microRNA information: hsa-miR-17-3p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-17-3p | miRbase |
| Accession: | MIMAT0000071 | miRbase |
| Precursor name: | hsa-mir-17 | miRbase |
| Precursor accession: | MI0000071 | miRbase |
| Symbol: | MIR17 | HGNC |
| RefSeq ID: | NR_029487 | GenBank |
| Sequence: | ACUGCAGUGAAGGCACUUGUAG |
Reported expression in cancers: hsa-miR-17-3p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-17-3p | bladder cancer | upregulation | "MicroRNA expression signatures of bladder cancer r ......" | 21464941 | RNA-Seq |
| hsa-miR-17-3p | breast cancer | downregulation | "Only four miRNAs including oncomiR miR-17 were dow ......" | 22562546 | |
| hsa-miR-17-3p | colon cancer | upregulation | "To present proof-of-principle application for empl ......" | 23741026 | qPCR; Microarray |
| hsa-miR-17-3p | colon cancer | upregulation | "The levels of CEA were measured by a radioimmunoas ......" | 26807240 | qPCR |
| hsa-miR-17-3p | colorectal cancer | upregulation | "Serum miRNAs were extracted from all subjects to a ......" | 27135244 | |
| hsa-miR-17-3p | gastric cancer | upregulation | "The most highly expressed miRNAs in gastric cancer ......" | 19175831 | |
| hsa-miR-17-3p | gastric cancer | upregulation | "In the analysis by real-time PCR-based miRNA array ......" | 22407237 | qPCR; Microarray |
| hsa-miR-17-3p | gastric cancer | upregulation | "We identified five miRNAs that were most consisten ......" | 24040025 | qPCR |
| hsa-miR-17-3p | head and neck cancer | downregulation | "Conversely decreased expressions of miR-153 miR-20 ......" | 25677760 | |
| hsa-miR-17-3p | lymphoma | upregulation | "Using quantitative real-time reverse transcription ......" | 26231295 | Reverse transcription PCR; qPCR |
| hsa-miR-17-3p | lymphoma | upregulation | "Therefore we investigated miR-17 miR-19a miR-19b m ......" | 27044389 |
Reported cancer pathway affected by hsa-miR-17-3p
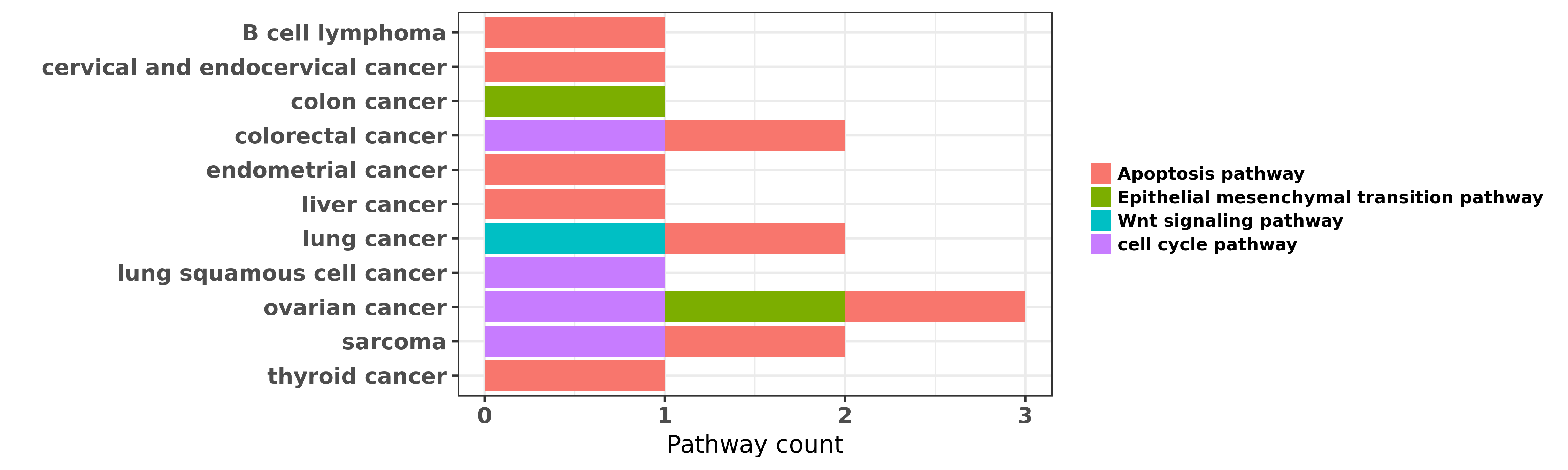
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-17-3p | B cell lymphoma | Apoptosis pathway | "The miR-17 approximately 92 cluster is frequently ......" | 20008931 | |
| hsa-miR-17-3p | cervical and endocervical cancer | Apoptosis pathway | "MiR 17 5p targets TP53INP1 and regulates cell prol ......" | 22730212 | |
| hsa-miR-17-3p | colon cancer | Epithelial mesenchymal transition pathway | "MicroRNA 17 induces epithelial mesenchymal transit ......" | 27278684 | Western blot |
| hsa-miR-17-3p | colorectal cancer | cell cycle pathway | "Up regulated miR 17 promotes cell proliferation tu ......" | 22132820 | |
| hsa-miR-17-3p | colorectal cancer | Apoptosis pathway | "Cell proliferation apoptosis and invasiveness were ......" | 26215320 | Flow cytometry; MTT assay; Western blot |
| hsa-miR-17-3p | endometrial cancer | Apoptosis pathway | "Bortezomib induces apoptosis of endometrial cancer ......" | 23716467 | |
| hsa-miR-17-3p | liver cancer | Apoptosis pathway | "Splicing regulator SLU7 preserves survival of hepa ......" | 26804174 | |
| hsa-miR-17-3p | lung cancer | Apoptosis pathway | "Apoptosis induction by antisense oligonucleotides ......" | 17384677 | |
| hsa-miR-17-3p | lung cancer | Apoptosis pathway | "MiR 16 targets Bcl 2 in paclitaxel resistant lung ......" | 25435430 | |
| hsa-miR-17-3p | lung cancer | Wnt signaling pathway | "miR 17 92/p38α Dysregulation Enhances Wnt Signali ......" | 27197183 | |
| hsa-miR-17-3p | lung squamous cell cancer | cell cycle pathway | "We found that the miR-17 family exerted important ......" | 24200043 | |
| hsa-miR-17-3p | ovarian cancer | cell cycle pathway | "MicroRNA 17 promotes normal ovarian cancer cells t ......" | 25510663 | Western blot |
| hsa-miR-17-3p | ovarian cancer | cell cycle pathway; Apoptosis pathway | "MiR 17 5p up regulates YES1 to modulate the cell c ......" | 25561420 | |
| hsa-miR-17-3p | ovarian cancer | Epithelial mesenchymal transition pathway | "MicroRNA 17 5p induces drug resistance and invasio ......" | 26500892 | MTT assay; Western blot |
| hsa-miR-17-3p | sarcoma | cell cycle pathway; Apoptosis pathway | "In our study we detected the six members of miR-17 ......" | 24824927 | Flow cytometry; Transwell assay |
| hsa-miR-17-3p | thyroid cancer | Apoptosis pathway | "Oncogenic role of miR 17 92 cluster in anaplastic ......" | 18429962 |
Reported cancer prognosis affected by hsa-miR-17-3p
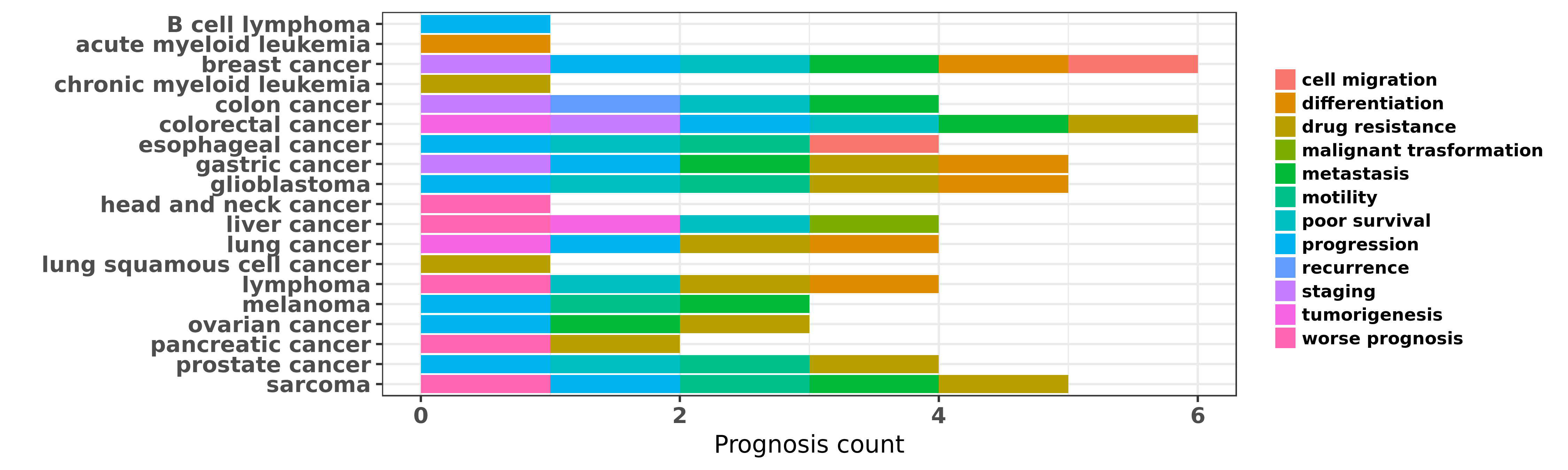
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-17-3p | B cell lymphoma | progression | "The High Expression of the microRNA 17 92 Cluster ......" | 25634356 | |
| hsa-miR-17-3p | acute myeloid leukemia | differentiation | "miR 17 deregulates a core RUNX1 miRNA mechanism of ......" | 25612891 | |
| hsa-miR-17-3p | breast cancer | cell migration | "miR 17 5p promotes human breast cancer cell migrat ......" | 20505989 | |
| hsa-miR-17-3p | breast cancer | differentiation | "In order to identify which miRNAs are involved in ......" | 22562546 | |
| hsa-miR-17-3p | breast cancer | progression | "Deregulated serum concentrations of circulating ce ......" | 23748853 | |
| hsa-miR-17-3p | breast cancer | poor survival | "Interestingly our data suggest that miRNA families ......" | 24398324 | |
| hsa-miR-17-3p | breast cancer | metastasis; progression | "Systematic analysis of metastasis associated genes ......" | 25001613 | |
| hsa-miR-17-3p | breast cancer | staging | "We employed qRT-PCR to determine expression level ......" | 27596294 | |
| hsa-miR-17-3p | chronic myeloid leukemia | drug resistance | "Expression of the miR 17 92 polycistron in chronic ......" | 17284533 | RNAi |
| hsa-miR-17-3p | chronic myeloid leukemia | drug resistance | "miR 17 in imatinib resistance and response to tyro ......" | 23818358 | |
| hsa-miR-17-3p | colon cancer | poor survival; staging | "Prognostic values of the miR 17 92 cluster and its ......" | 22065543 | |
| hsa-miR-17-3p | colon cancer | metastasis | "miR 21 miR 17 and miR 19a induced by phosphatase o ......" | 22677902 | RNAi |
| hsa-miR-17-3p | colon cancer | staging | "To present proof-of-principle application for empl ......" | 23741026 | |
| hsa-miR-17-3p | colon cancer | metastasis | "Nine dysregulated miRNA pairs fell into three miRN ......" | 25287248 | |
| hsa-miR-17-3p | colon cancer | staging | "microRNA 17 Is the Most Up Regulated Member of the ......" | 26465597 | |
| hsa-miR-17-3p | colon cancer | recurrence | "Serum expression levels of miR 17 miR 21 and miR 9 ......" | 26781797 | |
| hsa-miR-17-3p | colon cancer | staging | "Combined assays for serum carcinoembryonic antigen ......" | 26807240 | |
| hsa-miR-17-3p | colorectal cancer | progression; tumorigenesis | "Up regulated miR 17 promotes cell proliferation tu ......" | 22132820 | |
| hsa-miR-17-3p | colorectal cancer | progression | "Elevated oncofoetal miR 17 5p expression regulates ......" | 23250421 | |
| hsa-miR-17-3p | colorectal cancer | metastasis; drug resistance | "MicroRNA 17 5p promotes chemotherapeutic drug resi ......" | 24912422 | |
| hsa-miR-17-3p | colorectal cancer | staging; poor survival | "Using individual RT-qPCR verification in 175 stage ......" | 26250939 | |
| hsa-miR-17-3p | esophageal cancer | progression; poor survival | "MicroRNA 17 microRNA 18a and microRNA 19a are prog ......" | 24360091 | |
| hsa-miR-17-3p | esophageal cancer | cell migration; motility | "Here we evaluated microRNA expression in ESCC cell ......" | 27508097 | |
| hsa-miR-17-3p | gastric cancer | staging; metastasis; differentiation; progression | "Downregulation of serum miR 17 and miR 106b levels ......" | 25561770 | |
| hsa-miR-17-3p | gastric cancer | staging | "A total of 305 cases of diagnosed gastric adenocar ......" | 26168960 | |
| hsa-miR-17-3p | gastric cancer | metastasis | "miR 17 92 host gene uderexpressed in gastric cance ......" | 26837962 | |
| hsa-miR-17-3p | gastric cancer | drug resistance | "MicroRNA 17 5p promotes gastric cancer proliferati ......" | 27725906 | |
| hsa-miR-17-3p | glioblastoma | differentiation | "De repression of CTGF via the miR 17 92 cluster up ......" | 20305691 | Luciferase |
| hsa-miR-17-3p | glioblastoma | differentiation | "We find that miR-9 miR-9* referred to as miR-9/9* ......" | 21857646 | |
| hsa-miR-17-3p | glioblastoma | drug resistance; poor survival; motility | "Stress response of glioblastoma cells mediated by ......" | 23391506 | |
| hsa-miR-17-3p | glioblastoma | drug resistance; progression | "microRNA 17 regulates the expression of ATG7 and m ......" | 23792642 | Luciferase |
| hsa-miR-17-3p | head and neck cancer | worse prognosis | "Conversely decreased expressions of miR-153 miR-20 ......" | 25677760 | |
| hsa-miR-17-3p | liver cancer | malignant trasformation | "Elevated expression of the miR 17 92 polycistron a ......" | 18688024 | |
| hsa-miR-17-3p | liver cancer | worse prognosis | "High expression of serum miR 17 5p associated with ......" | 23108086 | |
| hsa-miR-17-3p | liver cancer | poor survival | "Mature miR 17 5p and passenger miR 17 3p induce he ......" | 23418359 | Colony formation |
| hsa-miR-17-3p | liver cancer | tumorigenesis | "In the present study we analyzed mCRP expression i ......" | 24297460 | Western blot |
| hsa-miR-17-3p | liver cancer | poor survival | "Splicing regulator SLU7 preserves survival of hepa ......" | 26804174 | |
| hsa-miR-17-3p | lung cancer | tumorigenesis | "Other significantly differentiated miR families in ......" | 23035181 | |
| hsa-miR-17-3p | lung cancer | differentiation | "Moreover the expression pattern of miR-17 miR-21 a ......" | 23036707 | |
| hsa-miR-17-3p | lung cancer | drug resistance | "miR 17 5p downregulation contributes to paclitaxel ......" | 24755562 | |
| hsa-miR-17-3p | lung cancer | drug resistance | "MiR 16 targets Bcl 2 in paclitaxel resistant lung ......" | 25435430 | |
| hsa-miR-17-3p | lung cancer | progression | "miR 17 92/p38α Dysregulation Enhances Wnt Signali ......" | 27197183 | |
| hsa-miR-17-3p | lung squamous cell cancer | drug resistance | "The regulatory and predictive functions of miR 17 ......" | 26482648 | |
| hsa-miR-17-3p | lung squamous cell cancer | drug resistance | "miR 17 5p down regulation contributes to erlotinib ......" | 27633093 | |
| hsa-miR-17-3p | lymphoma | drug resistance | "MicroRNA 17 92 significantly enhances radioresista ......" | 21040528 | |
| hsa-miR-17-3p | lymphoma | differentiation | "Overexpression of microRNAs from the miR 17 92 par ......" | 21698185 | |
| hsa-miR-17-3p | lymphoma | poor survival | "Using quantitative real-time reverse transcription ......" | 26231295 | |
| hsa-miR-17-3p | lymphoma | worse prognosis; poor survival | "miR 17 92 cluster components analysis in Burkitt l ......" | 27044389 | |
| hsa-miR-17-3p | melanoma | progression | "In situ analysis of melanoma samples using progres ......" | 23728176 | |
| hsa-miR-17-3p | melanoma | motility; metastasis | "miR 17 regulates melanoma cell motility by inhibit ......" | 26158900 | Cell migration assay; Luciferase |
| hsa-miR-17-3p | ovarian cancer | progression | "MiR 17 5p up regulates YES1 to modulate the cell c ......" | 25561420 | |
| hsa-miR-17-3p | ovarian cancer | drug resistance | "MicroRNA 17 5p induces drug resistance and invasio ......" | 26500892 | MTT assay; Western blot |
| hsa-miR-17-3p | ovarian cancer | drug resistance | "Evidence for miR 17 92 and miR 134 gene cluster re ......" | 27383301 | |
| hsa-miR-17-3p | ovarian cancer | metastasis | "miR 17 inhibits ovarian cancer cell peritoneal met ......" | 27499367 | |
| hsa-miR-17-3p | pancreatic cancer | worse prognosis | "MicroRNA miR 17 5p is overexpressed in pancreatic ......" | 20703102 | |
| hsa-miR-17-3p | pancreatic cancer | drug resistance | "The miR 17 92 cluster counteracts quiescence and c ......" | 25887381 | |
| hsa-miR-17-3p | pancreatic cancer | drug resistance | "GFRα2 prompts cell growth and chemoresistance thr ......" | 27400681 | |
| hsa-miR-17-3p | prostate cancer | motility; progression | "MicroRNA 17 3p is a prostate tumor suppressor in v ......" | 19771525 | |
| hsa-miR-17-3p | prostate cancer | poor survival | "Both mature miR 17 5p and passenger strand miR 17 ......" | 23990326 | Colony formation |
| hsa-miR-17-3p | prostate cancer | poor survival | "Non-responders to docetaxel and patients with shor ......" | 24714754 | |
| hsa-miR-17-3p | prostate cancer | drug resistance | "miR 17 92 plays an oncogenic role and conveys chem ......" | 26891588 | |
| hsa-miR-17-3p | prostate cancer | drug resistance | "Downregulation of miR 17 92a cluster promotes auto ......" | 27501757 | Luciferase |
| hsa-miR-17-3p | sarcoma | metastasis; progression | "miR 17 inhibitor suppressed osteosarcoma tumor gro ......" | 24462867 | |
| hsa-miR-17-3p | sarcoma | progression; worse prognosis | "Upregulation of microRNA 17 92 cluster associates ......" | 24645838 | |
| hsa-miR-17-3p | sarcoma | motility; drug resistance | "SS18 SSX regulated miR 17 promotes tumor growth of ......" | 24989082 | Luciferase |
Reported gene related to hsa-miR-17-3p
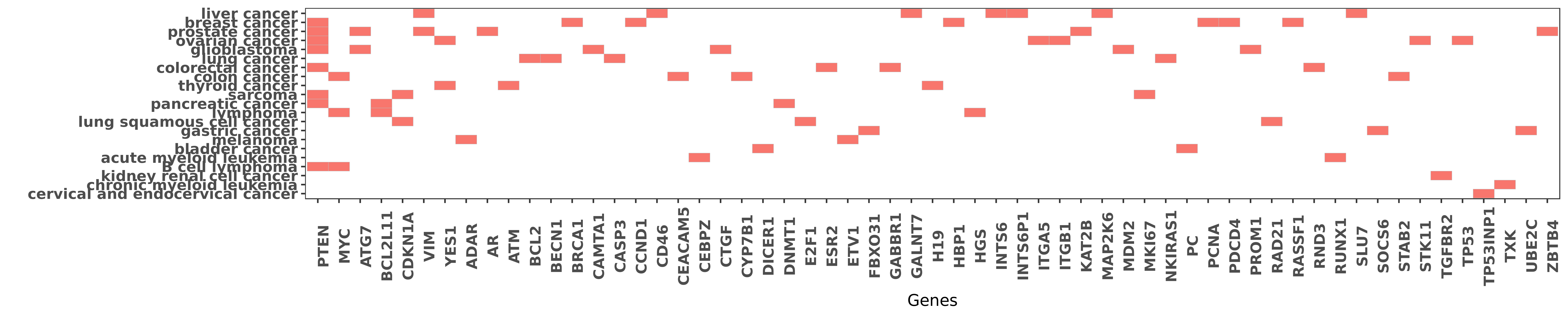
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-17-3p | B cell lymphoma | PTEN | "The High Expression of the microRNA 17 92 Cluster ......" | 25634356 |
| hsa-miR-17-3p | breast cancer | PTEN | "Bioinformatics analysis and folding energy calcula ......" | 26629823 |
| hsa-miR-17-3p | colorectal cancer | PTEN | "In addition ERβ expression significantly increase ......" | 26215320 |
| hsa-miR-17-3p | colorectal cancer | PTEN | "MicroRNA 17 5p promotes chemotherapeutic drug resi ......" | 24912422 |
| hsa-miR-17-3p | glioblastoma | PTEN | "We found that these phenotypes were the results of ......" | 23391506 |
| hsa-miR-17-3p | ovarian cancer | PTEN | "MicroRNA 17 5p induces drug resistance and invasio ......" | 26500892 |
| hsa-miR-17-3p | pancreatic cancer | PTEN | "GFRα2 prompts cell growth and chemoresistance thr ......" | 27400681 |
| hsa-miR-17-3p | prostate cancer | PTEN | "Resveratrol and pterostilbene epigenetically resto ......" | 26318586 |
| hsa-miR-17-3p | sarcoma | PTEN | "PTEN mRNA correlated inversely with miR-92a and me ......" | 23133552 |
| hsa-miR-17-3p | sarcoma | PTEN | "miR 17 inhibitor suppressed osteosarcoma tumor gro ......" | 24462867 |
| hsa-miR-17-3p | B cell lymphoma | MYC | "miR 17 92 fine tunes MYC expression and function t ......" | 26555894 |
| hsa-miR-17-3p | B cell lymphoma | MYC | "The miR-17 approximately 92 cluster is frequently ......" | 20008931 |
| hsa-miR-17-3p | colon cancer | MYC | "Butyrate inhibits pro proliferative miR 92a by dim ......" | 26463716 |
| hsa-miR-17-3p | colon cancer | MYC | "Using target prediction and anticorrelation to gen ......" | 23436804 |
| hsa-miR-17-3p | lymphoma | MYC | "Statistical analysis showed that higher expression ......" | 26231295 |
| hsa-miR-17-3p | lymphoma | MYC | "The Myc miR 17 92 axis amplifies B cell receptor s ......" | 24169826 |
| hsa-miR-17-3p | melanoma | ADAR | "We have recently shown that miR-17 directly target ......" | 24920276 |
| hsa-miR-17-3p | melanoma | ADAR | "We found that cancer cells silence ADAR1 by overex ......" | 23728176 |
| hsa-miR-17-3p | glioblastoma | ATG7 | "microRNA 17 regulates the expression of ATG7 and m ......" | 23792642 |
| hsa-miR-17-3p | prostate cancer | ATG7 | "Autophagy-related gene ATG7 was validated as a tar ......" | 27501757 |
| hsa-miR-17-3p | lymphoma | BCL2L11 | "We found that upregulated expression of miR-17 and ......" | 27044389 |
| hsa-miR-17-3p | pancreatic cancer | BCL2L11 | "miR 17 5p inhibitor enhances chemosensitivity to g ......" | 23001407 |
| hsa-miR-17-3p | lung cancer | BECN1 | "miR 17 5p downregulation contributes to paclitaxel ......" | 24755562 |
| hsa-miR-17-3p | lung cancer | BECN1 | "Previously utilizing miRNA arrays we reported that ......" | 25435430 |
| hsa-miR-17-3p | lung squamous cell cancer | CDKN1A | "The low expressions of miR-17 and miR-92 families ......" | 26482648 |
| hsa-miR-17-3p | sarcoma | CDKN1A | "According to PicTar and Miranda algorithms which p ......" | 24989082 |
| hsa-miR-17-3p | liver cancer | VIM | "In contrast the passenger strand miR-17-3p repress ......" | 23418359 |
| hsa-miR-17-3p | prostate cancer | VIM | "The lack of miR-17-3p expression correlated with a ......" | 19771525 |
| hsa-miR-17-3p | ovarian cancer | YES1 | "MiR 17 5p up regulates YES1 to modulate the cell c ......" | 25561420 |
| hsa-miR-17-3p | thyroid cancer | YES1 | "Long noncoding RNA H19 competitively binds miR 17 ......" | 27093644 |
| hsa-miR-17-3p | prostate cancer | AR | "miR 17 5p targets the p300/CBP associated factor a ......" | 23095762 |
| hsa-miR-17-3p | thyroid cancer | ATM | "In 3 of 6 clinical ATC samples miR-17-3p and miR-1 ......" | 18429962 |
| hsa-miR-17-3p | lung cancer | BCL2 | "Combined overexpression of miR-16 and miR-17 great ......" | 25435430 |
| hsa-miR-17-3p | breast cancer | BRCA1 | "In the target in vitro assay we observed that miR- ......" | 19048628 |
| hsa-miR-17-3p | glioblastoma | CAMTA1 | "We identify CAMTA1 as an miR-9/9* and miR-17 targe ......" | 21857646 |
| hsa-miR-17-3p | lung cancer | CASP3 | "Moreover in this report we showed that the combine ......" | 25435430 |
| hsa-miR-17-3p | breast cancer | CCND1 | "Bioinformatics Prediction and In Vitro Analysis Re ......" | 26431674 |
| hsa-miR-17-3p | liver cancer | CD46 | "The present study suggests that CD46 plays an impo ......" | 24297460 |
| hsa-miR-17-3p | colon cancer | CEACAM5 | "Combined assays for serum carcinoembryonic antigen ......" | 26807240 |
| hsa-miR-17-3p | acute myeloid leukemia | CEBPZ | "miR 17 deregulates a core RUNX1 miRNA mechanism of ......" | 25612891 |
| hsa-miR-17-3p | glioblastoma | CTGF | "De repression of CTGF via the miR 17 92 cluster up ......" | 20305691 |
| hsa-miR-17-3p | colon cancer | CYP7B1 | "MicroRNA 17 induces epithelial mesenchymal transit ......" | 27278684 |
| hsa-miR-17-3p | bladder cancer | DICER1 | "microRNA 18a a member of the oncogenic miR 17 92 c ......" | 21935572 |
| hsa-miR-17-3p | pancreatic cancer | DNMT1 | "DNMT1 Inhibition Reprograms Pancreatic Cancer Stem ......" | 27261509 |
| hsa-miR-17-3p | lung squamous cell cancer | E2F1 | "We proposed a model for the miR-17 family E2F1 and ......" | 24200043 |
| hsa-miR-17-3p | colorectal cancer | ESR2 | "mRNA levels of ER beta ERβ and miR-17 were quanti ......" | 26215320 |
| hsa-miR-17-3p | melanoma | ETV1 | "miR 17 regulates melanoma cell motility by inhibit ......" | 26158900 |
| hsa-miR-17-3p | gastric cancer | FBXO31 | "F box protein FBXO31 is down regulated in gastric ......" | 25115392 |
| hsa-miR-17-3p | colorectal cancer | GABBR1 | "In conclusion miR-106a/b miR-20a/b and miR-17 cont ......" | 27230463 |
| hsa-miR-17-3p | liver cancer | GALNT7 | "In contrast the passenger strand miR-17-3p repress ......" | 23418359 |
| hsa-miR-17-3p | thyroid cancer | H19 | "Long noncoding RNA H19 competitively binds miR 17 ......" | 27093644 |
| hsa-miR-17-3p | breast cancer | HBP1 | "miR 17 5p promotes human breast cancer cell migrat ......" | 20505989 |
| hsa-miR-17-3p | lymphoma | HGS | "Expression of miR-17 and miR-106a was detected in ......" | 21953646 |
| hsa-miR-17-3p | liver cancer | INTS6 | "Pseudogene INTS6P1 regulates its cognate gene INTS ......" | 25686840 |
| hsa-miR-17-3p | liver cancer | INTS6P1 | "Pseudogene INTS6P1 regulates its cognate gene INTS ......" | 25686840 |
| hsa-miR-17-3p | ovarian cancer | ITGA5 | "miR 17 inhibits ovarian cancer cell peritoneal met ......" | 27499367 |
| hsa-miR-17-3p | ovarian cancer | ITGB1 | "miR 17 inhibits ovarian cancer cell peritoneal met ......" | 27499367 |
| hsa-miR-17-3p | prostate cancer | KAT2B | "miR 17 5p targets the p300/CBP associated factor a ......" | 23095762 |
| hsa-miR-17-3p | liver cancer | MAP2K6 | "miR 17 5p Promotes migration of human hepatocellul ......" | 20209605 |
| hsa-miR-17-3p | glioblastoma | MDM2 | "Finally we demonstrated that miR-17 could repress ......" | 23391506 |
| hsa-miR-17-3p | sarcoma | MKI67 | "Tumor volume formed in mice in vivo was significan ......" | 24989082 |
| hsa-miR-17-3p | lung cancer | NKIRAS1 | "Besides a statistically significant negative corre ......" | 23156677 |
| hsa-miR-17-3p | bladder cancer | PC | "Long Noncoding RNA GCASPC a Target of miR 17 3p Ne ......" | 27450454 |
| hsa-miR-17-3p | breast cancer | PCNA | "Bioinformatics Prediction and In Vitro Analysis Re ......" | 26431674 |
| hsa-miR-17-3p | breast cancer | PDCD4 | "Bioinformatics analysis and folding energy calcula ......" | 26629823 |
| hsa-miR-17-3p | glioblastoma | PROM1 | "Ectopic expression of miR-17 was found to facilita ......" | 23391506 |
| hsa-miR-17-3p | lung squamous cell cancer | RAD21 | "The low expressions of miR-17 and miR-92 families ......" | 26482648 |
| hsa-miR-17-3p | breast cancer | RASSF1 | "RASSF1A resulted more frequently methylated in men ......" | 23160909 |
| hsa-miR-17-3p | colorectal cancer | RND3 | "Up regulated miR 17 promotes cell proliferation tu ......" | 22132820 |
| hsa-miR-17-3p | acute myeloid leukemia | RUNX1 | "miR 17 deregulates a core RUNX1 miRNA mechanism of ......" | 25612891 |
| hsa-miR-17-3p | liver cancer | SLU7 | "Interestingly altered splicing of miR-17-92 and do ......" | 26804174 |
| hsa-miR-17-3p | gastric cancer | SOCS6 | "miR 17 5p promotes proliferation by targeting SOCS ......" | 24801601 |
| hsa-miR-17-3p | colon cancer | STAB2 | "Nine dysregulated miRNA pairs fell into three miRN ......" | 25287248 |
| hsa-miR-17-3p | ovarian cancer | STK11 | "MicroRNA 17 promotes normal ovarian cancer cells t ......" | 25510663 |
| hsa-miR-17-3p | kidney renal cell cancer | TGFBR2 | "We demonstrate that miR-17 overexpression interfer ......" | 25011053 |
| hsa-miR-17-3p | ovarian cancer | TP53 | "MicroRNA 17 promotes normal ovarian cancer cells t ......" | 25510663 |
| hsa-miR-17-3p | cervical and endocervical cancer | TP53INP1 | "MiR 17 5p targets TP53INP1 and regulates cell prol ......" | 22730212 |
| hsa-miR-17-3p | chronic myeloid leukemia | TXK | "miR 17 in imatinib resistance and response to tyro ......" | 23818358 |
| hsa-miR-17-3p | gastric cancer | UBE2C | "We found that both miR-17 and miR-20a miR-17/20a t ......" | 25760688 |
| hsa-miR-17-3p | prostate cancer | ZBTB4 | "Induction of the transcriptional repressor ZBTB4 i ......" | 22752225 |
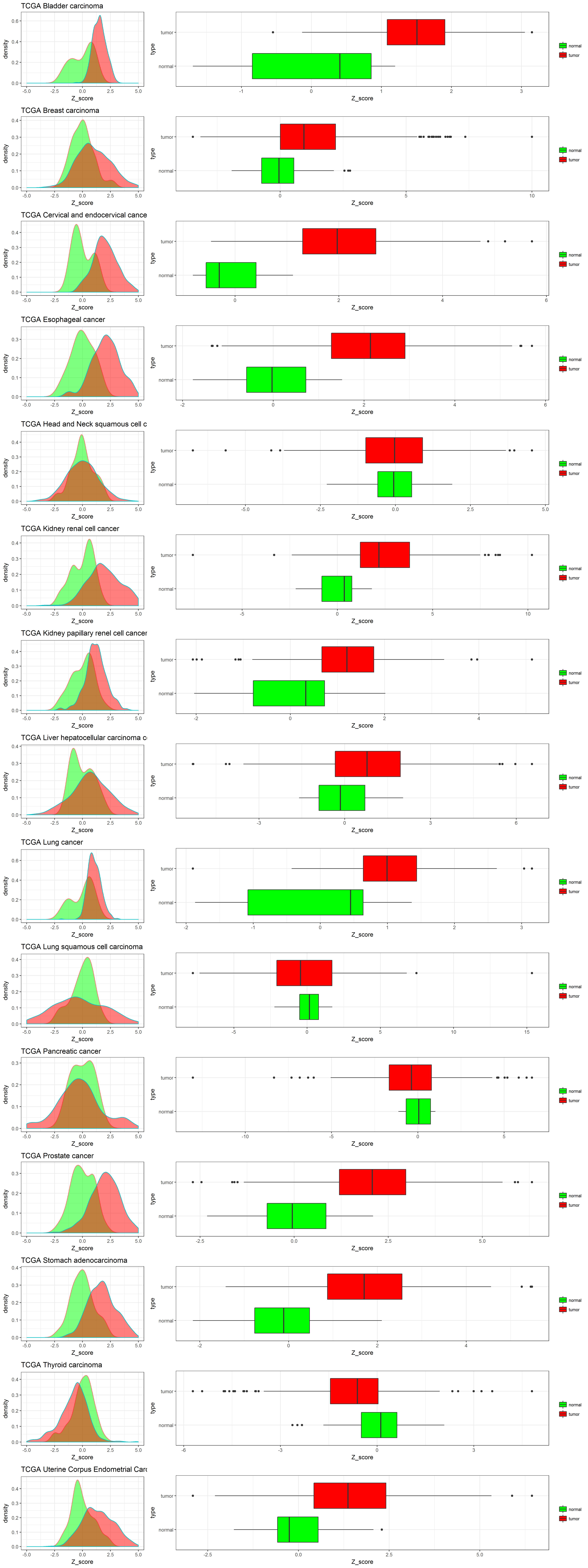
Expression profile in cancer corhorts:
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-17-3p | HBP1 | 13 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LIHC; LUSC; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate; MirTarget | TCGA BLCA -0.076; TCGA BRCA -0.219; TCGA CESC -0.182; TCGA ESCA -0.201; TCGA HNSC -0.161; TCGA KIRC -0.082; TCGA KIRP -0.073; TCGA LIHC -0.17; TCGA LUSC -0.077; TCGA PRAD -0.078; TCGA SARC -0.122; TCGA STAD -0.124; TCGA UCEC -0.091 |
hsa-miR-17-3p | HOXB2 | 14 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.443; TCGA BRCA -0.547; TCGA CESC -0.453; TCGA COAD -0.436; TCGA HNSC -0.362; TCGA KIRC -0.248; TCGA KIRP -0.372; TCGA LIHC -0.118; TCGA LUAD -0.324; TCGA LUSC -0.269; TCGA PRAD -0.638; TCGA SARC -0.233; TCGA STAD -0.43; TCGA UCEC -0.156 |
hsa-miR-17-3p | P2RX6 | 11 cancers: BLCA; BRCA; ESCA; HNSC; KIRP; LUAD; LUSC; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.311; TCGA BRCA -0.158; TCGA ESCA -0.398; TCGA HNSC -0.471; TCGA KIRP -0.641; TCGA LUAD -0.478; TCGA LUSC -0.248; TCGA PRAD -0.319; TCGA SARC -0.483; TCGA STAD -0.479; TCGA UCEC -0.202 |
hsa-miR-17-3p | PRKCA | 10 cancers: BLCA; CESC; HNSC; KIRC; KIRP; LUAD; PRAD; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.163; TCGA CESC -0.25; TCGA HNSC -0.215; TCGA KIRC -0.553; TCGA KIRP -0.17; TCGA LUAD -0.221; TCGA PRAD -0.467; TCGA THCA -0.519; TCGA STAD -0.147; TCGA UCEC -0.285 |
hsa-miR-17-3p | PRMT2 | 9 cancers: BLCA; COAD; ESCA; LUAD; LUSC; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.098; TCGA COAD -0.068; TCGA ESCA -0.165; TCGA LUAD -0.08; TCGA LUSC -0.075; TCGA PRAD -0.05; TCGA SARC -0.085; TCGA STAD -0.247; TCGA UCEC -0.068 |
hsa-miR-17-3p | SELE | 12 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; LIHC; OV; PRAD; THCA; UCEC | miRNAWalker2 validate | TCGA BLCA -0.781; TCGA BRCA -0.149; TCGA CESC -0.718; TCGA COAD -0.353; TCGA HNSC -0.333; TCGA KIRC -0.598; TCGA KIRP -1.194; TCGA LIHC -0.613; TCGA OV -0.275; TCGA PRAD -0.769; TCGA THCA -0.407; TCGA UCEC -0.458 |
hsa-miR-17-3p | STIM1 | 14 cancers: BLCA; CESC; COAD; ESCA; HNSC; LGG; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.112; TCGA CESC -0.121; TCGA COAD -0.105; TCGA ESCA -0.254; TCGA HNSC -0.135; TCGA LGG -0.124; TCGA LUAD -0.071; TCGA LUSC -0.19; TCGA PAAD -0.139; TCGA PRAD -0.124; TCGA SARC -0.249; TCGA THCA -0.178; TCGA STAD -0.221; TCGA UCEC -0.266 |
hsa-miR-17-3p | VIM | 13 cancers: BLCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | miRNAWalker2 validate; miRNATAP | TCGA BLCA -0.482; TCGA CESC -0.328; TCGA COAD -0.407; TCGA ESCA -0.461; TCGA HNSC -0.348; TCGA LIHC -0.107; TCGA LUAD -0.342; TCGA LUSC -0.297; TCGA OV -0.193; TCGA PAAD -0.271; TCGA PRAD -0.276; TCGA STAD -0.422; TCGA UCEC -0.255 |
hsa-miR-17-3p | ZNF423 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; LUAD; PAAD; PRAD; THCA; STAD; UCEC | miRNAWalker2 validate; MirTarget; miRNATAP | TCGA BLCA -0.466; TCGA BRCA -0.531; TCGA CESC -0.468; TCGA COAD -0.42; TCGA ESCA -0.609; TCGA HNSC -0.508; TCGA LGG -0.099; TCGA LIHC -0.273; TCGA LUAD -0.46; TCGA PAAD -0.635; TCGA PRAD -0.558; TCGA THCA -0.243; TCGA STAD -0.586; TCGA UCEC -0.291 |
hsa-miR-17-3p | PCYT1B | 13 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LUAD; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.388; TCGA CESC -0.796; TCGA COAD -0.602; TCGA ESCA -0.619; TCGA HNSC -0.38; TCGA KIRC -0.456; TCGA LGG -0.105; TCGA LUAD -0.284; TCGA PAAD -0.929; TCGA PRAD -0.907; TCGA SARC -0.643; TCGA STAD -0.534; TCGA UCEC -0.301 |
hsa-miR-17-3p | LRCH2 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; PAAD; PRAD; STAD | MirTarget | TCGA BLCA -0.689; TCGA BRCA -0.326; TCGA CESC -0.985; TCGA COAD -0.754; TCGA ESCA -0.865; TCGA HNSC -0.414; TCGA KIRC -0.36; TCGA KIRP -0.881; TCGA LIHC -0.143; TCGA LUAD -0.302; TCGA PAAD -0.656; TCGA PRAD -0.707; TCGA STAD -0.666 |
hsa-miR-17-3p | WWTR1 | 10 cancers: BLCA; COAD; ESCA; HNSC; KIRP; LIHC; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.372; TCGA COAD -0.51; TCGA ESCA -0.519; TCGA HNSC -0.148; TCGA KIRP -0.323; TCGA LIHC -0.375; TCGA PRAD -0.496; TCGA THCA -0.222; TCGA STAD -0.516; TCGA UCEC -0.155 |
hsa-miR-17-3p | ITIH5 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.921; TCGA BRCA -0.421; TCGA CESC -0.255; TCGA COAD -0.531; TCGA ESCA -0.587; TCGA HNSC -0.186; TCGA KIRC -0.234; TCGA KIRP -1.175; TCGA LIHC -0.463; TCGA LUAD -0.491; TCGA PAAD -0.697; TCGA PRAD -0.618; TCGA SARC -0.528; TCGA STAD -0.98; TCGA UCEC -0.287 |
hsa-miR-17-3p | INPP5A | 11 cancers: BLCA; BRCA; COAD; ESCA; KIRP; LGG; LUSC; OV; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.327; TCGA BRCA -0.133; TCGA COAD -0.078; TCGA ESCA -0.201; TCGA KIRP -0.104; TCGA LGG -0.084; TCGA LUSC -0.111; TCGA OV -0.166; TCGA SARC -0.245; TCGA STAD -0.349; TCGA UCEC -0.08 |
hsa-miR-17-3p | SUCLG2 | 10 cancers: BLCA; BRCA; KIRC; KIRP; LGG; LIHC; LUSC; SARC; THCA; STAD | MirTarget | TCGA BLCA -0.106; TCGA BRCA -0.211; TCGA KIRC -0.554; TCGA KIRP -0.153; TCGA LGG -0.088; TCGA LIHC -0.134; TCGA LUSC -0.091; TCGA SARC -0.103; TCGA THCA -0.152; TCGA STAD -0.104 |
hsa-miR-17-3p | CYTH3 | 10 cancers: BLCA; COAD; ESCA; HNSC; KIRP; LIHC; LUAD; PAAD; THCA; STAD | MirTarget | TCGA BLCA -0.13; TCGA COAD -0.116; TCGA ESCA -0.234; TCGA HNSC -0.141; TCGA KIRP -0.261; TCGA LIHC -0.22; TCGA LUAD -0.189; TCGA PAAD -0.292; TCGA THCA -0.17; TCGA STAD -0.231 |
hsa-miR-17-3p | SNX1 | 9 cancers: BLCA; BRCA; ESCA; KIRC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.123; TCGA BRCA -0.216; TCGA ESCA -0.183; TCGA KIRC -0.101; TCGA PAAD -0.141; TCGA PRAD -0.119; TCGA SARC -0.095; TCGA STAD -0.201; TCGA UCEC -0.075 |
hsa-miR-17-3p | KIAA0232 | 11 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LIHC; PRAD; SARC; STAD | MirTarget | TCGA BLCA -0.163; TCGA BRCA -0.358; TCGA CESC -0.182; TCGA ESCA -0.162; TCGA HNSC -0.148; TCGA KIRC -0.295; TCGA KIRP -0.162; TCGA LIHC -0.102; TCGA PRAD -0.089; TCGA SARC -0.107; TCGA STAD -0.069 |
hsa-miR-17-3p | CPEB3 | 12 cancers: BLCA; BRCA; CESC; ESCA; KIRC; KIRP; LGG; LIHC; PAAD; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.239; TCGA BRCA -0.371; TCGA CESC -0.248; TCGA ESCA -0.357; TCGA KIRC -0.469; TCGA KIRP -0.328; TCGA LGG -0.108; TCGA LIHC -0.359; TCGA PAAD -0.238; TCGA THCA -0.124; TCGA STAD -0.128; TCGA UCEC -0.123 |
hsa-miR-17-3p | ANGPTL1 | 17 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; OV; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -1.313; TCGA BRCA -0.644; TCGA CESC -1.15; TCGA COAD -1.505; TCGA ESCA -1.557; TCGA HNSC -0.551; TCGA KIRC -1.399; TCGA KIRP -1.402; TCGA LIHC -0.505; TCGA LUAD -0.365; TCGA LUSC -0.315; TCGA OV -0.347; TCGA PAAD -0.961; TCGA PRAD -0.871; TCGA SARC -0.482; TCGA STAD -1.518; TCGA UCEC -0.532 |
hsa-miR-17-3p | CDO1 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -1.005; TCGA BRCA -0.451; TCGA CESC -1.004; TCGA COAD -0.936; TCGA ESCA -1.326; TCGA HNSC -0.483; TCGA KIRC -0.892; TCGA KIRP -0.566; TCGA LGG -0.102; TCGA LUAD -0.413; TCGA LUSC -0.43; TCGA PAAD -0.584; TCGA PRAD -0.492; TCGA STAD -1.214; TCGA UCEC -0.594 |
hsa-miR-17-3p | CSDE1 | 10 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; KIRP; LIHC; PRAD; THCA; STAD | MirTarget; miRNATAP | TCGA BLCA -0.116; TCGA BRCA -0.059; TCGA ESCA -0.134; TCGA HNSC -0.068; TCGA KIRC -0.149; TCGA KIRP -0.126; TCGA LIHC -0.095; TCGA PRAD -0.087; TCGA THCA -0.081; TCGA STAD -0.14 |
hsa-miR-17-3p | DOK6 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.946; TCGA BRCA -0.389; TCGA CESC -0.708; TCGA COAD -0.605; TCGA ESCA -0.625; TCGA HNSC -0.473; TCGA KIRC -0.393; TCGA KIRP -0.358; TCGA LUAD -0.197; TCGA PAAD -0.827; TCGA PRAD -0.309; TCGA STAD -0.5; TCGA UCEC -0.337 |
hsa-miR-17-3p | KITLG | 12 cancers: BLCA; BRCA; COAD; ESCA; KIRC; KIRP; LGG; LUSC; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.393; TCGA BRCA -0.553; TCGA COAD -0.254; TCGA ESCA -0.338; TCGA KIRC -0.588; TCGA KIRP -0.832; TCGA LGG -0.22; TCGA LUSC -0.224; TCGA PRAD -0.52; TCGA THCA -0.31; TCGA STAD -0.203; TCGA UCEC -0.185 |
hsa-miR-17-3p | FOXP1 | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; SARC; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.213; TCGA BRCA -0.417; TCGA CESC -0.172; TCGA COAD -0.172; TCGA ESCA -0.232; TCGA HNSC -0.112; TCGA KIRC -0.282; TCGA KIRP -0.15; TCGA SARC -0.18; TCGA UCEC -0.259 |
hsa-miR-17-3p | DNAJC18 | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LUAD; PAAD; PRAD; STAD | MirTarget | TCGA BLCA -0.254; TCGA BRCA -0.16; TCGA COAD -0.209; TCGA ESCA -0.462; TCGA HNSC -0.137; TCGA KIRP -0.149; TCGA LUAD -0.105; TCGA PAAD -0.344; TCGA PRAD -0.312; TCGA STAD -0.464 |
hsa-miR-17-3p | PNMA1 | 13 cancers: BLCA; BRCA; COAD; ESCA; KIRC; KIRP; LGG; OV; PAAD; PRAD; SARC; THCA; STAD | MirTarget | TCGA BLCA -0.259; TCGA BRCA -0.103; TCGA COAD -0.41; TCGA ESCA -0.261; TCGA KIRC -0.167; TCGA KIRP -0.165; TCGA LGG -0.091; TCGA OV -0.113; TCGA PAAD -0.253; TCGA PRAD -0.411; TCGA SARC -0.177; TCGA THCA -0.096; TCGA STAD -0.332 |
hsa-miR-17-3p | TSHZ3 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.768; TCGA BRCA -0.352; TCGA CESC -0.364; TCGA COAD -0.576; TCGA ESCA -0.785; TCGA HNSC -0.454; TCGA KIRP -0.502; TCGA LIHC -0.27; TCGA LUAD -0.371; TCGA LUSC -0.244; TCGA PAAD -0.305; TCGA PRAD -0.407; TCGA STAD -0.635; TCGA UCEC -0.528 |
hsa-miR-17-3p | SH3PXD2A | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.4; TCGA BRCA -0.089; TCGA COAD -0.209; TCGA ESCA -0.217; TCGA HNSC -0.178; TCGA KIRP -0.27; TCGA LUAD -0.071; TCGA LUSC -0.189; TCGA PAAD -0.265; TCGA PRAD -0.423; TCGA STAD -0.172; TCGA UCEC -0.105 |
hsa-miR-17-3p | TNS1 | 16 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -1.145; TCGA BRCA -0.429; TCGA CESC -0.371; TCGA COAD -0.768; TCGA ESCA -1.037; TCGA HNSC -0.402; TCGA KIRP -0.405; TCGA LIHC -0.107; TCGA LUAD -0.235; TCGA LUSC -0.217; TCGA PAAD -0.591; TCGA PRAD -0.811; TCGA SARC -0.782; TCGA THCA -0.127; TCGA STAD -1.051; TCGA UCEC -0.435 |
hsa-miR-17-3p | IQSEC1 | 13 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.094; TCGA BRCA -0.159; TCGA COAD -0.139; TCGA ESCA -0.281; TCGA HNSC -0.274; TCGA KIRC -0.089; TCGA KIRP -0.361; TCGA LUAD -0.19; TCGA LUSC -0.138; TCGA PAAD -0.141; TCGA THCA -0.212; TCGA STAD -0.168; TCGA UCEC -0.185 |
hsa-miR-17-3p | ADAMTS15 | 13 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PRAD; SARC; STAD | MirTarget; miRNATAP | TCGA BLCA -0.8; TCGA BRCA -0.969; TCGA COAD -0.385; TCGA ESCA -1.016; TCGA HNSC -0.767; TCGA KIRC -0.784; TCGA KIRP -0.486; TCGA LIHC -0.416; TCGA LUAD -0.415; TCGA LUSC -0.502; TCGA PRAD -0.691; TCGA SARC -0.444; TCGA STAD -0.553 |
hsa-miR-17-3p | TOX | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; PRAD; STAD | MirTarget | TCGA BLCA -0.298; TCGA CESC -0.32; TCGA COAD -0.477; TCGA ESCA -0.602; TCGA HNSC -0.365; TCGA KIRC -0.737; TCGA KIRP -0.618; TCGA PRAD -0.316; TCGA STAD -0.697 |
hsa-miR-17-3p | SORCS2 | 16 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.781; TCGA BRCA -0.418; TCGA CESC -0.417; TCGA COAD -0.54; TCGA ESCA -0.883; TCGA HNSC -0.806; TCGA KIRC -0.525; TCGA KIRP -0.861; TCGA LGG -0.121; TCGA LUAD -0.35; TCGA LUSC -0.545; TCGA OV -0.313; TCGA PAAD -0.528; TCGA PRAD -0.601; TCGA STAD -0.589; TCGA UCEC -0.288 |
hsa-miR-17-3p | GCOM1 | 13 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.638; TCGA BRCA -0.207; TCGA CESC -0.351; TCGA ESCA -0.518; TCGA HNSC -0.341; TCGA KIRC -1.203; TCGA KIRP -1.118; TCGA LIHC -0.228; TCGA LUAD -0.364; TCGA PRAD -0.654; TCGA SARC -0.396; TCGA STAD -0.097; TCGA UCEC -0.534 |
hsa-miR-17-3p | CDK17 | 9 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; PAAD; PRAD | MirTarget; miRNATAP | TCGA BLCA -0.104; TCGA BRCA -0.309; TCGA CESC -0.109; TCGA COAD -0.172; TCGA ESCA -0.294; TCGA HNSC -0.064; TCGA LGG -0.058; TCGA PAAD -0.173; TCGA PRAD -0.074 |
hsa-miR-17-3p | ANKRD50 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; LUSC; PRAD; THCA; STAD | MirTarget; miRNATAP | TCGA BLCA -0.245; TCGA BRCA -0.288; TCGA CESC -0.189; TCGA COAD -0.229; TCGA ESCA -0.289; TCGA HNSC -0.172; TCGA KIRC -0.121; TCGA LIHC -0.238; TCGA LUSC -0.118; TCGA PRAD -0.205; TCGA THCA -0.107; TCGA STAD -0.141 |
hsa-miR-17-3p | ZFHX4 | 9 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; PAAD; PRAD; STAD | MirTarget; miRNATAP | TCGA BLCA -0.678; TCGA BRCA -0.506; TCGA CESC -0.574; TCGA COAD -1.002; TCGA ESCA -0.747; TCGA KIRC -0.399; TCGA PAAD -0.397; TCGA PRAD -0.252; TCGA STAD -0.727 |
hsa-miR-17-3p | ARHGEF12 | 11 cancers: BLCA; BRCA; CESC; ESCA; KIRC; KIRP; LIHC; PRAD; SARC; THCA; UCEC | MirTarget | TCGA BLCA -0.121; TCGA BRCA -0.337; TCGA CESC -0.123; TCGA ESCA -0.087; TCGA KIRC -0.322; TCGA KIRP -0.277; TCGA LIHC -0.209; TCGA PRAD -0.165; TCGA SARC -0.112; TCGA THCA -0.148; TCGA UCEC -0.076 |
hsa-miR-17-3p | RAB8B | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; OV; PRAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.124; TCGA BRCA -0.223; TCGA CESC -0.116; TCGA COAD -0.201; TCGA ESCA -0.278; TCGA HNSC -0.129; TCGA LIHC -0.117; TCGA LUAD -0.18; TCGA LUSC -0.136; TCGA OV -0.1; TCGA PRAD -0.159; TCGA UCEC -0.097 |
hsa-miR-17-3p | IRS1 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.354; TCGA BRCA -0.443; TCGA CESC -0.339; TCGA COAD -0.273; TCGA ESCA -0.3; TCGA HNSC -0.211; TCGA KIRC -0.42; TCGA LUAD -0.162; TCGA LUSC -0.375; TCGA PRAD -0.23; TCGA SARC -0.286; TCGA STAD -0.231; TCGA UCEC -0.247 |
hsa-miR-17-3p | CPNE8 | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; LIHC; LUSC; PAAD; PRAD; STAD | MirTarget | TCGA BLCA -0.463; TCGA CESC -0.292; TCGA COAD -0.224; TCGA ESCA -0.312; TCGA HNSC -0.136; TCGA LIHC -0.182; TCGA LUSC -0.197; TCGA PAAD -0.349; TCGA PRAD -0.454; TCGA STAD -0.226 |
hsa-miR-17-3p | MBNL1 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; LIHC; LUAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.248; TCGA BRCA -0.119; TCGA CESC -0.095; TCGA COAD -0.142; TCGA ESCA -0.258; TCGA LIHC -0.094; TCGA LUAD -0.1; TCGA PRAD -0.238; TCGA SARC -0.35; TCGA STAD -0.188; TCGA UCEC -0.087 |
hsa-miR-17-3p | MAP4 | 14 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LGG; LIHC; LUAD; LUSC; PRAD; SARC; THCA; STAD | MirTarget; miRNATAP | TCGA BLCA -0.122; TCGA BRCA -0.074; TCGA COAD -0.123; TCGA ESCA -0.243; TCGA HNSC -0.236; TCGA KIRC -0.171; TCGA LGG -0.097; TCGA LIHC -0.109; TCGA LUAD -0.062; TCGA LUSC -0.108; TCGA PRAD -0.078; TCGA SARC -0.142; TCGA THCA -0.076; TCGA STAD -0.202 |
hsa-miR-17-3p | MAGEH1 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; PAAD; SARC; STAD | MirTarget | TCGA BLCA -0.328; TCGA BRCA -0.058; TCGA CESC -0.598; TCGA COAD -0.336; TCGA ESCA -0.422; TCGA HNSC -0.379; TCGA KIRC -0.152; TCGA KIRP -0.272; TCGA LUAD -0.126; TCGA PAAD -0.44; TCGA SARC -0.101; TCGA STAD -0.494 |
hsa-miR-17-3p | HBEGF | 10 cancers: BLCA; ESCA; KIRC; KIRP; LIHC; LUAD; LUSC; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.268; TCGA ESCA -0.254; TCGA KIRC -0.199; TCGA KIRP -0.423; TCGA LIHC -0.253; TCGA LUAD -0.358; TCGA LUSC -0.325; TCGA PRAD -0.159; TCGA STAD -0.212; TCGA UCEC -0.137 |
hsa-miR-17-3p | EBF1 | 16 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LGG; LUAD; LUSC; OV; PAAD; PRAD; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.71; TCGA BRCA -0.462; TCGA CESC -0.754; TCGA COAD -0.478; TCGA ESCA -0.519; TCGA HNSC -0.355; TCGA KIRP -0.682; TCGA LGG -0.109; TCGA LUAD -0.189; TCGA LUSC -0.257; TCGA OV -0.218; TCGA PAAD -0.399; TCGA PRAD -0.407; TCGA THCA -0.186; TCGA STAD -0.374; TCGA UCEC -0.37 |
hsa-miR-17-3p | FZD4 | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LIHC; LUAD; PAAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.256; TCGA BRCA -0.489; TCGA COAD -0.344; TCGA ESCA -0.419; TCGA HNSC -0.269; TCGA LIHC -0.252; TCGA LUAD -0.104; TCGA PAAD -0.349; TCGA THCA -0.298; TCGA STAD -0.362; TCGA UCEC -0.161 |
hsa-miR-17-3p | MFN2 | 10 cancers: BLCA; BRCA; ESCA; KIRC; KIRP; LIHC; LUSC; PRAD; THCA; STAD | MirTarget | TCGA BLCA -0.085; TCGA BRCA -0.078; TCGA ESCA -0.173; TCGA KIRC -0.216; TCGA KIRP -0.121; TCGA LIHC -0.104; TCGA LUSC -0.088; TCGA PRAD -0.235; TCGA THCA -0.114; TCGA STAD -0.218 |
hsa-miR-17-3p | DPT | 16 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -1.613; TCGA BRCA -0.493; TCGA CESC -1.126; TCGA COAD -1.102; TCGA ESCA -1.561; TCGA HNSC -0.617; TCGA KIRC -0.749; TCGA KIRP -0.764; TCGA LIHC -0.37; TCGA LUAD -0.59; TCGA LUSC -0.588; TCGA OV -0.501; TCGA PAAD -0.681; TCGA PRAD -0.709; TCGA STAD -1.413; TCGA UCEC -0.796 |
hsa-miR-17-3p | TUB | 14 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LUAD; PAAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.452; TCGA CESC -0.352; TCGA COAD -0.578; TCGA ESCA -0.819; TCGA HNSC -0.222; TCGA KIRC -0.401; TCGA LGG -0.154; TCGA LUAD -0.168; TCGA PAAD -0.557; TCGA PRAD -0.147; TCGA SARC -0.294; TCGA THCA -0.18; TCGA STAD -0.762; TCGA UCEC -0.197 |
hsa-miR-17-3p | DCLK1 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; OV; PAAD; PRAD; SARC; STAD | MirTarget; miRNATAP | TCGA BLCA -0.744; TCGA BRCA -0.645; TCGA CESC -0.933; TCGA COAD -0.874; TCGA ESCA -0.959; TCGA HNSC -0.468; TCGA LIHC -0.199; TCGA OV -0.195; TCGA PAAD -0.446; TCGA PRAD -0.537; TCGA SARC -0.203; TCGA STAD -0.862 |
hsa-miR-17-3p | EHD3 | 16 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.112; TCGA BRCA -0.115; TCGA CESC -0.376; TCGA COAD -0.31; TCGA ESCA -0.314; TCGA HNSC -0.237; TCGA KIRC -0.528; TCGA KIRP -0.762; TCGA LIHC -0.208; TCGA LUAD -0.099; TCGA LUSC -0.221; TCGA PAAD -0.355; TCGA PRAD -0.216; TCGA THCA -0.194; TCGA STAD -0.266; TCGA UCEC -0.101 |
hsa-miR-17-3p | FXYD6 | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LIHC; LUAD; PAAD; PRAD | MirTarget | TCGA BLCA -1.004; TCGA CESC -0.621; TCGA COAD -0.779; TCGA ESCA -0.688; TCGA HNSC -0.392; TCGA KIRP -0.646; TCGA LIHC -0.179; TCGA LUAD -0.272; TCGA PAAD -0.631; TCGA PRAD -0.522 |
hsa-miR-17-3p | COL12A1 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; OV; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.285; TCGA BRCA -0.449; TCGA CESC -0.362; TCGA COAD -0.366; TCGA ESCA -0.404; TCGA HNSC -0.53; TCGA KIRC -0.326; TCGA KIRP -0.544; TCGA LIHC -0.356; TCGA LUAD -0.46; TCGA LUSC -0.585; TCGA OV -0.223; TCGA UCEC -0.153 |
hsa-miR-17-3p | SASH1 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LGG; LIHC; PAAD; STAD; UCEC | MirTarget | TCGA BLCA -0.206; TCGA BRCA -0.335; TCGA CESC -0.297; TCGA COAD -0.189; TCGA ESCA -0.333; TCGA HNSC -0.206; TCGA KIRP -0.271; TCGA LGG -0.146; TCGA LIHC -0.154; TCGA PAAD -0.441; TCGA STAD -0.198; TCGA UCEC -0.086 |
hsa-miR-17-3p | NFASC | 15 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; PAAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.631; TCGA CESC -0.392; TCGA COAD -0.521; TCGA ESCA -1.128; TCGA HNSC -0.192; TCGA KIRC -0.693; TCGA KIRP -1.015; TCGA LIHC -0.21; TCGA LUAD -0.325; TCGA PAAD -0.676; TCGA PRAD -0.468; TCGA SARC -0.991; TCGA THCA -0.325; TCGA STAD -0.956; TCGA UCEC -0.641 |
hsa-miR-17-3p | HIPK3 | 11 cancers: BLCA; BRCA; COAD; ESCA; KIRC; KIRP; LIHC; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.315; TCGA BRCA -0.443; TCGA COAD -0.128; TCGA ESCA -0.41; TCGA KIRC -0.576; TCGA KIRP -0.548; TCGA LIHC -0.375; TCGA PRAD -0.532; TCGA SARC -0.256; TCGA STAD -0.205; TCGA UCEC -0.362 |
hsa-miR-17-3p | SCN4A | 9 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; PRAD; SARC; STAD | mirMAP | TCGA BLCA -0.226; TCGA BRCA -0.483; TCGA COAD -0.398; TCGA ESCA -0.436; TCGA HNSC -0.743; TCGA KIRP -0.449; TCGA PRAD -0.508; TCGA SARC -0.282; TCGA STAD -0.452 |
hsa-miR-17-3p | AGPAT4 | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; PAAD; PRAD; STAD | mirMAP | TCGA BLCA -0.195; TCGA CESC -0.245; TCGA COAD -0.537; TCGA ESCA -0.294; TCGA HNSC -0.311; TCGA KIRC -0.142; TCGA LUAD -0.252; TCGA PAAD -0.499; TCGA PRAD -0.439; TCGA STAD -0.239 |
hsa-miR-17-3p | LZTS1 | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; OV; PRAD; THCA; UCEC | mirMAP | TCGA BLCA -0.204; TCGA CESC -0.532; TCGA COAD -0.302; TCGA ESCA -0.242; TCGA HNSC -0.306; TCGA OV -0.2; TCGA PRAD -0.27; TCGA THCA -0.304; TCGA UCEC -0.131 |
hsa-miR-17-3p | NTN1 | 10 cancers: BLCA; COAD; ESCA; KIRC; KIRP; LUAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.599; TCGA COAD -0.682; TCGA ESCA -0.854; TCGA KIRC -0.706; TCGA KIRP -1.038; TCGA LUAD -0.243; TCGA PRAD -0.721; TCGA SARC -0.525; TCGA STAD -1.085; TCGA UCEC -0.318 |
hsa-miR-17-3p | CA12 | 12 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; LUSC; PRAD; THCA; UCEC | mirMAP | TCGA BLCA -0.255; TCGA BRCA -1.013; TCGA CESC -0.362; TCGA ESCA -0.669; TCGA HNSC -0.32; TCGA KIRC -0.58; TCGA KIRP -0.372; TCGA LGG -0.279; TCGA LUSC -0.572; TCGA PRAD -0.477; TCGA THCA -0.512; TCGA UCEC -0.565 |
hsa-miR-17-3p | FOSL2 | 13 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.132; TCGA BRCA -0.082; TCGA COAD -0.113; TCGA ESCA -0.333; TCGA HNSC -0.131; TCGA KIRP -0.307; TCGA LIHC -0.363; TCGA LUAD -0.12; TCGA LUSC -0.136; TCGA PRAD -0.383; TCGA SARC -0.152; TCGA STAD -0.226; TCGA UCEC -0.093 |
hsa-miR-17-3p | PGR | 17 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.827; TCGA BRCA -1.409; TCGA CESC -0.977; TCGA COAD -0.944; TCGA ESCA -1.257; TCGA HNSC -0.429; TCGA KIRC -0.819; TCGA KIRP -1.037; TCGA LGG -0.207; TCGA LIHC -0.625; TCGA LUAD -0.599; TCGA LUSC -0.43; TCGA PAAD -0.919; TCGA PRAD -0.904; TCGA SARC -0.966; TCGA STAD -1.029; TCGA UCEC -0.625 |
hsa-miR-17-3p | SSH1 | 11 cancers: BLCA; BRCA; COAD; ESCA; KIRP; LIHC; LUAD; LUSC; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.133; TCGA BRCA -0.09; TCGA COAD -0.124; TCGA ESCA -0.172; TCGA KIRP -0.112; TCGA LIHC -0.057; TCGA LUAD -0.092; TCGA LUSC -0.109; TCGA PRAD -0.189; TCGA STAD -0.106; TCGA UCEC -0.083 |
hsa-miR-17-3p | RASSF2 | 14 cancers: BLCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; LUAD; OV; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.215; TCGA CESC -0.356; TCGA COAD -0.426; TCGA ESCA -0.401; TCGA HNSC -0.239; TCGA LGG -0.133; TCGA LIHC -0.139; TCGA LUAD -0.321; TCGA OV -0.167; TCGA PAAD -0.332; TCGA PRAD -0.275; TCGA SARC -0.236; TCGA STAD -0.379; TCGA UCEC -0.236 |
hsa-miR-17-3p | LONP2 | 12 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUSC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.11; TCGA BRCA -0.257; TCGA ESCA -0.122; TCGA HNSC -0.08; TCGA KIRC -0.28; TCGA KIRP -0.137; TCGA LGG -0.061; TCGA LIHC -0.228; TCGA LUSC -0.143; TCGA THCA -0.213; TCGA STAD -0.062; TCGA UCEC -0.096 |
hsa-miR-17-3p | SLC1A2 | 10 cancers: BLCA; BRCA; COAD; ESCA; KIRP; LGG; LIHC; LUAD; STAD; UCEC | mirMAP | TCGA BLCA -0.352; TCGA BRCA -0.546; TCGA COAD -0.356; TCGA ESCA -0.591; TCGA KIRP -0.655; TCGA LGG -0.41; TCGA LIHC -0.512; TCGA LUAD -0.226; TCGA STAD -0.276; TCGA UCEC -0.287 |
hsa-miR-17-3p | IL1R1 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; STAD; UCEC | mirMAP | TCGA BLCA -0.457; TCGA BRCA -0.264; TCGA CESC -0.479; TCGA COAD -0.545; TCGA ESCA -0.694; TCGA HNSC -0.462; TCGA KIRC -0.294; TCGA KIRP -0.25; TCGA LIHC -0.391; TCGA LUAD -0.291; TCGA LUSC -0.423; TCGA PAAD -0.357; TCGA STAD -0.347; TCGA UCEC -0.324 |
hsa-miR-17-3p | C1orf21 | 13 cancers: BLCA; BRCA; CESC; COAD; KIRC; KIRP; LGG; LIHC; PAAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.322; TCGA BRCA -0.441; TCGA CESC -0.233; TCGA COAD -0.178; TCGA KIRC -0.25; TCGA KIRP -0.424; TCGA LGG -0.07; TCGA LIHC -0.331; TCGA PAAD -0.319; TCGA SARC -0.288; TCGA THCA -0.218; TCGA STAD -0.258; TCGA UCEC -0.235 |
hsa-miR-17-3p | KCNJ5 | 13 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; STAD | mirMAP | TCGA BLCA -0.364; TCGA BRCA -0.155; TCGA COAD -0.373; TCGA ESCA -0.541; TCGA HNSC -0.31; TCGA KIRC -0.457; TCGA KIRP -0.742; TCGA LIHC -0.279; TCGA LUAD -0.398; TCGA LUSC -0.445; TCGA PAAD -1.037; TCGA PRAD -0.706; TCGA STAD -0.45 |
hsa-miR-17-3p | PDLIM2 | 11 cancers: BLCA; COAD; KIRP; LIHC; LUAD; LUSC; OV; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.319; TCGA COAD -0.189; TCGA KIRP -0.321; TCGA LIHC -0.129; TCGA LUAD -0.406; TCGA LUSC -0.185; TCGA OV -0.176; TCGA PRAD -0.129; TCGA SARC -0.111; TCGA STAD -0.287; TCGA UCEC -0.164 |
hsa-miR-17-3p | WDFY4 | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.577; TCGA CESC -0.468; TCGA COAD -0.389; TCGA ESCA -0.441; TCGA HNSC -0.248; TCGA LUAD -0.247; TCGA LUSC -0.378; TCGA PRAD -0.381; TCGA STAD -0.252; TCGA UCEC -0.265 |
hsa-miR-17-3p | TMEM127 | 16 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; OV; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.123; TCGA BRCA -0.16; TCGA COAD -0.086; TCGA ESCA -0.092; TCGA HNSC -0.09; TCGA KIRC -0.14; TCGA KIRP -0.26; TCGA LIHC -0.142; TCGA LUAD -0.08; TCGA LUSC -0.137; TCGA OV -0.112; TCGA PRAD -0.184; TCGA SARC -0.089; TCGA THCA -0.171; TCGA STAD -0.1; TCGA UCEC -0.078 |
hsa-miR-17-3p | ENTPD1 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; OV; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.39; TCGA BRCA -0.094; TCGA CESC -0.189; TCGA COAD -0.242; TCGA ESCA -0.245; TCGA HNSC -0.096; TCGA KIRP -0.131; TCGA LUAD -0.201; TCGA LUSC -0.159; TCGA OV -0.086; TCGA PAAD -0.282; TCGA PRAD -0.173; TCGA SARC -0.396; TCGA STAD -0.194; TCGA UCEC -0.119 |
hsa-miR-17-3p | KIAA1644 | 16 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -1.285; TCGA BRCA -0.135; TCGA CESC -0.402; TCGA COAD -0.808; TCGA ESCA -1.017; TCGA HNSC -0.645; TCGA KIRP -0.743; TCGA LGG -0.248; TCGA LIHC -0.382; TCGA LUAD -0.367; TCGA LUSC -0.322; TCGA OV -0.36; TCGA PAAD -1.065; TCGA PRAD -0.914; TCGA STAD -0.987; TCGA UCEC -0.71 |
hsa-miR-17-3p | FTO | 13 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.203; TCGA BRCA -0.211; TCGA COAD -0.115; TCGA ESCA -0.154; TCGA HNSC -0.128; TCGA LIHC -0.183; TCGA LUAD -0.112; TCGA LUSC -0.099; TCGA PAAD -0.146; TCGA PRAD -0.227; TCGA THCA -0.18; TCGA STAD -0.142; TCGA UCEC -0.193 |
hsa-miR-17-3p | NFIC | 14 cancers: BLCA; BRCA; COAD; ESCA; KIRC; KIRP; LIHC; LUAD; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.28; TCGA BRCA -0.274; TCGA COAD -0.249; TCGA ESCA -0.44; TCGA KIRC -0.22; TCGA KIRP -0.464; TCGA LIHC -0.265; TCGA LUAD -0.106; TCGA PAAD -0.438; TCGA PRAD -0.275; TCGA SARC -0.259; TCGA THCA -0.321; TCGA STAD -0.393; TCGA UCEC -0.478 |
hsa-miR-17-3p | DST | 12 cancers: BLCA; BRCA; COAD; ESCA; KIRC; KIRP; LUAD; LUSC; PRAD; SARC; STAD; UCEC | mirMAP; miRNATAP | TCGA BLCA -0.309; TCGA BRCA -0.332; TCGA COAD -0.256; TCGA ESCA -0.427; TCGA KIRC -0.27; TCGA KIRP -0.171; TCGA LUAD -0.333; TCGA LUSC -0.256; TCGA PRAD -0.549; TCGA SARC -0.264; TCGA STAD -0.442; TCGA UCEC -0.302 |
hsa-miR-17-3p | PLXDC1 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.215; TCGA BRCA -0.225; TCGA CESC -0.456; TCGA COAD -0.198; TCGA ESCA -0.278; TCGA HNSC -0.376; TCGA KIRP -0.257; TCGA LUAD -0.225; TCGA LUSC -0.276; TCGA PAAD -0.258; TCGA PRAD -0.12; TCGA STAD -0.178; TCGA UCEC -0.362 |
hsa-miR-17-3p | ZYG11B | 9 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; KIRP; LIHC; PRAD; STAD | mirMAP | TCGA BLCA -0.075; TCGA BRCA -0.161; TCGA ESCA -0.183; TCGA HNSC -0.094; TCGA KIRC -0.278; TCGA KIRP -0.172; TCGA LIHC -0.224; TCGA PRAD -0.221; TCGA STAD -0.067 |
hsa-miR-17-3p | UBXN10 | 10 cancers: BLCA; BRCA; HNSC; KIRP; LIHC; LUAD; LUSC; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.291; TCGA BRCA -0.754; TCGA HNSC -0.357; TCGA KIRP -0.282; TCGA LIHC -0.591; TCGA LUAD -0.22; TCGA LUSC -0.361; TCGA PRAD -0.992; TCGA STAD -0.417; TCGA UCEC -0.207 |
hsa-miR-17-3p | DUSP19 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; KIRP; LIHC; LUAD; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.336; TCGA BRCA -0.199; TCGA CESC -0.192; TCGA COAD -0.201; TCGA ESCA -0.433; TCGA KIRC -0.238; TCGA KIRP -0.112; TCGA LIHC -0.128; TCGA LUAD -0.142; TCGA PAAD -0.381; TCGA PRAD -0.415; TCGA SARC -0.338; TCGA THCA -0.213; TCGA STAD -0.227; TCGA UCEC -0.139 |
hsa-miR-17-3p | GALNT10 | 15 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; OV; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.217; TCGA BRCA -0.527; TCGA COAD -0.093; TCGA ESCA -0.257; TCGA HNSC -0.292; TCGA KIRC -0.327; TCGA KIRP -0.3; TCGA LIHC -0.163; TCGA LUAD -0.266; TCGA LUSC -0.335; TCGA OV -0.154; TCGA PRAD -0.135; TCGA THCA -0.377; TCGA STAD -0.175; TCGA UCEC -0.18 |
hsa-miR-17-3p | SCN3B | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.613; TCGA BRCA -0.421; TCGA CESC -0.54; TCGA COAD -0.645; TCGA ESCA -0.753; TCGA HNSC -0.523; TCGA KIRC -0.317; TCGA KIRP -0.3; TCGA LGG -0.305; TCGA LUAD -0.346; TCGA PAAD -0.771; TCGA PRAD -0.506; TCGA STAD -0.563; TCGA UCEC -0.38 |
hsa-miR-17-3p | SLFN5 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; LIHC; LUAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.16; TCGA BRCA -0.189; TCGA CESC -0.27; TCGA COAD -0.291; TCGA ESCA -0.218; TCGA LIHC -0.392; TCGA LUAD -0.092; TCGA PRAD -0.529; TCGA SARC -0.257; TCGA STAD -0.14; TCGA UCEC -0.136 |
hsa-miR-17-3p | SAMD14 | 9 cancers: BLCA; CESC; HNSC; LGG; LUAD; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.355; TCGA CESC -0.274; TCGA HNSC -0.299; TCGA LGG -0.086; TCGA LUAD -0.268; TCGA PRAD -0.485; TCGA THCA -0.347; TCGA STAD -0.333; TCGA UCEC -0.136 |
hsa-miR-17-3p | PLEKHA2 | 9 cancers: BLCA; CESC; ESCA; KIRP; LIHC; LUAD; LUSC; PRAD; SARC | mirMAP | TCGA BLCA -0.185; TCGA CESC -0.165; TCGA ESCA -0.113; TCGA KIRP -0.164; TCGA LIHC -0.102; TCGA LUAD -0.193; TCGA LUSC -0.163; TCGA PRAD -0.58; TCGA SARC -0.242 |
hsa-miR-17-3p | KCNK3 | 13 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LGG; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.911; TCGA CESC -0.374; TCGA COAD -1.003; TCGA ESCA -0.92; TCGA HNSC -0.471; TCGA KIRP -0.512; TCGA LGG -0.261; TCGA LUSC -0.377; TCGA PAAD -0.832; TCGA PRAD -0.964; TCGA SARC -1.149; TCGA STAD -1.391; TCGA UCEC -0.467 |
hsa-miR-17-3p | PRND | 9 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUAD; LUSC; STAD; UCEC | mirMAP | TCGA BLCA -0.82; TCGA BRCA -0.641; TCGA COAD -0.883; TCGA ESCA -0.602; TCGA HNSC -0.388; TCGA LUAD -0.562; TCGA LUSC -0.33; TCGA STAD -0.481; TCGA UCEC -0.723 |
hsa-miR-17-3p | LRRC15 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; OV; PRAD; UCEC | mirMAP | TCGA BLCA -0.753; TCGA BRCA -0.353; TCGA CESC -0.623; TCGA COAD -0.617; TCGA ESCA -0.808; TCGA HNSC -0.897; TCGA KIRC -0.451; TCGA KIRP -0.591; TCGA LIHC -0.449; TCGA LUAD -0.543; TCGA LUSC -0.692; TCGA OV -0.491; TCGA PRAD -0.556; TCGA UCEC -0.342 |
hsa-miR-17-3p | IL16 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.415; TCGA BRCA -0.155; TCGA CESC -0.345; TCGA COAD -0.273; TCGA ESCA -0.497; TCGA HNSC -0.207; TCGA LUAD -0.382; TCGA LUSC -0.362; TCGA PRAD -0.222; TCGA SARC -0.249; TCGA STAD -0.449; TCGA UCEC -0.271 |
hsa-miR-17-3p | SH3RF3 | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.569; TCGA BRCA -0.098; TCGA COAD -0.199; TCGA ESCA -0.4; TCGA HNSC -0.48; TCGA LIHC -0.576; TCGA LUAD -0.17; TCGA LUSC -0.22; TCGA THCA -0.206; TCGA STAD -0.3; TCGA UCEC -0.223 |
hsa-miR-17-3p | GNG7 | 15 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; OV; PAAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.684; TCGA CESC -0.655; TCGA COAD -0.458; TCGA ESCA -0.78; TCGA HNSC -0.307; TCGA KIRC -0.362; TCGA KIRP -0.404; TCGA LGG -0.087; TCGA LUAD -0.375; TCGA LUSC -0.257; TCGA OV -0.233; TCGA PAAD -0.623; TCGA SARC -0.212; TCGA STAD -0.877; TCGA UCEC -0.371 |
hsa-miR-17-3p | RNF152 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.224; TCGA BRCA -0.141; TCGA CESC -0.326; TCGA COAD -0.312; TCGA ESCA -0.621; TCGA HNSC -0.365; TCGA KIRC -0.743; TCGA LIHC -0.134; TCGA LUAD -0.137; TCGA PAAD -0.365; TCGA PRAD -0.612; TCGA SARC -0.164; TCGA STAD -0.449; TCGA UCEC -0.139 |
hsa-miR-17-3p | MYO1D | 9 cancers: BLCA; BRCA; HNSC; KIRC; KIRP; LUSC; SARC; THCA; UCEC | mirMAP; miRNATAP | TCGA BLCA -0.098; TCGA BRCA -0.088; TCGA HNSC -0.129; TCGA KIRC -0.337; TCGA KIRP -0.23; TCGA LUSC -0.096; TCGA SARC -0.219; TCGA THCA -0.136; TCGA UCEC -0.089 |
hsa-miR-17-3p | ADAMTSL1 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.719; TCGA BRCA -0.341; TCGA CESC -0.585; TCGA COAD -0.507; TCGA ESCA -1.157; TCGA HNSC -0.36; TCGA KIRC -0.566; TCGA KIRP -1.053; TCGA LIHC -0.331; TCGA LUAD -0.388; TCGA LUSC -0.259; TCGA PAAD -0.539; TCGA PRAD -0.292; TCGA STAD -0.727; TCGA UCEC -0.548 |
hsa-miR-17-3p | LYNX1 | 13 cancers: BLCA; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.555; TCGA COAD -0.658; TCGA ESCA -0.7; TCGA HNSC -0.547; TCGA KIRC -0.45; TCGA KIRP -0.848; TCGA LGG -0.21; TCGA LUAD -0.441; TCGA LUSC -0.338; TCGA PRAD -0.357; TCGA SARC -0.673; TCGA STAD -0.921; TCGA UCEC -0.401 |
hsa-miR-17-3p | OLFML2A | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.301; TCGA BRCA -0.259; TCGA COAD -0.319; TCGA ESCA -0.305; TCGA HNSC -0.216; TCGA KIRP -0.322; TCGA LUAD -0.173; TCGA LUSC -0.342; TCGA PAAD -0.275; TCGA PRAD -0.544; TCGA STAD -0.275; TCGA UCEC -0.371 |
hsa-miR-17-3p | PIP5K1C | 13 cancers: BLCA; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; OV; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.172; TCGA COAD -0.17; TCGA ESCA -0.371; TCGA HNSC -0.081; TCGA KIRP -0.139; TCGA LUAD -0.124; TCGA LUSC -0.082; TCGA OV -0.087; TCGA PRAD -0.129; TCGA SARC -0.189; TCGA THCA -0.194; TCGA STAD -0.348; TCGA UCEC -0.238 |
hsa-miR-17-3p | MAPT | 10 cancers: BLCA; BRCA; COAD; ESCA; LUAD; PAAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.249; TCGA BRCA -0.907; TCGA COAD -0.795; TCGA ESCA -0.759; TCGA LUAD -0.244; TCGA PAAD -0.563; TCGA SARC -0.414; TCGA THCA -0.518; TCGA STAD -0.936; TCGA UCEC -0.241 |
hsa-miR-17-3p | C15orf52 | 11 cancers: BLCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.449; TCGA CESC -0.312; TCGA COAD -0.236; TCGA ESCA -0.38; TCGA HNSC -0.255; TCGA LIHC -0.159; TCGA LUAD -0.133; TCGA PRAD -0.183; TCGA SARC -0.881; TCGA STAD -0.573; TCGA UCEC -0.155 |
hsa-miR-17-3p | PTPRT | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; LGG; LUAD; PAAD; SARC; STAD | mirMAP | TCGA BLCA -0.38; TCGA BRCA -1.321; TCGA CESC -0.762; TCGA COAD -0.323; TCGA ESCA -0.726; TCGA KIRC -0.984; TCGA LGG -0.334; TCGA LUAD -0.566; TCGA PAAD -0.883; TCGA SARC -0.604; TCGA STAD -0.71 |
hsa-miR-17-3p | TECPR2 | 17 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUSC; OV; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.172; TCGA BRCA -0.16; TCGA CESC -0.09; TCGA COAD -0.164; TCGA ESCA -0.292; TCGA HNSC -0.134; TCGA KIRC -0.085; TCGA KIRP -0.087; TCGA LIHC -0.159; TCGA LUSC -0.143; TCGA OV -0.083; TCGA PAAD -0.28; TCGA PRAD -0.145; TCGA SARC -0.149; TCGA THCA -0.127; TCGA STAD -0.319; TCGA UCEC -0.177 |
hsa-miR-17-3p | SPN | 9 cancers: BLCA; HNSC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.275; TCGA HNSC -0.22; TCGA LUAD -0.52; TCGA LUSC -0.26; TCGA PAAD -0.495; TCGA PRAD -0.315; TCGA SARC -0.224; TCGA STAD -0.203; TCGA UCEC -0.253 |
hsa-miR-17-3p | MYO5A | 14 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; THCA; STAD | mirMAP | TCGA BLCA -0.187; TCGA BRCA -0.146; TCGA COAD -0.38; TCGA ESCA -0.589; TCGA HNSC -0.232; TCGA KIRC -0.192; TCGA KIRP -0.357; TCGA LIHC -0.126; TCGA LUAD -0.243; TCGA LUSC -0.211; TCGA PAAD -0.506; TCGA PRAD -0.169; TCGA THCA -0.133; TCGA STAD -0.208 |
hsa-miR-17-3p | NTRK2 | 16 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; KIRP; LGG; LIHC; LUAD; OV; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.986; TCGA BRCA -0.451; TCGA CESC -1.09; TCGA COAD -0.977; TCGA ESCA -0.694; TCGA KIRC -0.897; TCGA KIRP -1.058; TCGA LGG -0.267; TCGA LIHC -0.453; TCGA LUAD -0.462; TCGA OV -0.293; TCGA PAAD -0.661; TCGA PRAD -0.789; TCGA SARC -0.613; TCGA STAD -0.544; TCGA UCEC -0.55 |
hsa-miR-17-3p | RGAG4 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; KIRP; LGG; LIHC; PAAD; PRAD; STAD | miRNATAP | TCGA BLCA -0.675; TCGA BRCA -0.375; TCGA CESC -0.46; TCGA COAD -0.516; TCGA ESCA -0.585; TCGA KIRC -0.367; TCGA KIRP -0.506; TCGA LGG -0.095; TCGA LIHC -0.34; TCGA PAAD -0.667; TCGA PRAD -0.438; TCGA STAD -0.765 |
hsa-miR-17-3p | AKT3 | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; PAAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.394; TCGA BRCA -0.105; TCGA COAD -0.52; TCGA ESCA -0.521; TCGA HNSC -0.168; TCGA KIRC -0.202; TCGA LIHC -0.35; TCGA LUAD -0.151; TCGA PAAD -0.519; TCGA SARC -0.208; TCGA STAD -0.478; TCGA UCEC -0.464 |
hsa-miR-17-3p | CAMK2G | 10 cancers: BLCA; KIRC; KIRP; LGG; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.18; TCGA KIRC -0.168; TCGA KIRP -0.115; TCGA LGG -0.189; TCGA PAAD -0.181; TCGA PRAD -0.283; TCGA SARC -0.353; TCGA THCA -0.114; TCGA STAD -0.303; TCGA UCEC -0.05 |
hsa-miR-17-3p | RORA | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.295; TCGA BRCA -0.226; TCGA CESC -0.433; TCGA COAD -0.383; TCGA ESCA -0.729; TCGA HNSC -0.392; TCGA LGG -0.098; TCGA LIHC -0.325; TCGA LUAD -0.168; TCGA LUSC -0.171; TCGA PAAD -0.367; TCGA PRAD -0.269; TCGA STAD -0.304; TCGA UCEC -0.236 |
hsa-miR-17-3p | GFRA1 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -1.192; TCGA BRCA -1.249; TCGA CESC -1.02; TCGA COAD -0.927; TCGA ESCA -1.406; TCGA HNSC -0.568; TCGA KIRC -0.738; TCGA KIRP -0.585; TCGA LIHC -0.395; TCGA LUAD -0.562; TCGA LUSC -0.3; TCGA PAAD -0.687; TCGA PRAD -0.599; TCGA STAD -1.574; TCGA UCEC -0.223 |
hsa-miR-17-3p | ATXN1L | 11 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; KIRP; LIHC; PRAD; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.086; TCGA BRCA -0.138; TCGA ESCA -0.17; TCGA HNSC -0.13; TCGA KIRC -0.294; TCGA KIRP -0.144; TCGA LIHC -0.184; TCGA PRAD -0.222; TCGA THCA -0.128; TCGA STAD -0.15; TCGA UCEC -0.109 |
hsa-miR-17-3p | KCNMA1 | 16 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -1.003; TCGA BRCA -0.655; TCGA CESC -0.683; TCGA COAD -1.146; TCGA ESCA -1.689; TCGA HNSC -0.462; TCGA LGG -0.132; TCGA LIHC -0.318; TCGA LUAD -0.201; TCGA LUSC -0.415; TCGA PAAD -0.78; TCGA PRAD -0.749; TCGA SARC -0.557; TCGA THCA -0.277; TCGA STAD -1.351; TCGA UCEC -0.534 |
hsa-miR-17-3p | USP9X | 9 cancers: BLCA; BRCA; HNSC; KIRC; KIRP; LIHC; OV; PRAD; THCA | miRNATAP | TCGA BLCA -0.098; TCGA BRCA -0.167; TCGA HNSC -0.131; TCGA KIRC -0.279; TCGA KIRP -0.193; TCGA LIHC -0.094; TCGA OV -0.086; TCGA PRAD -0.207; TCGA THCA -0.127 |
hsa-miR-17-3p | SLC2A10 | 12 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; STAD; UCEC | miRNATAP | TCGA BLCA -0.23; TCGA BRCA -0.457; TCGA CESC -0.43; TCGA COAD -0.331; TCGA HNSC -0.357; TCGA KIRC -0.369; TCGA KIRP -0.33; TCGA LIHC -0.18; TCGA LUAD -0.363; TCGA LUSC -0.322; TCGA STAD -0.262; TCGA UCEC -0.189 |
hsa-miR-17-3p | WNK1 | 9 cancers: BLCA; BRCA; CESC; ESCA; KIRC; KIRP; PRAD; THCA; UCEC | miRNATAP | TCGA BLCA -0.129; TCGA BRCA -0.091; TCGA CESC -0.133; TCGA ESCA -0.228; TCGA KIRC -0.615; TCGA KIRP -0.571; TCGA PRAD -0.11; TCGA THCA -0.11; TCGA UCEC -0.064 |
hsa-miR-17-3p | NR3C1 | 10 cancers: BLCA; BRCA; COAD; ESCA; LIHC; PAAD; PRAD; SARC; THCA; STAD | miRNATAP | TCGA BLCA -0.32; TCGA BRCA -0.296; TCGA COAD -0.462; TCGA ESCA -0.514; TCGA LIHC -0.16; TCGA PAAD -0.399; TCGA PRAD -0.55; TCGA SARC -0.099; TCGA THCA -0.106; TCGA STAD -0.43 |
hsa-miR-17-3p | CLIP3 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.806; TCGA BRCA -0.159; TCGA CESC -0.627; TCGA COAD -0.695; TCGA ESCA -0.842; TCGA HNSC -0.458; TCGA KIRC -0.233; TCGA KIRP -0.45; TCGA LUAD -0.244; TCGA PAAD -0.701; TCGA PRAD -0.592; TCGA SARC -0.237; TCGA STAD -0.895; TCGA UCEC -0.363 |
hsa-miR-17-3p | OLFM1 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; LGG; LIHC; LUAD; OV; PAAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.798; TCGA BRCA -0.412; TCGA CESC -0.627; TCGA COAD -0.689; TCGA ESCA -0.513; TCGA LGG -0.192; TCGA LIHC -0.361; TCGA LUAD -0.462; TCGA OV -0.413; TCGA PAAD -0.683; TCGA SARC -0.419; TCGA STAD -0.741; TCGA UCEC -0.496 |
hsa-miR-17-3p | SHISA9 | 9 cancers: BRCA; ESCA; LGG; LIHC; OV; PAAD; PRAD; THCA; STAD | MirTarget; miRNATAP | TCGA BRCA -0.462; TCGA ESCA -0.919; TCGA LGG -0.305; TCGA LIHC -0.38; TCGA OV -0.37; TCGA PAAD -0.693; TCGA PRAD -0.732; TCGA THCA -0.612; TCGA STAD -1.073 |
hsa-miR-17-3p | TOB2 | 10 cancers: BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LUSC; SARC; STAD; UCEC | MirTarget | TCGA BRCA -0.126; TCGA COAD -0.064; TCGA ESCA -0.181; TCGA HNSC -0.081; TCGA KIRC -0.118; TCGA KIRP -0.195; TCGA LUSC -0.096; TCGA SARC -0.075; TCGA STAD -0.137; TCGA UCEC -0.083 |
hsa-miR-17-3p | SLC9A7 | 9 cancers: BRCA; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; OV; THCA | miRNATAP | TCGA BRCA -0.19; TCGA ESCA -0.259; TCGA HNSC -0.156; TCGA KIRC -0.394; TCGA KIRP -0.568; TCGA LIHC -0.34; TCGA LUAD -0.216; TCGA OV -0.111; TCGA THCA -0.324 |
hsa-miR-17-3p | EPB41L1 | 9 cancers: BRCA; CESC; KIRC; LUAD; LUSC; PRAD; SARC; THCA; STAD | miRNATAP | TCGA BRCA -0.158; TCGA CESC -0.334; TCGA KIRC -0.364; TCGA LUAD -0.075; TCGA LUSC -0.171; TCGA PRAD -0.195; TCGA SARC -0.249; TCGA THCA -0.136; TCGA STAD -0.134 |
hsa-miR-17-3p | TANC2 | 11 cancers: BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUSC; PRAD; THCA; STAD | miRNATAP | TCGA BRCA -0.307; TCGA COAD -0.483; TCGA ESCA -0.281; TCGA HNSC -0.226; TCGA KIRC -0.159; TCGA KIRP -0.282; TCGA LIHC -0.249; TCGA LUSC -0.107; TCGA PRAD -0.101; TCGA THCA -0.165; TCGA STAD -0.136 |
hsa-miR-17-3p | TTC9 | 9 cancers: BRCA; CESC; COAD; KIRC; LIHC; LUAD; LUSC; OV; UCEC | miRNATAP | TCGA BRCA -0.17; TCGA CESC -0.382; TCGA COAD -0.263; TCGA KIRC -0.226; TCGA LIHC -0.797; TCGA LUAD -0.193; TCGA LUSC -0.428; TCGA OV -0.234; TCGA UCEC -0.131 |
hsa-miR-17-3p | ASTN1 | 12 cancers: CESC; COAD; ESCA; KIRC; LGG; LIHC; LUAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA CESC -0.929; TCGA COAD -0.969; TCGA ESCA -1.5; TCGA KIRC -0.825; TCGA LGG -0.098; TCGA LIHC -0.28; TCGA LUAD -0.268; TCGA PRAD -0.704; TCGA SARC -0.465; TCGA THCA -0.622; TCGA STAD -1.292; TCGA UCEC -0.367 |
hsa-miR-17-3p | STOX2 | 10 cancers: CESC; COAD; ESCA; KIRC; LIHC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA CESC -0.306; TCGA COAD -0.394; TCGA ESCA -0.547; TCGA KIRC -0.171; TCGA LIHC -0.271; TCGA PAAD -0.545; TCGA PRAD -0.869; TCGA SARC -0.424; TCGA STAD -0.708; TCGA UCEC -0.159 |
hsa-miR-17-3p | NOS1 | 9 cancers: COAD; ESCA; HNSC; KIRC; KIRP; OV; PAAD; PRAD; STAD | mirMAP | TCGA COAD -0.738; TCGA ESCA -1.339; TCGA HNSC -0.399; TCGA KIRC -2.095; TCGA KIRP -1.578; TCGA OV -0.368; TCGA PAAD -0.994; TCGA PRAD -0.986; TCGA STAD -1.322 |
hsa-miR-17-3p | IGSF3 | 9 cancers: ESCA; HNSC; KIRC; KIRP; LGG; LUSC; PRAD; SARC; STAD | mirMAP | TCGA ESCA -0.246; TCGA HNSC -0.121; TCGA KIRC -0.498; TCGA KIRP -0.288; TCGA LGG -0.225; TCGA LUSC -0.211; TCGA PRAD -0.22; TCGA SARC -0.28; TCGA STAD -0.212 |
Enriched cancer pathways of putative targets