanda | TCGA BLCA -0.143; TCGA CESC -0.105; TCGA ESCA -0.358; TCGA HNSC -0.16; TCGA LUAD -0.111; TCGA LUSC -0.134; TCGA PRAD -0.074; TCGA SARC -0.185; TCGA STAD -0.308; TCGA UCEC -0.061 |
hsa-miR-140-5p | UBR5 | 13 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LIHC; LUAD; LUSC; SARC; STAD; UCEC | MirTarget; miRanda | TCGA BLCA -0.08; TCGA CESC -0.099; TCGA COAD -0.152; TCGA ESCA -0.262; TCGA HNSC -0.146; TCGA KIRC -0.119; TCGA LGG -0.061; TCGA LIHC -0.135; TCGA LUAD -0.171; TCGA LUSC -0.214; TCGA SARC -0.152; TCGA STAD -0.149; TCGA UCEC -0.113 |
hsa-miR-140-5p | SHROOM3 | 9 cancers: BLCA; BRCA; KIRC; KIRP; LUAD; PAAD; SARC; THCA; UCEC | MirTarget; PITA; miRanda; miRNATAP | TCGA BLCA -0.198; TCGA BRCA -0.286; TCGA KIRC -0.654; TCGA KIRP -0.185; TCGA LUAD -0.161; TCGA PAAD -0.365; TCGA SARC -0.998; TCGA THCA -0.136; TCGA UCEC -0.116 |
hsa-miR-140-5p | WDFY3 | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; PAAD; PRAD; STAD | MirTarget; PITA; miRanda | TCGA BLCA -0.153; TCGA CESC -0.118; TCGA COAD -0.15; TCGA ESCA -0.174; TCGA HNSC -0.108; TCGA KIRC -0.128; TCGA LUAD -0.095; TCGA PAAD -0.151; TCGA PRAD -0.121; TCGA STAD -0.193 |
hsa-miR-140-5p | PKN2 | 11 cancers: BLCA; CESC; HNSC; KIRP; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget; miRanda | TCGA BLCA -0.094; TCGA CESC -0.1; TCGA HNSC -0.107; TCGA KIRP -0.081; TCGA LUAD -0.107; TCGA LUSC -0.096; TCGA PAAD -0.154; TCGA PRAD -0.186; TCGA SARC -0.107; TCGA STAD -0.191; TCGA UCEC -0.08 |
hsa-miR-140-5p | ANKFY1 | 9 cancers: BLCA; ESCA; HNSC; KIRC; LGG; LUAD; PRAD; THCA; STAD | MirTarget; PITA; miRanda | TCGA BLCA -0.087; TCGA ESCA -0.141; TCGA HNSC -0.094; TCGA KIRC -0.129; TCGA LGG -0.143; TCGA LUAD -0.06; TCGA PRAD -0.152; TCGA THCA -0.202; TCGA STAD -0.066 |
hsa-miR-140-5p | ARIH1 | 13 cancers: BLCA; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD | MirTarget; miRanda | TCGA BLCA -0.116; TCGA COAD -0.073; TCGA ESCA -0.113; TCGA HNSC -0.056; TCGA KIRC -0.084; TCGA KIRP -0.061; TCGA LUAD -0.118; TCGA LUSC -0.087; TCGA PAAD -0.099; TCGA PRAD -0.1; TCGA SARC -0.111; TCGA THCA -0.078; TCGA STAD -0.161 |
hsa-miR-140-5p | SLC2A1 | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LUSC; PAAD; PRAD; THCA; UCEC | MirTarget; miRanda; miRNATAP | TCGA BLCA -0.27; TCGA BRCA -0.159; TCGA CESC -0.354; TCGA ESCA -0.969; TCGA HNSC -0.642; TCGA LUSC -1.229; TCGA PAAD -0.668; TCGA PRAD -0.103; TCGA THCA -0.271; TCGA UCEC -0.35 |
hsa-miR-140-5p | NCKAP1 | 14 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget; PITA; miRanda | TCGA BLCA -0.205; TCGA CESC -0.135; TCGA COAD -0.097; TCGA ESCA -0.239; TCGA HNSC -0.207; TCGA KIRC -0.168; TCGA LUAD -0.134; TCGA LUSC -0.276; TCGA PAAD -0.326; TCGA PRAD -0.168; TCGA SARC -0.268; TCGA THCA -0.199; TCGA STAD -0.242; TCGA UCEC -0.135 |
hsa-miR-140-5p | NUCKS1 | 9 cancers: BLCA; ESCA; KIRC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget; miRanda; miRNATAP | TCGA BLCA -0.113; TCGA ESCA -0.152; TCGA KIRC -0.107; TCGA LUAD -0.11; TCGA LUSC -0.162; TCGA PAAD -0.108; TCGA PRAD -0.096; TCGA STAD -0.209; TCGA UCEC -0.151 |
hsa-miR-140-5p | ZMYM4 | 9 cancers: BLCA; CESC; COAD; KIRP; LUAD; LUSC; PRAD; STAD; UCEC | MirTarget; miRanda | TCGA BLCA -0.074; TCGA CESC -0.071; TCGA COAD -0.09; TCGA KIRP -0.063; TCGA LUAD -0.092; TCGA LUSC -0.081; TCGA PRAD -0.081; TCGA STAD -0.171; TCGA UCEC -0.079 |
hsa-miR-140-5p | LAMC1 | 11 cancers: BLCA; COAD; HNSC; KIRP; LUAD; LUSC; PAAD; SARC; THCA; STAD; UCEC | MirTarget; PITA; miRanda; miRNATAP | TCGA BLCA -0.188; TCGA COAD -0.219; TCGA HNSC -0.148; TCGA KIRP -0.306; TCGA LUAD -0.098; TCGA LUSC -0.326; TCGA PAAD -0.186; TCGA SARC -0.092; TCGA THCA -0.183; TCGA STAD -0.262; TCGA UCEC -0.31 |
hsa-miR-140-5p | FZD6 | 14 cancers: BLCA; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget; miRanda | TCGA BLCA -0.138; TCGA COAD -0.277; TCGA ESCA -0.77; TCGA HNSC -0.299; TCGA KIRC -0.182; TCGA KIRP -0.172; TCGA LUAD -0.099; TCGA LUSC -0.323; TCGA PAAD -0.267; TCGA PRAD -0.12; TCGA SARC -0.344; TCGA THCA -0.255; TCGA STAD -0.227; TCGA UCEC -0.221 |
hsa-miR-140-5p | BTBD7 | 12 cancers: BLCA; CESC; ESCA; HNSC; KIRP; LGG; LUAD; LUSC; PAAD; PRAD; SARC; STAD | PITA | TCGA BLCA -0.167; TCGA CESC -0.114; TCGA ESCA -0.222; TCGA HNSC -0.214; TCGA KIRP -0.206; TCGA LGG -0.063; TCGA LUAD -0.133; TCGA LUSC -0.107; TCGA PAAD -0.127; TCGA PRAD -0.149; TCGA SARC -0.124; TCGA STAD -0.136 |
hsa-miR-140-5p | RGMB | 10 cancers: BLCA; BRCA; ESCA; KIRP; LGG; LUAD; PRAD; SARC; THCA; STAD | PITA; miRanda; miRNATAP | TCGA BLCA -0.16; TCGA BRCA -0.103; TCGA ESCA -0.309; TCGA KIRP -0.093; TCGA LGG -0.216; TCGA LUAD -0.101; TCGA PRAD -0.223; TCGA SARC -0.138; TCGA THCA -0.115; TCGA STAD -0.444 |
hsa-miR-140-5p | PPP2R3A | 13 cancers: BLCA; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | PITA; miRanda; miRNATAP | TCGA BLCA -0.163; TCGA ESCA -0.505; TCGA HNSC -0.212; TCGA KIRC -0.229; TCGA KIRP -0.147; TCGA LUAD -0.116; TCGA LUSC -0.232; TCGA PAAD -0.216; TCGA PRAD -0.191; TCGA SARC -0.121; TCGA THCA -0.185; TCGA STAD -0.42; TCGA UCEC -0.228 |
hsa-miR-140-5p | DAG1 | 9 cancers: BLCA; BRCA; KIRC; LUAD; LUSC; PAAD; SARC; THCA; UCEC | PITA; miRanda; miRNATAP | TCGA BLCA -0.124; TCGA BRCA -0.063; TCGA KIRC -0.285; TCGA LUAD -0.117; TCGA LUSC -0.107; TCGA PAAD -0.286; TCGA SARC -0.135; TCGA THCA -0.179; TCGA UCEC -0.113 |
hsa-miR-140-5p | CDYL | 11 cancers: BLCA; ESCA; HNSC; KIRC; LGG; LUAD; LUSC; PRAD; THCA; STAD; UCEC | PITA; miRanda | TCGA BLCA -0.094; TCGA ESCA -0.208; TCGA HNSC -0.147; TCGA KIRC -0.134; TCGA LGG -0.131; TCGA LUAD -0.071; TCGA LUSC -0.272; TCGA PRAD -0.063; TCGA THCA -0.201; TCGA STAD -0.134; TCGA UCEC -0.182 |
hsa-miR-140-5p | BAZ2B | 10 cancers: BLCA; COAD; HNSC; KIRP; LGG; LUAD; PRAD; SARC; THCA; STAD | PITA; miRanda; miRNATAP | TCGA BLCA -0.266; TCGA COAD -0.146; TCGA HNSC -0.096; TCGA KIRP -0.117; TCGA LGG -0.066; TCGA LUAD -0.136; TCGA PRAD -0.163; TCGA SARC -0.145; TCGA THCA -0.095; TCGA STAD -0.262 |
hsa-miR-140-5p | PLEKHG3 | 12 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LIHC; LUSC; PAAD; PRAD; SARC; THCA; STAD | PITA; miRanda; miRNATAP | TCGA BLCA -0.343; TCGA BRCA -0.181; TCGA ESCA -0.519; TCGA HNSC -0.23; TCGA KIRC -0.422; TCGA LIHC -0.204; TCGA LUSC -0.445; TCGA PAAD -0.257; TCGA PRAD -0.263; TCGA SARC -0.412; TCGA THCA -0.094; TCGA STAD -0.211 |
hsa-miR-140-5p | CUEDC1 | 10 cancers: BLCA; BRCA; ESCA; KIRC; KIRP; LGG; PAAD; PRAD; SARC; STAD | PITA; miRanda | TCGA BLCA -0.202; TCGA BRCA -0.19; TCGA ESCA -0.278; TCGA KIRC -0.295; TCGA KIRP -0.087; TCGA LGG -0.051; TCGA PAAD -0.204; TCGA PRAD -0.107; TCGA SARC -0.13; TCGA STAD -0.222 |
hsa-miR-140-5p | TEAD1 | 9 cancers: BLCA; ESCA; KIRC; LGG; PAAD; PRAD; SARC; THCA; STAD | PITA; miRanda; miRNATAP | TCGA BLCA -0.212; TCGA ESCA -0.181; TCGA KIRC -0.122; TCGA LGG -0.115; TCGA PAAD -0.172; TCGA PRAD -0.224; TCGA SARC -0.233; TCGA THCA -0.201; TCGA STAD -0.479 |
hsa-miR-140-5p | SNX27 | 9 cancers: BLCA; BRCA; ESCA; LUAD; LUSC; PRAD; SARC; THCA; STAD | PITA; miRanda; miRNATAP | TCGA BLCA -0.127; TCGA BRCA -0.056; TCGA ESCA -0.121; TCGA LUAD -0.174; TCGA LUSC -0.08; TCGA PRAD -0.061; TCGA SARC -0.071; TCGA THCA -0.162; TCGA STAD -0.255 |
hsa-miR-140-5p | MIPOL1 | 11 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; SARC; STAD | PITA; miRanda; miRNATAP | TCGA BLCA -0.234; TCGA BRCA -0.178; TCGA CESC -0.315; TCGA ESCA -0.396; TCGA HNSC -0.208; TCGA KIRC -0.39; TCGA LUAD -0.133; TCGA LUSC -0.213; TCGA PAAD -0.365; TCGA SARC -0.482; TCGA STAD -0.336 |
hsa-miR-140-5p | ACACA | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LUAD; LUSC; SARC; STAD | miRanda | TCGA BLCA -0.166; TCGA BRCA -0.07; TCGA CESC -0.125; TCGA ESCA -0.198; TCGA HNSC -0.13; TCGA KIRC -0.28; TCGA LUAD -0.124; TCGA LUSC -0.187; TCGA SARC -0.141; TCGA STAD -0.129 |
hsa-miR-140-5p | VEZF1 | 10 cancers: BLCA; BRCA; COAD; ESCA; KIRC; KIRP; LUAD; PRAD; SARC; STAD | miRanda | TCGA BLCA -0.137; TCGA BRCA -0.059; TCGA COAD -0.123; TCGA ESCA -0.13; TCGA KIRC -0.133; TCGA KIRP -0.15; TCGA LUAD -0.1; TCGA PRAD -0.169; TCGA SARC -0.113; TCGA STAD -0.181 |
hsa-miR-140-5p | LPP | 9 cancers: BLCA; COAD; ESCA; HNSC; LUAD; LUSC; PRAD; SARC; STAD | miRanda | TCGA BLCA -0.453; TCGA COAD -0.177; TCGA ESCA -0.226; TCGA HNSC -0.223; TCGA LUAD -0.116; TCGA LUSC -0.214; TCGA PRAD -0.481; TCGA SARC -0.699; TCGA STAD -0.639 |
hsa-miR-140-5p | ASH1L | 9 cancers: BLCA; COAD; ESCA; KIRC; LUAD; LUSC; PRAD; SARC; STAD | miRanda | TCGA BLCA -0.26; TCGA COAD -0.113; TCGA ESCA -0.165; TCGA KIRC -0.129; TCGA LUAD -0.212; TCGA LUSC -0.167; TCGA PRAD -0.179; TCGA SARC -0.232; TCGA STAD -0.223 |
hsa-miR-140-5p | SLC24A1 | 9 cancers: BLCA; BRCA; KIRC; KIRP; LIHC; LUAD; PRAD; SARC; STAD | miRanda | TCGA BLCA -0.138; TCGA BRCA -0.078; TCGA KIRC -0.267; TCGA KIRP -0.144; TCGA LIHC -0.238; TCGA LUAD -0.093; TCGA PRAD -0.137; TCGA SARC -0.11; TCGA STAD -0.119 |
hsa-miR-140-5p | PTK2 | 13 cancers: BLCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; SARC; THCA; STAD; UCEC | miRanda | TCGA BLCA -0.097; TCGA CESC -0.117; TCGA COAD -0.165; TCGA ESCA -0.217; TCGA HNSC -0.224; TCGA LIHC -0.143; TCGA LUAD -0.162; TCGA LUSC -0.218; TCGA PAAD -0.173; TCGA SARC -0.107; TCGA THCA -0.088; TCGA STAD -0.069; TCGA UCEC -0.136 |
hsa-miR-140-5p | PLEKHA5 | 11 cancers: BLCA; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; THCA; STAD | miRanda | TCGA BLCA -0.211; TCGA COAD -0.155; TCGA ESCA -0.2; TCGA HNSC -0.122; TCGA LIHC -0.148; TCGA LUAD -0.163; TCGA LUSC -0.22; TCGA PAAD -0.397; TCGA PRAD -0.166; TCGA THCA -0.346; TCGA STAD -0.337 |
hsa-miR-140-5p | RBM41 | 9 cancers: BLCA; HNSC; KIRP; LUAD; LUSC; PAAD; SARC; STAD; UCEC | miRanda | TCGA BLCA -0.078; TCGA HNSC -0.131; TCGA KIRP -0.273; TCGA LUAD -0.105; TCGA LUSC -0.205; TCGA PAAD -0.164; TCGA SARC -0.117; TCGA STAD -0.32; TCGA UCEC -0.09 |
hsa-miR-140-5p | KCTD3 | 9 cancers: BLCA; BRCA; KIRP; LGG; LUSC; PAAD; PRAD; SARC; STAD | miRanda | TCGA BLCA -0.136; TCGA BRCA -0.213; TCGA KIRP -0.112; TCGA LGG -0.067; TCGA LUSC -0.112; TCGA PAAD -0.226; TCGA PRAD -0.09; TCGA SARC -0.191; TCGA STAD -0.106 |
hsa-miR-140-5p | EGLN3 | 9 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LGG; LUSC; PAAD; SARC | miRanda | TCGA BLCA -0.395; TCGA BRCA -0.31; TCGA CESC -0.473; TCGA ESCA -0.745; TCGA HNSC -0.324; TCGA LGG -0.108; TCGA LUSC -0.746; TCGA PAAD -0.983; TCGA SARC -0.415 |
hsa-miR-140-5p | STAU2 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; KIRP; LUAD; LUSC; SARC; THCA; STAD; UCEC | miRanda | TCGA BLCA -0.08; TCGA BRCA -0.071; TCGA CESC -0.163; TCGA COAD -0.123; TCGA ESCA -0.211; TCGA KIRC -0.163; TCGA KIRP -0.095; TCGA LUAD -0.202; TCGA LUSC -0.106; TCGA SARC -0.082; TCGA THCA -0.084; TCGA STAD -0.172; TCGA UCEC -0.249 |
hsa-miR-140-5p | SMARCC2 | 10 cancers: BLCA; BRCA; CESC; KIRP; LGG; LUAD; LUSC; SARC; THCA; STAD | miRanda | TCGA BLCA -0.101; TCGA BRCA -0.08; TCGA CESC -0.076; TCGA KIRP -0.155; TCGA LGG -0.067; TCGA LUAD -0.119; TCGA LUSC -0.084; TCGA SARC -0.059; TCGA THCA -0.091; TCGA STAD -0.057 |
hsa-miR-140-5p | BRD1 | 10 cancers: BLCA; ESCA; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; SARC; STAD | miRanda | TCGA BLCA -0.081; TCGA ESCA -0.15; TCGA HNSC -0.097; TCGA KIRP -0.087; TCGA LGG -0.079; TCGA LIHC -0.146; TCGA LUAD -0.09; TCGA LUSC -0.118; TCGA SARC -0.092; TCGA STAD -0.12 |
hsa-miR-140-5p | LRBA | 9 cancers: BLCA; BRCA; CESC; HNSC; KIRC; LIHC; LUAD; SARC; THCA | miRanda | TCGA BLCA -0.166; TCGA BRCA -0.209; TCGA CESC -0.126; TCGA HNSC -0.095; TCGA KIRC -0.473; TCGA LIHC -0.134; TCGA LUAD -0.159; TCGA SARC -0.102; TCGA THCA -0.11 |
hsa-miR-140-5p | CDK8 | 9 cancers: BLCA; COAD; KIRC; LGG; LUAD; LUSC; PRAD; SARC; STAD | miRanda | TCGA BLCA -0.148; TCGA COAD -0.183; TCGA KIRC -0.146; TCGA LGG -0.122; TCGA LUAD -0.07; TCGA LUSC -0.234; TCGA PRAD -0.161; TCGA SARC -0.18; TCGA STAD -0.156 |
hsa-miR-140-5p | TOM1L1 | 9 cancers: BLCA; BRCA; CESC; HNSC; KIRC; LUAD; LUSC; PAAD; SARC | miRanda | TCGA BLCA -0.154; TCGA BRCA -0.254; TCGA CESC -0.274; TCGA HNSC -0.16; TCGA KIRC -0.393; TCGA LUAD -0.14; TCGA LUSC -0.134; TCGA PAAD -0.274; TCGA SARC -0.516 |
hsa-miR-140-5p | FAM131A | 10 cancers: BLCA; BRCA; HNSC; KIRC; KIRP; LUSC; SARC; THCA; STAD; UCEC | miRanda | TCGA BLCA -0.116; TCGA BRCA -0.086; TCGA HNSC -0.133; TCGA KIRC -0.115; TCGA KIRP -0.103; TCGA LUSC -0.259; TCGA SARC -0.313; TCGA THCA -0.131; TCGA STAD -0.151; TCGA UCEC -0.147 |
hsa-miR-140-5p | SNX16 | 10 cancers: BLCA; CESC; HNSC; LGG; LUAD; LUSC; SARC; THCA; STAD; UCEC | miRanda | TCGA BLCA -0.087; TCGA CESC -0.152; TCGA HNSC -0.149; TCGA LGG -0.075; TCGA LUAD -0.161; TCGA LUSC -0.102; TCGA SARC -0.24; TCGA THCA -0.241; TCGA STAD -0.113; TCGA UCEC -0.189 |
hsa-miR-140-5p | PLA2G12A | 10 cancers: BLCA; BRCA; CESC; KIRC; LIHC; LUAD; SARC; THCA; STAD; UCEC | miRanda | TCGA BLCA -0.148; TCGA BRCA -0.112; TCGA CESC -0.084; TCGA KIRC -0.189; TCGA LIHC -0.148; TCGA LUAD -0.098; TCGA SARC -0.126; TCGA THCA -0.085; TCGA STAD -0.182; TCGA UCEC -0.083 |
hsa-miR-140-5p | DDX42 | 9 cancers: BLCA; CESC; ESCA; KIRP; LIHC; LUAD; LUSC; SARC; STAD | miRanda | TCGA BLCA -0.07; TCGA CESC -0.074; TCGA ESCA -0.108; TCGA KIRP -0.077; TCGA LIHC -0.114; TCGA LUAD -0.118; TCGA LUSC -0.097; TCGA SARC -0.073; TCGA STAD -0.096 |
hsa-miR-140-5p | LNX2 | 11 cancers: BLCA; BRCA; CESC; HNSC; KIRC; LUAD; PAAD; PRAD; THCA; STAD; UCEC | miRanda | TCGA BLCA -0.181; TCGA BRCA -0.131; TCGA CESC -0.119; TCGA HNSC -0.2; TCGA KIRC -0.18; TCGA LUAD -0.146; TCGA PAAD -0.176; TCGA PRAD -0.112; TCGA THCA -0.107; TCGA STAD -0.115; TCGA UCEC -0.116 |
hsa-miR-140-5p | MPP7 | 10 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LUAD; PRAD; SARC; THCA; STAD | miRanda | TCGA BLCA -0.414; TCGA BRCA -0.253; TCGA ESCA -0.396; TCGA HNSC -0.181; TCGA KIRC -1.137; TCGA LUAD -0.212; TCGA PRAD -0.179; TCGA SARC -0.935; TCGA THCA -0.406; TCGA STAD -0.609 |
hsa-miR-140-5p | ARHGEF4 | 11 cancers: BLCA; ESCA; HNSC; KIRC; LGG; LIHC; LUSC; PAAD; PRAD; SARC; STAD | miRanda | TCGA BLCA -0.481; TCGA ESCA -1.059; TCGA HNSC -0.437; TCGA KIRC -0.408; TCGA LGG -0.091; TCGA LIHC -0.657; TCGA LUSC -0.477; TCGA PAAD -0.305; TCGA PRAD -0.286; TCGA SARC -0.459; TCGA STAD -0.66 |
hsa-miR-140-5p | GLS2 | 10 cancers: BLCA; BRCA; CESC; KIRC; KIRP; LUAD; PRAD; SARC; THCA; STAD | miRanda | TCGA BLCA -0.267; TCGA BRCA -0.358; TCGA CESC -0.316; TCGA KIRC -0.84; TCGA KIRP -0.439; TCGA LUAD -0.193; TCGA PRAD -0.356; TCGA SARC -0.173; TCGA THCA -0.742; TCGA STAD -0.137 |
hsa-miR-140-5p | PHLDB2 | 9 cancers: BLCA; COAD; ESCA; HNSC; KIRC; PRAD; SARC; THCA; STAD | miRanda | TCGA BLCA -0.318; TCGA COAD -0.486; TCGA ESCA -0.591; TCGA HNSC -0.401; TCGA KIRC -0.158; TCGA PRAD -0.165; TCGA SARC -0.406; TCGA THCA -0.223; TCGA STAD -0.44 |
hsa-miR-140-5p | AGAP1 | 11 cancers: BLCA; BRCA; CESC; HNSC; LIHC; LUAD; LUSC; PAAD; SARC; THCA; STAD | miRanda | TCGA BLCA -0.124; TCGA BRCA -0.055; TCGA CESC -0.128; TCGA HNSC -0.116; TCGA LIHC -0.221; TCGA LUAD -0.163; TCGA LUSC -0.12; TCGA PAAD -0.333; TCGA SARC -0.093; TCGA THCA -0.129; TCGA STAD -0.179 |
hsa-miR-140-5p | ZMYND8 | 11 cancers: BLCA; BRCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; SARC; THCA; STAD | miRanda | TCGA BLCA -0.091; TCGA BRCA -0.128; TCGA HNSC -0.127; TCGA KIRC -0.148; TCGA KIRP -0.132; TCGA LUAD -0.113; TCGA LUSC -0.162; TCGA PAAD -0.169; TCGA SARC -0.186; TCGA THCA -0.115; TCGA STAD -0.125 |
hsa-miR-140-5p | PPL | 11 cancers: BLCA; ESCA; KIRC; KIRP; LUAD; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRanda | TCGA BLCA -0.299; TCGA ESCA -0.441; TCGA KIRC -0.487; TCGA KIRP -0.473; TCGA LUAD -0.134; TCGA PAAD -1.001; TCGA PRAD -0.257; TCGA SARC -0.623; TCGA THCA -0.618; TCGA STAD -0.438; TCGA UCEC -0.263 |
hsa-miR-140-5p | UBXN7 | 11 cancers: BLCA; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | miRanda | TCGA BLCA -0.137; TCGA COAD -0.295; TCGA ESCA -0.286; TCGA HNSC -0.284; TCGA KIRC -0.21; TCGA LUAD -0.099; TCGA LUSC -0.443; TCGA PRAD -0.255; TCGA SARC -0.146; TCGA STAD -0.107; TCGA UCEC -0.155 |
hsa-miR-140-5p | KLF5 | 10 cancers: BLCA; ESCA; HNSC; KIRC; KIRP; LUSC; PAAD; PRAD; THCA; UCEC | miRanda | TCGA BLCA -0.216; TCGA ESCA -0.316; TCGA HNSC -0.279; TCGA KIRC -0.579; TCGA KIRP -0.507; TCGA LUSC -0.247; TCGA PAAD -0.715; TCGA PRAD -0.366; TCGA THCA -0.316; TCGA UCEC -0.19 |
hsa-miR-140-5p | TMEM67 | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LGG; LUSC; PAAD; PRAD; STAD; UCEC | miRanda | TCGA BLCA -0.108; TCGA BRCA -0.063; TCGA COAD -0.285; TCGA ESCA -0.287; TCGA HNSC -0.193; TCGA KIRP -0.237; TCGA LGG -0.097; TCGA LUSC -0.137; TCGA PAAD -0.297; TCGA PRAD -0.131; TCGA STAD -0.279; TCGA UCEC -0.127 |
hsa-miR-140-5p | STXBP4 | 9 cancers: BLCA; CESC; ESCA; HNSC; KIRC; LUAD; PRAD; SARC; STAD | miRanda | TCGA BLCA -0.203; TCGA CESC -0.125; TCGA ESCA -0.207; TCGA HNSC -0.148; TCGA KIRC -0.267; TCGA LUAD -0.089; TCGA PRAD -0.202; TCGA SARC -0.202; TCGA STAD -0.278 |
hsa-miR-140-5p | BLZF1 | 9 cancers: BLCA; ESCA; HNSC; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRanda | TCGA BLCA -0.08; TCGA ESCA -0.185; TCGA HNSC -0.112; TCGA LUSC -0.14; TCGA PAAD -0.167; TCGA PRAD -0.149; TCGA SARC -0.081; TCGA STAD -0.185; TCGA UCEC -0.173 |
hsa-miR-140-5p | STON2 | 9 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LGG; LUSC; STAD; UCEC | miRanda | TCGA BLCA -0.23; TCGA BRCA -0.168; TCGA CESC -0.3; TCGA ESCA -0.753; TCGA HNSC -0.36; TCGA LGG -0.216; TCGA LUSC -0.46; TCGA STAD -0.176; TCGA UCEC -0.169 |
hsa-miR-140-5p | KCTD11 | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRP; LUSC; PAAD; PRAD; THCA | miRanda | TCGA BLCA -0.209; TCGA BRCA -0.115; TCGA CESC -0.238; TCGA ESCA -0.389; TCGA HNSC -0.295; TCGA KIRP -0.19; TCGA LUSC -0.263; TCGA PAAD -0.209; TCGA PRAD -0.082; TCGA THCA -0.069 |
hsa-miR-140-5p | PTPRF | 11 cancers: BLCA; CESC; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; UCEC | miRanda | TCGA BLCA -0.162; TCGA CESC -0.14; TCGA ESCA -0.347; TCGA HNSC -0.144; TCGA LUAD -0.089; TCGA LUSC -0.348; TCGA PAAD -0.387; TCGA PRAD -0.137; TCGA SARC -0.561; TCGA THCA -0.434; TCGA UCEC -0.158 |
hsa-miR-140-5p | MKRN1 | 11 cancers: BLCA; BRCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; SARC; THCA; UCEC | miRanda | TCGA BLCA -0.064; TCGA BRCA -0.051; TCGA HNSC -0.063; TCGA KIRC -0.073; TCGA KIRP -0.156; TCGA LGG -0.088; TCGA LUAD -0.088; TCGA LUSC -0.131; TCGA SARC -0.135; TCGA THCA -0.073; TCGA UCEC -0.075 |
hsa-miR-140-5p | FAT1 | 9 cancers: BLCA; COAD; ESCA; HNSC; KIRP; LGG; LUSC; PAAD; PRAD | miRanda | TCGA BLCA -0.161; TCGA COAD -0.165; TCGA ESCA -0.301; TCGA HNSC -0.308; TCGA KIRP -0.268; TCGA LGG -0.118; TCGA LUSC -0.338; TCGA PAAD -0.682; TCGA PRAD -0.193 |
hsa-miR-140-5p | TM9SF3 | 9 cancers: BLCA; KIRC; KIRP; LIHC; LUAD; PAAD; PRAD; THCA; UCEC | miRanda | TCGA BLCA -0.087; TCGA KIRC -0.102; TCGA KIRP -0.074; TCGA LIHC -0.107; TCGA LUAD -0.107; TCGA PAAD -0.229; TCGA PRAD -0.084; TCGA THCA -0.079; TCGA UCEC -0.094 |
hsa-miR-140-5p | TAOK1 | 9 cancers: BLCA; COAD; ESCA; HNSC; LUAD; LUSC; PRAD; SARC; STAD | miRanda | TCGA BLCA -0.245; TCGA COAD -0.369; TCGA ESCA -0.221; TCGA HNSC -0.25; TCGA LUAD -0.138; TCGA LUSC -0.185; TCGA PRAD -0.3; TCGA SARC -0.262; TCGA STAD -0.239 |
hsa-miR-140-5p | UEVLD | 9 cancers: BLCA; COAD; ESCA; HNSC; KIRC; PAAD; PRAD; SARC; STAD | miRanda | TCGA BLCA -0.093; TCGA COAD -0.157; TCGA ESCA -0.174; TCGA HNSC -0.148; TCGA KIRC -0.264; TCGA PAAD -0.184; TCGA PRAD -0.183; TCGA SARC -0.092; TCGA STAD -0.134 |
hsa-miR-140-5p | RCOR3 | 9 cancers: BLCA; BRCA; KIRP; LUAD; LUSC; PRAD; SARC; THCA; STAD | miRanda | TCGA BLCA -0.148; TCGA BRCA -0.105; TCGA KIRP -0.076; TCGA LUAD -0.096; TCGA LUSC -0.092; TCGA PRAD -0.134; TCGA SARC -0.193; TCGA THCA -0.105; TCGA STAD -0.227 |
hsa-miR-140-5p | PPP1CB | 12 cancers: BLCA; CESC; ESCA; HNSC; KIRP; LUAD; LUSC; PRAD; SARC; THCA; STAD; UCEC | miRanda | TCGA BLCA -0.166; TCGA CESC -0.11; TCGA ESCA -0.24; TCGA HNSC -0.124; TCGA KIRP -0.059; TCGA LUAD -0.114; TCGA LUSC -0.148; TCGA PRAD -0.137; TCGA SARC -0.2; TCGA THCA -0.193; TCGA STAD -0.253; TCGA UCEC -0.173 |
hsa-miR-140-5p | SBF2 | 13 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LUSC; PAAD; PRAD; SARC; THCA; STAD | miRanda | TCGA BLCA -0.236; TCGA CESC -0.136; TCGA COAD -0.087; TCGA ESCA -0.176; TCGA HNSC -0.127; TCGA KIRC -0.147; TCGA LGG -0.092; TCGA LUSC -0.142; TCGA PAAD -0.21; TCGA PRAD -0.112; TCGA SARC -0.176; TCGA THCA -0.116; TCGA STAD -0.306 |
hsa-miR-140-5p | EPN2 | 12 cancers: BLCA; BRCA; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; SARC; THCA; STAD | miRanda | TCGA BLCA -0.18; TCGA BRCA -0.117; TCGA ESCA -0.43; TCGA HNSC -0.065; TCGA KIRP -0.189; TCGA LIHC -0.185; TCGA LUAD -0.154; TCGA LUSC -0.13; TCGA PAAD -0.126; TCGA SARC -0.417; TCGA THCA -0.17; TCGA STAD -0.231 |
hsa-miR-140-5p | CAPN1 | 9 cancers: BLCA; BRCA; KIRC; KIRP; LIHC; LUSC; PAAD; PRAD; THCA | miRanda; miRNATAP | TCGA BLCA -0.091; TCGA BRCA -0.125; TCGA KIRC -0.216; TCGA KIRP -0.25; TCGA LIHC -0.15; TCGA LUSC -0.193; TCGA PAAD -0.241; TCGA PRAD -0.131; TCGA THCA -0.136 |
hsa-miR-140-5p | ZBTB41 | 9 cancers: BLCA; BRCA; ESCA; HNSC; LUAD; LUSC; THCA; STAD; UCEC | miRanda | TCGA BLCA -0.103; TCGA BRCA -0.124; TCGA ESCA -0.378; TCGA HNSC -0.142; TCGA LUAD -0.13; TCGA LUSC -0.273; TCGA THCA -0.103; TCGA STAD -0.211; TCGA UCEC -0.156 |
hsa-miR-140-5p | CHD8 | 9 cancers: BLCA; ESCA; HNSC; LIHC; LUAD; LUSC; PRAD; SARC; STAD | miRanda | TCGA BLCA -0.073; TCGA ESCA -0.158; TCGA HNSC -0.109; TCGA LIHC -0.105; TCGA LUAD -0.121; TCGA LUSC -0.128; TCGA PRAD -0.061; TCGA SARC -0.071; TCGA STAD -0.065 |
hsa-miR-140-5p | EGFR | 10 cancers: BLCA; ESCA; HNSC; LGG; LUSC; PAAD; PRAD; SARC; THCA; STAD | miRanda | TCGA BLCA -0.29; TCGA ESCA -0.643; TCGA HNSC -0.396; TCGA LGG -0.331; TCGA LUSC -0.503; TCGA PAAD -0.498; TCGA PRAD -0.414; TCGA SARC -0.189; TCGA THCA -0.236; TCGA STAD -0.289 |
hsa-miR-140-5p | GBAS | 9 cancers: BLCA; KIRC; LGG; LIHC; LUSC; PAAD; SARC; STAD; UCEC | miRanda | TCGA BLCA -0.084; TCGA KIRC -0.387; TCGA LGG -0.101; TCGA LIHC -0.281; TCGA LUSC -0.152; TCGA PAAD -0.129; TCGA SARC -0.214; TCGA STAD -0.187; TCGA UCEC -0.11 |
hsa-miR-140-5p | CPNE3 | 10 cancers: BLCA; COAD; ESCA; HNSC; KIRC; LGG; LUAD; SARC; STAD; UCEC | miRanda | TCGA BLCA -0.088; TCGA COAD -0.154; TCGA ESCA -0.217; TCGA HNSC -0.119; TCGA KIRC -0.19; TCGA LGG -0.08; TCGA LUAD -0.118; TCGA SARC -0.097; TCGA STAD -0.173; TCGA UCEC -0.102 |
hsa-miR-140-5p | SMARCC1 | 9 cancers: BLCA; KIRC; LUAD; LUSC; PAAD; SARC; THCA; STAD; UCEC | miRanda | TCGA BLCA -0.096; TCGA KIRC -0.251; TCGA LUAD -0.123; TCGA LUSC -0.301; TCGA PAAD -0.11; TCGA SARC -0.12; TCGA THCA -0.113; TCGA STAD -0.078; TCGA UCEC -0.173 |
hsa-miR-140-5p | MEGF9 | 9 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; PRAD; SARC; THCA; STAD | miRanda | TCGA BLCA -0.173; TCGA BRCA -0.152; TCGA ESCA -0.393; TCGA HNSC -0.113; TCGA KIRC -0.347; TCGA PRAD -0.194; TCGA SARC -0.137; TCGA THCA -0.425; TCGA STAD -0.194 |
hsa-miR-140-5p | TMEM106B | 12 cancers: BLCA; COAD; HNSC; KIRC; LGG; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRanda | TCGA BLCA -0.119; TCGA COAD -0.123; TCGA HNSC -0.086; TCGA KIRC -0.248; TCGA LGG -0.105; TCGA LUSC -0.134; TCGA PAAD -0.146; TCGA PRAD -0.313; TCGA SARC -0.085; TCGA THCA -0.193; TCGA STAD -0.291; TCGA UCEC -0.12 |
hsa-miR-140-5p | MAN1A2 | 9 cancers: BLCA; COAD; HNSC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | miRanda | TCGA BLCA -0.112; TCGA COAD -0.177; TCGA HNSC -0.112; TCGA LUAD -0.117; TCGA LUSC -0.122; TCGA PRAD -0.261; TCGA SARC -0.192; TCGA STAD -0.132; TCGA UCEC -0.103 |
hsa-miR-140-5p | OXR1 | 13 cancers: BLCA; CESC; COAD; ESCA; KIRC; KIRP; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD | miRanda | TCGA BLCA -0.164; TCGA CESC -0.129; TCGA COAD -0.241; TCGA ESCA -0.208; TCGA KIRC -0.218; TCGA KIRP -0.407; TCGA LUAD -0.079; TCGA LUSC -0.128; TCGA PAAD -0.168; TCGA PRAD -0.304; TCGA SARC -0.158; TCGA THCA -0.155; TCGA STAD -0.27 |
hsa-miR-140-5p | ZNF770 | 10 cancers: BLCA; ESCA; HNSC; KIRC; LUAD; LUSC; SARC; THCA; STAD; UCEC | miRanda | TCGA BLCA -0.175; TCGA ESCA -0.317; TCGA HNSC -0.197; TCGA KIRC -0.13; TCGA LUAD -0.102; TCGA LUSC -0.213; TCGA SARC -0.088; TCGA THCA -0.147; TCGA STAD -0.257; TCGA UCEC -0.196 |
hsa-miR-140-5p | WDR26 | 9 cancers: BLCA; ESCA; LGG; LUAD; LUSC; SARC; THCA; STAD; UCEC | miRanda | TCGA BLCA -0.066; TCGA ESCA -0.171; TCGA LGG -0.054; TCGA LUAD -0.075; TCGA LUSC -0.078; TCGA SARC -0.077; TCGA THCA -0.16; TCGA STAD -0.221; TCGA UCEC -0.071 |
hsa-miR-140-5p | MPP5 | 9 cancers: BLCA; COAD; HNSC; KIRC; LUAD; PAAD; PRAD; STAD; UCEC | miRanda | TCGA BLCA -0.116; TCGA COAD -0.095; TCGA HNSC -0.13; TCGA KIRC -0.428; TCGA LUAD -0.104; TCGA PAAD -0.236; TCGA PRAD -0.252; TCGA STAD -0.134; TCGA UCEC -0.111 |
hsa-miR-140-5p | VEZT | 12 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; SARC; STAD | miRanda | TCGA BLCA -0.077; TCGA BRCA -0.072; TCGA CESC -0.083; TCGA ESCA -0.157; TCGA HNSC -0.095; TCGA KIRP -0.134; TCGA LIHC -0.146; TCGA LUAD -0.12; TCGA LUSC -0.092; TCGA PAAD -0.136; TCGA SARC -0.134; TCGA STAD -0.128 |
hsa-miR-140-5p | THSD4 | 10 cancers: BLCA; BRCA; COAD; KIRC; LIHC; PAAD; PRAD; SARC; THCA; STAD | miRanda | TCGA BLCA -0.22; TCGA BRCA -0.592; TCGA COAD -0.319; TCGA KIRC -0.782; TCGA LIHC -0.373; TCGA PAAD -0.286; TCGA PRAD -0.422; TCGA SARC -0.613; TCGA THCA -0.202; TCGA STAD -0.251 |
hsa-miR-140-5p | ZHX1 | 9 cancers: BLCA; COAD; ESCA; KIRC; LGG; LUAD; PRAD; SARC; STAD | miRNATAP | TCGA BLCA -0.144; TCGA COAD -0.146; TCGA ESCA -0.272; TCGA KIRC -0.154; TCGA LGG -0.072; TCGA LUAD -0.141; TCGA PRAD -0.087; TCGA SARC -0.15; TCGA STAD -0.157 |
hsa-miR-140-5p | NCS1 | 9 cancers: BLCA; ESCA; HNSC; KIRC; LIHC; LUSC; PRAD; SARC; STAD | miRNATAP | TCGA BLCA -0.273; TCGA ESCA -0.593; TCGA HNSC -0.122; TCGA KIRC -0.321; TCGA LIHC -0.326; TCGA LUSC -0.42; TCGA PRAD -0.255; TCGA SARC -0.337; TCGA STAD -0.555 |
hsa-miR-140-5p | PSEN1 | 9 cancers: BRCA; CESC; ESCA; HNSC; KIRC; LUAD; PAAD; THCA; UCEC | MirTarget; miRanda | TCGA BRCA -0.052; TCGA CESC -0.075; TCGA ESCA -0.116; TCGA HNSC -0.162; TCGA KIRC -0.156; TCGA LUAD -0.066; TCGA PAAD -0.214; TCGA THCA -0.087; TCGA UCEC -0.069 |
hsa-miR-140-5p | IGSF3 | 13 cancers: BRCA; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget; PITA; miRanda; miRNATAP | TCGA BRCA -0.111; TCGA ESCA -0.596; TCGA HNSC -0.318; TCGA KIRC -0.332; TCGA KIRP -0.127; TCGA LUAD -0.236; TCGA LUSC -0.399; TCGA PAAD -0.27; TCGA PRAD -0.261; TCGA SARC -0.309; TCGA THCA -0.351; TCGA STAD -0.23; TCGA UCEC -0.334 |
hsa-miR-140-5p | CD46 | 10 cancers: BRCA; COAD; KIRC; LUAD; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRanda | TCGA BRCA -0.137; TCGA COAD -0.202; TCGA KIRC -0.253; TCGA LUAD -0.155; TCGA PAAD -0.226; TCGA PRAD -0.234; TCGA SARC -0.202; TCGA THCA -0.171; TCGA STAD -0.117; TCGA UCEC -0.095 |
hsa-miR-140-5p | EVPL | 11 cancers: BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LUAD; PAAD; SARC; THCA; UCEC | miRanda | TCGA BRCA -0.415; TCGA CESC -0.281; TCGA ESCA -0.536; TCGA HNSC -0.217; TCGA KIRC -0.669; TCGA KIRP -0.388; TCGA LUAD -0.17; TCGA PAAD -0.784; TCGA SARC -0.313; TCGA THCA -0.398; TCGA UCEC -0.368 |
hsa-miR-140-5p | BRD8 | 9 cancers: BRCA; KIRC; KIRP; LGG; LUAD; SARC; THCA; STAD; UCEC | miRanda | TCGA BRCA -0.109; TCGA KIRC -0.105; TCGA KIRP -0.061; TCGA LGG -0.067; TCGA LUAD -0.073; TCGA SARC -0.134; TCGA THCA -0.077; TCGA STAD -0.102; TCGA UCEC -0.056 |
hsa-miR-140-5p | AFTPH | 9 cancers: BRCA; CESC; HNSC; KIRC; LIHC; LUAD; LUSC; SARC; UCEC | miRanda; miRNATAP | TCGA BRCA -0.062; TCGA CESC -0.069; TCGA HNSC -0.095; TCGA KIRC -0.195; TCGA LIHC -0.078; TCGA LUAD -0.107; TCGA LUSC -0.087; TCGA SARC -0.069; TCGA UCEC -0.142 |
hsa-miR-140-5p | RFC5 | 9 cancers: BRCA; CESC; KIRP; LGG; LIHC; LUAD; LUSC; THCA; UCEC | miRanda | TCGA BRCA -0.061; TCGA CESC -0.115; TCGA KIRP -0.227; TCGA LGG -0.108; TCGA LIHC -0.117; TCGA LUAD -0.138; TCGA LUSC -0.352; TCGA THCA -0.119; TCGA UCEC -0.115 |
hsa-miR-140-5p | METTL2A | 13 cancers: BRCA; CESC; COAD; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; THCA; UCEC | miRanda | TCGA BRCA -0.077; TCGA CESC -0.132; TCGA COAD -0.129; TCGA HNSC -0.069; TCGA KIRC -0.114; TCGA KIRP -0.104; TCGA LGG -0.086; TCGA LIHC -0.122; TCGA LUAD -0.071; TCGA LUSC -0.207; TCGA PAAD -0.12; TCGA THCA -0.056; TCGA UCEC -0.145 |
hsa-miR-140-5p | POGZ | 9 cancers: BRCA; CESC; KIRP; LIHC; LUAD; LUSC; SARC; STAD; UCEC | miRanda | TCGA BRCA -0.061; TCGA CESC -0.086; TCGA KIRP -0.163; TCGA LIHC -0.111; TCGA LUAD -0.176; TCGA LUSC -0.146; TCGA SARC -0.086; TCGA STAD -0.185; TCGA UCEC -0.062 |
hsa-miR-140-5p | ANAPC7 | 10 cancers: BRCA; ESCA; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; SARC; UCEC | miRanda | TCGA BRCA -0.08; TCGA ESCA -0.144; TCGA KIRC -0.084; TCGA KIRP -0.076; TCGA LGG -0.05; TCGA LIHC -0.089; TCGA LUAD -0.06; TCGA LUSC -0.17; TCGA SARC -0.065; TCGA UCEC -0.065 |
hsa-miR-140-5p | RBM39 | 9 cancers: BRCA; CESC; KIRP; LGG; LIHC; LUAD; LUSC; STAD; UCEC | miRanda; miRNATAP | TCGA BRCA -0.074; TCGA CESC -0.063; TCGA KIRP -0.191; TCGA LGG -0.078; TCGA LIHC -0.11; TCGA LUAD -0.069; TCGA LUSC -0.163; TCGA STAD -0.063; TCGA UCEC -0.105 |
hsa-miR-140-5p | SRP9 | 9 cancers: BRCA; KIRC; LGG; LIHC; LUAD; LUSC; PAAD; THCA; UCEC | miRanda | TCGA BRCA -0.15; TCGA KIRC -0.126; TCGA LGG -0.078; TCGA LIHC -0.095; TCGA LUAD -0.065; TCGA LUSC -0.131; TCGA PAAD -0.092; TCGA THCA -0.059; TCGA UCEC -0.148 |
hsa-miR-140-5p | L2HGDH | 10 cancers: BRCA; CESC; ESCA; KIRC; LUAD; LUSC; PAAD; SARC; STAD; UCEC | miRanda | TCGA BRCA -0.093; TCGA CESC -0.131; TCGA ESCA -0.198; TCGA KIRC -0.497; TCGA LUAD -0.129; TCGA LUSC -0.323; TCGA PAAD -0.156; TCGA SARC -0.213; TCGA STAD -0.11; TCGA UCEC -0.21 |
hsa-miR-140-5p | KIAA0907 | 11 cancers: BRCA; CESC; KIRP; LGG; LIHC; LUAD; LUSC; SARC; THCA; STAD; UCEC | miRanda | TCGA BRCA -0.096; TCGA CESC -0.141; TCGA KIRP -0.319; TCGA LGG -0.086; TCGA LIHC -0.156; TCGA LUAD -0.101; TCGA LUSC -0.376; TCGA SARC -0.068; TCGA THCA -0.179; TCGA STAD -0.166; TCGA UCEC -0.233 |
hsa-miR-140-5p | MAP7 | 9 cancers: BRCA; CESC; HNSC; KIRC; LUAD; LUSC; PAAD; THCA; UCEC | miRanda | TCGA BRCA -0.178; TCGA CESC -0.203; TCGA HNSC -0.313; TCGA KIRC -0.41; TCGA LUAD -0.225; TCGA LUSC -0.369; TCGA PAAD -0.257; TCGA THCA -0.105; TCGA UCEC -0.368 |
hsa-miR-140-5p | CHD4 | 9 cancers: BRCA; CESC; HNSC; KIRC; LUAD; LUSC; THCA; STAD; UCEC | miRanda | TCGA BRCA -0.067; TCGA CESC -0.079; TCGA HNSC -0.091; TCGA KIRC -0.099; TCGA LUAD -0.113; TCGA LUSC -0.124; TCGA THCA -0.227; TCGA STAD -0.056; TCGA UCEC -0.117 |
hsa-miR-140-5p | TANC2 | 13 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; THCA; STAD; UCEC | miRanda; miRNATAP | TCGA BRCA -0.173; TCGA CESC -0.172; TCGA COAD -0.349; TCGA ESCA -0.577; TCGA HNSC -0.256; TCGA KIRP -0.174; TCGA LIHC -0.243; TCGA LUAD -0.253; TCGA LUSC -0.134; TCGA PAAD -0.34; TCGA THCA -0.213; TCGA STAD -0.255; TCGA UCEC -0.16 |
hsa-miR-140-5p | PIP4K2C | 11 cancers: BRCA; CESC; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; THCA; UCEC | miRanda | TCGA BRCA -0.176; TCGA CESC -0.082; TCGA ESCA -0.134; TCGA HNSC -0.205; TCGA KIRC -0.511; TCGA LUAD -0.077; TCGA LUSC -0.199; TCGA PAAD -0.223; TCGA PRAD -0.123; TCGA THCA -0.109; TCGA UCEC -0.103 |
hsa-miR-140-5p | FAM134A | 9 cancers: BRCA; HNSC; LIHC; LUAD; LUSC; PAAD; SARC; THCA; UCEC | miRanda | TCGA BRCA -0.101; TCGA HNSC -0.069; TCGA LIHC -0.099; TCGA LUAD -0.063; TCGA LUSC -0.09; TCGA PAAD -0.148; TCGA SARC -0.075; TCGA THCA -0.169; TCGA UCEC -0.086 |
hsa-miR-140-5p | FAM199X | 10 cancers: BRCA; COAD; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | miRanda | TCGA BRCA -0.095; TCGA COAD -0.124; TCGA HNSC -0.113; TCGA KIRC -0.228; TCGA LUAD -0.109; TCGA LUSC -0.284; TCGA PAAD -0.207; TCGA PRAD -0.099; TCGA STAD -0.124; TCGA UCEC -0.246 |
hsa-miR-140-5p | DNAH14 | 10 cancers: BRCA; CESC; COAD; KIRC; KIRP; LIHC; LUSC; THCA; STAD; UCEC | miRanda | TCGA BRCA -0.213; TCGA CESC -0.468; TCGA COAD -0.385; TCGA KIRC -0.24; TCGA KIRP -0.245; TCGA LIHC -0.225; TCGA LUSC -0.559; TCGA THCA -0.3; TCGA STAD -0.305; TCGA UCEC -0.203 |
hsa-miR-140-5p | BCL7A | 10 cancers: BRCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; SARC; THCA | miRanda | TCGA BRCA -0.079; TCGA HNSC -0.128; TCGA KIRC -0.334; TCGA KIRP -0.098; TCGA LGG -0.168; TCGA LIHC -0.213; TCGA LUAD -0.159; TCGA LUSC -0.228; TCGA SARC -0.111; TCGA THCA -0.234 |
hsa-miR-140-5p | MARVELD2 | 9 cancers: BRCA; CESC; HNSC; KIRC; KIRP; LIHC; LUAD; PAAD; UCEC | miRanda | TCGA BRCA -0.272; TCGA CESC -0.222; TCGA HNSC -0.159; TCGA KIRC -0.965; TCGA KIRP -0.209; TCGA LIHC -0.397; TCGA LUAD -0.217; TCGA PAAD -0.55; TCGA UCEC -0.167 |
hsa-miR-140-5p | PARD6B | 10 cancers: BRCA; CESC; HNSC; KIRC; LIHC; LUAD; PAAD; SARC; THCA; UCEC | miRanda | TCGA BRCA -0.524; TCGA CESC -0.173; TCGA HNSC -0.143; TCGA KIRC -0.46; TCGA LIHC -0.25; TCGA LUAD -0.169; TCGA PAAD -0.412; TCGA SARC -0.465; TCGA THCA -0.269; TCGA UCEC -0.232 |
hsa-miR-140-5p | MAL2 | 10 cancers: BRCA; CESC; HNSC; KIRC; KIRP; LIHC; LUAD; PAAD; THCA; UCEC | miRanda | TCGA BRCA -0.211; TCGA CESC -0.29; TCGA HNSC -0.267; TCGA KIRC -1.254; TCGA KIRP -0.636; TCGA LIHC -0.834; TCGA LUAD -0.199; TCGA PAAD -0.73; TCGA THCA -0.271; TCGA UCEC -0.632 |
hsa-miR-140-5p | CREB3L4 | 9 cancers: BRCA; CESC; KIRP; LIHC; LUAD; LUSC; PAAD; SARC; THCA | miRanda | TCGA BRCA -0.565; TCGA CESC -0.153; TCGA KIRP -0.253; TCGA LIHC -0.151; TCGA LUAD -0.134; TCGA LUSC -0.203; TCGA PAAD -0.228; TCGA SARC -0.134; TCGA THCA -0.089 |
hsa-miR-140-5p | SLC44A1 | 9 cancers: BRCA; CESC; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; THCA | miRanda | TCGA BRCA -0.061; TCGA CESC -0.092; TCGA ESCA -0.287; TCGA HNSC -0.155; TCGA KIRC -0.268; TCGA LUAD -0.11; TCGA LUSC -0.255; TCGA PAAD -0.395; TCGA THCA -0.179 |
hsa-miR-140-5p | CCDC43 | 10 cancers: BRCA; CESC; ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; THCA; UCEC | miRanda | TCGA BRCA -0.057; TCGA CESC -0.107; TCGA ESCA -0.109; TCGA HNSC -0.07; TCGA KIRP -0.08; TCGA LUAD -0.116; TCGA LUSC -0.152; TCGA PAAD -0.133; TCGA THCA -0.105; TCGA UCEC -0.11 |
hsa-miR-140-5p | MYO19 | 13 cancers: BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; SARC; THCA | miRanda | TCGA BRCA -0.098; TCGA CESC -0.168; TCGA ESCA -0.192; TCGA HNSC -0.086; TCGA KIRC -0.216; TCGA KIRP -0.179; TCGA LGG -0.112; TCGA LIHC -0.219; TCGA LUAD -0.11; TCGA LUSC -0.381; TCGA PAAD -0.155; TCGA SARC -0.156; TCGA THCA -0.157 |
hsa-miR-140-5p | MNAT1 | 9 cancers: BRCA; CESC; ESCA; HNSC; LGG; LUAD; LUSC; PAAD; UCEC | miRanda | TCGA BRCA -0.093; TCGA CESC -0.114; TCGA ESCA -0.368; TCGA HNSC -0.173; TCGA LGG -0.102; TCGA LUAD -0.079; TCGA LUSC -0.25; TCGA PAAD -0.217; TCGA UCEC -0.113 |
hsa-miR-140-5p | RAB15 | 9 cancers: BRCA; HNSC; KIRC; LUAD; LUSC; PAAD; SARC; THCA; UCEC | miRanda | TCGA BRCA -0.182; TCGA HNSC -0.151; TCGA KIRC -0.285; TCGA LUAD -0.17; TCGA LUSC -0.183; TCGA PAAD -0.185; TCGA SARC -0.28; TCGA THCA -0.341; TCGA UCEC -0.151 |
hsa-miR-140-5p | RTKN | 9 cancers: BRCA; CESC; HNSC; LIHC; LUAD; LUSC; PAAD; THCA; UCEC | miRanda | TCGA BRCA -0.139; TCGA CESC -0.129; TCGA HNSC -0.208; TCGA LIHC -0.224; TCGA LUAD -0.112; TCGA LUSC -0.399; TCGA PAAD -0.197; TCGA THCA -0.367; TCGA UCEC -0.174 |
hsa-miR-140-5p | ZBTB9 | 9 cancers: BRCA; CESC; HNSC; KIRC; LGG; LIHC; LUAD; LUSC; UCEC | miRanda | TCGA BRCA -0.1; TCGA CESC -0.087; TCGA HNSC -0.105; TCGA KIRC -0.17; TCGA LGG -0.051; TCGA LIHC -0.156; TCGA LUAD -0.078; TCGA LUSC -0.218; TCGA UCEC -0.149 |
hsa-miR-140-5p | LRRC1 | 11 cancers: BRCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; THCA; UCEC | miRanda | TCGA BRCA -0.144; TCGA HNSC -0.213; TCGA KIRC -0.309; TCGA KIRP -0.172; TCGA LGG -0.128; TCGA LIHC -0.624; TCGA LUAD -0.141; TCGA LUSC -0.27; TCGA PAAD -0.274; TCGA THCA -0.132; TCGA UCEC -0.18 |
hsa-miR-140-5p | PANK3 | 9 cancers: BRCA; COAD; ESCA; HNSC; KIRC; LUSC; PRAD; STAD; UCEC | miRanda | TCGA BRCA -0.142; TCGA COAD -0.158; TCGA ESCA -0.17; TCGA HNSC -0.144; TCGA KIRC -0.282; TCGA LUSC -0.212; TCGA PRAD -0.136; TCGA STAD -0.098; TCGA UCEC -0.207 |
hsa-miR-140-5p | FKBP4 | 9 cancers: BRCA; CESC; ESCA; KIRC; LIHC; LUAD; LUSC; PAAD; UCEC | miRanda; miRNATAP | TCGA BRCA -0.358; TCGA CESC -0.135; TCGA ESCA -0.231; TCGA KIRC -0.324; TCGA LIHC -0.155; TCGA LUAD -0.137; TCGA LUSC -0.427; TCGA PAAD -0.325; TCGA UCEC -0.257 |
hsa-miR-140-5p | CCDC120 | 10 cancers: BRCA; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; SARC; THCA; UCEC | miRanda | TCGA BRCA -0.146; TCGA KIRP -0.128; TCGA LGG -0.185; TCGA LIHC -0.282; TCGA LUAD -0.133; TCGA LUSC -0.161; TCGA PAAD -0.275; TCGA SARC -0.234; TCGA THCA -0.238; TCGA UCEC -0.18 |
hsa-miR-140-5p | IPO7 | 10 cancers: CESC; COAD; KIRC; LGG; LUAD; LUSC; PAAD; SARC; THCA; STAD | MirTarget; miRanda; miRNATAP | TCGA CESC -0.11; TCGA COAD -0.117; TCGA KIRC -0.126; TCGA LGG -0.097; TCGA LUAD -0.091; TCGA LUSC -0.154; TCGA PAAD -0.133; TCGA SARC -0.086; TCGA THCA -0.199; TCGA STAD -0.257 |
hsa-miR-140-5p | MYO10 | 9 cancers: CESC; HNSC; KIRC; LGG; LIHC; LUAD; LUSC; PAAD; UCEC | MirTarget; miRanda; miRNATAP | TCGA CESC -0.19; TCGA HNSC -0.272; TCGA KIRC -0.391; TCGA LGG -0.074; TCGA LIHC -0.314; TCGA LUAD -0.167; TCGA LUSC -0.198; TCGA PAAD -0.27; TCGA UCEC -0.254 |
hsa-miR-140-5p | STK3 | 15 cancers: CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | PITA; miRanda | TCGA CESC -0.164; TCGA COAD -0.227; TCGA ESCA -0.323; TCGA HNSC -0.177; TCGA KIRC -0.159; TCGA KIRP -0.071; TCGA LIHC -0.112; TCGA LUAD -0.129; TCGA LUSC -0.197; TCGA PAAD -0.293; TCGA PRAD -0.072; TCGA SARC -0.173; TCGA THCA -0.11; TCGA STAD -0.151; TCGA UCEC -0.175 |
hsa-miR-140-5p | INSIG2 | 9 cancers: CESC; HNSC; KIRP; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRanda | TCGA CESC -0.1; TCGA HNSC -0.118; TCGA KIRP -0.128; TCGA PAAD -0.272; TCGA PRAD -0.098; TCGA SARC -0.081; TCGA THCA -0.08; TCGA STAD -0.101; TCGA UCEC -0.129 |
hsa-miR-140-5p | PHF6 | 9 cancers: CESC; KIRC; KIRP; LGG; LUAD; LUSC; THCA; STAD; UCEC | miRanda | TCGA CESC -0.102; TCGA KIRC -0.251; TCGA KIRP -0.109; TCGA LGG -0.108; TCGA LUAD -0.102; TCGA LUSC -0.235; TCGA THCA -0.077; TCGA STAD -0.155; TCGA UCEC -0.209 |
hsa-miR-140-5p | RAD21 | 10 cancers: CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PRAD; SARC; UCEC | miRanda | TCGA CESC -0.145; TCGA COAD -0.12; TCGA ESCA -0.267; TCGA HNSC -0.111; TCGA KIRC -0.09; TCGA LUAD -0.182; TCGA LUSC -0.171; TCGA PRAD -0.099; TCGA SARC -0.08; TCGA UCEC -0.262 |
hsa-miR-140-5p | POGK | 10 cancers: CESC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; THCA; STAD; UCEC | miRanda | TCGA CESC -0.102; TCGA KIRC -0.101; TCGA KIRP -0.131; TCGA LIHC -0.12; TCGA LUAD -0.135; TCGA LUSC -0.172; TCGA PAAD -0.101; TCGA THCA -0.139; TCGA STAD -0.083; TCGA UCEC -0.161 |
hsa-miR-140-5p | YWHAQ | 9 cancers: CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; STAD; UCEC | miRanda | TCGA CESC -0.098; TCGA COAD -0.132; TCGA ESCA -0.247; TCGA HNSC -0.144; TCGA LIHC -0.114; TCGA LUAD -0.065; TCGA LUSC -0.192; TCGA STAD -0.073; TCGA UCEC -0.099 |
hsa-miR-140-5p | NMD3 | 9 cancers: CESC; COAD; ESCA; HNSC; LGG; LUSC; PAAD; THCA; UCEC | miRanda | TCGA CESC -0.129; TCGA COAD -0.106; TCGA ESCA -0.176; TCGA HNSC -0.15; TCGA LGG -0.103; TCGA LUSC -0.256; TCGA PAAD -0.188; TCGA THCA -0.101; TCGA UCEC -0.134 |
hsa-miR-140-5p | INTS8 | 10 cancers: CESC; COAD; ESCA; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; UCEC | miRanda | TCGA CESC -0.118; TCGA COAD -0.127; TCGA ESCA -0.155; TCGA HNSC -0.109; TCGA KIRP -0.179; TCGA LGG -0.081; TCGA LIHC -0.169; TCGA LUAD -0.119; TCGA LUSC -0.281; TCGA UCEC -0.187 |
hsa-miR-140-5p | CXADR | 12 cancers: CESC; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; PRAD; SARC; THCA; UCEC | miRanda | TCGA CESC -0.192; TCGA COAD -0.336; TCGA ESCA -0.288; TCGA HNSC -0.405; TCGA KIRP -0.425; TCGA LUAD -0.243; TCGA LUSC -0.425; TCGA PAAD -0.431; TCGA PRAD -0.317; TCGA SARC -0.579; TCGA THCA -0.178; TCGA UCEC -0.243 |
hsa-miR-140-5p | UGGT2 | 9 cancers: CESC; COAD; KIRP; LGG; LUAD; PAAD; THCA; STAD; UCEC | miRanda | TCGA CESC -0.122; TCGA COAD -0.181; TCGA KIRP -0.149; TCGA LGG -0.102; TCGA LUAD -0.073; TCGA PAAD -0.213; TCGA THCA -0.156; TCGA STAD -0.224; TCGA UCEC -0.12 |
hsa-miR-140-5p | FAM60A | 12 cancers: CESC; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; SARC; THCA; UCEC | miRanda | TCGA CESC -0.13; TCGA HNSC -0.098; TCGA KIRC -0.403; TCGA KIRP -0.215; TCGA LGG -0.187; TCGA LIHC -0.254; TCGA LUAD -0.126; TCGA LUSC -0.359; TCGA PAAD -0.172; TCGA SARC -0.168; TCGA THCA -0.252; TCGA UCEC -0.268 |
hsa-miR-140-5p | ZNF623 | 9 cancers: CESC; COAD; ESCA; HNSC; LUAD; LUSC; SARC; STAD; UCEC | miRanda | TCGA CESC -0.164; TCGA COAD -0.264; TCGA ESCA -0.244; TCGA HNSC -0.14; TCGA LUAD -0.164; TCGA LUSC -0.367; TCGA SARC -0.116; TCGA STAD -0.103; TCGA UCEC -0.152 |
hsa-miR-140-5p | ZDHHC5 | 12 cancers: CESC; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; THCA; UCEC | miRanda | TCGA CESC -0.11; TCGA ESCA -0.204; TCGA HNSC -0.16; TCGA KIRC -0.063; TCGA KIRP -0.102; TCGA LIHC -0.083; TCGA LUAD -0.061; TCGA LUSC -0.137; TCGA PAAD -0.201; TCGA PRAD -0.081; TCGA THCA -0.179; TCGA UCEC -0.067 |
hsa-miR-140-5p | DDX52 | 13 cancers: CESC; COAD; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PRAD; THCA; STAD; UCEC | miRanda | TCGA CESC -0.102; TCGA COAD -0.108; TCGA HNSC -0.103; TCGA KIRC -0.234; TCGA KIRP -0.171; TCGA LGG -0.084; TCGA LIHC -0.085; TCGA LUAD -0.075; TCGA LUSC -0.182; TCGA PRAD -0.052; TCGA THCA -0.238; TCGA STAD -0.077; TCGA UCEC -0.254 |
hsa-miR-140-5p | UBA6 | 9 cancers: CESC; ESCA; HNSC; KIRP; LUSC; PAAD; THCA; STAD; UCEC | miRanda | TCGA CESC -0.135; TCGA ESCA -0.197; TCGA HNSC -0.113; TCGA KIRP -0.08; TCGA LUSC -0.234; TCGA PAAD -0.14; TCGA THCA -0.059; TCGA STAD -0.159; TCGA UCEC -0.086 |
hsa-miR-140-5p | ESYT3 | 9 cancers: CESC; ESCA; KIRC; LIHC; LUAD; PAAD; SARC; STAD; UCEC | miRanda | TCGA CESC -0.38; TCGA ESCA -0.483; TCGA KIRC -0.698; TCGA LIHC -0.551; TCGA LUAD -0.227; TCGA PAAD -0.374; TCGA SARC -0.321; TCGA STAD -0.662; TCGA UCEC -0.274 |
hsa-miR-140-5p | YY1 | 9 cancers: CESC; ESCA; HNSC; LUAD; LUSC; PAAD; SARC; THCA; UCEC | mirMAP | TCGA CESC -0.108; TCGA ESCA -0.218; TCGA HNSC -0.199; TCGA LUAD -0.098; TCGA LUSC -0.215; TCGA PAAD -0.145; TCGA SARC -0.065; TCGA THCA -0.14; TCGA UCEC -0.15 |
hsa-miR-140-5p | CALU | 9 cancers: COAD; HNSC; KIRP; LUSC; PAAD; PRAD; THCA; STAD; UCEC | PITA; miRanda | TCGA COAD -0.195; TCGA HNSC -0.118; TCGA KIRP -0.135; TCGA LUSC -0.268; TCGA PAAD -0.266; TCGA PRAD -0.132; TCGA THCA -0.081; TCGA STAD -0.093; TCGA UCEC -0.077 |
hsa-miR-140-5p | ZBTB33 | 9 cancers: COAD; HNSC; KIRP; LGG; LUAD; LUSC; PRAD; THCA; UCEC | miRanda | TCGA COAD -0.143; TCGA HNSC -0.093; TCGA KIRP -0.069; TCGA LGG -0.063; TCGA LUAD -0.055; TCGA LUSC -0.205; TCGA PRAD -0.065; TCGA THCA -0.243; TCGA UCEC -0.145 |
hsa-miR-140-5p | RNF2 | 9 cancers: COAD; ESCA; HNSC; KIRP; LUAD; LUSC; THCA; STAD; UCEC | miRanda | TCGA COAD -0.105; TCGA ESCA -0.193; TCGA HNSC -0.173; TCGA KIRP -0.082; TCGA LUAD -0.068; TCGA LUSC -0.274; TCGA THCA -0.077; TCGA STAD -0.155; TCGA UCEC -0.151 |
hsa-miR-140-5p | RRM2B | 9 cancers: COAD; ESCA; KIRC; LUAD; PRAD; SARC; THCA; STAD; UCEC | miRanda | TCGA COAD -0.176; TCGA ESCA -0.345; TCGA KIRC -0.141; TCGA LUAD -0.084; TCGA PRAD -0.101; TCGA SARC -0.078; TCGA THCA -0.105; TCGA STAD -0.136; TCGA UCEC -0.191 |
hsa-miR-140-5p | SLC35F5 | 10 cancers: COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; STAD; UCEC | miRanda | TCGA COAD -0.186; TCGA ESCA -0.124; TCGA HNSC -0.17; TCGA KIRC -0.33; TCGA KIRP -0.181; TCGA LUAD -0.088; TCGA LUSC -0.247; TCGA PAAD -0.208; TCGA STAD -0.074; TCGA UCEC -0.262 |
hsa-miR-140-5p | LRRC57 | 12 cancers: COAD; ESCA; HNSC; KIRC; LIHC; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRanda | TCGA COAD -0.097; TCGA ESCA -0.161; TCGA HNSC -0.13; TCGA KIRC -0.281; TCGA LIHC -0.132; TCGA LUSC -0.136; TCGA PAAD -0.182; TCGA PRAD -0.062; TCGA SARC -0.059; TCGA THCA -0.082; TCGA STAD -0.153; TCGA UCEC -0.068 |
hsa-miR-140-5p | DHX36 | 10 cancers: COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | miRanda | TCGA COAD -0.156; TCGA ESCA -0.302; TCGA HNSC -0.168; TCGA KIRC -0.164; TCGA LUAD -0.061; TCGA LUSC -0.3; TCGA PAAD -0.105; TCGA PRAD -0.08; TCGA STAD -0.152; TCGA UCEC -0.156 |
hsa-miR-140-5p | IRF6 | 10 cancers: ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; UCEC | MirTarget | TCGA ESCA -0.685; TCGA HNSC -0.305; TCGA KIRC -0.979; TCGA LUAD -0.141; TCGA LUSC -0.667; TCGA PAAD -0.643; TCGA PRAD -0.218; TCGA SARC -1.134; TCGA THCA -0.115; TCGA UCEC -0.219 |
hsa-miR-140-5p | NCBP2 | 12 cancers: ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; THCA; STAD; UCEC | MirTarget; miRanda | TCGA ESCA -0.158; TCGA HNSC -0.139; TCGA KIRC -0.103; TCGA KIRP -0.091; TCGA LGG -0.118; TCGA LIHC -0.099; TCGA LUAD -0.058; TCGA LUSC -0.366; TCGA PAAD -0.086; TCGA THCA -0.113; TCGA STAD -0.112; TCGA UCEC -0.244 |
hsa-miR-140-5p | GPR161 | 9 cancers: ESCA; HNSC; LGG; LUSC; PAAD; PRAD; SARC; THCA; STAD | PITA; miRanda; miRNATAP | TCGA ESCA -0.48; TCGA HNSC -0.144; TCGA LGG -0.077; TCGA LUSC -0.427; TCGA PAAD -0.283; TCGA PRAD -0.403; TCGA SARC -0.333; TCGA THCA -0.262; TCGA STAD -0.19 |
hsa-miR-140-5p | GJB3 | 9 cancers: ESCA; HNSC; KIRC; KIRP; LUSC; PAAD; PRAD; THCA; UCEC | miRanda | TCGA ESCA -0.606; TCGA HNSC -0.301; TCGA KIRC -0.612; TCGA KIRP -1.397; TCGA LUSC -0.773; TCGA PAAD -0.709; TCGA PRAD -0.708; TCGA THCA -1.316; TCGA UCEC -0.501 |
hsa-miR-140-5p | BRAP | 9 cancers: ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; THCA; STAD; UCEC | miRanda | TCGA ESCA -0.098; TCGA HNSC -0.058; TCGA KIRP -0.064; TCGA LIHC -0.081; TCGA LUAD -0.12; TCGA LUSC -0.106; TCGA THCA -0.058; TCGA STAD -0.061; TCGA UCEC -0.099 |
hsa-miR-140-5p | GFM1 | 10 cancers: ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRanda | TCGA ESCA -0.266; TCGA HNSC -0.125; TCGA KIRC -0.368; TCGA LUAD -0.097; TCGA LUSC -0.284; TCGA PAAD -0.102; TCGA PRAD -0.113; TCGA SARC -0.077; TCGA STAD -0.068; TCGA UCEC -0.205 |
hsa-miR-140-5p | ZNF639 | 10 cancers: ESCA; HNSC; KIRP; LGG; LUAD; LUSC; SARC; THCA; STAD; UCEC | miRanda | TCGA ESCA -0.191; TCGA HNSC -0.128; TCGA KIRP -0.069; TCGA LGG -0.099; TCGA LUAD -0.085; TCGA LUSC -0.285; TCGA SARC -0.054; TCGA THCA -0.053; TCGA STAD -0.152; TCGA UCEC -0.144 |
hsa-miR-140-5p | ZMYM1 | 9 cancers: ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PRAD; SARC; STAD | miRanda | TCGA ESCA -0.125; TCGA HNSC -0.096; TCGA KIRC -0.09; TCGA KIRP -0.165; TCGA LUAD -0.079; TCGA LUSC -0.241; TCGA PRAD -0.128; TCGA SARC -0.172; TCGA STAD -0.283 |
hsa-miR-140-5p | CPSF2 | 9 cancers: ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | miRanda | TCGA ESCA -0.134; TCGA HNSC -0.149; TCGA KIRC -0.129; TCGA LUAD -0.096; TCGA LUSC -0.125; TCGA PAAD -0.098; TCGA PRAD -0.071; TCGA STAD -0.073; TCGA UCEC -0.097 |
hsa-miR-140-5p | NAA30 | 9 cancers: ESCA; HNSC; KIRC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | mirMAP | TCGA ESCA -0.251; TCGA HNSC -0.156; TCGA KIRC -0.125; TCGA LUAD -0.124; TCGA LUSC -0.15; TCGA PRAD -0.053; TCGA SARC -0.071; TCGA STAD -0.166; TCGA UCEC -0.147 |
hsa-miR-140-5p | PURB | 9 cancers: ESCA; HNSC; KIRP; LUAD; LUSC; SARC; THCA; STAD; UCEC | miRNATAP | TCGA ESCA -0.159; TCGA HNSC -0.105; TCGA KIRP -0.168; TCGA LUAD -0.064; TCGA LUSC -0.149; TCGA SARC -0.094; TCGA THCA -0.059; TCGA STAD -0.295; TCGA UCEC -0.157 |
hsa-miR-140-5p | FGD6 | 9 cancers: HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD | miRanda | TCGA HNSC -0.127; TCGA KIRP -0.227; TCGA LIHC -0.27; TCGA LUAD -0.101; TCGA LUSC -0.226; TCGA PAAD -0.314; TCGA PRAD -0.166; TCGA SARC -0.286; TCGA STAD -0.293 |
Enriched cancer pathways of putative targets