microRNA information: hsa-miR-15b-3p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-15b-3p | miRbase |
| Accession: | MIMAT0004586 | miRbase |
| Precursor name: | hsa-mir-15b | miRbase |
| Precursor accession: | MI0000438 | miRbase |
| Symbol: | MIR15B | HGNC |
| RefSeq ID: | NR_029663 | GenBank |
| Sequence: | CGAAUCAUUAUUUGCUGCUCUA |
Reported expression in cancers: hsa-miR-15b-3p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-15b-3p | cervical and endocervical cancer | upregulation | "Vote-counting analysis showed that up-regulation w ......" | 25920605 | |
| hsa-miR-15b-3p | colon cancer | downregulation | "miR-15 microRNA 15 and miR-16 are frequently delet ......" | 22574716 | |
| hsa-miR-15b-3p | colorectal cancer | downregulation | "The in vivo significance of expression of miR-16/1 ......" | 21390519 | qPCR |
| hsa-miR-15b-3p | colorectal cancer | upregulation | "Similarly to miR-195 the members of the same miR f ......" | 22710713 | |
| hsa-miR-15b-3p | colorectal cancer | upregulation | "Quantitative reverse-transcription PCR was used to ......" | 23267864 | Reverse transcription PCR |
| hsa-miR-15b-3p | colorectal cancer | downregulation | "Expression of miR-15b and miR-135b was significant ......" | 25557290 | |
| hsa-miR-15b-3p | endometrial cancer | deregulation | "Compared with the miRNAs in the control endometriu ......" | 22904162 | |
| hsa-miR-15b-3p | esophageal cancer | upregulation | "This study was designed to detect novel microRNAs ......" | 25117812 | Microarray |
| hsa-miR-15b-3p | gastric cancer | downregulation | "Among the downregulated microRNAs are miR-15b and ......" | 18449891 | qPCR |
| hsa-miR-15b-3p | liver cancer | downregulation | "In this study we experimentally demonstrated throu ......" | 23487264 | |
| hsa-miR-15b-3p | lymphoma | upregulation | "Identification of miR 15b as a transformation rela ......" | 26676972 | Microarray; in situ hybridization |
| hsa-miR-15b-3p | melanoma | deregulation | "MicroRNA 15b represents an independent prognostic ......" | 19830692 | qPCR |
| hsa-miR-15b-3p | pancreatic cancer | upregulation | "Profiling of 95 microRNAs in pancreatic cancer cel ......" | 19030927 | qPCR |
| hsa-miR-15b-3p | pancreatic cancer | downregulation | "Moreover plasmatic miR-15b down-regulation was det ......" | 24892299 | |
| hsa-miR-15b-3p | prostate cancer | downregulation | "We demonstrated that miR-15 and miR-16 are downreg ......" | 21532615 |
Reported cancer pathway affected by hsa-miR-15b-3p
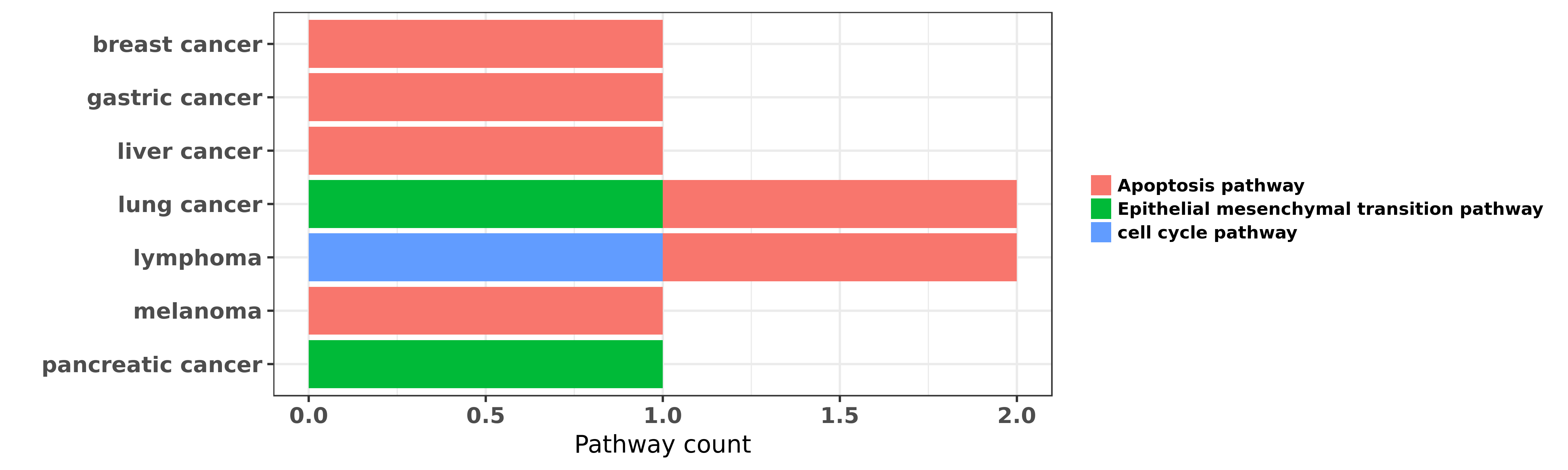
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-15b-3p | breast cancer | Apoptosis pathway | "The miR 15 family enhances the radiosensitivity of ......" | 25594541 | Western blot; Flow cytometry; Luciferase |
| hsa-miR-15b-3p | breast cancer | Apoptosis pathway | "We conclude that combination of Etoposide or C-10 ......" | 26551008 | |
| hsa-miR-15b-3p | gastric cancer | Apoptosis pathway | "miR 15b and miR 16 modulate multidrug resistance b ......" | 18449891 | Luciferase |
| hsa-miR-15b-3p | liver cancer | Apoptosis pathway | "High expression of microRNA 15b predicts a low ris ......" | 19956871 | |
| hsa-miR-15b-3p | liver cancer | Apoptosis pathway | "miR 15b 5p induces endoplasmic reticulum stress an ......" | 26023735 | |
| hsa-miR-15b-3p | lung cancer | Apoptosis pathway | "Dynamically expressed microRNA 15b modulates the a ......" | 23517578 | Flow cytometry |
| hsa-miR-15b-3p | lung cancer | Epithelial mesenchymal transition pathway | "miR 15b regulates cisplatin resistance and metasta ......" | 25721211 | |
| hsa-miR-15b-3p | lung cancer | Apoptosis pathway | "Specifically following HDAC inhibition MYC which n ......" | 26915294 | |
| hsa-miR-15b-3p | lymphoma | cell cycle pathway; Apoptosis pathway | "Identification of miR 15b as a transformation rela ......" | 26676972 | |
| hsa-miR-15b-3p | melanoma | Apoptosis pathway | "MicroRNA 15b represents an independent prognostic ......" | 19830692 | |
| hsa-miR-15b-3p | pancreatic cancer | Epithelial mesenchymal transition pathway | "miR 15b promotes epithelial mesenchymal transition ......" | 26166038 |
Reported cancer prognosis affected by hsa-miR-15b-3p
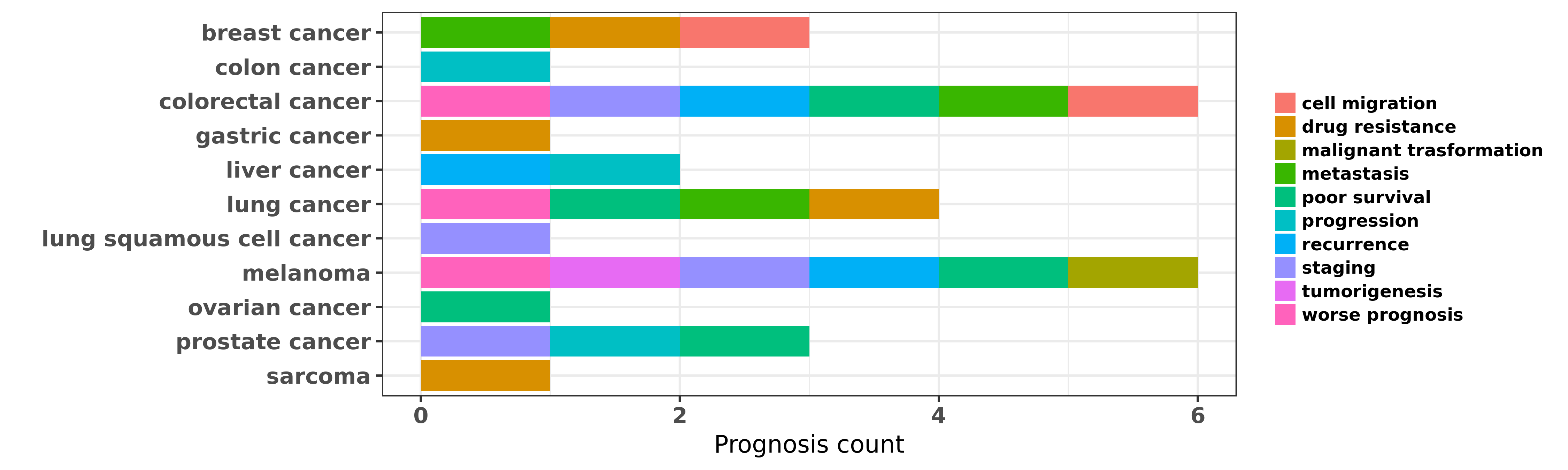
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-15b-3p | breast cancer | drug resistance | "The miR 15 family enhances the radiosensitivity of ......" | 25594541 | Western blot; Flow cytometry; Luciferase |
| hsa-miR-15b-3p | breast cancer | cell migration; metastasis | "EGF induces microRNAs that target suppressors of c ......" | 25783158 | |
| hsa-miR-15b-3p | colon cancer | progression | "miR-15 microRNA 15 and miR-16 are frequently delet ......" | 22574716 | Colony formation; Western blot |
| hsa-miR-15b-3p | colorectal cancer | staging | "A panel of 8 plasma miRNAs miR-532-3p miR-331 miR- ......" | 24022433 | |
| hsa-miR-15b-3p | colorectal cancer | metastasis | "Expression of miR-15b and miR-135b was significant ......" | 25557290 | |
| hsa-miR-15b-3p | colorectal cancer | metastasis; cell migration; recurrence; poor survival; worse prognosis | "Inhibition of miR 15b decreases cell migration and ......" | 26743779 | Western blot; Colony formation |
| hsa-miR-15b-3p | gastric cancer | drug resistance | "miR 15b and miR 16 modulate multidrug resistance b ......" | 18449891 | Luciferase |
| hsa-miR-15b-3p | liver cancer | recurrence | "High expression of microRNA 15b predicts a low ris ......" | 19956871 | |
| hsa-miR-15b-3p | liver cancer | recurrence | "miR 15b 5p induces endoplasmic reticulum stress an ......" | 26023735 | |
| hsa-miR-15b-3p | liver cancer | progression | "A panel of ten cancer-associated miRNAs was analyz ......" | 26264553 | |
| hsa-miR-15b-3p | lung cancer | metastasis; drug resistance; worse prognosis | "miR 15b regulates cisplatin resistance and metasta ......" | 25721211 | |
| hsa-miR-15b-3p | lung cancer | poor survival | "Specifically following HDAC inhibition MYC which n ......" | 26915294 | |
| hsa-miR-15b-3p | lung cancer | poor survival | "Tissues were microdissected to obtain >90% tumor o ......" | 27081085 | |
| hsa-miR-15b-3p | lung squamous cell cancer | staging | "However the study demonstrated a correlation betwe ......" | 25384507 | |
| hsa-miR-15b-3p | melanoma | malignant trasformation; poor survival; recurrence; tumorigenesis; worse prognosis | "MicroRNA 15b represents an independent prognostic ......" | 19830692 | |
| hsa-miR-15b-3p | melanoma | staging; poor survival; recurrence | "We recently reported our array-based discovery of ......" | 25155861 | |
| hsa-miR-15b-3p | ovarian cancer | poor survival | "WNT7A Regulation by miR 15b in Ovarian Cancer; In ......" | 27195958 | Luciferase |
| hsa-miR-15b-3p | prostate cancer | progression; poor survival; staging | "We demonstrated that miR-15 and miR-16 are downreg ......" | 21532615 | |
| hsa-miR-15b-3p | sarcoma | drug resistance | "In addition higher expression of miR-451 and miR-1 ......" | 22350417 |
Reported gene related to hsa-miR-15b-3p
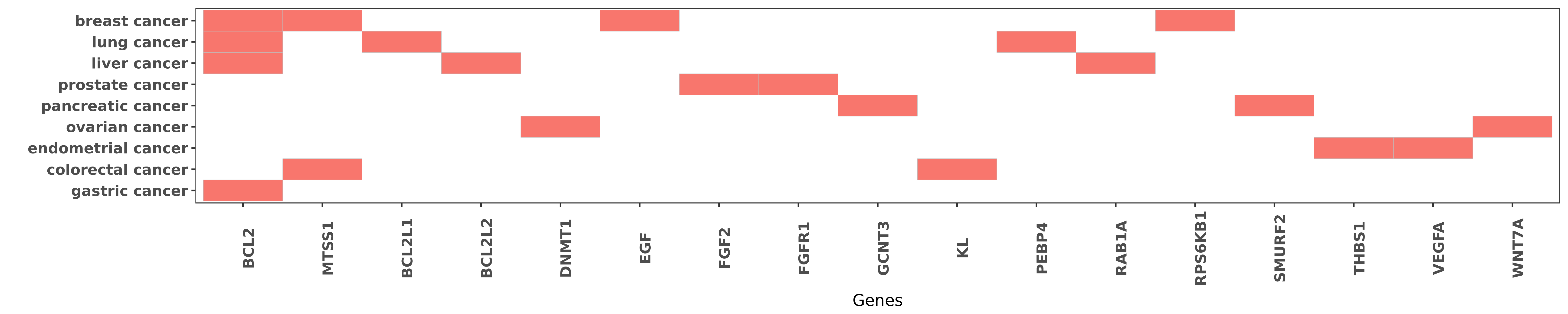
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-15b-3p | breast cancer | BCL2 | "MicroRNAs miRNAs encoded by the miR-15 cluster are ......" | 25594541 |
| hsa-miR-15b-3p | gastric cancer | BCL2 | "miR 15b and miR 16 modulate multidrug resistance b ......" | 18449891 |
| hsa-miR-15b-3p | liver cancer | BCL2 | "MiR 15b mediates liver cancer cells proliferation ......" | 26884837 |
| hsa-miR-15b-3p | lung cancer | BCL2 | "As a result transcript levels of the tumor-suppres ......" | 26915294 |
| hsa-miR-15b-3p | breast cancer | MTSS1 | "EGF induces microRNAs that target suppressors of c ......" | 25783158 |
| hsa-miR-15b-3p | colorectal cancer | MTSS1 | "High plasma miR-15b level and negative MTSS1 and K ......" | 26743779 |
| hsa-miR-15b-3p | lung cancer | BCL2L1 | "As a result transcript levels of the tumor-suppres ......" | 26915294 |
| hsa-miR-15b-3p | liver cancer | BCL2L2 | "Bcl-w was identified as a target molecule regulate ......" | 19956871 |
| hsa-miR-15b-3p | ovarian cancer | DNMT1 | "Treatment with decitabine dose-dependently increas ......" | 27195958 |
| hsa-miR-15b-3p | breast cancer | EGF | "EGF induces microRNAs that target suppressors of c ......" | 25783158 |
| hsa-miR-15b-3p | prostate cancer | FGF2 | "Such downregulation of miR-15 and miR-16 in cancer ......" | 21532615 |
| hsa-miR-15b-3p | prostate cancer | FGFR1 | "Such downregulation of miR-15 and miR-16 in cancer ......" | 21532615 |
| hsa-miR-15b-3p | pancreatic cancer | GCNT3 | "Expression of microRNA 15b and the glycosyltransfe ......" | 24892299 |
| hsa-miR-15b-3p | colorectal cancer | KL | "High plasma miR-15b level and negative MTSS1 and K ......" | 26743779 |
| hsa-miR-15b-3p | lung cancer | PEBP4 | "miR 15b regulates cisplatin resistance and metasta ......" | 25721211 |
| hsa-miR-15b-3p | liver cancer | RAB1A | "miR 15b 5p induces endoplasmic reticulum stress an ......" | 26023735 |
| hsa-miR-15b-3p | breast cancer | RPS6KB1 | "In addition the dual luciferase reporter assays in ......" | 25261849 |
| hsa-miR-15b-3p | pancreatic cancer | SMURF2 | "miR 15b promotes epithelial mesenchymal transition ......" | 26166038 |
| hsa-miR-15b-3p | endometrial cancer | THBS1 | "TaqMan qRT-PCR was used to assess the expression o ......" | 22904162 |
| hsa-miR-15b-3p | endometrial cancer | VEGFA | "TaqMan qRT-PCR was used to assess the expression o ......" | 22904162 |
| hsa-miR-15b-3p | ovarian cancer | WNT7A | "WNT7A Regulation by miR 15b in Ovarian Cancer; In ......" | 27195958 |
Expression profile in cancer corhorts:

Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-15b-3p | NAV2 | 11 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; PRAD; THCA; STAD | MirTarget | TCGA BLCA -0.175; TCGA BRCA -0.166; TCGA CESC -0.15; TCGA ESCA -0.173; TCGA HNSC -0.118; TCGA KIRC -0.361; TCGA KIRP -0.198; TCGA LGG -0.104; TCGA PRAD -0.52; TCGA THCA -0.164; TCGA STAD -0.347 |
hsa-miR-15b-3p | GPC6 | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUSC; OV; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.891; TCGA CESC -0.554; TCGA COAD -0.889; TCGA ESCA -0.609; TCGA HNSC -0.624; TCGA LUSC -0.596; TCGA OV -0.248; TCGA PRAD -0.151; TCGA STAD -0.382; TCGA UCEC -0.243 |
hsa-miR-15b-3p | NID1 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUSC; PRAD; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.33; TCGA BRCA -0.075; TCGA CESC -0.296; TCGA COAD -0.39; TCGA ESCA -0.246; TCGA HNSC -0.218; TCGA LUSC -0.187; TCGA PRAD -0.324; TCGA THCA -0.155; TCGA STAD -0.28; TCGA UCEC -0.344 |
hsa-miR-15b-3p | UNC80 | 9 cancers: BLCA; ESCA; KIRC; LGG; OV; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.338; TCGA ESCA -0.439; TCGA KIRC -0.208; TCGA LGG -0.208; TCGA OV -0.502; TCGA PAAD -1.125; TCGA PRAD -0.21; TCGA STAD -0.723; TCGA UCEC -0.374 |
hsa-miR-15b-3p | ABL1 | 9 cancers: BLCA; BRCA; CESC; ESCA; LGG; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.226; TCGA BRCA -0.051; TCGA CESC -0.13; TCGA ESCA -0.152; TCGA LGG -0.055; TCGA SARC -0.082; TCGA THCA -0.058; TCGA STAD -0.216; TCGA UCEC -0.173 |
hsa-miR-15b-3p | COL4A4 | 12 cancers: BLCA; COAD; ESCA; KIRC; KIRP; LGG; LUAD; LUSC; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.593; TCGA COAD -0.525; TCGA ESCA -0.642; TCGA KIRC -0.703; TCGA KIRP -0.321; TCGA LGG -0.092; TCGA LUAD -0.387; TCGA LUSC -0.65; TCGA PRAD -0.371; TCGA SARC -0.232; TCGA STAD -0.444; TCGA UCEC -0.385 |
hsa-miR-15b-3p | CROT | 13 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LUAD; LUSC; OV; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.112; TCGA BRCA -0.33; TCGA CESC -0.153; TCGA HNSC -0.171; TCGA KIRC -0.33; TCGA KIRP -0.089; TCGA LUAD -0.135; TCGA LUSC -0.135; TCGA OV -0.123; TCGA PRAD -0.119; TCGA SARC -0.108; TCGA STAD -0.221; TCGA UCEC -0.191 |
hsa-miR-15b-3p | CHL1 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; OV; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.675; TCGA BRCA -0.591; TCGA CESC -0.578; TCGA COAD -1.193; TCGA ESCA -0.532; TCGA HNSC -0.725; TCGA KIRC -1.212; TCGA KIRP -0.487; TCGA OV -0.328; TCGA PRAD -0.377; TCGA STAD -0.521; TCGA UCEC -0.326 |
hsa-miR-15b-3p | ST6GALNAC3 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; LUSC; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.374; TCGA BRCA -0.182; TCGA CESC -0.426; TCGA COAD -0.335; TCGA ESCA -0.39; TCGA HNSC -0.275; TCGA KIRC -0.4; TCGA LIHC -0.38; TCGA LUSC -0.393; TCGA PRAD -0.291; TCGA STAD -0.277; TCGA UCEC -0.395 |
hsa-miR-15b-3p | MFAP5 | 11 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -1.407; TCGA CESC -0.542; TCGA COAD -1.42; TCGA ESCA -1.152; TCGA HNSC -0.864; TCGA LUAD -0.326; TCGA LUSC -0.659; TCGA PRAD -0.475; TCGA SARC -0.452; TCGA STAD -0.744; TCGA UCEC -0.64 |
hsa-miR-15b-3p | SEMA3C | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; OV; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.151; TCGA BRCA -0.412; TCGA CESC -0.351; TCGA COAD -0.266; TCGA ESCA -0.361; TCGA HNSC -0.5; TCGA LUAD -0.199; TCGA LUSC -0.357; TCGA OV -0.179; TCGA PRAD -0.454; TCGA SARC -0.307; TCGA STAD -0.529; TCGA UCEC -0.342 |
hsa-miR-15b-3p | TIMP2 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.685; TCGA BRCA -0.118; TCGA CESC -0.345; TCGA COAD -0.794; TCGA ESCA -0.431; TCGA HNSC -0.481; TCGA LUAD -0.244; TCGA LUSC -0.567; TCGA PAAD -0.149; TCGA PRAD -0.303; TCGA SARC -0.088; TCGA STAD -0.409; TCGA UCEC -0.413 |
hsa-miR-15b-3p | TMEM47 | 18 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; OV; PAAD; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.377; TCGA BRCA -0.21; TCGA CESC -0.314; TCGA COAD -0.805; TCGA ESCA -0.748; TCGA HNSC -0.501; TCGA KIRC -0.157; TCGA KIRP -0.206; TCGA LGG -0.154; TCGA LIHC -0.431; TCGA LUAD -0.244; TCGA LUSC -0.549; TCGA OV -0.179; TCGA PAAD -0.326; TCGA PRAD -0.313; TCGA SARC -0.331; TCGA STAD -0.686; TCGA UCEC -0.539 |
hsa-miR-15b-3p | FAM110B | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; OV; PAAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.627; TCGA BRCA -0.18; TCGA CESC -0.468; TCGA COAD -0.804; TCGA ESCA -0.431; TCGA HNSC -0.34; TCGA KIRC -0.308; TCGA KIRP -0.191; TCGA LGG -0.35; TCGA LUAD -0.136; TCGA OV -0.173; TCGA PAAD -0.284; TCGA THCA -0.143; TCGA STAD -0.45; TCGA UCEC -0.33 |
hsa-miR-15b-3p | MACROD2 | 12 cancers: BLCA; BRCA; COAD; ESCA; KIRC; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.215; TCGA BRCA -0.223; TCGA COAD -0.379; TCGA ESCA -0.497; TCGA KIRC -0.228; TCGA LUAD -0.281; TCGA LUSC -0.744; TCGA OV -0.303; TCGA PAAD -0.255; TCGA PRAD -0.316; TCGA STAD -0.365; TCGA UCEC -0.175 |
hsa-miR-15b-3p | COL4A3BP | 12 cancers: BLCA; BRCA; CESC; ESCA; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.078; TCGA BRCA -0.143; TCGA CESC -0.085; TCGA ESCA -0.15; TCGA LIHC -0.157; TCGA LUAD -0.109; TCGA LUSC -0.173; TCGA PAAD -0.133; TCGA PRAD -0.131; TCGA SARC -0.152; TCGA STAD -0.157; TCGA UCEC -0.089 |
hsa-miR-15b-3p | MAN1A1 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.248; TCGA BRCA -0.133; TCGA CESC -0.42; TCGA COAD -0.158; TCGA ESCA -0.334; TCGA HNSC -0.463; TCGA KIRC -0.356; TCGA LUSC -0.456; TCGA PAAD -0.267; TCGA PRAD -0.351; TCGA SARC -0.175; TCGA STAD -0.105; TCGA UCEC -0.394 |
hsa-miR-15b-3p | PABPC5 | 13 cancers: BLCA; BRCA; COAD; ESCA; KIRC; LGG; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.735; TCGA BRCA -0.274; TCGA COAD -0.8; TCGA ESCA -0.639; TCGA KIRC -0.259; TCGA LGG -0.339; TCGA LUAD -0.224; TCGA LUSC -0.581; TCGA OV -0.357; TCGA PAAD -0.464; TCGA PRAD -0.332; TCGA STAD -0.758; TCGA UCEC -0.824 |
hsa-miR-15b-3p | NEK7 | 11 cancers: BLCA; COAD; ESCA; LGG; LUAD; LUSC; OV; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.106; TCGA COAD -0.162; TCGA ESCA -0.112; TCGA LGG -0.077; TCGA LUAD -0.099; TCGA LUSC -0.208; TCGA OV -0.12; TCGA PRAD -0.12; TCGA SARC -0.117; TCGA STAD -0.194; TCGA UCEC -0.17 |
hsa-miR-15b-3p | TMEM200B | 9 cancers: BLCA; BRCA; COAD; ESCA; LUAD; LUSC; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.38; TCGA BRCA -0.214; TCGA COAD -1.037; TCGA ESCA -0.265; TCGA LUAD -0.271; TCGA LUSC -0.353; TCGA PRAD -0.592; TCGA STAD -0.135; TCGA UCEC -0.914 |
hsa-miR-15b-3p | NALCN | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LUAD; LUSC; PAAD; STAD; UCEC | MirTarget | TCGA BLCA -0.602; TCGA BRCA -0.327; TCGA CESC -0.5; TCGA COAD -1.502; TCGA ESCA -1.013; TCGA HNSC -0.576; TCGA LGG -0.221; TCGA LUAD -0.538; TCGA LUSC -0.696; TCGA PAAD -0.709; TCGA STAD -0.639; TCGA UCEC -0.437 |
hsa-miR-15b-3p | CWF19L2 | 9 cancers: BLCA; ESCA; KIRC; KIRP; LGG; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.067; TCGA ESCA -0.188; TCGA KIRC -0.167; TCGA KIRP -0.085; TCGA LGG -0.076; TCGA PRAD -0.099; TCGA SARC -0.12; TCGA STAD -0.241; TCGA UCEC -0.153 |
hsa-miR-15b-3p | SLC7A2 | 9 cancers: BLCA; BRCA; COAD; ESCA; LUSC; PAAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.218; TCGA BRCA -0.556; TCGA COAD -0.649; TCGA ESCA -0.489; TCGA LUSC -0.413; TCGA PAAD -0.465; TCGA THCA -0.239; TCGA STAD -0.474; TCGA UCEC -0.532 |
hsa-miR-15b-3p | SARM1 | 9 cancers: BLCA; BRCA; ESCA; HNSC; LGG; LUAD; PAAD; STAD; UCEC | mirMAP | TCGA BLCA -0.132; TCGA BRCA -0.142; TCGA ESCA -0.267; TCGA HNSC -0.14; TCGA LGG -0.108; TCGA LUAD -0.169; TCGA PAAD -0.215; TCGA STAD -0.117; TCGA UCEC -0.093 |
hsa-miR-15b-3p | IGF1 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -1.142; TCGA BRCA -0.538; TCGA CESC -0.729; TCGA COAD -1.372; TCGA ESCA -0.969; TCGA HNSC -0.6; TCGA LIHC -0.318; TCGA LUAD -0.294; TCGA LUSC -0.643; TCGA PAAD -0.546; TCGA PRAD -0.27; TCGA STAD -0.796; TCGA UCEC -0.39 |
hsa-miR-15b-3p | SNX1 | 12 cancers: BLCA; BRCA; CESC; ESCA; KIRC; LIHC; LUAD; LUSC; PAAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.084; TCGA BRCA -0.121; TCGA CESC -0.066; TCGA ESCA -0.089; TCGA KIRC -0.082; TCGA LIHC -0.061; TCGA LUAD -0.058; TCGA LUSC -0.067; TCGA PAAD -0.077; TCGA SARC -0.09; TCGA STAD -0.169; TCGA UCEC -0.127 |
hsa-miR-15b-3p | H6PD | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LIHC; LUSC; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.118; TCGA BRCA -0.146; TCGA CESC -0.122; TCGA ESCA -0.184; TCGA HNSC -0.115; TCGA LIHC -0.103; TCGA LUSC -0.103; TCGA PRAD -0.097; TCGA STAD -0.146; TCGA UCEC -0.165 |
hsa-miR-15b-3p | KIAA2022 | 13 cancers: BLCA; BRCA; COAD; ESCA; KIRC; KIRP; LGG; LUAD; PAAD; PRAD; SARC; STAD; UCEC | mirMAP; miRNATAP | TCGA BLCA -0.81; TCGA BRCA -0.127; TCGA COAD -1.349; TCGA ESCA -1.075; TCGA KIRC -1.213; TCGA KIRP -0.436; TCGA LGG -0.113; TCGA LUAD -0.405; TCGA PAAD -0.594; TCGA PRAD -0.321; TCGA SARC -0.323; TCGA STAD -1.219; TCGA UCEC -0.305 |
hsa-miR-15b-3p | NGFR | 9 cancers: BLCA; BRCA; COAD; LIHC; LUAD; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.744; TCGA BRCA -0.662; TCGA COAD -1.185; TCGA LIHC -0.51; TCGA LUAD -0.248; TCGA PAAD -0.543; TCGA PRAD -0.551; TCGA STAD -1.008; TCGA UCEC -0.406 |
hsa-miR-15b-3p | CALCRL | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.294; TCGA BRCA -0.156; TCGA CESC -0.384; TCGA COAD -0.61; TCGA ESCA -0.282; TCGA HNSC -0.251; TCGA LGG -0.427; TCGA LIHC -0.315; TCGA LUAD -0.245; TCGA LUSC -0.373; TCGA PAAD -0.451; TCGA PRAD -0.241; TCGA SARC -0.173; TCGA STAD -0.271; TCGA UCEC -0.503 |
hsa-miR-15b-3p | CAMK2A | 11 cancers: BLCA; CESC; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -1.067; TCGA CESC -0.372; TCGA ESCA -0.506; TCGA HNSC -0.949; TCGA KIRC -0.754; TCGA LIHC -0.441; TCGA LUAD -0.328; TCGA LUSC -0.83; TCGA PRAD -0.534; TCGA STAD -0.859; TCGA UCEC -0.841 |
hsa-miR-15b-3p | GLI2 | 9 cancers: BLCA; BRCA; COAD; LUAD; OV; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.699; TCGA BRCA -0.287; TCGA COAD -0.77; TCGA LUAD -0.191; TCGA OV -0.249; TCGA PRAD -0.415; TCGA SARC -0.17; TCGA STAD -0.404; TCGA UCEC -0.412 |
hsa-miR-15b-3p | RUNX1T1 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | mirMAP; miRNATAP | TCGA BLCA -0.894; TCGA BRCA -0.427; TCGA CESC -0.875; TCGA COAD -1.042; TCGA ESCA -0.811; TCGA HNSC -0.553; TCGA KIRC -0.257; TCGA LGG -0.252; TCGA LUAD -0.215; TCGA LUSC -0.7; TCGA PAAD -0.418; TCGA PRAD -0.528; TCGA STAD -0.839; TCGA UCEC -0.703 |
hsa-miR-15b-3p | CYLD | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.156; TCGA CESC -0.1; TCGA COAD -0.115; TCGA ESCA -0.157; TCGA HNSC -0.134; TCGA LIHC -0.103; TCGA LUAD -0.101; TCGA LUSC -0.242; TCGA PRAD -0.103; TCGA SARC -0.084; TCGA STAD -0.159; TCGA UCEC -0.158 |
hsa-miR-15b-3p | ARHGAP28 | 9 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; LGG; STAD; UCEC | mirMAP | TCGA BLCA -0.276; TCGA BRCA -0.073; TCGA CESC -0.436; TCGA COAD -0.569; TCGA HNSC -0.422; TCGA KIRC -0.189; TCGA LGG -0.2; TCGA STAD -0.284; TCGA UCEC -0.252 |
hsa-miR-15b-3p | WDR7 | 11 cancers: BLCA; COAD; ESCA; HNSC; KIRC; LIHC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.069; TCGA COAD -0.114; TCGA ESCA -0.151; TCGA HNSC -0.077; TCGA KIRC -0.053; TCGA LIHC -0.093; TCGA PAAD -0.133; TCGA PRAD -0.152; TCGA SARC -0.084; TCGA STAD -0.124; TCGA UCEC -0.073 |
hsa-miR-15b-3p | FMO2 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.914; TCGA BRCA -0.455; TCGA CESC -0.899; TCGA COAD -1.147; TCGA ESCA -0.744; TCGA HNSC -0.716; TCGA LGG -0.131; TCGA LIHC -0.705; TCGA LUAD -0.547; TCGA LUSC -0.727; TCGA PAAD -0.607; TCGA PRAD -0.627; TCGA SARC -1.016; TCGA STAD -0.811; TCGA UCEC -0.745 |
hsa-miR-15b-3p | ABLIM1 | 10 cancers: BLCA; BRCA; HNSC; KIRC; KIRP; LGG; LIHC; PRAD; SARC; UCEC | mirMAP | TCGA BLCA -0.185; TCGA BRCA -0.081; TCGA HNSC -0.208; TCGA KIRC -0.282; TCGA KIRP -0.117; TCGA LGG -0.155; TCGA LIHC -0.126; TCGA PRAD -0.146; TCGA SARC -0.298; TCGA UCEC -0.129 |
hsa-miR-15b-3p | NFATC4 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUSC; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.427; TCGA BRCA -0.238; TCGA CESC -0.333; TCGA COAD -0.27; TCGA ESCA -0.402; TCGA HNSC -0.308; TCGA KIRP -0.211; TCGA LUSC -0.144; TCGA PRAD -0.22; TCGA STAD -0.529; TCGA UCEC -0.235 |
hsa-miR-15b-3p | CHRDL1 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -1.24; TCGA BRCA -0.559; TCGA CESC -1.2; TCGA COAD -2.088; TCGA ESCA -1.657; TCGA HNSC -0.74; TCGA KIRC -0.693; TCGA LUAD -0.483; TCGA LUSC -0.846; TCGA PAAD -0.714; TCGA PRAD -0.92; TCGA SARC -0.431; TCGA STAD -1.441; TCGA UCEC -1.035 |
hsa-miR-15b-3p | ZC3H12B | 16 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; OV; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.334; TCGA BRCA -0.223; TCGA CESC -0.299; TCGA ESCA -0.513; TCGA HNSC -0.135; TCGA KIRC -0.341; TCGA KIRP -0.262; TCGA LGG -0.226; TCGA LUAD -0.109; TCGA LUSC -0.255; TCGA OV -0.143; TCGA PAAD -0.197; TCGA PRAD -0.379; TCGA SARC -0.169; TCGA STAD -0.422; TCGA UCEC -0.421 |
hsa-miR-15b-3p | DRP2 | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; LGG; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.343; TCGA CESC -0.157; TCGA COAD -0.352; TCGA ESCA -0.278; TCGA HNSC -0.276; TCGA LGG -0.137; TCGA PRAD -0.363; TCGA STAD -0.569; TCGA UCEC -0.213 |
hsa-miR-15b-3p | NFAT5 | 14 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; OV; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.105; TCGA BRCA -0.089; TCGA CESC -0.126; TCGA ESCA -0.152; TCGA HNSC -0.085; TCGA KIRC -0.202; TCGA KIRP -0.162; TCGA LUAD -0.057; TCGA LUSC -0.136; TCGA OV -0.094; TCGA PRAD -0.139; TCGA SARC -0.099; TCGA STAD -0.213; TCGA UCEC -0.177 |
hsa-miR-15b-3p | CRTC1 | 10 cancers: BLCA; BRCA; ESCA; KIRC; KIRP; LGG; OV; PAAD; THCA; STAD | mirMAP | TCGA BLCA -0.129; TCGA BRCA -0.134; TCGA ESCA -0.144; TCGA KIRC -0.194; TCGA KIRP -0.185; TCGA LGG -0.137; TCGA OV -0.126; TCGA PAAD -0.236; TCGA THCA -0.112; TCGA STAD -0.105 |
hsa-miR-15b-3p | KANK1 | 15 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.271; TCGA BRCA -0.067; TCGA CESC -0.317; TCGA ESCA -0.195; TCGA HNSC -0.314; TCGA KIRC -0.112; TCGA LGG -0.128; TCGA LIHC -0.165; TCGA LUAD -0.096; TCGA LUSC -0.119; TCGA PAAD -0.199; TCGA PRAD -0.348; TCGA SARC -0.355; TCGA STAD -0.235; TCGA UCEC -0.202 |
hsa-miR-15b-3p | ASPA | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.768; TCGA BRCA -0.498; TCGA CESC -0.581; TCGA COAD -0.713; TCGA ESCA -1.1; TCGA LIHC -0.24; TCGA LUAD -0.38; TCGA LUSC -0.769; TCGA PAAD -0.514; TCGA PRAD -1.048; TCGA SARC -0.925; TCGA STAD -0.698; TCGA UCEC -0.825 |
hsa-miR-15b-3p | INPP4B | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUSC; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.159; TCGA BRCA -0.443; TCGA CESC -0.242; TCGA COAD -0.223; TCGA ESCA -0.24; TCGA HNSC -0.321; TCGA LIHC -0.209; TCGA LUSC -0.482; TCGA SARC -0.154; TCGA STAD -0.23; TCGA UCEC -0.234 |
hsa-miR-15b-3p | GAB1 | 12 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LUAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.172; TCGA BRCA -0.105; TCGA CESC -0.144; TCGA ESCA -0.237; TCGA HNSC -0.227; TCGA KIRC -0.179; TCGA KIRP -0.106; TCGA LUAD -0.112; TCGA PRAD -0.126; TCGA SARC -0.118; TCGA STAD -0.337; TCGA UCEC -0.277 |
hsa-miR-15b-3p | MDGA1 | 10 cancers: BLCA; BRCA; COAD; KIRC; LGG; LUAD; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.403; TCGA BRCA -0.171; TCGA COAD -0.588; TCGA KIRC -0.133; TCGA LGG -0.067; TCGA LUAD -0.317; TCGA PAAD -0.339; TCGA PRAD -0.167; TCGA STAD -0.4; TCGA UCEC -0.259 |
hsa-miR-15b-3p | SOBP | 14 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.69; TCGA BRCA -0.195; TCGA COAD -0.427; TCGA ESCA -0.58; TCGA HNSC -0.432; TCGA KIRC -0.201; TCGA KIRP -0.162; TCGA LGG -0.108; TCGA LUAD -0.182; TCGA LUSC -0.321; TCGA PAAD -0.259; TCGA PRAD -0.337; TCGA STAD -0.706; TCGA UCEC -0.322 |
hsa-miR-15b-3p | PHACTR2 | 11 cancers: BLCA; CESC; ESCA; HNSC; LIHC; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.091; TCGA CESC -0.131; TCGA ESCA -0.177; TCGA HNSC -0.175; TCGA LIHC -0.104; TCGA LUSC -0.267; TCGA PAAD -0.17; TCGA PRAD -0.281; TCGA SARC -0.16; TCGA STAD -0.157; TCGA UCEC -0.16 |
hsa-miR-15b-3p | CDH6 | 11 cancers: BLCA; BRCA; CESC; COAD; HNSC; LUAD; LUSC; PAAD; PRAD; THCA; STAD | mirMAP | TCGA BLCA -0.215; TCGA BRCA -0.176; TCGA CESC -0.328; TCGA COAD -0.432; TCGA HNSC -0.125; TCGA LUAD -0.252; TCGA LUSC -0.654; TCGA PAAD -0.295; TCGA PRAD -0.257; TCGA THCA -0.498; TCGA STAD -0.182 |
hsa-miR-15b-3p | CPEB4 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LIHC; LUAD; LUSC; PAAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.229; TCGA BRCA -0.126; TCGA CESC -0.23; TCGA COAD -0.189; TCGA ESCA -0.254; TCGA HNSC -0.352; TCGA KIRC -0.243; TCGA LGG -0.059; TCGA LIHC -0.272; TCGA LUAD -0.08; TCGA LUSC -0.341; TCGA PAAD -0.145; TCGA SARC -0.201; TCGA STAD -0.161; TCGA UCEC -0.241 |
hsa-miR-15b-3p | STAM2 | 12 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LIHC; LUSC; OV; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.052; TCGA BRCA -0.084; TCGA ESCA -0.12; TCGA HNSC -0.072; TCGA KIRC -0.168; TCGA LIHC -0.089; TCGA LUSC -0.102; TCGA OV -0.061; TCGA PRAD -0.092; TCGA SARC -0.088; TCGA STAD -0.068; TCGA UCEC -0.14 |
hsa-miR-15b-3p | IGFBP5 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; OV; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.726; TCGA BRCA -0.126; TCGA CESC -0.456; TCGA COAD -0.848; TCGA ESCA -0.604; TCGA HNSC -0.46; TCGA KIRC -0.333; TCGA LIHC -0.334; TCGA OV -0.188; TCGA PAAD -0.292; TCGA PRAD -0.331; TCGA SARC -0.277; TCGA STAD -0.49; TCGA UCEC -0.498 |
hsa-miR-15b-3p | RIMS3 | 9 cancers: BLCA; BRCA; ESCA; HNSC; LUAD; LUSC; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.217; TCGA BRCA -0.097; TCGA ESCA -0.467; TCGA HNSC -0.277; TCGA LUAD -0.137; TCGA LUSC -0.432; TCGA PRAD -0.405; TCGA STAD -0.563; TCGA UCEC -0.231 |
hsa-miR-15b-3p | CNR1 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.431; TCGA BRCA -0.34; TCGA CESC -0.534; TCGA COAD -1.279; TCGA ESCA -0.954; TCGA HNSC -0.224; TCGA KIRP -0.563; TCGA LUSC -0.488; TCGA PAAD -0.566; TCGA PRAD -0.504; TCGA SARC -0.552; TCGA STAD -1.157; TCGA UCEC -0.403 |
hsa-miR-15b-3p | SGIP1 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.443; TCGA BRCA -0.088; TCGA CESC -0.535; TCGA COAD -0.633; TCGA ESCA -0.548; TCGA HNSC -0.34; TCGA KIRC -0.376; TCGA KIRP -0.508; TCGA LUAD -0.285; TCGA LUSC -0.675; TCGA PAAD -0.332; TCGA PRAD -0.139; TCGA SARC -0.304; TCGA STAD -0.497; TCGA UCEC -0.435 |
hsa-miR-15b-3p | KLF12 | 13 cancers: BLCA; BRCA; COAD; ESCA; KIRC; KIRP; LGG; LIHC; LUSC; OV; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.312; TCGA BRCA -0.055; TCGA COAD -0.515; TCGA ESCA -0.277; TCGA KIRC -0.131; TCGA KIRP -0.123; TCGA LGG -0.091; TCGA LIHC -0.209; TCGA LUSC -0.14; TCGA OV -0.121; TCGA PRAD -0.171; TCGA STAD -0.417; TCGA UCEC -0.164 |
hsa-miR-15b-3p | RASSF8 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUSC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.286; TCGA BRCA -0.12; TCGA CESC -0.237; TCGA COAD -0.777; TCGA ESCA -0.31; TCGA HNSC -0.157; TCGA KIRC -0.374; TCGA KIRP -0.111; TCGA LUSC -0.187; TCGA PAAD -0.331; TCGA PRAD -0.203; TCGA STAD -0.682; TCGA UCEC -0.511 |
hsa-miR-15b-3p | ATXN1 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | mirMAP; miRNATAP | TCGA BLCA -0.328; TCGA BRCA -0.102; TCGA CESC -0.179; TCGA COAD -0.238; TCGA ESCA -0.171; TCGA HNSC -0.215; TCGA LUAD -0.106; TCGA LUSC -0.121; TCGA PRAD -0.272; TCGA SARC -0.114; TCGA STAD -0.287; TCGA UCEC -0.233 |
hsa-miR-15b-3p | FOSB | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LIHC; LUSC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.512; TCGA BRCA -0.656; TCGA COAD -0.426; TCGA ESCA -0.456; TCGA HNSC -0.238; TCGA KIRC -0.344; TCGA LIHC -0.46; TCGA LUSC -0.694; TCGA PAAD -0.408; TCGA PRAD -0.538; TCGA STAD -0.312; TCGA UCEC -0.57 |
hsa-miR-15b-3p | CD93 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.385; TCGA BRCA -0.187; TCGA CESC -0.413; TCGA COAD -0.456; TCGA ESCA -0.312; TCGA HNSC -0.255; TCGA LIHC -0.119; TCGA LUAD -0.214; TCGA LUSC -0.65; TCGA PAAD -0.295; TCGA PRAD -0.164; TCGA SARC -0.226; TCGA STAD -0.165; TCGA UCEC -0.403 |
hsa-miR-15b-3p | PTPRB | 16 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; OV; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.244; TCGA BRCA -0.352; TCGA CESC -0.441; TCGA COAD -0.169; TCGA ESCA -0.511; TCGA HNSC -0.294; TCGA KIRC -0.242; TCGA LIHC -0.459; TCGA LUAD -0.209; TCGA LUSC -0.615; TCGA OV -0.104; TCGA PAAD -0.236; TCGA PRAD -0.168; TCGA SARC -0.352; TCGA STAD -0.255; TCGA UCEC -0.491 |
hsa-miR-15b-3p | TNFRSF19 | 13 cancers: BLCA; BRCA; COAD; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; OV; PAAD; PRAD; UCEC | mirMAP | TCGA BLCA -0.371; TCGA BRCA -0.185; TCGA COAD -0.787; TCGA HNSC -0.368; TCGA KIRC -0.226; TCGA KIRP -0.333; TCGA LIHC -0.42; TCGA LUAD -0.13; TCGA LUSC -0.29; TCGA OV -0.167; TCGA PAAD -0.329; TCGA PRAD -0.166; TCGA UCEC -0.142 |
hsa-miR-15b-3p | MAP1B | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.676; TCGA BRCA -0.113; TCGA CESC -0.214; TCGA COAD -0.775; TCGA ESCA -0.336; TCGA HNSC -0.263; TCGA LIHC -0.169; TCGA PAAD -0.492; TCGA PRAD -0.429; TCGA SARC -0.147; TCGA STAD -0.828; TCGA UCEC -0.322 |
hsa-miR-15b-3p | TMTC1 | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.377; TCGA BRCA -0.355; TCGA COAD -0.754; TCGA ESCA -0.296; TCGA HNSC -0.258; TCGA KIRC -0.25; TCGA PAAD -0.293; TCGA PRAD -0.439; TCGA SARC -0.156; TCGA STAD -0.688; TCGA UCEC -0.185 |
hsa-miR-15b-3p | KIAA0513 | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.429; TCGA CESC -0.161; TCGA COAD -0.188; TCGA ESCA -0.336; TCGA HNSC -0.229; TCGA LIHC -0.11; TCGA LUAD -0.115; TCGA LUSC -0.436; TCGA PRAD -0.283; TCGA SARC -0.083; TCGA STAD -0.225; TCGA UCEC -0.236 |
hsa-miR-15b-3p | RAB30 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.247; TCGA BRCA -0.173; TCGA CESC -0.131; TCGA COAD -0.294; TCGA ESCA -0.147; TCGA HNSC -0.149; TCGA KIRC -0.124; TCGA LIHC -0.142; TCGA SARC -0.157; TCGA STAD -0.171; TCGA UCEC -0.082 |
hsa-miR-15b-3p | ENTPD1 | 12 cancers: BLCA; CESC; COAD; ESCA; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.369; TCGA CESC -0.176; TCGA COAD -0.351; TCGA ESCA -0.277; TCGA LUAD -0.107; TCGA LUSC -0.302; TCGA PAAD -0.13; TCGA PRAD -0.083; TCGA SARC -0.214; TCGA THCA -0.319; TCGA STAD -0.276; TCGA UCEC -0.257 |
hsa-miR-15b-3p | FGF2 | 17 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; OV; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.698; TCGA BRCA -0.417; TCGA CESC -0.251; TCGA COAD -0.874; TCGA ESCA -0.602; TCGA KIRC -0.217; TCGA KIRP -0.309; TCGA LGG -0.138; TCGA LIHC -0.499; TCGA LUAD -0.159; TCGA LUSC -0.222; TCGA OV -0.159; TCGA PRAD -0.599; TCGA SARC -0.27; TCGA THCA -0.225; TCGA STAD -0.67; TCGA UCEC -0.724 |
hsa-miR-15b-3p | FRMD6 | 9 cancers: BLCA; BRCA; CESC; COAD; HNSC; LUAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.294; TCGA BRCA -0.272; TCGA CESC -0.222; TCGA COAD -0.805; TCGA HNSC -0.119; TCGA LUAD -0.134; TCGA PRAD -0.634; TCGA STAD -0.573; TCGA UCEC -0.557 |
hsa-miR-15b-3p | IGF1R | 11 cancers: BLCA; BRCA; KIRC; KIRP; LGG; OV; PAAD; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.139; TCGA BRCA -0.198; TCGA KIRC -0.135; TCGA KIRP -0.076; TCGA LGG -0.111; TCGA OV -0.221; TCGA PAAD -0.149; TCGA PRAD -0.153; TCGA THCA -0.098; TCGA STAD -0.258; TCGA UCEC -0.196 |
hsa-miR-15b-3p | NTRK3 | 17 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LUAD; LUSC; OV; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.98; TCGA BRCA -0.477; TCGA CESC -0.913; TCGA COAD -1.234; TCGA ESCA -1.239; TCGA HNSC -0.595; TCGA KIRC -0.494; TCGA LGG -0.078; TCGA LUAD -0.51; TCGA LUSC -0.782; TCGA OV -0.368; TCGA PAAD -0.312; TCGA PRAD -0.832; TCGA SARC -0.458; TCGA THCA -0.238; TCGA STAD -0.967; TCGA UCEC -0.621 |
hsa-miR-15b-3p | FTO | 14 cancers: BLCA; BRCA; CESC; COAD; HNSC; LIHC; LUAD; LUSC; OV; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.093; TCGA BRCA -0.129; TCGA CESC -0.065; TCGA COAD -0.148; TCGA HNSC -0.094; TCGA LIHC -0.124; TCGA LUAD -0.104; TCGA LUSC -0.09; TCGA OV -0.1; TCGA PAAD -0.079; TCGA PRAD -0.151; TCGA SARC -0.094; TCGA STAD -0.222; TCGA UCEC -0.144 |
hsa-miR-15b-3p | RGS5 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.565; TCGA BRCA -0.29; TCGA CESC -0.426; TCGA COAD -0.617; TCGA ESCA -0.492; TCGA HNSC -0.282; TCGA LIHC -0.193; TCGA LUAD -0.314; TCGA LUSC -0.519; TCGA PAAD -0.394; TCGA PRAD -0.132; TCGA SARC -0.296; TCGA THCA -0.233; TCGA STAD -0.512; TCGA UCEC -0.447 |
hsa-miR-15b-3p | ST6GAL2 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LGG; LIHC; LUAD; OV; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.627; TCGA BRCA -0.208; TCGA CESC -0.555; TCGA COAD -0.977; TCGA ESCA -0.432; TCGA HNSC -0.577; TCGA KIRP -0.321; TCGA LGG -0.277; TCGA LIHC -0.441; TCGA LUAD -0.249; TCGA OV -0.395; TCGA PRAD -0.402; TCGA STAD -0.903; TCGA UCEC -0.384 |
hsa-miR-15b-3p | GULP1 | 9 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; OV; STAD; UCEC | mirMAP | TCGA BLCA -0.459; TCGA BRCA -0.272; TCGA COAD -0.703; TCGA ESCA -0.348; TCGA HNSC -0.342; TCGA KIRC -0.391; TCGA OV -0.32; TCGA STAD -0.338; TCGA UCEC -0.369 |
hsa-miR-15b-3p | SETD7 | 12 cancers: BLCA; BRCA; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.188; TCGA BRCA -0.088; TCGA ESCA -0.098; TCGA HNSC -0.16; TCGA LGG -0.05; TCGA LIHC -0.134; TCGA LUAD -0.067; TCGA LUSC -0.105; TCGA PRAD -0.079; TCGA SARC -0.26; TCGA STAD -0.308; TCGA UCEC -0.424 |
hsa-miR-15b-3p | TACC1 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.353; TCGA BRCA -0.169; TCGA CESC -0.269; TCGA COAD -0.155; TCGA ESCA -0.302; TCGA KIRC -0.196; TCGA LIHC -0.122; TCGA LUAD -0.171; TCGA LUSC -0.284; TCGA PAAD -0.147; TCGA PRAD -0.299; TCGA SARC -0.247; TCGA STAD -0.446; TCGA UCEC -0.53 |
hsa-miR-15b-3p | NTRK2 | 11 cancers: BLCA; BRCA; COAD; KIRC; LGG; LUAD; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.789; TCGA BRCA -0.563; TCGA COAD -1.037; TCGA KIRC -0.883; TCGA LGG -0.275; TCGA LUAD -0.278; TCGA PAAD -0.626; TCGA PRAD -0.579; TCGA SARC -0.507; TCGA STAD -0.742; TCGA UCEC -0.511 |
hsa-miR-15b-3p | MKX | 13 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LGG; LIHC; PAAD; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.856; TCGA BRCA -0.223; TCGA COAD -0.471; TCGA ESCA -0.465; TCGA HNSC -0.224; TCGA KIRC -0.209; TCGA LGG -0.609; TCGA LIHC -0.532; TCGA PAAD -0.227; TCGA PRAD -0.498; TCGA THCA -0.545; TCGA STAD -0.952; TCGA UCEC -0.211 |
hsa-miR-15b-3p | DLG2 | 16 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; OV; PAAD; STAD; UCEC | mirMAP | TCGA BLCA -0.771; TCGA BRCA -0.363; TCGA CESC -0.704; TCGA COAD -1.281; TCGA ESCA -1.061; TCGA HNSC -0.706; TCGA KIRC -0.645; TCGA KIRP -0.159; TCGA LGG -0.076; TCGA LIHC -0.37; TCGA LUAD -0.274; TCGA LUSC -0.554; TCGA OV -0.492; TCGA PAAD -0.666; TCGA STAD -0.997; TCGA UCEC -0.593 |
hsa-miR-15b-3p | EDNRA | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.63; TCGA BRCA -0.179; TCGA CESC -0.527; TCGA COAD -0.536; TCGA ESCA -0.169; TCGA HNSC -0.403; TCGA LUAD -0.326; TCGA LUSC -0.58; TCGA PAAD -0.264; TCGA PRAD -0.571; TCGA THCA -0.127; TCGA STAD -0.512; TCGA UCEC -0.591 |
hsa-miR-15b-3p | GFRA1 | 16 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; OV; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.974; TCGA BRCA -0.699; TCGA CESC -0.987; TCGA COAD -1.258; TCGA ESCA -1.118; TCGA HNSC -0.899; TCGA LGG -0.371; TCGA LIHC -0.69; TCGA LUAD -0.612; TCGA LUSC -0.662; TCGA OV -0.305; TCGA PAAD -0.368; TCGA PRAD -0.658; TCGA SARC -0.353; TCGA STAD -1.039; TCGA UCEC -0.769 |
hsa-miR-15b-3p | DST | 12 cancers: BLCA; BRCA; COAD; HNSC; KIRC; LGG; LIHC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.23; TCGA BRCA -0.342; TCGA COAD -0.286; TCGA HNSC -0.104; TCGA KIRC -0.1; TCGA LGG -0.117; TCGA LIHC -0.137; TCGA PAAD -0.128; TCGA PRAD -0.421; TCGA SARC -0.182; TCGA STAD -0.504; TCGA UCEC -0.389 |
hsa-miR-15b-3p | MR1 | 9 cancers: BLCA; COAD; LIHC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.077; TCGA COAD -0.289; TCGA LIHC -0.103; TCGA LUAD -0.136; TCGA LUSC -0.192; TCGA PRAD -0.388; TCGA SARC -0.114; TCGA STAD -0.15; TCGA UCEC -0.123 |
hsa-miR-15b-3p | SCN3A | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.374; TCGA BRCA -0.674; TCGA CESC -0.659; TCGA COAD -0.765; TCGA ESCA -0.623; TCGA HNSC -0.186; TCGA KIRC -0.759; TCGA KIRP -0.455; TCGA LGG -0.287; TCGA PAAD -0.55; TCGA PRAD -0.226; TCGA SARC -0.482; TCGA STAD -0.453; TCGA UCEC -0.35 |
hsa-miR-15b-3p | NCAM2 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.794; TCGA BRCA -0.398; TCGA CESC -0.728; TCGA COAD -1.408; TCGA ESCA -0.816; TCGA HNSC -0.497; TCGA KIRC -0.427; TCGA LGG -0.112; TCGA LUAD -0.388; TCGA LUSC -0.571; TCGA PAAD -0.44; TCGA PRAD -0.279; TCGA STAD -0.85; TCGA UCEC -0.364 |
hsa-miR-15b-3p | PDE1C | 14 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.886; TCGA BRCA -0.427; TCGA COAD -0.422; TCGA ESCA -0.783; TCGA HNSC -0.232; TCGA KIRC -0.333; TCGA LUAD -0.194; TCGA LUSC -0.507; TCGA PAAD -0.311; TCGA PRAD -0.876; TCGA SARC -0.28; TCGA THCA -0.469; TCGA STAD -0.636; TCGA UCEC -0.456 |
hsa-miR-15b-3p | ADAMTS5 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LGG; LUSC; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.428; TCGA BRCA -0.323; TCGA CESC -0.536; TCGA COAD -0.423; TCGA ESCA -0.494; TCGA HNSC -0.385; TCGA KIRP -0.266; TCGA LGG -0.212; TCGA LUSC -0.421; TCGA PRAD -0.446; TCGA STAD -0.233; TCGA UCEC -0.535 |
hsa-miR-15b-3p | CYP2U1 | 15 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.29; TCGA BRCA -0.134; TCGA COAD -0.318; TCGA ESCA -0.391; TCGA HNSC -0.138; TCGA KIRC -0.133; TCGA LGG -0.066; TCGA LIHC -0.248; TCGA LUAD -0.182; TCGA LUSC -0.189; TCGA PAAD -0.242; TCGA PRAD -0.113; TCGA SARC -0.242; TCGA STAD -0.451; TCGA UCEC -0.281 |
hsa-miR-15b-3p | NMNAT2 | 9 cancers: BLCA; BRCA; COAD; LGG; LUSC; PAAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.195; TCGA BRCA -0.106; TCGA COAD -0.461; TCGA LGG -0.276; TCGA LUSC -0.582; TCGA PAAD -0.567; TCGA SARC -0.362; TCGA STAD -0.563; TCGA UCEC -0.238 |
hsa-miR-15b-3p | SKI | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; STAD; UCEC | mirMAP | TCGA BLCA -0.105; TCGA BRCA -0.05; TCGA CESC -0.175; TCGA COAD -0.071; TCGA ESCA -0.09; TCGA HNSC -0.087; TCGA KIRP -0.076; TCGA LUAD -0.121; TCGA LUSC -0.19; TCGA STAD -0.15; TCGA UCEC -0.105 |
hsa-miR-15b-3p | PRKAA2 | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.478; TCGA BRCA -0.246; TCGA COAD -0.952; TCGA ESCA -0.614; TCGA HNSC -0.437; TCGA KIRC -0.231; TCGA KIRP -0.231; TCGA PRAD -0.088; TCGA SARC -0.337; TCGA STAD -1.152; TCGA UCEC -0.126 |
hsa-miR-15b-3p | ANTXR2 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.634; TCGA BRCA -0.167; TCGA CESC -0.379; TCGA COAD -0.312; TCGA ESCA -0.348; TCGA HNSC -0.24; TCGA LIHC -0.235; TCGA LUAD -0.181; TCGA LUSC -0.482; TCGA PRAD -0.347; TCGA SARC -0.174; TCGA STAD -0.575; TCGA UCEC -0.674 |
hsa-miR-15b-3p | CCDC141 | 10 cancers: BLCA; BRCA; ESCA; HNSC; LGG; LUAD; LUSC; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.503; TCGA BRCA -0.164; TCGA ESCA -0.446; TCGA HNSC -0.522; TCGA LGG -0.157; TCGA LUAD -0.262; TCGA LUSC -0.6; TCGA SARC -0.346; TCGA STAD -0.214; TCGA UCEC -0.418 |
hsa-miR-15b-3p | PRICKLE2 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.628; TCGA BRCA -0.35; TCGA CESC -0.305; TCGA COAD -0.688; TCGA ESCA -0.659; TCGA HNSC -0.283; TCGA KIRC -0.076; TCGA LGG -0.088; TCGA LUAD -0.22; TCGA LUSC -0.441; TCGA PAAD -0.305; TCGA PRAD -0.6; TCGA SARC -0.156; TCGA STAD -0.85; TCGA UCEC -0.505 |
hsa-miR-15b-3p | APBB2 | 16 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; OV; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.216; TCGA BRCA -0.158; TCGA CESC -0.18; TCGA COAD -0.103; TCGA ESCA -0.183; TCGA HNSC -0.16; TCGA KIRC -0.064; TCGA KIRP -0.106; TCGA LGG -0.073; TCGA LIHC -0.163; TCGA LUAD -0.097; TCGA LUSC -0.324; TCGA OV -0.089; TCGA SARC -0.123; TCGA STAD -0.198; TCGA UCEC -0.098 |
hsa-miR-15b-3p | HEYL | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; STAD; UCEC | mirMAP | TCGA BLCA -0.269; TCGA BRCA -0.05; TCGA CESC -0.332; TCGA COAD -0.525; TCGA ESCA -0.24; TCGA HNSC -0.239; TCGA LUAD -0.239; TCGA LUSC -0.5; TCGA PAAD -0.475; TCGA STAD -0.333; TCGA UCEC -0.368 |
hsa-miR-15b-3p | HHIP | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PRAD; STAD | mirMAP | TCGA BLCA -0.548; TCGA CESC -0.383; TCGA COAD -1.049; TCGA ESCA -0.56; TCGA HNSC -0.411; TCGA KIRC -0.527; TCGA KIRP -0.41; TCGA LIHC -0.499; TCGA LUAD -0.649; TCGA LUSC -0.905; TCGA PRAD -0.716; TCGA STAD -0.306 |
hsa-miR-15b-3p | BEND7 | 12 cancers: BLCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.26; TCGA HNSC -0.332; TCGA KIRC -0.222; TCGA KIRP -0.119; TCGA LGG -0.155; TCGA LIHC -0.147; TCGA LUAD -0.153; TCGA LUSC -0.218; TCGA PAAD -0.286; TCGA SARC -0.266; TCGA STAD -0.156; TCGA UCEC -0.231 |
hsa-miR-15b-3p | COL4A3 | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.598; TCGA BRCA -0.13; TCGA COAD -0.511; TCGA ESCA -0.946; TCGA HNSC -0.303; TCGA KIRC -0.814; TCGA KIRP -0.6; TCGA LUAD -0.556; TCGA LUSC -0.901; TCGA PRAD -0.224; TCGA STAD -0.632; TCGA UCEC -0.407 |
hsa-miR-15b-3p | CA5B | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; KIRP; LUAD; STAD; UCEC | mirMAP | TCGA BLCA -0.129; TCGA BRCA -0.162; TCGA CESC -0.106; TCGA COAD -0.201; TCGA ESCA -0.328; TCGA KIRC -0.109; TCGA KIRP -0.121; TCGA LUAD -0.149; TCGA STAD -0.369; TCGA UCEC -0.205 |
hsa-miR-15b-3p | ANTXR1 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.467; TCGA BRCA -0.09; TCGA CESC -0.312; TCGA COAD -1.018; TCGA ESCA -0.268; TCGA HNSC -0.283; TCGA LGG -0.071; TCGA LIHC -0.214; TCGA LUAD -0.259; TCGA LUSC -0.287; TCGA PRAD -0.324; TCGA STAD -0.374; TCGA UCEC -0.356 |
hsa-miR-15b-3p | NLGN1 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.523; TCGA BRCA -0.422; TCGA CESC -0.317; TCGA COAD -1.462; TCGA ESCA -0.718; TCGA HNSC -0.216; TCGA LGG -0.117; TCGA PAAD -0.684; TCGA PRAD -0.356; TCGA STAD -1.141; TCGA UCEC -0.439 |
hsa-miR-15b-3p | NLGN2 | 10 cancers: BLCA; BRCA; COAD; KIRP; LGG; LIHC; PAAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.152; TCGA BRCA -0.121; TCGA COAD -0.521; TCGA KIRP -0.211; TCGA LGG -0.159; TCGA LIHC -0.136; TCGA PAAD -0.215; TCGA THCA -0.09; TCGA STAD -0.431; TCGA UCEC -0.138 |
hsa-miR-15b-3p | LONRF2 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; PAAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.875; TCGA BRCA -0.423; TCGA CESC -0.759; TCGA COAD -1.83; TCGA ESCA -0.903; TCGA HNSC -0.291; TCGA KIRC -0.773; TCGA KIRP -0.379; TCGA LUAD -0.236; TCGA PAAD -0.657; TCGA SARC -0.587; TCGA THCA -0.275; TCGA STAD -1.098; TCGA UCEC -0.482 |
hsa-miR-15b-3p | PAQR8 | 9 cancers: BLCA; CESC; ESCA; KIRC; KIRP; LGG; LUSC; PRAD; UCEC | mirMAP | TCGA BLCA -0.132; TCGA CESC -0.158; TCGA ESCA -0.256; TCGA KIRC -0.181; TCGA KIRP -0.285; TCGA LGG -0.084; TCGA LUSC -0.174; TCGA PRAD -0.232; TCGA UCEC -0.233 |
hsa-miR-15b-3p | MCC | 10 cancers: BLCA; BRCA; COAD; KIRC; LUAD; OV; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.15; TCGA BRCA -0.269; TCGA COAD -0.723; TCGA KIRC -0.189; TCGA LUAD -0.2; TCGA OV -0.132; TCGA PAAD -0.209; TCGA PRAD -0.607; TCGA STAD -0.54; TCGA UCEC -0.34 |
hsa-miR-15b-3p | BCL2 | 11 cancers: BLCA; BRCA; CESC; ESCA; KIRP; LIHC; OV; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.323; TCGA BRCA -0.311; TCGA CESC -0.161; TCGA ESCA -0.238; TCGA KIRP -0.124; TCGA LIHC -0.104; TCGA OV -0.193; TCGA PAAD -0.191; TCGA PRAD -0.334; TCGA STAD -0.398; TCGA UCEC -0.413 |
hsa-miR-15b-3p | PTEN | 14 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LIHC; LUAD; LUSC; OV; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.111; TCGA BRCA -0.099; TCGA CESC -0.102; TCGA ESCA -0.084; TCGA HNSC -0.15; TCGA LIHC -0.176; TCGA LUAD -0.061; TCGA LUSC -0.253; TCGA OV -0.126; TCGA PAAD -0.073; TCGA PRAD -0.218; TCGA SARC -0.084; TCGA STAD -0.14; TCGA UCEC -0.207 |
hsa-miR-15b-3p | LRRC15 | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PRAD; UCEC | mirMAP | TCGA BLCA -0.846; TCGA CESC -0.278; TCGA COAD -0.905; TCGA ESCA -0.303; TCGA HNSC -0.544; TCGA LUAD -0.33; TCGA LUSC -0.449; TCGA PRAD -0.193; TCGA UCEC -0.333 |
hsa-miR-15b-3p | NEGR1 | 16 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LUAD; LUSC; OV; PAAD; PRAD; SARC; STAD; UCEC | mirMAP; miRNATAP | TCGA BLCA -1.245; TCGA BRCA -0.338; TCGA CESC -0.492; TCGA COAD -1.078; TCGA ESCA -1.123; TCGA HNSC -0.725; TCGA KIRC -0.432; TCGA LGG -0.187; TCGA LUAD -0.471; TCGA LUSC -0.751; TCGA OV -0.248; TCGA PAAD -0.564; TCGA PRAD -0.572; TCGA SARC -0.539; TCGA STAD -1.028; TCGA UCEC -0.753 |
hsa-miR-15b-3p | IL16 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.479; TCGA BRCA -0.123; TCGA CESC -0.269; TCGA COAD -0.296; TCGA ESCA -0.316; TCGA HNSC -0.13; TCGA LUAD -0.197; TCGA LUSC -0.512; TCGA PRAD -0.113; TCGA SARC -0.207; TCGA STAD -0.167; TCGA UCEC -0.297 |
hsa-miR-15b-3p | ZHX3 | 15 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; OV; PAAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.17; TCGA BRCA -0.109; TCGA CESC -0.121; TCGA ESCA -0.299; TCGA HNSC -0.123; TCGA KIRC -0.336; TCGA KIRP -0.128; TCGA LIHC -0.075; TCGA LUAD -0.077; TCGA LUSC -0.163; TCGA OV -0.103; TCGA PAAD -0.18; TCGA SARC -0.156; TCGA STAD -0.293; TCGA UCEC -0.124 |
hsa-miR-15b-3p | CHD9 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; LGG; LIHC; LUAD; LUSC; OV; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.108; TCGA BRCA -0.095; TCGA CESC -0.124; TCGA COAD -0.137; TCGA ESCA -0.118; TCGA KIRC -0.112; TCGA LGG -0.055; TCGA LIHC -0.242; TCGA LUAD -0.116; TCGA LUSC -0.158; TCGA OV -0.108; TCGA PRAD -0.167; TCGA STAD -0.224; TCGA UCEC -0.245 |
hsa-miR-15b-3p | FAM26E | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.505; TCGA BRCA -0.113; TCGA CESC -0.277; TCGA COAD -0.795; TCGA ESCA -0.431; TCGA HNSC -0.448; TCGA LIHC -0.196; TCGA LUAD -0.158; TCGA LUSC -0.514; TCGA PAAD -0.196; TCGA PRAD -0.208; TCGA SARC -0.254; TCGA STAD -0.295; TCGA UCEC -0.418 |
hsa-miR-15b-3p | SLITRK4 | 11 cancers: BLCA; CESC; ESCA; HNSC; KIRC; KIRP; PAAD; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.413; TCGA CESC -0.869; TCGA ESCA -0.709; TCGA HNSC -0.992; TCGA KIRC -0.192; TCGA KIRP -0.282; TCGA PAAD -0.443; TCGA PRAD -0.628; TCGA THCA -0.197; TCGA STAD -0.73; TCGA UCEC -0.342 |
hsa-miR-15b-3p | GIPC3 | 14 cancers: BLCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.128; TCGA CESC -0.365; TCGA COAD -0.354; TCGA ESCA -0.348; TCGA HNSC -0.116; TCGA LIHC -0.207; TCGA LUAD -0.165; TCGA LUSC -0.41; TCGA PAAD -0.227; TCGA PRAD -0.192; TCGA SARC -0.236; TCGA THCA -0.25; TCGA STAD -0.173; TCGA UCEC -0.307 |
hsa-miR-15b-3p | GREM2 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; LUSC; OV; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -1.03; TCGA BRCA -0.64; TCGA CESC -0.581; TCGA COAD -1.524; TCGA ESCA -1.39; TCGA HNSC -0.996; TCGA KIRC -0.766; TCGA LIHC -0.526; TCGA LUSC -0.785; TCGA OV -0.311; TCGA PAAD -0.657; TCGA PRAD -0.346; TCGA STAD -1.206; TCGA UCEC -0.763 |
hsa-miR-15b-3p | CACNB4 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.727; TCGA BRCA -0.34; TCGA CESC -0.208; TCGA COAD -0.499; TCGA ESCA -0.382; TCGA HNSC -0.246; TCGA KIRC -0.464; TCGA KIRP -0.367; TCGA LIHC -0.287; TCGA LUAD -0.145; TCGA PAAD -0.284; TCGA PRAD -0.452; TCGA SARC -0.283; TCGA STAD -0.496; TCGA UCEC -0.352 |
hsa-miR-15b-3p | SATB1 | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LUAD; OV; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.144; TCGA CESC -0.325; TCGA COAD -0.338; TCGA ESCA -0.281; TCGA HNSC -0.261; TCGA KIRC -0.198; TCGA LGG -0.153; TCGA LUAD -0.131; TCGA OV -0.202; TCGA PRAD -0.353; TCGA STAD -0.445; TCGA UCEC -0.447 |
hsa-miR-15b-3p | GRIN2A | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; LUAD; LUSC; OV; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.733; TCGA BRCA -0.368; TCGA CESC -0.431; TCGA COAD -1.136; TCGA ESCA -1.025; TCGA LUAD -0.65; TCGA LUSC -0.517; TCGA OV -0.496; TCGA PAAD -0.286; TCGA PRAD -0.756; TCGA SARC -0.519; TCGA STAD -1.088; TCGA UCEC -0.601 |
hsa-miR-15b-3p | TEAD1 | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; PAAD; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.23; TCGA BRCA -0.082; TCGA COAD -0.171; TCGA ESCA -0.129; TCGA HNSC -0.087; TCGA KIRC -0.107; TCGA PAAD -0.108; TCGA PRAD -0.271; TCGA THCA -0.086; TCGA STAD -0.455; TCGA UCEC -0.144 |
hsa-miR-15b-3p | TLR5 | 12 cancers: BLCA; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; OV; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.27; TCGA COAD -0.333; TCGA ESCA -0.209; TCGA HNSC -0.147; TCGA KIRC -0.228; TCGA LUAD -0.302; TCGA LUSC -0.233; TCGA OV -0.14; TCGA PRAD -0.324; TCGA SARC -0.29; TCGA STAD -0.234; TCGA UCEC -0.203 |
hsa-miR-15b-3p | TCF4 | 13 cancers: BLCA; BRCA; CESC; COAD; HNSC; LGG; LIHC; LUAD; OV; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.416; TCGA BRCA -0.178; TCGA CESC -0.236; TCGA COAD -0.672; TCGA HNSC -0.225; TCGA LGG -0.07; TCGA LIHC -0.179; TCGA LUAD -0.148; TCGA OV -0.124; TCGA PAAD -0.215; TCGA PRAD -0.275; TCGA STAD -0.371; TCGA UCEC -0.359 |
hsa-miR-15b-3p | LRP10 | 11 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LUSC; PRAD; SARC; THCA; UCEC | mirMAP | TCGA BLCA -0.18; TCGA BRCA -0.148; TCGA CESC -0.141; TCGA ESCA -0.127; TCGA HNSC -0.201; TCGA KIRC -0.178; TCGA LUSC -0.258; TCGA PRAD -0.077; TCGA SARC -0.141; TCGA THCA -0.111; TCGA UCEC -0.069 |
hsa-miR-15b-3p | DNM3 | 17 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LIHC; LUAD; LUSC; OV; PAAD; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.297; TCGA BRCA -0.15; TCGA CESC -0.403; TCGA COAD -0.577; TCGA ESCA -0.39; TCGA HNSC -0.285; TCGA KIRC -0.385; TCGA LGG -0.204; TCGA LIHC -0.19; TCGA LUAD -0.286; TCGA LUSC -0.452; TCGA OV -0.184; TCGA PAAD -0.256; TCGA PRAD -0.215; TCGA THCA -0.104; TCGA STAD -0.301; TCGA UCEC -0.315 |
hsa-miR-15b-3p | RYR2 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.952; TCGA BRCA -0.114; TCGA CESC -0.474; TCGA COAD -0.901; TCGA ESCA -1.011; TCGA LUAD -0.401; TCGA LUSC -0.491; TCGA PAAD -0.314; TCGA PRAD -0.316; TCGA SARC -0.408; TCGA STAD -0.984; TCGA UCEC -0.337 |
hsa-miR-15b-3p | GABBR1 | 10 cancers: BLCA; BRCA; ESCA; KIRP; LGG; LUAD; OV; PAAD; SARC; UCEC | mirMAP | TCGA BLCA -0.172; TCGA BRCA -0.145; TCGA ESCA -0.293; TCGA KIRP -0.528; TCGA LGG -0.353; TCGA LUAD -0.185; TCGA OV -0.177; TCGA PAAD -0.17; TCGA SARC -0.166; TCGA UCEC -0.35 |
hsa-miR-15b-3p | TMEM170B | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LGG; PAAD; STAD; UCEC | mirMAP | TCGA BLCA -0.129; TCGA BRCA -0.056; TCGA COAD -0.239; TCGA ESCA -0.294; TCGA HNSC -0.195; TCGA KIRC -0.08; TCGA LGG -0.119; TCGA PAAD -0.246; TCGA STAD -0.387; TCGA UCEC -0.096 |
hsa-miR-15b-3p | DOK6 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.826; TCGA BRCA -0.22; TCGA CESC -0.568; TCGA COAD -0.782; TCGA ESCA -0.406; TCGA HNSC -0.411; TCGA LGG -0.187; TCGA LUAD -0.169; TCGA LUSC -0.258; TCGA OV -0.206; TCGA PAAD -0.558; TCGA PRAD -0.191; TCGA STAD -0.539; TCGA UCEC -0.535 |
hsa-miR-15b-3p | UBE2QL1 | 14 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LIHC; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.461; TCGA CESC -0.28; TCGA COAD -0.96; TCGA ESCA -0.368; TCGA HNSC -0.467; TCGA KIRC -0.385; TCGA LGG -0.122; TCGA LIHC -0.862; TCGA PAAD -0.423; TCGA PRAD -0.531; TCGA SARC -0.367; TCGA THCA -0.32; TCGA STAD -0.579; TCGA UCEC -0.251 |
hsa-miR-15b-3p | CCPG1 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; OV; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.292; TCGA BRCA -0.089; TCGA CESC -0.116; TCGA COAD -0.207; TCGA ESCA -0.284; TCGA HNSC -0.159; TCGA KIRC -0.115; TCGA LIHC -0.22; TCGA LUAD -0.112; TCGA LUSC -0.27; TCGA OV -0.073; TCGA PRAD -0.16; TCGA SARC -0.195; TCGA STAD -0.164; TCGA UCEC -0.214 |
hsa-miR-15b-3p | SSBP2 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; OV; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.166; TCGA BRCA -0.151; TCGA CESC -0.154; TCGA COAD -0.543; TCGA ESCA -0.197; TCGA HNSC -0.11; TCGA KIRP -0.124; TCGA LUAD -0.208; TCGA OV -0.13; TCGA PAAD -0.254; TCGA PRAD -0.2; TCGA SARC -0.111; TCGA STAD -0.579; TCGA UCEC -0.199 |
hsa-miR-15b-3p | PPARGC1A | 13 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; KIRP; LIHC; LUSC; OV; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.883; TCGA BRCA -0.168; TCGA ESCA -0.666; TCGA HNSC -0.756; TCGA KIRC -0.729; TCGA KIRP -0.367; TCGA LIHC -0.43; TCGA LUSC -0.337; TCGA OV -0.329; TCGA PRAD -0.864; TCGA SARC -0.434; TCGA STAD -0.422; TCGA UCEC -0.393 |
hsa-miR-15b-3p | KLHL14 | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; KIRP; LUSC; PRAD; STAD | miRNATAP | TCGA BLCA -0.488; TCGA BRCA -0.325; TCGA CESC -0.706; TCGA COAD -0.828; TCGA ESCA -0.477; TCGA KIRC -0.998; TCGA KIRP -0.608; TCGA LUSC -0.313; TCGA PRAD -0.525; TCGA STAD -0.505 |
hsa-miR-15b-3p | BMPER | 14 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.433; TCGA BRCA -0.479; TCGA COAD -0.425; TCGA ESCA -0.536; TCGA HNSC -0.332; TCGA KIRP -0.344; TCGA LIHC -0.487; TCGA LUAD -0.35; TCGA LUSC -0.385; TCGA OV -0.235; TCGA PAAD -0.331; TCGA PRAD -0.763; TCGA STAD -0.923; TCGA UCEC -0.7 |
hsa-miR-15b-3p | RERG | 16 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.65; TCGA BRCA -0.377; TCGA CESC -0.564; TCGA COAD -1.074; TCGA ESCA -0.391; TCGA HNSC -0.368; TCGA KIRC -0.172; TCGA KIRP -0.158; TCGA LIHC -0.168; TCGA LUAD -0.386; TCGA LUSC -0.332; TCGA PAAD -0.451; TCGA PRAD -0.383; TCGA SARC -0.427; TCGA STAD -0.985; TCGA UCEC -0.927 |
hsa-miR-15b-3p | MEGF9 | 9 cancers: BRCA; COAD; KIRC; LUAD; PAAD; PRAD; THCA; STAD; UCEC | MirTarget; mirMAP | TCGA BRCA -0.158; TCGA COAD -0.088; TCGA KIRC -0.327; TCGA LUAD -0.101; TCGA PAAD -0.109; TCGA PRAD -0.099; TCGA THCA -0.31; TCGA STAD -0.187; TCGA UCEC -0.115 |
hsa-miR-15b-3p | DCAF5 | 13 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRP; LGG; LIHC; LUSC; PAAD; SARC; STAD; UCEC | MirTarget | TCGA BRCA -0.095; TCGA CESC -0.083; TCGA COAD -0.059; TCGA ESCA -0.125; TCGA HNSC -0.144; TCGA KIRP -0.059; TCGA LGG -0.081; TCGA LIHC -0.064; TCGA LUSC -0.086; TCGA PAAD -0.054; TCGA SARC -0.105; TCGA STAD -0.125; TCGA UCEC -0.051 |
hsa-miR-15b-3p | ADAMTS19 | 10 cancers: BRCA; CESC; COAD; ESCA; KIRC; LGG; OV; PRAD; STAD; UCEC | MirTarget | TCGA BRCA -0.329; TCGA CESC -0.763; TCGA COAD -0.56; TCGA ESCA -0.41; TCGA KIRC -0.971; TCGA LGG -0.288; TCGA OV -0.603; TCGA PRAD -0.261; TCGA STAD -0.409; TCGA UCEC -0.575 |
hsa-miR-15b-3p | LZTFL1 | 9 cancers: BRCA; ESCA; KIRC; KIRP; LUAD; LUSC; OV; STAD; UCEC | MirTarget | TCGA BRCA -0.103; TCGA ESCA -0.083; TCGA KIRC -0.241; TCGA KIRP -0.081; TCGA LUAD -0.062; TCGA LUSC -0.129; TCGA OV -0.09; TCGA STAD -0.12; TCGA UCEC -0.155 |
hsa-miR-15b-3p | CFTR | 10 cancers: BRCA; CESC; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; PRAD; THCA | mirMAP | TCGA BRCA -0.254; TCGA CESC -0.504; TCGA HNSC -0.443; TCGA KIRC -0.962; TCGA KIRP -0.522; TCGA LUAD -0.494; TCGA LUSC -0.806; TCGA PAAD -1.048; TCGA PRAD -0.892; TCGA THCA -0.352 |
hsa-miR-15b-3p | F7 | 11 cancers: BRCA; CESC; ESCA; KIRC; KIRP; LGG; LIHC; PAAD; PRAD; THCA; UCEC | mirMAP | TCGA BRCA -0.588; TCGA CESC -0.367; TCGA ESCA -0.389; TCGA KIRC -0.667; TCGA KIRP -0.305; TCGA LGG -0.263; TCGA LIHC -0.436; TCGA PAAD -0.65; TCGA PRAD -0.385; TCGA THCA -0.421; TCGA UCEC -0.337 |
hsa-miR-15b-3p | AFF4 | 12 cancers: BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUSC; OV; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BRCA -0.076; TCGA CESC -0.101; TCGA COAD -0.074; TCGA ESCA -0.158; TCGA HNSC -0.107; TCGA LIHC -0.181; TCGA LUSC -0.106; TCGA OV -0.076; TCGA PRAD -0.083; TCGA SARC -0.076; TCGA STAD -0.155; TCGA UCEC -0.093 |
hsa-miR-15b-3p | LIN7A | 9 cancers: BRCA; CESC; ESCA; KIRC; KIRP; LIHC; PAAD; STAD; UCEC | mirMAP | TCGA BRCA -0.492; TCGA CESC -0.36; TCGA ESCA -0.347; TCGA KIRC -0.287; TCGA KIRP -0.259; TCGA LIHC -0.173; TCGA PAAD -0.243; TCGA STAD -0.206; TCGA UCEC -0.412 |
hsa-miR-15b-3p | SEMA5A | 9 cancers: BRCA; CESC; ESCA; HNSC; KIRC; LUAD; LUSC; PRAD; UCEC | mirMAP | TCGA BRCA -0.395; TCGA CESC -0.534; TCGA ESCA -0.254; TCGA HNSC -0.246; TCGA KIRC -0.22; TCGA LUAD -0.349; TCGA LUSC -0.592; TCGA PRAD -0.433; TCGA UCEC -0.277 |
hsa-miR-15b-3p | PTPRE | 9 cancers: BRCA; HNSC; LGG; LUAD; LUSC; PRAD; SARC; THCA; UCEC | mirMAP | TCGA BRCA -0.136; TCGA HNSC -0.146; TCGA LGG -0.09; TCGA LUAD -0.301; TCGA LUSC -0.462; TCGA PRAD -0.153; TCGA SARC -0.158; TCGA THCA -0.301; TCGA UCEC -0.098 |
hsa-miR-15b-3p | FGD4 | 10 cancers: BRCA; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PRAD; SARC | mirMAP | TCGA BRCA -0.07; TCGA ESCA -0.391; TCGA HNSC -0.145; TCGA KIRC -0.09; TCGA KIRP -0.116; TCGA LIHC -0.266; TCGA LUAD -0.126; TCGA LUSC -0.34; TCGA PRAD -0.146; TCGA SARC -0.284 |
hsa-miR-15b-3p | PPP1R9A | 9 cancers: BRCA; ESCA; HNSC; KIRC; KIRP; LGG; OV; THCA; STAD | mirMAP | TCGA BRCA -0.093; TCGA ESCA -0.747; TCGA HNSC -0.518; TCGA KIRC -0.28; TCGA KIRP -0.175; TCGA LGG -0.103; TCGA OV -0.081; TCGA THCA -0.118; TCGA STAD -0.555 |
hsa-miR-15b-3p | MARCH8 | 11 cancers: BRCA; CESC; ESCA; KIRC; LGG; LIHC; LUSC; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BRCA -0.178; TCGA CESC -0.174; TCGA ESCA -0.109; TCGA KIRC -0.144; TCGA LGG -0.216; TCGA LIHC -0.201; TCGA LUSC -0.118; TCGA PRAD -0.19; TCGA SARC -0.173; TCGA STAD -0.12; TCGA UCEC -0.174 |
hsa-miR-15b-3p | ZIK1 | 11 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUSC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BRCA -0.119; TCGA CESC -0.411; TCGA COAD -0.619; TCGA ESCA -0.321; TCGA HNSC -0.293; TCGA KIRC -0.256; TCGA LUSC -0.346; TCGA PAAD -0.292; TCGA PRAD -0.228; TCGA STAD -0.473; TCGA UCEC -0.349 |
hsa-miR-15b-3p | OPCML | 9 cancers: BRCA; COAD; ESCA; KIRC; KIRP; LGG; LUAD; STAD; UCEC | mirMAP | TCGA BRCA -0.26; TCGA COAD -0.743; TCGA ESCA -0.797; TCGA KIRC -0.48; TCGA KIRP -0.382; TCGA LGG -0.182; TCGA LUAD -0.37; TCGA STAD -0.483; TCGA UCEC -0.702 |
hsa-miR-15b-3p | LUZP2 | 9 cancers: BRCA; COAD; HNSC; LGG; LIHC; LUAD; PRAD; STAD; UCEC | mirMAP | TCGA BRCA -0.849; TCGA COAD -0.699; TCGA HNSC -0.438; TCGA LGG -0.503; TCGA LIHC -0.655; TCGA LUAD -0.55; TCGA PRAD -0.227; TCGA STAD -0.599; TCGA UCEC -0.227 |
hsa-miR-15b-3p | ACADSB | 10 cancers: BRCA; CESC; ESCA; KIRC; KIRP; LGG; LIHC; SARC; STAD; UCEC | mirMAP | TCGA BRCA -0.354; TCGA CESC -0.181; TCGA ESCA -0.191; TCGA KIRC -0.552; TCGA KIRP -0.112; TCGA LGG -0.12; TCGA LIHC -0.338; TCGA SARC -0.158; TCGA STAD -0.125; TCGA UCEC -0.119 |
hsa-miR-15b-3p | ZNF655 | 9 cancers: BRCA; CESC; HNSC; KIRP; LIHC; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BRCA -0.128; TCGA CESC -0.123; TCGA HNSC -0.074; TCGA KIRP -0.174; TCGA LIHC -0.075; TCGA PRAD -0.516; TCGA SARC -0.101; TCGA STAD -0.22; TCGA UCEC -0.133 |
hsa-miR-15b-3p | ZNF69 | 10 cancers: BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; OV; THCA | mirMAP | TCGA BRCA -0.082; TCGA CESC -0.306; TCGA ESCA -0.354; TCGA HNSC -0.245; TCGA KIRC -0.204; TCGA KIRP -0.223; TCGA LUAD -0.237; TCGA LUSC -0.422; TCGA OV -0.222; TCGA THCA -0.063 |
hsa-miR-15b-3p | ZNF10 | 11 cancers: BRCA; CESC; ESCA; KIRC; KIRP; LGG; OV; PAAD; SARC; STAD; UCEC | mirMAP | TCGA BRCA -0.096; TCGA CESC -0.131; TCGA ESCA -0.332; TCGA KIRC -0.262; TCGA KIRP -0.181; TCGA LGG -0.052; TCGA OV -0.125; TCGA PAAD -0.229; TCGA SARC -0.142; TCGA STAD -0.331; TCGA UCEC -0.149 |
hsa-miR-15b-3p | ZNF225 | 9 cancers: BRCA; CESC; COAD; ESCA; KIRC; KIRP; SARC; STAD; UCEC | mirMAP | TCGA BRCA -0.068; TCGA CESC -0.132; TCGA COAD -0.07; TCGA ESCA -0.126; TCGA KIRC -0.069; TCGA KIRP -0.061; TCGA SARC -0.083; TCGA STAD -0.121; TCGA UCEC -0.066 |
hsa-miR-15b-3p | ESRRG | 9 cancers: BRCA; ESCA; HNSC; KIRC; KIRP; OV; PAAD; PRAD; STAD | miRNATAP | TCGA BRCA -0.368; TCGA ESCA -0.728; TCGA HNSC -0.637; TCGA KIRC -1.083; TCGA KIRP -0.281; TCGA OV -0.501; TCGA PAAD -0.541; TCGA PRAD -0.276; TCGA STAD -0.336 |
hsa-miR-15b-3p | KIDINS220 | 10 cancers: CESC; COAD; KIRC; LGG; LUAD; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA CESC -0.112; TCGA COAD -0.111; TCGA KIRC -0.146; TCGA LGG -0.06; TCGA LUAD -0.051; TCGA PAAD -0.136; TCGA PRAD -0.065; TCGA SARC -0.114; TCGA STAD -0.17; TCGA UCEC -0.108 |
hsa-miR-15b-3p | INSR | 9 cancers: CESC; ESCA; HNSC; LIHC; PAAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA CESC -0.215; TCGA ESCA -0.222; TCGA HNSC -0.111; TCGA LIHC -0.207; TCGA PAAD -0.117; TCGA SARC -0.2; TCGA THCA -0.237; TCGA STAD -0.108; TCGA UCEC -0.261 |
hsa-miR-15b-3p | KCTD21 | 10 cancers: CESC; COAD; HNSC; LGG; LIHC; LUAD; OV; SARC; THCA; STAD | mirMAP | TCGA CESC -0.08; TCGA COAD -0.1; TCGA HNSC -0.17; TCGA LGG -0.096; TCGA LIHC -0.211; TCGA LUAD -0.064; TCGA OV -0.095; TCGA SARC -0.188; TCGA THCA -0.091; TCGA STAD -0.101 |
hsa-miR-15b-3p | ZNF585B | 9 cancers: CESC; COAD; ESCA; HNSC; KIRC; KIRP; PAAD; PRAD; STAD | mirMAP | TCGA CESC -0.282; TCGA COAD -0.137; TCGA ESCA -0.347; TCGA HNSC -0.153; TCGA KIRC -0.123; TCGA KIRP -0.149; TCGA PAAD -0.226; TCGA PRAD -0.102; TCGA STAD -0.333 |
hsa-miR-15b-3p | MFAP3L | 9 cancers: COAD; HNSC; KIRC; LIHC; LUAD; LUSC; PAAD; PRAD; STAD | mirMAP; miRNATAP | TCGA COAD -0.232; TCGA HNSC -0.35; TCGA KIRC -0.469; TCGA LIHC -0.423; TCGA LUAD -0.137; TCGA LUSC -0.399; TCGA PAAD -0.261; TCGA PRAD -0.22; TCGA STAD -0.374 |
hsa-miR-15b-3p | MTSS1 | 9 cancers: HNSC; KIRC; KIRP; LGG; LIHC; LUAD; PRAD; STAD; UCEC | mirMAP | TCGA HNSC -0.094; TCGA KIRC -0.192; TCGA KIRP -0.226; TCGA LGG -0.124; TCGA LIHC -0.195; TCGA LUAD -0.193; TCGA PRAD -0.115; TCGA STAD -0.229; TCGA UCEC -0.099 |
Enriched cancer pathways of putative targets