microRNA information: hsa-miR-16-5p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-16-5p | miRbase |
| Accession: | MIMAT0000069 | miRbase |
| Precursor name: | hsa-mir-16-1 | miRbase |
| Precursor accession: | MI0000070 | miRbase |
| Symbol: | MIR16-1 | HGNC |
| RefSeq ID: | NR_029486 | GenBank |
| Sequence: | UAGCAGCACGUAAAUAUUGGCG |
Reported expression in cancers: hsa-miR-16-5p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-16-5p | B cell lymphoma | downregulation | "We measured the levels of miRNAs miR-15a miR-16-1 ......" | 21987025 | Reverse transcription PCR; qPCR |
| hsa-miR-16-5p | bladder cancer | downregulation | "MicroRNA-16 miR-16 has been demonstrated to regula ......" | 23991964 | |
| hsa-miR-16-5p | bladder cancer | upregulation | "Identification and validation of reference genes f ......" | 25738263 | qPCR; RNA-Seq |
| hsa-miR-16-5p | breast cancer | upregulation | "Western blot and QPCR were used to determine the e ......" | 22260523 | qPCR |
| hsa-miR-16-5p | breast cancer | upregulation | "miR-16 which is located in 13q14 chromosome plays ......" | 25672252 | |
| hsa-miR-16-5p | cervical and endocervical cancer | deregulation | "Using miRNA array analyses for age-matched normal ......" | 18596939 | Northern blot; Microarray |
| hsa-miR-16-5p | chronic myeloid leukemia | deregulation | "Some onco-miRNAs were found to be downregulated mi ......" | 25833191 | |
| hsa-miR-16-5p | colon cancer | downregulation | "miR-15 microRNA 15 and miR-16 are frequently delet ......" | 22574716 | |
| hsa-miR-16-5p | colorectal cancer | downregulation | "The in vivo significance of expression of miR-16/1 ......" | 21390519 | qPCR |
| hsa-miR-16-5p | colorectal cancer | downregulation | "miR-16 has been reported to be downregulated and t ......" | 23380758 | |
| hsa-miR-16-5p | colorectal cancer | downregulation | "The aim of this study was to detect the expression ......" | 24045965 | Reverse transcription PCR; qPCR |
| hsa-miR-16-5p | colorectal cancer | downregulation | "The aim of this study was to reveal the associatio ......" | 25435873 | qPCR |
| hsa-miR-16-5p | esophageal cancer | upregulation | "Plasma levels of miR-16 miR-21 miR-22 miR-126 miR- ......" | 26221263 | qPCR |
| hsa-miR-16-5p | gastric cancer | upregulation | "We found that miR-16 and miR-21 were upregulated u ......" | 21081469 | |
| hsa-miR-16-5p | gastric cancer | upregulation | "miRNA expression was determined using a custom-des ......" | 22112324 | Microarray |
| hsa-miR-16-5p | gastric cancer | upregulation | "High expression of miR 16 and miR 451 predicating ......" | 27605261 | Microarray; in situ hybridization |
| hsa-miR-16-5p | head and neck cancer | deregulation | "A global miRNA profiling was done on 51 formalin-f ......" | 20145181 | Reverse transcription PCR |
| hsa-miR-16-5p | kidney renal cell cancer | upregulation | "The present study aimed to determine the expressio ......" | 25815587 | qPCR |
| hsa-miR-16-5p | liver cancer | upregulation | "Up-regulation of miR-15a miR-16 miR-27b miR-30b mi ......" | 24314246 | qPCR |
| hsa-miR-16-5p | liver cancer | downregulation | "Expression of serum miR 16 let 7f and miR 21 in pa ......" | 24697119 | Reverse transcription PCR; qPCR |
| hsa-miR-16-5p | liver cancer | deregulation | "Interestingly miR-195 miR-25 and miR-16 were up-re ......" | 25519019 | |
| hsa-miR-16-5p | lung squamous cell cancer | upregulation | "Global miRNA microarray expression profiling based ......" | 23774211 | qPCR; Microarray |
| hsa-miR-16-5p | lung squamous cell cancer | downregulation | "MicroRNAs play important roles in the development ......" | 23954293 | |
| hsa-miR-16-5p | lung squamous cell cancer | deregulation | "Identification of serum miRNAs by nano quantum dot ......" | 26695145 | Microarray; qPCR |
| hsa-miR-16-5p | lymphoma | upregulation | "The major findings were: the detection of a panel ......" | 19945163 | |
| hsa-miR-16-5p | lymphoma | upregulation | "The expression levels of miR-16 were examined by T ......" | 24447552 | qPCR |
| hsa-miR-16-5p | lymphoma | upregulation | "Expression of miR 16 in patients with T lymphoblas ......" | 24598659 | |
| hsa-miR-16-5p | ovarian cancer | downregulation | "MiR 15a and MiR 16 control Bmi 1 expression in ova ......" | 19903841 | |
| hsa-miR-16-5p | prostate cancer | deregulation | "This study aimed to investigate the microRNA miRNA ......" | 19676045 | qPCR; Microarray |
| hsa-miR-16-5p | prostate cancer | downregulation | "Control of tumor and microenvironment cross talk b ......" | 21532615 | |
| hsa-miR-16-5p | prostate cancer | upregulation | "Next generation sequencing technology was applied ......" | 21980368 | RNA-Seq |
| hsa-miR-16-5p | sarcoma | downregulation | "The signature includes high expression of miR-181a ......" | 22350417 | qPCR |
| hsa-miR-16-5p | sarcoma | downregulation | "Similar analysis in mouse metastatic sarcomas reve ......" | 26044957 | |
| hsa-miR-16-5p | thyroid cancer | downregulation | "We also find that the expression of miR-195 is dow ......" | 26527888 |
Reported cancer pathway affected by hsa-miR-16-5p
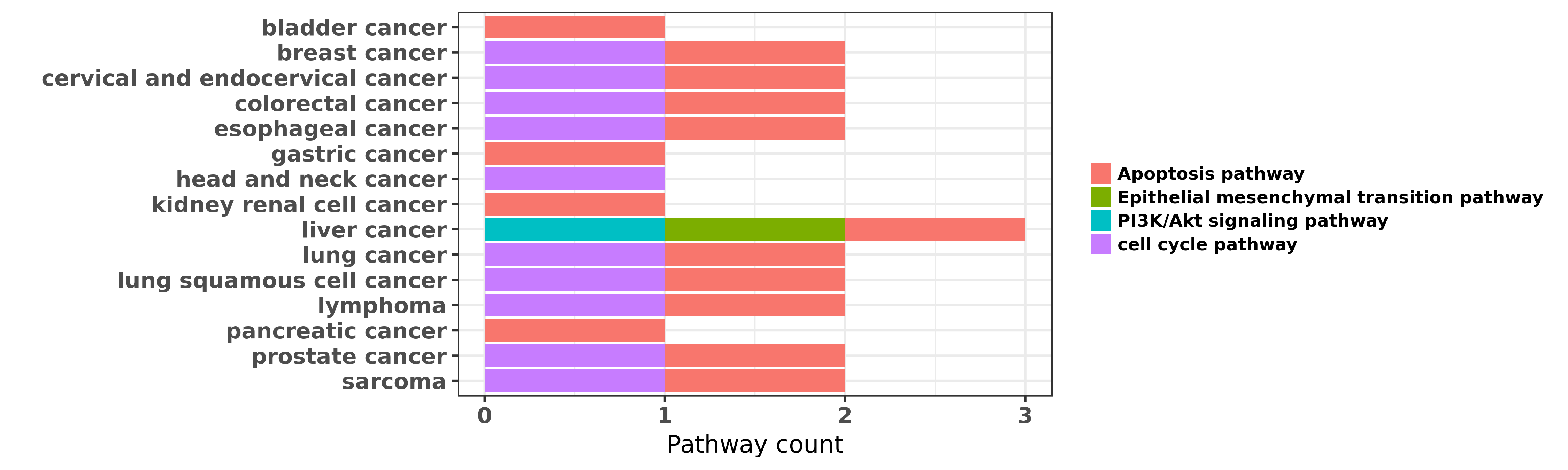
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-16-5p | bladder cancer | Apoptosis pathway | "MicroRNA-16 miR-16 has been demonstrated to regula ......" | 23991964 | |
| hsa-miR-16-5p | bladder cancer | Apoptosis pathway | "Artesunate induces apoptosis of bladder cancer cel ......" | 25196524 | Flow cytometry; MTT assay |
| hsa-miR-16-5p | breast cancer | cell cycle pathway | "Downregulation of the tumor suppressor miR 16 via ......" | 22583478 | Western blot |
| hsa-miR-16-5p | breast cancer | cell cycle pathway; Apoptosis pathway | "miR-16 which is located in 13q14 chromosome plays ......" | 25672252 | MTT assay |
| hsa-miR-16-5p | breast cancer | Apoptosis pathway | "Here we showed that overexpression of miR-16 promo ......" | 26993770 | |
| hsa-miR-16-5p | breast cancer | Apoptosis pathway | "Renilla luciferase reporter assay was performed to ......" | 27596816 | Luciferase; MTT assay; Western blot; Wound Healing Assay |
| hsa-miR-16-5p | cervical and endocervical cancer | Apoptosis pathway; cell cycle pathway | "MicroRNA miR 16 1 regulates CCNE1 cyclin E1 gene e ......" | 26629104 | |
| hsa-miR-16-5p | colorectal cancer | Apoptosis pathway | "miR-16 has been reported to be downregulated and t ......" | 23380758 | |
| hsa-miR-16-5p | colorectal cancer | cell cycle pathway | "Clinicopathologic features clinical outcomes and s ......" | 26934556 | |
| hsa-miR-16-5p | colorectal cancer | Apoptosis pathway | "Effective Targeting Survivin Caspase 3 and MicroRN ......" | 27055667 | |
| hsa-miR-16-5p | esophageal cancer | Apoptosis pathway; cell cycle pathway | "MiR 16 induced the suppression of cell apoptosis w ......" | 24852767 | Western blot; Luciferase |
| hsa-miR-16-5p | gastric cancer | Apoptosis pathway | "miR 15b and miR 16 modulate multidrug resistance b ......" | 18449891 | Luciferase |
| hsa-miR-16-5p | gastric cancer | Apoptosis pathway | "In order to identify miRNA signatures for gastric ......" | 22112324 | |
| hsa-miR-16-5p | head and neck cancer | cell cycle pathway | "A global miRNA profiling was done on 51 formalin-f ......" | 20145181 | Flow cytometry |
| hsa-miR-16-5p | kidney renal cell cancer | Apoptosis pathway | "The present study aimed to determine the expressio ......" | 25815587 | |
| hsa-miR-16-5p | kidney renal cell cancer | Apoptosis pathway | "Herein we sought to investigate the impact of gemc ......" | 25833690 | Western blot; MTT assay |
| hsa-miR-16-5p | liver cancer | Apoptosis pathway | "P glycoprotein enhances radiation induced apoptoti ......" | 21336967 | |
| hsa-miR-16-5p | liver cancer | Apoptosis pathway; PI3K/Akt signaling pathway; Epithelial mesenchymal transition pathway | "miR-16 is known to be abnormally expressed in hepa ......" | 26499886 | MTT assay; Western blot |
| hsa-miR-16-5p | lung cancer | Apoptosis pathway | "MiR 16 targets Bcl 2 in paclitaxel resistant lung ......" | 25435430 | |
| hsa-miR-16-5p | lung cancer | Apoptosis pathway; cell cycle pathway | "Effect of mir 16 on proliferation and apoptosis in ......" | 26064212 | Flow cytometry; Cell Proliferation Assay; Luciferase |
| hsa-miR-16-5p | lung squamous cell cancer | cell cycle pathway; Apoptosis pathway | "miR 15a and miR 16 are implicated in cell cycle re ......" | 19549910 | RNAi |
| hsa-miR-16-5p | lung squamous cell cancer | Apoptosis pathway | "MicroRNA 16 1 Inhibits Tumor Cell Proliferation an ......" | 27712591 | MTT assay; Western blot |
| hsa-miR-16-5p | lymphoma | cell cycle pathway; Apoptosis pathway | "MicroRNA profiles were correlated with correspondi ......" | 21960592 | |
| hsa-miR-16-5p | lymphoma | Apoptosis pathway | "On screening for upstream regulators of BMI1 we fo ......" | 23686310 | |
| hsa-miR-16-5p | lymphoma | Apoptosis pathway | "Here we demonstrate that a noncoding regulatory RN ......" | 26640145 | |
| hsa-miR-16-5p | pancreatic cancer | Apoptosis pathway | "The overall objective of our investigation was to ......" | 22966344 | Luciferase |
| hsa-miR-16-5p | prostate cancer | Apoptosis pathway | "The miR 15a miR 16 1 cluster controls prostate can ......" | 18931683 | |
| hsa-miR-16-5p | prostate cancer | cell cycle pathway | "Here we show that microRNA miR-16 which is express ......" | 19738602 | |
| hsa-miR-16-5p | sarcoma | cell cycle pathway; Apoptosis pathway | "miR 15a and miR 16 1 downregulate CCND1 and induce ......" | 22922827 | |
| hsa-miR-16-5p | sarcoma | cell cycle pathway | "c Myc Represses Tumor Suppressive microRNAs let 7a ......" | 26393798 |
Reported cancer prognosis affected by hsa-miR-16-5p

| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-16-5p | breast cancer | malignant trasformation | "Real-time quantitative PCR RQ-PCR is a sensitive a ......" | 18718003 | |
| hsa-miR-16-5p | breast cancer | drug resistance | "MiRNA microarray analysis identified 299 and 226 m ......" | 21399894 | |
| hsa-miR-16-5p | breast cancer | tumorigenesis | "Downregulation of the tumor suppressor miR 16 via ......" | 22583478 | Western blot |
| hsa-miR-16-5p | breast cancer | poor survival | "Two miRNA signatures miR-16 155 125b 374a and miR- ......" | 23405235 | |
| hsa-miR-16-5p | breast cancer | poor survival | "miR-16 which is located in 13q14 chromosome plays ......" | 25672252 | MTT assay |
| hsa-miR-16-5p | breast cancer | metastasis; progression | "Aberrant plasma levels of circulating miR 16 miR 1 ......" | 26033453 | |
| hsa-miR-16-5p | breast cancer | cell migration; motility | "To address this question miRNA expression profiles ......" | 26684238 | |
| hsa-miR-16-5p | breast cancer | metastasis | "miR 16 5p Is a Stably Expressed Housekeeping Micro ......" | 26821018 | |
| hsa-miR-16-5p | breast cancer | staging; metastasis; drug resistance | "Here we showed that overexpression of miR-16 promo ......" | 26993770 | |
| hsa-miR-16-5p | cervical and endocervical cancer | tumorigenesis | "MicroRNA miR 16 1 regulates CCNE1 cyclin E1 gene e ......" | 26629104 | |
| hsa-miR-16-5p | cervical and endocervical cancer | tumorigenesis | "Serum miRNAs panel miR 16 2* miR 195 miR 2861 miR ......" | 26656154 | |
| hsa-miR-16-5p | colon cancer | progression; poor survival | "miR-15 microRNA 15 and miR-16 are frequently delet ......" | 22574716 | Colony formation; Western blot |
| hsa-miR-16-5p | colon cancer | drug resistance; poor survival | "miR 15a and miR 16 modulate drug resistance by tar ......" | 25623762 | Western blot |
| hsa-miR-16-5p | colorectal cancer | differentiation | "miR-16 has been reported to be downregulated and t ......" | 23380758 | |
| hsa-miR-16-5p | colorectal cancer | staging; metastasis; differentiation; recurrence; progression; worse prognosis | "The aim of this study was to detect the expression ......" | 24045965 | |
| hsa-miR-16-5p | colorectal cancer | cell migration | "AP4 downregulation by p53 was mediated indirectly ......" | 24285725 | |
| hsa-miR-16-5p | colorectal cancer | worse prognosis; staging; metastasis; poor survival; progression | "The aim of this study was to reveal the associatio ......" | 25435873 | |
| hsa-miR-16-5p | colorectal cancer | progression | "Of these 3 upregulated let-7b miR-1290 and miR-126 ......" | 27698796 | |
| hsa-miR-16-5p | esophageal cancer | cell migration | "MiR 16 induced the suppression of cell apoptosis w ......" | 24852767 | Western blot; Luciferase |
| hsa-miR-16-5p | esophageal cancer | progression; poor survival | "Plasma levels of miR-16 miR-21 miR-22 miR-126 miR- ......" | 26221263 | |
| hsa-miR-16-5p | gastric cancer | drug resistance | "miR 15b and miR 16 modulate multidrug resistance b ......" | 18449891 | Luciferase |
| hsa-miR-16-5p | gastric cancer | drug resistance | "In order to identify miRNA signatures for gastric ......" | 22112324 | |
| hsa-miR-16-5p | gastric cancer | progression; staging; differentiation | "Circulating MiR 16 5p and MiR 19b 3p as Two Novel ......" | 25897338 | |
| hsa-miR-16-5p | gastric cancer | drug resistance; staging | "MiR 16 mediates trastuzumab and lapatinib response ......" | 27157613 | |
| hsa-miR-16-5p | gastric cancer | worse prognosis; staging; poor survival; tumor size; metastasis | "High expression of miR 16 and miR 451 predicating ......" | 27605261 | |
| hsa-miR-16-5p | kidney renal cell cancer | malignant trasformation | "We performed genome-wide expression profiling of m ......" | 19228262 | |
| hsa-miR-16-5p | kidney renal cell cancer | tumorigenesis | "The present study aimed to determine the expressio ......" | 25815587 | |
| hsa-miR-16-5p | liver cancer | staging | "RNA was extracted from the sera and miR-21 as well ......" | 22066022 | |
| hsa-miR-16-5p | liver cancer | worse prognosis | "miRNAs are small non-coding RNAs which target comp ......" | 24496037 | |
| hsa-miR-16-5p | liver cancer | tumor size; recurrence | "Expression of serum miR 16 let 7f and miR 21 in pa ......" | 24697119 | |
| hsa-miR-16-5p | liver cancer | progression | "We analysed the expression of mature miRNA-122 miR ......" | 26133725 | |
| hsa-miR-16-5p | liver cancer | metastasis; cell migration | "miR-16 is known to be abnormally expressed in hepa ......" | 26499886 | MTT assay; Western blot |
| hsa-miR-16-5p | lung cancer | drug resistance | "MiR 16 targets Bcl 2 in paclitaxel resistant lung ......" | 25435430 | |
| hsa-miR-16-5p | lung cancer | tumorigenesis | "Lung tumorigenesis induced by human vascular endot ......" | 25912305 | Western blot |
| hsa-miR-16-5p | lung squamous cell cancer | drug resistance | "miR 15a and miR 16 are implicated in cell cycle re ......" | 19549910 | RNAi |
| hsa-miR-16-5p | lung squamous cell cancer | poor survival | "The clinical and functional significance of RNA-in ......" | 23349018 | RNAi |
| hsa-miR-16-5p | lung squamous cell cancer | poor survival | "Global miRNA microarray expression profiling based ......" | 23774211 | |
| hsa-miR-16-5p | lung squamous cell cancer | motility; progression; cell migration | "Downregulation of miR 16 promotes growth and motil ......" | 23954293 | Colony formation |
| hsa-miR-16-5p | lung squamous cell cancer | staging | "However the study demonstrated a correlation betwe ......" | 25384507 | |
| hsa-miR-16-5p | lung squamous cell cancer | progression | "MicroRNA 16 1 Inhibits Tumor Cell Proliferation an ......" | 27712591 | MTT assay; Western blot |
| hsa-miR-16-5p | lymphoma | differentiation | "MicroRNA profiles were correlated with correspondi ......" | 21960592 | |
| hsa-miR-16-5p | lymphoma | tumor size; progression | "On screening for upstream regulators of BMI1 we fo ......" | 23686310 | |
| hsa-miR-16-5p | lymphoma | worse prognosis; poor survival | "To study the expression of miR-16 and bcl-2 in T l ......" | 24447552 | |
| hsa-miR-16-5p | lymphoma | worse prognosis; poor survival | "Expression of miR 16 in patients with T lymphoblas ......" | 24598659 | |
| hsa-miR-16-5p | lymphoma | staging; progression | "Herein we used a global quantitative real-time pol ......" | 25503151 | |
| hsa-miR-16-5p | lymphoma | staging; progression; drug resistance | "Here we demonstrate that a noncoding regulatory RN ......" | 26640145 | |
| hsa-miR-16-5p | melanoma | staging; metastasis | "Our analyses identified a 4-microRNA miR-150-5p mi ......" | 26089374 | |
| hsa-miR-16-5p | melanoma | staging; poor survival; worse prognosis | "In a hospital based case-control study we found a ......" | 26829037 | |
| hsa-miR-16-5p | ovarian cancer | drug resistance | "MiR 15a and MiR 16 control Bmi 1 expression in ova ......" | 19903841 | |
| hsa-miR-16-5p | ovarian cancer | poor survival; recurrence | "Three miRNAs hsa-miR-16 hsa-miR-22* and ebv-miR-BH ......" | 23554906 | |
| hsa-miR-16-5p | ovarian cancer | progression; drug resistance; poor survival | "Here we report that multiple mechanisms that promo ......" | 26918603 | |
| hsa-miR-16-5p | pancreatic cancer | poor survival | "The overall objective of our investigation was to ......" | 22966344 | Luciferase |
| hsa-miR-16-5p | prostate cancer | poor survival; staging | "The miR 15a miR 16 1 cluster controls prostate can ......" | 18931683 | |
| hsa-miR-16-5p | prostate cancer | metastasis; staging | "Here we show that microRNA miR-16 which is express ......" | 19738602 | |
| hsa-miR-16-5p | prostate cancer | malignant trasformation | "Real time quantitative PCR qPCR is the method of c ......" | 20890088 | |
| hsa-miR-16-5p | prostate cancer | progression; poor survival; staging | "Control of tumor and microenvironment cross talk b ......" | 21532615 | |
| hsa-miR-16-5p | prostate cancer | metastasis | "Next generation sequencing technology was applied ......" | 21980368 | |
| hsa-miR-16-5p | sarcoma | metastasis | "Similar analysis in mouse metastatic sarcomas reve ......" | 26044957 | |
| hsa-miR-16-5p | sarcoma | progression | "c Myc Represses Tumor Suppressive microRNAs let 7a ......" | 26393798 | |
| hsa-miR-16-5p | sarcoma | differentiation | "GLIPR1 inhibits the proliferation and induces the ......" | 27460987 | |
| hsa-miR-16-5p | thyroid cancer | metastasis | "These miRNAs and mRNAs were further validated by q ......" | 24064622 | Luciferase |
Reported gene related to hsa-miR-16-5p

| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-16-5p | colon cancer | BCL2 | "miR 15a and miR 16 modulate drug resistance by tar ......" | 25623762 |
| hsa-miR-16-5p | gastric cancer | BCL2 | "miR 15b and miR 16 modulate multidrug resistance b ......" | 18449891 |
| hsa-miR-16-5p | liver cancer | BCL2 | "P glycoprotein enhances radiation induced apoptoti ......" | 21336967 |
| hsa-miR-16-5p | lung cancer | BCL2 | "We demonstrated that anti-apoptotic protein Bcl-2 ......" | 25435430 |
| hsa-miR-16-5p | lymphoma | BCL2 | "To study the expression of miR-16 and bcl-2 in T l ......" | 24447552 |
| hsa-miR-16-5p | lymphoma | BCL2 | "The miR-16 expression correlated with BCL-2 protei ......" | 24598659 |
| hsa-miR-16-5p | pancreatic cancer | BCL2 | "The overall objective of our investigation was to ......" | 22966344 |
| hsa-miR-16-5p | prostate cancer | BCL2 | "MicroRNAs miRNAs are noncoding small RNAs that rep ......" | 18931683 |
| hsa-miR-16-5p | breast cancer | BMI1 | "Our results suggest that overexpression of miR-15a ......" | 27596816 |
| hsa-miR-16-5p | lymphoma | BMI1 | "Finally we found that a proteasome inhibitor borte ......" | 23686310 |
| hsa-miR-16-5p | ovarian cancer | BMI1 | "Significant correlations between low expression of ......" | 26918603 |
| hsa-miR-16-5p | bladder cancer | CCND1 | "At the molecular level our results further reveale ......" | 23991964 |
| hsa-miR-16-5p | lymphoma | CCND1 | "Truncation in CCND1 mRNA alters miR 16 1 regulatio ......" | 18483394 |
| hsa-miR-16-5p | sarcoma | CCND1 | "CCND1 has been found to be a target of miR-15a and ......" | 22922827 |
| hsa-miR-16-5p | breast cancer | CCNE1 | "In contrast miR-16 blocks HuR-mediated upregulatio ......" | 25830480 |
| hsa-miR-16-5p | breast cancer | CCNE1 | "A search for miR-16 targets showed that the CCNE1 ......" | 22583478 |
| hsa-miR-16-5p | cervical and endocervical cancer | CCNE1 | "Thus in the present brief communication we determi ......" | 26629104 |
| hsa-miR-16-5p | bladder cancer | PCNA | "At the molecular level our results further reveale ......" | 23991964 |
| hsa-miR-16-5p | breast cancer | PCNA | "In contrast miR-16 blocks HuR-mediated upregulatio ......" | 25830480 |
| hsa-miR-16-5p | breast cancer | PCNA | "Target genes of miR-16 were searched through a bio ......" | 22583478 |
| hsa-miR-16-5p | colorectal cancer | TP53 | "In the present study we sought to investigate the ......" | 23380758 |
| hsa-miR-16-5p | lung squamous cell cancer | TP53 | "p53 regulates the maturation process of miR-16 and ......" | 21400525 |
| hsa-miR-16-5p | lymphoma | TP53 | "Alternatively in CTCL cells lacking functional p53 ......" | 26640145 |
| hsa-miR-16-5p | liver cancer | AFP | "The combination of serum miR-16 with serum alpha f ......" | 26133725 |
| hsa-miR-16-5p | liver cancer | AFP | "The combination of miR-16 AFP AFP-L3% and DCP yiel ......" | 21278583 |
| hsa-miR-16-5p | colorectal cancer | CASP3 | "Effective Targeting Survivin Caspase 3 and MicroRN ......" | 27055667 |
| hsa-miR-16-5p | lung cancer | CASP3 | "Moreover in this report we showed that the combine ......" | 25435430 |
| hsa-miR-16-5p | breast cancer | MYC | "miR-16 expression was studied by RT-qPCR in cancer ......" | 22583478 |
| hsa-miR-16-5p | gastric cancer | MYC | "TZ and lapatinib ability to block extracellular si ......" | 27157613 |
| hsa-miR-16-5p | bladder cancer | PTGS2 | "Artesunate induces apoptosis of bladder cancer cel ......" | 25196524 |
| hsa-miR-16-5p | liver cancer | PTGS2 | "MiR-16 and miR-101 levels do not correlate with CO ......" | 24759835 |
| hsa-miR-16-5p | lung cancer | VEGFA | "We further demonstrated that the intranasal admini ......" | 25912305 |
| hsa-miR-16-5p | lymphoma | VEGFA | "First we observed that ALK expression leads to dow ......" | 21778999 |
| hsa-miR-16-5p | bladder cancer | AGRP | "ART efficiently inhibited orthotopic tumor growth ......" | 25196524 |
| hsa-miR-16-5p | lymphoma | ALK | "First we observed that ALK expression leads to dow ......" | 21778999 |
| hsa-miR-16-5p | ovarian cancer | ATP7B | "Additionally epithelial to mesenchymal transition ......" | 26918603 |
| hsa-miR-16-5p | lung cancer | BECN1 | "Combined overexpression of miR-16 and miR-17 great ......" | 25435430 |
| hsa-miR-16-5p | colon cancer | CCNB1 | "The levels of miR-15a and miR-16-1 in colon cancer ......" | 22574716 |
| hsa-miR-16-5p | prostate cancer | CDK1 | "Cell model studies indicate that miR-16 likely sup ......" | 19738602 |
| hsa-miR-16-5p | prostate cancer | CDK2 | "Cell model studies indicate that miR-16 likely sup ......" | 19738602 |
| hsa-miR-16-5p | lung cancer | CLDN2 | "Quercetin Decreases Claudin 2 Expression Mediated ......" | 26061016 |
| hsa-miR-16-5p | breast cancer | ELAVL1 | "Loss of repression of HuR translation by miR 16 ma ......" | 20626035 |
| hsa-miR-16-5p | liver cancer | F2 | "The expression of serum miR-16 is down-regulated i ......" | 24697119 |
| hsa-miR-16-5p | breast cancer | FASN | "Both miR-15a and miR-16-1 contributes to inhibitin ......" | 27713175 |
| hsa-miR-16-5p | prostate cancer | FGF2 | "Such downregulation of miR-15 and miR-16 in cancer ......" | 21532615 |
| hsa-miR-16-5p | prostate cancer | FGFR1 | "Such downregulation of miR-15 and miR-16 in cancer ......" | 21532615 |
| hsa-miR-16-5p | gastric cancer | FUBP1 | "Supporting the clinical relevance of our results w ......" | 27157613 |
| hsa-miR-16-5p | sarcoma | GLIPR1 | "GLIPR1 inhibits the proliferation and induces the ......" | 27460987 |
| hsa-miR-16-5p | bladder cancer | GLS2 | "The RNA levels of urothelial carcinoma-associated ......" | 26373319 |
| hsa-miR-16-5p | lung squamous cell cancer | HDGF | "Downregulation of miR 16 promotes growth and motil ......" | 23954293 |
| hsa-miR-16-5p | gastric cancer | HGF | "Direct targeting of HGF by miR 16 regulates prolif ......" | 27683052 |
| hsa-miR-16-5p | ovarian cancer | HGS | "Significant correlations between low expression of ......" | 26918603 |
| hsa-miR-16-5p | prostate cancer | HPRT1 | "The levels of miR-16 and RNU1A-1 were reliably mea ......" | 21539977 |
| hsa-miR-16-5p | sarcoma | IGF1R | "Furthermore we confirmed that IGF1R is a direct ta ......" | 23507142 |
| hsa-miR-16-5p | breast cancer | IKBKB | "Furthermore IκB kinase β IKBKB was identified as ......" | 26993770 |
| hsa-miR-16-5p | colorectal cancer | KDR | "The expression levels of two target genes Myb and ......" | 26934556 |
| hsa-miR-16-5p | kidney renal cell cancer | LARP6 | "The impact of miR-16 on cell proliferation migrati ......" | 25815587 |
| hsa-miR-16-5p | colorectal cancer | MYB | "The expression levels of two target genes Myb and ......" | 26934556 |
| hsa-miR-16-5p | gastric cancer | NFASC | "NF κB targets miR 16 and miR 21 in gastric cancer ......" | 21081469 |
| hsa-miR-16-5p | liver cancer | PGP | "RHepG2 cells the multidrug resistant subline of hu ......" | 21336967 |
| hsa-miR-16-5p | gastric cancer | PIK3CD | "TZ and lapatinib ability to block extracellular si ......" | 27157613 |
| hsa-miR-16-5p | lung cancer | PPM1D | "The relative activity of renila luciferase was det ......" | 26064212 |
| hsa-miR-16-5p | gastric cancer | PTGER4 | "EP2 or EP4 siRNA or antagonists impaired the nicot ......" | 21081469 |
| hsa-miR-16-5p | thyroid cancer | RAF1 | "Besides we identify that Raf1 is a direct target o ......" | 26527888 |
| hsa-miR-16-5p | esophageal cancer | RECK | "Aberrant expression level of miR-16 could suppress ......" | 24852767 |
| hsa-miR-16-5p | prostate cancer | RNU6-6P | "In this study the expression of four putative refe ......" | 20890088 |
| hsa-miR-16-5p | bladder cancer | ROS1 | "Together our results revealed that urothelial carc ......" | 26373319 |
| hsa-miR-16-5p | pancreatic cancer | SCARNA17 | "Four internal controls were utilized for RT-qPCR r ......" | 26499892 |
| hsa-miR-16-5p | lung squamous cell cancer | SCLC1 | "In SCLC the miR-16 in the miRNA-drug target-drug n ......" | 26273338 |
| hsa-miR-16-5p | colon cancer | SLC33A1 | "The CCK8 test showed that at 1 5 25 and 125 µg/ml ......" | 25623762 |
| hsa-miR-16-5p | pancreatic cancer | SNORD15A | "Four internal controls were utilized for RT-qPCR r ......" | 26499892 |
| hsa-miR-16-5p | prostate cancer | SNORD7 | "In this study the expression of four putative refe ......" | 20890088 |
| hsa-miR-16-5p | esophageal cancer | SOX6 | "Aberrant expression level of miR-16 could suppress ......" | 24852767 |
| hsa-miR-16-5p | breast cancer | STAT3 | "miR-16 expression was studied by RT-qPCR in cancer ......" | 22583478 |
| hsa-miR-16-5p | liver cancer | TECRL | "P glycoprotein enhances radiation induced apoptoti ......" | 21336967 |
| hsa-miR-16-5p | lymphoma | TSPYL2 | "First we examined the expression of miR-16 in prim ......" | 26640145 |
| hsa-miR-16-5p | bladder cancer | UCA1 | "Long non coding RNA UCA1 promotes glutamine metabo ......" | 26373319 |
| hsa-miR-16-5p | liver cancer | XIAP | "On the other hand ectopic mdr1 expression enhanced ......" | 21336967 |
| hsa-miR-16-5p | breast cancer | YARS | "Two miRNA signatures miR-16 155 125b 374a and miR- ......" | 23405235 |
Expression profile in cancer corhorts:

Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-16-5p | ACTN1 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.469; TCGA BRCA -0.123; TCGA CESC -0.196; TCGA COAD -0.251; TCGA ESCA -0.427; TCGA HNSC -0.385; TCGA LUAD -0.196; TCGA LUSC -0.555; TCGA PAAD -0.351; TCGA PRAD -0.406; TCGA SARC -0.387; TCGA STAD -0.496; TCGA UCEC -0.211 |
hsa-miR-16-5p | ACTN4 | 9 cancers: BLCA; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; THCA; UCEC | miRNAWalker2 validate | TCGA BLCA -0.123; TCGA HNSC -0.117; TCGA KIRC -0.071; TCGA LUAD -0.098; TCGA LUSC -0.158; TCGA PAAD -0.217; TCGA PRAD -0.109; TCGA THCA -0.08; TCGA UCEC -0.079 |
hsa-miR-16-5p | ACVR2A | 10 cancers: BLCA; BRCA; KIRC; KIRP; LGG; OV; PAAD; SARC; STAD; UCEC | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | TCGA BLCA -0.096; TCGA BRCA -0.182; TCGA KIRC -0.225; TCGA KIRP -0.109; TCGA LGG -0.072; TCGA OV -0.197; TCGA PAAD -0.186; TCGA SARC -0.172; TCGA STAD -0.204; TCGA UCEC -0.222 |
hsa-miR-16-5p | AHNAK2 | 13 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.999; TCGA BRCA -0.118; TCGA COAD -0.666; TCGA ESCA -0.874; TCGA HNSC -0.727; TCGA KIRP -0.475; TCGA LUAD -0.471; TCGA LUSC -0.747; TCGA PAAD -1.337; TCGA PRAD -0.666; TCGA SARC -0.491; TCGA STAD -1.143; TCGA UCEC -0.545 |
hsa-miR-16-5p | AKAP13 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.282; TCGA BRCA -0.066; TCGA CESC -0.163; TCGA COAD -0.173; TCGA ESCA -0.333; TCGA HNSC -0.205; TCGA LUAD -0.108; TCGA LUSC -0.288; TCGA PAAD -0.225; TCGA PRAD -0.12; TCGA UCEC -0.251 |
hsa-miR-16-5p | AKT3 | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; LUAD; PRAD; STAD; UCEC | miRNAWalker2 validate; miRTarBase; miRNATAP | TCGA BLCA -0.528; TCGA BRCA -0.137; TCGA CESC -0.31; TCGA COAD -0.746; TCGA ESCA -0.474; TCGA KIRC -0.175; TCGA LUAD -0.27; TCGA PRAD -0.188; TCGA STAD -0.568; TCGA UCEC -0.739 |
hsa-miR-16-5p | ANAPC16 | 9 cancers: BLCA; KIRC; KIRP; LGG; LIHC; OV; PRAD; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.098; TCGA KIRC -0.156; TCGA KIRP -0.071; TCGA LGG -0.127; TCGA LIHC -0.079; TCGA OV -0.11; TCGA PRAD -0.066; TCGA STAD -0.133; TCGA UCEC -0.113 |
hsa-miR-16-5p | APLNR | 9 cancers: BLCA; BRCA; CESC; COAD; HNSC; LIHC; LUSC; THCA; UCEC | miRNAWalker2 validate; MirTarget | TCGA BLCA -0.397; TCGA BRCA -0.164; TCGA CESC -0.456; TCGA COAD -0.502; TCGA HNSC -0.351; TCGA LIHC -0.376; TCGA LUSC -0.504; TCGA THCA -0.274; TCGA UCEC -0.277 |
hsa-miR-16-5p | APP | 12 cancers: BLCA; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | miRNAWalker2 validate; miRNATAP | TCGA BLCA -0.178; TCGA COAD -0.127; TCGA ESCA -0.192; TCGA HNSC -0.134; TCGA KIRC -0.335; TCGA KIRP -0.127; TCGA LUAD -0.187; TCGA LUSC -0.26; TCGA PAAD -0.201; TCGA PRAD -0.357; TCGA STAD -0.063; TCGA UCEC -0.145 |
hsa-miR-16-5p | ARL10 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRP; LUAD; LUSC; OV; PAAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.338; TCGA BRCA -0.138; TCGA CESC -0.259; TCGA COAD -0.672; TCGA ESCA -0.385; TCGA KIRP -0.18; TCGA LUAD -0.244; TCGA LUSC -0.196; TCGA OV -0.142; TCGA PAAD -0.263; TCGA PRAD -0.376; TCGA SARC -0.246; TCGA STAD -0.49; TCGA UCEC -0.255 |
hsa-miR-16-5p | ARL2 | 9 cancers: BLCA; BRCA; HNSC; KIRC; LIHC; OV; THCA; STAD; UCEC | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | TCGA BLCA -0.163; TCGA BRCA -0.152; TCGA HNSC -0.099; TCGA KIRC -0.22; TCGA LIHC -0.299; TCGA OV -0.179; TCGA THCA -0.119; TCGA STAD -0.072; TCGA UCEC -0.138 |
hsa-miR-16-5p | ARL3 | 10 cancers: BLCA; BRCA; KIRC; KIRP; LGG; LIHC; OV; THCA; STAD; UCEC | miRNAWalker2 validate; MirTarget | TCGA BLCA -0.094; TCGA BRCA -0.067; TCGA KIRC -0.239; TCGA KIRP -0.18; TCGA LGG -0.214; TCGA LIHC -0.149; TCGA OV -0.173; TCGA THCA -0.149; TCGA STAD -0.118; TCGA UCEC -0.156 |
hsa-miR-16-5p | ARMCX2 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUSC; PRAD; THCA; STAD; UCEC | miRNAWalker2 validate; MirTarget; miRNATAP | TCGA BLCA -0.213; TCGA BRCA -0.192; TCGA CESC -0.35; TCGA COAD -0.495; TCGA ESCA -0.429; TCGA HNSC -0.341; TCGA KIRC -0.289; TCGA LUSC -0.254; TCGA PRAD -0.546; TCGA THCA -0.208; TCGA STAD -0.535; TCGA UCEC -0.412 |
hsa-miR-16-5p | ASH1L | 10 cancers: BLCA; COAD; ESCA; KIRC; KIRP; LUAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate; MirTarget; miRNATAP | TCGA BLCA -0.123; TCGA COAD -0.1; TCGA ESCA -0.167; TCGA KIRC -0.225; TCGA KIRP -0.092; TCGA LUAD -0.118; TCGA PRAD -0.165; TCGA SARC -0.248; TCGA STAD -0.204; TCGA UCEC -0.168 |
hsa-miR-16-5p | ATF7 | 13 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.145; TCGA CESC -0.066; TCGA COAD -0.09; TCGA ESCA -0.163; TCGA HNSC -0.118; TCGA KIRP -0.112; TCGA LUAD -0.109; TCGA LUSC -0.073; TCGA PAAD -0.253; TCGA PRAD -0.091; TCGA SARC -0.171; TCGA STAD -0.22; TCGA UCEC -0.084 |
hsa-miR-16-5p | AXIN2 | 14 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; UCEC | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | TCGA BLCA -0.751; TCGA BRCA -0.25; TCGA CESC -0.696; TCGA HNSC -0.516; TCGA KIRC -0.444; TCGA KIRP -0.371; TCGA LGG -0.178; TCGA LIHC -0.493; TCGA LUAD -0.2; TCGA LUSC -0.315; TCGA PAAD -0.334; TCGA PRAD -0.339; TCGA SARC -0.35; TCGA UCEC -0.291 |
hsa-miR-16-5p | BNC2 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -1.182; TCGA BRCA -0.313; TCGA CESC -0.796; TCGA COAD -1.088; TCGA ESCA -1.327; TCGA HNSC -0.679; TCGA LUAD -0.314; TCGA LUSC -0.839; TCGA PAAD -0.986; TCGA PRAD -0.535; TCGA STAD -0.898; TCGA UCEC -1.053 |
hsa-miR-16-5p | BTG2 | 9 cancers: BLCA; BRCA; ESCA; KIRC; LUSC; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate; MirTarget; miRNATAP | TCGA BLCA -0.322; TCGA BRCA -0.275; TCGA ESCA -0.415; TCGA KIRC -0.604; TCGA LUSC -0.171; TCGA PRAD -0.298; TCGA SARC -0.231; TCGA STAD -0.267; TCGA UCEC -0.326 |
hsa-miR-16-5p | BTRC | 17 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; OV; PRAD; THCA; STAD; UCEC | miRNAWalker2 validate; MirTarget; miRNATAP | TCGA BLCA -0.072; TCGA BRCA -0.077; TCGA CESC -0.13; TCGA COAD -0.091; TCGA ESCA -0.147; TCGA HNSC -0.14; TCGA KIRC -0.171; TCGA KIRP -0.074; TCGA LGG -0.121; TCGA LIHC -0.171; TCGA LUAD -0.107; TCGA LUSC -0.065; TCGA OV -0.103; TCGA PRAD -0.089; TCGA THCA -0.065; TCGA STAD -0.133; TCGA UCEC -0.112 |
hsa-miR-16-5p | CACNA2D1 | 9 cancers: BLCA; BRCA; COAD; ESCA; LUAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -1.089; TCGA BRCA -0.217; TCGA COAD -1.444; TCGA ESCA -1.061; TCGA LUAD -0.35; TCGA PRAD -1.224; TCGA SARC -0.717; TCGA STAD -0.797; TCGA UCEC -0.776 |
hsa-miR-16-5p | CACNB2 | 9 cancers: BLCA; CESC; ESCA; KIRP; LUSC; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.972; TCGA CESC -0.543; TCGA ESCA -0.99; TCGA KIRP -0.398; TCGA LUSC -0.305; TCGA PRAD -0.319; TCGA SARC -0.656; TCGA STAD -0.955; TCGA UCEC -0.632 |
hsa-miR-16-5p | CADM1 | 10 cancers: BLCA; CESC; COAD; HNSC; KIRC; LIHC; LUAD; OV; THCA; STAD | miRNAWalker2 validate; miRTarBase; miRNATAP | TCGA BLCA -0.38; TCGA CESC -0.396; TCGA COAD -0.828; TCGA HNSC -0.328; TCGA KIRC -0.486; TCGA LIHC -0.351; TCGA LUAD -0.231; TCGA OV -0.203; TCGA THCA -0.248; TCGA STAD -0.319 |
hsa-miR-16-5p | CALU | 11 cancers: BLCA; COAD; ESCA; HNSC; LUSC; OV; PAAD; PRAD; THCA; STAD; UCEC | miRNAWalker2 validate; MirTarget | TCGA BLCA -0.243; TCGA COAD -0.231; TCGA ESCA -0.182; TCGA HNSC -0.2; TCGA LUSC -0.268; TCGA OV -0.064; TCGA PAAD -0.418; TCGA PRAD -0.236; TCGA THCA -0.19; TCGA STAD -0.106; TCGA UCEC -0.172 |
hsa-miR-16-5p | CAMK2G | 10 cancers: BLCA; CESC; ESCA; KIRC; LGG; LIHC; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.19; TCGA CESC -0.083; TCGA ESCA -0.183; TCGA KIRC -0.094; TCGA LGG -0.245; TCGA LIHC -0.136; TCGA PRAD -0.378; TCGA SARC -0.349; TCGA STAD -0.436; TCGA UCEC -0.17 |
hsa-miR-16-5p | CAPZA2 | 11 cancers: BLCA; ESCA; HNSC; KIRP; LUSC; OV; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate; MirTarget | TCGA BLCA -0.151; TCGA ESCA -0.137; TCGA HNSC -0.152; TCGA KIRP -0.112; TCGA LUSC -0.107; TCGA OV -0.095; TCGA PRAD -0.157; TCGA SARC -0.144; TCGA THCA -0.106; TCGA STAD -0.158; TCGA UCEC -0.231 |
hsa-miR-16-5p | CCND2 | 9 cancers: BLCA; BRCA; CESC; ESCA; KIRP; LUAD; PRAD; STAD; UCEC | miRNAWalker2 validate; miRNATAP | TCGA BLCA -0.625; TCGA BRCA -0.126; TCGA CESC -0.251; TCGA ESCA -0.666; TCGA KIRP -0.352; TCGA LUAD -0.135; TCGA PRAD -0.256; TCGA STAD -0.516; TCGA UCEC -0.598 |
hsa-miR-16-5p | CCPG1 | 13 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LUSC; OV; PAAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.293; TCGA CESC -0.172; TCGA COAD -0.13; TCGA ESCA -0.346; TCGA HNSC -0.113; TCGA KIRC -0.142; TCGA LUSC -0.304; TCGA OV -0.091; TCGA PAAD -0.275; TCGA PRAD -0.356; TCGA SARC -0.158; TCGA STAD -0.131; TCGA UCEC -0.26 |
hsa-miR-16-5p | CD55 | 9 cancers: BLCA; HNSC; KIRC; LUAD; LUSC; OV; PAAD; THCA; UCEC | miRNAWalker2 validate | TCGA BLCA -0.171; TCGA HNSC -0.229; TCGA KIRC -0.276; TCGA LUAD -0.2; TCGA LUSC -0.287; TCGA OV -0.143; TCGA PAAD -0.359; TCGA THCA -0.317; TCGA UCEC -0.25 |
hsa-miR-16-5p | CDK6 | 11 cancers: BLCA; CESC; HNSC; KIRP; LIHC; LUSC; OV; PAAD; PRAD; THCA; UCEC | miRNAWalker2 validate; miRTarBase | TCGA BLCA -0.211; TCGA CESC -0.233; TCGA HNSC -0.39; TCGA KIRP -0.166; TCGA LIHC -0.498; TCGA LUSC -0.198; TCGA OV -0.304; TCGA PAAD -0.231; TCGA PRAD -0.176; TCGA THCA -0.184; TCGA UCEC -0.422 |
hsa-miR-16-5p | CLIP2 | 9 cancers: BLCA; BRCA; CESC; KIRP; LGG; LUAD; LUSC; THCA; UCEC | miRNAWalker2 validate | TCGA BLCA -0.25; TCGA BRCA -0.086; TCGA CESC -0.222; TCGA KIRP -0.155; TCGA LGG -0.07; TCGA LUAD -0.198; TCGA LUSC -0.224; TCGA THCA -0.227; TCGA UCEC -0.181 |
hsa-miR-16-5p | CNN3 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.249; TCGA BRCA -0.083; TCGA CESC -0.218; TCGA COAD -0.296; TCGA ESCA -0.363; TCGA HNSC -0.188; TCGA KIRC -0.095; TCGA LUAD -0.116; TCGA LUSC -0.43; TCGA PAAD -0.268; TCGA PRAD -0.389; TCGA THCA -0.184; TCGA STAD -0.267; TCGA UCEC -0.268 |
hsa-miR-16-5p | COL4A2 | 10 cancers: BLCA; CESC; COAD; LIHC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.379; TCGA CESC -0.366; TCGA COAD -0.46; TCGA LIHC -0.201; TCGA LUAD -0.325; TCGA LUSC -0.419; TCGA PAAD -0.469; TCGA PRAD -0.169; TCGA STAD -0.283; TCGA UCEC -0.182 |
hsa-miR-16-5p | CREB3L2 | 10 cancers: BLCA; ESCA; HNSC; KIRP; LUAD; LUSC; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.226; TCGA ESCA -0.272; TCGA HNSC -0.266; TCGA KIRP -0.127; TCGA LUAD -0.201; TCGA LUSC -0.316; TCGA PRAD -0.134; TCGA SARC -0.13; TCGA STAD -0.183; TCGA UCEC -0.244 |
hsa-miR-16-5p | CSDE1 | 12 cancers: BLCA; CESC; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate; MirTarget; miRNATAP | TCGA BLCA -0.151; TCGA CESC -0.095; TCGA ESCA -0.222; TCGA HNSC -0.085; TCGA KIRC -0.143; TCGA LUAD -0.063; TCGA LUSC -0.111; TCGA PAAD -0.104; TCGA PRAD -0.108; TCGA SARC -0.118; TCGA STAD -0.222; TCGA UCEC -0.147 |
hsa-miR-16-5p | CSGALNACT1 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LUAD; LUSC; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.476; TCGA BRCA -0.136; TCGA CESC -0.259; TCGA COAD -0.285; TCGA ESCA -0.48; TCGA HNSC -0.281; TCGA KIRC -0.295; TCGA LGG -0.122; TCGA LUAD -0.332; TCGA LUSC -0.442; TCGA PRAD -0.212; TCGA SARC -0.629; TCGA STAD -0.193; TCGA UCEC -0.449 |
hsa-miR-16-5p | DIXDC1 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; OV; PAAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate; MirTarget | TCGA BLCA -0.843; TCGA BRCA -0.196; TCGA CESC -0.468; TCGA COAD -0.551; TCGA ESCA -0.93; TCGA HNSC -0.349; TCGA KIRP -0.118; TCGA LUAD -0.237; TCGA LUSC -0.397; TCGA OV -0.126; TCGA PAAD -0.447; TCGA PRAD -0.524; TCGA SARC -0.606; TCGA STAD -0.804; TCGA UCEC -0.656 |
hsa-miR-16-5p | DLC1 | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUSC; PRAD; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.433; TCGA BRCA -0.283; TCGA CESC -0.597; TCGA COAD -0.501; TCGA ESCA -0.593; TCGA HNSC -0.433; TCGA LUSC -0.686; TCGA PRAD -0.143; TCGA STAD -0.468; TCGA UCEC -0.481 |
hsa-miR-16-5p | DMD | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -1.028; TCGA BRCA -0.206; TCGA CESC -0.481; TCGA COAD -0.628; TCGA ESCA -0.798; TCGA LUAD -0.224; TCGA LUSC -0.263; TCGA PAAD -0.467; TCGA PRAD -0.613; TCGA SARC -0.604; TCGA STAD -0.989; TCGA UCEC -0.823 |
hsa-miR-16-5p | DNAJB4 | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUSC; OV; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.342; TCGA BRCA -0.071; TCGA COAD -0.313; TCGA ESCA -0.419; TCGA HNSC -0.256; TCGA LUSC -0.221; TCGA OV -0.087; TCGA PRAD -0.472; TCGA SARC -0.17; TCGA STAD -0.452; TCGA UCEC -0.53 |
hsa-miR-16-5p | EPC1 | 9 cancers: BLCA; COAD; ESCA; LGG; LUAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.129; TCGA COAD -0.109; TCGA ESCA -0.196; TCGA LGG -0.127; TCGA LUAD -0.065; TCGA PRAD -0.234; TCGA SARC -0.19; TCGA STAD -0.142; TCGA UCEC -0.14 |
hsa-miR-16-5p | FAM69A | 11 cancers: BLCA; COAD; ESCA; HNSC; LGG; LUAD; LUSC; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.281; TCGA COAD -0.112; TCGA ESCA -0.306; TCGA HNSC -0.176; TCGA LGG -0.157; TCGA LUAD -0.167; TCGA LUSC -0.198; TCGA PRAD -0.367; TCGA SARC -0.285; TCGA STAD -0.247; TCGA UCEC -0.44 |
hsa-miR-16-5p | FAT3 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; LGG; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate; MirTarget; miRNATAP | TCGA BLCA -0.795; TCGA BRCA -0.531; TCGA CESC -0.584; TCGA COAD -0.831; TCGA ESCA -1.409; TCGA KIRC -0.435; TCGA LGG -0.228; TCGA LUAD -0.417; TCGA LUSC -0.462; TCGA PAAD -0.538; TCGA PRAD -1.199; TCGA SARC -1.064; TCGA STAD -1.335; TCGA UCEC -0.425 |
hsa-miR-16-5p | FBXO3 | 11 cancers: BLCA; COAD; ESCA; HNSC; KIRC; KIRP; OV; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.088; TCGA COAD -0.068; TCGA ESCA -0.156; TCGA HNSC -0.157; TCGA KIRC -0.19; TCGA KIRP -0.104; TCGA OV -0.077; TCGA PRAD -0.154; TCGA SARC -0.201; TCGA STAD -0.169; TCGA UCEC -0.201 |
hsa-miR-16-5p | FBXW11 | 11 cancers: BLCA; COAD; ESCA; KIRC; LUAD; OV; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.105; TCGA COAD -0.055; TCGA ESCA -0.152; TCGA KIRC -0.147; TCGA LUAD -0.082; TCGA OV -0.078; TCGA PRAD -0.102; TCGA SARC -0.129; TCGA THCA -0.092; TCGA STAD -0.094; TCGA UCEC -0.158 |
hsa-miR-16-5p | FGF2 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; KIRP; LGG; LUSC; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate; MirTarget; miRNATAP | TCGA BLCA -0.791; TCGA BRCA -0.335; TCGA CESC -0.382; TCGA COAD -0.781; TCGA ESCA -0.877; TCGA KIRC -0.243; TCGA KIRP -0.439; TCGA LGG -0.238; TCGA LUSC -0.244; TCGA PRAD -0.555; TCGA SARC -0.38; TCGA THCA -0.221; TCGA STAD -0.677; TCGA UCEC -0.918 |
hsa-miR-16-5p | FLNA | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.822; TCGA BRCA -0.173; TCGA COAD -0.818; TCGA ESCA -0.828; TCGA HNSC -0.135; TCGA LUAD -0.291; TCGA LUSC -0.444; TCGA PAAD -0.645; TCGA PRAD -0.745; TCGA SARC -0.503; TCGA STAD -0.94; TCGA UCEC -0.459 |
hsa-miR-16-5p | GABARAPL1 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LIHC; OV; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.261; TCGA BRCA -0.122; TCGA CESC -0.207; TCGA COAD -0.217; TCGA ESCA -0.339; TCGA HNSC -0.167; TCGA KIRC -0.706; TCGA LGG -0.105; TCGA LIHC -0.217; TCGA OV -0.13; TCGA PRAD -0.262; TCGA SARC -0.25; TCGA THCA -0.113; TCGA STAD -0.364; TCGA UCEC -0.436 |
hsa-miR-16-5p | GNAL | 14 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; OV; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate; miRNATAP | TCGA BLCA -1.076; TCGA BRCA -0.402; TCGA COAD -0.619; TCGA ESCA -0.729; TCGA HNSC -0.267; TCGA KIRC -0.38; TCGA KIRP -0.326; TCGA LGG -0.421; TCGA LUAD -0.237; TCGA OV -0.177; TCGA PRAD -0.793; TCGA SARC -0.965; TCGA STAD -0.913; TCGA UCEC -0.561 |
hsa-miR-16-5p | HTRA1 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.601; TCGA BRCA -0.422; TCGA CESC -0.286; TCGA COAD -0.577; TCGA ESCA -0.378; TCGA HNSC -0.472; TCGA LIHC -0.276; TCGA LUAD -0.293; TCGA LUSC -0.549; TCGA OV -0.26; TCGA PAAD -0.544; TCGA PRAD -0.18; TCGA STAD -0.261; TCGA UCEC -0.299 |
hsa-miR-16-5p | IDS | 11 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PRAD; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.159; TCGA CESC -0.218; TCGA COAD -0.159; TCGA ESCA -0.387; TCGA HNSC -0.177; TCGA LUAD -0.188; TCGA LUSC -0.196; TCGA PRAD -0.089; TCGA THCA -0.117; TCGA STAD -0.314; TCGA UCEC -0.25 |
hsa-miR-16-5p | KCND3 | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -1.113; TCGA BRCA -0.412; TCGA COAD -0.936; TCGA ESCA -1.459; TCGA HNSC -0.24; TCGA KIRC -0.835; TCGA LUAD -0.417; TCGA LUSC -0.753; TCGA PRAD -0.725; TCGA SARC -0.976; TCGA STAD -0.521; TCGA UCEC -0.633 |
hsa-miR-16-5p | KIDINS220 | 13 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.115; TCGA CESC -0.116; TCGA COAD -0.155; TCGA ESCA -0.133; TCGA HNSC -0.141; TCGA KIRC -0.139; TCGA LUAD -0.142; TCGA LUSC -0.087; TCGA PAAD -0.197; TCGA PRAD -0.106; TCGA SARC -0.173; TCGA STAD -0.218; TCGA UCEC -0.211 |
hsa-miR-16-5p | KIF1B | 9 cancers: BLCA; ESCA; HNSC; KIRC; LGG; PRAD; THCA; STAD; UCEC | miRNAWalker2 validate; miRNATAP | TCGA BLCA -0.11; TCGA ESCA -0.194; TCGA HNSC -0.232; TCGA KIRC -0.178; TCGA LGG -0.112; TCGA PRAD -0.11; TCGA THCA -0.116; TCGA STAD -0.244; TCGA UCEC -0.106 |
hsa-miR-16-5p | LAMB1 | 13 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; LUAD; LUSC; OV; PAAD; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.101; TCGA BRCA -0.149; TCGA CESC -0.31; TCGA COAD -0.284; TCGA HNSC -0.183; TCGA KIRC -0.136; TCGA KIRP -0.456; TCGA LUAD -0.245; TCGA LUSC -0.442; TCGA OV -0.165; TCGA PAAD -0.389; TCGA STAD -0.153; TCGA UCEC -0.235 |
hsa-miR-16-5p | LAMC1 | 16 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; OV; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate; MirTarget | TCGA BLCA -0.297; TCGA CESC -0.144; TCGA COAD -0.373; TCGA ESCA -0.222; TCGA HNSC -0.12; TCGA KIRP -0.159; TCGA LIHC -0.156; TCGA LUAD -0.117; TCGA LUSC -0.259; TCGA OV -0.09; TCGA PAAD -0.515; TCGA PRAD -0.108; TCGA SARC -0.197; TCGA THCA -0.15; TCGA STAD -0.346; TCGA UCEC -0.132 |
hsa-miR-16-5p | LONRF2 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; LUAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate; mirMAP | TCGA BLCA -1.19; TCGA BRCA -0.248; TCGA CESC -1.087; TCGA COAD -1.816; TCGA ESCA -1.229; TCGA KIRC -1.101; TCGA LUAD -0.337; TCGA SARC -1.074; TCGA THCA -0.459; TCGA STAD -1.124; TCGA UCEC -0.663 |
hsa-miR-16-5p | MACF1 | 13 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.257; TCGA CESC -0.211; TCGA COAD -0.259; TCGA ESCA -0.353; TCGA HNSC -0.283; TCGA KIRC -0.17; TCGA LUAD -0.201; TCGA LUSC -0.426; TCGA PRAD -0.247; TCGA SARC -0.387; TCGA THCA -0.127; TCGA STAD -0.383; TCGA UCEC -0.319 |
hsa-miR-16-5p | MAP4 | 13 cancers: BLCA; COAD; ESCA; HNSC; KIRC; LGG; LUAD; LUSC; PAAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.184; TCGA COAD -0.102; TCGA ESCA -0.318; TCGA HNSC -0.231; TCGA KIRC -0.086; TCGA LGG -0.063; TCGA LUAD -0.134; TCGA LUSC -0.267; TCGA PAAD -0.241; TCGA SARC -0.125; TCGA THCA -0.105; TCGA STAD -0.21; TCGA UCEC -0.114 |
hsa-miR-16-5p | MEGF8 | 12 cancers: BLCA; BRCA; COAD; ESCA; KIRC; LIHC; LUAD; LUSC; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.114; TCGA BRCA -0.16; TCGA COAD -0.124; TCGA ESCA -0.233; TCGA KIRC -0.167; TCGA LIHC -0.164; TCGA LUAD -0.114; TCGA LUSC -0.158; TCGA SARC -0.122; TCGA THCA -0.082; TCGA STAD -0.142; TCGA UCEC -0.116 |
hsa-miR-16-5p | MRC2 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.483; TCGA BRCA -0.376; TCGA CESC -0.329; TCGA COAD -0.743; TCGA ESCA -0.664; TCGA HNSC -0.262; TCGA LUAD -0.338; TCGA LUSC -0.667; TCGA PAAD -0.779; TCGA PRAD -0.125; TCGA STAD -0.303; TCGA UCEC -0.332 |
hsa-miR-16-5p | NAV2 | 11 cancers: BLCA; BRCA; COAD; ESCA; KIRC; LGG; LUAD; LUSC; PAAD; PRAD; STAD | miRNAWalker2 validate | TCGA BLCA -0.208; TCGA BRCA -0.117; TCGA COAD -0.164; TCGA ESCA -0.327; TCGA KIRC -0.484; TCGA LGG -0.135; TCGA LUAD -0.169; TCGA LUSC -0.195; TCGA PAAD -0.253; TCGA PRAD -0.473; TCGA STAD -0.454 |
hsa-miR-16-5p | NOTCH2 | 14 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate; MirTarget | TCGA BLCA -0.149; TCGA CESC -0.2; TCGA COAD -0.279; TCGA ESCA -0.39; TCGA HNSC -0.179; TCGA KIRP -0.2; TCGA LUAD -0.181; TCGA LUSC -0.277; TCGA PAAD -0.497; TCGA PRAD -0.154; TCGA SARC -0.133; TCGA THCA -0.157; TCGA STAD -0.258; TCGA UCEC -0.126 |
hsa-miR-16-5p | NPR3 | 9 cancers: BLCA; COAD; HNSC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.634; TCGA COAD -1.184; TCGA HNSC -0.308; TCGA LUAD -0.633; TCGA LUSC -0.388; TCGA PAAD -1.021; TCGA PRAD -0.545; TCGA STAD -0.878; TCGA UCEC -0.409 |
hsa-miR-16-5p | NRP1 | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.167; TCGA CESC -0.339; TCGA COAD -0.546; TCGA ESCA -0.395; TCGA HNSC -0.25; TCGA KIRP -0.132; TCGA LUAD -0.295; TCGA LUSC -0.561; TCGA PAAD -0.373; TCGA THCA -0.14; TCGA STAD -0.168; TCGA UCEC -0.318 |
hsa-miR-16-5p | NRXN1 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; LGG; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -1.307; TCGA BRCA -0.477; TCGA CESC -0.406; TCGA COAD -1.608; TCGA ESCA -1.597; TCGA KIRC -0.652; TCGA LGG -0.35; TCGA PRAD -0.474; TCGA SARC -0.58; TCGA STAD -1.399; TCGA UCEC -0.706 |
hsa-miR-16-5p | PCDHGB4 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PRAD; SARC; UCEC | miRNAWalker2 validate | TCGA BLCA -0.462; TCGA BRCA -0.239; TCGA CESC -0.522; TCGA COAD -0.464; TCGA ESCA -0.35; TCGA HNSC -0.604; TCGA KIRC -0.708; TCGA LUAD -0.328; TCGA LUSC -0.408; TCGA PRAD -0.418; TCGA SARC -0.536; TCGA UCEC -0.541 |
hsa-miR-16-5p | PDE4D | 9 cancers: BLCA; COAD; ESCA; KIRC; KIRP; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.48; TCGA COAD -0.143; TCGA ESCA -0.466; TCGA KIRC -0.114; TCGA KIRP -0.099; TCGA PRAD -0.431; TCGA SARC -0.456; TCGA STAD -0.375; TCGA UCEC -0.355 |
hsa-miR-16-5p | PDK4 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; OV; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate; MirTarget; miRNATAP | TCGA BLCA -0.761; TCGA BRCA -0.449; TCGA CESC -0.707; TCGA COAD -0.324; TCGA ESCA -1.065; TCGA HNSC -0.627; TCGA LUAD -0.338; TCGA LUSC -0.511; TCGA OV -0.212; TCGA PRAD -0.434; TCGA SARC -0.91; TCGA STAD -0.923; TCGA UCEC -0.757 |
hsa-miR-16-5p | PDLIM5 | 11 cancers: BLCA; CESC; ESCA; HNSC; LGG; LUAD; LUSC; PAAD; SARC; STAD; UCEC | miRNAWalker2 validate; miRNATAP | TCGA BLCA -0.34; TCGA CESC -0.214; TCGA ESCA -0.153; TCGA HNSC -0.265; TCGA LGG -0.277; TCGA LUAD -0.196; TCGA LUSC -0.252; TCGA PAAD -0.355; TCGA SARC -0.456; TCGA STAD -0.189; TCGA UCEC -0.304 |
hsa-miR-16-5p | PHKB | 11 cancers: BLCA; CESC; ESCA; HNSC; KIRC; LGG; LIHC; OV; PRAD; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.162; TCGA CESC -0.082; TCGA ESCA -0.116; TCGA HNSC -0.168; TCGA KIRC -0.284; TCGA LGG -0.052; TCGA LIHC -0.108; TCGA OV -0.1; TCGA PRAD -0.139; TCGA STAD -0.07; TCGA UCEC -0.118 |
hsa-miR-16-5p | PHLDB2 | 14 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.403; TCGA BRCA -0.151; TCGA COAD -0.975; TCGA ESCA -0.41; TCGA HNSC -0.216; TCGA KIRC -0.216; TCGA KIRP -0.205; TCGA LUAD -0.227; TCGA LUSC -0.284; TCGA PRAD -0.317; TCGA SARC -0.43; TCGA THCA -0.242; TCGA STAD -0.694; TCGA UCEC -0.726 |
hsa-miR-16-5p | PLEC | 9 cancers: BLCA; BRCA; HNSC; LGG; LIHC; LUAD; LUSC; PAAD; STAD | miRNAWalker2 validate | TCGA BLCA -0.107; TCGA BRCA -0.096; TCGA HNSC -0.298; TCGA LGG -0.227; TCGA LIHC -0.165; TCGA LUAD -0.207; TCGA LUSC -0.407; TCGA PAAD -0.257; TCGA STAD -0.117 |
hsa-miR-16-5p | PLSCR4 | 13 cancers: BLCA; BRCA; COAD; ESCA; KIRP; LGG; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate; MirTarget | TCGA BLCA -0.624; TCGA BRCA -0.268; TCGA COAD -0.253; TCGA ESCA -0.59; TCGA KIRP -0.139; TCGA LGG -0.149; TCGA LUAD -0.27; TCGA LUSC -0.249; TCGA PAAD -0.406; TCGA PRAD -0.403; TCGA SARC -0.271; TCGA STAD -0.656; TCGA UCEC -0.754 |
hsa-miR-16-5p | PRKAB2 | 16 cancers: BLCA; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; OV; PAAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate; MirTarget | TCGA BLCA -0.213; TCGA COAD -0.159; TCGA ESCA -0.323; TCGA HNSC -0.249; TCGA KIRC -0.238; TCGA KIRP -0.069; TCGA LGG -0.054; TCGA LIHC -0.189; TCGA LUAD -0.098; TCGA LUSC -0.116; TCGA OV -0.14; TCGA PAAD -0.235; TCGA PRAD -0.189; TCGA SARC -0.325; TCGA STAD -0.39; TCGA UCEC -0.207 |
hsa-miR-16-5p | PRKAR1A | 9 cancers: BLCA; COAD; ESCA; HNSC; KIRC; LUSC; PRAD; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.064; TCGA COAD -0.083; TCGA ESCA -0.115; TCGA HNSC -0.096; TCGA KIRC -0.166; TCGA LUSC -0.144; TCGA PRAD -0.158; TCGA STAD -0.111; TCGA UCEC -0.275 |
hsa-miR-16-5p | PRKAR2A | 10 cancers: BLCA; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PRAD; SARC; STAD | miRNAWalker2 validate | TCGA BLCA -0.207; TCGA COAD -0.161; TCGA ESCA -0.208; TCGA HNSC -0.233; TCGA KIRC -0.496; TCGA LUAD -0.162; TCGA LUSC -0.216; TCGA PRAD -0.472; TCGA SARC -0.318; TCGA STAD -0.149 |
hsa-miR-16-5p | PRNP | 11 cancers: BLCA; BRCA; COAD; HNSC; LUAD; LUSC; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.216; TCGA BRCA -0.101; TCGA COAD -0.323; TCGA HNSC -0.173; TCGA LUAD -0.106; TCGA LUSC -0.368; TCGA PRAD -0.283; TCGA SARC -0.17; TCGA THCA -0.164; TCGA STAD -0.462; TCGA UCEC -0.481 |
hsa-miR-16-5p | PTGS1 | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; STAD | miRNAWalker2 validate | TCGA BLCA -1.272; TCGA BRCA -0.069; TCGA COAD -0.585; TCGA ESCA -0.826; TCGA HNSC -0.3; TCGA LUAD -0.275; TCGA LUSC -0.715; TCGA PAAD -0.293; TCGA PRAD -0.7; TCGA STAD -0.374 |
hsa-miR-16-5p | PTGS2 | 9 cancers: BLCA; BRCA; KIRC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | miRNAWalker2 validate; miRTarBase | TCGA BLCA -0.555; TCGA BRCA -0.31; TCGA KIRC -0.541; TCGA LUAD -0.558; TCGA LUSC -0.37; TCGA PAAD -0.605; TCGA PRAD -0.547; TCGA STAD -0.362; TCGA UCEC -0.321 |
hsa-miR-16-5p | PTPRT | 12 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; LGG; LUAD; OV; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.507; TCGA BRCA -0.774; TCGA CESC -0.69; TCGA COAD -0.884; TCGA HNSC -0.392; TCGA KIRC -1.24; TCGA LGG -0.635; TCGA LUAD -0.524; TCGA OV -0.575; TCGA SARC -0.597; TCGA STAD -0.462; TCGA UCEC -0.659 |
hsa-miR-16-5p | PYGB | 9 cancers: BLCA; HNSC; KIRC; KIRP; LGG; LUSC; PRAD; SARC; THCA | miRNAWalker2 validate | TCGA BLCA -0.291; TCGA HNSC -0.281; TCGA KIRC -0.191; TCGA KIRP -0.16; TCGA LGG -0.116; TCGA LUSC -0.144; TCGA PRAD -0.307; TCGA SARC -0.315; TCGA THCA -0.152 |
hsa-miR-16-5p | RAB23 | 12 cancers: BLCA; CESC; COAD; ESCA; KIRP; LUSC; OV; PAAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.663; TCGA CESC -0.221; TCGA COAD -0.36; TCGA ESCA -0.461; TCGA KIRP -0.101; TCGA LUSC -0.208; TCGA OV -0.139; TCGA PAAD -0.472; TCGA PRAD -0.338; TCGA SARC -0.361; TCGA STAD -0.674; TCGA UCEC -0.492 |
hsa-miR-16-5p | RAB30 | 11 cancers: BLCA; COAD; ESCA; HNSC; KIRC; LUSC; OV; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.241; TCGA COAD -0.359; TCGA ESCA -0.317; TCGA HNSC -0.21; TCGA KIRC -0.126; TCGA LUSC -0.159; TCGA OV -0.186; TCGA PRAD -0.214; TCGA SARC -0.333; TCGA STAD -0.126; TCGA UCEC -0.132 |
hsa-miR-16-5p | RBMS1 | 9 cancers: BLCA; COAD; ESCA; HNSC; LUSC; PAAD; PRAD; STAD; UCEC | miRNAWalker2 validate; miRNATAP | TCGA BLCA -0.088; TCGA COAD -0.745; TCGA ESCA -0.234; TCGA HNSC -0.144; TCGA LUSC -0.194; TCGA PAAD -0.354; TCGA PRAD -0.231; TCGA STAD -0.434; TCGA UCEC -0.378 |
hsa-miR-16-5p | RBMS3 | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.687; TCGA BRCA -0.403; TCGA COAD -0.79; TCGA ESCA -0.776; TCGA HNSC -0.538; TCGA LUAD -0.3; TCGA LUSC -0.463; TCGA OV -0.257; TCGA PAAD -0.601; TCGA PRAD -0.604; TCGA STAD -0.778; TCGA UCEC -0.266 |
hsa-miR-16-5p | RECK | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; SARC; STAD; UCEC | miRNAWalker2 validate; miRNATAP | TCGA BLCA -0.591; TCGA BRCA -0.351; TCGA CESC -0.522; TCGA COAD -0.628; TCGA ESCA -0.87; TCGA HNSC -0.545; TCGA KIRC -0.104; TCGA LUAD -0.248; TCGA LUSC -0.489; TCGA PAAD -0.354; TCGA SARC -0.199; TCGA STAD -0.563; TCGA UCEC -0.524 |
hsa-miR-16-5p | RIMS3 | 11 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LUSC; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.216; TCGA BRCA -0.228; TCGA ESCA -0.623; TCGA HNSC -0.246; TCGA KIRC -0.201; TCGA LUSC -0.402; TCGA PRAD -0.493; TCGA SARC -0.513; TCGA THCA -0.18; TCGA STAD -0.55; TCGA UCEC -0.412 |
hsa-miR-16-5p | RNF217 | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LUSC; PAAD; PRAD; STAD | miRNAWalker2 validate; MirTarget | TCGA BLCA -0.35; TCGA BRCA -0.102; TCGA COAD -0.308; TCGA ESCA -0.588; TCGA HNSC -0.397; TCGA KIRP -0.119; TCGA LUSC -0.189; TCGA PAAD -0.593; TCGA PRAD -0.207; TCGA STAD -0.576 |
hsa-miR-16-5p | RTN4 | 12 cancers: BLCA; CESC; HNSC; KIRC; LUSC; OV; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.14; TCGA CESC -0.086; TCGA HNSC -0.289; TCGA KIRC -0.165; TCGA LUSC -0.21; TCGA OV -0.079; TCGA PAAD -0.143; TCGA PRAD -0.226; TCGA SARC -0.086; TCGA THCA -0.085; TCGA STAD -0.22; TCGA UCEC -0.235 |
hsa-miR-16-5p | RUNDC3B | 10 cancers: BLCA; BRCA; CESC; ESCA; KIRP; LUSC; OV; PRAD; STAD; UCEC | miRNAWalker2 validate; MirTarget | TCGA BLCA -0.314; TCGA BRCA -0.362; TCGA CESC -0.457; TCGA ESCA -0.483; TCGA KIRP -0.298; TCGA LUSC -0.265; TCGA OV -0.151; TCGA PRAD -0.271; TCGA STAD -0.466; TCGA UCEC -0.637 |
hsa-miR-16-5p | RUNX1T1 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate; miRNATAP | TCGA BLCA -1.135; TCGA BRCA -0.446; TCGA CESC -0.959; TCGA COAD -1.156; TCGA ESCA -1.17; TCGA HNSC -0.702; TCGA KIRC -0.352; TCGA LGG -0.303; TCGA LUAD -0.38; TCGA LUSC -0.922; TCGA PAAD -0.634; TCGA PRAD -0.495; TCGA SARC -0.483; TCGA STAD -0.853; TCGA UCEC -0.749 |
hsa-miR-16-5p | SEMA4C | 9 cancers: BLCA; BRCA; COAD; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.16; TCGA BRCA -0.106; TCGA COAD -0.297; TCGA PAAD -0.263; TCGA PRAD -0.125; TCGA SARC -0.211; TCGA THCA -0.117; TCGA STAD -0.282; TCGA UCEC -0.17 |
hsa-miR-16-5p | SIPA1L2 | 9 cancers: BLCA; CESC; ESCA; HNSC; LUAD; OV; PAAD; THCA; STAD | miRNAWalker2 validate; miRNATAP | TCGA BLCA -0.202; TCGA CESC -0.29; TCGA ESCA -0.332; TCGA HNSC -0.358; TCGA LUAD -0.239; TCGA OV -0.166; TCGA PAAD -0.277; TCGA THCA -0.206; TCGA STAD -0.388 |
hsa-miR-16-5p | SIRT4 | 9 cancers: BLCA; BRCA; COAD; KIRC; LGG; LIHC; OV; STAD; UCEC | miRNAWalker2 validate; MirTarget | TCGA BLCA -0.193; TCGA BRCA -0.081; TCGA COAD -0.203; TCGA KIRC -0.223; TCGA LGG -0.158; TCGA LIHC -0.32; TCGA OV -0.112; TCGA STAD -0.265; TCGA UCEC -0.097 |
hsa-miR-16-5p | SMAD7 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRP; LGG; LUAD; LUSC; OV; UCEC | miRNAWalker2 validate; MirTarget; miRNATAP | TCGA BLCA -0.269; TCGA BRCA -0.064; TCGA CESC -0.11; TCGA COAD -0.291; TCGA ESCA -0.18; TCGA KIRP -0.171; TCGA LGG -0.115; TCGA LUAD -0.146; TCGA LUSC -0.48; TCGA OV -0.127; TCGA UCEC -0.289 |
hsa-miR-16-5p | SOCS2 | 10 cancers: BLCA; BRCA; CESC; ESCA; KIRC; LUSC; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.227; TCGA BRCA -0.565; TCGA CESC -0.379; TCGA ESCA -0.516; TCGA KIRC -0.399; TCGA LUSC -0.243; TCGA PRAD -0.118; TCGA SARC -0.496; TCGA STAD -0.277; TCGA UCEC -0.765 |
hsa-miR-16-5p | SOCS5 | 14 cancers: BLCA; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; OV; PAAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.158; TCGA COAD -0.137; TCGA ESCA -0.13; TCGA HNSC -0.135; TCGA KIRC -0.075; TCGA KIRP -0.072; TCGA LUAD -0.106; TCGA LUSC -0.081; TCGA OV -0.111; TCGA PAAD -0.214; TCGA PRAD -0.096; TCGA SARC -0.125; TCGA STAD -0.296; TCGA UCEC -0.254 |
hsa-miR-16-5p | SON | 10 cancers: BLCA; COAD; ESCA; KIRC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate; miRNATAP | TCGA BLCA -0.076; TCGA COAD -0.071; TCGA ESCA -0.114; TCGA KIRC -0.125; TCGA LUAD -0.059; TCGA LUSC -0.119; TCGA PRAD -0.138; TCGA SARC -0.135; TCGA STAD -0.122; TCGA UCEC -0.081 |
hsa-miR-16-5p | SPRED1 | 9 cancers: BLCA; CESC; HNSC; LUAD; LUSC; OV; PAAD; PRAD; UCEC | miRNAWalker2 validate; miRNATAP | TCGA BLCA -0.184; TCGA CESC -0.107; TCGA HNSC -0.131; TCGA LUAD -0.179; TCGA LUSC -0.28; TCGA OV -0.117; TCGA PAAD -0.347; TCGA PRAD -0.12; TCGA UCEC -0.293 |
hsa-miR-16-5p | SPRYD3 | 15 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; OV; PRAD; SARC; THCA; UCEC | miRNAWalker2 validate; MirTarget; miRNATAP | TCGA BLCA -0.225; TCGA BRCA -0.16; TCGA CESC -0.188; TCGA ESCA -0.246; TCGA HNSC -0.228; TCGA KIRC -0.19; TCGA KIRP -0.101; TCGA LIHC -0.21; TCGA LUAD -0.202; TCGA LUSC -0.238; TCGA OV -0.073; TCGA PRAD -0.196; TCGA SARC -0.203; TCGA THCA -0.174; TCGA UCEC -0.192 |
hsa-miR-16-5p | STK33 | 9 cancers: BLCA; CESC; COAD; KIRC; KIRP; LGG; PRAD; STAD; UCEC | miRNAWalker2 validate; MirTarget; miRNATAP | TCGA BLCA -0.563; TCGA CESC -0.558; TCGA COAD -0.962; TCGA KIRC -0.846; TCGA KIRP -0.453; TCGA LGG -0.094; TCGA PRAD -0.545; TCGA STAD -0.527; TCGA UCEC -0.296 |
hsa-miR-16-5p | SYT11 | 10 cancers: BLCA; CESC; COAD; ESCA; LUAD; LUSC; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.552; TCGA CESC -0.317; TCGA COAD -0.726; TCGA ESCA -0.467; TCGA LUAD -0.118; TCGA LUSC -0.299; TCGA PRAD -0.386; TCGA SARC -0.237; TCGA STAD -0.501; TCGA UCEC -0.368 |
hsa-miR-16-5p | TES | 13 cancers: BLCA; COAD; ESCA; HNSC; KIRP; LUSC; OV; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.274; TCGA COAD -0.156; TCGA ESCA -0.238; TCGA HNSC -0.154; TCGA KIRP -0.452; TCGA LUSC -0.146; TCGA OV -0.097; TCGA PAAD -0.215; TCGA PRAD -0.275; TCGA SARC -0.41; TCGA THCA -0.084; TCGA STAD -0.227; TCGA UCEC -0.208 |
hsa-miR-16-5p | TFPI | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; OV; PAAD; PRAD; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.244; TCGA BRCA -0.247; TCGA CESC -0.374; TCGA COAD -0.307; TCGA ESCA -0.292; TCGA HNSC -0.215; TCGA KIRP -0.205; TCGA OV -0.184; TCGA PAAD -0.661; TCGA PRAD -0.277; TCGA STAD -0.261; TCGA UCEC -0.621 |
hsa-miR-16-5p | TIMP3 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.593; TCGA BRCA -0.253; TCGA CESC -0.588; TCGA COAD -0.807; TCGA ESCA -1.151; TCGA HNSC -0.785; TCGA KIRC -0.349; TCGA LGG -0.26; TCGA LIHC -0.239; TCGA LUAD -0.464; TCGA LUSC -1.002; TCGA PAAD -0.636; TCGA PRAD -0.326; TCGA STAD -0.51; TCGA UCEC -0.869 |
hsa-miR-16-5p | TMEM100 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LUAD; OV; PAAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate; MirTarget | TCGA BLCA -1.18; TCGA BRCA -0.49; TCGA CESC -0.649; TCGA COAD -1.091; TCGA ESCA -1.308; TCGA HNSC -0.757; TCGA LGG -0.652; TCGA LUAD -0.321; TCGA OV -0.744; TCGA PAAD -0.482; TCGA PRAD -0.44; TCGA SARC -0.606; TCGA STAD -0.965; TCGA UCEC -0.546 |
hsa-miR-16-5p | TMEM109 | 9 cancers: BLCA; CESC; COAD; LIHC; LUAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.158; TCGA CESC -0.113; TCGA COAD -0.081; TCGA LIHC -0.126; TCGA LUAD -0.112; TCGA PRAD -0.126; TCGA SARC -0.071; TCGA STAD -0.118; TCGA UCEC -0.202 |
hsa-miR-16-5p | TMEM43 | 13 cancers: BLCA; CESC; COAD; ESCA; KIRP; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate; MirTarget | TCGA BLCA -0.113; TCGA CESC -0.11; TCGA COAD -0.114; TCGA ESCA -0.242; TCGA KIRP -0.264; TCGA LUAD -0.086; TCGA LUSC -0.147; TCGA PAAD -0.333; TCGA PRAD -0.108; TCGA SARC -0.213; TCGA THCA -0.127; TCGA STAD -0.321; TCGA UCEC -0.324 |
hsa-miR-16-5p | TNRC6B | 11 cancers: BLCA; CESC; ESCA; KIRC; KIRP; LGG; LUAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate; miRNATAP | TCGA BLCA -0.061; TCGA CESC -0.09; TCGA ESCA -0.189; TCGA KIRC -0.096; TCGA KIRP -0.245; TCGA LGG -0.084; TCGA LUAD -0.095; TCGA PRAD -0.085; TCGA SARC -0.17; TCGA STAD -0.212; TCGA UCEC -0.088 |
hsa-miR-16-5p | TUBA1A | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUSC; OV; PRAD; THCA | miRNAWalker2 validate; MirTarget; miRNATAP | TCGA BLCA -0.437; TCGA BRCA -0.063; TCGA CESC -0.248; TCGA COAD -0.452; TCGA ESCA -0.449; TCGA HNSC -0.318; TCGA KIRP -0.125; TCGA LUSC -0.297; TCGA OV -0.197; TCGA PRAD -0.221; TCGA THCA -0.139 |
hsa-miR-16-5p | VASN | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; OV; PAAD; THCA; STAD | miRNAWalker2 validate | TCGA BLCA -0.454; TCGA BRCA -0.199; TCGA CESC -0.226; TCGA COAD -0.467; TCGA ESCA -0.4; TCGA HNSC -0.35; TCGA KIRP -0.135; TCGA LUAD -0.162; TCGA LUSC -0.714; TCGA OV -0.13; TCGA PAAD -0.352; TCGA THCA -0.286; TCGA STAD -0.207 |
hsa-miR-16-5p | VCL | 13 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.372; TCGA CESC -0.172; TCGA COAD -0.293; TCGA ESCA -0.432; TCGA HNSC -0.209; TCGA KIRC -0.238; TCGA LUAD -0.237; TCGA LUSC -0.356; TCGA PAAD -0.449; TCGA PRAD -0.597; TCGA SARC -0.434; TCGA STAD -0.499; TCGA UCEC -0.294 |
hsa-miR-16-5p | VIM | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUSC; OV; PAAD; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.47; TCGA BRCA -0.16; TCGA CESC -0.472; TCGA COAD -0.602; TCGA ESCA -0.503; TCGA HNSC -0.238; TCGA LUSC -0.584; TCGA OV -0.123; TCGA PAAD -0.226; TCGA STAD -0.164; TCGA UCEC -0.269 |
hsa-miR-16-5p | XYLT1 | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | miRNAWalker2 validate; mirMAP | TCGA BLCA -0.448; TCGA BRCA -0.095; TCGA COAD -0.181; TCGA ESCA -0.567; TCGA HNSC -0.312; TCGA KIRP -0.148; TCGA LUAD -0.479; TCGA LUSC -0.563; TCGA PAAD -0.339; TCGA PRAD -0.163; TCGA STAD -0.253; TCGA UCEC -0.673 |
hsa-miR-16-5p | ZFHX3 | 11 cancers: BLCA; CESC; COAD; HNSC; KIRC; LUAD; OV; PAAD; PRAD; SARC; STAD | miRNAWalker2 validate; MirTarget; miRNATAP | TCGA BLCA -0.193; TCGA CESC -0.158; TCGA COAD -0.282; TCGA HNSC -0.194; TCGA KIRC -0.161; TCGA LUAD -0.177; TCGA OV -0.103; TCGA PAAD -0.223; TCGA PRAD -0.218; TCGA SARC -0.151; TCGA STAD -0.377 |
hsa-miR-16-5p | ZYX | 13 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate; miRTarBase; miRNATAP | TCGA BLCA -0.298; TCGA BRCA -0.168; TCGA CESC -0.168; TCGA ESCA -0.22; TCGA HNSC -0.164; TCGA KIRP -0.092; TCGA LUAD -0.15; TCGA LUSC -0.416; TCGA PAAD -0.232; TCGA PRAD -0.107; TCGA SARC -0.143; TCGA STAD -0.204; TCGA UCEC -0.207 |
hsa-miR-16-5p | FGFR1 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; OV; PAAD; PRAD; STAD | miRTarBase; miRNATAP | TCGA BLCA -0.873; TCGA BRCA -0.184; TCGA CESC -0.564; TCGA COAD -0.71; TCGA ESCA -0.647; TCGA HNSC -0.389; TCGA KIRC -0.148; TCGA LUAD -0.247; TCGA LUSC -0.234; TCGA OV -0.282; TCGA PAAD -0.3; TCGA PRAD -0.293; TCGA STAD -0.537 |
hsa-miR-16-5p | MASP1 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRP; LGG; OV; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.87; TCGA BRCA -0.765; TCGA CESC -0.785; TCGA COAD -1.768; TCGA ESCA -0.689; TCGA KIRP -0.377; TCGA LGG -0.446; TCGA OV -0.346; TCGA PAAD -0.648; TCGA PRAD -0.956; TCGA SARC -1.014; TCGA STAD -0.606; TCGA UCEC -1.377 |
hsa-miR-16-5p | DCAF8 | 11 cancers: BLCA; ESCA; HNSC; KIRC; LGG; LIHC; OV; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.071; TCGA ESCA -0.135; TCGA HNSC -0.085; TCGA KIRC -0.09; TCGA LGG -0.053; TCGA LIHC -0.098; TCGA OV -0.057; TCGA PRAD -0.072; TCGA SARC -0.196; TCGA STAD -0.099; TCGA UCEC -0.062 |
hsa-miR-16-5p | ADCY5 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; LUAD; OV; PAAD; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -1.471; TCGA BRCA -0.723; TCGA CESC -0.405; TCGA COAD -1.475; TCGA ESCA -1.838; TCGA KIRC -0.44; TCGA LUAD -0.429; TCGA OV -0.318; TCGA PAAD -0.64; TCGA PRAD -0.827; TCGA SARC -1.269; TCGA STAD -1.19; TCGA UCEC -0.369 |
hsa-miR-16-5p | REEP1 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; OV; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -1.14; TCGA BRCA -0.382; TCGA CESC -0.396; TCGA COAD -0.721; TCGA ESCA -0.736; TCGA HNSC -0.474; TCGA OV -0.234; TCGA PRAD -0.59; TCGA SARC -0.787; TCGA STAD -1.023; TCGA UCEC -0.627 |
hsa-miR-16-5p | EGLN1 | 9 cancers: BLCA; ESCA; HNSC; LGG; LIHC; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.072; TCGA ESCA -0.128; TCGA HNSC -0.095; TCGA LGG -0.098; TCGA LIHC -0.185; TCGA PRAD -0.092; TCGA SARC -0.113; TCGA STAD -0.157; TCGA UCEC -0.109 |
hsa-miR-16-5p | CRIM1 | 13 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.221; TCGA BRCA -0.196; TCGA COAD -0.174; TCGA ESCA -0.295; TCGA HNSC -0.231; TCGA KIRC -0.181; TCGA LUAD -0.264; TCGA LUSC -0.27; TCGA PAAD -0.339; TCGA PRAD -0.33; TCGA SARC -0.317; TCGA STAD -0.306; TCGA UCEC -0.421 |
hsa-miR-16-5p | EDA | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LGG; LUAD; OV; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.379; TCGA BRCA -0.163; TCGA COAD -0.513; TCGA ESCA -0.708; TCGA HNSC -0.471; TCGA KIRC -0.261; TCGA LGG -0.095; TCGA LUAD -0.259; TCGA OV -0.215; TCGA SARC -0.317; TCGA STAD -0.296; TCGA UCEC -0.198 |
hsa-miR-16-5p | DZIP1 | 10 cancers: BLCA; BRCA; COAD; ESCA; KIRC; LUAD; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.371; TCGA BRCA -0.107; TCGA COAD -0.826; TCGA ESCA -0.359; TCGA KIRC -0.208; TCGA LUAD -0.22; TCGA PAAD -0.473; TCGA PRAD -0.292; TCGA STAD -0.661; TCGA UCEC -0.436 |
hsa-miR-16-5p | THRB | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LUSC; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.196; TCGA BRCA -0.321; TCGA COAD -0.807; TCGA ESCA -0.714; TCGA HNSC -0.451; TCGA KIRC -1.029; TCGA LUSC -0.302; TCGA SARC -0.466; TCGA STAD -0.572; TCGA UCEC -0.587 |
hsa-miR-16-5p | NBR1 | 11 cancers: BLCA; CESC; HNSC; KIRC; KIRP; LGG; LUAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.074; TCGA CESC -0.07; TCGA HNSC -0.081; TCGA KIRC -0.127; TCGA KIRP -0.113; TCGA LGG -0.053; TCGA LUAD -0.073; TCGA PRAD -0.17; TCGA SARC -0.177; TCGA STAD -0.098; TCGA UCEC -0.217 |
hsa-miR-16-5p | RAB11FIP2 | 15 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; OV; PAAD; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.179; TCGA CESC -0.219; TCGA COAD -0.212; TCGA ESCA -0.266; TCGA HNSC -0.117; TCGA KIRC -0.298; TCGA KIRP -0.274; TCGA LUAD -0.094; TCGA LUSC -0.203; TCGA OV -0.215; TCGA PAAD -0.151; TCGA PRAD -0.276; TCGA SARC -0.139; TCGA STAD -0.257; TCGA UCEC -0.52 |
hsa-miR-16-5p | SLC25A12 | 9 cancers: BLCA; BRCA; COAD; ESCA; KIRC; KIRP; PRAD; SARC; STAD | MirTarget | TCGA BLCA -0.231; TCGA BRCA -0.061; TCGA COAD -0.135; TCGA ESCA -0.268; TCGA KIRC -0.264; TCGA KIRP -0.141; TCGA PRAD -0.21; TCGA SARC -0.256; TCGA STAD -0.347 |
hsa-miR-16-5p | CNTNAP1 | 11 cancers: BLCA; CESC; COAD; ESCA; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.565; TCGA CESC -0.285; TCGA COAD -0.893; TCGA ESCA -0.766; TCGA LUAD -0.194; TCGA LUSC -0.396; TCGA PAAD -0.434; TCGA PRAD -0.293; TCGA SARC -0.251; TCGA STAD -0.604; TCGA UCEC -0.209 |
hsa-miR-16-5p | PIK3R1 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LUAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.203; TCGA BRCA -0.185; TCGA CESC -0.233; TCGA COAD -0.19; TCGA ESCA -0.405; TCGA HNSC -0.138; TCGA LGG -0.122; TCGA LUAD -0.147; TCGA PRAD -0.329; TCGA SARC -0.384; TCGA STAD -0.332; TCGA UCEC -0.156 |
hsa-miR-16-5p | TGFBR3 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.536; TCGA BRCA -0.274; TCGA CESC -0.399; TCGA COAD -0.222; TCGA ESCA -0.679; TCGA HNSC -0.288; TCGA KIRC -0.443; TCGA LUAD -0.217; TCGA PRAD -0.44; TCGA SARC -0.424; TCGA STAD -0.707; TCGA UCEC -0.61 |
hsa-miR-16-5p | SEMA3A | 9 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUSC; OV; PRAD; STAD | MirTarget | TCGA BLCA -0.732; TCGA BRCA -0.138; TCGA COAD -0.665; TCGA ESCA -0.589; TCGA HNSC -0.404; TCGA LUSC -0.369; TCGA OV -0.216; TCGA PRAD -0.894; TCGA STAD -0.674 |
hsa-miR-16-5p | KLHDC8B | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; PAAD; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.181; TCGA BRCA -0.161; TCGA CESC -0.17; TCGA COAD -0.387; TCGA ESCA -0.246; TCGA HNSC -0.206; TCGA LGG -0.106; TCGA LIHC -0.17; TCGA PAAD -0.221; TCGA PRAD -0.133; TCGA THCA -0.109; TCGA STAD -0.344; TCGA UCEC -0.298 |
hsa-miR-16-5p | THSD4 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.355; TCGA BRCA -0.49; TCGA CESC -0.328; TCGA COAD -0.318; TCGA ESCA -0.589; TCGA HNSC -0.443; TCGA KIRC -0.884; TCGA LGG -0.143; TCGA LUAD -0.211; TCGA LUSC -0.417; TCGA PAAD -0.707; TCGA PRAD -0.65; TCGA SARC -0.504; TCGA STAD -0.426; TCGA UCEC -0.257 |
hsa-miR-16-5p | UBL3 | 10 cancers: BLCA; ESCA; HNSC; KIRC; LGG; LUSC; OV; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.228; TCGA ESCA -0.298; TCGA HNSC -0.226; TCGA KIRC -0.363; TCGA LGG -0.113; TCGA LUSC -0.207; TCGA OV -0.166; TCGA PRAD -0.099; TCGA STAD -0.137; TCGA UCEC -0.428 |
hsa-miR-16-5p | SETD3 | 11 cancers: BLCA; CESC; ESCA; HNSC; KIRC; LGG; LUAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.127; TCGA CESC -0.07; TCGA ESCA -0.163; TCGA HNSC -0.195; TCGA KIRC -0.178; TCGA LGG -0.065; TCGA LUAD -0.058; TCGA SARC -0.085; TCGA THCA -0.073; TCGA STAD -0.115; TCGA UCEC -0.152 |
hsa-miR-16-5p | ZBTB46 | 10 cancers: BLCA; BRCA; COAD; ESCA; KIRP; LGG; LUAD; LUSC; THCA; UCEC | MirTarget | TCGA BLCA -0.157; TCGA BRCA -0.1; TCGA COAD -0.304; TCGA ESCA -0.378; TCGA KIRP -0.139; TCGA LGG -0.081; TCGA LUAD -0.136; TCGA LUSC -0.425; TCGA THCA -0.124; TCGA UCEC -0.33 |
hsa-miR-16-5p | PTPN14 | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; STAD | MirTarget | TCGA BLCA -0.094; TCGA BRCA -0.101; TCGA COAD -0.195; TCGA ESCA -0.228; TCGA HNSC -0.17; TCGA KIRC -0.333; TCGA LUAD -0.17; TCGA LUSC -0.238; TCGA PAAD -0.605; TCGA PRAD -0.391; TCGA STAD -0.318 |
hsa-miR-16-5p | SAMD4A | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.63; TCGA BRCA -0.143; TCGA CESC -0.285; TCGA COAD -0.45; TCGA ESCA -0.643; TCGA HNSC -0.279; TCGA LUAD -0.235; TCGA LUSC -0.706; TCGA PRAD -0.235; TCGA SARC -0.244; TCGA STAD -0.611; TCGA UCEC -0.508 |
hsa-miR-16-5p | FERMT2 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.803; TCGA BRCA -0.156; TCGA CESC -0.539; TCGA COAD -0.962; TCGA ESCA -0.92; TCGA HNSC -0.477; TCGA KIRC -0.226; TCGA KIRP -0.131; TCGA LUAD -0.117; TCGA LUSC -0.471; TCGA PAAD -0.447; TCGA PRAD -0.469; TCGA SARC -0.383; TCGA STAD -0.811; TCGA UCEC -0.439 |
hsa-miR-16-5p | MAP3K3 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PRAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.224; TCGA BRCA -0.091; TCGA CESC -0.122; TCGA COAD -0.28; TCGA ESCA -0.298; TCGA HNSC -0.071; TCGA KIRC -0.119; TCGA KIRP -0.065; TCGA LUAD -0.103; TCGA LUSC -0.166; TCGA PRAD -0.078; TCGA STAD -0.243; TCGA UCEC -0.243 |
hsa-miR-16-5p | TSPYL2 | 9 cancers: BLCA; BRCA; CESC; COAD; ESCA; LGG; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.215; TCGA BRCA -0.182; TCGA CESC -0.111; TCGA COAD -0.136; TCGA ESCA -0.261; TCGA LGG -0.108; TCGA SARC -0.297; TCGA STAD -0.391; TCGA UCEC -0.157 |
hsa-miR-16-5p | MFAP5 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; STAD; UCEC | MirTarget | TCGA BLCA -1.787; TCGA BRCA -0.147; TCGA CESC -0.962; TCGA COAD -1.242; TCGA ESCA -1.648; TCGA HNSC -1.245; TCGA LUAD -0.592; TCGA LUSC -0.95; TCGA PAAD -1.52; TCGA STAD -0.719; TCGA UCEC -0.735 |
hsa-miR-16-5p | BVES | 9 cancers: BLCA; COAD; ESCA; KIRC; OV; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.686; TCGA COAD -0.964; TCGA ESCA -1.14; TCGA KIRC -0.369; TCGA OV -0.268; TCGA PRAD -0.518; TCGA SARC -0.38; TCGA STAD -1.018; TCGA UCEC -0.55 |
hsa-miR-16-5p | SCN3A | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.357; TCGA BRCA -0.676; TCGA CESC -0.619; TCGA COAD -0.775; TCGA ESCA -0.64; TCGA HNSC -0.334; TCGA KIRC -1.198; TCGA KIRP -0.402; TCGA LGG -0.182; TCGA SARC -0.931; TCGA STAD -0.447; TCGA UCEC -0.442 |
hsa-miR-16-5p | DUSP3 | 13 cancers: BLCA; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.276; TCGA COAD -0.088; TCGA ESCA -0.326; TCGA HNSC -0.206; TCGA KIRC -0.07; TCGA LIHC -0.141; TCGA LUAD -0.061; TCGA LUSC -0.099; TCGA PAAD -0.179; TCGA PRAD -0.295; TCGA SARC -0.245; TCGA STAD -0.346; TCGA UCEC -0.171 |
hsa-miR-16-5p | PAPPA | 14 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LGG; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.311; TCGA BRCA -0.244; TCGA COAD -0.654; TCGA ESCA -0.517; TCGA HNSC -0.574; TCGA KIRC -1.076; TCGA LGG -0.308; TCGA LUAD -0.328; TCGA LUSC -0.912; TCGA PAAD -0.381; TCGA PRAD -0.402; TCGA SARC -0.366; TCGA STAD -0.225; TCGA UCEC -0.993 |
hsa-miR-16-5p | SSBP2 | 9 cancers: BLCA; BRCA; COAD; ESCA; OV; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.212; TCGA BRCA -0.12; TCGA COAD -0.387; TCGA ESCA -0.369; TCGA OV -0.092; TCGA PRAD -0.176; TCGA SARC -0.242; TCGA STAD -0.555; TCGA UCEC -0.271 |
hsa-miR-16-5p | ZNF662 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.375; TCGA BRCA -0.312; TCGA CESC -0.526; TCGA COAD -0.616; TCGA ESCA -0.897; TCGA HNSC -0.473; TCGA KIRC -0.51; TCGA KIRP -0.556; TCGA LUSC -0.33; TCGA PAAD -0.334; TCGA PRAD -0.432; TCGA SARC -0.349; TCGA STAD -0.714; TCGA UCEC -0.65 |
hsa-miR-16-5p | RORA | 11 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LGG; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.328; TCGA CESC -0.284; TCGA COAD -0.547; TCGA ESCA -0.599; TCGA HNSC -0.37; TCGA KIRP -0.251; TCGA LGG -0.093; TCGA PRAD -0.113; TCGA SARC -0.231; TCGA STAD -0.449; TCGA UCEC -0.339 |
hsa-miR-16-5p | DYNC1I1 | 9 cancers: BLCA; CESC; COAD; HNSC; LUAD; OV; PRAD; SARC; STAD | MirTarget; miRNATAP | TCGA BLCA -0.692; TCGA CESC -0.694; TCGA COAD -1.045; TCGA HNSC -0.586; TCGA LUAD -0.266; TCGA OV -0.341; TCGA PRAD -0.705; TCGA SARC -0.62; TCGA STAD -0.866 |
hsa-miR-16-5p | CSRNP1 | 9 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LUSC; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.364; TCGA BRCA -0.34; TCGA ESCA -0.485; TCGA HNSC -0.128; TCGA KIRC -0.247; TCGA LUSC -0.354; TCGA PRAD -0.232; TCGA STAD -0.174; TCGA UCEC -0.145 |
hsa-miR-16-5p | NUAK1 | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; LUAD; LUSC; PAAD; STAD; UCEC | MirTarget | TCGA BLCA -0.198; TCGA BRCA -0.22; TCGA CESC -0.286; TCGA COAD -0.611; TCGA ESCA -0.219; TCGA LUAD -0.415; TCGA LUSC -0.568; TCGA PAAD -0.677; TCGA STAD -0.201; TCGA UCEC -0.4 |
hsa-miR-16-5p | UBE4B | 9 cancers: BLCA; ESCA; HNSC; KIRC; LUSC; PRAD; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.09; TCGA ESCA -0.182; TCGA HNSC -0.117; TCGA KIRC -0.16; TCGA LUSC -0.119; TCGA PRAD -0.131; TCGA THCA -0.12; TCGA STAD -0.186; TCGA UCEC -0.134 |
hsa-miR-16-5p | PCDH17 | 10 cancers: BLCA; CESC; COAD; HNSC; LUAD; LUSC; OV; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.202; TCGA CESC -0.503; TCGA COAD -0.493; TCGA HNSC -0.286; TCGA LUAD -0.177; TCGA LUSC -0.35; TCGA OV -0.147; TCGA THCA -0.243; TCGA STAD -0.195; TCGA UCEC -0.422 |
hsa-miR-16-5p | ASB1 | 9 cancers: BLCA; BRCA; COAD; ESCA; KIRC; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.103; TCGA BRCA -0.151; TCGA COAD -0.11; TCGA ESCA -0.1; TCGA KIRC -0.275; TCGA PRAD -0.206; TCGA SARC -0.228; TCGA STAD -0.208; TCGA UCEC -0.142 |
hsa-miR-16-5p | PPM1L | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; PRAD; SARC; STAD | MirTarget | TCGA BLCA -0.419; TCGA CESC -0.247; TCGA COAD -0.258; TCGA ESCA -0.483; TCGA HNSC -0.399; TCGA KIRC -0.425; TCGA PRAD -0.528; TCGA SARC -0.33; TCGA STAD -0.367 |
hsa-miR-16-5p | TNFAIP8L3 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LUAD; LUSC; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.756; TCGA BRCA -0.088; TCGA CESC -0.37; TCGA COAD -0.561; TCGA ESCA -0.777; TCGA HNSC -0.437; TCGA LGG -0.127; TCGA LUAD -0.207; TCGA LUSC -0.213; TCGA PRAD -0.392; TCGA STAD -0.665; TCGA UCEC -1.312 |
hsa-miR-16-5p | ZDHHC15 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUSC; PAAD; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.809; TCGA BRCA -0.202; TCGA CESC -0.81; TCGA COAD -1.002; TCGA ESCA -1.301; TCGA HNSC -0.608; TCGA KIRC -1.101; TCGA KIRP -0.233; TCGA LUSC -0.343; TCGA PAAD -0.314; TCGA PRAD -0.603; TCGA THCA -0.083; TCGA STAD -0.836; TCGA UCEC -0.784 |
hsa-miR-16-5p | PCDHAC2 | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUAD; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.316; TCGA CESC -0.578; TCGA COAD -0.602; TCGA ESCA -0.878; TCGA HNSC -0.521; TCGA LUAD -0.287; TCGA PRAD -0.573; TCGA THCA -0.436; TCGA STAD -0.566; TCGA UCEC -0.294 |
hsa-miR-16-5p | DSEL | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; OV; PAAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.48; TCGA BRCA -0.234; TCGA CESC -0.34; TCGA COAD -0.85; TCGA ESCA -0.735; TCGA HNSC -0.576; TCGA LUAD -0.221; TCGA LUSC -0.4; TCGA OV -0.191; TCGA PAAD -0.534; TCGA STAD -0.611; TCGA UCEC -0.542 |
hsa-miR-16-5p | RGMA | 11 cancers: BLCA; BRCA; COAD; ESCA; KIRC; LGG; LIHC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.853; TCGA BRCA -0.17; TCGA COAD -1.308; TCGA ESCA -0.991; TCGA KIRC -0.393; TCGA LGG -0.3; TCGA LIHC -0.341; TCGA PAAD -0.654; TCGA PRAD -0.307; TCGA STAD -1.233; TCGA UCEC -0.555 |
hsa-miR-16-5p | TRPC1 | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LGG; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.365; TCGA BRCA -0.209; TCGA COAD -0.486; TCGA ESCA -0.742; TCGA HNSC -0.386; TCGA KIRP -0.284; TCGA LGG -0.079; TCGA PRAD -0.286; TCGA STAD -0.746; TCGA UCEC -0.587 |
hsa-miR-16-5p | RABGAP1 | 10 cancers: BLCA; COAD; ESCA; KIRC; KIRP; LGG; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.188; TCGA COAD -0.079; TCGA ESCA -0.264; TCGA KIRC -0.142; TCGA KIRP -0.08; TCGA LGG -0.125; TCGA PRAD -0.113; TCGA SARC -0.246; TCGA STAD -0.305; TCGA UCEC -0.114 |
hsa-miR-16-5p | AMOTL1 | 16 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LGG; LUAD; OV; PAAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.438; TCGA BRCA -0.257; TCGA CESC -0.219; TCGA COAD -0.928; TCGA ESCA -0.454; TCGA HNSC -0.184; TCGA KIRP -0.165; TCGA LGG -0.133; TCGA LUAD -0.236; TCGA OV -0.171; TCGA PAAD -0.421; TCGA PRAD -0.157; TCGA SARC -0.322; TCGA THCA -0.214; TCGA STAD -0.655; TCGA UCEC -0.324 |
hsa-miR-16-5p | PTHLH | 10 cancers: BLCA; BRCA; CESC; COAD; HNSC; OV; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.371; TCGA BRCA -0.465; TCGA CESC -0.381; TCGA COAD -0.818; TCGA HNSC -0.563; TCGA OV -0.285; TCGA PAAD -0.579; TCGA PRAD -0.229; TCGA STAD -0.481; TCGA UCEC -0.24 |
hsa-miR-16-5p | GALNT13 | 11 cancers: BLCA; CESC; COAD; HNSC; KIRC; LGG; OV; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.611; TCGA CESC -0.808; TCGA COAD -1.04; TCGA HNSC -1.073; TCGA KIRC -0.385; TCGA LGG -0.596; TCGA OV -0.408; TCGA PRAD -0.729; TCGA SARC -0.825; TCGA STAD -0.855; TCGA UCEC -0.816 |
hsa-miR-16-5p | MKX | 14 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LGG; LIHC; OV; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -1.076; TCGA BRCA -0.237; TCGA COAD -0.518; TCGA ESCA -0.892; TCGA HNSC -0.327; TCGA KIRC -0.458; TCGA LGG -0.976; TCGA LIHC -0.424; TCGA OV -0.218; TCGA PAAD -0.712; TCGA PRAD -0.415; TCGA SARC -0.448; TCGA STAD -1.029; TCGA UCEC -0.337 |
hsa-miR-16-5p | LRRN3 | 9 cancers: BLCA; BRCA; COAD; ESCA; LGG; OV; PRAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.395; TCGA BRCA -0.518; TCGA COAD -0.548; TCGA ESCA -0.302; TCGA LGG -0.198; TCGA OV -0.167; TCGA PRAD -0.587; TCGA STAD -0.449; TCGA UCEC -0.249 |
hsa-miR-16-5p | LSM11 | 11 cancers: BLCA; CESC; COAD; ESCA; KIRP; LIHC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.333; TCGA CESC -0.098; TCGA COAD -0.078; TCGA ESCA -0.259; TCGA KIRP -0.148; TCGA LIHC -0.111; TCGA PAAD -0.183; TCGA PRAD -0.263; TCGA SARC -0.379; TCGA STAD -0.229; TCGA UCEC -0.186 |
hsa-miR-16-5p | MYLK | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -1.274; TCGA BRCA -0.387; TCGA CESC -0.738; TCGA COAD -1.15; TCGA ESCA -1.184; TCGA HNSC -0.461; TCGA LUAD -0.449; TCGA LUSC -0.639; TCGA PAAD -0.751; TCGA PRAD -0.943; TCGA SARC -0.949; TCGA STAD -1.218; TCGA UCEC -0.982 |
hsa-miR-16-5p | GHR | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LUAD; OV; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.877; TCGA BRCA -0.35; TCGA CESC -0.562; TCGA COAD -0.807; TCGA ESCA -0.698; TCGA HNSC -0.713; TCGA KIRC -0.254; TCGA LGG -0.219; TCGA LUAD -0.371; TCGA OV -0.243; TCGA PRAD -0.515; TCGA SARC -0.366; TCGA STAD -0.858; TCGA UCEC -0.808 |
hsa-miR-16-5p | TPM2 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -1.062; TCGA BRCA -0.438; TCGA CESC -0.324; TCGA COAD -0.827; TCGA ESCA -0.92; TCGA HNSC -0.562; TCGA LUAD -0.234; TCGA LUSC -0.642; TCGA PAAD -0.596; TCGA PRAD -0.616; TCGA SARC -0.52; TCGA STAD -0.949; TCGA UCEC -0.514 |
hsa-miR-16-5p | ENAH | 12 cancers: BLCA; COAD; HNSC; LGG; LUAD; LUSC; OV; PAAD; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.229; TCGA COAD -0.252; TCGA HNSC -0.237; TCGA LGG -0.134; TCGA LUAD -0.1; TCGA LUSC -0.22; TCGA OV -0.097; TCGA PAAD -0.29; TCGA PRAD -0.079; TCGA SARC -0.207; TCGA STAD -0.407; TCGA UCEC -0.129 |
hsa-miR-16-5p | TBC1D20 | 9 cancers: BLCA; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; STAD; UCEC | MirTarget | TCGA BLCA -0.062; TCGA COAD -0.132; TCGA ESCA -0.097; TCGA HNSC -0.13; TCGA LIHC -0.07; TCGA LUAD -0.057; TCGA LUSC -0.104; TCGA STAD -0.157; TCGA UCEC -0.052 |
hsa-miR-16-5p | ITGA10 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.321; TCGA BRCA -0.52; TCGA CESC -0.392; TCGA COAD -0.395; TCGA ESCA -0.619; TCGA HNSC -0.415; TCGA LIHC -0.27; TCGA LUAD -0.234; TCGA LUSC -0.271; TCGA THCA -0.259; TCGA STAD -0.416; TCGA UCEC -0.188 |
hsa-miR-16-5p | UBXN10 | 10 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; KIRP; LUSC; PRAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.339; TCGA BRCA -0.571; TCGA ESCA -1; TCGA HNSC -0.579; TCGA KIRC -0.244; TCGA KIRP -0.323; TCGA LUSC -0.594; TCGA PRAD -1.006; TCGA STAD -0.61; TCGA UCEC -0.286 |
hsa-miR-16-5p | SESN1 | 10 cancers: BLCA; BRCA; CESC; ESCA; LGG; LUSC; OV; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.124; TCGA BRCA -0.202; TCGA CESC -0.167; TCGA ESCA -0.232; TCGA LGG -0.099; TCGA LUSC -0.16; TCGA OV -0.188; TCGA SARC -0.177; TCGA STAD -0.144; TCGA UCEC -0.258 |
hsa-miR-16-5p | ANKS1A | 11 cancers: BLCA; ESCA; KIRC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.189; TCGA ESCA -0.249; TCGA KIRC -0.407; TCGA LUAD -0.169; TCGA LUSC -0.13; TCGA PAAD -0.116; TCGA PRAD -0.137; TCGA SARC -0.304; TCGA THCA -0.076; TCGA STAD -0.264; TCGA UCEC -0.088 |
hsa-miR-16-5p | CCDC6 | 11 cancers: BLCA; CESC; ESCA; HNSC; LGG; LUAD; PAAD; PRAD; SARC; THCA; STAD | MirTarget; miRNATAP | TCGA BLCA -0.136; TCGA CESC -0.084; TCGA ESCA -0.142; TCGA HNSC -0.147; TCGA LGG -0.054; TCGA LUAD -0.099; TCGA PAAD -0.205; TCGA PRAD -0.083; TCGA SARC -0.171; TCGA THCA -0.071; TCGA STAD -0.163 |
hsa-miR-16-5p | MYRIP | 9 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.693; TCGA BRCA -0.484; TCGA CESC -0.439; TCGA ESCA -0.876; TCGA HNSC -0.46; TCGA KIRC -0.485; TCGA SARC -0.751; TCGA STAD -0.627; TCGA UCEC -0.211 |
hsa-miR-16-5p | BACH2 | 9 cancers: BLCA; BRCA; CESC; COAD; KIRC; LUAD; OV; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.524; TCGA BRCA -0.296; TCGA CESC -0.237; TCGA COAD -0.686; TCGA KIRC -0.356; TCGA LUAD -0.206; TCGA OV -0.197; TCGA STAD -0.341; TCGA UCEC -0.611 |
hsa-miR-16-5p | TSPAN5 | 13 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; LUAD; LUSC; OV; PRAD; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.182; TCGA BRCA -0.347; TCGA CESC -0.402; TCGA COAD -0.381; TCGA HNSC -0.34; TCGA KIRC -0.59; TCGA LUAD -0.216; TCGA LUSC -0.212; TCGA OV -0.192; TCGA PRAD -0.5; TCGA THCA -0.335; TCGA STAD -0.135; TCGA UCEC -0.572 |
hsa-miR-16-5p | PSKH1 | 14 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; LIHC; LUAD; LUSC; OV; PRAD; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.109; TCGA BRCA -0.126; TCGA CESC -0.138; TCGA COAD -0.069; TCGA HNSC -0.25; TCGA KIRC -0.307; TCGA LIHC -0.326; TCGA LUAD -0.108; TCGA LUSC -0.202; TCGA OV -0.135; TCGA PRAD -0.105; TCGA THCA -0.111; TCGA STAD -0.092; TCGA UCEC -0.157 |
hsa-miR-16-5p | EYA4 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.625; TCGA BRCA -0.183; TCGA CESC -0.55; TCGA COAD -0.806; TCGA ESCA -0.746; TCGA HNSC -0.668; TCGA KIRC -1.813; TCGA LUAD -0.424; TCGA LUSC -0.83; TCGA OV -0.486; TCGA PAAD -0.762; TCGA PRAD -0.763; TCGA STAD -0.596; TCGA UCEC -0.435 |
hsa-miR-16-5p | SEPT2 | 10 cancers: BLCA; ESCA; HNSC; LGG; OV; PAAD; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.112; TCGA ESCA -0.101; TCGA HNSC -0.08; TCGA LGG -0.071; TCGA OV -0.104; TCGA PAAD -0.164; TCGA PRAD -0.091; TCGA SARC -0.196; TCGA STAD -0.107; TCGA UCEC -0.14 |
hsa-miR-16-5p | ZNF704 | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.328; TCGA CESC -0.674; TCGA COAD -0.281; TCGA ESCA -0.332; TCGA HNSC -0.542; TCGA KIRC -0.265; TCGA LUAD -0.374; TCGA PAAD -0.626; TCGA PRAD -0.191; TCGA SARC -0.417; TCGA STAD -0.456; TCGA UCEC -0.231 |
hsa-miR-16-5p | PLEKHM3 | 9 cancers: BLCA; COAD; ESCA; HNSC; LUSC; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.114; TCGA COAD -0.157; TCGA ESCA -0.188; TCGA HNSC -0.142; TCGA LUSC -0.125; TCGA PRAD -0.261; TCGA SARC -0.235; TCGA STAD -0.378; TCGA UCEC -0.271 |
hsa-miR-16-5p | RELN | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUSC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -1.023; TCGA BRCA -0.794; TCGA CESC -0.55; TCGA COAD -1.485; TCGA ESCA -1.287; TCGA HNSC -0.831; TCGA KIRC -0.758; TCGA KIRP -0.957; TCGA LIHC -0.763; TCGA LUSC -0.662; TCGA STAD -0.792; TCGA UCEC -0.779 |
hsa-miR-16-5p | COBLL1 | 9 cancers: BLCA; ESCA; HNSC; KIRC; KIRP; LUAD; PAAD; PRAD; SARC | MirTarget; miRNATAP | TCGA BLCA -0.172; TCGA ESCA -0.292; TCGA HNSC -0.236; TCGA KIRC -1.04; TCGA KIRP -0.357; TCGA LUAD -0.134; TCGA PAAD -0.191; TCGA PRAD -0.105; TCGA SARC -0.704 |
hsa-miR-16-5p | TMEM55A | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; OV; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.264; TCGA CESC -0.205; TCGA COAD -0.169; TCGA ESCA -0.19; TCGA HNSC -0.227; TCGA KIRC -0.219; TCGA LUAD -0.169; TCGA LUSC -0.257; TCGA OV -0.093; TCGA PRAD -0.131; TCGA STAD -0.57; TCGA UCEC -0.458 |
hsa-miR-16-5p | NRBP1 | 9 cancers: BLCA; CESC; COAD; HNSC; LIHC; LUAD; OV; PAAD; THCA | MirTarget; miRNATAP | TCGA BLCA -0.07; TCGA CESC -0.066; TCGA COAD -0.114; TCGA HNSC -0.091; TCGA LIHC -0.121; TCGA LUAD -0.073; TCGA OV -0.06; TCGA PAAD -0.139; TCGA THCA -0.06 |
hsa-miR-16-5p | SYNE1 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.597; TCGA BRCA -0.104; TCGA CESC -0.421; TCGA COAD -0.79; TCGA ESCA -0.982; TCGA HNSC -0.238; TCGA KIRC -0.355; TCGA LUAD -0.176; TCGA LUSC -0.593; TCGA PRAD -0.28; TCGA SARC -0.371; TCGA STAD -0.595; TCGA UCEC -0.75 |
hsa-miR-16-5p | PLXNA2 | 9 cancers: BLCA; ESCA; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD | MirTarget; miRNATAP | TCGA BLCA -0.169; TCGA ESCA -0.273; TCGA LIHC -0.174; TCGA LUAD -0.187; TCGA LUSC -0.345; TCGA PAAD -0.22; TCGA PRAD -0.205; TCGA SARC -0.261; TCGA STAD -0.109 |
hsa-miR-16-5p | BCL2L2 | 11 cancers: BLCA; BRCA; COAD; ESCA; KIRC; LUAD; LUSC; PRAD; SARC; THCA; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.205; TCGA BRCA -0.072; TCGA COAD -0.117; TCGA ESCA -0.348; TCGA KIRC -0.338; TCGA LUAD -0.101; TCGA LUSC -0.123; TCGA PRAD -0.241; TCGA SARC -0.371; TCGA THCA -0.102; TCGA UCEC -0.294 |
hsa-miR-16-5p | ENTPD1 | 12 cancers: BLCA; CESC; COAD; ESCA; LIHC; LUAD; LUSC; PAAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.381; TCGA CESC -0.195; TCGA COAD -0.362; TCGA ESCA -0.325; TCGA LIHC -0.095; TCGA LUAD -0.081; TCGA LUSC -0.267; TCGA PAAD -0.212; TCGA SARC -0.3; TCGA THCA -0.222; TCGA STAD -0.276; TCGA UCEC -0.302 |
hsa-miR-16-5p | HAS2 | 11 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LUSC; PAAD; PRAD; UCEC | MirTarget | TCGA BLCA -0.285; TCGA CESC -0.401; TCGA COAD -0.6; TCGA ESCA -0.436; TCGA HNSC -0.258; TCGA KIRC -0.381; TCGA LGG -0.228; TCGA LUSC -0.592; TCGA PAAD -0.444; TCGA PRAD -0.269; TCGA UCEC -0.26 |
hsa-miR-16-5p | ZFHX4 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; PAAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.894; TCGA BRCA -0.316; TCGA CESC -0.929; TCGA COAD -1.359; TCGA ESCA -0.643; TCGA HNSC -0.529; TCGA KIRC -0.384; TCGA LGG -0.116; TCGA PAAD -0.959; TCGA STAD -0.84; TCGA UCEC -0.417 |
hsa-miR-16-5p | ARHGEF12 | 14 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.089; TCGA CESC -0.136; TCGA COAD -0.064; TCGA ESCA -0.152; TCGA HNSC -0.194; TCGA KIRC -0.223; TCGA LUAD -0.163; TCGA LUSC -0.132; TCGA PAAD -0.247; TCGA PRAD -0.116; TCGA SARC -0.192; TCGA THCA -0.134; TCGA STAD -0.169; TCGA UCEC -0.148 |
hsa-miR-16-5p | DYNC1LI2 | 13 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.124; TCGA CESC -0.113; TCGA COAD -0.056; TCGA ESCA -0.235; TCGA HNSC -0.172; TCGA KIRP -0.103; TCGA LUAD -0.093; TCGA LUSC -0.2; TCGA PAAD -0.158; TCGA PRAD -0.182; TCGA SARC -0.153; TCGA STAD -0.264; TCGA UCEC -0.209 |
hsa-miR-16-5p | CCDC149 | 11 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.249; TCGA BRCA -0.108; TCGA ESCA -0.312; TCGA HNSC -0.212; TCGA KIRC -0.187; TCGA LUAD -0.135; TCGA LUSC -0.172; TCGA PRAD -0.125; TCGA SARC -0.107; TCGA STAD -0.254; TCGA UCEC -0.172 |
hsa-miR-16-5p | NCS1 | 9 cancers: BLCA; COAD; ESCA; KIRC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.442; TCGA COAD -0.363; TCGA ESCA -0.43; TCGA KIRC -0.297; TCGA PAAD -0.213; TCGA PRAD -0.338; TCGA SARC -0.321; TCGA STAD -0.762; TCGA UCEC -0.286 |
hsa-miR-16-5p | NFAT5 | 12 cancers: BLCA; CESC; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.089; TCGA CESC -0.092; TCGA ESCA -0.186; TCGA HNSC -0.15; TCGA KIRC -0.337; TCGA KIRP -0.333; TCGA LUAD -0.09; TCGA LUSC -0.23; TCGA PRAD -0.148; TCGA SARC -0.098; TCGA STAD -0.271; TCGA UCEC -0.23 |
hsa-miR-16-5p | RSPO3 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -1.298; TCGA BRCA -0.303; TCGA CESC -0.887; TCGA COAD -1.181; TCGA ESCA -1.978; TCGA HNSC -0.607; TCGA KIRC -0.512; TCGA LGG -0.447; TCGA LUAD -0.293; TCGA LUSC -0.675; TCGA PAAD -0.536; TCGA PRAD -0.626; TCGA SARC -0.825; TCGA STAD -0.857; TCGA UCEC -0.93 |
hsa-miR-16-5p | ARHGAP20 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -1.069; TCGA BRCA -0.477; TCGA CESC -0.519; TCGA COAD -0.759; TCGA ESCA -1.299; TCGA HNSC -0.263; TCGA LUAD -0.293; TCGA LUSC -0.434; TCGA PAAD -0.348; TCGA PRAD -0.743; TCGA SARC -0.816; TCGA THCA -0.266; TCGA STAD -0.75; TCGA UCEC -0.951 |
hsa-miR-16-5p | PAFAH1B1 | 12 cancers: BLCA; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PRAD; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.117; TCGA COAD -0.081; TCGA ESCA -0.168; TCGA HNSC -0.134; TCGA KIRC -0.133; TCGA KIRP -0.144; TCGA LUAD -0.086; TCGA LUSC -0.093; TCGA PRAD -0.202; TCGA THCA -0.06; TCGA STAD -0.134; TCGA UCEC -0.13 |
hsa-miR-16-5p | SNRK | 11 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.119; TCGA CESC -0.107; TCGA COAD -0.099; TCGA ESCA -0.256; TCGA HNSC -0.079; TCGA KIRC -0.13; TCGA LUAD -0.082; TCGA LUSC -0.107; TCGA SARC -0.136; TCGA STAD -0.214; TCGA UCEC -0.267 |
hsa-miR-16-5p | BMX | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.713; TCGA BRCA -0.663; TCGA CESC -0.758; TCGA COAD -0.719; TCGA ESCA -0.793; TCGA HNSC -0.612; TCGA LUAD -0.396; TCGA SARC -0.543; TCGA STAD -0.407; TCGA UCEC -0.377 |
hsa-miR-16-5p | KCNAB1 | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUAD; OV; PAAD; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.404; TCGA CESC -0.188; TCGA COAD -0.33; TCGA ESCA -0.32; TCGA HNSC -0.238; TCGA LUAD -0.173; TCGA OV -0.151; TCGA PAAD -0.344; TCGA PRAD -0.791; TCGA SARC -0.299; TCGA STAD -0.408; TCGA UCEC -0.446 |
hsa-miR-16-5p | HPSE2 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; LGG; LUAD; LUSC; OV; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -2.091; TCGA BRCA -0.738; TCGA CESC -0.659; TCGA COAD -0.543; TCGA ESCA -2.325; TCGA KIRC -1.36; TCGA LGG -1.121; TCGA LUAD -0.673; TCGA LUSC -0.405; TCGA OV -0.387; TCGA PRAD -1.149; TCGA STAD -0.907; TCGA UCEC -0.767 |
hsa-miR-16-5p | SKI | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.157; TCGA BRCA -0.06; TCGA CESC -0.104; TCGA COAD -0.085; TCGA ESCA -0.201; TCGA HNSC -0.111; TCGA LUAD -0.181; TCGA LUSC -0.334; TCGA PAAD -0.186; TCGA STAD -0.186; TCGA UCEC -0.157 |
hsa-miR-16-5p | PPM1K | 9 cancers: BLCA; CESC; COAD; ESCA; KIRC; LGG; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.267; TCGA CESC -0.22; TCGA COAD -0.165; TCGA ESCA -0.347; TCGA KIRC -0.479; TCGA LGG -0.169; TCGA PRAD -0.237; TCGA STAD -0.371; TCGA UCEC -0.406 |
hsa-miR-16-5p | USP44 | 10 cancers: BLCA; BRCA; ESCA; KIRC; KIRP; OV; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.32; TCGA BRCA -0.341; TCGA ESCA -0.707; TCGA KIRC -1.397; TCGA KIRP -0.618; TCGA OV -0.323; TCGA PRAD -0.432; TCGA SARC -0.478; TCGA STAD -0.766; TCGA UCEC -0.531 |
hsa-miR-16-5p | PARVA | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.374; TCGA BRCA -0.071; TCGA CESC -0.198; TCGA COAD -0.201; TCGA ESCA -0.464; TCGA HNSC -0.234; TCGA LUAD -0.195; TCGA LUSC -0.253; TCGA PAAD -0.591; TCGA PRAD -0.361; TCGA SARC -0.263; TCGA THCA -0.126; TCGA STAD -0.531; TCGA UCEC -0.231 |
hsa-miR-16-5p | DPH3 | 9 cancers: BLCA; ESCA; KIRC; LIHC; LUSC; OV; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.077; TCGA ESCA -0.118; TCGA KIRC -0.201; TCGA LIHC -0.091; TCGA LUSC -0.098; TCGA OV -0.096; TCGA PRAD -0.164; TCGA STAD -0.072; TCGA UCEC -0.188 |
hsa-miR-16-5p | VAT1 | 13 cancers: BLCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; OV; PAAD; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.112; TCGA CESC -0.208; TCGA COAD -0.115; TCGA ESCA -0.212; TCGA HNSC -0.326; TCGA LIHC -0.124; TCGA LUAD -0.085; TCGA LUSC -0.236; TCGA OV -0.101; TCGA PAAD -0.193; TCGA THCA -0.112; TCGA STAD -0.12; TCGA UCEC -0.145 |
hsa-miR-16-5p | KIAA2022 | 10 cancers: BLCA; COAD; ESCA; KIRC; LGG; OV; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -1.039; TCGA COAD -1.383; TCGA ESCA -1.602; TCGA KIRC -1.832; TCGA LGG -0.145; TCGA OV -0.24; TCGA PRAD -0.425; TCGA SARC -1.017; TCGA STAD -1.214; TCGA UCEC -0.477 |
hsa-miR-16-5p | WISP1 | 11 cancers: BLCA; CESC; COAD; ESCA; KIRC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.494; TCGA CESC -0.599; TCGA COAD -0.831; TCGA ESCA -0.533; TCGA KIRC -0.307; TCGA LUAD -0.278; TCGA LUSC -0.633; TCGA PAAD -0.812; TCGA PRAD -0.24; TCGA STAD -0.251; TCGA UCEC -0.509 |
hsa-miR-16-5p | ANK2 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -1.062; TCGA BRCA -0.35; TCGA CESC -0.749; TCGA COAD -1.109; TCGA ESCA -1.47; TCGA HNSC -0.632; TCGA KIRC -0.586; TCGA LUAD -0.19; TCGA LUSC -0.271; TCGA PRAD -0.542; TCGA SARC -0.434; TCGA STAD -1.025; TCGA UCEC -0.636 |
hsa-miR-16-5p | SGCA | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -1.922; TCGA BRCA -0.5; TCGA CESC -0.827; TCGA COAD -1.525; TCGA ESCA -2.256; TCGA HNSC -1.049; TCGA LIHC -0.462; TCGA LUAD -0.514; TCGA LUSC -0.936; TCGA PRAD -0.643; TCGA SARC -1.194; TCGA STAD -1.138; TCGA UCEC -1.157 |
hsa-miR-16-5p | UBR3 | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; OV; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.124; TCGA CESC -0.085; TCGA COAD -0.059; TCGA ESCA -0.167; TCGA HNSC -0.216; TCGA KIRC -0.176; TCGA LUAD -0.088; TCGA OV -0.073; TCGA PRAD -0.254; TCGA SARC -0.22; TCGA STAD -0.187; TCGA UCEC -0.129 |
hsa-miR-16-5p | SLC7A2 | 9 cancers: BLCA; BRCA; COAD; ESCA; LUSC; SARC; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.294; TCGA BRCA -0.501; TCGA COAD -0.546; TCGA ESCA -0.479; TCGA LUSC -0.351; TCGA SARC -0.474; TCGA THCA -0.384; TCGA STAD -0.531; TCGA UCEC -0.683 |
hsa-miR-16-5p | COL4A3BP | 9 cancers: BLCA; BRCA; ESCA; LUAD; LUSC; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.102; TCGA BRCA -0.067; TCGA ESCA -0.187; TCGA LUAD -0.09; TCGA LUSC -0.125; TCGA PRAD -0.25; TCGA SARC -0.161; TCGA STAD -0.161; TCGA UCEC -0.118 |
hsa-miR-16-5p | FZD4 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; STAD; UCEC | MirTarget | TCGA BLCA -0.16; TCGA BRCA -0.244; TCGA CESC -0.228; TCGA COAD -0.386; TCGA ESCA -0.494; TCGA HNSC -0.276; TCGA KIRP -0.229; TCGA LUAD -0.145; TCGA LUSC -0.379; TCGA STAD -0.306; TCGA UCEC -0.375 |
hsa-miR-16-5p | PNMAL2 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; KIRP; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.715; TCGA BRCA -0.369; TCGA CESC -0.282; TCGA COAD -0.88; TCGA ESCA -0.351; TCGA KIRC -0.242; TCGA KIRP -0.253; TCGA LUAD -0.399; TCGA LUSC -0.387; TCGA OV -0.217; TCGA PAAD -0.341; TCGA PRAD -0.311; TCGA STAD -0.465; TCGA UCEC -0.536 |
hsa-miR-16-5p | MEOX2 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -1.297; TCGA BRCA -0.483; TCGA CESC -0.82; TCGA COAD -1.343; TCGA ESCA -0.72; TCGA HNSC -0.498; TCGA LUAD -0.352; TCGA LUSC -1.019; TCGA PAAD -0.866; TCGA PRAD -0.284; TCGA STAD -0.906; TCGA UCEC -1.014 |
hsa-miR-16-5p | PID1 | 16 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LUAD; LUSC; OV; PAAD; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.911; TCGA BRCA -0.333; TCGA CESC -0.459; TCGA COAD -0.53; TCGA ESCA -0.514; TCGA HNSC -0.298; TCGA KIRC -0.238; TCGA LGG -0.25; TCGA LUAD -0.308; TCGA LUSC -0.325; TCGA OV -0.456; TCGA PAAD -0.462; TCGA PRAD -0.279; TCGA SARC -0.495; TCGA STAD -0.54; TCGA UCEC -0.912 |
hsa-miR-16-5p | COL24A1 | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LUSC; PAAD; STAD | MirTarget; miRNATAP | TCGA BLCA -0.306; TCGA CESC -0.401; TCGA COAD -0.825; TCGA ESCA -0.406; TCGA HNSC -0.675; TCGA KIRC -0.409; TCGA LUSC -0.453; TCGA PAAD -0.82; TCGA STAD -0.593 |
hsa-miR-16-5p | ADAMTSL3 | 12 cancers: BLCA; CESC; COAD; ESCA; KIRC; KIRP; LUAD; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -1.291; TCGA CESC -0.419; TCGA COAD -1.39; TCGA ESCA -1.191; TCGA KIRC -0.268; TCGA KIRP -0.354; TCGA LUAD -0.277; TCGA PAAD -0.482; TCGA PRAD -0.696; TCGA SARC -0.438; TCGA STAD -0.861; TCGA UCEC -0.749 |
hsa-miR-16-5p | MBNL2 | 11 cancers: BLCA; CESC; ESCA; HNSC; LUSC; OV; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.24; TCGA CESC -0.236; TCGA ESCA -0.343; TCGA HNSC -0.265; TCGA LUSC -0.296; TCGA OV -0.147; TCGA PAAD -0.179; TCGA PRAD -0.474; TCGA SARC -0.214; TCGA STAD -0.291; TCGA UCEC -0.355 |
hsa-miR-16-5p | FAM189A1 | 9 cancers: BLCA; BRCA; ESCA; HNSC; KIRP; LUAD; LUSC; OV; STAD | MirTarget; miRNATAP | TCGA BLCA -0.793; TCGA BRCA -0.608; TCGA ESCA -0.734; TCGA HNSC -0.809; TCGA KIRP -0.519; TCGA LUAD -0.417; TCGA LUSC -0.519; TCGA OV -0.35; TCGA STAD -0.892 |
hsa-miR-16-5p | SEL1L | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; SARC; UCEC | MirTarget | TCGA BLCA -0.103; TCGA CESC -0.078; TCGA COAD -0.136; TCGA ESCA -0.189; TCGA HNSC -0.152; TCGA KIRC -0.156; TCGA LUAD -0.092; TCGA LUSC -0.182; TCGA PAAD -0.258; TCGA PRAD -0.146; TCGA SARC -0.144; TCGA UCEC -0.153 |
hsa-miR-16-5p | ZMAT3 | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUAD; PAAD; PRAD; STAD | MirTarget | TCGA BLCA -0.288; TCGA CESC -0.166; TCGA COAD -0.397; TCGA ESCA -0.184; TCGA HNSC -0.175; TCGA LUAD -0.109; TCGA PAAD -0.285; TCGA PRAD -0.337; TCGA STAD -0.251 |
hsa-miR-16-5p | ITPR1 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUSC; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.46; TCGA BRCA -0.289; TCGA CESC -0.342; TCGA COAD -0.611; TCGA ESCA -0.971; TCGA HNSC -0.165; TCGA KIRC -0.38; TCGA LUSC -0.342; TCGA PRAD -0.763; TCGA SARC -0.521; TCGA STAD -0.426; TCGA UCEC -0.638 |
hsa-miR-16-5p | SMPD1 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; PRAD; THCA; STAD | MirTarget | TCGA BLCA -0.22; TCGA BRCA -0.167; TCGA CESC -0.103; TCGA COAD -0.189; TCGA ESCA -0.252; TCGA HNSC -0.219; TCGA KIRC -0.223; TCGA LIHC -0.159; TCGA LUAD -0.13; TCGA LUSC -0.282; TCGA PRAD -0.194; TCGA THCA -0.128; TCGA STAD -0.08 |
hsa-miR-16-5p | COL12A1 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.359; TCGA BRCA -0.134; TCGA CESC -0.507; TCGA COAD -0.785; TCGA ESCA -0.553; TCGA HNSC -0.583; TCGA KIRC -0.191; TCGA LUAD -0.602; TCGA LUSC -1.213; TCGA PAAD -1.054; TCGA STAD -0.324; TCGA UCEC -0.345 |
hsa-miR-16-5p | KY | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.939; TCGA BRCA -0.648; TCGA CESC -0.324; TCGA COAD -0.421; TCGA ESCA -0.681; TCGA HNSC -0.644; TCGA KIRC -0.55; TCGA PRAD -1.27; TCGA SARC -0.58; TCGA STAD -0.953; TCGA UCEC -0.712 |
hsa-miR-16-5p | SEMA3D | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LUAD; LUSC; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.878; TCGA BRCA -0.587; TCGA CESC -0.575; TCGA COAD -1.243; TCGA ESCA -0.616; TCGA HNSC -0.591; TCGA KIRC -0.408; TCGA LGG -0.496; TCGA LUAD -0.367; TCGA LUSC -0.347; TCGA PRAD -0.774; TCGA SARC -0.721; TCGA STAD -0.702; TCGA UCEC -0.371 |
hsa-miR-16-5p | NFASC | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | MirTarget; mirMAP | TCGA BLCA -0.591; TCGA CESC -0.311; TCGA COAD -0.583; TCGA ESCA -1.03; TCGA HNSC -0.441; TCGA KIRC -0.593; TCGA LUAD -0.305; TCGA LUSC -0.271; TCGA PRAD -0.342; TCGA SARC -1.041; TCGA STAD -0.923; TCGA UCEC -0.665 |
hsa-miR-16-5p | ZFYVE1 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LUAD; LUSC; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.086; TCGA BRCA -0.099; TCGA CESC -0.102; TCGA COAD -0.113; TCGA ESCA -0.253; TCGA HNSC -0.208; TCGA KIRC -0.223; TCGA LGG -0.075; TCGA LUAD -0.098; TCGA LUSC -0.183; TCGA PRAD -0.129; TCGA SARC -0.141; TCGA STAD -0.137; TCGA UCEC -0.093 |
hsa-miR-16-5p | KCNA6 | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LGG; LUAD; OV; STAD; UCEC | MirTarget | TCGA BLCA -0.652; TCGA BRCA -0.551; TCGA CESC -0.607; TCGA ESCA -1.325; TCGA HNSC -0.71; TCGA LGG -0.159; TCGA LUAD -0.246; TCGA OV -0.365; TCGA STAD -0.716; TCGA UCEC -0.473 |
hsa-miR-16-5p | PCDHA3 | 9 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; LUAD; THCA; UCEC | MirTarget | TCGA BLCA -0.622; TCGA BRCA -0.551; TCGA CESC -0.83; TCGA COAD -0.937; TCGA ESCA -0.817; TCGA KIRC -0.561; TCGA LUAD -0.253; TCGA THCA -0.318; TCGA UCEC -0.756 |
hsa-miR-16-5p | PEX12 | 12 cancers: BLCA; CESC; COAD; HNSC; KIRC; KIRP; LGG; OV; PRAD; SARC; THCA; UCEC | MirTarget | TCGA BLCA -0.11; TCGA CESC -0.093; TCGA COAD -0.136; TCGA HNSC -0.101; TCGA KIRC -0.196; TCGA KIRP -0.174; TCGA LGG -0.093; TCGA OV -0.107; TCGA PRAD -0.165; TCGA SARC -0.296; TCGA THCA -0.147; TCGA UCEC -0.225 |
hsa-miR-16-5p | RET | 9 cancers: BLCA; COAD; ESCA; HNSC; KIRC; LGG; LUAD; PRAD; STAD | MirTarget; miRNATAP | TCGA BLCA -0.285; TCGA COAD -0.654; TCGA ESCA -0.934; TCGA HNSC -0.353; TCGA KIRC -0.54; TCGA LGG -0.535; TCGA LUAD -0.336; TCGA PRAD -0.537; TCGA STAD -0.615 |
hsa-miR-16-5p | ANKRD29 | 9 cancers: BLCA; BRCA; COAD; ESCA; LIHC; OV; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.912; TCGA BRCA -0.371; TCGA COAD -0.344; TCGA ESCA -0.698; TCGA LIHC -0.535; TCGA OV -0.218; TCGA SARC -0.507; TCGA STAD -0.51; TCGA UCEC -0.725 |
hsa-miR-16-5p | FAT4 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; OV; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.826; TCGA BRCA -0.383; TCGA CESC -0.604; TCGA COAD -0.901; TCGA ESCA -0.93; TCGA HNSC -0.424; TCGA KIRP -0.212; TCGA LUAD -0.342; TCGA LUSC -0.63; TCGA OV -0.222; TCGA PAAD -0.626; TCGA PRAD -0.619; TCGA SARC -0.515; TCGA STAD -0.581; TCGA UCEC -0.35 |
hsa-miR-16-5p | HSPG2 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.483; TCGA BRCA -0.253; TCGA CESC -0.257; TCGA COAD -0.453; TCGA ESCA -0.44; TCGA HNSC -0.177; TCGA LIHC -0.149; TCGA LUAD -0.365; TCGA LUSC -0.621; TCGA PAAD -0.557; TCGA PRAD -0.192; TCGA SARC -0.364; TCGA THCA -0.175; TCGA STAD -0.316; TCGA UCEC -0.193 |
hsa-miR-16-5p | TACC1 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.467; TCGA BRCA -0.131; TCGA CESC -0.25; TCGA COAD -0.191; TCGA ESCA -0.511; TCGA KIRC -0.283; TCGA LUAD -0.234; TCGA LUSC -0.203; TCGA PAAD -0.217; TCGA PRAD -0.416; TCGA SARC -0.383; TCGA STAD -0.497; TCGA UCEC -0.6 |
hsa-miR-16-5p | STON1 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LUAD; LUSC; PAAD; PRAD; SARC; THCA; UCEC | MirTarget | TCGA BLCA -0.817; TCGA BRCA -0.111; TCGA CESC -0.435; TCGA COAD -1.065; TCGA ESCA -0.871; TCGA HNSC -0.184; TCGA KIRC -0.273; TCGA LGG -0.395; TCGA LUAD -0.399; TCGA LUSC -0.254; TCGA PAAD -0.664; TCGA PRAD -0.342; TCGA SARC -0.59; TCGA THCA -0.172; TCGA UCEC -0.883 |
hsa-miR-16-5p | EZH1 | 12 cancers: BLCA; BRCA; CESC; ESCA; KIRC; KIRP; LGG; LUAD; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.159; TCGA BRCA -0.127; TCGA CESC -0.178; TCGA ESCA -0.273; TCGA KIRC -0.227; TCGA KIRP -0.133; TCGA LGG -0.058; TCGA LUAD -0.072; TCGA PRAD -0.111; TCGA SARC -0.34; TCGA STAD -0.35; TCGA UCEC -0.259 |
hsa-miR-16-5p | ISLR | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; OV; PAAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -1.189; TCGA BRCA -0.342; TCGA CESC -0.736; TCGA COAD -0.928; TCGA ESCA -1.004; TCGA HNSC -0.704; TCGA KIRC -0.602; TCGA LUAD -0.419; TCGA LUSC -0.899; TCGA OV -0.265; TCGA PAAD -0.841; TCGA SARC -0.327; TCGA STAD -0.58; TCGA UCEC -0.62 |
hsa-miR-16-5p | AGPAT4 | 12 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; LUSC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.157; TCGA BRCA -0.151; TCGA CESC -0.26; TCGA COAD -0.412; TCGA HNSC -0.449; TCGA KIRC -0.307; TCGA KIRP -0.41; TCGA LUSC -0.468; TCGA PAAD -0.219; TCGA PRAD -0.18; TCGA STAD -0.401; TCGA UCEC -0.151 |
hsa-miR-16-5p | H6PD | 9 cancers: BLCA; BRCA; ESCA; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.133; TCGA BRCA -0.158; TCGA ESCA -0.276; TCGA LUAD -0.088; TCGA LUSC -0.221; TCGA PAAD -0.288; TCGA PRAD -0.08; TCGA STAD -0.162; TCGA UCEC -0.196 |
hsa-miR-16-5p | NTN1 | 12 cancers: BLCA; BRCA; COAD; ESCA; KIRC; LGG; LUAD; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.763; TCGA BRCA -0.126; TCGA COAD -0.673; TCGA ESCA -0.971; TCGA KIRC -0.737; TCGA LGG -0.158; TCGA LUAD -0.271; TCGA PAAD -0.372; TCGA PRAD -0.465; TCGA SARC -0.791; TCGA STAD -0.949; TCGA UCEC -0.23 |
hsa-miR-16-5p | ADCY2 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LGG; LUAD; OV; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -1.189; TCGA BRCA -0.322; TCGA CESC -0.6; TCGA COAD -1.083; TCGA ESCA -1.724; TCGA HNSC -0.922; TCGA KIRP -0.538; TCGA LGG -0.44; TCGA LUAD -0.31; TCGA OV -0.893; TCGA PAAD -0.467; TCGA PRAD -0.42; TCGA SARC -1.153; TCGA STAD -1.028; TCGA UCEC -0.842 |
hsa-miR-16-5p | IGSF9B | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; KIRP; LUAD; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.963; TCGA BRCA -0.347; TCGA CESC -0.375; TCGA COAD -0.489; TCGA ESCA -0.874; TCGA KIRC -0.286; TCGA KIRP -0.344; TCGA LUAD -0.429; TCGA PAAD -0.49; TCGA PRAD -0.545; TCGA SARC -1.272; TCGA STAD -0.601; TCGA UCEC -0.778 |
hsa-miR-16-5p | COL4A4 | 9 cancers: BLCA; COAD; ESCA; KIRC; KIRP; LUAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.636; TCGA COAD -0.295; TCGA ESCA -0.501; TCGA KIRC -0.982; TCGA KIRP -0.183; TCGA LUAD -0.437; TCGA PRAD -0.371; TCGA STAD -0.44; TCGA UCEC -0.539 |
hsa-miR-16-5p | GNAO1 | 12 cancers: BLCA; COAD; ESCA; LGG; LUAD; LUSC; OV; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -1.04; TCGA COAD -1.266; TCGA ESCA -1.01; TCGA LGG -0.12; TCGA LUAD -0.263; TCGA LUSC -0.24; TCGA OV -0.345; TCGA PRAD -0.546; TCGA SARC -0.834; TCGA THCA -0.296; TCGA STAD -1.05; TCGA UCEC -0.564 |
hsa-miR-16-5p | PITPNM2 | 9 cancers: BLCA; ESCA; HNSC; KIRP; LGG; LUSC; PRAD; SARC; STAD | mirMAP | TCGA BLCA -0.159; TCGA ESCA -0.379; TCGA HNSC -0.18; TCGA KIRP -0.08; TCGA LGG -0.095; TCGA LUSC -0.148; TCGA PRAD -0.102; TCGA SARC -0.23; TCGA STAD -0.262 |
hsa-miR-16-5p | TRIOBP | 9 cancers: BLCA; BRCA; CESC; ESCA; KIRP; PAAD; SARC; THCA; UCEC | mirMAP | TCGA BLCA -0.083; TCGA BRCA -0.146; TCGA CESC -0.139; TCGA ESCA -0.16; TCGA KIRP -0.235; TCGA PAAD -0.152; TCGA SARC -0.147; TCGA THCA -0.123; TCGA UCEC -0.109 |
hsa-miR-16-5p | ZNF516 | 11 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; LGG; OV; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.276; TCGA BRCA -0.154; TCGA CESC -0.156; TCGA COAD -0.178; TCGA HNSC -0.117; TCGA KIRC -0.122; TCGA LGG -0.072; TCGA OV -0.141; TCGA PRAD -0.656; TCGA STAD -0.315; TCGA UCEC -0.267 |
hsa-miR-16-5p | SEC14L5 | 9 cancers: BLCA; ESCA; KIRC; KIRP; LIHC; OV; PRAD; SARC; STAD | mirMAP | TCGA BLCA -0.274; TCGA ESCA -0.453; TCGA KIRC -0.493; TCGA KIRP -0.164; TCGA LIHC -0.395; TCGA OV -0.248; TCGA PRAD -0.204; TCGA SARC -0.588; TCGA STAD -0.298 |
hsa-miR-16-5p | NOVA2 | 9 cancers: BLCA; BRCA; CESC; COAD; HNSC; LIHC; LUAD; LUSC; UCEC | mirMAP | TCGA BLCA -0.127; TCGA BRCA -0.171; TCGA CESC -0.418; TCGA COAD -0.37; TCGA HNSC -0.174; TCGA LIHC -0.29; TCGA LUAD -0.188; TCGA LUSC -0.394; TCGA UCEC -0.242 |
hsa-miR-16-5p | PRX | 16 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.339; TCGA BRCA -0.521; TCGA CESC -0.212; TCGA COAD -0.222; TCGA ESCA -0.445; TCGA HNSC -0.161; TCGA KIRC -0.256; TCGA LIHC -0.311; TCGA LUAD -0.199; TCGA LUSC -0.3; TCGA PAAD -0.272; TCGA PRAD -0.121; TCGA SARC -0.334; TCGA THCA -0.118; TCGA STAD -0.306; TCGA UCEC -0.492 |
hsa-miR-16-5p | ZMIZ1 | 11 cancers: BLCA; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; STAD; UCEC | mirMAP | TCGA BLCA -0.106; TCGA COAD -0.139; TCGA ESCA -0.175; TCGA HNSC -0.188; TCGA KIRC -0.134; TCGA KIRP -0.139; TCGA LGG -0.155; TCGA LUAD -0.182; TCGA LUSC -0.174; TCGA STAD -0.2; TCGA UCEC -0.236 |
hsa-miR-16-5p | VWF | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.332; TCGA BRCA -0.308; TCGA CESC -0.479; TCGA COAD -0.522; TCGA ESCA -0.416; TCGA HNSC -0.253; TCGA LIHC -0.312; TCGA LUAD -0.345; TCGA LUSC -0.429; TCGA SARC -0.196; TCGA STAD -0.151; TCGA UCEC -0.26 |
hsa-miR-16-5p | C1orf21 | 10 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; OV; PAAD; SARC; STAD; UCEC | mirMAP; miRNATAP | TCGA BLCA -0.35; TCGA BRCA -0.338; TCGA ESCA -0.3; TCGA HNSC -0.194; TCGA KIRC -0.19; TCGA OV -0.173; TCGA PAAD -0.229; TCGA SARC -0.167; TCGA STAD -0.383; TCGA UCEC -0.314 |
hsa-miR-16-5p | PDLIM2 | 9 cancers: BLCA; BRCA; CESC; COAD; HNSC; LGG; LUAD; LUSC; SARC | mirMAP | TCGA BLCA -0.375; TCGA BRCA -0.179; TCGA CESC -0.207; TCGA COAD -0.263; TCGA HNSC -0.334; TCGA LGG -0.054; TCGA LUAD -0.192; TCGA LUSC -0.424; TCGA SARC -0.141 |
hsa-miR-16-5p | SNX21 | 12 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LIHC; LUSC; PAAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.17; TCGA BRCA -0.169; TCGA CESC -0.156; TCGA ESCA -0.245; TCGA HNSC -0.328; TCGA KIRC -0.148; TCGA LIHC -0.154; TCGA LUSC -0.125; TCGA PAAD -0.135; TCGA SARC -0.25; TCGA STAD -0.234; TCGA UCEC -0.168 |
hsa-miR-16-5p | PMEPA1 | 11 cancers: BLCA; BRCA; CESC; COAD; HNSC; LUAD; LUSC; OV; PAAD; THCA; UCEC | mirMAP | TCGA BLCA -0.406; TCGA BRCA -0.125; TCGA CESC -0.259; TCGA COAD -0.329; TCGA HNSC -0.432; TCGA LUAD -0.372; TCGA LUSC -0.975; TCGA OV -0.284; TCGA PAAD -0.633; TCGA THCA -0.229; TCGA UCEC -0.272 |
hsa-miR-16-5p | VPS13B | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.083; TCGA CESC -0.164; TCGA COAD -0.189; TCGA ESCA -0.249; TCGA HNSC -0.103; TCGA LIHC -0.12; TCGA LUAD -0.124; TCGA LUSC -0.146; TCGA PRAD -0.132; TCGA SARC -0.233; TCGA STAD -0.16; TCGA UCEC -0.224 |
hsa-miR-16-5p | MTSS1L | 12 cancers: BLCA; CESC; COAD; HNSC; LGG; LUAD; LUSC; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.119; TCGA CESC -0.224; TCGA COAD -0.145; TCGA HNSC -0.131; TCGA LGG -0.198; TCGA LUAD -0.202; TCGA LUSC -0.173; TCGA PRAD -0.082; TCGA SARC -0.246; TCGA THCA -0.152; TCGA STAD -0.38; TCGA UCEC -0.2 |
hsa-miR-16-5p | KIAA0513 | 10 cancers: BLCA; ESCA; HNSC; KIRC; KIRP; LIHC; LUSC; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.401; TCGA ESCA -0.465; TCGA HNSC -0.296; TCGA KIRC -0.136; TCGA KIRP -0.069; TCGA LIHC -0.165; TCGA LUSC -0.471; TCGA PRAD -0.415; TCGA STAD -0.25; TCGA UCEC -0.124 |
hsa-miR-16-5p | KIAA1614 | 12 cancers: BLCA; BRCA; COAD; ESCA; KIRP; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.552; TCGA BRCA -0.318; TCGA COAD -0.452; TCGA ESCA -0.46; TCGA KIRP -0.147; TCGA LUAD -0.274; TCGA LUSC -0.22; TCGA PAAD -0.426; TCGA PRAD -0.579; TCGA SARC -0.23; TCGA STAD -0.507; TCGA UCEC -0.242 |
hsa-miR-16-5p | DAB2IP | 11 cancers: BLCA; BRCA; ESCA; KIRC; KIRP; LUAD; PAAD; PRAD; SARC; THCA; UCEC | mirMAP | TCGA BLCA -0.094; TCGA BRCA -0.189; TCGA ESCA -0.16; TCGA KIRC -0.221; TCGA KIRP -0.092; TCGA LUAD -0.129; TCGA PAAD -0.198; TCGA PRAD -0.11; TCGA SARC -0.197; TCGA THCA -0.204; TCGA UCEC -0.11 |
hsa-miR-16-5p | KIAA1644 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LUSC; OV; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -1.465; TCGA BRCA -0.271; TCGA CESC -0.688; TCGA COAD -1.192; TCGA ESCA -0.938; TCGA HNSC -0.559; TCGA LGG -0.371; TCGA LUSC -0.807; TCGA OV -0.378; TCGA PAAD -0.409; TCGA PRAD -0.801; TCGA STAD -0.983; TCGA UCEC -1.084 |
hsa-miR-16-5p | WTIP | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; LUSC; OV; PAAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.151; TCGA BRCA -0.384; TCGA CESC -0.201; TCGA COAD -0.575; TCGA ESCA -0.359; TCGA HNSC -0.251; TCGA KIRC -0.354; TCGA LIHC -0.233; TCGA LUSC -0.425; TCGA OV -0.135; TCGA PAAD -0.535; TCGA THCA -0.207; TCGA STAD -0.314; TCGA UCEC -0.211 |
hsa-miR-16-5p | WNT9A | 10 cancers: BLCA; BRCA; COAD; ESCA; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.528; TCGA BRCA -0.294; TCGA COAD -0.883; TCGA ESCA -0.767; TCGA LUAD -0.266; TCGA LUSC -0.488; TCGA PAAD -0.376; TCGA PRAD -0.19; TCGA STAD -1.03; TCGA UCEC -0.336 |
hsa-miR-16-5p | LPP | 9 cancers: BLCA; COAD; ESCA; HNSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP; miRNATAP | TCGA BLCA -0.522; TCGA COAD -0.349; TCGA ESCA -0.455; TCGA HNSC -0.165; TCGA PAAD -0.61; TCGA PRAD -0.611; TCGA SARC -0.518; TCGA STAD -0.796; TCGA UCEC -0.198 |
hsa-miR-16-5p | SCN2B | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -1.105; TCGA BRCA -1.167; TCGA CESC -0.755; TCGA COAD -1.618; TCGA ESCA -1.377; TCGA HNSC -1.072; TCGA KIRC -1.69; TCGA LUAD -0.654; TCGA LUSC -0.871; TCGA PAAD -0.913; TCGA PRAD -0.743; TCGA SARC -0.718; TCGA STAD -1.294; TCGA UCEC -0.969 |
hsa-miR-16-5p | PLXDC1 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; STAD; UCEC | mirMAP | TCGA BLCA -0.233; TCGA BRCA -0.271; TCGA CESC -0.446; TCGA COAD -0.452; TCGA ESCA -0.341; TCGA HNSC -0.381; TCGA LIHC -0.258; TCGA LUAD -0.157; TCGA LUSC -0.471; TCGA PAAD -0.326; TCGA STAD -0.208; TCGA UCEC -0.395 |
hsa-miR-16-5p | KLHL21 | 10 cancers: BLCA; BRCA; ESCA; KIRC; LUSC; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.206; TCGA BRCA -0.082; TCGA ESCA -0.211; TCGA KIRC -0.45; TCGA LUSC -0.141; TCGA PRAD -0.203; TCGA SARC -0.173; TCGA THCA -0.091; TCGA STAD -0.27; TCGA UCEC -0.171 |
hsa-miR-16-5p | SDC3 | 9 cancers: BLCA; BRCA; COAD; ESCA; LGG; LUSC; PRAD; THCA; STAD | mirMAP | TCGA BLCA -0.279; TCGA BRCA -0.115; TCGA COAD -0.209; TCGA ESCA -0.244; TCGA LGG -0.104; TCGA LUSC -0.192; TCGA PRAD -0.143; TCGA THCA -0.186; TCGA STAD -0.347 |
hsa-miR-16-5p | PTCHD1 | 9 cancers: BLCA; BRCA; CESC; COAD; ESCA; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.779; TCGA BRCA -0.242; TCGA CESC -0.342; TCGA COAD -1.077; TCGA ESCA -1.014; TCGA PRAD -0.876; TCGA SARC -1.099; TCGA STAD -1.591; TCGA UCEC -0.796 |
hsa-miR-16-5p | SLC39A13 | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LUAD; LUSC; THCA; STAD | mirMAP | TCGA BLCA -0.106; TCGA BRCA -0.162; TCGA CESC -0.142; TCGA ESCA -0.128; TCGA HNSC -0.124; TCGA KIRC -0.233; TCGA LUAD -0.099; TCGA LUSC -0.324; TCGA THCA -0.097; TCGA STAD -0.101 |
hsa-miR-16-5p | GFRA2 | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUSC; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.614; TCGA BRCA -0.338; TCGA CESC -0.664; TCGA COAD -0.591; TCGA ESCA -0.845; TCGA HNSC -0.3; TCGA LUSC -0.405; TCGA PRAD -0.217; TCGA STAD -0.577; TCGA UCEC -0.564 |
hsa-miR-16-5p | AR | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP; miRNATAP | TCGA BLCA -0.634; TCGA BRCA -0.548; TCGA CESC -0.745; TCGA COAD -1.025; TCGA ESCA -1.317; TCGA HNSC -0.677; TCGA LUAD -0.361; TCGA LUSC -0.743; TCGA PAAD -1.103; TCGA PRAD -0.267; TCGA SARC -0.617; TCGA STAD -0.985; TCGA UCEC -0.374 |
hsa-miR-16-5p | SHE | 11 cancers: BLCA; BRCA; COAD; ESCA; LUAD; LUSC; OV; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.497; TCGA BRCA -0.302; TCGA COAD -0.676; TCGA ESCA -0.497; TCGA LUAD -0.258; TCGA LUSC -0.418; TCGA OV -0.149; TCGA PRAD -0.218; TCGA SARC -0.315; TCGA STAD -0.393; TCGA UCEC -0.689 |
hsa-miR-16-5p | KCNK3 | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; LGG; LUSC; OV; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.993; TCGA CESC -0.381; TCGA COAD -1.172; TCGA ESCA -1.027; TCGA HNSC -0.364; TCGA LGG -0.407; TCGA LUSC -0.612; TCGA OV -0.391; TCGA PRAD -1.004; TCGA SARC -1.061; TCGA STAD -1.103; TCGA UCEC -0.876 |
hsa-miR-16-5p | SYNPO | 12 cancers: BLCA; BRCA; COAD; ESCA; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.448; TCGA BRCA -0.194; TCGA COAD -0.372; TCGA ESCA -0.503; TCGA LUAD -0.332; TCGA LUSC -0.565; TCGA PAAD -0.339; TCGA PRAD -0.226; TCGA SARC -0.356; TCGA THCA -0.126; TCGA STAD -0.369; TCGA UCEC -0.384 |
hsa-miR-16-5p | ZHX3 | 12 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LUAD; LUSC; OV; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.264; TCGA BRCA -0.139; TCGA CESC -0.228; TCGA ESCA -0.508; TCGA HNSC -0.303; TCGA KIRC -0.527; TCGA LUAD -0.191; TCGA LUSC -0.271; TCGA OV -0.127; TCGA SARC -0.393; TCGA STAD -0.368; TCGA UCEC -0.222 |
hsa-miR-16-5p | SH3PXD2B | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.316; TCGA BRCA -0.093; TCGA CESC -0.173; TCGA COAD -0.307; TCGA ESCA -0.378; TCGA HNSC -0.253; TCGA LUAD -0.238; TCGA LUSC -0.356; TCGA PAAD -0.706; TCGA PRAD -0.349; TCGA THCA -0.174; TCGA STAD -0.335; TCGA UCEC -0.124 |
hsa-miR-16-5p | MLXIP | 10 cancers: BLCA; ESCA; HNSC; KIRC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.276; TCGA ESCA -0.206; TCGA HNSC -0.199; TCGA KIRC -0.086; TCGA LUAD -0.206; TCGA LUSC -0.21; TCGA PRAD -0.143; TCGA SARC -0.273; TCGA STAD -0.159; TCGA UCEC -0.088 |
hsa-miR-16-5p | HHIPL1 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.453; TCGA BRCA -0.27; TCGA CESC -0.424; TCGA COAD -0.803; TCGA ESCA -0.551; TCGA HNSC -0.416; TCGA LGG -0.078; TCGA LIHC -0.214; TCGA LUAD -0.238; TCGA LUSC -0.314; TCGA PAAD -0.509; TCGA PRAD -0.298; TCGA STAD -0.296; TCGA UCEC -0.163 |
hsa-miR-16-5p | MEX3B | 9 cancers: BLCA; CESC; COAD; LGG; OV; PAAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.16; TCGA CESC -0.332; TCGA COAD -0.581; TCGA LGG -0.269; TCGA OV -0.195; TCGA PAAD -0.336; TCGA SARC -0.188; TCGA STAD -0.182; TCGA UCEC -0.279 |
hsa-miR-16-5p | KIRREL | 14 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUAD; LUSC; OV; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.52; TCGA BRCA -0.305; TCGA COAD -0.854; TCGA ESCA -0.361; TCGA HNSC -0.627; TCGA LUAD -0.429; TCGA LUSC -0.569; TCGA OV -0.113; TCGA PAAD -0.838; TCGA PRAD -0.656; TCGA SARC -0.455; TCGA THCA -0.32; TCGA STAD -0.341; TCGA UCEC -0.33 |
hsa-miR-16-5p | MKL2 | 11 cancers: BLCA; BRCA; COAD; ESCA; KIRC; KIRP; LUAD; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.079; TCGA BRCA -0.073; TCGA COAD -0.097; TCGA ESCA -0.163; TCGA KIRC -0.098; TCGA KIRP -0.136; TCGA LUAD -0.196; TCGA PAAD -0.22; TCGA PRAD -0.126; TCGA STAD -0.215; TCGA UCEC -0.191 |
hsa-miR-16-5p | C15orf52 | 16 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.498; TCGA BRCA -0.196; TCGA CESC -0.328; TCGA COAD -0.325; TCGA ESCA -0.695; TCGA HNSC -0.439; TCGA KIRC -0.315; TCGA KIRP -0.241; TCGA LGG -0.13; TCGA LUAD -0.19; TCGA LUSC -0.478; TCGA PAAD -0.44; TCGA PRAD -0.235; TCGA SARC -0.624; TCGA STAD -0.85; TCGA UCEC -0.403 |
hsa-miR-16-5p | LDOC1L | 13 cancers: BLCA; COAD; ESCA; KIRC; KIRP; LGG; LUAD; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.193; TCGA COAD -0.159; TCGA ESCA -0.185; TCGA KIRC -0.237; TCGA KIRP -0.11; TCGA LGG -0.06; TCGA LUAD -0.187; TCGA PAAD -0.178; TCGA PRAD -0.166; TCGA SARC -0.197; TCGA THCA -0.157; TCGA STAD -0.113; TCGA UCEC -0.202 |
hsa-miR-16-5p | KIF13B | 12 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; SARC; THCA | mirMAP | TCGA BLCA -0.134; TCGA BRCA -0.352; TCGA CESC -0.194; TCGA ESCA -0.275; TCGA HNSC -0.224; TCGA KIRC -0.609; TCGA LUAD -0.264; TCGA LUSC -0.245; TCGA PAAD -0.398; TCGA PRAD -0.188; TCGA SARC -0.203; TCGA THCA -0.077 |
hsa-miR-16-5p | CYS1 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -1.074; TCGA BRCA -0.55; TCGA CESC -0.62; TCGA COAD -1; TCGA ESCA -0.9; TCGA HNSC -0.83; TCGA KIRC -0.985; TCGA KIRP -0.328; TCGA LGG -0.252; TCGA LUAD -0.617; TCGA LUSC -0.872; TCGA PAAD -1.105; TCGA PRAD -0.3; TCGA STAD -0.942; TCGA UCEC -1.157 |
hsa-miR-16-5p | BTBD19 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.323; TCGA BRCA -0.405; TCGA CESC -0.533; TCGA COAD -0.485; TCGA ESCA -0.524; TCGA HNSC -0.344; TCGA KIRP -0.208; TCGA LUAD -0.293; TCGA LUSC -0.908; TCGA PAAD -0.414; TCGA SARC -0.219; TCGA STAD -0.366; TCGA UCEC -0.465 |
hsa-miR-16-5p | DCLK1 | 11 cancers: BLCA; CESC; COAD; ESCA; HNSC; LGG; LUAD; OV; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.927; TCGA CESC -0.748; TCGA COAD -1.299; TCGA ESCA -0.914; TCGA HNSC -0.868; TCGA LGG -0.133; TCGA LUAD -0.211; TCGA OV -0.188; TCGA PRAD -0.371; TCGA STAD -0.84; TCGA UCEC -0.757 |
hsa-miR-16-5p | LRP6 | 12 cancers: BLCA; CESC; COAD; KIRC; LGG; LUAD; OV; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.088; TCGA CESC -0.184; TCGA COAD -0.087; TCGA KIRC -0.304; TCGA LGG -0.064; TCGA LUAD -0.129; TCGA OV -0.112; TCGA PAAD -0.254; TCGA PRAD -0.136; TCGA SARC -0.154; TCGA STAD -0.11; TCGA UCEC -0.137 |
hsa-miR-16-5p | CPEB2 | 9 cancers: BLCA; BRCA; ESCA; HNSC; LUSC; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.221; TCGA BRCA -0.141; TCGA ESCA -0.498; TCGA HNSC -0.403; TCGA LUSC -0.315; TCGA PRAD -0.161; TCGA SARC -0.316; TCGA STAD -0.291; TCGA UCEC -0.301 |
hsa-miR-16-5p | TLL1 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.777; TCGA BRCA -0.465; TCGA CESC -0.355; TCGA COAD -1.018; TCGA ESCA -0.86; TCGA HNSC -0.605; TCGA LGG -0.314; TCGA LUAD -0.271; TCGA LUSC -0.541; TCGA PAAD -0.751; TCGA PRAD -0.37; TCGA SARC -0.367; TCGA STAD -0.575; TCGA UCEC -0.715 |
hsa-miR-16-5p | IGF1R | 15 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; OV; PAAD; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.186; TCGA BRCA -0.126; TCGA COAD -0.147; TCGA ESCA -0.308; TCGA HNSC -0.134; TCGA KIRC -0.223; TCGA KIRP -0.102; TCGA LGG -0.113; TCGA LUAD -0.128; TCGA LUSC -0.199; TCGA OV -0.16; TCGA PAAD -0.276; TCGA THCA -0.122; TCGA STAD -0.295; TCGA UCEC -0.281 |
hsa-miR-16-5p | SEMA6D | 11 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LUAD; PRAD; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.489; TCGA BRCA -0.446; TCGA CESC -0.324; TCGA ESCA -0.385; TCGA HNSC -0.568; TCGA KIRC -1.296; TCGA LUAD -0.305; TCGA PRAD -0.589; TCGA THCA -0.348; TCGA STAD -0.443; TCGA UCEC -0.39 |
hsa-miR-16-5p | PHACTR2 | 11 cancers: BLCA; CESC; ESCA; HNSC; LGG; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.097; TCGA CESC -0.14; TCGA ESCA -0.274; TCGA HNSC -0.21; TCGA LGG -0.115; TCGA LUAD -0.105; TCGA LUSC -0.293; TCGA PAAD -0.348; TCGA PRAD -0.318; TCGA STAD -0.223; TCGA UCEC -0.222 |
hsa-miR-16-5p | PTGFR | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LUSC; PAAD; STAD; UCEC | miRNATAP | TCGA BLCA -1.232; TCGA BRCA -0.295; TCGA CESC -0.573; TCGA COAD -1.012; TCGA ESCA -0.867; TCGA HNSC -0.503; TCGA KIRC -0.869; TCGA LGG -0.297; TCGA LUSC -0.715; TCGA PAAD -0.372; TCGA STAD -0.709; TCGA UCEC -1.527 |
hsa-miR-16-5p | BACE1 | 16 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.295; TCGA BRCA -0.064; TCGA CESC -0.24; TCGA COAD -0.326; TCGA ESCA -0.347; TCGA HNSC -0.278; TCGA KIRC -0.116; TCGA KIRP -0.116; TCGA LIHC -0.167; TCGA LUAD -0.284; TCGA LUSC -0.247; TCGA PAAD -0.267; TCGA SARC -0.1; TCGA THCA -0.096; TCGA STAD -0.359; TCGA UCEC -0.218 |
hsa-miR-16-5p | RASSF8 | 13 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.445; TCGA CESC -0.256; TCGA COAD -0.978; TCGA ESCA -0.698; TCGA HNSC -0.172; TCGA KIRC -0.459; TCGA LUAD -0.161; TCGA LUSC -0.31; TCGA PAAD -0.684; TCGA PRAD -0.177; TCGA SARC -0.28; TCGA STAD -0.755; TCGA UCEC -0.762 |
hsa-miR-16-5p | PLXNC1 | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PRAD; UCEC | miRNATAP | TCGA BLCA -0.501; TCGA CESC -0.5; TCGA COAD -0.96; TCGA ESCA -0.71; TCGA HNSC -0.294; TCGA LUAD -0.215; TCGA LUSC -0.784; TCGA PRAD -0.321; TCGA UCEC -0.449 |
hsa-miR-16-5p | FGF7 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -1.286; TCGA BRCA -0.295; TCGA CESC -0.833; TCGA COAD -0.996; TCGA ESCA -1.408; TCGA HNSC -0.528; TCGA KIRC -0.562; TCGA LUAD -0.36; TCGA LUSC -0.615; TCGA PAAD -0.7; TCGA PRAD -0.636; TCGA SARC -0.689; TCGA STAD -0.821; TCGA UCEC -1.011 |
hsa-miR-16-5p | SGCD | 16 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LUAD; LUSC; OV; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -1.507; TCGA BRCA -0.416; TCGA CESC -0.84; TCGA COAD -1.223; TCGA ESCA -1.46; TCGA HNSC -1.241; TCGA KIRC -0.46; TCGA LGG -0.295; TCGA LUAD -0.565; TCGA LUSC -0.95; TCGA OV -0.386; TCGA PAAD -0.765; TCGA PRAD -0.379; TCGA SARC -0.387; TCGA STAD -0.887; TCGA UCEC -0.546 |
hsa-miR-16-5p | PGM2L1 | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LUSC; PAAD; THCA; UCEC | miRNATAP | TCGA BLCA -0.344; TCGA CESC -0.328; TCGA COAD -0.354; TCGA ESCA -0.334; TCGA HNSC -0.165; TCGA KIRP -0.232; TCGA LUSC -0.29; TCGA PAAD -0.467; TCGA THCA -0.283; TCGA UCEC -0.196 |
hsa-miR-16-5p | SH3BGRL2 | 10 cancers: BLCA; BRCA; CESC; HNSC; KIRC; LIHC; OV; PAAD; PRAD; UCEC | miRNATAP | TCGA BLCA -0.256; TCGA BRCA -0.327; TCGA CESC -0.264; TCGA HNSC -0.298; TCGA KIRC -0.211; TCGA LIHC -0.211; TCGA OV -0.308; TCGA PAAD -0.294; TCGA PRAD -0.389; TCGA UCEC -0.308 |
hsa-miR-16-5p | ESRRG | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LUAD; OV; PAAD; PRAD | miRNATAP | TCGA BLCA -0.303; TCGA BRCA -0.216; TCGA CESC -0.57; TCGA ESCA -0.696; TCGA HNSC -0.624; TCGA KIRC -1.411; TCGA LUAD -0.291; TCGA OV -0.355; TCGA PAAD -0.495; TCGA PRAD -0.581 |
hsa-miR-16-5p | PRKG1 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; OV; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -1.102; TCGA BRCA -0.249; TCGA CESC -0.853; TCGA COAD -0.738; TCGA ESCA -0.883; TCGA HNSC -0.666; TCGA LUAD -0.562; TCGA LUSC -0.844; TCGA OV -0.188; TCGA PAAD -1.023; TCGA PRAD -0.897; TCGA SARC -0.863; TCGA STAD -0.724; TCGA UCEC -0.918 |
hsa-miR-16-5p | KIF1C | 16 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.169; TCGA BRCA -0.161; TCGA CESC -0.096; TCGA ESCA -0.29; TCGA HNSC -0.339; TCGA KIRC -0.334; TCGA KIRP -0.088; TCGA LIHC -0.273; TCGA LUAD -0.144; TCGA LUSC -0.315; TCGA PAAD -0.31; TCGA PRAD -0.191; TCGA SARC -0.098; TCGA THCA -0.105; TCGA STAD -0.093; TCGA UCEC -0.079 |
hsa-miR-16-5p | CAPN6 | 9 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; OV; PRAD; STAD | miRNATAP | TCGA BLCA -0.897; TCGA BRCA -0.395; TCGA CESC -1.174; TCGA COAD -0.999; TCGA HNSC -0.605; TCGA KIRC -0.858; TCGA OV -0.717; TCGA PRAD -0.771; TCGA STAD -0.638 |
hsa-miR-16-5p | ZNF532 | 11 cancers: BLCA; CESC; COAD; KIRP; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.157; TCGA CESC -0.138; TCGA COAD -0.601; TCGA KIRP -0.105; TCGA LUAD -0.088; TCGA LUSC -0.126; TCGA PAAD -0.412; TCGA PRAD -0.28; TCGA SARC -0.152; TCGA STAD -0.367; TCGA UCEC -0.18 |
hsa-miR-16-5p | ADAMTS18 | 9 cancers: BLCA; BRCA; CESC; COAD; HNSC; LUAD; OV; THCA; STAD | miRNATAP | TCGA BLCA -0.582; TCGA BRCA -0.457; TCGA CESC -0.383; TCGA COAD -1.087; TCGA HNSC -0.371; TCGA LUAD -0.333; TCGA OV -0.62; TCGA THCA -0.526; TCGA STAD -0.245 |
hsa-miR-16-5p | ADAMTS5 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LUAD; LUSC; OV; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.558; TCGA BRCA -0.354; TCGA CESC -0.796; TCGA COAD -0.458; TCGA ESCA -0.677; TCGA HNSC -0.635; TCGA LGG -0.321; TCGA LUAD -0.231; TCGA LUSC -0.78; TCGA OV -0.196; TCGA PRAD -0.691; TCGA STAD -0.276; TCGA UCEC -0.69 |
hsa-miR-16-5p | SOS2 | 11 cancers: BLCA; CESC; ESCA; HNSC; KIRC; LGG; LUSC; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.091; TCGA CESC -0.118; TCGA ESCA -0.263; TCGA HNSC -0.154; TCGA KIRC -0.242; TCGA LGG -0.066; TCGA LUSC -0.173; TCGA PRAD -0.112; TCGA SARC -0.123; TCGA STAD -0.211; TCGA UCEC -0.218 |
hsa-miR-16-5p | ROR1 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.938; TCGA BRCA -0.146; TCGA CESC -0.405; TCGA COAD -0.697; TCGA ESCA -0.784; TCGA HNSC -0.491; TCGA LUAD -0.272; TCGA LUSC -0.785; TCGA PAAD -0.637; TCGA PRAD -0.819; TCGA SARC -0.524; TCGA STAD -0.39; TCGA UCEC -0.273 |
hsa-miR-16-5p | NLGN1 | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LGG; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.852; TCGA BRCA -0.405; TCGA COAD -1.235; TCGA ESCA -1.001; TCGA HNSC -0.374; TCGA KIRP -0.233; TCGA LGG -0.126; TCGA PRAD -0.223; TCGA SARC -0.945; TCGA STAD -1.185; TCGA UCEC -0.717 |
hsa-miR-16-5p | SLC20A2 | 11 cancers: BLCA; BRCA; HNSC; KIRC; LIHC; LUAD; OV; PAAD; PRAD; SARC; STAD | miRNATAP | TCGA BLCA -0.196; TCGA BRCA -0.085; TCGA HNSC -0.163; TCGA KIRC -0.529; TCGA LIHC -0.263; TCGA LUAD -0.119; TCGA OV -0.099; TCGA PAAD -0.288; TCGA PRAD -0.152; TCGA SARC -0.381; TCGA STAD -0.2 |
hsa-miR-16-5p | CRTC3 | 9 cancers: BLCA; COAD; ESCA; HNSC; LUAD; PAAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.202; TCGA COAD -0.103; TCGA ESCA -0.186; TCGA HNSC -0.075; TCGA LUAD -0.168; TCGA PAAD -0.137; TCGA SARC -0.274; TCGA STAD -0.23; TCGA UCEC -0.131 |
hsa-miR-16-5p | FGF9 | 10 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LUAD; OV; PAAD; STAD | miRNATAP | TCGA BLCA -0.884; TCGA BRCA -0.266; TCGA CESC -0.626; TCGA HNSC -0.267; TCGA KIRC -2.939; TCGA KIRP -0.787; TCGA LUAD -0.466; TCGA OV -0.423; TCGA PAAD -0.45; TCGA STAD -0.292 |
hsa-miR-16-5p | SYT3 | 9 cancers: BLCA; BRCA; HNSC; KIRC; KIRP; LGG; LIHC; LUSC; STAD | miRNATAP | TCGA BLCA -0.338; TCGA BRCA -0.426; TCGA HNSC -0.623; TCGA KIRC -0.751; TCGA KIRP -0.28; TCGA LGG -0.182; TCGA LIHC -0.552; TCGA LUSC -0.495; TCGA STAD -0.538 |
hsa-miR-16-5p | SCN4B | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.585; TCGA BRCA -0.635; TCGA CESC -0.489; TCGA COAD -0.86; TCGA ESCA -0.854; TCGA HNSC -0.545; TCGA LIHC -0.326; TCGA LUAD -0.468; TCGA LUSC -0.34; TCGA PAAD -0.626; TCGA PRAD -0.336; TCGA SARC -0.407; TCGA STAD -0.651; TCGA UCEC -0.814 |
hsa-miR-16-5p | RAPGEF2 | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; STAD; UCEC | miRNATAP | TCGA BLCA -0.077; TCGA CESC -0.119; TCGA COAD -0.089; TCGA ESCA -0.2; TCGA HNSC -0.186; TCGA KIRP -0.1; TCGA LUAD -0.141; TCGA LUSC -0.268; TCGA STAD -0.136; TCGA UCEC -0.141 |
hsa-miR-16-5p | FGF18 | 12 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LUAD; LUSC; OV; PAAD; SARC; STAD | miRNATAP | TCGA BLCA -0.717; TCGA BRCA -0.407; TCGA CESC -0.606; TCGA ESCA -0.545; TCGA HNSC -0.634; TCGA KIRC -0.347; TCGA LUAD -0.278; TCGA LUSC -0.485; TCGA OV -0.311; TCGA PAAD -0.608; TCGA SARC -0.36; TCGA STAD -0.369 |
hsa-miR-16-5p | LATS2 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.386; TCGA BRCA -0.085; TCGA CESC -0.127; TCGA COAD -0.322; TCGA ESCA -0.403; TCGA HNSC -0.249; TCGA LUAD -0.171; TCGA LUSC -0.425; TCGA PAAD -0.53; TCGA STAD -0.323; TCGA UCEC -0.313 |
hsa-miR-16-5p | NFE2L1 | 11 cancers: BLCA; CESC; ESCA; HNSC; KIRC; LUAD; PRAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.13; TCGA CESC -0.079; TCGA ESCA -0.155; TCGA HNSC -0.132; TCGA KIRC -0.085; TCGA LUAD -0.082; TCGA PRAD -0.189; TCGA SARC -0.181; TCGA THCA -0.114; TCGA STAD -0.06; TCGA UCEC -0.217 |
hsa-miR-16-5p | CALM1 | 9 cancers: BLCA; ESCA; HNSC; KIRC; LUAD; OV; PRAD; SARC; STAD | miRNATAP | TCGA BLCA -0.179; TCGA ESCA -0.191; TCGA HNSC -0.226; TCGA KIRC -0.133; TCGA LUAD -0.061; TCGA OV -0.083; TCGA PRAD -0.23; TCGA SARC -0.099; TCGA STAD -0.172 |
hsa-miR-16-5p | CACNB4 | 10 cancers: BLCA; BRCA; COAD; ESCA; KIRC; LIHC; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.797; TCGA BRCA -0.377; TCGA COAD -0.427; TCGA ESCA -0.465; TCGA KIRC -0.701; TCGA LIHC -0.528; TCGA PRAD -0.661; TCGA SARC -0.479; TCGA STAD -0.566; TCGA UCEC -0.412 |
hsa-miR-16-5p | SHROOM4 | 16 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; OV; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.298; TCGA BRCA -0.159; TCGA CESC -0.513; TCGA COAD -0.313; TCGA ESCA -0.493; TCGA HNSC -0.465; TCGA LIHC -0.233; TCGA LUAD -0.318; TCGA LUSC -0.515; TCGA OV -0.275; TCGA PAAD -0.337; TCGA PRAD -0.254; TCGA SARC -0.181; TCGA THCA -0.41; TCGA STAD -0.332; TCGA UCEC -0.345 |
hsa-miR-16-5p | SOX5 | 9 cancers: BLCA; CESC; COAD; HNSC; KIRC; LUAD; PRAD; SARC; STAD | miRNATAP | TCGA BLCA -0.745; TCGA CESC -0.68; TCGA COAD -0.627; TCGA HNSC -0.405; TCGA KIRC -0.456; TCGA LUAD -0.237; TCGA PRAD -0.63; TCGA SARC -0.582; TCGA STAD -0.511 |
hsa-miR-16-5p | USP2 | 9 cancers: BLCA; CESC; ESCA; HNSC; KIRC; LGG; LUSC; PRAD; STAD | miRNATAP | TCGA BLCA -0.36; TCGA CESC -0.233; TCGA ESCA -0.438; TCGA HNSC -0.966; TCGA KIRC -0.78; TCGA LGG -0.162; TCGA LUSC -0.344; TCGA PRAD -0.313; TCGA STAD -0.644 |
hsa-miR-16-5p | MRAS | 11 cancers: BLCA; COAD; ESCA; HNSC; KIRC; LIHC; LUSC; PAAD; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.454; TCGA COAD -0.769; TCGA ESCA -0.431; TCGA HNSC -0.303; TCGA KIRC -0.152; TCGA LIHC -0.33; TCGA LUSC -0.543; TCGA PAAD -0.332; TCGA PRAD -0.283; TCGA STAD -0.424; TCGA UCEC -0.294 |
hsa-miR-16-5p | SYDE1 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.394; TCGA BRCA -0.256; TCGA CESC -0.465; TCGA COAD -0.621; TCGA ESCA -0.479; TCGA HNSC -0.273; TCGA LUAD -0.295; TCGA LUSC -0.468; TCGA PAAD -0.481; TCGA PRAD -0.26; TCGA SARC -0.112; TCGA STAD -0.394; TCGA UCEC -0.422 |
hsa-miR-16-5p | FRY | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.472; TCGA BRCA -0.135; TCGA CESC -0.503; TCGA COAD -0.511; TCGA ESCA -0.764; TCGA HNSC -0.372; TCGA KIRC -0.568; TCGA KIRP -0.18; TCGA LGG -0.202; TCGA LUAD -0.316; TCGA PAAD -0.349; TCGA PRAD -0.164; TCGA SARC -0.669; TCGA STAD -0.367; TCGA UCEC -0.522 |
hsa-miR-16-5p | ADAMTS3 | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LUSC; PAAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.342; TCGA CESC -0.324; TCGA COAD -0.71; TCGA ESCA -0.393; TCGA HNSC -0.247; TCGA KIRP -0.381; TCGA LUSC -0.281; TCGA PAAD -0.407; TCGA STAD -0.311; TCGA UCEC -0.327 |
hsa-miR-16-5p | ZNF423 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.538; TCGA BRCA -0.497; TCGA CESC -0.69; TCGA COAD -0.687; TCGA ESCA -0.724; TCGA HNSC -0.716; TCGA LGG -0.174; TCGA LIHC -0.304; TCGA LUAD -0.402; TCGA LUSC -0.582; TCGA OV -0.459; TCGA PAAD -0.627; TCGA PRAD -0.432; TCGA STAD -0.736; TCGA UCEC -0.312 |
hsa-miR-16-5p | PTPRD | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LGG; LUAD; LUSC; OV; STAD; UCEC | miRNATAP | TCGA BLCA -1.138; TCGA CESC -0.962; TCGA COAD -0.404; TCGA ESCA -0.963; TCGA HNSC -0.942; TCGA KIRP -0.392; TCGA LGG -0.174; TCGA LUAD -0.369; TCGA LUSC -0.422; TCGA OV -0.304; TCGA STAD -0.468; TCGA UCEC -0.714 |
hsa-miR-16-5p | CHD5 | 9 cancers: BLCA; COAD; ESCA; KIRC; KIRP; PRAD; SARC; THCA; STAD | miRNATAP | TCGA BLCA -0.553; TCGA COAD -0.288; TCGA ESCA -1.426; TCGA KIRC -1.723; TCGA KIRP -0.632; TCGA PRAD -0.241; TCGA SARC -0.732; TCGA THCA -0.288; TCGA STAD -0.935 |
hsa-miR-16-5p | RARB | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; LUAD; PAAD; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.614; TCGA BRCA -0.115; TCGA CESC -0.374; TCGA COAD -0.522; TCGA ESCA -0.488; TCGA KIRC -0.419; TCGA LUAD -0.164; TCGA PAAD -0.278; TCGA PRAD -0.3; TCGA STAD -0.346; TCGA UCEC -0.257 |
hsa-miR-16-5p | PAM | 13 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; OV; PRAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.365; TCGA CESC -0.316; TCGA COAD -0.424; TCGA ESCA -0.498; TCGA HNSC -0.364; TCGA LUAD -0.128; TCGA LUSC -0.39; TCGA OV -0.276; TCGA PRAD -0.356; TCGA SARC -0.176; TCGA THCA -0.329; TCGA STAD -0.396; TCGA UCEC -0.131 |
hsa-miR-16-5p | SYDE2 | 11 cancers: BLCA; CESC; COAD; ESCA; KIRC; LGG; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.341; TCGA CESC -0.32; TCGA COAD -0.273; TCGA ESCA -0.485; TCGA KIRC -0.444; TCGA LGG -0.191; TCGA PAAD -0.242; TCGA PRAD -0.427; TCGA SARC -0.496; TCGA STAD -0.667; TCGA UCEC -0.266 |
hsa-miR-16-5p | TSC22D3 | 10 cancers: BLCA; BRCA; COAD; ESCA; LUSC; OV; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.333; TCGA BRCA -0.251; TCGA COAD -0.394; TCGA ESCA -0.589; TCGA LUSC -0.205; TCGA OV -0.179; TCGA PRAD -0.27; TCGA SARC -0.273; TCGA STAD -0.269; TCGA UCEC -0.356 |
hsa-miR-16-5p | CHD9 | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.142; TCGA CESC -0.13; TCGA COAD -0.176; TCGA ESCA -0.201; TCGA HNSC -0.166; TCGA KIRC -0.118; TCGA KIRP -0.116; TCGA LUAD -0.129; TCGA LUSC -0.227; TCGA PRAD -0.237; TCGA STAD -0.257; TCGA UCEC -0.287 |
hsa-miR-16-5p | PELI2 | 11 cancers: BLCA; BRCA; KIRC; KIRP; LGG; LUAD; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.514; TCGA BRCA -0.363; TCGA KIRC -0.498; TCGA KIRP -0.123; TCGA LGG -0.201; TCGA LUAD -0.165; TCGA PAAD -0.288; TCGA PRAD -0.185; TCGA SARC -0.314; TCGA STAD -0.14; TCGA UCEC -0.253 |
hsa-miR-16-5p | TRIM2 | 9 cancers: BLCA; CESC; HNSC; KIRC; KIRP; OV; PRAD; SARC; STAD | miRNATAP | TCGA BLCA -0.23; TCGA CESC -0.264; TCGA HNSC -0.228; TCGA KIRC -0.996; TCGA KIRP -0.321; TCGA OV -0.202; TCGA PRAD -0.414; TCGA SARC -0.185; TCGA STAD -0.115 |
hsa-miR-16-5p | SPAG7 | 9 cancers: BLCA; BRCA; HNSC; KIRC; KIRP; LIHC; OV; PRAD; UCEC | miRNATAP | TCGA BLCA -0.07; TCGA BRCA -0.124; TCGA HNSC -0.079; TCGA KIRC -0.142; TCGA KIRP -0.118; TCGA LIHC -0.254; TCGA OV -0.122; TCGA PRAD -0.1; TCGA UCEC -0.112 |
hsa-miR-16-5p | RASL12 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; OV; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -1.049; TCGA BRCA -0.297; TCGA CESC -0.535; TCGA COAD -0.711; TCGA ESCA -0.999; TCGA HNSC -0.341; TCGA LIHC -0.34; TCGA LUAD -0.22; TCGA LUSC -0.584; TCGA OV -0.214; TCGA PAAD -0.237; TCGA PRAD -0.666; TCGA SARC -0.968; TCGA STAD -0.485; TCGA UCEC -0.741 |
hsa-miR-16-5p | CNN1 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -2.104; TCGA BRCA -0.911; TCGA CESC -1.147; TCGA COAD -1.641; TCGA ESCA -2.034; TCGA HNSC -0.926; TCGA LIHC -0.401; TCGA LUAD -0.557; TCGA LUSC -1.008; TCGA PAAD -0.753; TCGA PRAD -0.837; TCGA SARC -1.38; TCGA STAD -1.527; TCGA UCEC -1.566 |
hsa-miR-16-5p | KALRN | 11 cancers: BLCA; CESC; ESCA; HNSC; LUAD; LUSC; PAAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.215; TCGA CESC -0.527; TCGA ESCA -0.559; TCGA HNSC -0.584; TCGA LUAD -0.189; TCGA LUSC -0.255; TCGA PAAD -0.327; TCGA SARC -0.492; TCGA THCA -0.21; TCGA STAD -0.232; TCGA UCEC -0.355 |
hsa-miR-16-5p | SIDT2 | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LIHC; LUAD; LUSC; PRAD; UCEC | miRNATAP | TCGA BLCA -0.205; TCGA BRCA -0.109; TCGA CESC -0.275; TCGA ESCA -0.254; TCGA HNSC -0.255; TCGA LIHC -0.177; TCGA LUAD -0.202; TCGA LUSC -0.265; TCGA PRAD -0.123; TCGA UCEC -0.135 |
hsa-miR-16-5p | CX3CL1 | 11 cancers: BLCA; BRCA; CESC; COAD; KIRP; LGG; LUAD; PRAD; SARC; THCA; UCEC | miRNATAP | TCGA BLCA -0.432; TCGA BRCA -0.321; TCGA CESC -0.282; TCGA COAD -0.344; TCGA KIRP -0.114; TCGA LGG -0.126; TCGA LUAD -0.346; TCGA PRAD -0.268; TCGA SARC -0.525; TCGA THCA -0.191; TCGA UCEC -0.274 |
hsa-miR-16-5p | SLC24A3 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.925; TCGA BRCA -0.227; TCGA CESC -0.585; TCGA COAD -1.008; TCGA ESCA -0.79; TCGA HNSC -0.885; TCGA KIRC -0.376; TCGA LGG -0.184; TCGA LUAD -0.596; TCGA LUSC -0.704; TCGA PAAD -0.277; TCGA PRAD -0.584; TCGA SARC -0.959; TCGA STAD -0.508; TCGA UCEC -0.994 |
hsa-miR-16-5p | ZC3H12B | 13 cancers: BLCA; BRCA; CESC; ESCA; KIRC; KIRP; LGG; LUAD; LUSC; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.335; TCGA BRCA -0.296; TCGA CESC -0.257; TCGA ESCA -0.409; TCGA KIRC -0.46; TCGA KIRP -0.275; TCGA LGG -0.221; TCGA LUAD -0.252; TCGA LUSC -0.219; TCGA PRAD -0.256; TCGA SARC -0.318; TCGA STAD -0.489; TCGA UCEC -0.543 |
hsa-miR-16-5p | ARHGEF9 | 9 cancers: BLCA; ESCA; HNSC; KIRC; LUAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.26; TCGA ESCA -0.313; TCGA HNSC -0.161; TCGA KIRC -0.124; TCGA LUAD -0.106; TCGA PRAD -0.29; TCGA SARC -0.286; TCGA STAD -0.23; TCGA UCEC -0.14 |
hsa-miR-16-5p | KBTBD4 | 9 cancers: BLCA; COAD; KIRC; LGG; LUAD; PRAD; SARC; THCA; UCEC | miRNATAP | TCGA BLCA -0.092; TCGA COAD -0.076; TCGA KIRC -0.273; TCGA LGG -0.053; TCGA LUAD -0.086; TCGA PRAD -0.267; TCGA SARC -0.281; TCGA THCA -0.163; TCGA UCEC -0.149 |
hsa-miR-16-5p | PPP3CB | 11 cancers: BLCA; CESC; ESCA; HNSC; KIRC; LGG; LUSC; OV; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.219; TCGA CESC -0.076; TCGA ESCA -0.205; TCGA HNSC -0.058; TCGA KIRC -0.134; TCGA LGG -0.091; TCGA LUSC -0.109; TCGA OV -0.104; TCGA PRAD -0.22; TCGA STAD -0.239; TCGA UCEC -0.19 |
hsa-miR-16-5p | CARD10 | 9 cancers: BRCA; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; THCA | miRNAWalker2 validate; MirTarget; miRNATAP | TCGA BRCA -0.261; TCGA HNSC -0.238; TCGA KIRP -0.121; TCGA LIHC -0.143; TCGA LUAD -0.324; TCGA LUSC -0.415; TCGA PAAD -0.288; TCGA PRAD -0.103; TCGA THCA -0.271 |
hsa-miR-16-5p | EFNB2 | 9 cancers: BRCA; HNSC; KIRP; LUAD; LUSC; OV; PAAD; THCA; UCEC | miRNAWalker2 validate; miRNATAP | TCGA BRCA -0.142; TCGA HNSC -0.268; TCGA KIRP -0.506; TCGA LUAD -0.15; TCGA LUSC -0.207; TCGA OV -0.311; TCGA PAAD -0.35; TCGA THCA -0.163; TCGA UCEC -0.298 |
hsa-miR-16-5p | EGFR | 11 cancers: BRCA; COAD; HNSC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BRCA -0.176; TCGA COAD -0.133; TCGA HNSC -0.329; TCGA LUAD -0.236; TCGA LUSC -0.243; TCGA PAAD -0.557; TCGA PRAD -0.333; TCGA SARC -0.377; TCGA THCA -0.193; TCGA STAD -0.348; TCGA UCEC -0.267 |
hsa-miR-16-5p | ITGA2 | 10 cancers: BRCA; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; THCA; UCEC | miRNAWalker2 validate; MirTarget; miRNATAP | TCGA BRCA -0.242; TCGA KIRC -0.459; TCGA KIRP -0.231; TCGA LIHC -0.251; TCGA LUAD -0.333; TCGA LUSC -0.405; TCGA PAAD -0.551; TCGA PRAD -0.532; TCGA THCA -0.488; TCGA UCEC -0.38 |
hsa-miR-16-5p | KIF5A | 9 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRC; THCA; STAD; UCEC | miRNAWalker2 validate; MirTarget; miRNATAP | TCGA BRCA -0.324; TCGA CESC -0.558; TCGA COAD -1.145; TCGA ESCA -0.974; TCGA HNSC -0.301; TCGA KIRC -1.394; TCGA THCA -0.44; TCGA STAD -0.866; TCGA UCEC -0.45 |
hsa-miR-16-5p | LUZP1 | 12 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | miRNAWalker2 validate; MirTarget; miRNATAP | TCGA BRCA -0.063; TCGA CESC -0.11; TCGA COAD -0.084; TCGA ESCA -0.165; TCGA HNSC -0.167; TCGA KIRC -0.12; TCGA LUAD -0.165; TCGA LUSC -0.254; TCGA PAAD -0.261; TCGA PRAD -0.245; TCGA STAD -0.101; TCGA UCEC -0.209 |
hsa-miR-16-5p | RPH3AL | 9 cancers: BRCA; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; OV; THCA | miRNAWalker2 validate; MirTarget | TCGA BRCA -0.283; TCGA KIRC -0.196; TCGA KIRP -0.305; TCGA LGG -0.064; TCGA LIHC -0.134; TCGA LUAD -0.153; TCGA LUSC -0.257; TCGA OV -0.238; TCGA THCA -0.114 |
hsa-miR-16-5p | SALL1 | 9 cancers: BRCA; CESC; COAD; HNSC; KIRC; KIRP; OV; PAAD; UCEC | miRNAWalker2 validate; MirTarget; miRNATAP | TCGA BRCA -0.452; TCGA CESC -0.947; TCGA COAD -0.354; TCGA HNSC -0.743; TCGA KIRC -0.356; TCGA KIRP -0.218; TCGA OV -0.836; TCGA PAAD -1.032; TCGA UCEC -0.351 |
hsa-miR-16-5p | USP53 | 11 cancers: BRCA; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate; miRNATAP | TCGA BRCA -0.151; TCGA ESCA -0.323; TCGA HNSC -0.115; TCGA KIRC -0.591; TCGA KIRP -0.156; TCGA LUAD -0.094; TCGA LUSC -0.204; TCGA PRAD -0.193; TCGA SARC -0.425; TCGA STAD -0.187; TCGA UCEC -0.218 |
hsa-miR-16-5p | WNT5A | 10 cancers: BRCA; CESC; COAD; HNSC; KIRC; KIRP; LUAD; OV; PAAD; UCEC | miRNAWalker2 validate | TCGA BRCA -0.443; TCGA CESC -0.29; TCGA COAD -0.347; TCGA HNSC -0.37; TCGA KIRC -0.292; TCGA KIRP -0.553; TCGA LUAD -0.269; TCGA OV -0.298; TCGA PAAD -0.807; TCGA UCEC -0.335 |
hsa-miR-16-5p | ATP7A | 10 cancers: BRCA; COAD; ESCA; HNSC; KIRC; LUAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BRCA -0.089; TCGA COAD -0.111; TCGA ESCA -0.166; TCGA HNSC -0.197; TCGA KIRC -0.123; TCGA LUAD -0.119; TCGA PRAD -0.097; TCGA SARC -0.11; TCGA STAD -0.125; TCGA UCEC -0.199 |
hsa-miR-16-5p | ASTN1 | 13 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LIHC; OV; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BRCA -0.656; TCGA CESC -0.89; TCGA COAD -1.119; TCGA ESCA -1.524; TCGA HNSC -0.523; TCGA KIRC -0.819; TCGA LGG -0.098; TCGA LIHC -0.292; TCGA OV -0.373; TCGA PRAD -0.834; TCGA THCA -0.366; TCGA STAD -1.231; TCGA UCEC -0.733 |
hsa-miR-16-5p | WWC1 | 9 cancers: BRCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; PAAD; THCA | MirTarget; miRNATAP | TCGA BRCA -0.191; TCGA HNSC -0.224; TCGA KIRC -0.396; TCGA KIRP -0.087; TCGA LGG -0.13; TCGA LUAD -0.102; TCGA LUSC -0.17; TCGA PAAD -0.323; TCGA THCA -0.147 |
hsa-miR-16-5p | PRTG | 11 cancers: BRCA; CESC; COAD; HNSC; KIRC; LGG; LUSC; OV; PAAD; STAD; UCEC | MirTarget | TCGA BRCA -0.439; TCGA CESC -0.767; TCGA COAD -0.829; TCGA HNSC -0.333; TCGA KIRC -1.101; TCGA LGG -0.253; TCGA LUSC -0.545; TCGA OV -0.427; TCGA PAAD -0.481; TCGA STAD -0.667; TCGA UCEC -0.743 |
hsa-miR-16-5p | DENND2C | 10 cancers: BRCA; CESC; COAD; HNSC; KIRC; LUAD; PAAD; PRAD; THCA; STAD | MirTarget | TCGA BRCA -0.129; TCGA CESC -0.307; TCGA COAD -0.165; TCGA HNSC -0.283; TCGA KIRC -0.145; TCGA LUAD -0.156; TCGA PAAD -0.53; TCGA PRAD -0.513; TCGA THCA -0.177; TCGA STAD -0.063 |
hsa-miR-16-5p | TBC1D16 | 10 cancers: BRCA; COAD; HNSC; KIRC; KIRP; LUAD; LUSC; OV; PAAD; THCA | MirTarget | TCGA BRCA -0.097; TCGA COAD -0.127; TCGA HNSC -0.395; TCGA KIRC -0.455; TCGA KIRP -0.153; TCGA LUAD -0.125; TCGA LUSC -0.276; TCGA OV -0.093; TCGA PAAD -0.23; TCGA THCA -0.223 |
hsa-miR-16-5p | SCN2A | 10 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; SARC; STAD; UCEC | MirTarget | TCGA BRCA -0.629; TCGA CESC -0.472; TCGA COAD -0.536; TCGA ESCA -0.585; TCGA HNSC -0.418; TCGA KIRC -1.85; TCGA KIRP -0.494; TCGA SARC -0.858; TCGA STAD -0.499; TCGA UCEC -0.485 |
hsa-miR-16-5p | DCAF5 | 11 cancers: BRCA; COAD; ESCA; HNSC; KIRP; LGG; LUSC; OV; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BRCA -0.063; TCGA COAD -0.064; TCGA ESCA -0.222; TCGA HNSC -0.227; TCGA KIRP -0.083; TCGA LGG -0.116; TCGA LUSC -0.113; TCGA OV -0.07; TCGA SARC -0.151; TCGA STAD -0.136; TCGA UCEC -0.058 |
hsa-miR-16-5p | UNC119B | 9 cancers: BRCA; KIRC; KIRP; LGG; LIHC; LUAD; OV; SARC; UCEC | MirTarget | TCGA BRCA -0.066; TCGA KIRC -0.275; TCGA KIRP -0.247; TCGA LGG -0.062; TCGA LIHC -0.172; TCGA LUAD -0.119; TCGA OV -0.159; TCGA SARC -0.165; TCGA UCEC -0.294 |
hsa-miR-16-5p | GRAMD3 | 10 cancers: BRCA; CESC; KIRC; LGG; LUAD; LUSC; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BRCA -0.306; TCGA CESC -0.208; TCGA KIRC -0.143; TCGA LGG -0.2; TCGA LUAD -0.153; TCGA LUSC -0.279; TCGA PRAD -0.074; TCGA THCA -0.201; TCGA STAD -0.146; TCGA UCEC -0.179 |
hsa-miR-16-5p | NDST1 | 10 cancers: BRCA; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; THCA; STAD; UCEC | mirMAP | TCGA BRCA -0.178; TCGA COAD -0.168; TCGA ESCA -0.183; TCGA HNSC -0.192; TCGA LIHC -0.206; TCGA LUAD -0.187; TCGA LUSC -0.337; TCGA THCA -0.149; TCGA STAD -0.145; TCGA UCEC -0.082 |
hsa-miR-16-5p | ABR | 9 cancers: BRCA; ESCA; HNSC; LGG; LUAD; LUSC; PRAD; THCA; UCEC | mirMAP | TCGA BRCA -0.109; TCGA ESCA -0.161; TCGA HNSC -0.108; TCGA LGG -0.084; TCGA LUAD -0.149; TCGA LUSC -0.132; TCGA PRAD -0.076; TCGA THCA -0.201; TCGA UCEC -0.141 |
hsa-miR-16-5p | ELFN2 | 9 cancers: BRCA; COAD; HNSC; KIRP; LGG; LIHC; OV; PAAD; THCA | mirMAP | TCGA BRCA -0.279; TCGA COAD -0.569; TCGA HNSC -0.352; TCGA KIRP -0.465; TCGA LGG -0.476; TCGA LIHC -0.929; TCGA OV -0.631; TCGA PAAD -0.532; TCGA THCA -0.797 |
hsa-miR-16-5p | TOM1L2 | 11 cancers: BRCA; ESCA; HNSC; KIRC; LGG; LIHC; LUAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BRCA -0.239; TCGA ESCA -0.329; TCGA HNSC -0.228; TCGA KIRC -0.384; TCGA LGG -0.13; TCGA LIHC -0.128; TCGA LUAD -0.17; TCGA PRAD -0.083; TCGA SARC -0.29; TCGA STAD -0.267; TCGA UCEC -0.176 |
hsa-miR-16-5p | MINK1 | 9 cancers: BRCA; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; PRAD; THCA | miRNATAP | TCGA BRCA -0.119; TCGA HNSC -0.128; TCGA KIRP -0.142; TCGA LGG -0.07; TCGA LIHC -0.098; TCGA LUAD -0.138; TCGA LUSC -0.169; TCGA PRAD -0.112; TCGA THCA -0.081 |
hsa-miR-16-5p | NYNRIN | 11 cancers: BRCA; CESC; COAD; HNSC; KIRC; LGG; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BRCA -0.123; TCGA CESC -0.334; TCGA COAD -0.31; TCGA HNSC -0.275; TCGA KIRC -0.513; TCGA LGG -0.088; TCGA PAAD -0.276; TCGA PRAD -0.468; TCGA SARC -0.244; TCGA STAD -0.304; TCGA UCEC -0.344 |
hsa-miR-16-5p | ZNF436 | 10 cancers: BRCA; CESC; COAD; ESCA; LUAD; PAAD; PRAD; THCA; STAD; UCEC | miRNATAP | TCGA BRCA -0.05; TCGA CESC -0.159; TCGA COAD -0.192; TCGA ESCA -0.216; TCGA LUAD -0.08; TCGA PAAD -0.234; TCGA PRAD -0.103; TCGA THCA -0.105; TCGA STAD -0.238; TCGA UCEC -0.155 |
hsa-miR-16-5p | VTCN1 | 10 cancers: BRCA; CESC; COAD; HNSC; KIRC; LGG; PAAD; PRAD; SARC; THCA | miRNATAP | TCGA BRCA -0.817; TCGA CESC -0.606; TCGA COAD -0.468; TCGA HNSC -0.526; TCGA KIRC -2.376; TCGA LGG -0.294; TCGA PAAD -1.518; TCGA PRAD -0.604; TCGA SARC -0.387; TCGA THCA -0.984 |
hsa-miR-16-5p | KIF16B | 12 cancers: BRCA; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; OV; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BRCA -0.105; TCGA COAD -0.132; TCGA ESCA -0.171; TCGA HNSC -0.229; TCGA KIRC -0.257; TCGA LUAD -0.143; TCGA LUSC -0.132; TCGA OV -0.076; TCGA PRAD -0.13; TCGA SARC -0.161; TCGA STAD -0.082; TCGA UCEC -0.091 |
hsa-miR-16-5p | BHLHE41 | 10 cancers: BRCA; CESC; COAD; ESCA; HNSC; LUSC; PRAD; THCA; STAD; UCEC | miRNATAP | TCGA BRCA -0.383; TCGA CESC -0.238; TCGA COAD -0.245; TCGA ESCA -0.479; TCGA HNSC -0.29; TCGA LUSC -0.707; TCGA PRAD -0.152; TCGA THCA -0.322; TCGA STAD -0.344; TCGA UCEC -0.202 |
hsa-miR-16-5p | GPR153 | 10 cancers: BRCA; ESCA; HNSC; KIRP; LGG; LUAD; LUSC; PAAD; STAD; UCEC | miRNATAP | TCGA BRCA -0.213; TCGA ESCA -0.369; TCGA HNSC -0.31; TCGA KIRP -0.367; TCGA LGG -0.224; TCGA LUAD -0.224; TCGA LUSC -0.534; TCGA PAAD -0.273; TCGA STAD -0.273; TCGA UCEC -0.308 |
hsa-miR-16-5p | CDS2 | 11 cancers: CESC; COAD; ESCA; KIRC; LGG; LIHC; OV; PRAD; SARC; THCA; STAD | miRNAWalker2 validate; mirMAP; miRNATAP | TCGA CESC -0.079; TCGA COAD -0.213; TCGA ESCA -0.095; TCGA KIRC -0.071; TCGA LGG -0.085; TCGA LIHC -0.117; TCGA OV -0.086; TCGA PRAD -0.117; TCGA SARC -0.118; TCGA THCA -0.056; TCGA STAD -0.242 |
hsa-miR-16-5p | EXD2 | 12 cancers: CESC; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA CESC -0.097; TCGA ESCA -0.189; TCGA HNSC -0.147; TCGA KIRC -0.242; TCGA KIRP -0.076; TCGA LGG -0.104; TCGA LUAD -0.076; TCGA PRAD -0.1; TCGA SARC -0.165; TCGA THCA -0.111; TCGA STAD -0.084; TCGA UCEC -0.104 |
hsa-miR-16-5p | FAM168A | 12 cancers: CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD | miRNAWalker2 validate | TCGA CESC -0.2; TCGA COAD -0.33; TCGA ESCA -0.375; TCGA HNSC -0.175; TCGA KIRC -0.27; TCGA LUAD -0.219; TCGA LUSC -0.168; TCGA PAAD -0.343; TCGA PRAD -0.334; TCGA SARC -0.222; TCGA THCA -0.194; TCGA STAD -0.148 |
hsa-miR-16-5p | MIB1 | 10 cancers: CESC; COAD; KIRC; LGG; LUAD; OV; PAAD; PRAD; STAD; UCEC | miRNAWalker2 validate; miRNATAP | TCGA CESC -0.09; TCGA COAD -0.094; TCGA KIRC -0.278; TCGA LGG -0.114; TCGA LUAD -0.101; TCGA OV -0.101; TCGA PAAD -0.215; TCGA PRAD -0.131; TCGA STAD -0.172; TCGA UCEC -0.235 |
hsa-miR-16-5p | MYO5A | 9 cancers: CESC; COAD; ESCA; HNSC; LUAD; LUSC; PRAD; STAD; UCEC | miRNAWalker2 validate; MirTarget | TCGA CESC -0.235; TCGA COAD -0.58; TCGA ESCA -0.422; TCGA HNSC -0.408; TCGA LUAD -0.15; TCGA LUSC -0.337; TCGA PRAD -0.109; TCGA STAD -0.248; TCGA UCEC -0.168 |
hsa-miR-16-5p | PPM1A | 11 cancers: CESC; ESCA; HNSC; KIRC; LGG; LUSC; OV; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA CESC -0.062; TCGA ESCA -0.249; TCGA HNSC -0.108; TCGA KIRC -0.289; TCGA LGG -0.079; TCGA LUSC -0.072; TCGA OV -0.084; TCGA PRAD -0.114; TCGA SARC -0.08; TCGA STAD -0.207; TCGA UCEC -0.185 |
hsa-miR-16-5p | MTMR3 | 13 cancers: CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA CESC -0.124; TCGA COAD -0.065; TCGA ESCA -0.192; TCGA HNSC -0.166; TCGA KIRC -0.149; TCGA KIRP -0.058; TCGA LGG -0.102; TCGA LUAD -0.144; TCGA PRAD -0.126; TCGA SARC -0.185; TCGA THCA -0.11; TCGA STAD -0.097; TCGA UCEC -0.11 |
hsa-miR-16-5p | RPS6KA6 | 11 cancers: CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; PRAD; SARC; STAD; UCEC | MirTarget | TCGA CESC -0.757; TCGA COAD -0.644; TCGA ESCA -1.177; TCGA HNSC -0.788; TCGA KIRC -2.166; TCGA KIRP -0.269; TCGA LGG -0.198; TCGA PRAD -0.194; TCGA SARC -0.522; TCGA STAD -0.943; TCGA UCEC -0.939 |
hsa-miR-16-5p | EPB41L1 | 10 cancers: CESC; KIRC; KIRP; LUAD; PAAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA CESC -0.21; TCGA KIRC -0.349; TCGA KIRP -0.138; TCGA LUAD -0.136; TCGA PAAD -0.406; TCGA PRAD -0.098; TCGA SARC -0.313; TCGA THCA -0.21; TCGA STAD -0.099; TCGA UCEC -0.137 |
hsa-miR-16-5p | ABTB2 | 10 cancers: CESC; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; THCA; STAD; UCEC | MirTarget | TCGA CESC -0.178; TCGA HNSC -0.336; TCGA KIRC -0.615; TCGA LUAD -0.257; TCGA LUSC -0.214; TCGA PAAD -0.327; TCGA PRAD -0.165; TCGA THCA -0.777; TCGA STAD -0.134; TCGA UCEC -0.19 |
hsa-miR-16-5p | HECTD1 | 9 cancers: CESC; ESCA; HNSC; KIRC; LUSC; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA CESC -0.092; TCGA ESCA -0.185; TCGA HNSC -0.189; TCGA KIRC -0.324; TCGA LUSC -0.105; TCGA PRAD -0.15; TCGA SARC -0.159; TCGA STAD -0.101; TCGA UCEC -0.091 |
hsa-miR-16-5p | IGF2R | 9 cancers: COAD; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; SARC; THCA | miRNAWalker2 validate; MirTarget | TCGA COAD -0.097; TCGA HNSC -0.124; TCGA KIRC -0.119; TCGA LUAD -0.14; TCGA LUSC -0.217; TCGA PAAD -0.189; TCGA PRAD -0.219; TCGA SARC -0.11; TCGA THCA -0.149 |
hsa-miR-16-5p | LATS1 | 10 cancers: COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate; MirTarget; miRNATAP | TCGA COAD -0.159; TCGA ESCA -0.118; TCGA HNSC -0.156; TCGA KIRC -0.231; TCGA LUAD -0.141; TCGA LUSC -0.181; TCGA PRAD -0.338; TCGA SARC -0.209; TCGA STAD -0.155; TCGA UCEC -0.114 |
hsa-miR-16-5p | SLC30A1 | 9 cancers: COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; SARC | miRNAWalker2 validate | TCGA COAD -0.122; TCGA ESCA -0.155; TCGA HNSC -0.178; TCGA KIRC -0.359; TCGA LUAD -0.132; TCGA LUSC -0.239; TCGA PAAD -0.303; TCGA PRAD -0.293; TCGA SARC -0.125 |
hsa-miR-16-5p | AFF4 | 9 cancers: COAD; ESCA; HNSC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA COAD -0.093; TCGA ESCA -0.277; TCGA HNSC -0.136; TCGA LUAD -0.108; TCGA LUSC -0.171; TCGA PRAD -0.166; TCGA SARC -0.177; TCGA STAD -0.214; TCGA UCEC -0.167 |
hsa-miR-16-5p | ATF6 | 9 cancers: COAD; HNSC; KIRC; LIHC; OV; PAAD; PRAD; SARC; UCEC | MirTarget | TCGA COAD -0.066; TCGA HNSC -0.121; TCGA KIRC -0.249; TCGA LIHC -0.113; TCGA OV -0.083; TCGA PAAD -0.135; TCGA PRAD -0.272; TCGA SARC -0.185; TCGA UCEC -0.112 |
hsa-miR-16-5p | TRAF6 | 9 cancers: COAD; ESCA; HNSC; KIRC; LUAD; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA COAD -0.088; TCGA ESCA -0.172; TCGA HNSC -0.086; TCGA KIRC -0.115; TCGA LUAD -0.067; TCGA PAAD -0.141; TCGA PRAD -0.13; TCGA STAD -0.133; TCGA UCEC -0.1 |
hsa-miR-16-5p | ATXN1L | 11 cancers: COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA COAD -0.101; TCGA ESCA -0.142; TCGA HNSC -0.101; TCGA KIRC -0.192; TCGA LUAD -0.159; TCGA LUSC -0.168; TCGA PAAD -0.133; TCGA PRAD -0.198; TCGA SARC -0.127; TCGA STAD -0.136; TCGA UCEC -0.205 |
hsa-miR-16-5p | DOCK9 | 9 cancers: ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA ESCA -0.267; TCGA HNSC -0.255; TCGA LUAD -0.253; TCGA LUSC -0.229; TCGA PAAD -0.212; TCGA PRAD -0.188; TCGA THCA -0.224; TCGA STAD -0.173; TCGA UCEC -0.198 |
hsa-miR-16-5p | PITPNM3 | 10 cancers: ESCA; HNSC; KIRC; KIRP; LUAD; OV; PRAD; SARC; STAD; UCEC | MirTarget | TCGA ESCA -0.451; TCGA HNSC -0.136; TCGA KIRC -0.737; TCGA KIRP -0.401; TCGA LUAD -0.289; TCGA OV -0.125; TCGA PRAD -0.507; TCGA SARC -0.674; TCGA STAD -0.681; TCGA UCEC -0.162 |
hsa-miR-16-5p | ARHGAP5 | 10 cancers: ESCA; HNSC; KIRC; KIRP; LGG; OV; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA ESCA -0.365; TCGA HNSC -0.112; TCGA KIRC -0.412; TCGA KIRP -0.129; TCGA LGG -0.103; TCGA OV -0.079; TCGA PRAD -0.096; TCGA SARC -0.133; TCGA STAD -0.209; TCGA UCEC -0.224 |
hsa-miR-16-5p | WASL | 11 cancers: ESCA; HNSC; KIRC; KIRP; LUSC; OV; PAAD; SARC; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA ESCA -0.128; TCGA HNSC -0.194; TCGA KIRC -0.143; TCGA KIRP -0.181; TCGA LUSC -0.107; TCGA OV -0.07; TCGA PAAD -0.165; TCGA SARC -0.115; TCGA THCA -0.115; TCGA STAD -0.083; TCGA UCEC -0.176 |
hsa-miR-16-5p | MAPK1 | 9 cancers: ESCA; HNSC; LGG; LUAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA ESCA -0.128; TCGA HNSC -0.094; TCGA LGG -0.074; TCGA LUAD -0.087; TCGA PRAD -0.164; TCGA SARC -0.122; TCGA THCA -0.157; TCGA STAD -0.082; TCGA UCEC -0.096 |
hsa-miR-16-5p | ZNRF3 | 11 cancers: HNSC; KIRC; KIRP; LGG; LIHC; LUAD; OV; PAAD; PRAD; SARC; THCA | miRNAWalker2 validate | TCGA HNSC -0.089; TCGA KIRC -0.428; TCGA KIRP -0.122; TCGA LGG -0.097; TCGA LIHC -0.253; TCGA LUAD -0.137; TCGA OV -0.091; TCGA PAAD -0.336; TCGA PRAD -0.351; TCGA SARC -0.136; TCGA THCA -0.284 |
hsa-miR-16-5p | CLDN12 | 9 cancers: HNSC; KIRC; KIRP; LUSC; OV; PRAD; SARC; THCA; UCEC | MirTarget; miRNATAP | TCGA HNSC -0.117; TCGA KIRC -0.102; TCGA KIRP -0.178; TCGA LUSC -0.107; TCGA OV -0.094; TCGA PRAD -0.083; TCGA SARC -0.127; TCGA THCA -0.152; TCGA UCEC -0.099 |
hsa-miR-16-5p | SMURF1 | 9 cancers: HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; SARC; THCA; UCEC | MirTarget; miRNATAP | TCGA HNSC -0.158; TCGA KIRP -0.109; TCGA LIHC -0.14; TCGA LUAD -0.113; TCGA LUSC -0.096; TCGA PAAD -0.191; TCGA SARC -0.095; TCGA THCA -0.12; TCGA UCEC -0.058 |
Enriched cancer pathways of putative targets