microRNA information: hsa-miR-185-3p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-185-3p | miRbase |
| Accession: | MIMAT0004611 | miRbase |
| Precursor name: | hsa-mir-185 | miRbase |
| Precursor accession: | MI0000482 | miRbase |
| Symbol: | MIR185 | HGNC |
| RefSeq ID: | NR_029706 | GenBank |
| Sequence: | AGGGGCUGGCUUUCCUCUGGUC |
Reported expression in cancers: hsa-miR-185-3p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-185-3p | breast cancer | downregulation | "In this study we found that miR-185 was strongly d ......" | 25319390 | |
| hsa-miR-185-3p | breast cancer | downregulation | "miR-185 has been demonstrated to be involved in th ......" | 25371748 | qPCR |
| hsa-miR-185-3p | colorectal cancer | deregulation | "miR 185 and miR 133b deregulation is associated wi ......" | 21573504 | qPCR |
| hsa-miR-185-3p | endometrial cancer | deregulation | "In endometrial cancer cells expression levels of m ......" | 26535032 | |
| hsa-miR-185-3p | esophageal cancer | downregulation | "To explore the role and mechanism of microRNA185 m ......" | 24025511 | qPCR |
| hsa-miR-185-3p | gastric cancer | deregulation | "The results from the miRNA microarray analysis wer ......" | 21475928 | qPCR; Microarray |
| hsa-miR-185-3p | gastric cancer | downregulation | "miR 185 is an independent prognosis factor and sup ......" | 24352663 | |
| hsa-miR-185-3p | gastric cancer | upregulation | "A total of 33 miRNAs were identified through the i ......" | 26059512 | Reverse transcription PCR; qPCR |
| hsa-miR-185-3p | liver cancer | upregulation | "Metastasis related miR 185 is a potential prognost ......" | 23648054 | Microarray |
| hsa-miR-185-3p | lung squamous cell cancer | downregulation | "Increasing evidence has shown that microRNAs play ......" | 26617940 | |
| hsa-miR-185-3p | prostate cancer | downregulation | "In this study miR-185 was found to be down-regulat ......" | 23417242 | |
| hsa-miR-185-3p | sarcoma | deregulation | "Hypoxic conditions were verified and miRNA express ......" | 24927770 | Microarray |
Reported cancer pathway affected by hsa-miR-185-3p

| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-185-3p | breast cancer | Apoptosis pathway | "MicroRNA 185 inhibits proliferation by targeting c ......" | 25371748 | Flow cytometry; MTT assay; Western blot; Luciferase |
| hsa-miR-185-3p | gastric cancer | Apoptosis pathway | "MicroRNA 185 regulates chemotherapeutic sensitivit ......" | 24763054 | |
| hsa-miR-185-3p | gastric cancer | cell cycle pathway; Apoptosis pathway | "TRIM29 functions as an oncogene in gastric cancer ......" | 26191199 | RNAi; Colony formation; Luciferase |
| hsa-miR-185-3p | kidney renal cell cancer | Apoptosis pathway | "MicroRNA 185 inhibits cell proliferation and induc ......" | 25700976 | Colony formation; Cell Proliferation Assay; Luciferase |
| hsa-miR-185-3p | liver cancer | Epithelial mesenchymal transition pathway | "MicroRNA 185 inhibits cell proliferation and epith ......" | 27212161 | Wound Healing Assay; Western blot; Luciferase |
| hsa-miR-185-3p | lung squamous cell cancer | cell cycle pathway | "MiR 107 and MiR 185 can induce cell cycle arrest i ......" | 19688090 | |
| hsa-miR-185-3p | ovarian cancer | Apoptosis pathway | "MiR 152 and miR 185 co contribute to ovarian cance ......" | 23318422 | |
| hsa-miR-185-3p | prostate cancer | cell cycle pathway | "MicroRNA 185 suppresses proliferation invasion mig ......" | 23417242 | Luciferase |
| hsa-miR-185-3p | prostate cancer | Apoptosis pathway | "MicroRNA 185 and 342 inhibit tumorigenicity and in ......" | 23951060 |
Reported cancer prognosis affected by hsa-miR-185-3p
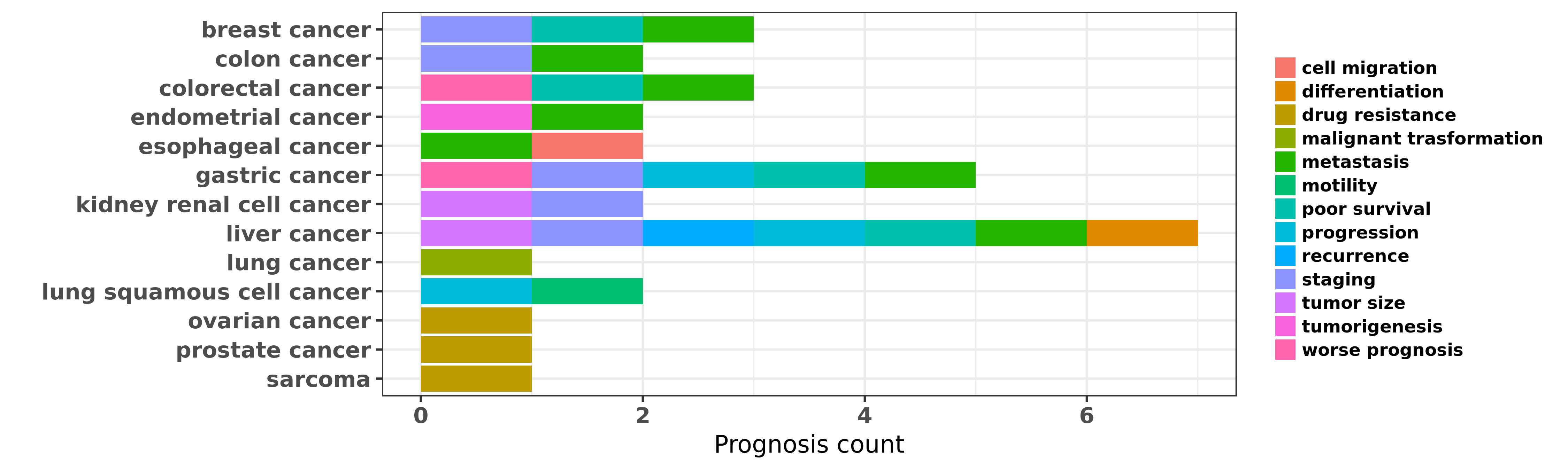
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-185-3p | breast cancer | staging; metastasis; poor survival | "miR 185 suppresses tumor proliferation by directly ......" | 25319390 | |
| hsa-miR-185-3p | breast cancer | metastasis | "RKIP suppresses the proliferation and metastasis o ......" | 27651238 | |
| hsa-miR-185-3p | colon cancer | metastasis | "MicroRNA 185 suppresses growth and invasion of col ......" | 25216407 | Western blot; MTT assay; Transwell assay |
| hsa-miR-185-3p | colon cancer | staging | "We assessed by qRT-PCR expression of 754 microRNAs ......" | 27485175 | |
| hsa-miR-185-3p | colorectal cancer | metastasis; poor survival | "miR 185 and miR 133b deregulation is associated wi ......" | 21573504 | |
| hsa-miR-185-3p | colorectal cancer | metastasis; worse prognosis | "STIM1 a direct target of microRNA 185 promotes tum ......" | 25531324 | |
| hsa-miR-185-3p | endometrial cancer | metastasis; tumorigenesis | "In endometrial cancer cells expression levels of m ......" | 26535032 | |
| hsa-miR-185-3p | esophageal cancer | cell migration; metastasis | "Plasma miR 185 is decreased in patients with esoph ......" | 26316588 | Luciferase |
| hsa-miR-185-3p | gastric cancer | metastasis; worse prognosis; staging; progression; poor survival | "miR 185 is an independent prognosis factor and sup ......" | 24352663 | |
| hsa-miR-185-3p | gastric cancer | staging | "A total of 33 miRNAs were identified through the i ......" | 26059512 | |
| hsa-miR-185-3p | kidney renal cell cancer | staging; tumor size | "MicroRNA 185 inhibits cell proliferation and induc ......" | 25700976 | Colony formation; Cell Proliferation Assay; Luciferase |
| hsa-miR-185-3p | liver cancer | staging; metastasis; recurrence; differentiation; tumor size; poor survival | "Metastasis related miR 185 is a potential prognost ......" | 23648054 | |
| hsa-miR-185-3p | liver cancer | metastasis; progression | "MicroRNA 185 inhibits cell proliferation and epith ......" | 27212161 | Wound Healing Assay; Western blot; Luciferase |
| hsa-miR-185-3p | lung cancer | malignant trasformation | "Diagnostic Value of Circulating Extracellular miR ......" | 24851110 | |
| hsa-miR-185-3p | lung squamous cell cancer | progression; motility | "MiR 185 acts as a tumor suppressor by targeting AK ......" | 26617940 | |
| hsa-miR-185-3p | ovarian cancer | drug resistance | "MiR 152 and miR 185 co contribute to ovarian cance ......" | 23318422 | |
| hsa-miR-185-3p | prostate cancer | drug resistance | "MiR 185 attenuates androgen receptor function in p ......" | 26940039 | Luciferase; Western blot |
| hsa-miR-185-3p | sarcoma | drug resistance | "Hypoxic conditions were verified and miRNA express ......" | 24927770 | Luciferase |
Reported gene related to hsa-miR-185-3p
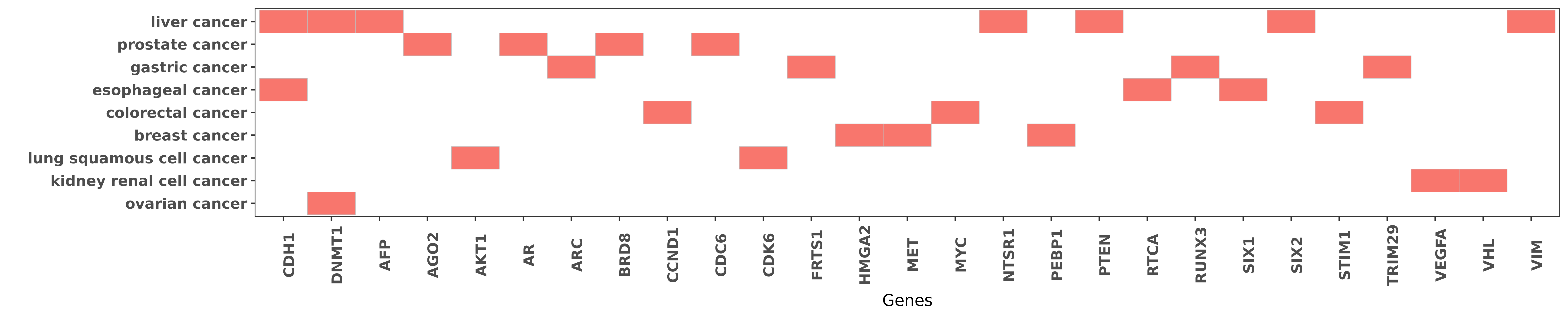
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-185-3p | prostate cancer | AR | "MicroRNA 185 downregulates androgen receptor expre ......" | 25673182 |
| hsa-miR-185-3p | prostate cancer | AR | "MMTV-Fluc reporter assays revealed that miR-185 ca ......" | 26940039 |
| hsa-miR-185-3p | prostate cancer | AR | "MicroRNA 185 suppresses proliferation invasion mig ......" | 23417242 |
| hsa-miR-185-3p | esophageal cancer | CDH1 | "After transfection of miR-185 inhibitor into KYSE3 ......" | 24025511 |
| hsa-miR-185-3p | liver cancer | CDH1 | "We also found that ectopic expression of miR-185 r ......" | 27212161 |
| hsa-miR-185-3p | liver cancer | DNMT1 | "miR-185 overexpression inhibited DNMT1 3' untransl ......" | 24911372 |
| hsa-miR-185-3p | ovarian cancer | DNMT1 | "MiR 152 and miR 185 co contribute to ovarian cance ......" | 23318422 |
| hsa-miR-185-3p | liver cancer | AFP | "There was no direct relationship between miR-185 a ......" | 23648054 |
| hsa-miR-185-3p | prostate cancer | AGO2 | "Putative miRNA response element MRE of miR-185 in ......" | 26940039 |
| hsa-miR-185-3p | lung squamous cell cancer | AKT1 | "MiR 185 acts as a tumor suppressor by targeting AK ......" | 26617940 |
| hsa-miR-185-3p | gastric cancer | ARC | "In elucidating the molecular mechanism by which mi ......" | 24763054 |
| hsa-miR-185-3p | prostate cancer | BRD8 | "Putative miRNA response element MRE of miR-185 in ......" | 26940039 |
| hsa-miR-185-3p | colorectal cancer | CCND1 | "Mechanically we demonstrated that miR-185 could su ......" | 27453420 |
| hsa-miR-185-3p | prostate cancer | CDC6 | "CDC6 one target of AR and an important regulatory ......" | 23417242 |
| hsa-miR-185-3p | lung squamous cell cancer | CDK6 | "Using miRNA-target prediction analyses and the arr ......" | 19688090 |
| hsa-miR-185-3p | gastric cancer | FRTS1 | "Moreover miR-185 might independently predict OS an ......" | 24352663 |
| hsa-miR-185-3p | breast cancer | HMGA2 | "RKIP suppresses the proliferation and metastasis o ......" | 27651238 |
| hsa-miR-185-3p | breast cancer | MET | "MicroRNA 185 inhibits proliferation by targeting c ......" | 25371748 |
| hsa-miR-185-3p | colorectal cancer | MYC | "Mechanically we demonstrated that miR-185 could su ......" | 27453420 |
| hsa-miR-185-3p | liver cancer | NTSR1 | "We classified 95 patients with early stage HCC int ......" | 23648054 |
| hsa-miR-185-3p | breast cancer | PEBP1 | "RKIP suppresses the proliferation and metastasis o ......" | 27651238 |
| hsa-miR-185-3p | liver cancer | PTEN | "These results provide novel evidence that miR-185 ......" | 24911372 |
| hsa-miR-185-3p | esophageal cancer | RTCA | "The xCELLigence RTCA MP system and Transwell assay ......" | 24025511 |
| hsa-miR-185-3p | gastric cancer | RUNX3 | "Finally we found that RUNX3 Runt-related transcrip ......" | 24763054 |
| hsa-miR-185-3p | esophageal cancer | SIX1 | "After transfection of miR-185 inhibitor into KYSE3 ......" | 24025511 |
| hsa-miR-185-3p | liver cancer | SIX2 | "MicroRNA 185 inhibits cell proliferation and epith ......" | 27212161 |
| hsa-miR-185-3p | colorectal cancer | STIM1 | "STIM1 a direct target of microRNA 185 promotes tum ......" | 25531324 |
| hsa-miR-185-3p | gastric cancer | TRIM29 | "TRIM29 functions as an oncogene in gastric cancer ......" | 26191199 |
| hsa-miR-185-3p | kidney renal cell cancer | VEGFA | "MicroRNA 185 inhibits cell proliferation and induc ......" | 25700976 |
| hsa-miR-185-3p | kidney renal cell cancer | VHL | "We used 786O A498 VHL inactivated and Caki-1 VHL i ......" | 25700976 |
| hsa-miR-185-3p | liver cancer | VIM | "We also found that ectopic expression of miR-185 r ......" | 27212161 |
Expression profile in cancer corhorts:
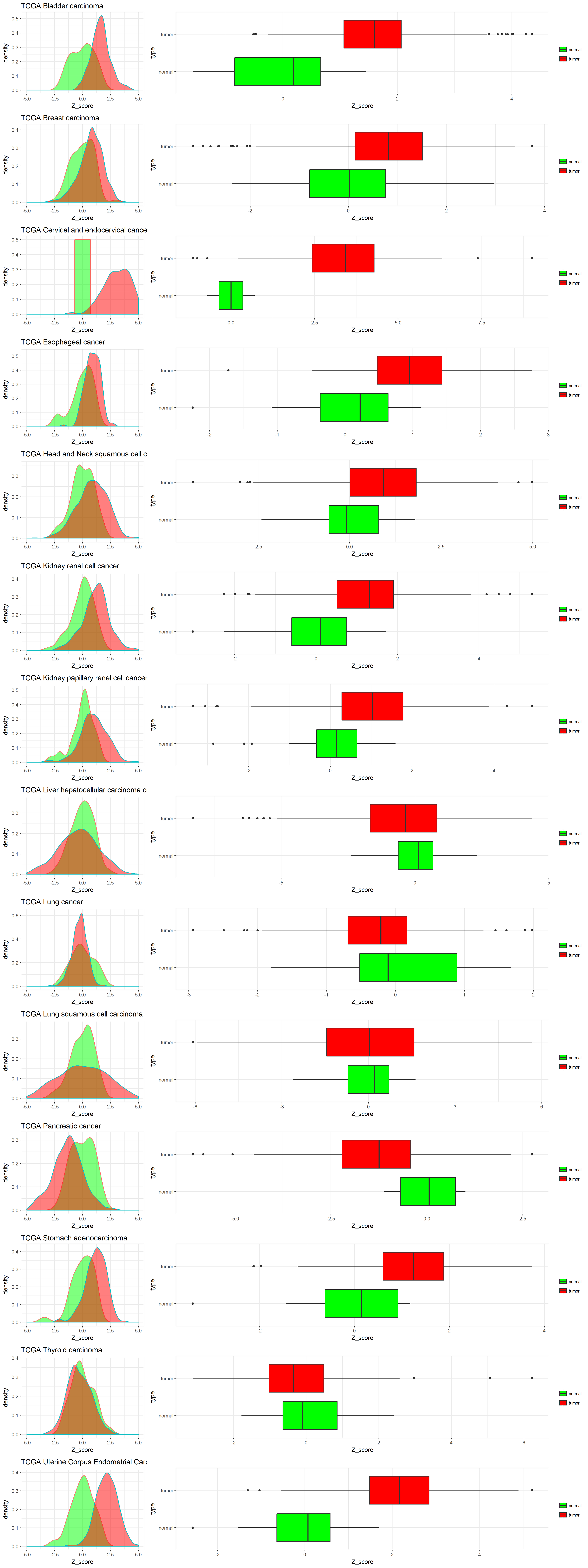
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-185-3p | LRRC32 | 9 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; LIHC; THCA; UCEC | MirTarget | TCGA BLCA -0.363; TCGA BRCA -0.202; TCGA CESC -0.273; TCGA COAD -0.305; TCGA ESCA -0.34; TCGA KIRC -0.143; TCGA LIHC -0.119; TCGA THCA -0.16; TCGA UCEC -0.258 |
hsa-miR-185-3p | TCF7L1 | 9 cancers: BLCA; COAD; KIRC; KIRP; LGG; OV; SARC; THCA; STAD | MirTarget | TCGA BLCA -0.265; TCGA COAD -0.297; TCGA KIRC -0.173; TCGA KIRP -0.186; TCGA LGG -0.148; TCGA OV -0.183; TCGA SARC -0.16; TCGA THCA -0.149; TCGA STAD -0.402 |
hsa-miR-185-3p | ATP1B2 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; KIRP; OV; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.268; TCGA BRCA -0.317; TCGA CESC -0.282; TCGA COAD -0.493; TCGA ESCA -0.522; TCGA KIRC -0.387; TCGA KIRP -0.188; TCGA OV -0.25; TCGA THCA -0.335; TCGA STAD -0.673; TCGA UCEC -0.539 |
hsa-miR-185-3p | PRELP | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; LUAD; LUSC; OV; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.828; TCGA BRCA -0.276; TCGA CESC -0.512; TCGA COAD -0.756; TCGA ESCA -0.956; TCGA KIRC -0.297; TCGA LUAD -0.159; TCGA LUSC -0.224; TCGA OV -0.247; TCGA SARC -0.958; TCGA THCA -0.366; TCGA STAD -0.911; TCGA UCEC -0.739 |
hsa-miR-185-3p | CORO2B | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; KIRP; LUAD; OV; STAD; UCEC | MirTarget | TCGA BLCA -0.269; TCGA BRCA -0.337; TCGA CESC -0.257; TCGA COAD -0.409; TCGA ESCA -0.472; TCGA KIRC -0.39; TCGA KIRP -0.216; TCGA LUAD -0.224; TCGA OV -0.284; TCGA STAD -0.477; TCGA UCEC -0.504 |
hsa-miR-185-3p | HSPA12A | 9 cancers: BLCA; BRCA; CESC; ESCA; KIRC; KIRP; LUAD; STAD; UCEC | MirTarget | TCGA BLCA -0.334; TCGA BRCA -0.117; TCGA CESC -0.168; TCGA ESCA -0.25; TCGA KIRC -0.179; TCGA KIRP -0.337; TCGA LUAD -0.174; TCGA STAD -0.381; TCGA UCEC -0.115 |
hsa-miR-185-3p | FAM131B | 9 cancers: BLCA; BRCA; CESC; COAD; KIRC; KIRP; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.276; TCGA BRCA -0.153; TCGA CESC -0.207; TCGA COAD -0.275; TCGA KIRC -0.111; TCGA KIRP -0.17; TCGA THCA -0.251; TCGA STAD -0.288; TCGA UCEC -0.431 |
hsa-miR-185-3p | FMOD | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; LUSC; OV; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.491; TCGA BRCA -0.376; TCGA CESC -0.258; TCGA COAD -0.269; TCGA ESCA -0.377; TCGA LUSC -0.179; TCGA OV -0.285; TCGA SARC -0.639; TCGA STAD -0.254; TCGA UCEC -0.438 |
hsa-miR-185-3p | MFGE8 | 10 cancers: BLCA; CESC; COAD; ESCA; LUAD; LUSC; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.269; TCGA CESC -0.119; TCGA COAD -0.282; TCGA ESCA -0.191; TCGA LUAD -0.097; TCGA LUSC -0.115; TCGA SARC -0.292; TCGA THCA -0.277; TCGA STAD -0.128; TCGA UCEC -0.113 |
hsa-miR-185-3p | ZCCHC24 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; LGG; LUAD; LUSC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.451; TCGA BRCA -0.326; TCGA CESC -0.223; TCGA COAD -0.293; TCGA ESCA -0.507; TCGA LGG -0.079; TCGA LUAD -0.089; TCGA LUSC -0.141; TCGA THCA -0.081; TCGA STAD -0.585; TCGA UCEC -0.406 |
hsa-miR-185-3p | ZBTB4 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; KIRP; LUAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.117; TCGA BRCA -0.178; TCGA CESC -0.086; TCGA COAD -0.122; TCGA ESCA -0.217; TCGA KIRC -0.12; TCGA KIRP -0.066; TCGA LUAD -0.077; TCGA SARC -0.116; TCGA STAD -0.339; TCGA UCEC -0.17 |
hsa-miR-185-3p | CTDSP2 | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; KIRP; OV; STAD; UCEC | MirTarget | TCGA BLCA -0.122; TCGA BRCA -0.098; TCGA CESC -0.074; TCGA COAD -0.059; TCGA ESCA -0.07; TCGA KIRC -0.07; TCGA KIRP -0.076; TCGA OV -0.078; TCGA STAD -0.094; TCGA UCEC -0.056 |
hsa-miR-185-3p | NFATC4 | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LUAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.271; TCGA BRCA -0.221; TCGA COAD -0.162; TCGA ESCA -0.315; TCGA HNSC -0.109; TCGA KIRP -0.311; TCGA LUAD -0.155; TCGA SARC -0.18; TCGA THCA -0.109; TCGA STAD -0.437; TCGA UCEC -0.213 |
hsa-miR-185-3p | PALM | 10 cancers: BLCA; BRCA; COAD; ESCA; KIRC; KIRP; LUAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.414; TCGA BRCA -0.36; TCGA COAD -0.379; TCGA ESCA -0.393; TCGA KIRC -0.361; TCGA KIRP -0.263; TCGA LUAD -0.131; TCGA THCA -0.331; TCGA STAD -0.452; TCGA UCEC -0.229 |
hsa-miR-185-3p | RHBDL3 | 9 cancers: BLCA; BRCA; COAD; ESCA; LGG; LUAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.203; TCGA BRCA -0.309; TCGA COAD -0.318; TCGA ESCA -0.301; TCGA LGG -0.119; TCGA LUAD -0.207; TCGA SARC -0.408; TCGA STAD -0.36; TCGA UCEC -0.184 |
hsa-miR-185-3p | PHYHIPL | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LGG; LIHC; SARC; STAD | MirTarget | TCGA BLCA -0.389; TCGA BRCA -0.387; TCGA COAD -0.563; TCGA ESCA -0.53; TCGA HNSC -0.263; TCGA KIRC -0.219; TCGA LGG -0.113; TCGA LIHC -0.252; TCGA SARC -0.556; TCGA STAD -0.494 |
hsa-miR-185-3p | RGMA | 13 cancers: BLCA; CESC; COAD; ESCA; KIRC; KIRP; LIHC; OV; PAAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.573; TCGA CESC -0.29; TCGA COAD -0.74; TCGA ESCA -0.412; TCGA KIRC -0.242; TCGA KIRP -0.224; TCGA LIHC -0.179; TCGA OV -0.149; TCGA PAAD -0.355; TCGA SARC -0.183; TCGA THCA -0.241; TCGA STAD -1.054; TCGA UCEC -0.398 |
hsa-miR-185-3p | CNN1 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; LIHC; LUAD; LUSC; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -1.146; TCGA BRCA -0.557; TCGA CESC -0.462; TCGA COAD -0.818; TCGA ESCA -1.091; TCGA KIRC -0.22; TCGA LIHC -0.259; TCGA LUAD -0.192; TCGA LUSC -0.261; TCGA SARC -1.444; TCGA THCA -0.334; TCGA STAD -1.454; TCGA UCEC -0.837 |
hsa-miR-185-3p | HSPA12B | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; LIHC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.318; TCGA BRCA -0.306; TCGA CESC -0.178; TCGA COAD -0.179; TCGA ESCA -0.244; TCGA KIRC -0.204; TCGA LIHC -0.156; TCGA THCA -0.108; TCGA STAD -0.149; TCGA UCEC -0.358 |
hsa-miR-185-3p | ILK | 9 cancers: BLCA; COAD; ESCA; HNSC; LUSC; PAAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.248; TCGA COAD -0.152; TCGA ESCA -0.12; TCGA HNSC -0.074; TCGA LUSC -0.086; TCGA PAAD -0.08; TCGA SARC -0.256; TCGA STAD -0.375; TCGA UCEC -0.128 |
hsa-miR-185-3p | ARHGEF17 | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; LUAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.201; TCGA BRCA -0.149; TCGA CESC -0.116; TCGA COAD -0.207; TCGA ESCA -0.276; TCGA LUAD -0.108; TCGA SARC -0.314; TCGA THCA -0.108; TCGA STAD -0.269; TCGA UCEC -0.158 |
hsa-miR-185-3p | SSPN | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; LUSC; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.311; TCGA BRCA -0.236; TCGA CESC -0.157; TCGA COAD -0.45; TCGA ESCA -0.488; TCGA LUSC -0.28; TCGA SARC -0.234; TCGA THCA -0.162; TCGA STAD -0.565; TCGA UCEC -0.371 |
hsa-miR-185-3p | CTF1 | 10 cancers: BLCA; BRCA; COAD; KIRC; KIRP; PAAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.189; TCGA BRCA -0.3; TCGA COAD -0.324; TCGA KIRC -0.265; TCGA KIRP -0.262; TCGA PAAD -0.194; TCGA SARC -0.455; TCGA THCA -0.245; TCGA STAD -0.529; TCGA UCEC -0.279 |
hsa-miR-185-3p | NFIX | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; LGG; LUAD; OV; PAAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.348; TCGA BRCA -0.134; TCGA CESC -0.19; TCGA COAD -0.075; TCGA ESCA -0.2; TCGA LGG -0.073; TCGA LUAD -0.177; TCGA OV -0.102; TCGA PAAD -0.115; TCGA SARC -0.182; TCGA THCA -0.088; TCGA STAD -0.344; TCGA UCEC -0.278 |
hsa-miR-185-3p | SLC27A1 | 10 cancers: BLCA; BRCA; HNSC; LUAD; LUSC; OV; PAAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.177; TCGA BRCA -0.242; TCGA HNSC -0.108; TCGA LUAD -0.206; TCGA LUSC -0.101; TCGA OV -0.114; TCGA PAAD -0.309; TCGA THCA -0.126; TCGA STAD -0.082; TCGA UCEC -0.135 |
hsa-miR-185-3p | ZBTB47 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LUAD; LUSC; PAAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.203; TCGA BRCA -0.147; TCGA CESC -0.144; TCGA COAD -0.123; TCGA ESCA -0.29; TCGA HNSC -0.155; TCGA LGG -0.06; TCGA LUAD -0.093; TCGA LUSC -0.094; TCGA PAAD -0.118; TCGA SARC -0.231; TCGA STAD -0.351; TCGA UCEC -0.145 |
hsa-miR-185-3p | MAPK4 | 9 cancers: BLCA; COAD; ESCA; KIRC; KIRP; OV; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.476; TCGA COAD -0.904; TCGA ESCA -1.094; TCGA KIRC -0.819; TCGA KIRP -0.329; TCGA OV -0.359; TCGA SARC -1.437; TCGA STAD -1.438; TCGA UCEC -0.374 |
hsa-miR-185-3p | CCL14 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.922; TCGA BRCA -0.683; TCGA CESC -0.701; TCGA COAD -0.722; TCGA ESCA -0.956; TCGA HNSC -0.341; TCGA KIRC -0.165; TCGA LUAD -0.183; TCGA SARC -0.367; TCGA STAD -0.879; TCGA UCEC -0.817 |
hsa-miR-185-3p | KY | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; LUAD; LUSC; OV; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.666; TCGA BRCA -0.515; TCGA CESC -0.357; TCGA COAD -0.295; TCGA ESCA -0.572; TCGA LUAD -0.23; TCGA LUSC -0.355; TCGA OV -0.269; TCGA SARC -0.766; TCGA STAD -0.956; TCGA UCEC -0.512 |
hsa-miR-185-3p | MTMR10 | 10 cancers: BLCA; BRCA; CESC; ESCA; KIRC; KIRP; LIHC; LUAD; STAD; UCEC | MirTarget | TCGA BLCA -0.095; TCGA BRCA -0.126; TCGA CESC -0.1; TCGA ESCA -0.078; TCGA KIRC -0.162; TCGA KIRP -0.136; TCGA LIHC -0.072; TCGA LUAD -0.061; TCGA STAD -0.184; TCGA UCEC -0.062 |
hsa-miR-185-3p | SLC9A3R2 | 10 cancers: BLCA; BRCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; THCA | mirMAP | TCGA BLCA -0.09; TCGA BRCA -0.128; TCGA HNSC -0.123; TCGA KIRC -0.236; TCGA KIRP -0.205; TCGA LIHC -0.125; TCGA LUAD -0.099; TCGA LUSC -0.108; TCGA PAAD -0.266; TCGA THCA -0.133 |
hsa-miR-185-3p | TNS1 | 9 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.671; TCGA BRCA -0.279; TCGA CESC -0.192; TCGA COAD -0.487; TCGA ESCA -0.723; TCGA KIRC -0.102; TCGA SARC -0.516; TCGA STAD -0.782; TCGA UCEC -0.415 |
hsa-miR-185-3p | CRTC1 | 11 cancers: BLCA; BRCA; ESCA; KIRC; KIRP; LUAD; OV; PAAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.101; TCGA BRCA -0.113; TCGA ESCA -0.145; TCGA KIRC -0.114; TCGA KIRP -0.181; TCGA LUAD -0.156; TCGA OV -0.095; TCGA PAAD -0.148; TCGA THCA -0.168; TCGA STAD -0.145; TCGA UCEC -0.059 |
hsa-miR-185-3p | VIPR2 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LUSC; OV; PAAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.736; TCGA BRCA -0.613; TCGA CESC -0.458; TCGA COAD -0.628; TCGA ESCA -1.052; TCGA HNSC -0.214; TCGA KIRC -0.255; TCGA KIRP -0.204; TCGA LGG -0.307; TCGA LUSC -0.288; TCGA OV -0.277; TCGA PAAD -0.485; TCGA SARC -1.02; TCGA STAD -1.079; TCGA UCEC -0.602 |
hsa-miR-185-3p | TSPAN11 | 10 cancers: BLCA; BRCA; COAD; ESCA; LGG; LUAD; LUSC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.283; TCGA BRCA -0.182; TCGA COAD -0.335; TCGA ESCA -0.448; TCGA LGG -0.141; TCGA LUAD -0.398; TCGA LUSC -0.227; TCGA THCA -0.266; TCGA STAD -0.371; TCGA UCEC -0.267 |
hsa-miR-185-3p | LDB3 | 13 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; OV; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.488; TCGA BRCA -0.203; TCGA COAD -0.801; TCGA ESCA -0.955; TCGA HNSC -0.38; TCGA KIRC -0.265; TCGA LIHC -0.199; TCGA LUAD -0.198; TCGA OV -0.126; TCGA SARC -0.97; TCGA THCA -0.2; TCGA STAD -1.38; TCGA UCEC -0.418 |
hsa-miR-185-3p | MXD4 | 12 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LGG; LUAD; LUSC; OV; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.106; TCGA BRCA -0.139; TCGA CESC -0.085; TCGA ESCA -0.127; TCGA HNSC -0.116; TCGA LGG -0.066; TCGA LUAD -0.102; TCGA LUSC -0.104; TCGA OV -0.121; TCGA THCA -0.104; TCGA STAD -0.131; TCGA UCEC -0.103 |
hsa-miR-185-3p | THRA | 10 cancers: BLCA; BRCA; CESC; KIRC; KIRP; LGG; OV; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.247; TCGA BRCA -0.125; TCGA CESC -0.089; TCGA KIRC -0.221; TCGA KIRP -0.142; TCGA LGG -0.069; TCGA OV -0.119; TCGA SARC -0.225; TCGA STAD -0.092; TCGA UCEC -0.225 |
hsa-miR-185-3p | PER2 | 9 cancers: BLCA; BRCA; ESCA; KIRP; LIHC; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.181; TCGA BRCA -0.25; TCGA ESCA -0.167; TCGA KIRP -0.128; TCGA LIHC -0.076; TCGA SARC -0.218; TCGA THCA -0.074; TCGA STAD -0.099; TCGA UCEC -0.15 |
hsa-miR-185-3p | SCN2B | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; OV; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.713; TCGA BRCA -0.918; TCGA CESC -0.499; TCGA COAD -0.79; TCGA ESCA -0.853; TCGA HNSC -0.281; TCGA KIRC -0.721; TCGA LUAD -0.176; TCGA LUSC -0.379; TCGA OV -0.3; TCGA SARC -0.782; TCGA THCA -0.37; TCGA STAD -1.077; TCGA UCEC -0.51 |
hsa-miR-185-3p | KCTD15 | 10 cancers: BLCA; CESC; COAD; KIRC; KIRP; LGG; LUAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.277; TCGA CESC -0.196; TCGA COAD -0.276; TCGA KIRC -0.158; TCGA KIRP -0.22; TCGA LGG -0.068; TCGA LUAD -0.139; TCGA SARC -0.359; TCGA STAD -0.314; TCGA UCEC -0.195 |
hsa-miR-185-3p | PLIN4 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; PAAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.339; TCGA BRCA -0.61; TCGA CESC -0.345; TCGA COAD -0.759; TCGA ESCA -0.825; TCGA HNSC -0.34; TCGA LUAD -0.37; TCGA PAAD -0.648; TCGA SARC -1.163; TCGA THCA -0.203; TCGA STAD -1.39; TCGA UCEC -0.461 |
hsa-miR-185-3p | PRIMA1 | 9 cancers: BLCA; BRCA; COAD; ESCA; KIRP; LUAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.621; TCGA BRCA -0.16; TCGA COAD -1.147; TCGA ESCA -0.751; TCGA KIRP -0.419; TCGA LUAD -0.233; TCGA SARC -0.903; TCGA STAD -1.558; TCGA UCEC -0.202 |
hsa-miR-185-3p | GNG7 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.439; TCGA BRCA -0.278; TCGA CESC -0.223; TCGA COAD -0.34; TCGA ESCA -0.667; TCGA HNSC -0.206; TCGA KIRC -0.23; TCGA KIRP -0.151; TCGA LIHC -0.236; TCGA LUAD -0.293; TCGA LUSC -0.216; TCGA SARC -0.344; TCGA STAD -0.664; TCGA UCEC -0.402 |
hsa-miR-185-3p | SCN4B | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; LIHC; LUAD; LUSC; OV; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.276; TCGA BRCA -0.53; TCGA CESC -0.359; TCGA COAD -0.468; TCGA ESCA -0.569; TCGA LIHC -0.171; TCGA LUAD -0.24; TCGA LUSC -0.214; TCGA OV -0.224; TCGA SARC -0.415; TCGA THCA -0.166; TCGA STAD -0.666; TCGA UCEC -0.531 |
hsa-miR-185-3p | LDLRAD2 | 12 cancers: BLCA; BRCA; ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.238; TCGA BRCA -0.117; TCGA ESCA -0.115; TCGA HNSC -0.137; TCGA KIRP -0.154; TCGA LUAD -0.094; TCGA LUSC -0.08; TCGA PAAD -0.246; TCGA SARC -0.198; TCGA THCA -0.212; TCGA STAD -0.146; TCGA UCEC -0.103 |
hsa-miR-185-3p | PRX | 12 cancers: BLCA; BRCA; ESCA; KIRC; KIRP; LIHC; LUAD; PAAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.203; TCGA BRCA -0.302; TCGA ESCA -0.221; TCGA KIRC -0.238; TCGA KIRP -0.142; TCGA LIHC -0.102; TCGA LUAD -0.127; TCGA PAAD -0.159; TCGA SARC -0.108; TCGA THCA -0.095; TCGA STAD -0.263; TCGA UCEC -0.235 |
hsa-miR-185-3p | NPTX1 | 11 cancers: BLCA; BRCA; COAD; ESCA; KIRC; LUAD; LUSC; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.414; TCGA BRCA -0.227; TCGA COAD -0.877; TCGA ESCA -0.625; TCGA KIRC -0.222; TCGA LUAD -0.27; TCGA LUSC -0.22; TCGA SARC -0.721; TCGA THCA -0.323; TCGA STAD -1.207; TCGA UCEC -0.358 |
hsa-miR-185-3p | RUNX1T1 | 9 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.552; TCGA BRCA -0.351; TCGA CESC -0.26; TCGA COAD -0.578; TCGA ESCA -0.666; TCGA KIRC -0.323; TCGA SARC -0.164; TCGA STAD -0.589; TCGA UCEC -0.617 |
hsa-miR-185-3p | VAMP2 | 13 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; OV; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.122; TCGA BRCA -0.192; TCGA CESC -0.08; TCGA ESCA -0.217; TCGA HNSC -0.161; TCGA KIRC -0.18; TCGA KIRP -0.15; TCGA LUAD -0.109; TCGA LUSC -0.078; TCGA OV -0.088; TCGA SARC -0.148; TCGA STAD -0.316; TCGA UCEC -0.212 |
hsa-miR-185-3p | DCHS1 | 9 cancers: BLCA; BRCA; CESC; COAD; ESCA; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.339; TCGA BRCA -0.136; TCGA CESC -0.161; TCGA COAD -0.152; TCGA ESCA -0.349; TCGA SARC -0.193; TCGA THCA -0.15; TCGA STAD -0.268; TCGA UCEC -0.286 |
hsa-miR-185-3p | APBB1 | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; OV; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.36; TCGA BRCA -0.168; TCGA CESC -0.18; TCGA COAD -0.338; TCGA ESCA -0.424; TCGA KIRC -0.083; TCGA OV -0.185; TCGA SARC -0.225; TCGA STAD -0.487; TCGA UCEC -0.269 |
hsa-miR-185-3p | EPN2 | 10 cancers: BRCA; CESC; KIRC; KIRP; LUAD; OV; SARC; THCA; STAD; UCEC | MirTarget | TCGA BRCA -0.065; TCGA CESC -0.145; TCGA KIRC -0.074; TCGA KIRP -0.15; TCGA LUAD -0.059; TCGA OV -0.074; TCGA SARC -0.217; TCGA THCA -0.054; TCGA STAD -0.189; TCGA UCEC -0.099 |
hsa-miR-185-3p | ZNF862 | 13 cancers: BRCA; CESC; ESCA; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; SARC; THCA; STAD; UCEC | MirTarget | TCGA BRCA -0.145; TCGA CESC -0.148; TCGA ESCA -0.235; TCGA HNSC -0.137; TCGA KIRP -0.211; TCGA LGG -0.071; TCGA LIHC -0.099; TCGA LUAD -0.163; TCGA LUSC -0.14; TCGA SARC -0.123; TCGA THCA -0.064; TCGA STAD -0.162; TCGA UCEC -0.171 |
hsa-miR-185-3p | MLLT6 | 10 cancers: BRCA; CESC; ESCA; KIRC; KIRP; LUAD; PAAD; SARC; THCA; UCEC | MirTarget; mirMAP | TCGA BRCA -0.068; TCGA CESC -0.088; TCGA ESCA -0.093; TCGA KIRC -0.095; TCGA KIRP -0.184; TCGA LUAD -0.103; TCGA PAAD -0.128; TCGA SARC -0.177; TCGA THCA -0.092; TCGA UCEC -0.063 |
hsa-miR-185-3p | SH2B1 | 13 cancers: BRCA; CESC; ESCA; HNSC; KIRP; LGG; LUAD; LUSC; OV; PAAD; SARC; THCA; UCEC | MirTarget; miRNATAP | TCGA BRCA -0.104; TCGA CESC -0.106; TCGA ESCA -0.073; TCGA HNSC -0.122; TCGA KIRP -0.146; TCGA LGG -0.061; TCGA LUAD -0.181; TCGA LUSC -0.148; TCGA OV -0.131; TCGA PAAD -0.127; TCGA SARC -0.151; TCGA THCA -0.08; TCGA UCEC -0.053 |
hsa-miR-185-3p | CROCC | 9 cancers: BRCA; CESC; KIRP; LGG; LUAD; OV; PAAD; THCA; UCEC | MirTarget | TCGA BRCA -0.121; TCGA CESC -0.179; TCGA KIRP -0.097; TCGA LGG -0.056; TCGA LUAD -0.202; TCGA OV -0.106; TCGA PAAD -0.289; TCGA THCA -0.136; TCGA UCEC -0.123 |
hsa-miR-185-3p | OSBPL7 | 9 cancers: BRCA; KIRP; LGG; LUAD; LUSC; PAAD; SARC; THCA; UCEC | mirMAP | TCGA BRCA -0.149; TCGA KIRP -0.137; TCGA LGG -0.084; TCGA LUAD -0.268; TCGA LUSC -0.182; TCGA PAAD -0.325; TCGA SARC -0.075; TCGA THCA -0.099; TCGA UCEC -0.095 |
hsa-miR-185-3p | SGSM2 | 12 cancers: BRCA; CESC; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; SARC; THCA; STAD; UCEC | mirMAP | TCGA BRCA -0.175; TCGA CESC -0.106; TCGA ESCA -0.162; TCGA HNSC -0.203; TCGA KIRP -0.133; TCGA LIHC -0.11; TCGA LUAD -0.206; TCGA LUSC -0.109; TCGA SARC -0.089; TCGA THCA -0.131; TCGA STAD -0.207; TCGA UCEC -0.055 |
hsa-miR-185-3p | ING5 | 10 cancers: BRCA; HNSC; KIRC; KIRP; LGG; LUAD; OV; PAAD; SARC; THCA | mirMAP | TCGA BRCA -0.103; TCGA HNSC -0.078; TCGA KIRC -0.091; TCGA KIRP -0.189; TCGA LGG -0.116; TCGA LUAD -0.122; TCGA OV -0.16; TCGA PAAD -0.091; TCGA SARC -0.142; TCGA THCA -0.066 |
hsa-miR-185-3p | SLC25A42 | 10 cancers: BRCA; CESC; COAD; ESCA; LIHC; LUAD; OV; SARC; STAD; UCEC | mirMAP | TCGA BRCA -0.09; TCGA CESC -0.113; TCGA COAD -0.105; TCGA ESCA -0.219; TCGA LIHC -0.148; TCGA LUAD -0.126; TCGA OV -0.106; TCGA SARC -0.097; TCGA STAD -0.244; TCGA UCEC -0.13 |
hsa-miR-185-3p | FLRT1 | 10 cancers: CESC; ESCA; KIRC; KIRP; LGG; LUAD; OV; PAAD; STAD; UCEC | MirTarget | TCGA CESC -0.169; TCGA ESCA -0.218; TCGA KIRC -0.595; TCGA KIRP -0.437; TCGA LGG -0.092; TCGA LUAD -0.215; TCGA OV -0.255; TCGA PAAD -0.219; TCGA STAD -0.613; TCGA UCEC -0.265 |
hsa-miR-185-3p | MAP2K7 | 11 cancers: CESC; HNSC; KIRP; LIHC; LUAD; LUSC; OV; PAAD; SARC; THCA; STAD | mirMAP | TCGA CESC -0.064; TCGA HNSC -0.056; TCGA KIRP -0.053; TCGA LIHC -0.074; TCGA LUAD -0.063; TCGA LUSC -0.056; TCGA OV -0.069; TCGA PAAD -0.147; TCGA SARC -0.086; TCGA THCA -0.082; TCGA STAD -0.057 |
Enriched cancer pathways of putative targets