microRNA information: hsa-miR-191-5p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-191-5p | miRbase |
| Accession: | MIMAT0000440 | miRbase |
| Precursor name: | hsa-mir-191 | miRbase |
| Precursor accession: | MI0000465 | miRbase |
| Symbol: | MIR191 | HGNC |
| RefSeq ID: | NR_029690 | GenBank |
| Sequence: | CAACGGAAUCCCAAAAGCAGCUG |
Reported expression in cancers: hsa-miR-191-5p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-191-5p | acute myeloid leukemia | deregulation | "After background subtraction and normalization usi ......" | 18187662 | Microarray; qPCR |
| hsa-miR-191-5p | bladder cancer | downregulation | "In the first - discovery - phase microarray cards ......" | 27468885 | Microarray; qPCR |
| hsa-miR-191-5p | breast cancer | deregulation | "Differential expression of miR 21 miR 125b and miR ......" | 22898264 | qPCR |
| hsa-miR-191-5p | colorectal cancer | upregulation | "miR 191 promotes tumorigenesis of human colorectal ......" | 25784653 | |
| hsa-miR-191-5p | gastric cancer | upregulation | "Here we found that the microRNA-191 miR-191 was at ......" | 21947487 | |
| hsa-miR-191-5p | gastric cancer | upregulation | "In the analysis by real-time PCR-based miRNA array ......" | 22407237 | qPCR; Microarray |
| hsa-miR-191-5p | liver cancer | upregulation | "hsa miR 191 is a candidate oncogene target for hep ......" | 20924108 | |
| hsa-miR-191-5p | liver cancer | upregulation | "Hypomethylation of the hsa miR 191 locus causes hi ......" | 21969817 | Northern blot; qPCR; RNA-Seq |
| hsa-miR-191-5p | lymphoma | upregulation | "Expression of microRNA 191 in T lymphoblastic leuk ......" | 27093985 | qPCR |
| hsa-miR-191-5p | melanoma | deregulation | "Furthermore low expression of miR-191 and high exp ......" | 20357817 |
Reported cancer pathway affected by hsa-miR-191-5p
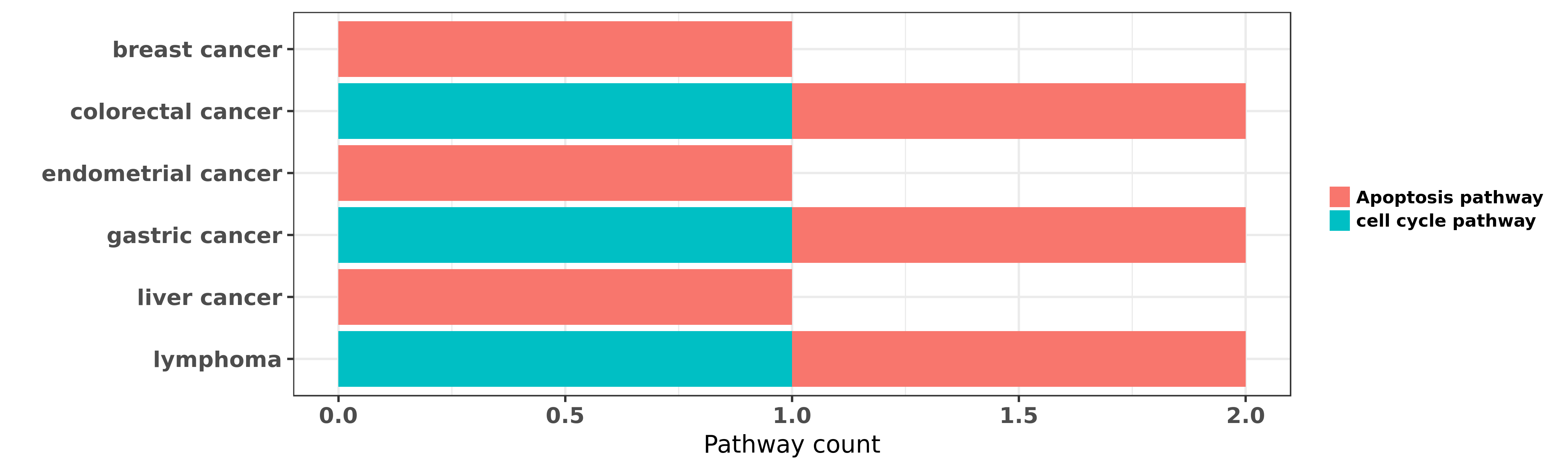
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-191-5p | breast cancer | Apoptosis pathway | "MiR-191/425 locus characterization revealed that t ......" | 23505378 | |
| hsa-miR-191-5p | colorectal cancer | cell cycle pathway; Apoptosis pathway | "miR 191 promotes tumorigenesis of human colorectal ......" | 25784653 | |
| hsa-miR-191-5p | endometrial cancer | Apoptosis pathway | "MiR 191 inhibits TNF α induced apoptosis of ovari ......" | 26191186 | Luciferase |
| hsa-miR-191-5p | gastric cancer | Apoptosis pathway | "MicroRNA 191 targets N deacetylase/N sulfotransfer ......" | 21947487 | Colony formation |
| hsa-miR-191-5p | gastric cancer | cell cycle pathway | "In this study the expression of miR-191 and miR-42 ......" | 24603541 | |
| hsa-miR-191-5p | liver cancer | Apoptosis pathway | "hsa miR 191 is a candidate oncogene target for hep ......" | 20924108 | |
| hsa-miR-191-5p | lymphoma | cell cycle pathway; Apoptosis pathway | "Expression of microRNA 191 in T lymphoblastic leuk ......" | 27093985 | Flow cytometry |
Reported cancer prognosis affected by hsa-miR-191-5p
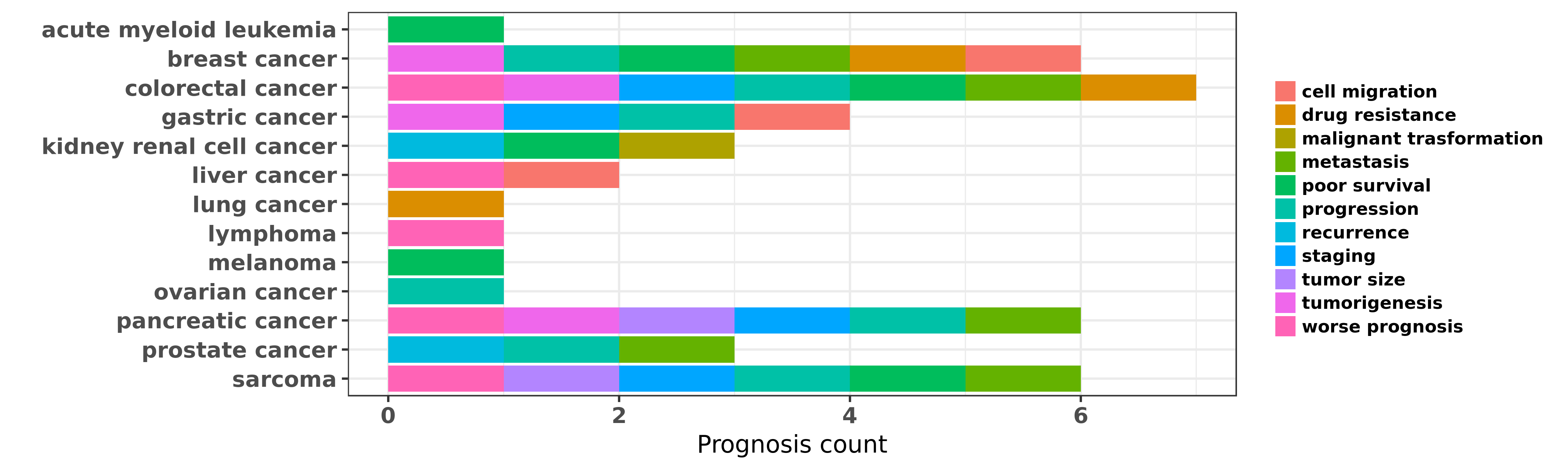
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-191-5p | acute myeloid leukemia | poor survival | "To determine whether miRNAs are associated with cy ......" | 18187662 | |
| hsa-miR-191-5p | acute myeloid leukemia | poor survival | "Finally reduction of the B subunit resulted in an ......" | 24858343 | |
| hsa-miR-191-5p | breast cancer | metastasis; tumorigenesis | "MiR-191/425 locus characterization revealed that t ......" | 23505378 | |
| hsa-miR-191-5p | breast cancer | poor survival; drug resistance; progression | "MicroRNA 191 an estrogen responsive microRNA funct ......" | 23542418 | |
| hsa-miR-191-5p | breast cancer | progression | "MicroRNA microArray analysis was performed compari ......" | 24626163 | |
| hsa-miR-191-5p | breast cancer | poor survival; cell migration; progression | "HIF inducible miR 191 promotes migration in breast ......" | 25867965 | |
| hsa-miR-191-5p | colorectal cancer | worse prognosis; progression; staging; metastasis; poor survival | "MicroRNA 191 correlates with poor prognosis of col ......" | 24050546 | |
| hsa-miR-191-5p | colorectal cancer | worse prognosis; progression; staging; metastasis; poor survival | "MicroRNA 191 correlates with poor prognosis of col ......" | 24195505 | |
| hsa-miR-191-5p | colorectal cancer | tumorigenesis; worse prognosis; progression; drug resistance | "miR 191 promotes tumorigenesis of human colorectal ......" | 25784653 | |
| hsa-miR-191-5p | gastric cancer | staging; progression; cell migration; tumorigenesis | "In this study the expression of miR-191 and miR-42 ......" | 24603541 | |
| hsa-miR-191-5p | kidney renal cell cancer | malignant trasformation | "Analysis of serum microRNAs miR 26a 2* miR 191 miR ......" | 22542158 | |
| hsa-miR-191-5p | kidney renal cell cancer | poor survival; recurrence | "In the survival analyses the expression levels of ......" | 25981392 | |
| hsa-miR-191-5p | liver cancer | worse prognosis; cell migration | "Hypomethylation of the hsa miR 191 locus causes hi ......" | 21969817 | Western blot |
| hsa-miR-191-5p | lung cancer | drug resistance | "Inhibition of miR 191 contributes to radiation res ......" | 25685068 | Western blot |
| hsa-miR-191-5p | lymphoma | worse prognosis | "Expression of microRNA 191 in T lymphoblastic leuk ......" | 27093985 | Flow cytometry |
| hsa-miR-191-5p | melanoma | poor survival | "Furthermore low expression of miR-191 and high exp ......" | 20357817 | |
| hsa-miR-191-5p | ovarian cancer | progression | "We identified an SNP SNP34091 in the 3'-UTR of MDM ......" | 21084273 | |
| hsa-miR-191-5p | pancreatic cancer | worse prognosis; staging; metastasis; tumor size | "The clinical significance and regulation mechanism ......" | 25119596 | Western blot; Luciferase |
| hsa-miR-191-5p | pancreatic cancer | progression; tumorigenesis | "MicroRNA 191 promotes pancreatic cancer progressio ......" | 25168367 | Luciferase |
| hsa-miR-191-5p | prostate cancer | recurrence | "Using the Cox regression test the risk of recurren ......" | 21255804 | |
| hsa-miR-191-5p | prostate cancer | metastasis; progression | "Computational predictions revealed that the rs4245 ......" | 25670033 | |
| hsa-miR-191-5p | sarcoma | worse prognosis; staging; metastasis; tumor size; poor survival; progression | "Increased expression of microRNA 191 as a potentia ......" | 26406942 |
Reported gene related to hsa-miR-191-5p
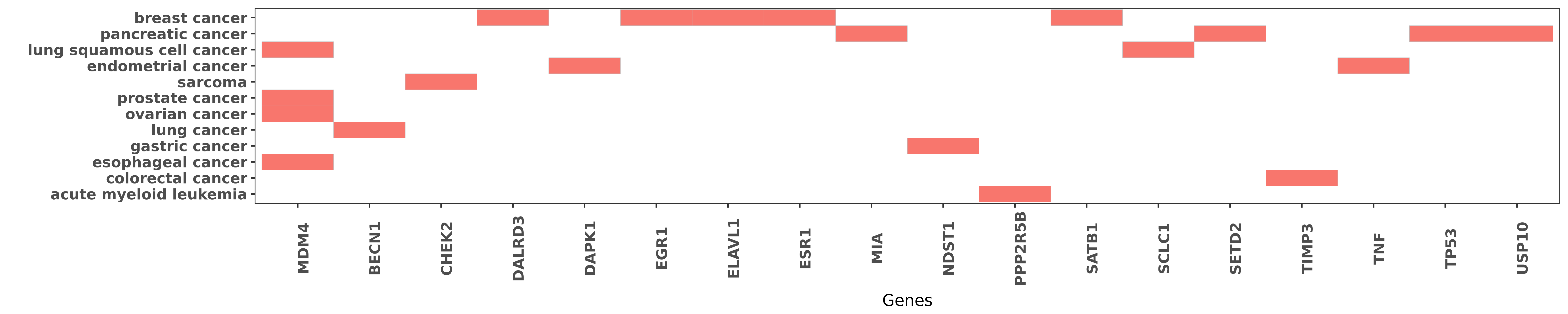
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-191-5p | esophageal cancer | MDM4 | "Association of a genetic variation in a miR 191 bi ......" | 23724042 |
| hsa-miR-191-5p | lung squamous cell cancer | MDM4 | "After co-tranfection of miRNAs and different allel ......" | 26274820 |
| hsa-miR-191-5p | ovarian cancer | MDM4 | "We identified an SNP SNP34091 in the 3'-UTR of MDM ......" | 21084273 |
| hsa-miR-191-5p | prostate cancer | MDM4 | "Herein we show using reporter gene assays and endo ......" | 25670033 |
| hsa-miR-191-5p | breast cancer | ESR1 | "MiR-191/425 locus characterization revealed that t ......" | 23505378 |
| hsa-miR-191-5p | breast cancer | ESR1 | "miR-191 is a direct estrogen receptor ER target an ......" | 23542418 |
| hsa-miR-191-5p | colorectal cancer | TIMP3 | "Furthermore we found that tissue inhibitor of meta ......" | 24195505 |
| hsa-miR-191-5p | colorectal cancer | TIMP3 | "Furthermore we found that tissue inhibitor of meta ......" | 24050546 |
| hsa-miR-191-5p | lung cancer | BECN1 | "Two binding sites between Beclin-1 and miR-191 sug ......" | 25685068 |
| hsa-miR-191-5p | sarcoma | CHEK2 | "MicroRNA 191 promotes osteosarcoma cells prolifera ......" | 25773391 |
| hsa-miR-191-5p | breast cancer | DALRD3 | "MiR-191/425 locus characterization revealed that t ......" | 23505378 |
| hsa-miR-191-5p | endometrial cancer | DAPK1 | "MiR 191 inhibits TNF α induced apoptosis of ovari ......" | 26191186 |
| hsa-miR-191-5p | breast cancer | EGR1 | "We demonstrated that miR-191 protects ERα positiv ......" | 23505378 |
| hsa-miR-191-5p | breast cancer | ELAVL1 | "We further established that miR-191 is a critical ......" | 25867965 |
| hsa-miR-191-5p | pancreatic cancer | MIA | "After hypoxic cultured for 6 and 12 h qRT-PCR and ......" | 25119596 |
| hsa-miR-191-5p | gastric cancer | NDST1 | "The N-deacetylase/N-sulfotransferase 1 NDST1 was c ......" | 21947487 |
| hsa-miR-191-5p | acute myeloid leukemia | PPP2R5B | "Finally reduction of the B subunit resulted in an ......" | 24858343 |
| hsa-miR-191-5p | breast cancer | SATB1 | "miR-191 and SATB1 show inverse correlation of expr ......" | 23542418 |
| hsa-miR-191-5p | lung squamous cell cancer | SCLC1 | "After co-tranfection of miRNAs and different allel ......" | 26274820 |
| hsa-miR-191-5p | pancreatic cancer | SETD2 | "The aim of study was to discuss the correlation an ......" | 25119596 |
| hsa-miR-191-5p | endometrial cancer | TNF | "MiR 191 inhibits TNF α induced apoptosis of ovari ......" | 26191186 |
| hsa-miR-191-5p | pancreatic cancer | TP53 | "At the molecular level bioinformatic prediction lu ......" | 25168367 |
| hsa-miR-191-5p | pancreatic cancer | USP10 | "MicroRNA 191 promotes pancreatic cancer progressio ......" | 25168367 |
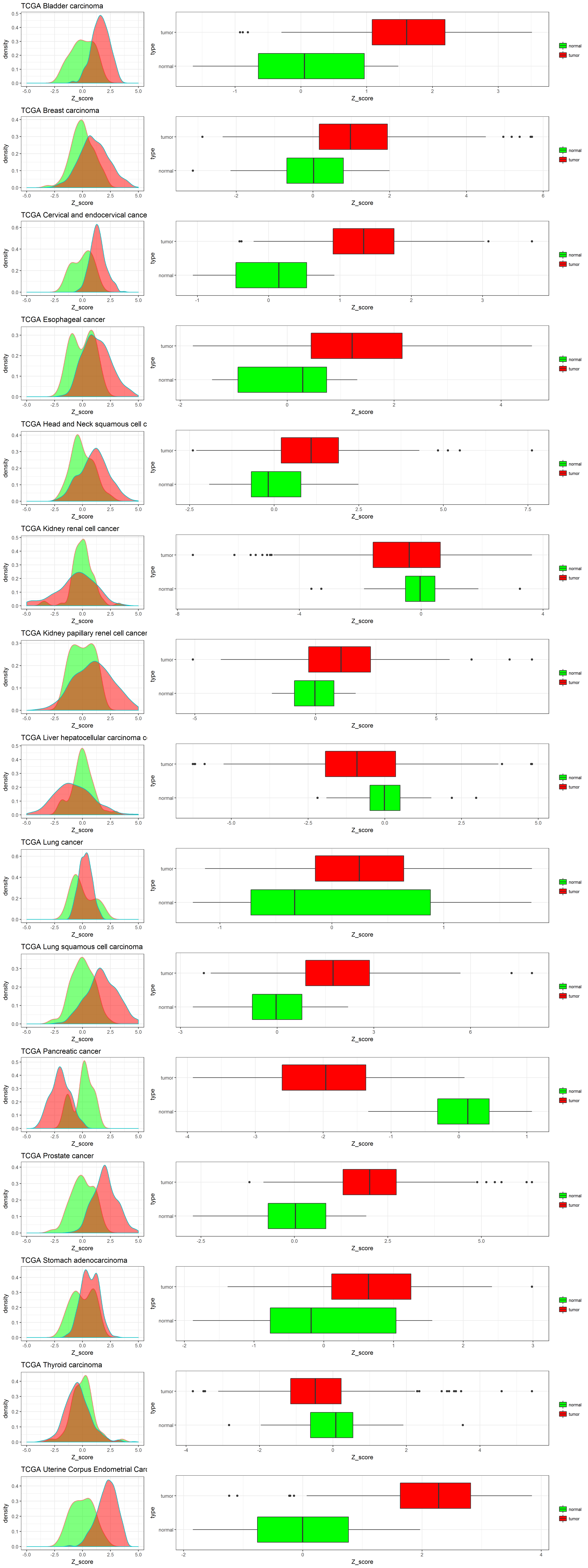
Expression profile in cancer corhorts:
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-191-5p | ATF6 | 12 cancers: BLCA; COAD; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; PRAD; SARC; THCA; UCEC | miRNAWalker2 validate | TCGA BLCA -0.068; TCGA COAD -0.085; TCGA HNSC -0.153; TCGA KIRP -0.241; TCGA LGG -0.072; TCGA LIHC -0.177; TCGA LUAD -0.097; TCGA LUSC -0.126; TCGA PRAD -0.259; TCGA SARC -0.146; TCGA THCA -0.097; TCGA UCEC -0.071 |
hsa-miR-191-5p | CDK6 | 10 cancers: BLCA; BRCA; CESC; HNSC; KIRP; LIHC; PAAD; PRAD; THCA; UCEC | miRNAWalker2 validate; miRTarBase | TCGA BLCA -0.712; TCGA BRCA -0.207; TCGA CESC -0.227; TCGA HNSC -0.304; TCGA KIRP -0.165; TCGA LIHC -0.29; TCGA PAAD -0.254; TCGA PRAD -0.141; TCGA THCA -0.164; TCGA UCEC -0.325 |
hsa-miR-191-5p | ENAH | 12 cancers: BLCA; COAD; ESCA; HNSC; LGG; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD | miRNAWalker2 validate | TCGA BLCA -0.255; TCGA COAD -0.121; TCGA ESCA -0.314; TCGA HNSC -0.195; TCGA LGG -0.202; TCGA LUAD -0.075; TCGA LUSC -0.122; TCGA PAAD -0.222; TCGA PRAD -0.189; TCGA SARC -0.252; TCGA THCA -0.086; TCGA STAD -0.31 |
hsa-miR-191-5p | MCFD2 | 11 cancers: BLCA; COAD; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.22; TCGA COAD -0.093; TCGA HNSC -0.166; TCGA KIRP -0.13; TCGA LIHC -0.105; TCGA LUAD -0.1; TCGA LUSC -0.16; TCGA PAAD -0.238; TCGA PRAD -0.202; TCGA STAD -0.122; TCGA UCEC -0.212 |
hsa-miR-191-5p | NDST1 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; OV; THCA; STAD; UCEC | miRTarBase; miRNATAP | TCGA BLCA -0.22; TCGA BRCA -0.101; TCGA CESC -0.274; TCGA COAD -0.18; TCGA ESCA -0.309; TCGA HNSC -0.262; TCGA KIRC -0.148; TCGA KIRP -0.078; TCGA LIHC -0.112; TCGA LUAD -0.102; TCGA LUSC -0.421; TCGA OV -0.223; TCGA THCA -0.062; TCGA STAD -0.103; TCGA UCEC -0.115 |
hsa-miR-191-5p | MAPRE2 | 11 cancers: BLCA; COAD; HNSC; LUAD; LUSC; OV; PRAD; SARC; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.421; TCGA COAD -0.158; TCGA HNSC -0.22; TCGA LUAD -0.074; TCGA LUSC -0.304; TCGA OV -0.118; TCGA PRAD -0.399; TCGA SARC -0.127; TCGA THCA -0.219; TCGA STAD -0.088; TCGA UCEC -0.188 |
hsa-miR-191-5p | CBFA2T3 | 9 cancers: BLCA; CESC; COAD; ESCA; KIRC; LUSC; OV; STAD; UCEC | MirTarget | TCGA BLCA -0.218; TCGA CESC -0.566; TCGA COAD -0.362; TCGA ESCA -0.527; TCGA KIRC -0.265; TCGA LUSC -0.316; TCGA OV -0.238; TCGA STAD -0.33; TCGA UCEC -0.267 |
hsa-miR-191-5p | SBDS | 15 cancers: BLCA; CESC; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; OV; PAAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.218; TCGA CESC -0.137; TCGA HNSC -0.124; TCGA KIRP -0.107; TCGA LGG -0.076; TCGA LIHC -0.102; TCGA LUAD -0.11; TCGA LUSC -0.214; TCGA OV -0.083; TCGA PAAD -0.108; TCGA PRAD -0.154; TCGA SARC -0.217; TCGA THCA -0.07; TCGA STAD -0.106; TCGA UCEC -0.334 |
hsa-miR-191-5p | CCPG1 | 13 cancers: BLCA; COAD; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.344; TCGA COAD -0.223; TCGA HNSC -0.093; TCGA KIRP -0.257; TCGA LIHC -0.126; TCGA LUAD -0.176; TCGA LUSC -0.39; TCGA PAAD -0.132; TCGA PRAD -0.258; TCGA SARC -0.117; TCGA THCA -0.084; TCGA STAD -0.091; TCGA UCEC -0.171 |
hsa-miR-191-5p | TJP1 | 14 cancers: BLCA; CESC; COAD; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.135; TCGA CESC -0.154; TCGA COAD -0.112; TCGA HNSC -0.294; TCGA KIRP -0.079; TCGA LGG -0.088; TCGA LIHC -0.111; TCGA LUAD -0.209; TCGA LUSC -0.365; TCGA PAAD -0.179; TCGA PRAD -0.074; TCGA SARC -0.129; TCGA STAD -0.234; TCGA UCEC -0.19 |
hsa-miR-191-5p | PRDM16 | 11 cancers: BLCA; BRCA; HNSC; KIRP; LIHC; LUAD; LUSC; OV; PRAD; SARC; UCEC | mirMAP | TCGA BLCA -0.382; TCGA BRCA -0.419; TCGA HNSC -0.312; TCGA KIRP -0.868; TCGA LIHC -0.368; TCGA LUAD -0.509; TCGA LUSC -0.672; TCGA OV -0.456; TCGA PRAD -0.378; TCGA SARC -0.878; TCGA UCEC -0.375 |
hsa-miR-191-5p | CUX1 | 11 cancers: BLCA; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; OV; PAAD; STAD; UCEC | mirMAP | TCGA BLCA -0.141; TCGA ESCA -0.214; TCGA HNSC -0.194; TCGA LGG -0.12; TCGA LIHC -0.212; TCGA LUAD -0.108; TCGA LUSC -0.142; TCGA OV -0.145; TCGA PAAD -0.142; TCGA STAD -0.102; TCGA UCEC -0.104 |
hsa-miR-191-5p | GAP43 | 9 cancers: BLCA; BRCA; COAD; ESCA; OV; PAAD; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.751; TCGA BRCA -0.264; TCGA COAD -1.6; TCGA ESCA -0.827; TCGA OV -0.378; TCGA PAAD -0.773; TCGA PRAD -0.316; TCGA STAD -0.841; TCGA UCEC -0.339 |
hsa-miR-191-5p | ZCCHC24 | 16 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; OV; PAAD; PRAD; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.656; TCGA BRCA -0.384; TCGA CESC -0.184; TCGA COAD -0.549; TCGA ESCA -0.64; TCGA HNSC -0.321; TCGA LGG -0.29; TCGA LIHC -0.206; TCGA LUAD -0.19; TCGA LUSC -0.713; TCGA OV -0.412; TCGA PAAD -0.453; TCGA PRAD -0.315; TCGA THCA -0.086; TCGA STAD -0.616; TCGA UCEC -0.551 |
hsa-miR-191-5p | EGR1 | 12 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.591; TCGA BRCA -0.517; TCGA CESC -0.351; TCGA ESCA -0.522; TCGA HNSC -0.178; TCGA KIRP -0.428; TCGA LUAD -0.243; TCGA LUSC -0.512; TCGA PAAD -0.356; TCGA PRAD -0.593; TCGA STAD -0.274; TCGA UCEC -0.439 |
hsa-miR-191-5p | TMOD2 | 14 cancers: BLCA; BRCA; CESC; COAD; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PRAD; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.247; TCGA BRCA -0.126; TCGA CESC -0.421; TCGA COAD -0.362; TCGA KIRC -0.264; TCGA KIRP -0.285; TCGA LGG -0.159; TCGA LIHC -0.25; TCGA LUAD -0.28; TCGA LUSC -0.269; TCGA PRAD -0.352; TCGA THCA -0.246; TCGA STAD -0.392; TCGA UCEC -0.527 |
hsa-miR-191-5p | ACSL1 | 9 cancers: BRCA; CESC; COAD; ESCA; HNSC; LUSC; OV; STAD; UCEC | miRNAWalker2 validate | TCGA BRCA -0.169; TCGA CESC -0.229; TCGA COAD -0.223; TCGA ESCA -0.363; TCGA HNSC -0.289; TCGA LUSC -0.257; TCGA OV -0.207; TCGA STAD -0.096; TCGA UCEC -0.175 |
hsa-miR-191-5p | MTSS1L | 12 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BRCA -0.149; TCGA CESC -0.273; TCGA COAD -0.118; TCGA ESCA -0.386; TCGA HNSC -0.107; TCGA KIRC -0.178; TCGA KIRP -0.197; TCGA LGG -0.237; TCGA PRAD -0.186; TCGA SARC -0.269; TCGA STAD -0.259; TCGA UCEC -0.077 |
hsa-miR-191-5p | MAPRE3 | 9 cancers: BRCA; CESC; ESCA; KIRP; LIHC; LUAD; LUSC; SARC; STAD | MirTarget; miRNATAP | TCGA BRCA -0.099; TCGA CESC -0.208; TCGA ESCA -0.334; TCGA KIRP -0.113; TCGA LIHC -0.194; TCGA LUAD -0.113; TCGA LUSC -0.14; TCGA SARC -0.238; TCGA STAD -0.225 |
hsa-miR-191-5p | DDX6 | 10 cancers: HNSC; KIRP; LGG; LIHC; LUAD; OV; PRAD; SARC; STAD; UCEC | MirTarget | TCGA HNSC -0.093; TCGA KIRP -0.115; TCGA LGG -0.124; TCGA LIHC -0.076; TCGA LUAD -0.133; TCGA OV -0.061; TCGA PRAD -0.054; TCGA SARC -0.077; TCGA STAD -0.151; TCGA UCEC -0.067 |
Enriched cancer pathways of putative targets