microRNA information: hsa-miR-181a-5p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-181a-5p | miRbase |
| Accession: | MIMAT0000256 | miRbase |
| Precursor name: | hsa-mir-181a-1 | miRbase |
| Precursor accession: | MI0000289 | miRbase |
| Symbol: | NA | HGNC |
| RefSeq ID: | NA | GenBank |
| Sequence: | AACAUUCAACGCUGUCGGUGAGU |
Reported expression in cancers: hsa-miR-181a-5p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-181a-5p | acute myeloid leukemia | upregulation | "This paper evaluated the association between micro ......" | 25646775 | RNA-Seq |
| hsa-miR-181a-5p | acute myeloid leukemia | upregulation | "Targeting the RAS/MAPK pathway with miR 181a in ac ......" | 27517749 | |
| hsa-miR-181a-5p | breast cancer | downregulation | "Increased expression of three of the most promisin ......" | 18708351 | Reverse transcription PCR; qPCR |
| hsa-miR-181a-5p | breast cancer | upregulation | "This was mediated by the micro miRNA family miR-18 ......" | 21102523 | |
| hsa-miR-181a-5p | breast cancer | upregulation | "TGF β upregulates miR 181a expression to promote ......" | 23241956 | |
| hsa-miR-181a-5p | breast cancer | downregulation | "RNA was extracted reverse transcribed and subjecte ......" | 24498016 | Microarray |
| hsa-miR-181a-5p | breast cancer | deregulation | "In this study we conducted miRNAs profile comparis ......" | 24788655 | Microarray; qPCR |
| hsa-miR-181a-5p | breast cancer | deregulation | "The expression profiles of miR-9 miR-21 miR-30a mi ......" | 25013435 | Reverse transcription PCR |
| hsa-miR-181a-5p | cervical and endocervical cancer | downregulation | "miR-181a has been reported to participate in tumor ......" | 27534652 | |
| hsa-miR-181a-5p | colorectal cancer | upregulation | "Recent studies have shown that miR-181a is dysregu ......" | 23023298 | qPCR |
| hsa-miR-181a-5p | gastric cancer | upregulation | "miRNA expression was determined using a custom-des ......" | 22112324 | Microarray |
| hsa-miR-181a-5p | gastric cancer | upregulation | "Genetic polymorphism at miR 181a binding site cont ......" | 22971574 | Reverse transcription PCR; Microarray |
| hsa-miR-181a-5p | gastric cancer | upregulation | "Hsa miR 181a 5p expression and effects on cell pro ......" | 23886199 | qPCR |
| hsa-miR-181a-5p | glioblastoma | deregulation | "In our study we examined by microarray the global ......" | 16039986 | Microarray |
| hsa-miR-181a-5p | head and neck cancer | deregulation | "A global miRNA profiling was performed on 12 sampl ......" | 21637912 | qPCR; Microarray |
| hsa-miR-181a-5p | liver cancer | upregulation | "Here using a global microarray-based miRNA profili ......" | 19585654 | Reverse transcription PCR; Microarray |
| hsa-miR-181a-5p | liver cancer | upregulation | "We demonstrated that conserved let-7 and miR-181 f ......" | 21352471 | qPCR; Microarray |
| hsa-miR-181a-5p | liver cancer | upregulation | "Up-regulation of miR-15a miR-16 miR-27b miR-30b mi ......" | 24314246 | qPCR |
| hsa-miR-181a-5p | liver cancer | upregulation | "Recent studies have found that miR-181a were dysre ......" | 24529171 | qPCR |
| hsa-miR-181a-5p | liver cancer | upregulation | "miR 181a mediates TGF β induced hepatocyte EMT an ......" | 24576072 | |
| hsa-miR-181a-5p | lung squamous cell cancer | deregulation | "Locked nucleic acids miRNA microarray expression p ......" | 20508945 | Microarray |
| hsa-miR-181a-5p | lung squamous cell cancer | deregulation | "In this study we analyzed the global expression pr ......" | 25524579 | |
| hsa-miR-181a-5p | lymphoma | upregulation | "The major findings were: the detection of a panel ......" | 19945163 | |
| hsa-miR-181a-5p | pancreatic cancer | downregulation | "The in vivo effect of miR-181a down-regulation on ......" | 26152285 | |
| hsa-miR-181a-5p | prostate cancer | upregulation | "In the present study the expression of miR-181 in ......" | 25187843 | |
| hsa-miR-181a-5p | sarcoma | downregulation | "Here we profiled miRNA expression of chondrosarcom ......" | 23940002 | qPCR; Microarray |
| hsa-miR-181a-5p | sarcoma | upregulation | "The expression of miR 181a 5p and miR 371b 5p in c ......" | 26214773 |
Reported cancer pathway affected by hsa-miR-181a-5p
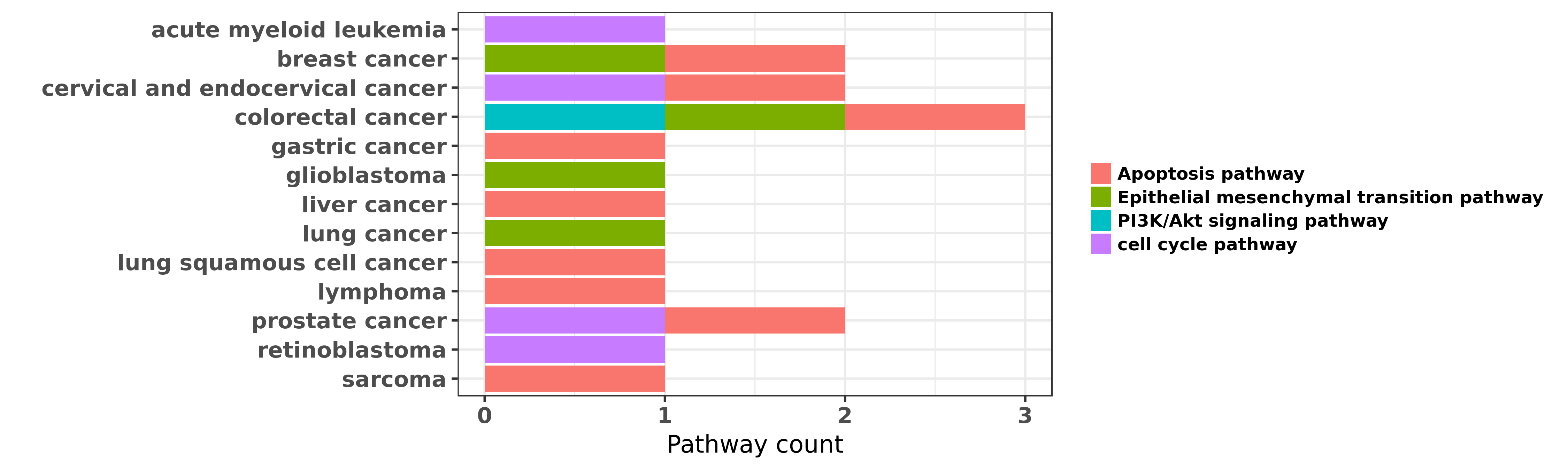
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-181a-5p | acute myeloid leukemia | cell cycle pathway | "miR 181a promotes G1/S transition and cell prolife ......" | 26113450 | Flow cytometry; Luciferase |
| hsa-miR-181a-5p | breast cancer | Epithelial mesenchymal transition pathway | "TGF β upregulates miR 181a expression to promote ......" | 23241956 | |
| hsa-miR-181a-5p | breast cancer | Apoptosis pathway | "The function role of miR 181a in chemosensitivity ......" | 24335172 | Western blot; Luciferase |
| hsa-miR-181a-5p | breast cancer | Apoptosis pathway | "Here we show that genotoxic treatments significant ......" | 26028030 | |
| hsa-miR-181a-5p | cervical and endocervical cancer | Apoptosis pathway | "MiR 181a confers resistance of cervical cancer to ......" | 22847611 | |
| hsa-miR-181a-5p | cervical and endocervical cancer | cell cycle pathway; Apoptosis pathway | "miR-181a has been reported to participate in tumor ......" | 27534652 | Western blot |
| hsa-miR-181a-5p | colorectal cancer | Epithelial mesenchymal transition pathway | "Expression profile was further assessed using quan ......" | 24755295 | Luciferase |
| hsa-miR-181a-5p | colorectal cancer | Apoptosis pathway; PI3K/Akt signaling pathway | "The emerging role of microRNA-181A miR-181A in CRC ......" | 26750720 | |
| hsa-miR-181a-5p | gastric cancer | Apoptosis pathway | "In order to identify miRNA signatures for gastric ......" | 22112324 | |
| hsa-miR-181a-5p | gastric cancer | Apoptosis pathway | "However it remains unknown whether miR-181a is inv ......" | 22581522 | Colony formation; Luciferase; Western blot |
| hsa-miR-181a-5p | gastric cancer | Apoptosis pathway | "Based on our previous experiments this study is to ......" | 24531888 | Western blot; Luciferase |
| hsa-miR-181a-5p | glioblastoma | Epithelial mesenchymal transition pathway | "We annotated 491 TCGA samples' miRNA expression pr ......" | 26283154 | |
| hsa-miR-181a-5p | liver cancer | Apoptosis pathway | "Functional analysis of miR 181a and Fas involved i ......" | 25449696 | |
| hsa-miR-181a-5p | lung cancer | Epithelial mesenchymal transition pathway | "The metastatic properties of the cells were assess ......" | 26323677 | Western blot; Cell migration assay; Wound Healing Assay; Luciferase |
| hsa-miR-181a-5p | lung squamous cell cancer | Apoptosis pathway | "In this study we analyzed the global expression pr ......" | 25524579 | |
| hsa-miR-181a-5p | lymphoma | Apoptosis pathway | "Bim down-regulation is posttranscriptionally regul ......" | 20841506 | |
| hsa-miR-181a-5p | prostate cancer | Apoptosis pathway | "MiR 181a contributes to bufalin induced apoptosis ......" | 24267199 | Western blot |
| hsa-miR-181a-5p | prostate cancer | cell cycle pathway | "In the present study the expression of miR-181 in ......" | 25187843 | |
| hsa-miR-181a-5p | retinoblastoma | cell cycle pathway | "Among them hsa-miR-373 hsa-miR-125b and hsa-miR-18 ......" | 26730174 | |
| hsa-miR-181a-5p | sarcoma | Apoptosis pathway | "MicroRNA 181a miR-181a was found dysregulated in a ......" | 23740615 | Western blot |
Reported cancer prognosis affected by hsa-miR-181a-5p
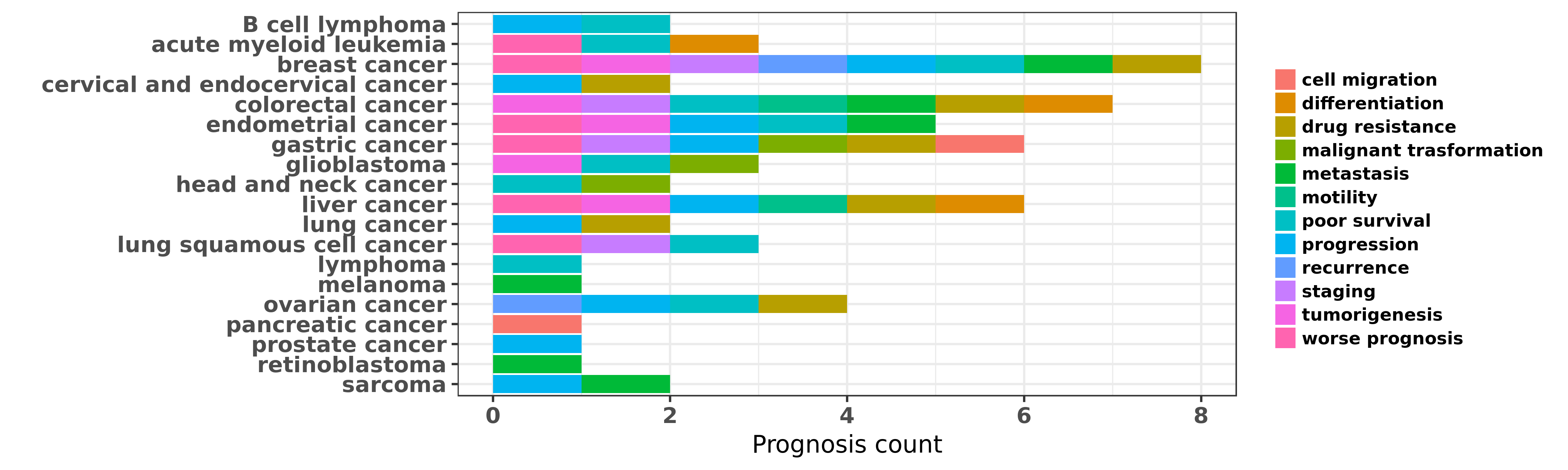
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-181a-5p | B cell lymphoma | poor survival; progression | "We measured the expression of each miRNA by quanti ......" | 21525173 | |
| hsa-miR-181a-5p | acute myeloid leukemia | differentiation | "The major features of these profiles were upregula ......" | 18809607 | |
| hsa-miR-181a-5p | acute myeloid leukemia | poor survival | "Prognostic significance of expression of a single ......" | 21079133 | |
| hsa-miR-181a-5p | acute myeloid leukemia | worse prognosis | "Recently we showed that increased miR-181a express ......" | 23100311 | |
| hsa-miR-181a-5p | acute myeloid leukemia | differentiation | "MiR 181 family: regulators of myeloid differentiat ......" | 25174404 | |
| hsa-miR-181a-5p | acute myeloid leukemia | worse prognosis; differentiation; poor survival | "The pathological role and prognostic impact of miR ......" | 25686674 | |
| hsa-miR-181a-5p | breast cancer | recurrence; poor survival; worse prognosis | "MicroRNA microarray analysis was performed to comp ......" | 21271219 | |
| hsa-miR-181a-5p | breast cancer | staging | "Decreased serum miR 181a is a potential new tool f ......" | 22692639 | |
| hsa-miR-181a-5p | breast cancer | metastasis; staging; poor survival | "TGF β upregulates miR 181a expression to promote ......" | 23241956 | |
| hsa-miR-181a-5p | breast cancer | progression | "MicroRNA-181 miR-181 is a multifaceted miRNA that ......" | 23524334 | |
| hsa-miR-181a-5p | breast cancer | drug resistance | "MiR 181a enhances drug sensitivity in mitoxantone ......" | 23780685 | Luciferase |
| hsa-miR-181a-5p | breast cancer | tumorigenesis | "The function role of miR 181a in chemosensitivity ......" | 24335172 | Western blot; Luciferase |
| hsa-miR-181a-5p | breast cancer | metastasis | "The expression of miRNAs in patients with primary ......" | 24846313 | |
| hsa-miR-181a-5p | breast cancer | metastasis; poor survival; drug resistance | "Here we show that genotoxic treatments significant ......" | 26028030 | |
| hsa-miR-181a-5p | breast cancer | poor survival | "In this multicenter study a 10-miRNA classifier in ......" | 27566954 | |
| hsa-miR-181a-5p | cervical and endocervical cancer | drug resistance | "MiR 181a confers resistance of cervical cancer to ......" | 22847611 | |
| hsa-miR-181a-5p | cervical and endocervical cancer | progression | "miR-181a has been reported to participate in tumor ......" | 27534652 | Western blot |
| hsa-miR-181a-5p | colorectal cancer | poor survival; drug resistance; differentiation | "miR 181a is associated with poor clinical outcome ......" | 24098024 | |
| hsa-miR-181a-5p | colorectal cancer | metastasis; staging; poor survival; motility | "Expression profile was further assessed using quan ......" | 24755295 | Luciferase |
| hsa-miR-181a-5p | colorectal cancer | metastasis; poor survival | "The emerging role of microRNA-181A miR-181A in CRC ......" | 26750720 | |
| hsa-miR-181a-5p | colorectal cancer | tumorigenesis | "It was found that miR-181a may be involved in the ......" | 27264420 | Luciferase |
| hsa-miR-181a-5p | endometrial cancer | poor survival; tumorigenesis; worse prognosis; metastasis; progression | "The aberrant expression of human microRNA-181a-1 h ......" | 25733820 | |
| hsa-miR-181a-5p | gastric cancer | drug resistance | "In order to identify miRNA signatures for gastric ......" | 22112324 | |
| hsa-miR-181a-5p | gastric cancer | malignant trasformation | "However it remains unknown whether miR-181a is inv ......" | 22581522 | Colony formation; Luciferase; Western blot |
| hsa-miR-181a-5p | gastric cancer | worse prognosis | "Genetic polymorphism at miR 181a binding site cont ......" | 22971574 | Luciferase |
| hsa-miR-181a-5p | gastric cancer | progression | "Hsa miR 181a 5p expression and effects on cell pro ......" | 23886199 | |
| hsa-miR-181a-5p | gastric cancer | staging; cell migration | "Here we show that miR-181a levels were significant ......" | 25706394 | |
| hsa-miR-181a-5p | gastric cancer | drug resistance | "MiR 181a suppresses autophagy and sensitizes gastr ......" | 26589846 | |
| hsa-miR-181a-5p | gastric cancer | drug resistance; progression | "Role of miR 27a miR 181a and miR 20b in gastric ca ......" | 26793992 | |
| hsa-miR-181a-5p | glioblastoma | malignant trasformation | "In our study we examined by microarray the global ......" | 16039986 | |
| hsa-miR-181a-5p | glioblastoma | tumorigenesis | "miR 181 subunits enhance the chemosensitivity of t ......" | 24573637 | |
| hsa-miR-181a-5p | glioblastoma | poor survival | "We annotated 491 TCGA samples' miRNA expression pr ......" | 26283154 | |
| hsa-miR-181a-5p | head and neck cancer | malignant trasformation | "A global miRNA profiling was performed on 12 sampl ......" | 21637912 | |
| hsa-miR-181a-5p | head and neck cancer | poor survival | "Our findings showed that significant elevated expr ......" | 25677760 | |
| hsa-miR-181a-5p | liver cancer | differentiation | "Here using a global microarray-based miRNA profili ......" | 19585654 | |
| hsa-miR-181a-5p | liver cancer | motility; drug resistance | "We evaluated the expression of microRNA in human H ......" | 21352471 | |
| hsa-miR-181a-5p | liver cancer | drug resistance | "The cell viability MTT assay was used to detect dr ......" | 23229111 | MTT assay |
| hsa-miR-181a-5p | liver cancer | worse prognosis | "Expression of serum microRNAs miR 222 miR 181 miR ......" | 24124720 | |
| hsa-miR-181a-5p | liver cancer | progression | "Recent studies have found that miR-181a were dysre ......" | 24529171 | Western blot; Luciferase; Colony formation |
| hsa-miR-181a-5p | liver cancer | drug resistance | "miR 181a mediates TGF β induced hepatocyte EMT an ......" | 24576072 | |
| hsa-miR-181a-5p | liver cancer | motility; tumorigenesis | "MiR 181a 5p is downregulated in hepatocellular car ......" | 25058462 | Luciferase |
| hsa-miR-181a-5p | lung cancer | progression; drug resistance | "The metastatic properties of the cells were assess ......" | 26323677 | Western blot; Cell migration assay; Wound Healing Assay; Luciferase |
| hsa-miR-181a-5p | lung squamous cell cancer | worse prognosis; staging; poor survival | "Deregulated expression of miR 21 miR 143 and miR 1 ......" | 20363096 | |
| hsa-miR-181a-5p | lymphoma | poor survival | "Bim down-regulation is posttranscriptionally regul ......" | 20841506 | |
| hsa-miR-181a-5p | melanoma | metastasis | "Circulating T cell natural killer NK natural kille ......" | 24370793 | Flow cytometry |
| hsa-miR-181a-5p | ovarian cancer | recurrence; poor survival; drug resistance | "Here we show that miR-181a promotes TGF-β-mediate ......" | 24394555 | |
| hsa-miR-181a-5p | ovarian cancer | progression; poor survival; drug resistance | "miRNA profiling by microarray real-time PCR and im ......" | 26782955 | |
| hsa-miR-181a-5p | ovarian cancer | drug resistance | "MiR 181a upregulation is associated with epithelia ......" | 27249598 | Flow cytometry; MTT assay |
| hsa-miR-181a-5p | pancreatic cancer | cell migration | "LPS induced miR 181a promotes pancreatic cancer ce ......" | 24532253 | |
| hsa-miR-181a-5p | prostate cancer | progression | "In the present study the expression of miR-181 in ......" | 25187843 | |
| hsa-miR-181a-5p | retinoblastoma | metastasis | "Among them hsa-miR-373 hsa-miR-125b and hsa-miR-18 ......" | 26730174 | |
| hsa-miR-181a-5p | sarcoma | progression | "MicroRNA 181a miR-181a was found dysregulated in a ......" | 23740615 | Western blot |
| hsa-miR-181a-5p | sarcoma | metastasis; progression | "miR 181a Targets RGS16 to Promote Chondrosarcoma G ......" | 26013170 |
Reported gene related to hsa-miR-181a-5p
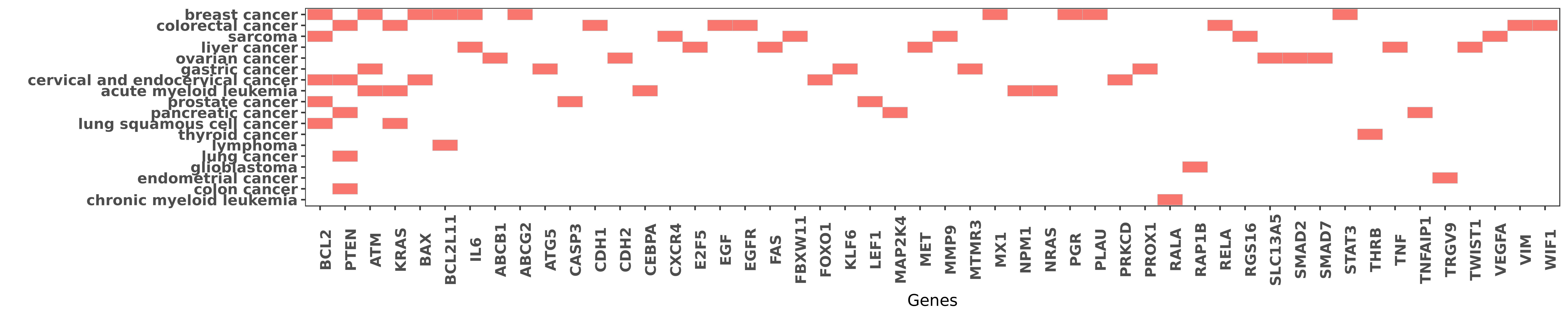
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-181a-5p | cervical and endocervical cancer | PTEN | "From our results down-regulation of miR-181a incre ......" | 27534652 |
| hsa-miR-181a-5p | colon cancer | PTEN | "miR-181a performs this function by inhibiting the ......" | 24685694 |
| hsa-miR-181a-5p | colorectal cancer | PTEN | "The expression of PTEN was regulated by IL-1β-sti ......" | 27264420 |
| hsa-miR-181a-5p | colorectal cancer | PTEN | "The expression of miR-181A and PTEN in CRC patient ......" | 26750720 |
| hsa-miR-181a-5p | lung cancer | PTEN | "Luciferase assays were performed to assess the abi ......" | 26323677 |
| hsa-miR-181a-5p | pancreatic cancer | PTEN | "LPS induced miR 181a promotes pancreatic cancer ce ......" | 24532253 |
| hsa-miR-181a-5p | acute myeloid leukemia | ATM | "MiR 181a Promotes Proliferation of Human Acute Mye ......" | 27150990 |
| hsa-miR-181a-5p | acute myeloid leukemia | ATM | "miR 181a promotes G1/S transition and cell prolife ......" | 26113450 |
| hsa-miR-181a-5p | breast cancer | ATM | "Ataxia telangiectasia mutated ATM a target gene of ......" | 21102523 |
| hsa-miR-181a-5p | breast cancer | ATM | "We report that miR-181a and miR-181b were overexpr ......" | 23656790 |
| hsa-miR-181a-5p | gastric cancer | ATM | "Ataxia-telangiectasia mutation ATM was predicted a ......" | 24531888 |
| hsa-miR-181a-5p | breast cancer | BCL2 | "The function role of miR 181a in chemosensitivity ......" | 24335172 |
| hsa-miR-181a-5p | cervical and endocervical cancer | BCL2 | "In addition inhibition of miR-181a promoted apopto ......" | 27534652 |
| hsa-miR-181a-5p | lung squamous cell cancer | BCL2 | "Moreover we also found miR-181 reduction was assoc ......" | 25524579 |
| hsa-miR-181a-5p | prostate cancer | BCL2 | "Bufalin was found to induce the expression of miR- ......" | 24267199 |
| hsa-miR-181a-5p | sarcoma | BCL2 | "The results from Western blotting indicated that m ......" | 23740615 |
| hsa-miR-181a-5p | acute myeloid leukemia | KRAS | "Here we report that miR-181a directly binds to 3'- ......" | 27517749 |
| hsa-miR-181a-5p | colorectal cancer | KRAS | "The KRAS mutational status was determined by pyros ......" | 24098024 |
| hsa-miR-181a-5p | lung squamous cell cancer | KRAS | "MiR 181a 5p inhibits cell proliferation and migrat ......" | 26124189 |
| hsa-miR-181a-5p | breast cancer | BAX | "We further identified BAX as a direct functional t ......" | 26028030 |
| hsa-miR-181a-5p | cervical and endocervical cancer | BAX | "In addition inhibition of miR-181a promoted apopto ......" | 27534652 |
| hsa-miR-181a-5p | breast cancer | BCL2L11 | "Mechanistically inactivation of miR-181a elevated ......" | 23241956 |
| hsa-miR-181a-5p | lymphoma | BCL2L11 | "Furthermore we found that cell adhesion-up-regulat ......" | 20841506 |
| hsa-miR-181a-5p | breast cancer | IL6 | "The upregulation of miR-181a was orchestrated by t ......" | 26028030 |
| hsa-miR-181a-5p | liver cancer | IL6 | "Knocking down IL-6 and Twist in HSCs significantly ......" | 21352471 |
| hsa-miR-181a-5p | sarcoma | VEGFA | "miR-181a is overexpressed in high-grade chondrosar ......" | 26013170 |
| hsa-miR-181a-5p | sarcoma | VEGFA | "miR-181a transfection of JJ cells doubled expressi ......" | 25106798 |
| hsa-miR-181a-5p | ovarian cancer | ABCB1 | "SKOV3 cells had increased P-gp expression after en ......" | 27249598 |
| hsa-miR-181a-5p | breast cancer | ABCG2 | "Luciferase activity assay showed that miR-181a mim ......" | 23780685 |
| hsa-miR-181a-5p | gastric cancer | ATG5 | "Then we indicated that ATG5 was a potential target ......" | 26589846 |
| hsa-miR-181a-5p | prostate cancer | CASP3 | "In prostate cancer PC-3cell line bufalin-induced a ......" | 24267199 |
| hsa-miR-181a-5p | colorectal cancer | CDH1 | "Ectopic expression of miR-181a suppressed the epit ......" | 24755295 |
| hsa-miR-181a-5p | ovarian cancer | CDH2 | "SKOV3/PTX cells had significantly higher expressio ......" | 27249598 |
| hsa-miR-181a-5p | acute myeloid leukemia | CEBPA | "Interestingly miR-181a expression was increased in ......" | 23100311 |
| hsa-miR-181a-5p | sarcoma | CXCR4 | "These data establish miR-181a as an oncomiR that p ......" | 26013170 |
| hsa-miR-181a-5p | liver cancer | E2F5 | "Mechanism investigation revealed that miR-181a inh ......" | 24529171 |
| hsa-miR-181a-5p | colorectal cancer | EGF | "However the role of miR-181a expression in CRC and ......" | 24098024 |
| hsa-miR-181a-5p | colorectal cancer | EGFR | "miR 181a is associated with poor clinical outcome ......" | 24098024 |
| hsa-miR-181a-5p | liver cancer | FAS | "Functional analysis of miR 181a and Fas involved i ......" | 25449696 |
| hsa-miR-181a-5p | sarcoma | FBXW11 | "First the expression of miR-181a in osteosarcoma c ......" | 23740615 |
| hsa-miR-181a-5p | cervical and endocervical cancer | FOXO1 | "From our results down-regulation of miR-181a incre ......" | 27534652 |
| hsa-miR-181a-5p | gastric cancer | KLF6 | "Site-directed mutagenesis and luciferase reporter ......" | 22581522 |
| hsa-miR-181a-5p | prostate cancer | LEF1 | "LEF1 targeting EMT in prostate cancer invasion is ......" | 26045991 |
| hsa-miR-181a-5p | pancreatic cancer | MAP2K4 | "LPS induced miR 181a promotes pancreatic cancer ce ......" | 24532253 |
| hsa-miR-181a-5p | liver cancer | MET | "MiR 181a 5p is downregulated in hepatocellular car ......" | 25058462 |
| hsa-miR-181a-5p | sarcoma | MMP9 | "The results from Western blotting indicated that m ......" | 23740615 |
| hsa-miR-181a-5p | gastric cancer | MTMR3 | "We further demonstrated that the rs12537CT genotyp ......" | 22971574 |
| hsa-miR-181a-5p | breast cancer | MX1 | "Overexpression of miR-181a down-regulated BCRP exp ......" | 23780685 |
| hsa-miR-181a-5p | acute myeloid leukemia | NPM1 | "The impact of miR-181a was most striking in poor m ......" | 21079133 |
| hsa-miR-181a-5p | acute myeloid leukemia | NRAS | "Here we report that miR-181a directly binds to 3'- ......" | 27517749 |
| hsa-miR-181a-5p | breast cancer | PGR | "In addition a transcriptome analysis revealed that ......" | 19826037 |
| hsa-miR-181a-5p | breast cancer | PLAU | "uPA mRNA was a direct target of miR-193a/b and miR ......" | 21779487 |
| hsa-miR-181a-5p | cervical and endocervical cancer | PRKCD | "MiR 181a confers resistance of cervical cancer to ......" | 22847611 |
| hsa-miR-181a-5p | gastric cancer | PROX1 | "In addition the ectopic expression of miR-181a in ......" | 25706394 |
| hsa-miR-181a-5p | chronic myeloid leukemia | RALA | "The delivery of miR-181a specifically inhibited th ......" | 26554155 |
| hsa-miR-181a-5p | glioblastoma | RAP1B | "miR 181 subunits enhance the chemosensitivity of t ......" | 24573637 |
| hsa-miR-181a-5p | colorectal cancer | RELA | "The expression levels of IL-1β NF-κB RelA and mi ......" | 27264420 |
| hsa-miR-181a-5p | sarcoma | RGS16 | "miR 181a Targets RGS16 to Promote Chondrosarcoma G ......" | 26013170 |
| hsa-miR-181a-5p | ovarian cancer | SLC13A5 | "It also confirms that concomitant analysis of P-Sm ......" | 26782955 |
| hsa-miR-181a-5p | ovarian cancer | SMAD2 | "Here we show that miR-181a promotes TGF-β-mediate ......" | 24394555 |
| hsa-miR-181a-5p | ovarian cancer | SMAD7 | "Here we show that miR-181a promotes TGF-β-mediate ......" | 24394555 |
| hsa-miR-181a-5p | breast cancer | STAT3 | "The upregulation of miR-181a was orchestrated by t ......" | 26028030 |
| hsa-miR-181a-5p | thyroid cancer | THRB | "We observed lower levels of THRB transcripts in ce ......" | 21159845 |
| hsa-miR-181a-5p | liver cancer | TNF | "TNF receptor superfamily member 6 Fas was further ......" | 25449696 |
| hsa-miR-181a-5p | pancreatic cancer | TNFAIP1 | "Targeting of miR-181a on TNFAIP1 in pancreatic can ......" | 26152285 |
| hsa-miR-181a-5p | endometrial cancer | TRGV9 | "To predict the targets of hsa-miR-181a ten differe ......" | 25733820 |
| hsa-miR-181a-5p | liver cancer | TWIST1 | "Knocking down IL-6 and Twist in HSCs significantly ......" | 21352471 |
| hsa-miR-181a-5p | colorectal cancer | VIM | "Ectopic expression of miR-181a suppressed the epit ......" | 24755295 |
| hsa-miR-181a-5p | colorectal cancer | WIF1 | "Additionally we identified WIF-1 as direct and fun ......" | 24755295 |
Expression profile in cancer corhorts:
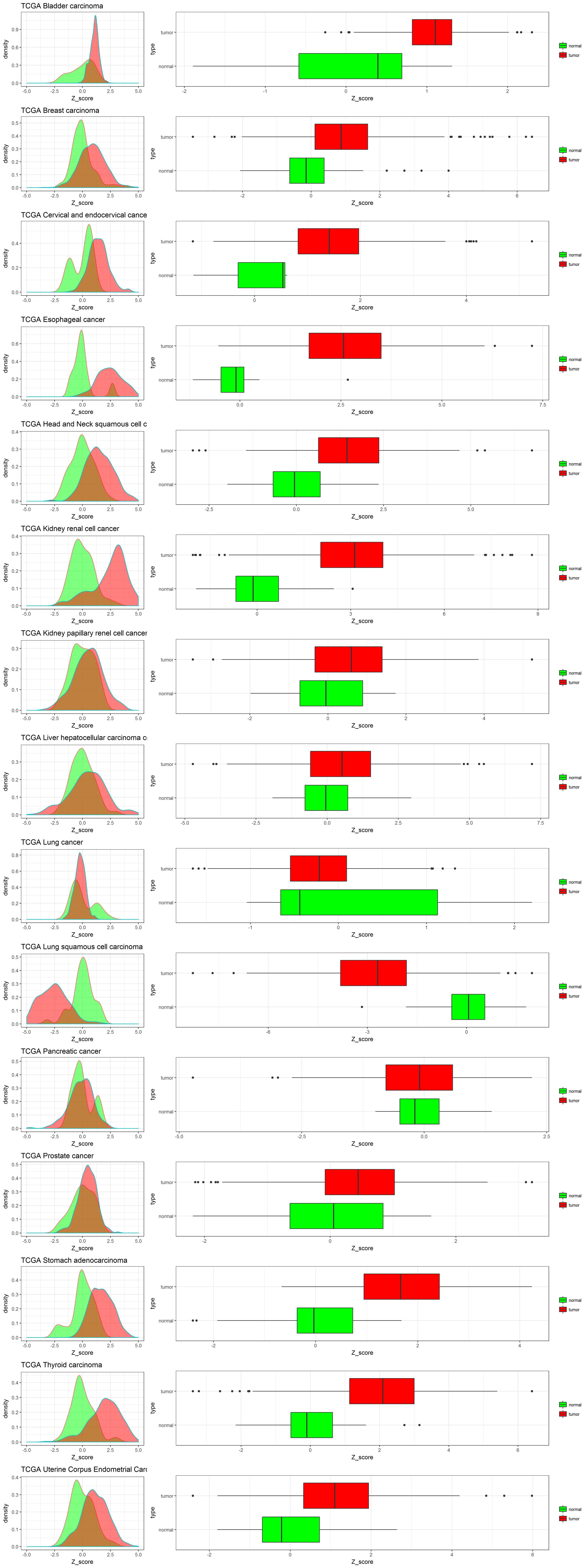
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-181a-5p | BAG2 | 12 cancers: BLCA; ESCA; KIRC; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD | miRNAWalker2 validate | TCGA BLCA -0.418; TCGA ESCA -0.3; TCGA KIRC -0.463; TCGA LGG -0.105; TCGA LIHC -0.296; TCGA LUAD -0.333; TCGA LUSC -0.246; TCGA PAAD -0.251; TCGA PRAD -0.133; TCGA SARC -0.524; TCGA THCA -0.209; TCGA STAD -0.346 |
hsa-miR-181a-5p | COPS2 | 9 cancers: BLCA; BRCA; CESC; HNSC; LIHC; LUAD; LUSC; PRAD; SARC | miRNAWalker2 validate; MirTarget | TCGA BLCA -0.118; TCGA BRCA -0.2; TCGA CESC -0.088; TCGA HNSC -0.059; TCGA LIHC -0.098; TCGA LUAD -0.143; TCGA LUSC -0.143; TCGA PRAD -0.087; TCGA SARC -0.102 |
hsa-miR-181a-5p | HUWE1 | 9 cancers: BLCA; BRCA; HNSC; KIRC; LUAD; LUSC; PRAD; SARC; THCA | miRNAWalker2 validate | TCGA BLCA -0.082; TCGA BRCA -0.088; TCGA HNSC -0.106; TCGA KIRC -0.224; TCGA LUAD -0.071; TCGA LUSC -0.068; TCGA PRAD -0.079; TCGA SARC -0.108; TCGA THCA -0.093 |
hsa-miR-181a-5p | KAT2B | 11 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LIHC; PAAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate; miRTarBase; MirTarget | TCGA BLCA -0.145; TCGA BRCA -0.326; TCGA ESCA -0.317; TCGA HNSC -0.301; TCGA KIRC -0.253; TCGA LIHC -0.427; TCGA PAAD -0.225; TCGA SARC -0.348; TCGA THCA -0.095; TCGA STAD -0.352; TCGA UCEC -0.135 |
hsa-miR-181a-5p | KCTD3 | 9 cancers: BLCA; BRCA; CESC; HNSC; LUAD; LUSC; PRAD; SARC; THCA | miRNAWalker2 validate | TCGA BLCA -0.191; TCGA BRCA -0.082; TCGA CESC -0.204; TCGA HNSC -0.2; TCGA LUAD -0.17; TCGA LUSC -0.191; TCGA PRAD -0.132; TCGA SARC -0.072; TCGA THCA -0.069 |
hsa-miR-181a-5p | MTMR12 | 11 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LIHC; OV; PAAD; SARC; STAD; UCEC | miRNAWalker2 validate; miRNATAP | TCGA BLCA -0.113; TCGA BRCA -0.101; TCGA ESCA -0.13; TCGA HNSC -0.239; TCGA KIRC -0.171; TCGA LIHC -0.082; TCGA OV -0.102; TCGA PAAD -0.171; TCGA SARC -0.212; TCGA STAD -0.12; TCGA UCEC -0.084 |
hsa-miR-181a-5p | PRDX3 | 10 cancers: BLCA; BRCA; COAD; KIRC; LIHC; LUAD; OV; PRAD; SARC; THCA | miRNAWalker2 validate; MirTarget | TCGA BLCA -0.104; TCGA BRCA -0.104; TCGA COAD -0.083; TCGA KIRC -0.288; TCGA LIHC -0.198; TCGA LUAD -0.177; TCGA OV -0.086; TCGA PRAD -0.064; TCGA SARC -0.061; TCGA THCA -0.081 |
hsa-miR-181a-5p | PTPN11 | 9 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LIHC; LUAD; SARC; THCA | miRNAWalker2 validate; miRTarBase; RAID | TCGA BLCA -0.185; TCGA BRCA -0.204; TCGA ESCA -0.081; TCGA HNSC -0.162; TCGA KIRC -0.142; TCGA LIHC -0.104; TCGA LUAD -0.205; TCGA SARC -0.099; TCGA THCA -0.106 |
hsa-miR-181a-5p | PTPRZ1 | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LUSC; PAAD; STAD | miRNAWalker2 validate | TCGA BLCA -0.535; TCGA CESC -1.032; TCGA COAD -0.396; TCGA ESCA -0.97; TCGA HNSC -0.286; TCGA KIRC -1.05; TCGA LUSC -1.571; TCGA PAAD -0.723; TCGA STAD -0.86 |
hsa-miR-181a-5p | SIK2 | 9 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LIHC; PAAD; THCA; STAD | miRNAWalker2 validate | TCGA BLCA -0.153; TCGA BRCA -0.342; TCGA ESCA -0.228; TCGA HNSC -0.154; TCGA KIRC -0.171; TCGA LIHC -0.159; TCGA PAAD -0.173; TCGA THCA -0.134; TCGA STAD -0.205 |
hsa-miR-181a-5p | TAB2 | 10 cancers: BLCA; BRCA; COAD; HNSC; KIRC; LGG; LIHC; LUAD; THCA; STAD | miRNAWalker2 validate | TCGA BLCA -0.109; TCGA BRCA -0.074; TCGA COAD -0.086; TCGA HNSC -0.074; TCGA KIRC -0.241; TCGA LGG -0.101; TCGA LIHC -0.225; TCGA LUAD -0.095; TCGA THCA -0.067; TCGA STAD -0.109 |
hsa-miR-181a-5p | TGFBR3 | 14 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; PAAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate; mirMAP; miRNATAP | TCGA BLCA -0.533; TCGA BRCA -0.522; TCGA COAD -0.263; TCGA ESCA -0.335; TCGA HNSC -0.481; TCGA KIRC -0.332; TCGA KIRP -0.227; TCGA LGG -0.111; TCGA LIHC -0.389; TCGA PAAD -0.255; TCGA SARC -0.297; TCGA THCA -0.275; TCGA STAD -0.604; TCGA UCEC -0.315 |
hsa-miR-181a-5p | TM9SF3 | 9 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; LUAD; PRAD | miRNAWalker2 validate; MirTarget; miRNATAP | TCGA BLCA -0.095; TCGA BRCA -0.104; TCGA CESC -0.084; TCGA COAD -0.08; TCGA HNSC -0.079; TCGA KIRC -0.154; TCGA KIRP -0.058; TCGA LUAD -0.158; TCGA PRAD -0.151 |
hsa-miR-181a-5p | TRIM2 | 10 cancers: BLCA; CESC; HNSC; KIRC; KIRP; LIHC; LUSC; PRAD; THCA; STAD | miRNAWalker2 validate; miRNATAP | TCGA BLCA -0.287; TCGA CESC -0.28; TCGA HNSC -0.398; TCGA KIRC -0.728; TCGA KIRP -0.388; TCGA LIHC -0.346; TCGA LUSC -0.327; TCGA PRAD -0.262; TCGA THCA -0.262; TCGA STAD -0.154 |
hsa-miR-181a-5p | ZNF148 | 9 cancers: BLCA; BRCA; COAD; HNSC; KIRC; LUAD; LUSC; PRAD; THCA | miRNAWalker2 validate; mirMAP | TCGA BLCA -0.116; TCGA BRCA -0.187; TCGA COAD -0.059; TCGA HNSC -0.13; TCGA KIRC -0.126; TCGA LUAD -0.154; TCGA LUSC -0.085; TCGA PRAD -0.095; TCGA THCA -0.087 |
hsa-miR-181a-5p | ZNF652 | 9 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; THCA | miRNAWalker2 validate | TCGA BLCA -0.141; TCGA BRCA -0.084; TCGA COAD -0.088; TCGA ESCA -0.153; TCGA HNSC -0.086; TCGA KIRC -0.106; TCGA KIRP -0.109; TCGA LIHC -0.145; TCGA THCA -0.172 |
hsa-miR-181a-5p | KPNA1 | 9 cancers: BLCA; BRCA; COAD; HNSC; KIRC; LIHC; LUAD; LUSC; SARC | MirTarget; miRNATAP | TCGA BLCA -0.079; TCGA BRCA -0.106; TCGA COAD -0.085; TCGA HNSC -0.136; TCGA KIRC -0.109; TCGA LIHC -0.138; TCGA LUAD -0.189; TCGA LUSC -0.235; TCGA SARC -0.119 |
hsa-miR-181a-5p | HLF | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; PAAD; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.91; TCGA BRCA -0.174; TCGA CESC -0.356; TCGA COAD -0.341; TCGA ESCA -0.818; TCGA HNSC -0.846; TCGA LIHC -0.832; TCGA PAAD -0.699; TCGA THCA -0.435; TCGA STAD -0.997; TCGA UCEC -0.363 |
hsa-miR-181a-5p | STXBP4 | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LGG; SARC; THCA; STAD | MirTarget | TCGA BLCA -0.235; TCGA BRCA -0.247; TCGA COAD -0.117; TCGA ESCA -0.153; TCGA HNSC -0.214; TCGA KIRC -0.33; TCGA LGG -0.107; TCGA SARC -0.091; TCGA THCA -0.089; TCGA STAD -0.096 |
hsa-miR-181a-5p | SIK3 | 10 cancers: BLCA; BRCA; COAD; ESCA; KIRC; LIHC; PAAD; SARC; THCA; STAD | MirTarget | TCGA BLCA -0.12; TCGA BRCA -0.224; TCGA COAD -0.076; TCGA ESCA -0.147; TCGA KIRC -0.203; TCGA LIHC -0.1; TCGA PAAD -0.252; TCGA SARC -0.053; TCGA THCA -0.133; TCGA STAD -0.216 |
hsa-miR-181a-5p | MLXIP | 10 cancers: BLCA; BRCA; ESCA; HNSC; LIHC; PAAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.272; TCGA BRCA -0.086; TCGA ESCA -0.117; TCGA HNSC -0.141; TCGA LIHC -0.104; TCGA PAAD -0.178; TCGA SARC -0.195; TCGA THCA -0.19; TCGA STAD -0.213; TCGA UCEC -0.091 |
hsa-miR-181a-5p | THRB | 10 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LIHC; SARC; THCA; STAD; UCEC | MirTarget; mirMAP; miRNATAP | TCGA BLCA -0.273; TCGA BRCA -0.328; TCGA ESCA -0.559; TCGA HNSC -0.435; TCGA KIRC -0.717; TCGA LIHC -0.297; TCGA SARC -0.363; TCGA THCA -0.253; TCGA STAD -0.587; TCGA UCEC -0.495 |
hsa-miR-181a-5p | UBL3 | 9 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LIHC; PAAD; PRAD; STAD | MirTarget; miRNATAP | TCGA BLCA -0.195; TCGA BRCA -0.2; TCGA ESCA -0.288; TCGA HNSC -0.213; TCGA KIRC -0.342; TCGA LIHC -0.235; TCGA PAAD -0.153; TCGA PRAD -0.171; TCGA STAD -0.207 |
hsa-miR-181a-5p | INPP5A | 9 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.217; TCGA BRCA -0.065; TCGA CESC -0.134; TCGA COAD -0.073; TCGA ESCA -0.207; TCGA HNSC -0.131; TCGA SARC -0.155; TCGA STAD -0.439; TCGA UCEC -0.152 |
hsa-miR-181a-5p | TNPO1 | 9 cancers: BLCA; BRCA; HNSC; KIRC; LIHC; LUAD; LUSC; PRAD; THCA | MirTarget | TCGA BLCA -0.124; TCGA BRCA -0.154; TCGA HNSC -0.08; TCGA KIRC -0.107; TCGA LIHC -0.08; TCGA LUAD -0.167; TCGA LUSC -0.136; TCGA PRAD -0.086; TCGA THCA -0.143 |
hsa-miR-181a-5p | CAMTA2 | 9 cancers: BLCA; ESCA; HNSC; KIRP; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.083; TCGA ESCA -0.177; TCGA HNSC -0.105; TCGA KIRP -0.145; TCGA PAAD -0.249; TCGA PRAD -0.051; TCGA SARC -0.078; TCGA STAD -0.242; TCGA UCEC -0.097 |
hsa-miR-181a-5p | SUCLG2 | 13 cancers: BLCA; BRCA; COAD; HNSC; KIRC; LGG; LIHC; LUAD; OV; PRAD; SARC; THCA; STAD | MirTarget | TCGA BLCA -0.125; TCGA BRCA -0.17; TCGA COAD -0.097; TCGA HNSC -0.126; TCGA KIRC -0.474; TCGA LGG -0.121; TCGA LIHC -0.335; TCGA LUAD -0.072; TCGA OV -0.109; TCGA PRAD -0.069; TCGA SARC -0.095; TCGA THCA -0.11; TCGA STAD -0.245 |
hsa-miR-181a-5p | LRRFIP1 | 11 cancers: BLCA; BRCA; CESC; HNSC; KIRC; LIHC; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.222; TCGA BRCA -0.18; TCGA CESC -0.1; TCGA HNSC -0.183; TCGA KIRC -0.187; TCGA LIHC -0.121; TCGA PRAD -0.128; TCGA SARC -0.18; TCGA THCA -0.134; TCGA STAD -0.064; TCGA UCEC -0.057 |
hsa-miR-181a-5p | PPP1R2 | 9 cancers: BLCA; CESC; HNSC; KIRC; LIHC; LUAD; LUSC; PRAD; SARC | MirTarget | TCGA BLCA -0.089; TCGA CESC -0.113; TCGA HNSC -0.112; TCGA KIRC -0.136; TCGA LIHC -0.085; TCGA LUAD -0.1; TCGA LUSC -0.201; TCGA PRAD -0.063; TCGA SARC -0.153 |
hsa-miR-181a-5p | IRS2 | 9 cancers: BLCA; BRCA; HNSC; KIRC; LIHC; LUAD; PAAD; SARC; STAD | MirTarget; miRNATAP | TCGA BLCA -0.182; TCGA BRCA -0.339; TCGA HNSC -0.158; TCGA KIRC -0.32; TCGA LIHC -0.42; TCGA LUAD -0.256; TCGA PAAD -0.157; TCGA SARC -0.17; TCGA STAD -0.323 |
hsa-miR-181a-5p | TAF9B | 10 cancers: BLCA; BRCA; CESC; COAD; KIRC; LIHC; PAAD; PRAD; SARC; STAD | MirTarget | TCGA BLCA -0.124; TCGA BRCA -0.116; TCGA CESC -0.127; TCGA COAD -0.174; TCGA KIRC -0.076; TCGA LIHC -0.171; TCGA PAAD -0.203; TCGA PRAD -0.057; TCGA SARC -0.249; TCGA STAD -0.212 |
hsa-miR-181a-5p | ZBTB4 | 11 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; KIRP; PAAD; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.22; TCGA BRCA -0.191; TCGA ESCA -0.311; TCGA HNSC -0.125; TCGA KIRC -0.221; TCGA KIRP -0.148; TCGA PAAD -0.149; TCGA PRAD -0.06; TCGA SARC -0.098; TCGA STAD -0.338; TCGA UCEC -0.151 |
hsa-miR-181a-5p | KANK1 | 11 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LIHC; OV; PAAD; SARC; STAD | MirTarget; miRNATAP | TCGA BLCA -0.346; TCGA BRCA -0.214; TCGA CESC -0.189; TCGA ESCA -0.404; TCGA HNSC -0.34; TCGA KIRC -0.133; TCGA LIHC -0.192; TCGA OV -0.131; TCGA PAAD -0.33; TCGA SARC -0.562; TCGA STAD -0.309 |
hsa-miR-181a-5p | ATXN7 | 9 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; KIRP; LIHC; SARC; THCA | MirTarget | TCGA BLCA -0.092; TCGA BRCA -0.208; TCGA ESCA -0.1; TCGA HNSC -0.108; TCGA KIRC -0.16; TCGA KIRP -0.11; TCGA LIHC -0.095; TCGA SARC -0.087; TCGA THCA -0.082 |
hsa-miR-181a-5p | API5 | 10 cancers: BLCA; BRCA; COAD; KIRC; LIHC; LUAD; LUSC; OV; PRAD; SARC | MirTarget | TCGA BLCA -0.057; TCGA BRCA -0.068; TCGA COAD -0.091; TCGA KIRC -0.171; TCGA LIHC -0.18; TCGA LUAD -0.101; TCGA LUSC -0.11; TCGA OV -0.086; TCGA PRAD -0.075; TCGA SARC -0.124 |
hsa-miR-181a-5p | ARIH1 | 9 cancers: BLCA; BRCA; CESC; HNSC; KIRC; LIHC; LUAD; LUSC; PRAD | MirTarget; mirMAP | TCGA BLCA -0.097; TCGA BRCA -0.172; TCGA CESC -0.084; TCGA HNSC -0.084; TCGA KIRC -0.1; TCGA LIHC -0.13; TCGA LUAD -0.066; TCGA LUSC -0.07; TCGA PRAD -0.05 |
hsa-miR-181a-5p | TTLL7 | 9 cancers: BLCA; BRCA; ESCA; LIHC; LUAD; PAAD; SARC; THCA; STAD | MirTarget | TCGA BLCA -0.468; TCGA BRCA -0.173; TCGA ESCA -0.576; TCGA LIHC -0.468; TCGA LUAD -0.477; TCGA PAAD -0.615; TCGA SARC -0.785; TCGA THCA -0.343; TCGA STAD -0.7 |
hsa-miR-181a-5p | TMF1 | 9 cancers: BLCA; BRCA; HNSC; KIRC; KIRP; LUAD; OV; SARC; THCA | MirTarget | TCGA BLCA -0.071; TCGA BRCA -0.235; TCGA HNSC -0.183; TCGA KIRC -0.199; TCGA KIRP -0.159; TCGA LUAD -0.063; TCGA OV -0.072; TCGA SARC -0.074; TCGA THCA -0.08 |
hsa-miR-181a-5p | G3BP2 | 9 cancers: BLCA; BRCA; HNSC; KIRC; LIHC; LUAD; PRAD; SARC; THCA | MirTarget; miRNATAP | TCGA BLCA -0.068; TCGA BRCA -0.124; TCGA HNSC -0.126; TCGA KIRC -0.218; TCGA LIHC -0.082; TCGA LUAD -0.065; TCGA PRAD -0.118; TCGA SARC -0.103; TCGA THCA -0.074 |
hsa-miR-181a-5p | TSC22D2 | 9 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; KIRP; LIHC; SARC; THCA | MirTarget | TCGA BLCA -0.103; TCGA BRCA -0.138; TCGA ESCA -0.164; TCGA HNSC -0.135; TCGA KIRC -0.211; TCGA KIRP -0.139; TCGA LIHC -0.193; TCGA SARC -0.099; TCGA THCA -0.179 |
hsa-miR-181a-5p | CLIP1 | 11 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LIHC; LUAD; SARC; THCA; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.272; TCGA BRCA -0.217; TCGA CESC -0.163; TCGA ESCA -0.193; TCGA HNSC -0.279; TCGA KIRC -0.152; TCGA LIHC -0.168; TCGA LUAD -0.157; TCGA SARC -0.297; TCGA THCA -0.091; TCGA UCEC -0.059 |
hsa-miR-181a-5p | LONRF1 | 9 cancers: BLCA; BRCA; CESC; HNSC; KIRP; OV; SARC; THCA; STAD | MirTarget; miRNATAP | TCGA BLCA -0.18; TCGA BRCA -0.121; TCGA CESC -0.14; TCGA HNSC -0.283; TCGA KIRP -0.107; TCGA OV -0.11; TCGA SARC -0.301; TCGA THCA -0.064; TCGA STAD -0.133 |
hsa-miR-181a-5p | MAP1B | 10 cancers: BLCA; BRCA; COAD; ESCA; LUAD; LUSC; PAAD; SARC; THCA; STAD | MirTarget; miRNATAP | TCGA BLCA -0.595; TCGA BRCA -0.125; TCGA COAD -0.243; TCGA ESCA -0.432; TCGA LUAD -0.237; TCGA LUSC -0.502; TCGA PAAD -0.4; TCGA SARC -0.418; TCGA THCA -0.217; TCGA STAD -0.484 |
hsa-miR-181a-5p | GHITM | 12 cancers: BLCA; BRCA; CESC; HNSC; KIRC; LIHC; LUAD; LUSC; OV; PRAD; THCA; STAD | MirTarget; miRNATAP | TCGA BLCA -0.109; TCGA BRCA -0.099; TCGA CESC -0.11; TCGA HNSC -0.146; TCGA KIRC -0.366; TCGA LIHC -0.236; TCGA LUAD -0.157; TCGA LUSC -0.134; TCGA OV -0.148; TCGA PRAD -0.052; TCGA THCA -0.179; TCGA STAD -0.109 |
hsa-miR-181a-5p | CBX7 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; OV; PAAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.537; TCGA BRCA -0.297; TCGA CESC -0.291; TCGA COAD -0.227; TCGA ESCA -0.517; TCGA HNSC -0.175; TCGA LIHC -0.275; TCGA OV -0.133; TCGA PAAD -0.487; TCGA SARC -0.557; TCGA STAD -0.721; TCGA UCEC -0.32 |
hsa-miR-181a-5p | UBE3C | 11 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; SARC; THCA | MirTarget; miRNATAP | TCGA BLCA -0.103; TCGA BRCA -0.102; TCGA ESCA -0.217; TCGA HNSC -0.158; TCGA KIRC -0.147; TCGA KIRP -0.095; TCGA LIHC -0.135; TCGA LUAD -0.124; TCGA LUSC -0.195; TCGA SARC -0.08; TCGA THCA -0.104 |
hsa-miR-181a-5p | PPP2R5E | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; SARC; THCA | MirTarget; mirMAP | TCGA BLCA -0.134; TCGA BRCA -0.173; TCGA COAD -0.127; TCGA ESCA -0.1; TCGA HNSC -0.192; TCGA KIRC -0.213; TCGA LUAD -0.175; TCGA LUSC -0.173; TCGA SARC -0.08; TCGA THCA -0.127 |
hsa-miR-181a-5p | TBL1X | 11 cancers: BLCA; HNSC; KIRC; LIHC; LUAD; LUSC; PAAD; SARC; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.275; TCGA HNSC -0.427; TCGA KIRC -0.095; TCGA LIHC -0.196; TCGA LUAD -0.13; TCGA LUSC -0.51; TCGA PAAD -0.318; TCGA SARC -0.221; TCGA THCA -0.224; TCGA STAD -0.247; TCGA UCEC -0.161 |
hsa-miR-181a-5p | MPP5 | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LIHC; PRAD; THCA; STAD | MirTarget | TCGA BLCA -0.113; TCGA BRCA -0.182; TCGA COAD -0.076; TCGA ESCA -0.122; TCGA HNSC -0.267; TCGA KIRC -0.494; TCGA LIHC -0.202; TCGA PRAD -0.207; TCGA THCA -0.087; TCGA STAD -0.077 |
hsa-miR-181a-5p | GOLGA1 | 9 cancers: BLCA; BRCA; ESCA; LIHC; LUSC; PAAD; SARC; THCA; STAD | MirTarget; miRNATAP | TCGA BLCA -0.086; TCGA BRCA -0.086; TCGA ESCA -0.109; TCGA LIHC -0.144; TCGA LUSC -0.078; TCGA PAAD -0.147; TCGA SARC -0.129; TCGA THCA -0.115; TCGA STAD -0.086 |
hsa-miR-181a-5p | MAMDC2 | 9 cancers: BLCA; BRCA; COAD; ESCA; KIRC; SARC; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.652; TCGA BRCA -0.483; TCGA COAD -0.532; TCGA ESCA -0.78; TCGA KIRC -0.817; TCGA SARC -0.803; TCGA THCA -0.288; TCGA STAD -1.333; TCGA UCEC -0.443 |
hsa-miR-181a-5p | LMO3 | 9 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; PAAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.917; TCGA BRCA -0.313; TCGA ESCA -0.597; TCGA HNSC -0.487; TCGA KIRC -1.481; TCGA PAAD -0.81; TCGA SARC -0.703; TCGA STAD -0.865; TCGA UCEC -0.517 |
hsa-miR-181a-5p | RPS6KB1 | 9 cancers: BLCA; BRCA; CESC; COAD; HNSC; LIHC; LUAD; LUSC; PRAD | MirTarget; miRNATAP | TCGA BLCA -0.056; TCGA BRCA -0.13; TCGA CESC -0.087; TCGA COAD -0.084; TCGA HNSC -0.087; TCGA LIHC -0.117; TCGA LUAD -0.126; TCGA LUSC -0.134; TCGA PRAD -0.059 |
hsa-miR-181a-5p | MPP7 | 9 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; PRAD; SARC; STAD | MirTarget | TCGA BLCA -0.42; TCGA BRCA -0.374; TCGA CESC -0.365; TCGA ESCA -0.521; TCGA HNSC -0.51; TCGA KIRC -1.095; TCGA PRAD -0.159; TCGA SARC -0.995; TCGA STAD -0.517 |
hsa-miR-181a-5p | SYNE1 | 9 cancers: BLCA; BRCA; ESCA; KIRC; PAAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.401; TCGA BRCA -0.305; TCGA ESCA -0.278; TCGA KIRC -0.169; TCGA PAAD -0.288; TCGA SARC -0.314; TCGA THCA -0.542; TCGA STAD -0.315; TCGA UCEC -0.144 |
hsa-miR-181a-5p | TAB3 | 9 cancers: BLCA; BRCA; CESC; HNSC; KIRC; PRAD; SARC; THCA; UCEC | MirTarget; mirMAP | TCGA BLCA -0.059; TCGA BRCA -0.164; TCGA CESC -0.098; TCGA HNSC -0.168; TCGA KIRC -0.269; TCGA PRAD -0.137; TCGA SARC -0.228; TCGA THCA -0.105; TCGA UCEC -0.061 |
hsa-miR-181a-5p | GPD1L | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; PAAD; PRAD; SARC; THCA; STAD | MirTarget; miRNATAP | TCGA BLCA -0.338; TCGA BRCA -0.353; TCGA CESC -0.164; TCGA COAD -0.106; TCGA ESCA -0.322; TCGA HNSC -0.551; TCGA KIRC -0.716; TCGA KIRP -0.121; TCGA PAAD -0.151; TCGA PRAD -0.083; TCGA SARC -0.343; TCGA THCA -0.306; TCGA STAD -0.346 |
hsa-miR-181a-5p | LMBRD2 | 9 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LIHC; LUAD; SARC; THCA | MirTarget; mirMAP | TCGA BLCA -0.186; TCGA BRCA -0.33; TCGA ESCA -0.143; TCGA HNSC -0.309; TCGA KIRC -0.331; TCGA LIHC -0.329; TCGA LUAD -0.185; TCGA SARC -0.228; TCGA THCA -0.322 |
hsa-miR-181a-5p | PLEKHA3 | 9 cancers: BLCA; BRCA; KIRC; LIHC; LUAD; OV; PRAD; SARC; STAD | MirTarget | TCGA BLCA -0.084; TCGA BRCA -0.126; TCGA KIRC -0.126; TCGA LIHC -0.208; TCGA LUAD -0.081; TCGA OV -0.086; TCGA PRAD -0.066; TCGA SARC -0.129; TCGA STAD -0.07 |
hsa-miR-181a-5p | KIAA2022 | 11 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; PAAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.801; TCGA BRCA -0.2; TCGA ESCA -1.089; TCGA HNSC -0.572; TCGA KIRC -1.619; TCGA PAAD -0.707; TCGA PRAD -0.162; TCGA SARC -0.526; TCGA THCA -0.592; TCGA STAD -1.228; TCGA UCEC -0.299 |
hsa-miR-181a-5p | CBLL1 | 9 cancers: BLCA; BRCA; COAD; HNSC; LUAD; LUSC; OV; SARC; THCA | MirTarget | TCGA BLCA -0.057; TCGA BRCA -0.103; TCGA COAD -0.137; TCGA HNSC -0.107; TCGA LUAD -0.13; TCGA LUSC -0.113; TCGA OV -0.06; TCGA SARC -0.077; TCGA THCA -0.139 |
hsa-miR-181a-5p | JMY | 12 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; KIRP; LIHC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.258; TCGA BRCA -0.196; TCGA ESCA -0.27; TCGA HNSC -0.176; TCGA KIRC -0.475; TCGA KIRP -0.166; TCGA LIHC -0.172; TCGA PAAD -0.25; TCGA PRAD -0.072; TCGA SARC -0.139; TCGA STAD -0.236; TCGA UCEC -0.096 |
hsa-miR-181a-5p | TEX2 | 11 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LIHC; LUSC; PAAD; PRAD; THCA; STAD | MirTarget | TCGA BLCA -0.177; TCGA BRCA -0.146; TCGA ESCA -0.263; TCGA HNSC -0.18; TCGA KIRC -0.212; TCGA LIHC -0.136; TCGA LUSC -0.138; TCGA PAAD -0.268; TCGA PRAD -0.095; TCGA THCA -0.262; TCGA STAD -0.119 |
hsa-miR-181a-5p | MAGI3 | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LIHC; SARC; THCA; STAD | MirTarget | TCGA BLCA -0.21; TCGA BRCA -0.164; TCGA CESC -0.193; TCGA ESCA -0.334; TCGA HNSC -0.321; TCGA KIRC -0.647; TCGA LIHC -0.138; TCGA SARC -0.153; TCGA THCA -0.124; TCGA STAD -0.164 |
hsa-miR-181a-5p | NNT | 9 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LIHC; PRAD; SARC; THCA | MirTarget; mirMAP | TCGA BLCA -0.116; TCGA BRCA -0.18; TCGA ESCA -0.197; TCGA HNSC -0.164; TCGA KIRC -0.551; TCGA LIHC -0.396; TCGA PRAD -0.166; TCGA SARC -0.197; TCGA THCA -0.32 |
hsa-miR-181a-5p | SLAIN2 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; PRAD; SARC; STAD | MirTarget | TCGA BLCA -0.178; TCGA BRCA -0.148; TCGA CESC -0.152; TCGA COAD -0.066; TCGA ESCA -0.17; TCGA HNSC -0.195; TCGA KIRC -0.112; TCGA LIHC -0.127; TCGA PRAD -0.097; TCGA SARC -0.128; TCGA STAD -0.138 |
hsa-miR-181a-5p | PAWR | 9 cancers: BLCA; BRCA; HNSC; KIRC; KIRP; LUAD; LUSC; PRAD; SARC | MirTarget | TCGA BLCA -0.107; TCGA BRCA -0.094; TCGA HNSC -0.112; TCGA KIRC -0.367; TCGA KIRP -0.184; TCGA LUAD -0.262; TCGA LUSC -0.304; TCGA PRAD -0.113; TCGA SARC -0.511 |
hsa-miR-181a-5p | SOS1 | 9 cancers: BLCA; BRCA; COAD; HNSC; KIRP; LIHC; LUAD; SARC; THCA | MirTarget; miRNATAP | TCGA BLCA -0.073; TCGA BRCA -0.16; TCGA COAD -0.06; TCGA HNSC -0.101; TCGA KIRP -0.098; TCGA LIHC -0.081; TCGA LUAD -0.105; TCGA SARC -0.055; TCGA THCA -0.11 |
hsa-miR-181a-5p | DCUN1D1 | 10 cancers: BLCA; BRCA; HNSC; KIRC; LIHC; LUAD; LUSC; OV; SARC; THCA | MirTarget; mirMAP | TCGA BLCA -0.105; TCGA BRCA -0.14; TCGA HNSC -0.183; TCGA KIRC -0.386; TCGA LIHC -0.111; TCGA LUAD -0.216; TCGA LUSC -0.497; TCGA OV -0.075; TCGA SARC -0.065; TCGA THCA -0.163 |
hsa-miR-181a-5p | ARMC8 | 11 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; LUAD; LUSC; SARC; THCA; STAD | MirTarget; miRNATAP | TCGA BLCA -0.079; TCGA BRCA -0.084; TCGA CESC -0.074; TCGA COAD -0.078; TCGA HNSC -0.054; TCGA KIRC -0.165; TCGA LUAD -0.087; TCGA LUSC -0.095; TCGA SARC -0.144; TCGA THCA -0.108; TCGA STAD -0.055 |
hsa-miR-181a-5p | KPNA4 | 10 cancers: BLCA; BRCA; COAD; HNSC; KIRC; LGG; LUAD; LUSC; SARC; THCA | MirTarget; miRNATAP | TCGA BLCA -0.066; TCGA BRCA -0.063; TCGA COAD -0.077; TCGA HNSC -0.101; TCGA KIRC -0.087; TCGA LGG -0.064; TCGA LUAD -0.23; TCGA LUSC -0.304; TCGA SARC -0.105; TCGA THCA -0.083 |
hsa-miR-181a-5p | NRXN1 | 10 cancers: BLCA; BRCA; COAD; ESCA; KIRC; KIRP; PAAD; PRAD; SARC; STAD | MirTarget; miRNATAP | TCGA BLCA -0.996; TCGA BRCA -0.223; TCGA COAD -0.873; TCGA ESCA -1.118; TCGA KIRC -1.169; TCGA KIRP -0.456; TCGA PAAD -1.346; TCGA PRAD -0.249; TCGA SARC -0.427; TCGA STAD -1.263 |
hsa-miR-181a-5p | NPEPPS | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; SARC | MirTarget; miRNATAP | TCGA BLCA -0.097; TCGA BRCA -0.173; TCGA COAD -0.076; TCGA ESCA -0.135; TCGA HNSC -0.282; TCGA KIRC -0.137; TCGA KIRP -0.101; TCGA LIHC -0.1; TCGA LUAD -0.082; TCGA LUSC -0.129; TCGA SARC -0.08 |
hsa-miR-181a-5p | WASL | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LIHC; LUAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.104; TCGA BRCA -0.117; TCGA CESC -0.124; TCGA COAD -0.116; TCGA ESCA -0.172; TCGA HNSC -0.183; TCGA KIRC -0.151; TCGA LGG -0.051; TCGA LIHC -0.133; TCGA LUAD -0.088; TCGA STAD -0.124; TCGA UCEC -0.054 |
hsa-miR-181a-5p | DENND4C | 9 cancers: BLCA; BRCA; ESCA; HNSC; KIRP; LIHC; OV; PRAD; SARC | MirTarget | TCGA BLCA -0.107; TCGA BRCA -0.275; TCGA ESCA -0.201; TCGA HNSC -0.162; TCGA KIRP -0.086; TCGA LIHC -0.101; TCGA OV -0.084; TCGA PRAD -0.107; TCGA SARC -0.136 |
hsa-miR-181a-5p | EXOC5 | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PRAD; SARC; THCA | MirTarget | TCGA BLCA -0.142; TCGA BRCA -0.134; TCGA COAD -0.08; TCGA ESCA -0.155; TCGA HNSC -0.143; TCGA LIHC -0.101; TCGA LUAD -0.174; TCGA LUSC -0.063; TCGA PRAD -0.059; TCGA SARC -0.068; TCGA THCA -0.103 |
hsa-miR-181a-5p | ZNF800 | 11 cancers: BLCA; BRCA; ESCA; HNSC; LGG; LIHC; LUAD; OV; PRAD; SARC; THCA | MirTarget; miRNATAP | TCGA BLCA -0.118; TCGA BRCA -0.22; TCGA ESCA -0.139; TCGA HNSC -0.205; TCGA LGG -0.072; TCGA LIHC -0.101; TCGA LUAD -0.107; TCGA OV -0.093; TCGA PRAD -0.086; TCGA SARC -0.058; TCGA THCA -0.119 |
hsa-miR-181a-5p | ARFGEF2 | 11 cancers: BLCA; BRCA; HNSC; KIRC; KIRP; LIHC; LUAD; OV; PRAD; SARC; THCA | MirTarget; miRNATAP | TCGA BLCA -0.094; TCGA BRCA -0.206; TCGA HNSC -0.231; TCGA KIRC -0.356; TCGA KIRP -0.17; TCGA LIHC -0.145; TCGA LUAD -0.097; TCGA OV -0.078; TCGA PRAD -0.116; TCGA SARC -0.105; TCGA THCA -0.108 |
hsa-miR-181a-5p | UBP1 | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; OV; PAAD; SARC | MirTarget; miRNATAP | TCGA BLCA -0.092; TCGA BRCA -0.123; TCGA COAD -0.067; TCGA ESCA -0.09; TCGA HNSC -0.088; TCGA KIRC -0.226; TCGA KIRP -0.138; TCGA OV -0.072; TCGA PAAD -0.069; TCGA SARC -0.169 |
hsa-miR-181a-5p | HIPK3 | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; SARC; THCA; STAD; UCEC | MirTarget; mirMAP | TCGA BLCA -0.496; TCGA BRCA -0.544; TCGA COAD -0.115; TCGA ESCA -0.325; TCGA HNSC -0.489; TCGA KIRC -0.375; TCGA SARC -0.336; TCGA THCA -0.305; TCGA STAD -0.199; TCGA UCEC -0.173 |
hsa-miR-181a-5p | LYRM1 | 10 cancers: BLCA; BRCA; HNSC; KIRC; LGG; LIHC; OV; PAAD; SARC; STAD | MirTarget | TCGA BLCA -0.078; TCGA BRCA -0.112; TCGA HNSC -0.196; TCGA KIRC -0.218; TCGA LGG -0.058; TCGA LIHC -0.267; TCGA OV -0.132; TCGA PAAD -0.173; TCGA SARC -0.209; TCGA STAD -0.126 |
hsa-miR-181a-5p | PPP2R3A | 9 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LUSC; STAD; UCEC | MirTarget | TCGA BLCA -0.215; TCGA BRCA -0.181; TCGA CESC -0.234; TCGA ESCA -0.396; TCGA HNSC -0.156; TCGA KIRC -0.322; TCGA LUSC -0.243; TCGA STAD -0.601; TCGA UCEC -0.127 |
hsa-miR-181a-5p | ZRANB1 | 10 cancers: BLCA; BRCA; CESC; HNSC; KIRC; LIHC; LUAD; PAAD; THCA; STAD | mirMAP | TCGA BLCA -0.141; TCGA BRCA -0.141; TCGA CESC -0.066; TCGA HNSC -0.098; TCGA KIRC -0.06; TCGA LIHC -0.102; TCGA LUAD -0.069; TCGA PAAD -0.107; TCGA THCA -0.059; TCGA STAD -0.095 |
hsa-miR-181a-5p | SNX1 | 9 cancers: BLCA; BRCA; CESC; ESCA; KIRC; LIHC; PRAD; SARC; STAD | mirMAP | TCGA BLCA -0.123; TCGA BRCA -0.159; TCGA CESC -0.128; TCGA ESCA -0.117; TCGA KIRC -0.052; TCGA LIHC -0.089; TCGA PRAD -0.058; TCGA SARC -0.103; TCGA STAD -0.197 |
hsa-miR-181a-5p | MYO9A | 10 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LIHC; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.355; TCGA BRCA -0.425; TCGA ESCA -0.299; TCGA HNSC -0.317; TCGA KIRC -0.189; TCGA LIHC -0.263; TCGA SARC -0.171; TCGA THCA -0.301; TCGA STAD -0.141; TCGA UCEC -0.135 |
hsa-miR-181a-5p | SEL1L | 9 cancers: BLCA; BRCA; COAD; HNSC; KIRC; LIHC; LUAD; PRAD; THCA | mirMAP; miRNATAP | TCGA BLCA -0.125; TCGA BRCA -0.203; TCGA COAD -0.109; TCGA HNSC -0.122; TCGA KIRC -0.206; TCGA LIHC -0.185; TCGA LUAD -0.18; TCGA PRAD -0.113; TCGA THCA -0.22 |
hsa-miR-181a-5p | AFF4 | 9 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LIHC; LUAD; SARC; THCA | mirMAP; miRNATAP | TCGA BLCA -0.167; TCGA BRCA -0.237; TCGA ESCA -0.151; TCGA HNSC -0.144; TCGA KIRC -0.089; TCGA LIHC -0.138; TCGA LUAD -0.06; TCGA SARC -0.091; TCGA THCA -0.061 |
hsa-miR-181a-5p | PIAS2 | 11 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; SARC; THCA | mirMAP | TCGA BLCA -0.172; TCGA BRCA -0.134; TCGA ESCA -0.146; TCGA HNSC -0.217; TCGA KIRC -0.212; TCGA LUAD -0.144; TCGA LUSC -0.227; TCGA PAAD -0.26; TCGA PRAD -0.139; TCGA SARC -0.115; TCGA THCA -0.209 |
hsa-miR-181a-5p | RAB18 | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; STAD | mirMAP | TCGA BLCA -0.057; TCGA BRCA -0.096; TCGA CESC -0.109; TCGA ESCA -0.202; TCGA HNSC -0.131; TCGA KIRC -0.139; TCGA LIHC -0.121; TCGA LUAD -0.139; TCGA LUSC -0.201; TCGA STAD -0.078 |
hsa-miR-181a-5p | MICALL1 | 9 cancers: BLCA; CESC; ESCA; KIRP; LUSC; PRAD; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.117; TCGA CESC -0.17; TCGA ESCA -0.375; TCGA KIRP -0.19; TCGA LUSC -0.445; TCGA PRAD -0.077; TCGA SARC -0.312; TCGA THCA -0.143; TCGA STAD -0.293 |
hsa-miR-181a-5p | DAAM1 | 9 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LIHC; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.323; TCGA BRCA -0.321; TCGA ESCA -0.323; TCGA HNSC -0.342; TCGA KIRC -0.451; TCGA LIHC -0.206; TCGA SARC -0.282; TCGA THCA -0.322; TCGA STAD -0.18 |
hsa-miR-181a-5p | ARHGAP5 | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LIHC; LUAD; PRAD; SARC | mirMAP | TCGA BLCA -0.205; TCGA BRCA -0.287; TCGA CESC -0.116; TCGA ESCA -0.141; TCGA HNSC -0.306; TCGA KIRC -0.283; TCGA LIHC -0.178; TCGA LUAD -0.078; TCGA PRAD -0.102; TCGA SARC -0.087 |
hsa-miR-181a-5p | AGFG2 | 12 cancers: BLCA; BRCA; HNSC; KIRC; KIRP; LGG; LIHC; OV; PAAD; PRAD; SARC; UCEC | mirMAP | TCGA BLCA -0.088; TCGA BRCA -0.062; TCGA HNSC -0.32; TCGA KIRC -0.186; TCGA KIRP -0.2; TCGA LGG -0.082; TCGA LIHC -0.186; TCGA OV -0.138; TCGA PAAD -0.179; TCGA PRAD -0.16; TCGA SARC -0.092; TCGA UCEC -0.096 |
hsa-miR-181a-5p | NCS1 | 10 cancers: BLCA; COAD; ESCA; KIRC; LUSC; PAAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.282; TCGA COAD -0.265; TCGA ESCA -0.325; TCGA KIRC -0.511; TCGA LUSC -0.418; TCGA PAAD -0.326; TCGA SARC -0.275; TCGA THCA -0.223; TCGA STAD -0.781; TCGA UCEC -0.131 |
hsa-miR-181a-5p | ASPA | 9 cancers: BLCA; BRCA; COAD; ESCA; LIHC; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.33; TCGA BRCA -0.528; TCGA COAD -0.318; TCGA ESCA -0.67; TCGA LIHC -0.353; TCGA SARC -0.378; TCGA THCA -0.49; TCGA STAD -0.677; TCGA UCEC -0.474 |
hsa-miR-181a-5p | RNF141 | 11 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; LIHC; OV; PRAD; SARC; THCA | mirMAP | TCGA BLCA -0.109; TCGA BRCA -0.161; TCGA CESC -0.145; TCGA COAD -0.149; TCGA HNSC -0.231; TCGA KIRC -0.233; TCGA LIHC -0.094; TCGA OV -0.065; TCGA PRAD -0.145; TCGA SARC -0.06; TCGA THCA -0.115 |
hsa-miR-181a-5p | LNPEP | 10 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LIHC; LUAD; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.247; TCGA BRCA -0.39; TCGA ESCA -0.216; TCGA HNSC -0.209; TCGA KIRC -0.418; TCGA LIHC -0.158; TCGA LUAD -0.129; TCGA SARC -0.2; TCGA THCA -0.281; TCGA STAD -0.147 |
hsa-miR-181a-5p | PCYOX1 | 12 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LIHC; OV; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.127; TCGA BRCA -0.301; TCGA ESCA -0.195; TCGA HNSC -0.287; TCGA KIRC -0.282; TCGA LIHC -0.271; TCGA OV -0.068; TCGA PAAD -0.121; TCGA PRAD -0.138; TCGA SARC -0.117; TCGA STAD -0.177; TCGA UCEC -0.063 |
hsa-miR-181a-5p | STX12 | 9 cancers: BLCA; BRCA; CESC; ESCA; KIRC; LIHC; PRAD; SARC; STAD | mirMAP | TCGA BLCA -0.124; TCGA BRCA -0.168; TCGA CESC -0.082; TCGA ESCA -0.275; TCGA KIRC -0.053; TCGA LIHC -0.145; TCGA PRAD -0.053; TCGA SARC -0.128; TCGA STAD -0.251 |
hsa-miR-181a-5p | NECAB1 | 9 cancers: BLCA; BRCA; COAD; ESCA; KIRC; PRAD; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.389; TCGA BRCA -0.25; TCGA COAD -0.403; TCGA ESCA -0.442; TCGA KIRC -0.237; TCGA PRAD -0.165; TCGA SARC -0.521; TCGA THCA -0.351; TCGA STAD -0.757 |
hsa-miR-181a-5p | GAN | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; PRAD; THCA | mirMAP | TCGA BLCA -0.202; TCGA BRCA -0.152; TCGA CESC -0.269; TCGA COAD -0.171; TCGA ESCA -0.203; TCGA HNSC -0.545; TCGA KIRC -0.172; TCGA LIHC -0.19; TCGA LUAD -0.25; TCGA LUSC -0.562; TCGA PRAD -0.117; TCGA THCA -0.232 |
hsa-miR-181a-5p | FAM63B | 9 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; KIRP; SARC; THCA; UCEC | mirMAP | TCGA BLCA -0.287; TCGA BRCA -0.399; TCGA ESCA -0.139; TCGA HNSC -0.291; TCGA KIRC -0.506; TCGA KIRP -0.326; TCGA SARC -0.294; TCGA THCA -0.291; TCGA UCEC -0.19 |
hsa-miR-181a-5p | IL6ST | 9 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LIHC; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.645; TCGA BRCA -0.838; TCGA ESCA -0.221; TCGA HNSC -0.541; TCGA KIRC -0.296; TCGA LIHC -0.323; TCGA SARC -0.283; TCGA THCA -0.381; TCGA STAD -0.165 |
hsa-miR-181a-5p | CLOCK | 10 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LIHC; LUAD; SARC; THCA; UCEC | mirMAP | TCGA BLCA -0.244; TCGA BRCA -0.336; TCGA ESCA -0.188; TCGA HNSC -0.346; TCGA KIRC -0.354; TCGA LIHC -0.201; TCGA LUAD -0.111; TCGA SARC -0.146; TCGA THCA -0.316; TCGA UCEC -0.093 |
hsa-miR-181a-5p | FBXO21 | 10 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; STAD | mirMAP | TCGA BLCA -0.089; TCGA BRCA -0.109; TCGA ESCA -0.145; TCGA HNSC -0.149; TCGA KIRC -0.337; TCGA LUAD -0.127; TCGA LUSC -0.056; TCGA PAAD -0.151; TCGA PRAD -0.062; TCGA STAD -0.081 |
hsa-miR-181a-5p | PPM1B | 9 cancers: BLCA; BRCA; COAD; HNSC; KIRC; LIHC; LUAD; PRAD; SARC | mirMAP; miRNATAP | TCGA BLCA -0.063; TCGA BRCA -0.107; TCGA COAD -0.084; TCGA HNSC -0.14; TCGA KIRC -0.21; TCGA LIHC -0.129; TCGA LUAD -0.11; TCGA PRAD -0.115; TCGA SARC -0.054 |
hsa-miR-181a-5p | NAA30 | 12 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; PAAD; PRAD; THCA; STAD | mirMAP | TCGA BLCA -0.111; TCGA BRCA -0.159; TCGA ESCA -0.151; TCGA HNSC -0.172; TCGA KIRC -0.219; TCGA LIHC -0.111; TCGA LUAD -0.19; TCGA LUSC -0.185; TCGA PAAD -0.088; TCGA PRAD -0.094; TCGA THCA -0.081; TCGA STAD -0.072 |
hsa-miR-181a-5p | RPRD1A | 11 cancers: BLCA; BRCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; PRAD; SARC; STAD | mirMAP | TCGA BLCA -0.158; TCGA BRCA -0.136; TCGA HNSC -0.175; TCGA KIRC -0.139; TCGA KIRP -0.092; TCGA LUAD -0.178; TCGA LUSC -0.146; TCGA PAAD -0.246; TCGA PRAD -0.128; TCGA SARC -0.15; TCGA STAD -0.136 |
hsa-miR-181a-5p | EXOC6B | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LUSC; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.247; TCGA BRCA -0.354; TCGA COAD -0.135; TCGA ESCA -0.245; TCGA HNSC -0.392; TCGA KIRC -0.339; TCGA LUSC -0.273; TCGA SARC -0.234; TCGA THCA -0.212; TCGA STAD -0.17 |
hsa-miR-181a-5p | RHBDD1 | 9 cancers: BLCA; BRCA; KIRC; LIHC; LUAD; PRAD; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.148; TCGA BRCA -0.154; TCGA KIRC -0.117; TCGA LIHC -0.1; TCGA LUAD -0.115; TCGA PRAD -0.106; TCGA SARC -0.15; TCGA THCA -0.078; TCGA STAD -0.071 |
hsa-miR-181a-5p | DIXDC1 | 11 cancers: BLCA; BRCA; COAD; ESCA; LIHC; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.55; TCGA BRCA -0.335; TCGA COAD -0.198; TCGA ESCA -0.424; TCGA LIHC -0.156; TCGA PAAD -0.165; TCGA PRAD -0.142; TCGA SARC -0.372; TCGA THCA -0.233; TCGA STAD -0.639; TCGA UCEC -0.144 |
hsa-miR-181a-5p | TMEM56 | 9 cancers: BLCA; BRCA; HNSC; KIRC; LIHC; PAAD; PRAD; SARC; STAD | mirMAP | TCGA BLCA -0.283; TCGA BRCA -0.288; TCGA HNSC -0.333; TCGA KIRC -0.579; TCGA LIHC -0.543; TCGA PAAD -0.384; TCGA PRAD -0.15; TCGA SARC -0.538; TCGA STAD -0.222 |
hsa-miR-181a-5p | C18orf25 | 9 cancers: BLCA; BRCA; CESC; HNSC; KIRP; LIHC; LUAD; LUSC; SARC | mirMAP | TCGA BLCA -0.067; TCGA BRCA -0.148; TCGA CESC -0.101; TCGA HNSC -0.212; TCGA KIRP -0.108; TCGA LIHC -0.069; TCGA LUAD -0.1; TCGA LUSC -0.065; TCGA SARC -0.232 |
hsa-miR-181a-5p | RMND5A | 9 cancers: BLCA; BRCA; CESC; HNSC; KIRC; LIHC; LUAD; LUSC; PRAD | mirMAP; miRNATAP | TCGA BLCA -0.078; TCGA BRCA -0.157; TCGA CESC -0.097; TCGA HNSC -0.316; TCGA KIRC -0.096; TCGA LIHC -0.21; TCGA LUAD -0.145; TCGA LUSC -0.308; TCGA PRAD -0.075 |
hsa-miR-181a-5p | CACNA2D1 | 9 cancers: BLCA; BRCA; ESCA; HNSC; LUAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.884; TCGA BRCA -0.714; TCGA ESCA -0.495; TCGA HNSC -0.48; TCGA LUAD -0.32; TCGA SARC -0.341; TCGA THCA -0.635; TCGA STAD -0.544; TCGA UCEC -0.447 |
hsa-miR-181a-5p | FBXO32 | 12 cancers: BLCA; CESC; COAD; ESCA; KIRC; LUAD; LUSC; OV; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.249; TCGA CESC -0.215; TCGA COAD -0.199; TCGA ESCA -0.367; TCGA KIRC -0.139; TCGA LUAD -0.248; TCGA LUSC -0.47; TCGA OV -0.254; TCGA PRAD -0.235; TCGA SARC -0.839; TCGA STAD -0.625; TCGA UCEC -0.273 |
hsa-miR-181a-5p | PRKAA2 | 10 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LUAD; PRAD; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.653; TCGA BRCA -0.426; TCGA ESCA -0.586; TCGA HNSC -0.743; TCGA KIRC -0.244; TCGA LUAD -0.311; TCGA PRAD -0.103; TCGA SARC -0.626; TCGA THCA -0.199; TCGA STAD -1.088 |
hsa-miR-181a-5p | PDLIM5 | 9 cancers: BLCA; BRCA; HNSC; KIRC; LIHC; LUSC; SARC; THCA; UCEC | mirMAP | TCGA BLCA -0.23; TCGA BRCA -0.312; TCGA HNSC -0.301; TCGA KIRC -0.13; TCGA LIHC -0.173; TCGA LUSC -0.149; TCGA SARC -0.532; TCGA THCA -0.175; TCGA UCEC -0.16 |
hsa-miR-181a-5p | HIPK1 | 9 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LIHC; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.184; TCGA BRCA -0.202; TCGA ESCA -0.155; TCGA HNSC -0.251; TCGA KIRC -0.14; TCGA LIHC -0.128; TCGA SARC -0.099; TCGA THCA -0.095; TCGA STAD -0.08 |
hsa-miR-181a-5p | PPM1L | 12 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; PAAD; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.528; TCGA BRCA -0.293; TCGA ESCA -0.435; TCGA HNSC -0.727; TCGA KIRC -0.399; TCGA LIHC -0.382; TCGA LUAD -0.177; TCGA LUSC -0.382; TCGA PAAD -0.24; TCGA SARC -0.318; TCGA THCA -0.727; TCGA STAD -0.363 |
hsa-miR-181a-5p | PTCHD1 | 10 cancers: BLCA; COAD; ESCA; HNSC; KIRP; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.755; TCGA COAD -0.652; TCGA ESCA -0.446; TCGA HNSC -0.558; TCGA KIRP -0.469; TCGA PAAD -0.455; TCGA PRAD -0.378; TCGA SARC -0.935; TCGA STAD -1.34; TCGA UCEC -0.251 |
hsa-miR-181a-5p | RIC3 | 10 cancers: BLCA; BRCA; COAD; ESCA; KIRC; KIRP; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.515; TCGA BRCA -0.361; TCGA COAD -0.363; TCGA ESCA -0.548; TCGA KIRC -1.21; TCGA KIRP -0.361; TCGA PAAD -0.745; TCGA PRAD -0.237; TCGA STAD -0.781; TCGA UCEC -0.522 |
hsa-miR-181a-5p | MAP3K2 | 9 cancers: BLCA; BRCA; HNSC; KIRC; LIHC; LUAD; LUSC; SARC; THCA | mirMAP | TCGA BLCA -0.139; TCGA BRCA -0.248; TCGA HNSC -0.125; TCGA KIRC -0.124; TCGA LIHC -0.223; TCGA LUAD -0.164; TCGA LUSC -0.084; TCGA SARC -0.136; TCGA THCA -0.208 |
hsa-miR-181a-5p | SMAD2 | 10 cancers: BLCA; BRCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; PRAD; STAD | mirMAP | TCGA BLCA -0.113; TCGA BRCA -0.115; TCGA HNSC -0.124; TCGA KIRC -0.111; TCGA KIRP -0.075; TCGA LUAD -0.101; TCGA LUSC -0.11; TCGA PAAD -0.066; TCGA PRAD -0.099; TCGA STAD -0.082 |
hsa-miR-181a-5p | YIPF6 | 9 cancers: BLCA; BRCA; HNSC; KIRC; LIHC; LUAD; LUSC; SARC; THCA | mirMAP | TCGA BLCA -0.065; TCGA BRCA -0.172; TCGA HNSC -0.176; TCGA KIRC -0.224; TCGA LIHC -0.208; TCGA LUAD -0.196; TCGA LUSC -0.121; TCGA SARC -0.085; TCGA THCA -0.208 |
hsa-miR-181a-5p | LYRM7 | 10 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LIHC; PAAD; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.175; TCGA BRCA -0.165; TCGA ESCA -0.123; TCGA HNSC -0.179; TCGA KIRC -0.092; TCGA LIHC -0.198; TCGA PAAD -0.25; TCGA SARC -0.064; TCGA THCA -0.216; TCGA STAD -0.085 |
hsa-miR-181a-5p | AKR1C1 | 10 cancers: BLCA; CESC; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; THCA; UCEC | mirMAP | TCGA BLCA -0.24; TCGA CESC -0.531; TCGA ESCA -0.979; TCGA HNSC -0.779; TCGA KIRC -0.23; TCGA LIHC -0.597; TCGA LUAD -0.708; TCGA LUSC -1.623; TCGA THCA -0.618; TCGA UCEC -0.432 |
hsa-miR-181a-5p | NHLRC2 | 9 cancers: BLCA; BRCA; HNSC; KIRC; LIHC; LUAD; PRAD; SARC; THCA | mirMAP | TCGA BLCA -0.198; TCGA BRCA -0.336; TCGA HNSC -0.256; TCGA KIRC -0.364; TCGA LIHC -0.129; TCGA LUAD -0.212; TCGA PRAD -0.131; TCGA SARC -0.096; TCGA THCA -0.3 |
hsa-miR-181a-5p | MAN1A2 | 10 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LIHC; LUAD; PRAD; SARC; THCA | mirMAP | TCGA BLCA -0.129; TCGA BRCA -0.223; TCGA ESCA -0.12; TCGA HNSC -0.245; TCGA KIRC -0.235; TCGA LIHC -0.165; TCGA LUAD -0.179; TCGA PRAD -0.105; TCGA SARC -0.118; TCGA THCA -0.137 |
hsa-miR-181a-5p | PEG3 | 11 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; KIRP; PAAD; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.563; TCGA BRCA -0.256; TCGA ESCA -0.513; TCGA HNSC -0.367; TCGA KIRC -0.826; TCGA KIRP -0.577; TCGA PAAD -0.629; TCGA PRAD -0.201; TCGA THCA -0.373; TCGA STAD -0.708; TCGA UCEC -0.398 |
hsa-miR-181a-5p | ZXDB | 9 cancers: BLCA; BRCA; HNSC; KIRC; LUAD; PRAD; SARC; THCA; UCEC | mirMAP | TCGA BLCA -0.147; TCGA BRCA -0.153; TCGA HNSC -0.098; TCGA KIRC -0.085; TCGA LUAD -0.137; TCGA PRAD -0.111; TCGA SARC -0.096; TCGA THCA -0.131; TCGA UCEC -0.068 |
hsa-miR-181a-5p | CAPZA2 | 9 cancers: BLCA; BRCA; COAD; HNSC; KIRC; LUAD; PRAD; SARC; UCEC | mirMAP | TCGA BLCA -0.17; TCGA BRCA -0.095; TCGA COAD -0.129; TCGA HNSC -0.062; TCGA KIRC -0.125; TCGA LUAD -0.168; TCGA PRAD -0.067; TCGA SARC -0.146; TCGA UCEC -0.083 |
hsa-miR-181a-5p | PPP1CB | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LIHC; LUAD; LUSC; OV; PRAD; SARC; STAD | mirMAP; miRNATAP | TCGA BLCA -0.209; TCGA BRCA -0.083; TCGA CESC -0.114; TCGA COAD -0.079; TCGA ESCA -0.171; TCGA HNSC -0.219; TCGA KIRC -0.14; TCGA LGG -0.078; TCGA LIHC -0.089; TCGA LUAD -0.169; TCGA LUSC -0.191; TCGA OV -0.085; TCGA PRAD -0.163; TCGA SARC -0.188; TCGA STAD -0.192 |
hsa-miR-181a-5p | CPEB4 | 9 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LIHC; SARC; THCA; STAD | miRNATAP | TCGA BLCA -0.341; TCGA BRCA -0.33; TCGA ESCA -0.181; TCGA HNSC -0.379; TCGA KIRC -0.408; TCGA LIHC -0.289; TCGA SARC -0.325; TCGA THCA -0.254; TCGA STAD -0.177 |
hsa-miR-181a-5p | TNRC6B | 10 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; KIRP; LUSC; PRAD; SARC; THCA | miRNATAP | TCGA BLCA -0.146; TCGA BRCA -0.158; TCGA ESCA -0.105; TCGA HNSC -0.143; TCGA KIRC -0.116; TCGA KIRP -0.15; TCGA LUSC -0.114; TCGA PRAD -0.094; TCGA SARC -0.064; TCGA THCA -0.139 |
hsa-miR-181a-5p | C2orf69 | 13 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; OV; PRAD; SARC; THCA | miRNATAP | TCGA BLCA -0.106; TCGA BRCA -0.205; TCGA COAD -0.148; TCGA ESCA -0.118; TCGA HNSC -0.202; TCGA KIRC -0.212; TCGA LIHC -0.272; TCGA LUAD -0.263; TCGA LUSC -0.182; TCGA OV -0.103; TCGA PRAD -0.094; TCGA SARC -0.075; TCGA THCA -0.196 |
hsa-miR-181a-5p | CALM1 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; PAAD; SARC; THCA; STAD | miRNATAP | TCGA BLCA -0.143; TCGA BRCA -0.082; TCGA CESC -0.11; TCGA COAD -0.099; TCGA ESCA -0.255; TCGA HNSC -0.148; TCGA KIRC -0.065; TCGA PAAD -0.287; TCGA SARC -0.081; TCGA THCA -0.068; TCGA STAD -0.285 |
hsa-miR-181a-5p | ADCY9 | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LIHC; PAAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.271; TCGA BRCA -0.222; TCGA COAD -0.111; TCGA ESCA -0.141; TCGA HNSC -0.117; TCGA LIHC -0.161; TCGA PAAD -0.257; TCGA SARC -0.264; TCGA THCA -0.197; TCGA STAD -0.315; TCGA UCEC -0.315 |
hsa-miR-181a-5p | KLF3 | 11 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PRAD; THCA | miRNATAP | TCGA BLCA -0.114; TCGA BRCA -0.182; TCGA CESC -0.137; TCGA HNSC -0.301; TCGA KIRC -0.079; TCGA KIRP -0.08; TCGA LIHC -0.097; TCGA LUAD -0.098; TCGA LUSC -0.129; TCGA PRAD -0.121; TCGA THCA -0.057 |
hsa-miR-181a-5p | MTMR9 | 9 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; KIRP; LUAD; SARC; THCA | miRNATAP | TCGA BLCA -0.153; TCGA BRCA -0.183; TCGA ESCA -0.15; TCGA HNSC -0.12; TCGA KIRC -0.071; TCGA KIRP -0.113; TCGA LUAD -0.092; TCGA SARC -0.07; TCGA THCA -0.057 |
hsa-miR-181a-5p | PPP1R12B | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.729; TCGA BRCA -0.266; TCGA COAD -0.337; TCGA ESCA -0.595; TCGA HNSC -0.196; TCGA KIRC -0.241; TCGA SARC -1.16; TCGA THCA -0.195; TCGA STAD -0.852; TCGA UCEC -0.337 |
hsa-miR-181a-5p | CACNB2 | 10 cancers: BLCA; BRCA; COAD; ESCA; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.706; TCGA BRCA -0.337; TCGA COAD -0.278; TCGA ESCA -0.509; TCGA PAAD -1.059; TCGA PRAD -0.135; TCGA SARC -0.485; TCGA THCA -0.143; TCGA STAD -0.612; TCGA UCEC -0.254 |
hsa-miR-181a-5p | AR | 9 cancers: BLCA; BRCA; ESCA; HNSC; LIHC; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.555; TCGA BRCA -0.74; TCGA ESCA -0.692; TCGA HNSC -0.529; TCGA LIHC -1.229; TCGA SARC -0.715; TCGA THCA -0.685; TCGA STAD -0.696; TCGA UCEC -0.336 |
hsa-miR-181a-5p | LPP | 13 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PRAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.592; TCGA BRCA -0.349; TCGA COAD -0.262; TCGA ESCA -0.224; TCGA HNSC -0.263; TCGA KIRC -0.16; TCGA LUAD -0.202; TCGA LUSC -0.119; TCGA PRAD -0.198; TCGA SARC -0.623; TCGA THCA -0.476; TCGA STAD -0.354; TCGA UCEC -0.263 |
hsa-miR-181a-5p | SYPL2 | 10 cancers: BLCA; BRCA; COAD; ESCA; KIRC; LIHC; PRAD; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.519; TCGA BRCA -0.159; TCGA COAD -0.323; TCGA ESCA -0.34; TCGA KIRC -0.572; TCGA LIHC -0.671; TCGA PRAD -0.215; TCGA THCA -0.342; TCGA STAD -0.62; TCGA UCEC -0.257 |
hsa-miR-181a-5p | MFSD6 | 9 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; PRAD; SARC | miRNATAP | TCGA BLCA -0.15; TCGA BRCA -0.18; TCGA CESC -0.139; TCGA ESCA -0.219; TCGA HNSC -0.474; TCGA KIRC -0.195; TCGA KIRP -0.232; TCGA PRAD -0.089; TCGA SARC -0.224 |
hsa-miR-181a-5p | PPARA | 13 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LIHC; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.299; TCGA BRCA -0.248; TCGA ESCA -0.17; TCGA HNSC -0.272; TCGA KIRC -0.351; TCGA LIHC -0.427; TCGA LUSC -0.189; TCGA PAAD -0.118; TCGA PRAD -0.13; TCGA SARC -0.133; TCGA THCA -0.213; TCGA STAD -0.234; TCGA UCEC -0.11 |
hsa-miR-181a-5p | MECP2 | 11 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; KIRP; OV; PAAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.114; TCGA BRCA -0.086; TCGA ESCA -0.127; TCGA HNSC -0.062; TCGA KIRC -0.117; TCGA KIRP -0.078; TCGA OV -0.085; TCGA PAAD -0.128; TCGA SARC -0.12; TCGA STAD -0.146; TCGA UCEC -0.06 |
hsa-miR-181a-5p | NR1D2 | 9 cancers: BLCA; BRCA; CESC; HNSC; KIRC; LIHC; OV; PRAD; SARC | miRNATAP | TCGA BLCA -0.106; TCGA BRCA -0.208; TCGA CESC -0.137; TCGA HNSC -0.186; TCGA KIRC -0.285; TCGA LIHC -0.186; TCGA OV -0.092; TCGA PRAD -0.111; TCGA SARC -0.13 |
hsa-miR-181a-5p | RGMA | 9 cancers: BLCA; ESCA; HNSC; LGG; LUSC; PAAD; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.594; TCGA ESCA -0.777; TCGA HNSC -0.372; TCGA LGG -0.188; TCGA LUSC -0.929; TCGA PAAD -0.392; TCGA THCA -0.315; TCGA STAD -1.002; TCGA UCEC -0.363 |
hsa-miR-181a-5p | PRIMA1 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; PAAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.987; TCGA BRCA -0.202; TCGA CESC -0.427; TCGA COAD -0.613; TCGA ESCA -0.76; TCGA HNSC -0.271; TCGA PAAD -0.527; TCGA SARC -0.752; TCGA THCA -1.213; TCGA STAD -1.653; TCGA UCEC -0.319 |
hsa-miR-181a-5p | RTF1 | 9 cancers: BLCA; BRCA; CESC; HNSC; KIRC; LUAD; LUSC; SARC; THCA | miRNATAP | TCGA BLCA -0.112; TCGA BRCA -0.125; TCGA CESC -0.092; TCGA HNSC -0.062; TCGA KIRC -0.085; TCGA LUAD -0.062; TCGA LUSC -0.076; TCGA SARC -0.1; TCGA THCA -0.096 |
hsa-miR-181a-5p | CAMK2G | 10 cancers: BLCA; CESC; COAD; HNSC; KIRC; KIRP; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.239; TCGA CESC -0.176; TCGA COAD -0.103; TCGA HNSC -0.106; TCGA KIRC -0.095; TCGA KIRP -0.135; TCGA SARC -0.286; TCGA THCA -0.093; TCGA STAD -0.28; TCGA UCEC -0.099 |
hsa-miR-181a-5p | PRKAG2 | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; PAAD; SARC; THCA; STAD | miRNATAP | TCGA BLCA -0.268; TCGA BRCA -0.156; TCGA COAD -0.109; TCGA ESCA -0.263; TCGA HNSC -0.093; TCGA KIRC -0.254; TCGA LIHC -0.407; TCGA LUAD -0.084; TCGA PAAD -0.156; TCGA SARC -0.493; TCGA THCA -0.124; TCGA STAD -0.34 |
hsa-miR-181a-5p | FAM102A | 9 cancers: BLCA; HNSC; KIRC; KIRP; LUSC; OV; SARC; THCA; STAD | miRNATAP | TCGA BLCA -0.105; TCGA HNSC -0.282; TCGA KIRC -0.508; TCGA KIRP -0.167; TCGA LUSC -0.392; TCGA OV -0.109; TCGA SARC -0.092; TCGA THCA -0.169; TCGA STAD -0.106 |
hsa-miR-181a-5p | ETF1 | 10 cancers: BLCA; BRCA; HNSC; KIRC; LIHC; LUAD; PRAD; SARC; THCA; STAD | miRNATAP | TCGA BLCA -0.126; TCGA BRCA -0.093; TCGA HNSC -0.099; TCGA KIRC -0.096; TCGA LIHC -0.064; TCGA LUAD -0.079; TCGA PRAD -0.058; TCGA SARC -0.113; TCGA THCA -0.096; TCGA STAD -0.063 |
hsa-miR-181a-5p | WSCD2 | 10 cancers: BLCA; COAD; ESCA; KIRC; LUSC; PAAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.504; TCGA COAD -0.451; TCGA ESCA -1.141; TCGA KIRC -1.346; TCGA LUSC -0.387; TCGA PAAD -0.479; TCGA SARC -0.424; TCGA THCA -1.487; TCGA STAD -1.395; TCGA UCEC -0.499 |
hsa-miR-181a-5p | ATP8B1 | 12 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PRAD; SARC; THCA | miRNATAP | TCGA BLCA -0.155; TCGA BRCA -0.324; TCGA CESC -0.226; TCGA ESCA -0.264; TCGA HNSC -0.497; TCGA KIRC -0.46; TCGA KIRP -0.223; TCGA LUAD -0.237; TCGA LUSC -0.3; TCGA PRAD -0.167; TCGA SARC -0.364; TCGA THCA -0.158 |
hsa-miR-181a-5p | CDC73 | 9 cancers: BLCA; BRCA; HNSC; LIHC; LUAD; LUSC; PRAD; SARC; THCA | miRNATAP | TCGA BLCA -0.115; TCGA BRCA -0.055; TCGA HNSC -0.064; TCGA LIHC -0.082; TCGA LUAD -0.119; TCGA LUSC -0.063; TCGA PRAD -0.076; TCGA SARC -0.093; TCGA THCA -0.126 |
hsa-miR-181a-5p | MAPT | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.523; TCGA BRCA -0.433; TCGA COAD -0.343; TCGA ESCA -0.751; TCGA HNSC -0.585; TCGA PAAD -0.908; TCGA PRAD -0.155; TCGA SARC -0.261; TCGA THCA -0.339; TCGA STAD -0.849; TCGA UCEC -0.168 |
hsa-miR-181a-5p | ARRDC4 | 9 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; PAAD; THCA; STAD | miRNATAP | TCGA BLCA -0.255; TCGA BRCA -0.248; TCGA CESC -0.323; TCGA COAD -0.167; TCGA ESCA -0.398; TCGA KIRC -0.25; TCGA PAAD -0.241; TCGA THCA -0.16; TCGA STAD -0.417 |
hsa-miR-181a-5p | EPS15 | 9 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LIHC; PRAD; THCA | miRNATAP | TCGA BLCA -0.052; TCGA BRCA -0.156; TCGA CESC -0.077; TCGA ESCA -0.104; TCGA HNSC -0.068; TCGA KIRC -0.074; TCGA LIHC -0.115; TCGA PRAD -0.058; TCGA THCA -0.06 |
hsa-miR-181a-5p | PDCD6IP | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; OV; PRAD | miRNATAP | TCGA BLCA -0.058; TCGA BRCA -0.143; TCGA CESC -0.084; TCGA COAD -0.062; TCGA ESCA -0.135; TCGA HNSC -0.186; TCGA KIRC -0.315; TCGA LIHC -0.113; TCGA OV -0.06; TCGA PRAD -0.079 |
hsa-miR-181a-5p | BCL6 | 9 cancers: BLCA; BRCA; CESC; HNSC; LIHC; LUSC; OV; SARC; THCA | miRNATAP | TCGA BLCA -0.238; TCGA BRCA -0.253; TCGA CESC -0.194; TCGA HNSC -0.274; TCGA LIHC -0.166; TCGA LUSC -0.104; TCGA OV -0.202; TCGA SARC -0.184; TCGA THCA -0.121 |
hsa-miR-181a-5p | AFTPH | 9 cancers: BRCA; CESC; HNSC; KIRC; LUAD; LUSC; OV; PRAD; SARC | miRNAWalker2 validate; MirTarget; miRNATAP | TCGA BRCA -0.114; TCGA CESC -0.09; TCGA HNSC -0.176; TCGA KIRC -0.106; TCGA LUAD -0.104; TCGA LUSC -0.132; TCGA OV -0.065; TCGA PRAD -0.078; TCGA SARC -0.081 |
hsa-miR-181a-5p | PROSC | 9 cancers: BRCA; ESCA; HNSC; KIRC; LIHC; LUAD; PRAD; SARC; STAD | miRNAWalker2 validate | TCGA BRCA -0.116; TCGA ESCA -0.172; TCGA HNSC -0.131; TCGA KIRC -0.18; TCGA LIHC -0.212; TCGA LUAD -0.13; TCGA PRAD -0.105; TCGA SARC -0.084; TCGA STAD -0.11 |
hsa-miR-181a-5p | STAG2 | 9 cancers: BRCA; COAD; HNSC; KIRC; LGG; LIHC; LUAD; OV; PRAD | miRNAWalker2 validate | TCGA BRCA -0.204; TCGA COAD -0.09; TCGA HNSC -0.151; TCGA KIRC -0.145; TCGA LGG -0.058; TCGA LIHC -0.136; TCGA LUAD -0.09; TCGA OV -0.075; TCGA PRAD -0.06 |
hsa-miR-181a-5p | TAF2 | 10 cancers: BRCA; CESC; HNSC; KIRC; KIRP; LUAD; LUSC; OV; SARC; THCA | miRNAWalker2 validate | TCGA BRCA -0.127; TCGA CESC -0.132; TCGA HNSC -0.132; TCGA KIRC -0.1; TCGA KIRP -0.11; TCGA LUAD -0.233; TCGA LUSC -0.228; TCGA OV -0.141; TCGA SARC -0.091; TCGA THCA -0.148 |
hsa-miR-181a-5p | TMLHE | 9 cancers: BRCA; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; THCA; STAD | MirTarget | TCGA BRCA -0.129; TCGA ESCA -0.107; TCGA HNSC -0.118; TCGA KIRC -0.212; TCGA LIHC -0.219; TCGA LUAD -0.163; TCGA LUSC -0.137; TCGA THCA -0.142; TCGA STAD -0.105 |
hsa-miR-181a-5p | MAPK1IP1L | 9 cancers: BRCA; COAD; HNSC; KIRC; LGG; LUAD; PRAD; SARC; THCA | MirTarget | TCGA BRCA -0.073; TCGA COAD -0.143; TCGA HNSC -0.095; TCGA KIRC -0.171; TCGA LGG -0.103; TCGA LUAD -0.084; TCGA PRAD -0.058; TCGA SARC -0.054; TCGA THCA -0.065 |
hsa-miR-181a-5p | USP33 | 10 cancers: BRCA; CESC; KIRC; KIRP; LIHC; LUAD; OV; PRAD; SARC; THCA | MirTarget; miRNATAP | TCGA BRCA -0.243; TCGA CESC -0.06; TCGA KIRC -0.089; TCGA KIRP -0.072; TCGA LIHC -0.088; TCGA LUAD -0.089; TCGA OV -0.071; TCGA PRAD -0.066; TCGA SARC -0.061; TCGA THCA -0.064 |
hsa-miR-181a-5p | ERLIN2 | 10 cancers: BRCA; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; PRAD; THCA; STAD | MirTarget | TCGA BRCA -0.159; TCGA ESCA -0.159; TCGA HNSC -0.153; TCGA KIRC -0.238; TCGA LIHC -0.209; TCGA LUAD -0.229; TCGA LUSC -0.246; TCGA PRAD -0.149; TCGA THCA -0.101; TCGA STAD -0.1 |
hsa-miR-181a-5p | NAA50 | 10 cancers: BRCA; COAD; HNSC; KIRC; LGG; LUAD; LUSC; OV; SARC; THCA | MirTarget; miRNATAP | TCGA BRCA -0.073; TCGA COAD -0.101; TCGA HNSC -0.167; TCGA KIRC -0.307; TCGA LGG -0.068; TCGA LUAD -0.262; TCGA LUSC -0.305; TCGA OV -0.076; TCGA SARC -0.072; TCGA THCA -0.073 |
hsa-miR-181a-5p | SFMBT1 | 9 cancers: BRCA; HNSC; KIRC; LIHC; LUAD; LUSC; OV; SARC; THCA | MirTarget; miRNATAP | TCGA BRCA -0.113; TCGA HNSC -0.104; TCGA KIRC -0.183; TCGA LIHC -0.116; TCGA LUAD -0.083; TCGA LUSC -0.079; TCGA OV -0.154; TCGA SARC -0.181; TCGA THCA -0.279 |
hsa-miR-181a-5p | CSNK2A2 | 9 cancers: BRCA; HNSC; LGG; LIHC; LUAD; LUSC; PRAD; STAD; UCEC | mirMAP | TCGA BRCA -0.079; TCGA HNSC -0.091; TCGA LGG -0.057; TCGA LIHC -0.063; TCGA LUAD -0.054; TCGA LUSC -0.251; TCGA PRAD -0.1; TCGA STAD -0.083; TCGA UCEC -0.053 |
hsa-miR-181a-5p | TMED5 | 9 cancers: BRCA; HNSC; KIRC; LGG; LIHC; LUAD; PRAD; SARC; THCA | mirMAP; miRNATAP | TCGA BRCA -0.219; TCGA HNSC -0.083; TCGA KIRC -0.206; TCGA LGG -0.066; TCGA LIHC -0.273; TCGA LUAD -0.096; TCGA PRAD -0.155; TCGA SARC -0.142; TCGA THCA -0.075 |
hsa-miR-181a-5p | FAM199X | 10 cancers: BRCA; HNSC; KIRC; LIHC; LUAD; LUSC; OV; PRAD; SARC; THCA | mirMAP | TCGA BRCA -0.153; TCGA HNSC -0.213; TCGA KIRC -0.348; TCGA LIHC -0.125; TCGA LUAD -0.201; TCGA LUSC -0.204; TCGA OV -0.092; TCGA PRAD -0.093; TCGA SARC -0.1; TCGA THCA -0.207 |
hsa-miR-181a-5p | SSR1 | 9 cancers: BRCA; COAD; KIRC; LGG; LUAD; LUSC; PAAD; PRAD; THCA | mirMAP | TCGA BRCA -0.07; TCGA COAD -0.129; TCGA KIRC -0.053; TCGA LGG -0.091; TCGA LUAD -0.184; TCGA LUSC -0.178; TCGA PAAD -0.138; TCGA PRAD -0.151; TCGA THCA -0.105 |
hsa-miR-181a-5p | TRMT5 | 10 cancers: BRCA; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; PRAD; THCA | mirMAP | TCGA BRCA -0.056; TCGA COAD -0.079; TCGA ESCA -0.098; TCGA HNSC -0.07; TCGA KIRC -0.101; TCGA LIHC -0.078; TCGA LUAD -0.099; TCGA LUSC -0.234; TCGA PRAD -0.093; TCGA THCA -0.06 |
hsa-miR-181a-5p | LIN7C | 10 cancers: BRCA; COAD; HNSC; KIRC; LIHC; LUAD; LUSC; OV; PRAD; SARC | mirMAP; miRNATAP | TCGA BRCA -0.119; TCGA COAD -0.108; TCGA HNSC -0.084; TCGA KIRC -0.08; TCGA LIHC -0.179; TCGA LUAD -0.11; TCGA LUSC -0.15; TCGA OV -0.064; TCGA PRAD -0.157; TCGA SARC -0.067 |
hsa-miR-181a-5p | CXADR | 9 cancers: BRCA; HNSC; KIRC; KIRP; LIHC; LUSC; PRAD; SARC; THCA | miRNATAP | TCGA BRCA -0.181; TCGA HNSC -0.464; TCGA KIRC -0.523; TCGA KIRP -0.328; TCGA LIHC -0.244; TCGA LUSC -0.288; TCGA PRAD -0.168; TCGA SARC -0.368; TCGA THCA -0.132 |
hsa-miR-181a-5p | GPD2 | 9 cancers: BRCA; CESC; COAD; HNSC; KIRC; LUAD; LUSC; OV; PRAD | miRNATAP | TCGA BRCA -0.22; TCGA CESC -0.157; TCGA COAD -0.087; TCGA HNSC -0.305; TCGA KIRC -0.193; TCGA LUAD -0.154; TCGA LUSC -0.336; TCGA OV -0.093; TCGA PRAD -0.129 |
hsa-miR-181a-5p | CDS2 | 9 cancers: BRCA; ESCA; KIRC; LUAD; LUSC; PAAD; SARC; THCA; STAD | miRNATAP | TCGA BRCA -0.089; TCGA ESCA -0.119; TCGA KIRC -0.078; TCGA LUAD -0.059; TCGA LUSC -0.121; TCGA PAAD -0.137; TCGA SARC -0.175; TCGA THCA -0.086; TCGA STAD -0.167 |
hsa-miR-181a-5p | TRIM9 | 9 cancers: BRCA; COAD; LUAD; LUSC; PAAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BRCA -0.129; TCGA COAD -0.374; TCGA LUAD -0.541; TCGA LUSC -0.506; TCGA PAAD -0.786; TCGA SARC -0.346; TCGA THCA -0.3; TCGA STAD -0.338; TCGA UCEC -0.23 |
hsa-miR-181a-5p | EPB41L4B | 10 cancers: BRCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; THCA | miRNATAP | TCGA BRCA -0.216; TCGA HNSC -0.232; TCGA KIRC -2.298; TCGA KIRP -0.47; TCGA LIHC -0.283; TCGA LUAD -0.094; TCGA LUSC -0.125; TCGA PAAD -0.541; TCGA PRAD -0.124; TCGA THCA -0.129 |
hsa-miR-181a-5p | ZNF396 | 9 cancers: BRCA; HNSC; KIRC; LIHC; PAAD; PRAD; SARC; THCA; STAD | miRNATAP | TCGA BRCA -0.289; TCGA HNSC -0.273; TCGA KIRC -0.192; TCGA LIHC -0.224; TCGA PAAD -0.416; TCGA PRAD -0.089; TCGA SARC -0.12; TCGA THCA -0.115; TCGA STAD -0.223 |
hsa-miR-181a-5p | RABGEF1 | 10 cancers: CESC; ESCA; HNSC; KIRC; KIRP; LIHC; LUSC; PAAD; SARC; STAD | MirTarget; miRNATAP | TCGA CESC -0.115; TCGA ESCA -0.189; TCGA HNSC -0.142; TCGA KIRC -0.122; TCGA KIRP -0.139; TCGA LIHC -0.078; TCGA LUSC -0.096; TCGA PAAD -0.066; TCGA SARC -0.072; TCGA STAD -0.063 |
hsa-miR-181a-5p | TXNL1 | 9 cancers: ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; THCA; STAD | mirMAP | TCGA ESCA -0.126; TCGA HNSC -0.126; TCGA LIHC -0.151; TCGA LUAD -0.144; TCGA LUSC -0.193; TCGA PAAD -0.16; TCGA PRAD -0.097; TCGA THCA -0.113; TCGA STAD -0.09 |
hsa-miR-181a-5p | MKRN1 | 9 cancers: ESCA; HNSC; KIRC; KIRP; LUSC; OV; PAAD; SARC; STAD | miRNATAP | TCGA ESCA -0.114; TCGA HNSC -0.112; TCGA KIRC -0.071; TCGA KIRP -0.077; TCGA LUSC -0.213; TCGA OV -0.119; TCGA PAAD -0.077; TCGA SARC -0.104; TCGA STAD -0.122 |
hsa-miR-181a-5p | AFG3L2 | 9 cancers: HNSC; KIRC; LIHC; LUAD; LUSC; OV; PRAD; THCA; STAD | MirTarget; miRNATAP | TCGA HNSC -0.175; TCGA KIRC -0.145; TCGA LIHC -0.15; TCGA LUAD -0.132; TCGA LUSC -0.282; TCGA OV -0.104; TCGA PRAD -0.066; TCGA THCA -0.215; TCGA STAD -0.129 |
Enriched cancer pathways of putative targets