microRNA information: hsa-miR-150-5p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-150-5p | miRbase |
| Accession: | MIMAT0000451 | miRbase |
| Precursor name: | hsa-mir-150 | miRbase |
| Precursor accession: | MI0000479 | miRbase |
| Symbol: | MIR150 | HGNC |
| RefSeq ID: | NR_029703 | GenBank |
| Sequence: | UCUCCCAACCCUUGUACCAGUG |
Reported expression in cancers: hsa-miR-150-5p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-150-5p | B cell lymphoma | deregulation | "A set of 11 miRNAs associated with the differentia ......" | 22936066 | qPCR |
| hsa-miR-150-5p | T cell leukemia | deregulation | "Specifically miR-150 and miR-223 were up-regulated ......" | 19246560 | |
| hsa-miR-150-5p | acute myeloid leukemia | deregulation | "Circulating miR 150 and miR 342 in plasma are nove ......" | 23391324 | Microarray; qPCR |
| hsa-miR-150-5p | colon cancer | downregulation | "Microarray analyses of miRNAs in exosome-enriched ......" | 24705249 | Microarray; qPCR |
| hsa-miR-150-5p | colorectal cancer | downregulation | "The study was divided into two phases: firstly qRT ......" | 25255814 | qPCR; Microarray |
| hsa-miR-150-5p | esophageal cancer | downregulation | "A public microarray database showed that the expre ......" | 23013135 | Microarray; qPCR |
| hsa-miR-150-5p | gastric cancer | upregulation | "In the analysis by real-time PCR-based miRNA array ......" | 22407237 | qPCR; Microarray |
| hsa-miR-150-5p | gastric cancer | upregulation | "From the set of the 29 microRNAs of interest we fo ......" | 27081844 | |
| hsa-miR-150-5p | liver cancer | downregulation | "MicroRNA-150 miR-150 is frequently dysregulated in ......" | 26871477 | |
| hsa-miR-150-5p | lung cancer | upregulation | "In the present study miR-150 was found to be signi ......" | 24456795 | qPCR |
| hsa-miR-150-5p | lung squamous cell cancer | downregulation | "Expression of miR 150 and miR 3940 5p is reduced i ......" | 23228962 | |
| hsa-miR-150-5p | lung squamous cell cancer | upregulation | "The levels of two mature miRNAs miR-143 and miR-15 ......" | 24286416 | qPCR |
| hsa-miR-150-5p | lymphoma | downregulation | "Using quantitative reverse transcription-polymeras ......" | 19177201 | Reverse transcription PCR; Microarray |
| hsa-miR-150-5p | lymphoma | downregulation | "Low expression of miR 150 in pediatric intestinal ......" | 24613688 | qPCR |
| hsa-miR-150-5p | ovarian cancer | upregulation | "We analyzed the miRNA expression profiles of prima ......" | 23554878 | qPCR |
| hsa-miR-150-5p | ovarian cancer | downregulation | "MicroRNA miR-150 has been reported to be dramatica ......" | 25090005 | qPCR |
| hsa-miR-150-5p | pancreatic cancer | downregulation | "The expression of miR-150-5p in pancreatic cancer ......" | 24246865 | qPCR |
| hsa-miR-150-5p | pancreatic cancer | downregulation | "Recently we identified miR-150 as a novel tumor su ......" | 24971005 | |
| hsa-miR-150-5p | pancreatic cancer | downregulation | "Limited research investigating the role of oncogen ......" | 25522282 | |
| hsa-miR-150-5p | prostate cancer | upregulation | "miR 150 is a factor of survival in prostate cancer ......" | 25778313 |
Reported cancer pathway affected by hsa-miR-150-5p
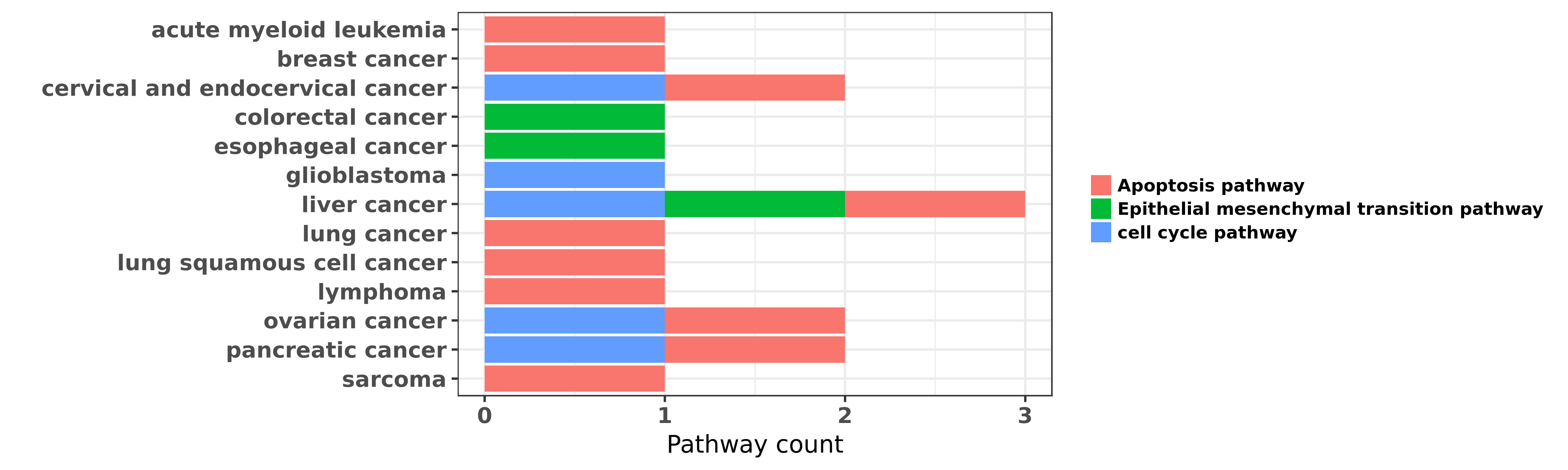
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-150-5p | acute myeloid leukemia | Apoptosis pathway | "Eradication of Acute Myeloid Leukemia with FLT3 Li ......" | 27280396 | |
| hsa-miR-150-5p | breast cancer | Apoptosis pathway | "miR 150 promotes human breast cancer growth and ma ......" | 24312495 | |
| hsa-miR-150-5p | cervical and endocervical cancer | Apoptosis pathway; cell cycle pathway | "microRNA 150 promotes cervical cancer cell growth ......" | 26715362 | Western blot; Flow cytometry; Luciferase |
| hsa-miR-150-5p | colorectal cancer | Epithelial mesenchymal transition pathway | "Wnt/β catenin pathway transactivates microRNA 150 ......" | 27285761 | |
| hsa-miR-150-5p | esophageal cancer | Epithelial mesenchymal transition pathway | "MiR 150 is associated with poor prognosis in esoph ......" | 23013135 | |
| hsa-miR-150-5p | glioblastoma | cell cycle pathway | "An unbiased functional microRNA screen identified ......" | 27399532 | |
| hsa-miR-150-5p | liver cancer | cell cycle pathway; Apoptosis pathway | "microRNA 150 inhibits human CD133 positive liver c ......" | 22025269 | Western blot |
| hsa-miR-150-5p | liver cancer | Epithelial mesenchymal transition pathway | "MicroRNA 150 suppresses cell proliferation and met ......" | 26871477 | |
| hsa-miR-150-5p | lung cancer | Apoptosis pathway | "miR 150 promotes the proliferation of lung cancer ......" | 23747308 | |
| hsa-miR-150-5p | lung cancer | Apoptosis pathway | "Jumonji domain containing 2A predicts prognosis an ......" | 26498874 | Western blot; MTT assay |
| hsa-miR-150-5p | lung squamous cell cancer | Apoptosis pathway | "Down regulation of miR 150 induces cell proliferat ......" | 24532468 | |
| hsa-miR-150-5p | lymphoma | Apoptosis pathway | "The role of microRNA 150 as a tumor suppressor in ......" | 21502955 | |
| hsa-miR-150-5p | ovarian cancer | cell cycle pathway; Apoptosis pathway | "We examined expression of miR-150 in ovarian cance ......" | 25986891 | Cell proliferation assay; Cell Proliferation Assay; Western blot |
| hsa-miR-150-5p | pancreatic cancer | Apoptosis pathway; cell cycle pathway | "MiR 150 5p inhibits the proliferation and promoted ......" | 24246865 | |
| hsa-miR-150-5p | pancreatic cancer | cell cycle pathway; Apoptosis pathway | "A decrease in miR 150 regulates the malignancy of ......" | 25522282 | |
| hsa-miR-150-5p | sarcoma | Apoptosis pathway | "MicroRNA 150 Inhibits Cell Invasion and Migration ......" | 26361218 | Western blot; Luciferase |
Reported cancer prognosis affected by hsa-miR-150-5p
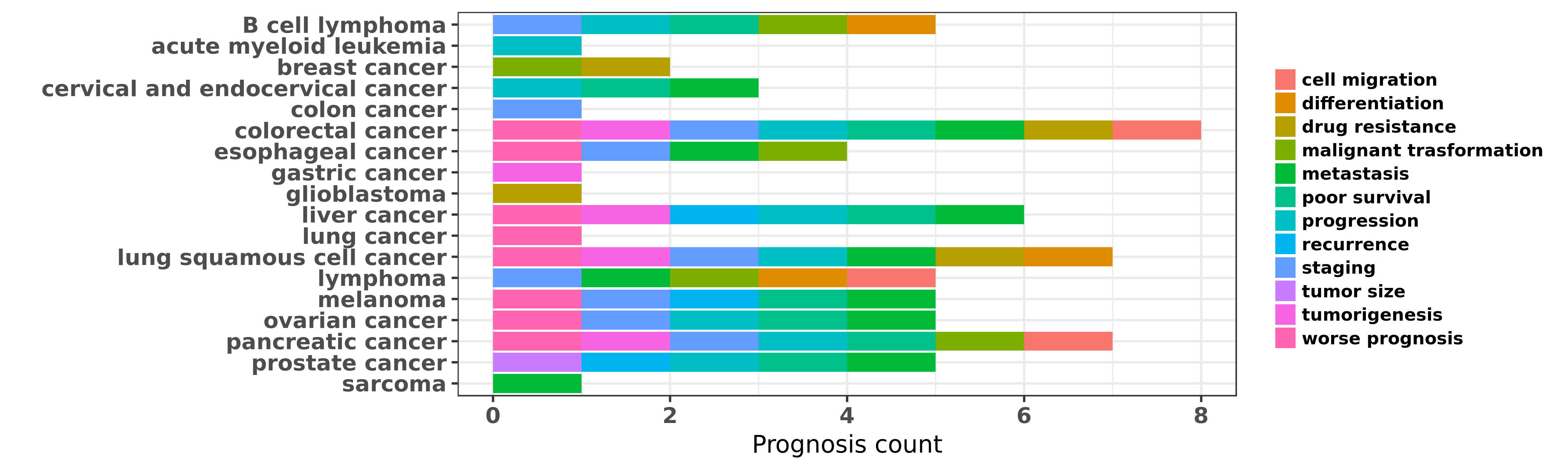
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-150-5p | B cell lymphoma | staging; differentiation; progression; poor survival | "A set of 11 miRNAs associated with the differentia ......" | 22936066 | |
| hsa-miR-150-5p | B cell lymphoma | malignant trasformation | "In this review we discuss miRNAs with essential fu ......" | 25541152 | |
| hsa-miR-150-5p | acute myeloid leukemia | progression | "Eradication of Acute Myeloid Leukemia with FLT3 Li ......" | 27280396 | |
| hsa-miR-150-5p | breast cancer | malignant trasformation | "miR 150 promotes human breast cancer growth and ma ......" | 24312495 | |
| hsa-miR-150-5p | breast cancer | drug resistance | "New miRNA-based drugs are also promising therapy f ......" | 26199650 | |
| hsa-miR-150-5p | cervical and endocervical cancer | poor survival; metastasis; progression | "microRNA 150 promotes cervical cancer cell growth ......" | 26715362 | Western blot; Flow cytometry; Luciferase |
| hsa-miR-150-5p | colon cancer | staging | "Microarray analyses of miRNAs in exosome-enriched ......" | 24705249 | |
| hsa-miR-150-5p | colorectal cancer | worse prognosis; poor survival; drug resistance; tumorigenesis | "miR 150 as a potential biomarker associated with p ......" | 22052060 | |
| hsa-miR-150-5p | colorectal cancer | cell migration; metastasis | "MiR 150 5p suppresses colorectal cancer cell migra ......" | 25124610 | Western blot; Luciferase |
| hsa-miR-150-5p | colorectal cancer | progression | "miR 150 functions as a tumour suppressor in human ......" | 25230975 | |
| hsa-miR-150-5p | colorectal cancer | progression; staging | "Circulating miRNAs miR 34a and miR 150 associated ......" | 25924769 | |
| hsa-miR-150-5p | colorectal cancer | drug resistance | "Wnt/β catenin pathway transactivates microRNA 150 ......" | 27285761 | |
| hsa-miR-150-5p | esophageal cancer | worse prognosis; staging; metastasis; malignant trasformation | "MiR 150 is associated with poor prognosis in esoph ......" | 23013135 | |
| hsa-miR-150-5p | gastric cancer | tumorigenesis | "MiR 150 promotes gastric cancer proliferation by n ......" | 20067763 | Luciferase |
| hsa-miR-150-5p | glioblastoma | drug resistance | "An unbiased functional microRNA screen identified ......" | 27399532 | |
| hsa-miR-150-5p | liver cancer | poor survival | "microRNA 150 inhibits human CD133 positive liver c ......" | 22025269 | Western blot |
| hsa-miR-150-5p | liver cancer | worse prognosis; recurrence; poor survival | "This study was conducted to detect the application ......" | 26215970 | |
| hsa-miR-150-5p | liver cancer | metastasis; progression; tumorigenesis; worse prognosis | "MicroRNA 150 suppresses cell proliferation and met ......" | 26871477 | |
| hsa-miR-150-5p | lung cancer | worse prognosis | "Jumonji domain containing 2A predicts prognosis an ......" | 26498874 | Western blot; MTT assay |
| hsa-miR-150-5p | lung squamous cell cancer | staging; tumorigenesis; differentiation | "Expression of miR 150 and miR 3940 5p is reduced i ......" | 23228962 | |
| hsa-miR-150-5p | lung squamous cell cancer | metastasis | "Altered miR 143 and miR 150 expressions in periphe ......" | 24286416 | |
| hsa-miR-150-5p | lung squamous cell cancer | progression | "Down regulation of miR 150 induces cell proliferat ......" | 24532468 | |
| hsa-miR-150-5p | lung squamous cell cancer | worse prognosis; staging; metastasis | "Increased expression of microRNA 150 is associated ......" | 25755784 | |
| hsa-miR-150-5p | lung squamous cell cancer | drug resistance | "Circulating miR 29a and miR 150 correlate with del ......" | 27117590 | |
| hsa-miR-150-5p | lymphoma | malignant trasformation | "The role of microRNA 150 as a tumor suppressor in ......" | 21502955 | |
| hsa-miR-150-5p | lymphoma | differentiation; staging | "Re expression of microRNA 150 induces EBV positive ......" | 23521217 | |
| hsa-miR-150-5p | lymphoma | staging; metastasis; cell migration | "MicroRNA 150 inhibits tumor invasion and metastasi ......" | 24385540 | |
| hsa-miR-150-5p | melanoma | metastasis | "Six microRNAs miR-9 miR-145 miR-150 miR-155 miR-20 ......" | 23863473 | |
| hsa-miR-150-5p | melanoma | staging; poor survival; recurrence | "We recently reported our array-based discovery of ......" | 25155861 | |
| hsa-miR-150-5p | melanoma | worse prognosis; staging | "Associations with prognosis were replicated for mi ......" | 25490969 | |
| hsa-miR-150-5p | melanoma | staging; metastasis | "Our analyses identified a 4-microRNA miR-150-5p mi ......" | 26089374 | |
| hsa-miR-150-5p | ovarian cancer | metastasis; poor survival | "We analyzed the miRNA expression profiles of prima ......" | 23554878 | |
| hsa-miR-150-5p | ovarian cancer | metastasis; worse prognosis; staging; progression; poor survival | "MicroRNA 150 predicts a favorable prognosis in pat ......" | 25090005 | Luciferase |
| hsa-miR-150-5p | pancreatic cancer | malignant trasformation | "MicroRNA 150 directly targets MUC4 and suppresses ......" | 21983127 | |
| hsa-miR-150-5p | pancreatic cancer | tumorigenesis; poor survival; progression | "A decrease in miR 150 regulates the malignancy of ......" | 25522282 | |
| hsa-miR-150-5p | pancreatic cancer | worse prognosis; poor survival | "The aim of this study was to investigate microRNAs ......" | 25906450 | |
| hsa-miR-150-5p | pancreatic cancer | staging | "At 25 weeks the miRNA microarray analysis revealed ......" | 26516699 | |
| hsa-miR-150-5p | pancreatic cancer | cell migration | "Hypoxia promotes C X C chemokine receptor type 4 e ......" | 26622579 | Luciferase; Cell migration assay |
| hsa-miR-150-5p | prostate cancer | poor survival; metastasis; recurrence; tumor size | "miR 150 is a factor of survival in prostate cancer ......" | 25778313 | Western blot |
| hsa-miR-150-5p | prostate cancer | metastasis; recurrence; progression | "MIR 150 promotes prostate cancer stem cell develop ......" | 26636522 | Western blot; Luciferase |
| hsa-miR-150-5p | sarcoma | metastasis | "MicroRNA 150 Inhibits Cell Invasion and Migration ......" | 26361218 | Western blot; Luciferase |
Reported gene related to hsa-miR-150-5p
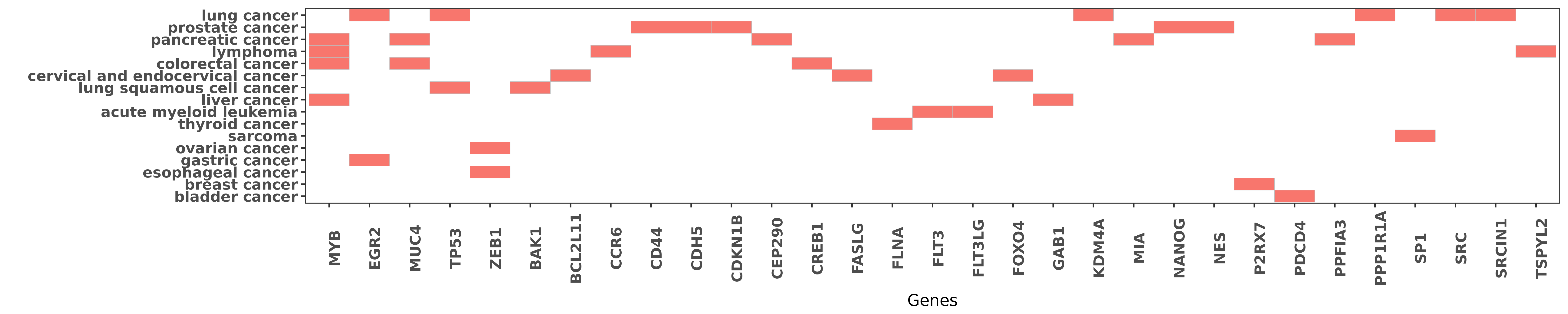
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-150-5p | colorectal cancer | MYB | "miR 150 functions as a tumour suppressor in human ......" | 25230975 |
| hsa-miR-150-5p | liver cancer | MYB | "We also show that miR-150 interacts with the 3'UTR ......" | 22025269 |
| hsa-miR-150-5p | lymphoma | MYB | "Re expression of microRNA 150 induces EBV positive ......" | 23521217 |
| hsa-miR-150-5p | lymphoma | MYB | "Moreover IHC and western blotting defined that c-M ......" | 24613688 |
| hsa-miR-150-5p | pancreatic cancer | MYB | "The associations of miR-150 c-Myb and MUC4 express ......" | 25522282 |
| hsa-miR-150-5p | pancreatic cancer | MYB | "Furthermore the addition of the miRNA inhibitor 2' ......" | 23675407 |
| hsa-miR-150-5p | colorectal cancer | MUC4 | "MiR 150 5p suppresses colorectal cancer cell migra ......" | 25124610 |
| hsa-miR-150-5p | pancreatic cancer | MUC4 | "MicroRNA 150 directly targets MUC4 and suppresses ......" | 21983127 |
| hsa-miR-150-5p | pancreatic cancer | MUC4 | "The associations of miR-150 c-Myb and MUC4 express ......" | 25522282 |
| hsa-miR-150-5p | pancreatic cancer | MUC4 | "Treatment of pancreatic cancer cells with miR-150- ......" | 24971005 |
| hsa-miR-150-5p | gastric cancer | EGR2 | "MiR 150 promotes gastric cancer proliferation by n ......" | 20067763 |
| hsa-miR-150-5p | lung cancer | EGR2 | "Here we demonstrate that miR-150 is aberrantly upr ......" | 23747308 |
| hsa-miR-150-5p | lung cancer | TP53 | "miR 150 promotes the proliferation of lung cancer ......" | 23747308 |
| hsa-miR-150-5p | lung squamous cell cancer | TP53 | "miR-150 and miR-3940-5p were found to be significa ......" | 23228962 |
| hsa-miR-150-5p | esophageal cancer | ZEB1 | "MiR 150 is associated with poor prognosis in esoph ......" | 23013135 |
| hsa-miR-150-5p | ovarian cancer | ZEB1 | "MicroRNA 150 predicts a favorable prognosis in pat ......" | 25090005 |
| hsa-miR-150-5p | lung squamous cell cancer | BAK1 | "Down regulation of miR 150 induces cell proliferat ......" | 24532468 |
| hsa-miR-150-5p | cervical and endocervical cancer | BCL2L11 | "Several cell cycle- and apoptosis-related genes Cy ......" | 26715362 |
| hsa-miR-150-5p | lymphoma | CCR6 | "MicroRNA 150 inhibits tumor invasion and metastasi ......" | 24385540 |
| hsa-miR-150-5p | prostate cancer | CD44 | "MiR-150 expression in CD144 or CD44 positive prima ......" | 26636522 |
| hsa-miR-150-5p | prostate cancer | CDH5 | "MiR-150 expression in CD144 or CD44 positive prima ......" | 26636522 |
| hsa-miR-150-5p | prostate cancer | CDKN1B | "MIR 150 promotes prostate cancer stem cell develop ......" | 26636522 |
| hsa-miR-150-5p | pancreatic cancer | CEP290 | "Hypoxia promotes C X C chemokine receptor type 4 e ......" | 26622579 |
| hsa-miR-150-5p | colorectal cancer | CREB1 | "Wnt/β catenin pathway transactivates microRNA 150 ......" | 27285761 |
| hsa-miR-150-5p | cervical and endocervical cancer | FASLG | "Several cell cycle- and apoptosis-related genes Cy ......" | 26715362 |
| hsa-miR-150-5p | thyroid cancer | FLNA | "Kaplan-Meier and multivariate analysis showed a si ......" | 24443580 |
| hsa-miR-150-5p | acute myeloid leukemia | FLT3 | "Eradication of Acute Myeloid Leukemia with FLT3 Li ......" | 27280396 |
| hsa-miR-150-5p | acute myeloid leukemia | FLT3LG | "Eradication of Acute Myeloid Leukemia with FLT3 Li ......" | 27280396 |
| hsa-miR-150-5p | cervical and endocervical cancer | FOXO4 | "microRNA 150 promotes cervical cancer cell growth ......" | 26715362 |
| hsa-miR-150-5p | liver cancer | GAB1 | "MicroRNA 150 suppresses cell proliferation and met ......" | 26871477 |
| hsa-miR-150-5p | lung cancer | KDM4A | "Finally the relationship between JMJD2A and miR-15 ......" | 26498874 |
| hsa-miR-150-5p | pancreatic cancer | MIA | "PANC-1 MIA PaCa-2BxPC-3 and AsPC-1 cells were tran ......" | 24246865 |
| hsa-miR-150-5p | prostate cancer | NANOG | "MiR-150 could directly target 3'UTR of p27 and dec ......" | 26636522 |
| hsa-miR-150-5p | prostate cancer | NES | "MiR-150 could directly target 3'UTR of p27 and dec ......" | 26636522 |
| hsa-miR-150-5p | breast cancer | P2RX7 | "miR 150 promotes human breast cancer growth and ma ......" | 24312495 |
| hsa-miR-150-5p | bladder cancer | PDCD4 | "miR 150 modulates cisplatin chemosensitivity and i ......" | 25287716 |
| hsa-miR-150-5p | pancreatic cancer | PPFIA3 | "Hypoxia promotes C X C chemokine receptor type 4 e ......" | 26622579 |
| hsa-miR-150-5p | lung cancer | PPP1R1A | "miR 150 promotes the proliferation and migration o ......" | 24456795 |
| hsa-miR-150-5p | sarcoma | SP1 | "A luciferase reporter assay displayed that miR-150 ......" | 26361218 |
| hsa-miR-150-5p | lung cancer | SRC | "miR 150 promotes the proliferation and migration o ......" | 24456795 |
| hsa-miR-150-5p | lung cancer | SRCIN1 | "Furthermore an inverse correlation between miR-150 ......" | 24456795 |
| hsa-miR-150-5p | lymphoma | TSPYL2 | "In this study we show that microRNA-150 miR-150 is ......" | 24385540 |
Expression profile in cancer corhorts:

Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-150-5p | VEGFA | 11 cancers: BLCA; BRCA; CESC; COAD; HNSC; LIHC; LUAD; LUSC; PAAD; THCA; UCEC | miRNAWalker2 validate; miRTarBase | TCGA BLCA -0.164; TCGA BRCA -0.151; TCGA CESC -0.19; TCGA COAD -0.105; TCGA HNSC -0.184; TCGA LIHC -0.115; TCGA LUAD -0.212; TCGA LUSC -0.217; TCGA PAAD -0.219; TCGA THCA -0.169; TCGA UCEC -0.153 |
hsa-miR-150-5p | FAM168B | 12 cancers: BLCA; CESC; COAD; ESCA; KIRC; LIHC; LUAD; LUSC; OV; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.052; TCGA CESC -0.088; TCGA COAD -0.057; TCGA ESCA -0.062; TCGA KIRC -0.133; TCGA LIHC -0.105; TCGA LUAD -0.081; TCGA LUSC -0.129; TCGA OV -0.075; TCGA SARC -0.145; TCGA STAD -0.143; TCGA UCEC -0.105 |
hsa-miR-150-5p | DVL3 | 9 cancers: BLCA; ESCA; HNSC; LGG; LUAD; LUSC; SARC; THCA; STAD | MirTarget | TCGA BLCA -0.079; TCGA ESCA -0.136; TCGA HNSC -0.065; TCGA LGG -0.113; TCGA LUAD -0.077; TCGA LUSC -0.221; TCGA SARC -0.081; TCGA THCA -0.051; TCGA STAD -0.057 |
hsa-miR-150-5p | KDM5B | 13 cancers: BLCA; BRCA; ESCA; HNSC; KIRP; LGG; LUAD; LUSC; OV; PAAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.095; TCGA BRCA -0.054; TCGA ESCA -0.069; TCGA HNSC -0.113; TCGA KIRP -0.057; TCGA LGG -0.176; TCGA LUAD -0.158; TCGA LUSC -0.179; TCGA OV -0.13; TCGA PAAD -0.058; TCGA SARC -0.139; TCGA STAD -0.102; TCGA UCEC -0.134 |
hsa-miR-150-5p | MARCH6 | 11 cancers: BLCA; CESC; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.063; TCGA CESC -0.08; TCGA HNSC -0.062; TCGA KIRC -0.091; TCGA KIRP -0.107; TCGA LIHC -0.074; TCGA LUAD -0.142; TCGA LUSC -0.154; TCGA SARC -0.123; TCGA STAD -0.069; TCGA UCEC -0.083 |
hsa-miR-150-5p | ZNF711 | 9 cancers: BLCA; KIRC; KIRP; LGG; OV; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.165; TCGA KIRC -0.249; TCGA KIRP -0.233; TCGA LGG -0.227; TCGA OV -0.134; TCGA SARC -0.485; TCGA THCA -0.089; TCGA STAD -0.12; TCGA UCEC -0.188 |
hsa-miR-150-5p | METAP1 | 9 cancers: BLCA; COAD; KIRC; LIHC; LUAD; LUSC; PRAD; SARC; STAD | MirTarget | TCGA BLCA -0.084; TCGA COAD -0.056; TCGA KIRC -0.062; TCGA LIHC -0.052; TCGA LUAD -0.093; TCGA LUSC -0.14; TCGA PRAD -0.066; TCGA SARC -0.081; TCGA STAD -0.064 |
hsa-miR-150-5p | WDR5B | 13 cancers: BLCA; BRCA; COAD; KIRC; KIRP; LGG; LIHC; LUSC; OV; PAAD; PRAD; THCA; STAD | MirTarget | TCGA BLCA -0.056; TCGA BRCA -0.092; TCGA COAD -0.081; TCGA KIRC -0.058; TCGA KIRP -0.084; TCGA LGG -0.081; TCGA LIHC -0.101; TCGA LUSC -0.067; TCGA OV -0.067; TCGA PAAD -0.051; TCGA PRAD -0.053; TCGA THCA -0.056; TCGA STAD -0.071 |
hsa-miR-150-5p | MEX3A | 12 cancers: BLCA; CESC; COAD; HNSC; KIRP; LGG; LUAD; LUSC; OV; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.19; TCGA CESC -0.258; TCGA COAD -0.211; TCGA HNSC -0.164; TCGA KIRP -0.116; TCGA LGG -0.495; TCGA LUAD -0.332; TCGA LUSC -0.364; TCGA OV -0.157; TCGA SARC -0.251; TCGA STAD -0.149; TCGA UCEC -0.224 |
hsa-miR-150-5p | ZMYM4 | 9 cancers: BLCA; CESC; COAD; KIRP; LGG; LUAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.063; TCGA CESC -0.064; TCGA COAD -0.051; TCGA KIRP -0.063; TCGA LGG -0.06; TCGA LUAD -0.079; TCGA SARC -0.068; TCGA STAD -0.08; TCGA UCEC -0.06 |
hsa-miR-150-5p | MANBAL | 13 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA | MirTarget | TCGA BLCA -0.053; TCGA BRCA -0.071; TCGA COAD -0.054; TCGA ESCA -0.073; TCGA HNSC -0.074; TCGA KIRC -0.055; TCGA LIHC -0.076; TCGA LUAD -0.095; TCGA LUSC -0.134; TCGA PAAD -0.141; TCGA PRAD -0.071; TCGA SARC -0.1; TCGA THCA -0.087 |
hsa-miR-150-5p | KIAA0895L | 15 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRP; LGG; LUAD; LUSC; OV; PAAD; PRAD; SARC; THCA; STAD | MirTarget | TCGA BLCA -0.161; TCGA BRCA -0.097; TCGA CESC -0.055; TCGA ESCA -0.077; TCGA HNSC -0.052; TCGA KIRP -0.204; TCGA LGG -0.23; TCGA LUAD -0.087; TCGA LUSC -0.152; TCGA OV -0.105; TCGA PAAD -0.092; TCGA PRAD -0.068; TCGA SARC -0.123; TCGA THCA -0.079; TCGA STAD -0.15 |
hsa-miR-150-5p | TRIM9 | 11 cancers: BLCA; BRCA; CESC; HNSC; KIRP; LGG; LUAD; LUSC; OV; SARC; UCEC | MirTarget | TCGA BLCA -0.15; TCGA BRCA -0.11; TCGA CESC -0.197; TCGA HNSC -0.204; TCGA KIRP -0.153; TCGA LGG -0.093; TCGA LUAD -0.264; TCGA LUSC -0.287; TCGA OV -0.118; TCGA SARC -0.331; TCGA UCEC -0.257 |
hsa-miR-150-5p | GK5 | 11 cancers: BLCA; CESC; COAD; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; SARC; STAD | MirTarget | TCGA BLCA -0.066; TCGA CESC -0.056; TCGA COAD -0.053; TCGA KIRC -0.175; TCGA KIRP -0.149; TCGA LIHC -0.073; TCGA LUAD -0.07; TCGA LUSC -0.15; TCGA PAAD -0.051; TCGA SARC -0.117; TCGA STAD -0.165 |
hsa-miR-150-5p | KCTD3 | 11 cancers: BLCA; BRCA; LGG; LIHC; LUAD; LUSC; PAAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.051; TCGA BRCA -0.116; TCGA LGG -0.093; TCGA LIHC -0.071; TCGA LUAD -0.114; TCGA LUSC -0.078; TCGA PAAD -0.091; TCGA SARC -0.115; TCGA THCA -0.077; TCGA STAD -0.14; TCGA UCEC -0.077 |
hsa-miR-150-5p | SUN1 | 14 cancers: BLCA; CESC; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; OV; PAAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.058; TCGA CESC -0.106; TCGA ESCA -0.062; TCGA HNSC -0.085; TCGA KIRC -0.062; TCGA KIRP -0.169; TCGA LIHC -0.086; TCGA LUAD -0.106; TCGA LUSC -0.156; TCGA OV -0.059; TCGA PAAD -0.134; TCGA SARC -0.108; TCGA STAD -0.187; TCGA UCEC -0.073 |
hsa-miR-150-5p | DGCR2 | 9 cancers: BLCA; BRCA; HNSC; LGG; LIHC; LUSC; PAAD; PRAD; THCA | mirMAP | TCGA BLCA -0.06; TCGA BRCA -0.056; TCGA HNSC -0.063; TCGA LGG -0.16; TCGA LIHC -0.066; TCGA LUSC -0.142; TCGA PAAD -0.08; TCGA PRAD -0.055; TCGA THCA -0.07 |
hsa-miR-150-5p | CA12 | 10 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; KIRP; LUSC; PRAD; SARC; THCA | mirMAP | TCGA BLCA -0.201; TCGA BRCA -0.269; TCGA ESCA -0.382; TCGA HNSC -0.176; TCGA KIRC -0.207; TCGA KIRP -0.22; TCGA LUSC -0.436; TCGA PRAD -0.167; TCGA SARC -0.176; TCGA THCA -0.433 |
hsa-miR-150-5p | TNPO2 | 9 cancers: BLCA; LIHC; LUSC; OV; PAAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.065; TCGA LIHC -0.06; TCGA LUSC -0.075; TCGA OV -0.096; TCGA PAAD -0.071; TCGA SARC -0.108; TCGA THCA -0.067; TCGA STAD -0.094; TCGA UCEC -0.05 |
hsa-miR-150-5p | CRTC1 | 11 cancers: BLCA; BRCA; CESC; KIRC; KIRP; LGG; OV; PRAD; SARC; THCA; UCEC | mirMAP | TCGA BLCA -0.083; TCGA BRCA -0.061; TCGA CESC -0.108; TCGA KIRC -0.157; TCGA KIRP -0.075; TCGA LGG -0.061; TCGA OV -0.09; TCGA PRAD -0.089; TCGA SARC -0.058; TCGA THCA -0.129; TCGA UCEC -0.053 |
hsa-miR-150-5p | SMAD5 | 11 cancers: BLCA; CESC; COAD; ESCA; KIRP; LGG; LUAD; OV; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.057; TCGA CESC -0.058; TCGA COAD -0.07; TCGA ESCA -0.074; TCGA KIRP -0.088; TCGA LGG -0.111; TCGA LUAD -0.08; TCGA OV -0.061; TCGA SARC -0.113; TCGA STAD -0.139; TCGA UCEC -0.096 |
hsa-miR-150-5p | FAM20B | 12 cancers: BLCA; BRCA; COAD; ESCA; KIRC; KIRP; LIHC; LUAD; LUSC; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.076; TCGA BRCA -0.067; TCGA COAD -0.054; TCGA ESCA -0.114; TCGA KIRC -0.116; TCGA KIRP -0.05; TCGA LIHC -0.078; TCGA LUAD -0.075; TCGA LUSC -0.116; TCGA SARC -0.081; TCGA STAD -0.166; TCGA UCEC -0.1 |
hsa-miR-150-5p | PLA2G12A | 11 cancers: BLCA; BRCA; CESC; COAD; KIRC; KIRP; LUAD; PAAD; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.1; TCGA BRCA -0.112; TCGA CESC -0.08; TCGA COAD -0.082; TCGA KIRC -0.148; TCGA KIRP -0.079; TCGA LUAD -0.109; TCGA PAAD -0.113; TCGA SARC -0.123; TCGA THCA -0.066; TCGA STAD -0.142 |
hsa-miR-150-5p | NDUFA5 | 12 cancers: BLCA; BRCA; COAD; KIRC; KIRP; LIHC; LUAD; PAAD; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.051; TCGA BRCA -0.082; TCGA COAD -0.074; TCGA KIRC -0.185; TCGA KIRP -0.097; TCGA LIHC -0.12; TCGA LUAD -0.085; TCGA PAAD -0.101; TCGA PRAD -0.098; TCGA THCA -0.08; TCGA STAD -0.07; TCGA UCEC -0.067 |
hsa-miR-150-5p | FAM155B | 11 cancers: BLCA; BRCA; HNSC; KIRC; KIRP; LUSC; OV; PRAD; SARC; THCA; UCEC | mirMAP | TCGA BLCA -0.255; TCGA BRCA -0.126; TCGA HNSC -0.202; TCGA KIRC -0.221; TCGA KIRP -0.07; TCGA LUSC -0.323; TCGA OV -0.168; TCGA PRAD -0.15; TCGA SARC -0.162; TCGA THCA -0.204; TCGA UCEC -0.144 |
hsa-miR-150-5p | MTSS1L | 9 cancers: BLCA; CESC; ESCA; HNSC; LGG; LUSC; PAAD; SARC; STAD | mirMAP | TCGA BLCA -0.109; TCGA CESC -0.081; TCGA ESCA -0.193; TCGA HNSC -0.067; TCGA LGG -0.088; TCGA LUSC -0.114; TCGA PAAD -0.119; TCGA SARC -0.221; TCGA STAD -0.22 |
hsa-miR-150-5p | GSTM3 | 13 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LUAD; LUSC; OV; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.251; TCGA BRCA -0.197; TCGA ESCA -0.235; TCGA HNSC -0.157; TCGA KIRC -0.531; TCGA LUAD -0.116; TCGA LUSC -0.291; TCGA OV -0.199; TCGA PRAD -0.117; TCGA SARC -0.132; TCGA THCA -0.079; TCGA STAD -0.21; TCGA UCEC -0.098 |
hsa-miR-150-5p | IGF1R | 16 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; OV; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.065; TCGA BRCA -0.142; TCGA CESC -0.13; TCGA ESCA -0.163; TCGA HNSC -0.143; TCGA KIRC -0.144; TCGA KIRP -0.108; TCGA LGG -0.114; TCGA LUAD -0.143; TCGA LUSC -0.213; TCGA OV -0.181; TCGA PRAD -0.131; TCGA SARC -0.146; TCGA THCA -0.068; TCGA STAD -0.1; TCGA UCEC -0.083 |
hsa-miR-150-5p | RAB40B | 9 cancers: BLCA; COAD; KIRC; LIHC; LUAD; LUSC; PAAD; SARC; THCA | mirMAP | TCGA BLCA -0.115; TCGA COAD -0.113; TCGA KIRC -0.23; TCGA LIHC -0.141; TCGA LUAD -0.137; TCGA LUSC -0.147; TCGA PAAD -0.141; TCGA SARC -0.059; TCGA THCA -0.097 |
hsa-miR-150-5p | PSD3 | 9 cancers: BLCA; BRCA; LIHC; LUAD; LUSC; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.119; TCGA BRCA -0.078; TCGA LIHC -0.081; TCGA LUAD -0.083; TCGA LUSC -0.087; TCGA SARC -0.127; TCGA THCA -0.06; TCGA STAD -0.097; TCGA UCEC -0.201 |
hsa-miR-150-5p | DPY19L4 | 14 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; OV; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.078; TCGA CESC -0.068; TCGA COAD -0.128; TCGA ESCA -0.129; TCGA HNSC -0.057; TCGA KIRC -0.157; TCGA KIRP -0.131; TCGA LUAD -0.101; TCGA LUSC -0.125; TCGA OV -0.05; TCGA SARC -0.084; TCGA THCA -0.073; TCGA STAD -0.168; TCGA UCEC -0.123 |
hsa-miR-150-5p | KSR2 | 9 cancers: BLCA; BRCA; CESC; KIRC; KIRP; OV; PRAD; THCA; UCEC | mirMAP | TCGA BLCA -0.207; TCGA BRCA -0.141; TCGA CESC -0.238; TCGA KIRC -0.388; TCGA KIRP -0.202; TCGA OV -0.129; TCGA PRAD -0.127; TCGA THCA -0.094; TCGA UCEC -0.102 |
hsa-miR-150-5p | PAWR | 13 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.052; TCGA BRCA -0.077; TCGA CESC -0.055; TCGA ESCA -0.093; TCGA HNSC -0.113; TCGA KIRC -0.13; TCGA KIRP -0.101; TCGA LUAD -0.152; TCGA LUSC -0.213; TCGA PRAD -0.057; TCGA SARC -0.304; TCGA STAD -0.077; TCGA UCEC -0.093 |
hsa-miR-150-5p | C5orf24 | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRP; LUAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.062; TCGA BRCA -0.057; TCGA CESC -0.074; TCGA COAD -0.08; TCGA ESCA -0.068; TCGA KIRP -0.053; TCGA LUAD -0.075; TCGA SARC -0.144; TCGA STAD -0.139; TCGA UCEC -0.093 |
hsa-miR-150-5p | ZNF623 | 12 cancers: BLCA; CESC; COAD; ESCA; KIRP; LGG; LIHC; LUAD; LUSC; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.058; TCGA CESC -0.058; TCGA COAD -0.066; TCGA ESCA -0.086; TCGA KIRP -0.088; TCGA LGG -0.054; TCGA LIHC -0.069; TCGA LUAD -0.099; TCGA LUSC -0.167; TCGA SARC -0.141; TCGA STAD -0.094; TCGA UCEC -0.107 |
hsa-miR-150-5p | SS18L1 | 14 cancers: BLCA; COAD; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; OV; PAAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.088; TCGA COAD -0.071; TCGA KIRC -0.148; TCGA KIRP -0.108; TCGA LGG -0.096; TCGA LIHC -0.149; TCGA LUAD -0.142; TCGA LUSC -0.125; TCGA OV -0.072; TCGA PAAD -0.109; TCGA SARC -0.099; TCGA THCA -0.053; TCGA STAD -0.168; TCGA UCEC -0.053 |
hsa-miR-150-5p | PCGF3 | 9 cancers: BLCA; BRCA; KIRP; LGG; LUAD; LUSC; OV; PAAD; SARC | mirMAP | TCGA BLCA -0.107; TCGA BRCA -0.064; TCGA KIRP -0.098; TCGA LGG -0.101; TCGA LUAD -0.075; TCGA LUSC -0.061; TCGA OV -0.053; TCGA PAAD -0.076; TCGA SARC -0.053 |
hsa-miR-150-5p | MAPT | 9 cancers: BLCA; BRCA; LGG; LIHC; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.179; TCGA BRCA -0.255; TCGA LGG -0.133; TCGA LIHC -0.18; TCGA PRAD -0.141; TCGA SARC -0.322; TCGA THCA -0.177; TCGA STAD -0.199; TCGA UCEC -0.113 |
hsa-miR-150-5p | TMEM201 | 9 cancers: BLCA; ESCA; KIRC; LIHC; LUAD; LUSC; OV; THCA; STAD | mirMAP | TCGA BLCA -0.06; TCGA ESCA -0.086; TCGA KIRC -0.123; TCGA LIHC -0.075; TCGA LUAD -0.06; TCGA LUSC -0.16; TCGA OV -0.056; TCGA THCA -0.067; TCGA STAD -0.061 |
hsa-miR-150-5p | C3orf18 | 10 cancers: BLCA; BRCA; CESC; KIRC; LIHC; LUAD; OV; SARC; THCA; UCEC | miRNATAP | TCGA BLCA -0.058; TCGA BRCA -0.127; TCGA CESC -0.166; TCGA KIRC -0.288; TCGA LIHC -0.133; TCGA LUAD -0.071; TCGA OV -0.101; TCGA SARC -0.089; TCGA THCA -0.086; TCGA UCEC -0.113 |
hsa-miR-150-5p | EDA | 9 cancers: BLCA; CESC; HNSC; KIRC; PRAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.157; TCGA CESC -0.149; TCGA HNSC -0.091; TCGA KIRC -0.152; TCGA PRAD -0.128; TCGA SARC -0.119; TCGA THCA -0.179; TCGA STAD -0.157; TCGA UCEC -0.19 |
hsa-miR-150-5p | NLGN2 | 9 cancers: BLCA; CESC; ESCA; HNSC; KIRP; LGG; LUSC; SARC; UCEC | miRNATAP | TCGA BLCA -0.083; TCGA CESC -0.081; TCGA ESCA -0.201; TCGA HNSC -0.101; TCGA KIRP -0.094; TCGA LGG -0.178; TCGA LUSC -0.151; TCGA SARC -0.156; TCGA UCEC -0.163 |
hsa-miR-150-5p | PFN2 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LUAD; LUSC; OV; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.106; TCGA BRCA -0.052; TCGA CESC -0.324; TCGA COAD -0.152; TCGA ESCA -0.287; TCGA HNSC -0.207; TCGA KIRC -0.293; TCGA LGG -0.103; TCGA LUAD -0.27; TCGA LUSC -0.389; TCGA OV -0.121; TCGA SARC -0.157; TCGA THCA -0.082; TCGA STAD -0.284; TCGA UCEC -0.104 |
hsa-miR-150-5p | GIGYF1 | 9 cancers: BLCA; KIRP; LGG; LIHC; LUSC; OV; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.051; TCGA KIRP -0.102; TCGA LGG -0.145; TCGA LIHC -0.072; TCGA LUSC -0.087; TCGA OV -0.053; TCGA THCA -0.067; TCGA STAD -0.069; TCGA UCEC -0.058 |
hsa-miR-150-5p | TCF12 | 9 cancers: BLCA; CESC; KIRP; LGG; LUAD; OV; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.064; TCGA CESC -0.104; TCGA KIRP -0.06; TCGA LGG -0.331; TCGA LUAD -0.064; TCGA OV -0.059; TCGA SARC -0.114; TCGA STAD -0.079; TCGA UCEC -0.076 |
hsa-miR-150-5p | ACVR1B | 9 cancers: BLCA; BRCA; COAD; KIRC; KIRP; LUAD; LUSC; OV; THCA | miRNATAP | TCGA BLCA -0.11; TCGA BRCA -0.064; TCGA COAD -0.076; TCGA KIRC -0.234; TCGA KIRP -0.072; TCGA LUAD -0.077; TCGA LUSC -0.08; TCGA OV -0.087; TCGA THCA -0.081 |
hsa-miR-150-5p | CLDN12 | 14 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; OV; PAAD; SARC; STAD | miRNATAP | TCGA BLCA -0.057; TCGA BRCA -0.098; TCGA CESC -0.073; TCGA COAD -0.076; TCGA HNSC -0.075; TCGA KIRC -0.087; TCGA KIRP -0.067; TCGA LIHC -0.054; TCGA LUAD -0.096; TCGA LUSC -0.155; TCGA OV -0.072; TCGA PAAD -0.207; TCGA SARC -0.134; TCGA STAD -0.167 |
hsa-miR-150-5p | SHISA4 | 11 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LIHC; LUAD; PRAD; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.101; TCGA BRCA -0.15; TCGA CESC -0.231; TCGA ESCA -0.226; TCGA HNSC -0.169; TCGA LIHC -0.25; TCGA LUAD -0.127; TCGA PRAD -0.076; TCGA THCA -0.058; TCGA STAD -0.101; TCGA UCEC -0.172 |
hsa-miR-150-5p | ZFYVE21 | 9 cancers: BLCA; ESCA; HNSC; KIRC; KIRP; LUSC; PAAD; PRAD; STAD | miRNATAP | TCGA BLCA -0.057; TCGA ESCA -0.077; TCGA HNSC -0.072; TCGA KIRC -0.112; TCGA KIRP -0.057; TCGA LUSC -0.105; TCGA PAAD -0.081; TCGA PRAD -0.064; TCGA STAD -0.085 |
hsa-miR-150-5p | CACNB3 | 11 cancers: BLCA; BRCA; CESC; KIRP; LUAD; LUSC; PAAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.138; TCGA BRCA -0.134; TCGA CESC -0.053; TCGA KIRP -0.099; TCGA LUAD -0.156; TCGA LUSC -0.088; TCGA PAAD -0.123; TCGA SARC -0.218; TCGA THCA -0.073; TCGA STAD -0.108; TCGA UCEC -0.08 |
hsa-miR-150-5p | BANF1 | 9 cancers: BRCA; COAD; ESCA; HNSC; LGG; LUAD; LUSC; PAAD; PRAD | MirTarget | TCGA BRCA -0.059; TCGA COAD -0.06; TCGA ESCA -0.096; TCGA HNSC -0.075; TCGA LGG -0.063; TCGA LUAD -0.097; TCGA LUSC -0.109; TCGA PAAD -0.063; TCGA PRAD -0.076 |
hsa-miR-150-5p | HN1L | 9 cancers: BRCA; COAD; ESCA; KIRP; LIHC; LUAD; LUSC; PAAD; SARC | MirTarget | TCGA BRCA -0.103; TCGA COAD -0.057; TCGA ESCA -0.094; TCGA KIRP -0.091; TCGA LIHC -0.109; TCGA LUAD -0.117; TCGA LUSC -0.132; TCGA PAAD -0.101; TCGA SARC -0.137 |
hsa-miR-150-5p | UBFD1 | 11 cancers: BRCA; CESC; LGG; LIHC; LUAD; LUSC; OV; PAAD; SARC; THCA; UCEC | MirTarget; miRNATAP | TCGA BRCA -0.07; TCGA CESC -0.057; TCGA LGG -0.056; TCGA LIHC -0.079; TCGA LUAD -0.095; TCGA LUSC -0.073; TCGA OV -0.051; TCGA PAAD -0.053; TCGA SARC -0.069; TCGA THCA -0.051; TCGA UCEC -0.07 |
hsa-miR-150-5p | NUBPL | 14 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUSC; OV; PAAD; SARC; STAD; UCEC | MirTarget | TCGA BRCA -0.065; TCGA CESC -0.078; TCGA COAD -0.087; TCGA ESCA -0.117; TCGA HNSC -0.067; TCGA KIRC -0.199; TCGA KIRP -0.066; TCGA LIHC -0.137; TCGA LUSC -0.094; TCGA OV -0.056; TCGA PAAD -0.065; TCGA SARC -0.079; TCGA STAD -0.145; TCGA UCEC -0.083 |
hsa-miR-150-5p | TUFT1 | 11 cancers: BRCA; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; PAAD; STAD; UCEC | MirTarget | TCGA BRCA -0.087; TCGA COAD -0.073; TCGA ESCA -0.157; TCGA HNSC -0.056; TCGA KIRC -0.213; TCGA LIHC -0.076; TCGA LUAD -0.177; TCGA LUSC -0.228; TCGA PAAD -0.171; TCGA STAD -0.128; TCGA UCEC -0.114 |
hsa-miR-150-5p | OTUD7B | 11 cancers: BRCA; CESC; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; SARC; STAD; UCEC | MirTarget | TCGA BRCA -0.096; TCGA CESC -0.061; TCGA HNSC -0.061; TCGA KIRC -0.056; TCGA KIRP -0.086; TCGA LIHC -0.062; TCGA LUAD -0.123; TCGA LUSC -0.098; TCGA SARC -0.149; TCGA STAD -0.109; TCGA UCEC -0.057 |
hsa-miR-150-5p | FAM84B | 9 cancers: BRCA; CESC; COAD; KIRC; KIRP; LGG; LUAD; PAAD; UCEC | MirTarget | TCGA BRCA -0.078; TCGA CESC -0.077; TCGA COAD -0.094; TCGA KIRC -0.228; TCGA KIRP -0.115; TCGA LGG -0.288; TCGA LUAD -0.07; TCGA PAAD -0.099; TCGA UCEC -0.129 |
hsa-miR-150-5p | MTA3 | 15 cancers: BRCA; CESC; COAD; HNSC; KIRC; LIHC; LUAD; LUSC; OV; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BRCA -0.077; TCGA CESC -0.081; TCGA COAD -0.058; TCGA HNSC -0.058; TCGA KIRC -0.08; TCGA LIHC -0.055; TCGA LUAD -0.127; TCGA LUSC -0.073; TCGA OV -0.078; TCGA PAAD -0.05; TCGA PRAD -0.062; TCGA SARC -0.131; TCGA THCA -0.057; TCGA STAD -0.088; TCGA UCEC -0.056 |
hsa-miR-150-5p | CAMK2B | 9 cancers: BRCA; KIRC; KIRP; LUSC; OV; PRAD; SARC; THCA; UCEC | mirMAP | TCGA BRCA -0.198; TCGA KIRC -0.298; TCGA KIRP -0.295; TCGA LUSC -0.239; TCGA OV -0.263; TCGA PRAD -0.156; TCGA SARC -0.231; TCGA THCA -0.165; TCGA UCEC -0.264 |
hsa-miR-150-5p | CLUAP1 | 11 cancers: BRCA; ESCA; KIRC; KIRP; LIHC; OV; PAAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BRCA -0.077; TCGA ESCA -0.117; TCGA KIRC -0.074; TCGA KIRP -0.116; TCGA LIHC -0.096; TCGA OV -0.073; TCGA PAAD -0.087; TCGA SARC -0.081; TCGA THCA -0.055; TCGA STAD -0.064; TCGA UCEC -0.054 |
hsa-miR-150-5p | HDAC5 | 9 cancers: BRCA; LGG; LIHC; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BRCA -0.052; TCGA LGG -0.07; TCGA LIHC -0.051; TCGA PAAD -0.084; TCGA PRAD -0.054; TCGA SARC -0.096; TCGA THCA -0.057; TCGA STAD -0.08; TCGA UCEC -0.063 |
hsa-miR-150-5p | MPP2 | 9 cancers: BRCA; CESC; KIRC; KIRP; LUSC; SARC; THCA; STAD; UCEC | mirMAP | TCGA BRCA -0.171; TCGA CESC -0.128; TCGA KIRC -0.36; TCGA KIRP -0.173; TCGA LUSC -0.158; TCGA SARC -0.372; TCGA THCA -0.16; TCGA STAD -0.159; TCGA UCEC -0.271 |
hsa-miR-150-5p | HS1BP3 | 9 cancers: BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; PRAD; STAD | mirMAP | TCGA BRCA -0.07; TCGA CESC -0.082; TCGA COAD -0.054; TCGA ESCA -0.099; TCGA HNSC -0.105; TCGA LIHC -0.156; TCGA LUAD -0.051; TCGA PRAD -0.056; TCGA STAD -0.068 |
hsa-miR-150-5p | VAPB | 10 cancers: BRCA; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; PAAD; STAD | mirMAP | TCGA BRCA -0.057; TCGA COAD -0.068; TCGA ESCA -0.07; TCGA HNSC -0.056; TCGA KIRC -0.112; TCGA LIHC -0.077; TCGA LUAD -0.124; TCGA LUSC -0.108; TCGA PAAD -0.057; TCGA STAD -0.097 |
hsa-miR-150-5p | KCTD15 | 10 cancers: BRCA; ESCA; LGG; LUSC; OV; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BRCA -0.074; TCGA ESCA -0.218; TCGA LGG -0.192; TCGA LUSC -0.241; TCGA OV -0.092; TCGA PRAD -0.052; TCGA SARC -0.293; TCGA THCA -0.108; TCGA STAD -0.142; TCGA UCEC -0.101 |
hsa-miR-150-5p | ZNF497 | 10 cancers: BRCA; KIRC; KIRP; LGG; LIHC; OV; PAAD; PRAD; SARC; THCA | mirMAP | TCGA BRCA -0.161; TCGA KIRC -0.116; TCGA KIRP -0.154; TCGA LGG -0.095; TCGA LIHC -0.113; TCGA OV -0.11; TCGA PAAD -0.117; TCGA PRAD -0.065; TCGA SARC -0.107; TCGA THCA -0.119 |
hsa-miR-150-5p | CCDC85C | 11 cancers: BRCA; KIRC; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD | mirMAP | TCGA BRCA -0.063; TCGA KIRC -0.158; TCGA LGG -0.106; TCGA LIHC -0.071; TCGA LUAD -0.096; TCGA LUSC -0.152; TCGA PAAD -0.141; TCGA PRAD -0.112; TCGA SARC -0.163; TCGA THCA -0.119; TCGA STAD -0.087 |
hsa-miR-150-5p | H1F0 | 13 cancers: BRCA; COAD; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; SARC; STAD; UCEC | miRNATAP | TCGA BRCA -0.15; TCGA COAD -0.091; TCGA HNSC -0.119; TCGA KIRC -0.339; TCGA KIRP -0.08; TCGA LGG -0.182; TCGA LIHC -0.079; TCGA LUAD -0.176; TCGA LUSC -0.285; TCGA PAAD -0.217; TCGA SARC -0.106; TCGA STAD -0.127; TCGA UCEC -0.09 |
hsa-miR-150-5p | MIB1 | 10 cancers: CESC; ESCA; KIRC; KIRP; LGG; LUAD; LUSC; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA CESC -0.077; TCGA ESCA -0.108; TCGA KIRC -0.113; TCGA KIRP -0.085; TCGA LGG -0.124; TCGA LUAD -0.1; TCGA LUSC -0.12; TCGA SARC -0.155; TCGA STAD -0.181; TCGA UCEC -0.119 |
hsa-miR-150-5p | ATRN | 12 cancers: CESC; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; OV; PAAD; THCA; UCEC | mirMAP | TCGA CESC -0.087; TCGA ESCA -0.089; TCGA HNSC -0.113; TCGA KIRC -0.18; TCGA KIRP -0.068; TCGA LIHC -0.06; TCGA LUAD -0.088; TCGA LUSC -0.215; TCGA OV -0.071; TCGA PAAD -0.062; TCGA THCA -0.072; TCGA UCEC -0.093 |
hsa-miR-150-5p | NAA30 | 10 cancers: CESC; COAD; ESCA; KIRC; KIRP; LUAD; LUSC; OV; STAD; UCEC | mirMAP | TCGA CESC -0.063; TCGA COAD -0.063; TCGA ESCA -0.058; TCGA KIRC -0.114; TCGA KIRP -0.079; TCGA LUAD -0.097; TCGA LUSC -0.105; TCGA OV -0.068; TCGA STAD -0.1; TCGA UCEC -0.091 |
hsa-miR-150-5p | IL17RD | 10 cancers: CESC; KIRC; KIRP; LGG; LUAD; LUSC; OV; SARC; STAD; UCEC | mirMAP | TCGA CESC -0.228; TCGA KIRC -0.245; TCGA KIRP -0.176; TCGA LGG -0.182; TCGA LUAD -0.114; TCGA LUSC -0.13; TCGA OV -0.171; TCGA SARC -0.325; TCGA STAD -0.308; TCGA UCEC -0.23 |
hsa-miR-150-5p | C16orf87 | 9 cancers: CESC; COAD; ESCA; HNSC; LGG; LUAD; LUSC; PRAD; STAD | mirMAP | TCGA CESC -0.082; TCGA COAD -0.054; TCGA ESCA -0.083; TCGA HNSC -0.093; TCGA LGG -0.173; TCGA LUAD -0.095; TCGA LUSC -0.123; TCGA PRAD -0.06; TCGA STAD -0.072 |
hsa-miR-150-5p | ZNF618 | 9 cancers: CESC; KIRC; KIRP; LUSC; OV; PAAD; SARC; THCA; UCEC | mirMAP | TCGA CESC -0.089; TCGA KIRC -0.127; TCGA KIRP -0.095; TCGA LUSC -0.069; TCGA OV -0.069; TCGA PAAD -0.068; TCGA SARC -0.089; TCGA THCA -0.058; TCGA UCEC -0.054 |
hsa-miR-150-5p | METTL2B | 9 cancers: CESC; KIRP; LGG; LIHC; LUAD; LUSC; OV; SARC; STAD | mirMAP | TCGA CESC -0.065; TCGA KIRP -0.083; TCGA LGG -0.06; TCGA LIHC -0.095; TCGA LUAD -0.096; TCGA LUSC -0.102; TCGA OV -0.053; TCGA SARC -0.064; TCGA STAD -0.069 |
hsa-miR-150-5p | QSOX2 | 9 cancers: CESC; ESCA; HNSC; LGG; LIHC; LUSC; OV; THCA; UCEC | mirMAP | TCGA CESC -0.093; TCGA ESCA -0.077; TCGA HNSC -0.077; TCGA LGG -0.139; TCGA LIHC -0.062; TCGA LUSC -0.149; TCGA OV -0.111; TCGA THCA -0.082; TCGA UCEC -0.134 |
hsa-miR-150-5p | PALM2 | 9 cancers: CESC; ESCA; HNSC; KIRP; LUAD; LUSC; OV; PAAD; SARC | mirMAP | TCGA CESC -0.247; TCGA ESCA -0.308; TCGA HNSC -0.146; TCGA KIRP -0.16; TCGA LUAD -0.147; TCGA LUSC -0.175; TCGA OV -0.123; TCGA PAAD -0.139; TCGA SARC -0.164 |
hsa-miR-150-5p | TADA1 | 10 cancers: COAD; KIRC; KIRP; LIHC; LUAD; LUSC; OV; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA COAD -0.08; TCGA KIRC -0.053; TCGA KIRP -0.057; TCGA LIHC -0.091; TCGA LUAD -0.115; TCGA LUSC -0.101; TCGA OV -0.087; TCGA SARC -0.126; TCGA STAD -0.064; TCGA UCEC -0.113 |
hsa-miR-150-5p | RBBP9 | 9 cancers: COAD; KIRC; KIRP; LGG; LIHC; OV; THCA; STAD; UCEC | mirMAP | TCGA COAD -0.063; TCGA KIRC -0.054; TCGA KIRP -0.054; TCGA LGG -0.091; TCGA LIHC -0.064; TCGA OV -0.062; TCGA THCA -0.052; TCGA STAD -0.128; TCGA UCEC -0.097 |
hsa-miR-150-5p | STAMBP | 9 cancers: COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; SARC; UCEC | mirMAP | TCGA COAD -0.109; TCGA ESCA -0.055; TCGA HNSC -0.054; TCGA LGG -0.07; TCGA LIHC -0.063; TCGA LUAD -0.085; TCGA LUSC -0.124; TCGA SARC -0.069; TCGA UCEC -0.055 |
hsa-miR-150-5p | PURB | 9 cancers: COAD; ESCA; KIRP; LIHC; LUAD; LUSC; SARC; STAD; UCEC | miRNATAP | TCGA COAD -0.059; TCGA ESCA -0.051; TCGA KIRP -0.113; TCGA LIHC -0.056; TCGA LUAD -0.068; TCGA LUSC -0.125; TCGA SARC -0.095; TCGA STAD -0.151; TCGA UCEC -0.106 |
hsa-miR-150-5p | GSK3B | 9 cancers: COAD; ESCA; KIRP; LIHC; LUAD; LUSC; SARC; STAD; UCEC | miRNATAP | TCGA COAD -0.059; TCGA ESCA -0.088; TCGA KIRP -0.067; TCGA LIHC -0.066; TCGA LUAD -0.066; TCGA LUSC -0.112; TCGA SARC -0.074; TCGA STAD -0.06; TCGA UCEC -0.065 |
hsa-miR-150-5p | ZNF326 | 9 cancers: COAD; KIRC; KIRP; LGG; LUAD; LUSC; SARC; STAD; UCEC | miRNATAP | TCGA COAD -0.058; TCGA KIRC -0.065; TCGA KIRP -0.086; TCGA LGG -0.079; TCGA LUAD -0.071; TCGA LUSC -0.141; TCGA SARC -0.052; TCGA STAD -0.106; TCGA UCEC -0.071 |
hsa-miR-150-5p | SUMO1 | 9 cancers: COAD; HNSC; KIRC; LIHC; LUAD; LUSC; PRAD; STAD; UCEC | miRNATAP | TCGA COAD -0.067; TCGA HNSC -0.05; TCGA KIRC -0.051; TCGA LIHC -0.071; TCGA LUAD -0.081; TCGA LUSC -0.081; TCGA PRAD -0.058; TCGA STAD -0.07; TCGA UCEC -0.088 |
hsa-miR-150-5p | CBX5 | 9 cancers: ESCA; KIRC; KIRP; LGG; LUAD; OV; SARC; THCA; UCEC | mirMAP | TCGA ESCA -0.075; TCGA KIRC -0.072; TCGA KIRP -0.083; TCGA LGG -0.074; TCGA LUAD -0.08; TCGA OV -0.07; TCGA SARC -0.15; TCGA THCA -0.058; TCGA UCEC -0.084 |
hsa-miR-150-5p | SH2D5 | 9 cancers: ESCA; HNSC; LIHC; LUSC; OV; PAAD; SARC; STAD; UCEC | mirMAP | TCGA ESCA -0.562; TCGA HNSC -0.252; TCGA LIHC -0.148; TCGA LUSC -0.802; TCGA OV -0.137; TCGA PAAD -0.281; TCGA SARC -0.219; TCGA STAD -0.374; TCGA UCEC -0.234 |
hsa-miR-150-5p | UBE2E3 | 10 cancers: ESCA; HNSC; KIRC; LGG; LIHC; LUAD; LUSC; OV; PRAD; UCEC | miRNATAP | TCGA ESCA -0.063; TCGA HNSC -0.097; TCGA KIRC -0.075; TCGA LGG -0.074; TCGA LIHC -0.062; TCGA LUAD -0.054; TCGA LUSC -0.137; TCGA OV -0.084; TCGA PRAD -0.114; TCGA UCEC -0.05 |
hsa-miR-150-5p | POGK | 9 cancers: KIRP; LGG; LIHC; LUAD; LUSC; OV; SARC; STAD; UCEC | MirTarget | TCGA KIRP -0.062; TCGA LGG -0.066; TCGA LIHC -0.055; TCGA LUAD -0.117; TCGA LUSC -0.091; TCGA OV -0.072; TCGA SARC -0.07; TCGA STAD -0.069; TCGA UCEC -0.103 |
Enriched cancer pathways of putative targets