microRNA information: hsa-miR-193a-3p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-193a-3p | miRbase |
| Accession: | MIMAT0000459 | miRbase |
| Precursor name: | hsa-mir-193a | miRbase |
| Precursor accession: | MI0000487 | miRbase |
| Symbol: | MIR193A | HGNC |
| RefSeq ID: | NR_029710 | GenBank |
| Sequence: | AACUGGCCUACAAAGUCCCAGU |
Reported expression in cancers: hsa-miR-193a-3p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-193a-3p | glioblastoma | deregulation | "Hypoxic signature of microRNAs in glioblastoma: in ......" | 25129238 | RNA-Seq |
| hsa-miR-193a-3p | lung cancer | downregulation | "The changes of EMT-related proteins were analyzed ......" | 22325218 | Microarray; qPCR |
| hsa-miR-193a-3p | lung squamous cell cancer | downregulation | "MicroRNA 193a 3p and 5p suppress the metastasis of ......" | 24469061 | |
| hsa-miR-193a-3p | prostate cancer | deregulation | "Upregulation of miR-122 miR-335 miR-184 miR-193 mi ......" | 23781281 | |
| hsa-miR-193a-3p | thyroid cancer | deregulation | "Deregulated miRNAs were confirmed by quantitative ......" | 24127332 | qPCR; in situ hybridization |
Reported cancer pathway affected by hsa-miR-193a-3p
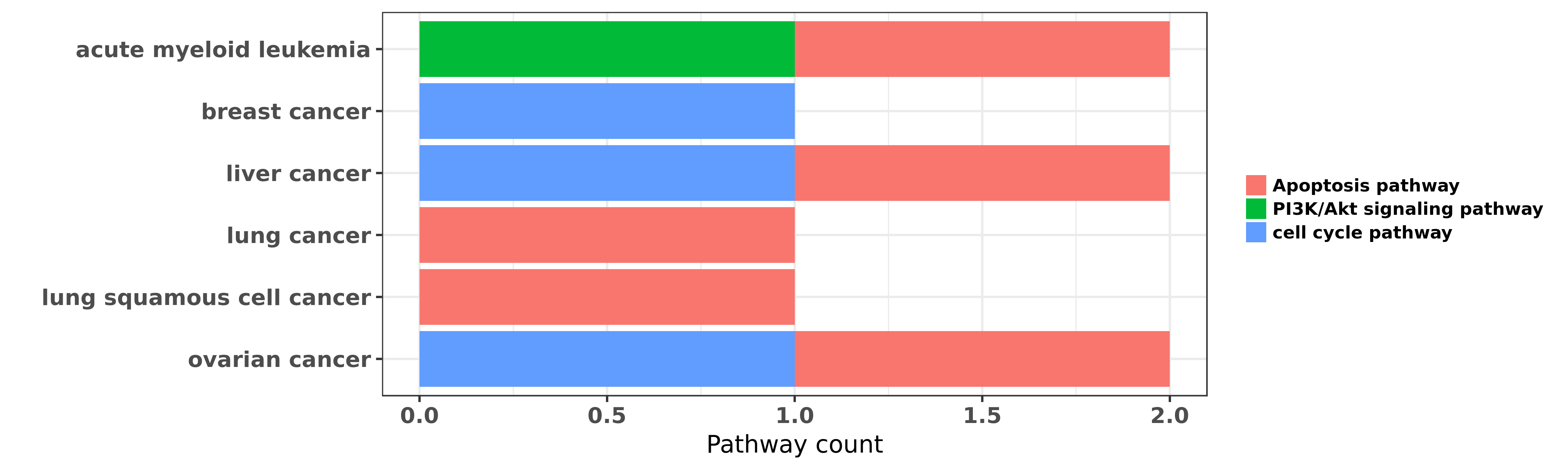
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-193a-3p | acute myeloid leukemia | Apoptosis pathway | "MicroRNA 193a represses c kit expression and funct ......" | 21399664 | Luciferase |
| hsa-miR-193a-3p | acute myeloid leukemia | Apoptosis pathway; PI3K/Akt signaling pathway | "Epigenetic silencing of microRNA 193a contributes ......" | 23223432 | |
| hsa-miR-193a-3p | acute myeloid leukemia | Apoptosis pathway | "Long non coding RNA HOTAIR modulates c KIT express ......" | 25979172 | Colony formation |
| hsa-miR-193a-3p | breast cancer | cell cycle pathway | "Finally our approach led us to identify and valida ......" | 22333974 | |
| hsa-miR-193a-3p | liver cancer | cell cycle pathway | "DNA methylation regulated miR 193a 3p dictates res ......" | 22117060 | |
| hsa-miR-193a-3p | liver cancer | Apoptosis pathway | "Effects of miR 193a and sorafenib on hepatocellula ......" | 24330766 | Luciferase; Western blot |
| hsa-miR-193a-3p | lung cancer | Apoptosis pathway | "miR 193a 3p functions as a tumor suppressor in lun ......" | 25391651 | |
| hsa-miR-193a-3p | lung squamous cell cancer | Apoptosis pathway | "Demethylation of miR 9 3 and miR 193a genes suppre ......" | 24356455 | Flow cytometry; Western blot |
| hsa-miR-193a-3p | ovarian cancer | Apoptosis pathway; cell cycle pathway | "Gain of function microRNA screens identify miR 193 ......" | 23588298 |
Reported cancer prognosis affected by hsa-miR-193a-3p
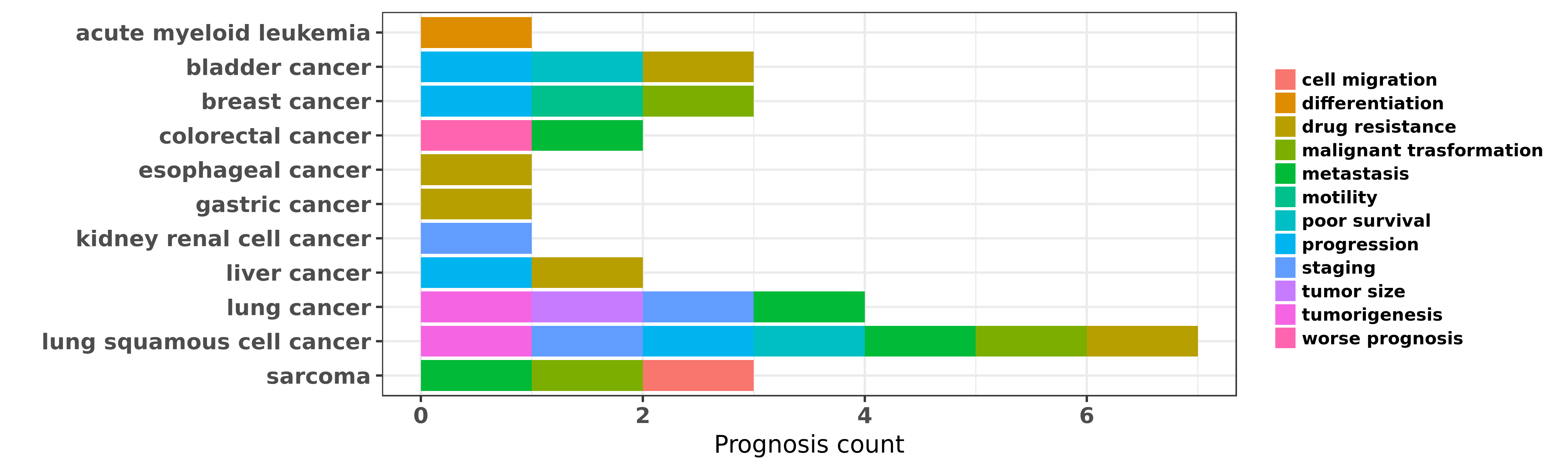
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-193a-3p | acute myeloid leukemia | differentiation | "MicroRNA 193a represses c kit expression and funct ......" | 21399664 | Luciferase |
| hsa-miR-193a-3p | acute myeloid leukemia | differentiation | "Epigenetic silencing of microRNA 193a contributes ......" | 23223432 | |
| hsa-miR-193a-3p | bladder cancer | drug resistance | "The DNA methylation regulated miR 193a 3p dictates ......" | 25188512 | |
| hsa-miR-193a-3p | bladder cancer | drug resistance | "miR 193a 3p regulates the multi drug resistance of ......" | 25311867 | |
| hsa-miR-193a-3p | bladder cancer | drug resistance | "MiR 193a 3p promotes the multi chemoresistance of ......" | 25444900 | |
| hsa-miR-193a-3p | bladder cancer | drug resistance | "The miR 193a 3p regulated PSEN1 gene suppresses th ......" | 25542424 | |
| hsa-miR-193a-3p | bladder cancer | progression; poor survival | "Of the eight most important progression-related mi ......" | 25990459 | |
| hsa-miR-193a-3p | bladder cancer | drug resistance | "The miR 193a 3p regulated ING5 gene activates the ......" | 25991669 | |
| hsa-miR-193a-3p | bladder cancer | drug resistance | "Focusing on the major obstacle regarding MIBC pati ......" | 27586262 | |
| hsa-miR-193a-3p | bladder cancer | drug resistance | "MiR 193a 5p Targets the Coding Region of AP 2α mR ......" | 27698912 | |
| hsa-miR-193a-3p | breast cancer | progression | "Finally our approach led us to identify and valida ......" | 22333974 | |
| hsa-miR-193a-3p | breast cancer | malignant trasformation | "In support of these findings in vitro functional s ......" | 24104550 | |
| hsa-miR-193a-3p | breast cancer | motility | "Arm Selection Preference of MicroRNA 193a Varies i ......" | 27307030 | |
| hsa-miR-193a-3p | colorectal cancer | metastasis; worse prognosis | "Downregulation of miR 193a 5p correlates with lymp ......" | 25232258 | |
| hsa-miR-193a-3p | esophageal cancer | drug resistance | "miR 193a 3p regulation of chemoradiation resistanc ......" | 26743123 | |
| hsa-miR-193a-3p | esophageal cancer | drug resistance | "MiR 193a 5p/ERBB2 act as concurrent chemoradiation ......" | 27203740 | |
| hsa-miR-193a-3p | gastric cancer | drug resistance | "Downregulation of microRNA 193 3p inhibits tumor p ......" | 26753960 | Western blot; Luciferase |
| hsa-miR-193a-3p | kidney renal cell cancer | staging | "Markedly dysregulated miRNAs in RCC cases were sub ......" | 25556603 | |
| hsa-miR-193a-3p | liver cancer | drug resistance; progression | "DNA methylation regulated miR 193a 3p dictates res ......" | 22117060 | |
| hsa-miR-193a-3p | lung cancer | tumorigenesis; staging; metastasis; tumor size | "Expression and clinicopathological significance of ......" | 26257582 | |
| hsa-miR-193a-3p | lung squamous cell cancer | staging; malignant trasformation | "Genome wide miRNA expression profiling identifies ......" | 22282464 | |
| hsa-miR-193a-3p | lung squamous cell cancer | progression; tumorigenesis; poor survival | "Demethylation of miR 9 3 and miR 193a genes suppre ......" | 24356455 | Flow cytometry; Western blot |
| hsa-miR-193a-3p | lung squamous cell cancer | metastasis | "MicroRNA 193a 3p and 5p suppress the metastasis of ......" | 24469061 | |
| hsa-miR-193a-3p | lung squamous cell cancer | metastasis; progression; drug resistance | "Quantitative proteomic analysis of the metastasis ......" | 25833338 | Western blot |
| hsa-miR-193a-3p | sarcoma | malignant trasformation | "Differential microRNA expression profiles between ......" | 24287458 | |
| hsa-miR-193a-3p | sarcoma | metastasis; cell migration | "MiR 193a 3p and miR 193a 5p suppress the metastasi ......" | 26913720 |
Reported gene related to hsa-miR-193a-3p
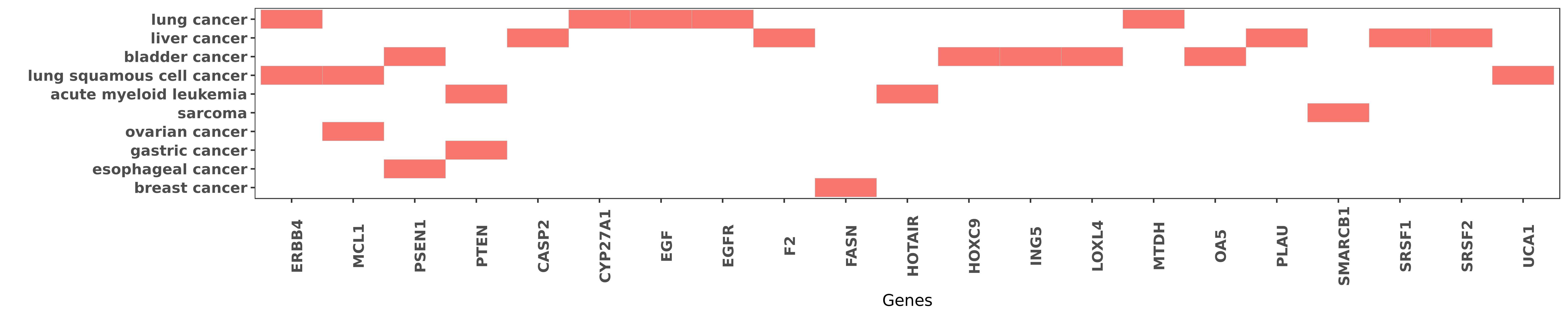
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-193a-3p | lung cancer | ERBB4 | "miR 193a 3p functions as a tumor suppressor in lun ......" | 25391651 |
| hsa-miR-193a-3p | lung squamous cell cancer | ERBB4 | "Moreover mechanistic investigations showed that UC ......" | 26655272 |
| hsa-miR-193a-3p | lung squamous cell cancer | MCL1 | "Treatment with 5-AzaC concomitantly upregulated ex ......" | 24356455 |
| hsa-miR-193a-3p | ovarian cancer | MCL1 | "We demonstrated that miR-193a decreased the amount ......" | 23588298 |
| hsa-miR-193a-3p | bladder cancer | PSEN1 | "The miR 193a 3p regulated PSEN1 gene suppresses th ......" | 25542424 |
| hsa-miR-193a-3p | esophageal cancer | PSEN1 | "miR 193a 3p regulation of chemoradiation resistanc ......" | 26743123 |
| hsa-miR-193a-3p | acute myeloid leukemia | PTEN | "Our study identifies miR-193a and PTEN as targets ......" | 23223432 |
| hsa-miR-193a-3p | gastric cancer | PTEN | "Downregulation of microRNA 193 3p inhibits tumor p ......" | 26753960 |
| hsa-miR-193a-3p | liver cancer | CASP2 | "This newly identified miR-193a-3p-SRSF2 axis highl ......" | 22117060 |
| hsa-miR-193a-3p | lung cancer | CYP27A1 | "Only miR-193 was associated with biochemical marke ......" | 23142363 |
| hsa-miR-193a-3p | lung cancer | EGF | "The results showed that miR-193a-3p level was decr ......" | 26257582 |
| hsa-miR-193a-3p | lung cancer | EGFR | "The results showed that miR-193a-3p level was decr ......" | 26257582 |
| hsa-miR-193a-3p | liver cancer | F2 | "We found miR-193a down-regulation in HCC respect t ......" | 24330766 |
| hsa-miR-193a-3p | breast cancer | FASN | "Profiling studies also exposed a rapid metformin-i ......" | 25213330 |
| hsa-miR-193a-3p | acute myeloid leukemia | HOTAIR | "Finally HOTAIR modulated c-KIT expression by compe ......" | 25979172 |
| hsa-miR-193a-3p | bladder cancer | HOXC9 | "MiR 193a 3p promotes the multi chemoresistance of ......" | 25444900 |
| hsa-miR-193a-3p | bladder cancer | ING5 | "The miR 193a 3p regulated ING5 gene activates the ......" | 25991669 |
| hsa-miR-193a-3p | bladder cancer | LOXL4 | "miR 193a 3p regulates the multi drug resistance of ......" | 25311867 |
| hsa-miR-193a-3p | lung cancer | MTDH | "Aberrant expression of miR-193a-3p and astrocyte e ......" | 26257582 |
| hsa-miR-193a-3p | bladder cancer | OA5 | "MiR 193a 5p Targets the Coding Region of AP 2α mR ......" | 27698912 |
| hsa-miR-193a-3p | liver cancer | PLAU | "The molecular interaction between miR-193a and uPA ......" | 24330766 |
| hsa-miR-193a-3p | sarcoma | SMARCB1 | "Differential microRNA expression profiles between ......" | 24287458 |
| hsa-miR-193a-3p | liver cancer | SRSF1 | "Genomic profiling at both DNA methylation and micr ......" | 22117060 |
| hsa-miR-193a-3p | liver cancer | SRSF2 | "DNA methylation regulated miR 193a 3p dictates res ......" | 22117060 |
| hsa-miR-193a-3p | lung squamous cell cancer | UCA1 | "LncRNA UCA1 exerts oncogenic functions in non smal ......" | 26655272 |
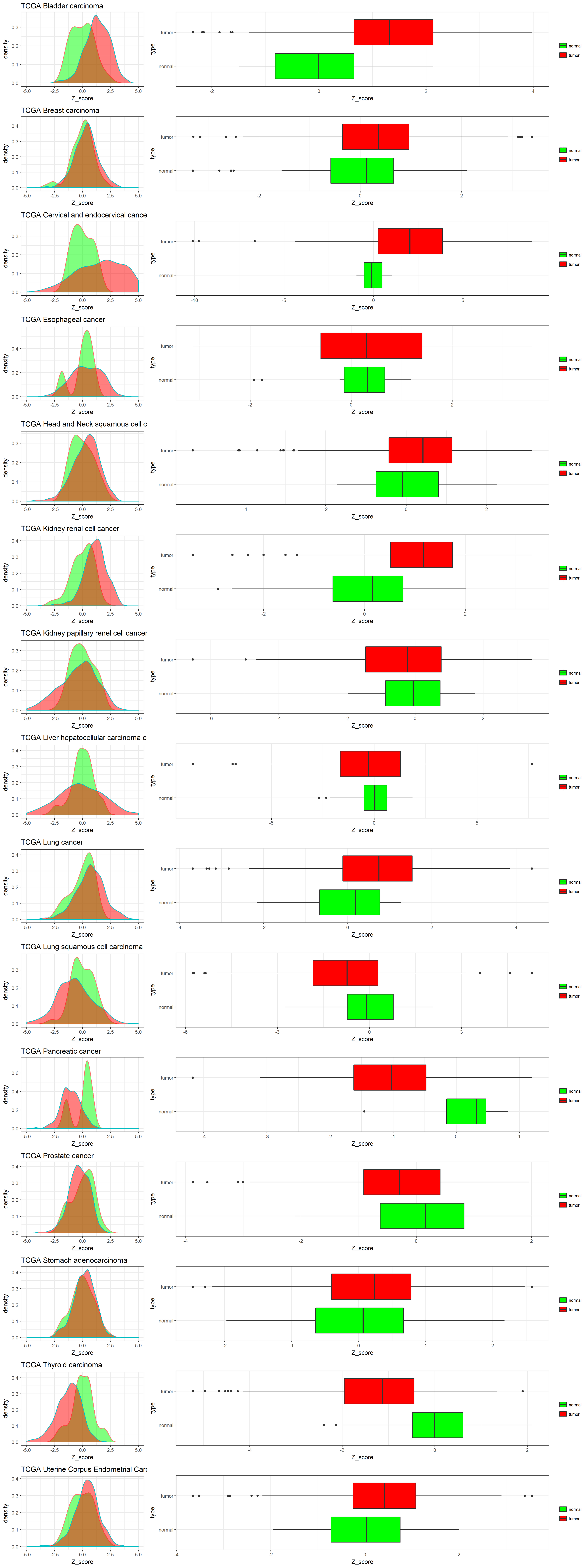
Expression profile in cancer corhorts:
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-193a-3p | TAPT1 | 13 cancers: BLCA; CESC; COAD; HNSC; KIRC; LGG; LIHC; LUAD; PAAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate; MirTarget; miRanda | TCGA BLCA -0.121; TCGA CESC -0.074; TCGA COAD -0.076; TCGA HNSC -0.056; TCGA KIRC -0.166; TCGA LGG -0.104; TCGA LIHC -0.105; TCGA LUAD -0.207; TCGA PAAD -0.123; TCGA PRAD -0.066; TCGA SARC -0.169; TCGA STAD -0.069; TCGA UCEC -0.064 |
hsa-miR-193a-3p | TGFBR3 | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LIHC; LUSC; SARC; STAD | MirTarget; miRanda; miRNATAP | TCGA BLCA -0.404; TCGA BRCA -0.375; TCGA CESC -0.216; TCGA ESCA -0.31; TCGA HNSC -0.56; TCGA KIRC -0.365; TCGA LIHC -0.192; TCGA LUSC -0.245; TCGA SARC -0.401; TCGA STAD -0.256 |
hsa-miR-193a-3p | ARHGEF12 | 13 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget; PITA; miRanda; miRNATAP | TCGA BLCA -0.138; TCGA CESC -0.192; TCGA COAD -0.106; TCGA ESCA -0.092; TCGA HNSC -0.096; TCGA KIRC -0.249; TCGA LIHC -0.098; TCGA LUAD -0.137; TCGA PRAD -0.076; TCGA SARC -0.109; TCGA THCA -0.073; TCGA STAD -0.168; TCGA UCEC -0.062 |
hsa-miR-193a-3p | ABI2 | 12 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LGG; LUAD; PAAD; PRAD; SARC; STAD | MirTarget; miRanda | TCGA BLCA -0.09; TCGA BRCA -0.063; TCGA CESC -0.21; TCGA HNSC -0.208; TCGA KIRC -0.196; TCGA KIRP -0.098; TCGA LGG -0.063; TCGA LUAD -0.065; TCGA PAAD -0.12; TCGA PRAD -0.1; TCGA SARC -0.11; TCGA STAD -0.14 |
hsa-miR-193a-3p | KIAA0430 | 11 cancers: BLCA; BRCA; CESC; HNSC; KIRC; LGG; LIHC; LUAD; SARC; STAD; UCEC | MirTarget; miRanda | TCGA BLCA -0.147; TCGA BRCA -0.053; TCGA CESC -0.104; TCGA HNSC -0.1; TCGA KIRC -0.06; TCGA LGG -0.07; TCGA LIHC -0.071; TCGA LUAD -0.085; TCGA SARC -0.118; TCGA STAD -0.119; TCGA UCEC -0.067 |
hsa-miR-193a-3p | ZNF562 | 9 cancers: BLCA; COAD; KIRC; KIRP; LIHC; PAAD; SARC; THCA; STAD | MirTarget; miRanda | TCGA BLCA -0.053; TCGA COAD -0.095; TCGA KIRC -0.15; TCGA KIRP -0.06; TCGA LIHC -0.085; TCGA PAAD -0.098; TCGA SARC -0.084; TCGA THCA -0.051; TCGA STAD -0.081 |
hsa-miR-193a-3p | WDR82 | 9 cancers: BLCA; ESCA; HNSC; KIRC; KIRP; LIHC; PAAD; SARC; STAD | MirTarget; miRanda; miRNATAP | TCGA BLCA -0.086; TCGA ESCA -0.075; TCGA HNSC -0.055; TCGA KIRC -0.069; TCGA KIRP -0.073; TCGA LIHC -0.078; TCGA PAAD -0.118; TCGA SARC -0.078; TCGA STAD -0.086 |
hsa-miR-193a-3p | NSF | 13 cancers: BLCA; CESC; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; THCA; UCEC | MirTarget; miRanda | TCGA BLCA -0.058; TCGA CESC -0.12; TCGA ESCA -0.164; TCGA HNSC -0.099; TCGA KIRC -0.084; TCGA KIRP -0.081; TCGA LIHC -0.093; TCGA LUAD -0.054; TCGA LUSC -0.085; TCGA PAAD -0.139; TCGA PRAD -0.139; TCGA THCA -0.074; TCGA UCEC -0.059 |
hsa-miR-193a-3p | JMY | 12 cancers: BLCA; CESC; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; PAAD; PRAD; SARC; STAD | MirTarget | TCGA BLCA -0.132; TCGA CESC -0.143; TCGA HNSC -0.104; TCGA KIRC -0.414; TCGA KIRP -0.081; TCGA LGG -0.089; TCGA LUAD -0.117; TCGA LUSC -0.094; TCGA PAAD -0.173; TCGA PRAD -0.062; TCGA SARC -0.238; TCGA STAD -0.199 |
hsa-miR-193a-3p | KIT | 10 cancers: BLCA; BRCA; CESC; HNSC; KIRC; LIHC; LUAD; PAAD; SARC; STAD | MirTarget; PITA; miRanda; miRNATAP | TCGA BLCA -0.489; TCGA BRCA -0.423; TCGA CESC -0.628; TCGA HNSC -0.488; TCGA KIRC -0.673; TCGA LIHC -0.332; TCGA LUAD -0.592; TCGA PAAD -0.393; TCGA SARC -0.238; TCGA STAD -0.292 |
hsa-miR-193a-3p | DCAF7 | 15 cancers: BLCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; UCEC | MirTarget; miRanda; miRNATAP | TCGA BLCA -0.073; TCGA CESC -0.152; TCGA ESCA -0.163; TCGA HNSC -0.147; TCGA KIRC -0.195; TCGA KIRP -0.178; TCGA LGG -0.093; TCGA LIHC -0.166; TCGA LUAD -0.076; TCGA LUSC -0.076; TCGA PAAD -0.145; TCGA PRAD -0.115; TCGA SARC -0.094; TCGA THCA -0.123; TCGA UCEC -0.052 |
hsa-miR-193a-3p | TMEM30A | 11 cancers: BLCA; CESC; COAD; HNSC; KIRC; LGG; LUAD; PRAD; SARC; THCA; STAD | MirTarget; PITA; miRanda; miRNATAP | TCGA BLCA -0.067; TCGA CESC -0.079; TCGA COAD -0.102; TCGA HNSC -0.064; TCGA KIRC -0.115; TCGA LGG -0.089; TCGA LUAD -0.098; TCGA PRAD -0.128; TCGA SARC -0.09; TCGA THCA -0.093; TCGA STAD -0.123 |
hsa-miR-193a-3p | ZMAT3 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.094; TCGA BRCA -0.091; TCGA CESC -0.208; TCGA COAD -0.225; TCGA ESCA -0.191; TCGA HNSC -0.27; TCGA KIRC -0.123; TCGA LIHC -0.209; TCGA SARC -0.147; TCGA THCA -0.375; TCGA STAD -0.116; TCGA UCEC -0.171 |
hsa-miR-193a-3p | BTRC | 10 cancers: BLCA; BRCA; KIRC; KIRP; LGG; LIHC; LUAD; PAAD; PRAD; STAD | MirTarget; miRanda | TCGA BLCA -0.12; TCGA BRCA -0.073; TCGA KIRC -0.194; TCGA KIRP -0.065; TCGA LGG -0.069; TCGA LIHC -0.149; TCGA LUAD -0.07; TCGA PAAD -0.108; TCGA PRAD -0.057; TCGA STAD -0.1 |
hsa-miR-193a-3p | CBX7 | 11 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LIHC; LUAD; PAAD; SARC; STAD; UCEC | MirTarget; miRanda; miRNATAP | TCGA BLCA -0.242; TCGA BRCA -0.272; TCGA ESCA -0.366; TCGA HNSC -0.318; TCGA KIRC -0.21; TCGA LIHC -0.123; TCGA LUAD -0.088; TCGA PAAD -0.223; TCGA SARC -0.379; TCGA STAD -0.255; TCGA UCEC -0.129 |
hsa-miR-193a-3p | FAT4 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; PAAD; SARC; STAD; UCEC | MirTarget; miRanda; miRNATAP | TCGA BLCA -0.247; TCGA BRCA -0.33; TCGA CESC -0.497; TCGA COAD -0.353; TCGA ESCA -0.289; TCGA HNSC -0.468; TCGA LIHC -0.344; TCGA LUAD -0.246; TCGA PAAD -0.474; TCGA SARC -0.233; TCGA STAD -0.353; TCGA UCEC -0.135 |
hsa-miR-193a-3p | PIKFYVE | 10 cancers: BLCA; CESC; HNSC; KIRC; KIRP; LGG; LUAD; PRAD; SARC; STAD | MirTarget | TCGA BLCA -0.111; TCGA CESC -0.221; TCGA HNSC -0.158; TCGA KIRC -0.243; TCGA KIRP -0.064; TCGA LGG -0.072; TCGA LUAD -0.056; TCGA PRAD -0.135; TCGA SARC -0.1; TCGA STAD -0.161 |
hsa-miR-193a-3p | IDE | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.115; TCGA CESC -0.162; TCGA COAD -0.106; TCGA ESCA -0.123; TCGA HNSC -0.15; TCGA KIRC -0.182; TCGA LIHC -0.177; TCGA LUAD -0.134; TCGA SARC -0.108; TCGA THCA -0.105; TCGA STAD -0.099; TCGA UCEC -0.126 |
hsa-miR-193a-3p | IGFBP5 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; LUSC; PAAD; SARC; STAD | PITA; miRanda; miRNATAP | TCGA BLCA -0.207; TCGA BRCA -0.154; TCGA CESC -0.701; TCGA COAD -0.291; TCGA ESCA -0.423; TCGA HNSC -0.648; TCGA KIRC -0.291; TCGA LIHC -0.26; TCGA LUSC -0.463; TCGA PAAD -0.316; TCGA SARC -0.408; TCGA STAD -0.263 |
hsa-miR-193a-3p | KIAA2022 | 11 cancers: BLCA; CESC; COAD; HNSC; KIRC; LGG; LUAD; PAAD; PRAD; SARC; STAD | PITA | TCGA BLCA -0.206; TCGA CESC -0.996; TCGA COAD -0.537; TCGA HNSC -0.704; TCGA KIRC -1.108; TCGA LGG -0.16; TCGA LUAD -0.254; TCGA PAAD -0.523; TCGA PRAD -0.166; TCGA SARC -0.706; TCGA STAD -0.521 |
hsa-miR-193a-3p | MYH11 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; SARC; STAD; UCEC | PITA; miRanda | TCGA BLCA -0.587; TCGA BRCA -0.512; TCGA CESC -0.568; TCGA COAD -0.598; TCGA ESCA -0.898; TCGA HNSC -0.835; TCGA LUAD -0.194; TCGA LUSC -0.542; TCGA SARC -1.336; TCGA STAD -0.783; TCGA UCEC -0.704 |
hsa-miR-193a-3p | CD34 | 10 cancers: BLCA; BRCA; CESC; COAD; HNSC; LIHC; LUAD; PAAD; SARC; STAD | PITA; miRanda | TCGA BLCA -0.095; TCGA BRCA -0.16; TCGA CESC -0.24; TCGA COAD -0.199; TCGA HNSC -0.177; TCGA LIHC -0.143; TCGA LUAD -0.1; TCGA PAAD -0.252; TCGA SARC -0.156; TCGA STAD -0.136 |
hsa-miR-193a-3p | NOVA1 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LUSC; PAAD; SARC; THCA | PITA; miRanda; miRNATAP | TCGA BLCA -0.481; TCGA BRCA -0.199; TCGA CESC -0.716; TCGA COAD -0.301; TCGA ESCA -0.665; TCGA HNSC -0.39; TCGA LGG -0.192; TCGA LUSC -0.259; TCGA PAAD -0.59; TCGA SARC -0.348; TCGA THCA -0.173 |
hsa-miR-193a-3p | SOS2 | 10 cancers: BLCA; CESC; COAD; KIRC; LGG; LUAD; PAAD; PRAD; SARC; STAD | PITA; miRanda; miRNATAP | TCGA BLCA -0.116; TCGA CESC -0.121; TCGA COAD -0.068; TCGA KIRC -0.27; TCGA LGG -0.085; TCGA LUAD -0.056; TCGA PAAD -0.111; TCGA PRAD -0.058; TCGA SARC -0.15; TCGA STAD -0.093 |
hsa-miR-193a-3p | ZSWIM5 | 10 cancers: BLCA; CESC; HNSC; KIRP; LGG; LUAD; PAAD; PRAD; THCA; UCEC | PITA; miRanda | TCGA BLCA -0.166; TCGA CESC -0.232; TCGA HNSC -0.3; TCGA KIRP -0.183; TCGA LGG -0.083; TCGA LUAD -0.189; TCGA PAAD -0.283; TCGA PRAD -0.085; TCGA THCA -0.208; TCGA UCEC -0.165 |
hsa-miR-193a-3p | CNOT6L | 10 cancers: BLCA; CESC; HNSC; KIRC; LIHC; LUAD; PRAD; SARC; STAD; UCEC | PITA | TCGA BLCA -0.108; TCGA CESC -0.09; TCGA HNSC -0.089; TCGA KIRC -0.132; TCGA LIHC -0.101; TCGA LUAD -0.086; TCGA PRAD -0.077; TCGA SARC -0.064; TCGA STAD -0.107; TCGA UCEC -0.069 |
hsa-miR-193a-3p | PTCH1 | 13 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LGG; LIHC; LUAD; LUSC; PAAD; STAD; UCEC | miRanda | TCGA BLCA -0.173; TCGA BRCA -0.12; TCGA CESC -0.293; TCGA ESCA -0.364; TCGA HNSC -0.522; TCGA KIRC -0.394; TCGA LGG -0.201; TCGA LIHC -0.213; TCGA LUAD -0.145; TCGA LUSC -0.299; TCGA PAAD -0.415; TCGA STAD -0.189; TCGA UCEC -0.155 |
hsa-miR-193a-3p | EPHA4 | 10 cancers: BLCA; BRCA; CESC; COAD; KIRC; LUAD; LUSC; THCA; STAD; UCEC | miRanda | TCGA BLCA -0.161; TCGA BRCA -0.201; TCGA CESC -0.355; TCGA COAD -0.525; TCGA KIRC -0.3; TCGA LUAD -0.279; TCGA LUSC -0.175; TCGA THCA -0.129; TCGA STAD -0.241; TCGA UCEC -0.199 |
hsa-miR-193a-3p | PDE1A | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; PAAD; SARC; STAD | miRanda | TCGA BLCA -0.162; TCGA BRCA -0.269; TCGA CESC -0.59; TCGA COAD -0.393; TCGA ESCA -0.278; TCGA HNSC -0.295; TCGA KIRC -0.585; TCGA LIHC -0.285; TCGA LUAD -0.153; TCGA PAAD -0.454; TCGA SARC -0.495; TCGA STAD -0.336 |
hsa-miR-193a-3p | TXNDC15 | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUAD; PAAD; SARC; STAD | miRanda | TCGA BLCA -0.12; TCGA CESC -0.084; TCGA COAD -0.109; TCGA ESCA -0.093; TCGA HNSC -0.106; TCGA LUAD -0.056; TCGA PAAD -0.224; TCGA SARC -0.075; TCGA STAD -0.102 |
hsa-miR-193a-3p | KIDINS220 | 12 cancers: BLCA; CESC; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; PAAD; PRAD; THCA; STAD | miRanda | TCGA BLCA -0.072; TCGA CESC -0.155; TCGA HNSC -0.116; TCGA KIRC -0.219; TCGA KIRP -0.089; TCGA LGG -0.081; TCGA LIHC -0.172; TCGA LUAD -0.14; TCGA PAAD -0.193; TCGA PRAD -0.071; TCGA THCA -0.08; TCGA STAD -0.119 |
hsa-miR-193a-3p | FHL1 | 10 cancers: BLCA; BRCA; COAD; ESCA; KIRP; LGG; LUAD; PAAD; SARC; STAD | miRanda | TCGA BLCA -0.186; TCGA BRCA -0.319; TCGA COAD -0.397; TCGA ESCA -0.411; TCGA KIRP -0.449; TCGA LGG -0.081; TCGA LUAD -0.153; TCGA PAAD -0.351; TCGA SARC -0.425; TCGA STAD -0.555 |
hsa-miR-193a-3p | NUMA1 | 9 cancers: BLCA; BRCA; CESC; HNSC; KIRC; LGG; PRAD; SARC; THCA | miRanda | TCGA BLCA -0.092; TCGA BRCA -0.062; TCGA CESC -0.09; TCGA HNSC -0.067; TCGA KIRC -0.092; TCGA LGG -0.134; TCGA PRAD -0.063; TCGA SARC -0.085; TCGA THCA -0.084 |
hsa-miR-193a-3p | MMRN1 | 10 cancers: BLCA; BRCA; CESC; COAD; HNSC; LIHC; LUAD; PAAD; SARC; STAD | miRanda | TCGA BLCA -0.424; TCGA BRCA -0.579; TCGA CESC -0.595; TCGA COAD -0.839; TCGA HNSC -0.349; TCGA LIHC -0.234; TCGA LUAD -0.3; TCGA PAAD -0.818; TCGA SARC -0.237; TCGA STAD -0.337 |
hsa-miR-193a-3p | MAPK10 | 12 cancers: BLCA; CESC; COAD; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; PAAD; SARC; STAD | miRanda; miRNATAP | TCGA BLCA -0.423; TCGA CESC -0.422; TCGA COAD -0.252; TCGA HNSC -0.494; TCGA KIRC -0.478; TCGA KIRP -0.149; TCGA LGG -0.09; TCGA LUAD -0.208; TCGA LUSC -0.364; TCGA PAAD -0.528; TCGA SARC -0.558; TCGA STAD -0.36 |
hsa-miR-193a-3p | EVI5L | 9 cancers: BLCA; BRCA; KIRC; LGG; LUSC; PAAD; SARC; THCA; UCEC | miRanda | TCGA BLCA -0.075; TCGA BRCA -0.061; TCGA KIRC -0.256; TCGA LGG -0.118; TCGA LUSC -0.074; TCGA PAAD -0.124; TCGA SARC -0.092; TCGA THCA -0.094; TCGA UCEC -0.064 |
hsa-miR-193a-3p | TNRC6C | 11 cancers: BLCA; CESC; ESCA; HNSC; KIRC; LGG; LIHC; LUAD; LUSC; PAAD; SARC | miRanda | TCGA BLCA -0.174; TCGA CESC -0.205; TCGA ESCA -0.157; TCGA HNSC -0.273; TCGA KIRC -0.086; TCGA LGG -0.117; TCGA LIHC -0.14; TCGA LUAD -0.114; TCGA LUSC -0.133; TCGA PAAD -0.321; TCGA SARC -0.086 |
hsa-miR-193a-3p | DGKH | 9 cancers: BLCA; ESCA; HNSC; KIRC; KIRP; LUSC; PRAD; SARC; STAD | miRanda | TCGA BLCA -0.203; TCGA ESCA -0.346; TCGA HNSC -0.134; TCGA KIRC -0.186; TCGA KIRP -0.214; TCGA LUSC -0.208; TCGA PRAD -0.119; TCGA SARC -0.132; TCGA STAD -0.15 |
hsa-miR-193a-3p | DPP8 | 11 cancers: BLCA; COAD; ESCA; HNSC; KIRC; LIHC; PAAD; PRAD; SARC; STAD; UCEC | miRanda | TCGA BLCA -0.096; TCGA COAD -0.169; TCGA ESCA -0.079; TCGA HNSC -0.164; TCGA KIRC -0.311; TCGA LIHC -0.129; TCGA PAAD -0.205; TCGA PRAD -0.107; TCGA SARC -0.11; TCGA STAD -0.109; TCGA UCEC -0.091 |
hsa-miR-193a-3p | KIAA2026 | 10 cancers: BLCA; CESC; KIRC; LGG; LIHC; LUAD; LUSC; PAAD; SARC; UCEC | miRanda | TCGA BLCA -0.071; TCGA CESC -0.15; TCGA KIRC -0.24; TCGA LGG -0.081; TCGA LIHC -0.118; TCGA LUAD -0.073; TCGA LUSC -0.14; TCGA PAAD -0.176; TCGA SARC -0.093; TCGA UCEC -0.055 |
hsa-miR-193a-3p | GPATCH8 | 10 cancers: BLCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; LUSC; SARC; STAD | miRanda | TCGA BLCA -0.05; TCGA CESC -0.081; TCGA ESCA -0.075; TCGA HNSC -0.082; TCGA KIRC -0.116; TCGA KIRP -0.063; TCGA LGG -0.067; TCGA LUSC -0.053; TCGA SARC -0.058; TCGA STAD -0.054 |
hsa-miR-193a-3p | FAM172A | 10 cancers: BLCA; BRCA; CESC; HNSC; LIHC; LUAD; PAAD; PRAD; SARC; STAD | miRanda | TCGA BLCA -0.194; TCGA BRCA -0.075; TCGA CESC -0.164; TCGA HNSC -0.167; TCGA LIHC -0.089; TCGA LUAD -0.095; TCGA PAAD -0.183; TCGA PRAD -0.085; TCGA SARC -0.143; TCGA STAD -0.166 |
hsa-miR-193a-3p | CCNDBP1 | 9 cancers: BLCA; BRCA; COAD; HNSC; KIRC; LIHC; LUAD; PRAD; SARC | miRanda | TCGA BLCA -0.119; TCGA BRCA -0.065; TCGA COAD -0.133; TCGA HNSC -0.067; TCGA KIRC -0.076; TCGA LIHC -0.088; TCGA LUAD -0.144; TCGA PRAD -0.059; TCGA SARC -0.11 |
hsa-miR-193a-3p | TRIM68 | 9 cancers: BLCA; BRCA; CESC; HNSC; KIRC; LUAD; PAAD; SARC; UCEC | miRanda | TCGA BLCA -0.119; TCGA BRCA -0.096; TCGA CESC -0.303; TCGA HNSC -0.124; TCGA KIRC -0.229; TCGA LUAD -0.151; TCGA PAAD -0.122; TCGA SARC -0.057; TCGA UCEC -0.081 |
hsa-miR-193a-3p | FAM131B | 9 cancers: BLCA; BRCA; CESC; HNSC; LGG; PAAD; SARC; THCA; UCEC | miRanda | TCGA BLCA -0.228; TCGA BRCA -0.19; TCGA CESC -0.281; TCGA HNSC -0.307; TCGA LGG -0.096; TCGA PAAD -0.315; TCGA SARC -0.285; TCGA THCA -0.112; TCGA UCEC -0.296 |
hsa-miR-193a-3p | KANK2 | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; LUAD; PAAD; SARC; STAD; UCEC | miRanda | TCGA BLCA -0.134; TCGA BRCA -0.172; TCGA CESC -0.214; TCGA COAD -0.232; TCGA ESCA -0.293; TCGA LUAD -0.063; TCGA PAAD -0.192; TCGA SARC -0.413; TCGA STAD -0.309; TCGA UCEC -0.195 |
hsa-miR-193a-3p | CYFIP2 | 12 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LGG; LUAD; PAAD; PRAD; SARC; THCA | miRanda | TCGA BLCA -0.183; TCGA BRCA -0.15; TCGA CESC -0.225; TCGA HNSC -0.234; TCGA KIRC -0.637; TCGA KIRP -0.108; TCGA LGG -0.1; TCGA LUAD -0.243; TCGA PAAD -0.454; TCGA PRAD -0.101; TCGA SARC -0.358; TCGA THCA -0.118 |
hsa-miR-193a-3p | ABCA9 | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LUAD; PAAD; STAD; UCEC | miRanda | TCGA BLCA -0.524; TCGA BRCA -0.516; TCGA CESC -0.787; TCGA ESCA -0.52; TCGA HNSC -0.47; TCGA KIRC -0.492; TCGA LUAD -0.173; TCGA PAAD -0.648; TCGA STAD -0.434; TCGA UCEC -0.338 |
hsa-miR-193a-3p | ZNF181 | 9 cancers: BLCA; CESC; HNSC; KIRC; KIRP; LUAD; PAAD; SARC; STAD | miRanda | TCGA BLCA -0.107; TCGA CESC -0.117; TCGA HNSC -0.096; TCGA KIRC -0.139; TCGA KIRP -0.06; TCGA LUAD -0.07; TCGA PAAD -0.247; TCGA SARC -0.15; TCGA STAD -0.097 |
hsa-miR-193a-3p | MPPED2 | 10 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; LIHC; LUAD; PAAD; STAD | miRanda | TCGA BLCA -0.61; TCGA BRCA -0.144; TCGA CESC -1.313; TCGA COAD -0.37; TCGA HNSC -0.475; TCGA KIRC -0.919; TCGA LIHC -0.474; TCGA LUAD -0.145; TCGA PAAD -0.335; TCGA STAD -0.237 |
hsa-miR-193a-3p | LATS1 | 9 cancers: BLCA; CESC; COAD; HNSC; KIRC; LGG; PAAD; PRAD; STAD | miRanda | TCGA BLCA -0.113; TCGA CESC -0.232; TCGA COAD -0.112; TCGA HNSC -0.225; TCGA KIRC -0.365; TCGA LGG -0.062; TCGA PAAD -0.209; TCGA PRAD -0.122; TCGA STAD -0.154 |
hsa-miR-193a-3p | RTF1 | 9 cancers: BLCA; CESC; COAD; HNSC; LGG; LUAD; PAAD; SARC; UCEC | miRanda | TCGA BLCA -0.082; TCGA CESC -0.061; TCGA COAD -0.063; TCGA HNSC -0.077; TCGA LGG -0.058; TCGA LUAD -0.069; TCGA PAAD -0.093; TCGA SARC -0.106; TCGA UCEC -0.09 |
hsa-miR-193a-3p | PTPN21 | 9 cancers: BLCA; BRCA; COAD; KIRC; KIRP; LIHC; PAAD; STAD; UCEC | miRanda | TCGA BLCA -0.128; TCGA BRCA -0.164; TCGA COAD -0.075; TCGA KIRC -0.429; TCGA KIRP -0.138; TCGA LIHC -0.154; TCGA PAAD -0.167; TCGA STAD -0.166; TCGA UCEC -0.122 |
hsa-miR-193a-3p | RAPGEF2 | 10 cancers: BLCA; CESC; HNSC; KIRC; KIRP; LIHC; LUAD; PAAD; STAD; UCEC | miRanda | TCGA BLCA -0.118; TCGA CESC -0.17; TCGA HNSC -0.095; TCGA KIRC -0.17; TCGA KIRP -0.087; TCGA LIHC -0.106; TCGA LUAD -0.14; TCGA PAAD -0.109; TCGA STAD -0.137; TCGA UCEC -0.053 |
hsa-miR-193a-3p | MLLT3 | 11 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC | miRanda | TCGA BLCA -0.151; TCGA BRCA -0.123; TCGA CESC -0.278; TCGA ESCA -0.241; TCGA HNSC -0.146; TCGA KIRC -0.138; TCGA KIRP -0.071; TCGA LGG -0.105; TCGA LIHC -0.243; TCGA LUAD -0.154; TCGA LUSC -0.18 |
hsa-miR-193a-3p | ACTG2 | 9 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; SARC; STAD; UCEC | miRanda | TCGA BLCA -0.286; TCGA BRCA -0.206; TCGA CESC -0.509; TCGA COAD -0.547; TCGA ESCA -0.429; TCGA HNSC -0.379; TCGA SARC -0.847; TCGA STAD -0.77; TCGA UCEC -0.66 |
hsa-miR-193a-3p | APC | 9 cancers: BLCA; CESC; COAD; HNSC; KIRC; LUAD; PAAD; SARC; STAD | miRanda | TCGA BLCA -0.069; TCGA CESC -0.103; TCGA COAD -0.15; TCGA HNSC -0.16; TCGA KIRC -0.107; TCGA LUAD -0.121; TCGA PAAD -0.256; TCGA SARC -0.101; TCGA STAD -0.164 |
hsa-miR-193a-3p | BMX | 9 cancers: BLCA; BRCA; CESC; COAD; HNSC; LUAD; SARC; STAD; UCEC | miRanda | TCGA BLCA -0.361; TCGA BRCA -0.485; TCGA CESC -0.491; TCGA COAD -0.432; TCGA HNSC -0.469; TCGA LUAD -0.279; TCGA SARC -0.272; TCGA STAD -0.263; TCGA UCEC -0.298 |
hsa-miR-193a-3p | SYNRG | 12 cancers: BLCA; CESC; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; PAAD; PRAD; STAD; UCEC | miRanda | TCGA BLCA -0.057; TCGA CESC -0.118; TCGA HNSC -0.109; TCGA KIRC -0.171; TCGA KIRP -0.057; TCGA LGG -0.056; TCGA LIHC -0.063; TCGA LUAD -0.06; TCGA PAAD -0.142; TCGA PRAD -0.079; TCGA STAD -0.073; TCGA UCEC -0.061 |
hsa-miR-193a-3p | SLC40A1 | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LIHC; LUAD; SARC; THCA | miRanda | TCGA BLCA -0.314; TCGA BRCA -0.368; TCGA CESC -0.558; TCGA ESCA -0.333; TCGA HNSC -0.412; TCGA KIRC -0.113; TCGA LIHC -0.269; TCGA LUAD -0.243; TCGA SARC -0.297; TCGA THCA -0.174 |
hsa-miR-193a-3p | PDE4D | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; PAAD; SARC; STAD; UCEC | miRanda | TCGA BLCA -0.366; TCGA BRCA -0.141; TCGA COAD -0.283; TCGA ESCA -0.361; TCGA HNSC -0.318; TCGA KIRC -0.152; TCGA LIHC -0.352; TCGA LUAD -0.347; TCGA PAAD -0.204; TCGA SARC -0.344; TCGA STAD -0.247; TCGA UCEC -0.224 |
hsa-miR-193a-3p | CCNT2 | 9 cancers: BLCA; CESC; KIRC; LGG; LUAD; LUSC; PRAD; SARC; STAD | miRanda | TCGA BLCA -0.127; TCGA CESC -0.124; TCGA KIRC -0.071; TCGA LGG -0.062; TCGA LUAD -0.078; TCGA LUSC -0.069; TCGA PRAD -0.089; TCGA SARC -0.079; TCGA STAD -0.079 |
hsa-miR-193a-3p | SOX5 | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUSC; PAAD; SARC | miRanda; miRNATAP | TCGA BLCA -0.245; TCGA BRCA -0.17; TCGA CESC -0.921; TCGA COAD -0.283; TCGA ESCA -0.45; TCGA HNSC -0.747; TCGA KIRC -0.379; TCGA LUSC -0.378; TCGA PAAD -0.515; TCGA SARC -0.338 |
hsa-miR-193a-3p | ARMCX3 | 10 cancers: BLCA; CESC; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; THCA; UCEC | miRanda | TCGA BLCA -0.058; TCGA CESC -0.376; TCGA ESCA -0.261; TCGA HNSC -0.392; TCGA KIRC -0.161; TCGA LUAD -0.055; TCGA LUSC -0.093; TCGA PAAD -0.114; TCGA THCA -0.163; TCGA UCEC -0.092 |
hsa-miR-193a-3p | BDP1 | 9 cancers: BLCA; CESC; KIRC; LGG; LUAD; PAAD; PRAD; SARC; STAD | miRanda | TCGA BLCA -0.07; TCGA CESC -0.142; TCGA KIRC -0.079; TCGA LGG -0.055; TCGA LUAD -0.101; TCGA PAAD -0.144; TCGA PRAD -0.093; TCGA SARC -0.118; TCGA STAD -0.115 |
hsa-miR-193a-3p | ZSCAN29 | 9 cancers: BLCA; CESC; HNSC; KIRC; KIRP; LGG; LUAD; PAAD; SARC | miRanda | TCGA BLCA -0.058; TCGA CESC -0.116; TCGA HNSC -0.056; TCGA KIRC -0.067; TCGA KIRP -0.057; TCGA LGG -0.08; TCGA LUAD -0.053; TCGA PAAD -0.111; TCGA SARC -0.098 |
hsa-miR-193a-3p | TP53INP1 | 11 cancers: BLCA; CESC; COAD; HNSC; KIRP; LUAD; LUSC; PRAD; SARC; THCA; UCEC | miRanda | TCGA BLCA -0.153; TCGA CESC -0.227; TCGA COAD -0.157; TCGA HNSC -0.081; TCGA KIRP -0.103; TCGA LUAD -0.178; TCGA LUSC -0.096; TCGA PRAD -0.083; TCGA SARC -0.222; TCGA THCA -0.131; TCGA UCEC -0.077 |
hsa-miR-193a-3p | ZMIZ1 | 10 cancers: BLCA; COAD; HNSC; KIRC; KIRP; LGG; LUAD; PAAD; STAD; UCEC | miRanda | TCGA BLCA -0.218; TCGA COAD -0.098; TCGA HNSC -0.172; TCGA KIRC -0.122; TCGA KIRP -0.135; TCGA LGG -0.158; TCGA LUAD -0.066; TCGA PAAD -0.161; TCGA STAD -0.125; TCGA UCEC -0.08 |
hsa-miR-193a-3p | ARHGEF6 | 10 cancers: BLCA; BRCA; CESC; COAD; HNSC; LIHC; LUAD; PAAD; STAD; UCEC | miRanda | TCGA BLCA -0.181; TCGA BRCA -0.116; TCGA CESC -0.239; TCGA COAD -0.336; TCGA HNSC -0.251; TCGA LIHC -0.269; TCGA LUAD -0.109; TCGA PAAD -0.227; TCGA STAD -0.256; TCGA UCEC -0.093 |
hsa-miR-193a-3p | PRKACB | 11 cancers: BLCA; CESC; HNSC; KIRC; KIRP; LIHC; PAAD; PRAD; SARC; STAD; UCEC | miRanda | TCGA BLCA -0.185; TCGA CESC -0.257; TCGA HNSC -0.121; TCGA KIRC -0.152; TCGA KIRP -0.09; TCGA LIHC -0.139; TCGA PAAD -0.215; TCGA PRAD -0.079; TCGA SARC -0.107; TCGA STAD -0.215; TCGA UCEC -0.113 |
hsa-miR-193a-3p | DPY19L1 | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LIHC; LUSC; PRAD; THCA; UCEC | miRanda | TCGA BLCA -0.062; TCGA BRCA -0.079; TCGA COAD -0.087; TCGA ESCA -0.194; TCGA HNSC -0.16; TCGA LIHC -0.156; TCGA LUSC -0.157; TCGA PRAD -0.08; TCGA THCA -0.158; TCGA UCEC -0.098 |
hsa-miR-193a-3p | CELF2 | 9 cancers: BLCA; BRCA; CESC; HNSC; LIHC; LUAD; SARC; STAD; UCEC | miRanda | TCGA BLCA -0.18; TCGA BRCA -0.191; TCGA CESC -0.306; TCGA HNSC -0.272; TCGA LIHC -0.222; TCGA LUAD -0.259; TCGA SARC -0.217; TCGA STAD -0.352; TCGA UCEC -0.331 |
hsa-miR-193a-3p | SNX18 | 9 cancers: BLCA; COAD; KIRC; LUAD; PAAD; SARC; THCA; STAD; UCEC | miRanda | TCGA BLCA -0.116; TCGA COAD -0.131; TCGA KIRC -0.217; TCGA LUAD -0.131; TCGA PAAD -0.149; TCGA SARC -0.189; TCGA THCA -0.059; TCGA STAD -0.085; TCGA UCEC -0.06 |
hsa-miR-193a-3p | PGM5 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; PAAD; SARC; STAD; UCEC | miRanda | TCGA BLCA -0.354; TCGA BRCA -0.482; TCGA CESC -0.87; TCGA COAD -0.403; TCGA ESCA -0.623; TCGA HNSC -0.394; TCGA KIRC -0.468; TCGA LIHC -0.577; TCGA LUAD -0.326; TCGA PAAD -0.489; TCGA SARC -0.75; TCGA STAD -0.593; TCGA UCEC -0.444 |
hsa-miR-193a-3p | RAPGEF6 | 9 cancers: BLCA; CESC; COAD; HNSC; KIRC; LUAD; PAAD; SARC; STAD | miRanda | TCGA BLCA -0.106; TCGA CESC -0.26; TCGA COAD -0.146; TCGA HNSC -0.239; TCGA KIRC -0.156; TCGA LUAD -0.166; TCGA PAAD -0.355; TCGA SARC -0.236; TCGA STAD -0.097 |
hsa-miR-193a-3p | AKAP6 | 10 cancers: BLCA; BRCA; COAD; KIRC; KIRP; LGG; LIHC; PAAD; SARC; STAD | miRanda | TCGA BLCA -0.235; TCGA BRCA -0.149; TCGA COAD -0.23; TCGA KIRC -0.48; TCGA KIRP -0.154; TCGA LGG -0.196; TCGA LIHC -0.217; TCGA PAAD -0.495; TCGA SARC -0.315; TCGA STAD -0.466 |
hsa-miR-193a-3p | NRXN3 | 10 cancers: BLCA; BRCA; CESC; HNSC; LUAD; LUSC; PAAD; SARC; THCA; STAD | miRanda | TCGA BLCA -0.679; TCGA BRCA -0.152; TCGA CESC -0.821; TCGA HNSC -0.715; TCGA LUAD -0.317; TCGA LUSC -0.495; TCGA PAAD -0.457; TCGA SARC -0.348; TCGA THCA -0.12; TCGA STAD -0.527 |
hsa-miR-193a-3p | ALDH6A1 | 11 cancers: BLCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; PAAD; SARC | miRanda | TCGA BLCA -0.097; TCGA CESC -0.246; TCGA ESCA -0.244; TCGA HNSC -0.306; TCGA KIRC -1.134; TCGA KIRP -0.297; TCGA LGG -0.108; TCGA LIHC -0.315; TCGA LUAD -0.253; TCGA PAAD -0.232; TCGA SARC -0.102 |
hsa-miR-193a-3p | ESR1 | 10 cancers: BLCA; CESC; COAD; KIRC; LIHC; PAAD; PRAD; SARC; THCA; STAD | miRanda | TCGA BLCA -0.154; TCGA CESC -0.511; TCGA COAD -0.327; TCGA KIRC -0.32; TCGA LIHC -0.411; TCGA PAAD -0.409; TCGA PRAD -0.186; TCGA SARC -0.319; TCGA THCA -0.326; TCGA STAD -0.303 |
hsa-miR-193a-3p | THSD7B | 10 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD | miRanda | TCGA BLCA -0.328; TCGA BRCA -0.469; TCGA CESC -0.351; TCGA COAD -0.543; TCGA HNSC -0.719; TCGA KIRC -0.284; TCGA LUAD -0.322; TCGA LUSC -0.453; TCGA PAAD -0.63; TCGA PRAD -0.221 |
hsa-miR-193a-3p | FAM179B | 9 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LIHC; PAAD; SARC | miRanda | TCGA BLCA -0.127; TCGA BRCA -0.067; TCGA CESC -0.141; TCGA HNSC -0.152; TCGA KIRC -0.269; TCGA KIRP -0.097; TCGA LIHC -0.148; TCGA PAAD -0.208; TCGA SARC -0.101 |
hsa-miR-193a-3p | ZNF720 | 9 cancers: BLCA; CESC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; SARC | miRanda | TCGA BLCA -0.074; TCGA CESC -0.157; TCGA KIRC -0.124; TCGA KIRP -0.086; TCGA LGG -0.079; TCGA LIHC -0.148; TCGA LUAD -0.115; TCGA LUSC -0.072; TCGA SARC -0.066 |
hsa-miR-193a-3p | LONP2 | 9 cancers: BLCA; CESC; ESCA; HNSC; KIRC; KIRP; LUAD; THCA; STAD | miRanda | TCGA BLCA -0.11; TCGA CESC -0.094; TCGA ESCA -0.147; TCGA HNSC -0.067; TCGA KIRC -0.223; TCGA KIRP -0.091; TCGA LUAD -0.064; TCGA THCA -0.114; TCGA STAD -0.094 |
hsa-miR-193a-3p | FBXL17 | 10 cancers: BLCA; KIRC; LGG; LIHC; LUAD; PAAD; PRAD; SARC; STAD; UCEC | miRanda | TCGA BLCA -0.202; TCGA KIRC -0.188; TCGA LGG -0.053; TCGA LIHC -0.088; TCGA LUAD -0.083; TCGA PAAD -0.143; TCGA PRAD -0.068; TCGA SARC -0.082; TCGA STAD -0.107; TCGA UCEC -0.063 |
hsa-miR-193a-3p | PBRM1 | 10 cancers: BLCA; CESC; COAD; HNSC; KIRC; KIRP; LUAD; PRAD; SARC; STAD | miRanda | TCGA BLCA -0.102; TCGA CESC -0.115; TCGA COAD -0.053; TCGA HNSC -0.071; TCGA KIRC -0.211; TCGA KIRP -0.086; TCGA LUAD -0.066; TCGA PRAD -0.081; TCGA SARC -0.121; TCGA STAD -0.158 |
hsa-miR-193a-3p | DOPEY1 | 10 cancers: BLCA; BRCA; CESC; KIRC; KIRP; LGG; LUAD; PAAD; SARC; STAD | miRanda | TCGA BLCA -0.163; TCGA BRCA -0.052; TCGA CESC -0.174; TCGA KIRC -0.163; TCGA KIRP -0.057; TCGA LGG -0.12; TCGA LUAD -0.052; TCGA PAAD -0.22; TCGA SARC -0.112; TCGA STAD -0.126 |
hsa-miR-193a-3p | C7 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; PAAD; SARC; STAD; UCEC | miRanda | TCGA BLCA -0.363; TCGA BRCA -0.725; TCGA CESC -1.017; TCGA COAD -0.57; TCGA ESCA -0.894; TCGA HNSC -0.883; TCGA KIRC -0.923; TCGA LUAD -0.465; TCGA PAAD -0.59; TCGA SARC -0.523; TCGA STAD -0.54; TCGA UCEC -0.406 |
hsa-miR-193a-3p | CX3CR1 | 9 cancers: BLCA; BRCA; CESC; COAD; HNSC; LIHC; LUAD; PAAD; STAD | miRanda | TCGA BLCA -0.23; TCGA BRCA -0.369; TCGA CESC -0.227; TCGA COAD -0.201; TCGA HNSC -0.245; TCGA LIHC -0.261; TCGA LUAD -0.266; TCGA PAAD -0.466; TCGA STAD -0.23 |
hsa-miR-193a-3p | NCOR1 | 12 cancers: BLCA; CESC; COAD; HNSC; KIRC; KIRP; LIHC; LUAD; PAAD; SARC; THCA; UCEC | miRanda | TCGA BLCA -0.095; TCGA CESC -0.133; TCGA COAD -0.132; TCGA HNSC -0.159; TCGA KIRC -0.181; TCGA KIRP -0.138; TCGA LIHC -0.142; TCGA LUAD -0.095; TCGA PAAD -0.228; TCGA SARC -0.211; TCGA THCA -0.051; TCGA UCEC -0.107 |
hsa-miR-193a-3p | TET2 | 10 cancers: BLCA; CESC; COAD; HNSC; KIRC; LGG; LUAD; LUSC; PRAD; STAD | miRanda | TCGA BLCA -0.119; TCGA CESC -0.237; TCGA COAD -0.141; TCGA HNSC -0.117; TCGA KIRC -0.121; TCGA LGG -0.081; TCGA LUAD -0.083; TCGA LUSC -0.08; TCGA PRAD -0.086; TCGA STAD -0.105 |
hsa-miR-193a-3p | UTRN | 13 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; LUAD; PAAD; PRAD; SARC; STAD; UCEC | miRanda | TCGA BLCA -0.275; TCGA BRCA -0.137; TCGA CESC -0.402; TCGA COAD -0.138; TCGA HNSC -0.39; TCGA KIRC -0.157; TCGA KIRP -0.114; TCGA LUAD -0.103; TCGA PAAD -0.225; TCGA PRAD -0.163; TCGA SARC -0.198; TCGA STAD -0.245; TCGA UCEC -0.117 |
hsa-miR-193a-3p | ROBO2 | 11 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; PAAD; STAD | miRanda | TCGA BLCA -0.328; TCGA BRCA -0.324; TCGA CESC -0.66; TCGA ESCA -0.375; TCGA HNSC -0.638; TCGA KIRC -0.479; TCGA LIHC -0.405; TCGA LUAD -0.341; TCGA LUSC -0.322; TCGA PAAD -0.39; TCGA STAD -0.275 |
hsa-miR-193a-3p | SBF2 | 14 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LGG; LIHC; LUSC; PRAD; SARC; THCA; STAD; UCEC | miRanda | TCGA BLCA -0.152; TCGA BRCA -0.066; TCGA CESC -0.15; TCGA HNSC -0.083; TCGA KIRC -0.236; TCGA KIRP -0.124; TCGA LGG -0.085; TCGA LIHC -0.19; TCGA LUSC -0.07; TCGA PRAD -0.107; TCGA SARC -0.111; TCGA THCA -0.115; TCGA STAD -0.09; TCGA UCEC -0.069 |
hsa-miR-193a-3p | BBS1 | 11 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; PAAD; SARC | miRanda | TCGA BLCA -0.134; TCGA BRCA -0.061; TCGA CESC -0.162; TCGA ESCA -0.108; TCGA HNSC -0.116; TCGA KIRC -0.296; TCGA KIRP -0.122; TCGA LIHC -0.104; TCGA LUAD -0.082; TCGA PAAD -0.155; TCGA SARC -0.147 |
hsa-miR-193a-3p | RUFY2 | 11 cancers: BLCA; BRCA; CESC; KIRC; KIRP; LGG; LIHC; LUAD; PRAD; STAD; UCEC | miRanda | TCGA BLCA -0.126; TCGA BRCA -0.061; TCGA CESC -0.076; TCGA KIRC -0.069; TCGA KIRP -0.069; TCGA LGG -0.065; TCGA LIHC -0.143; TCGA LUAD -0.117; TCGA PRAD -0.054; TCGA STAD -0.1; TCGA UCEC -0.059 |
hsa-miR-193a-3p | ATP9B | 13 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; LUAD; PAAD; PRAD; SARC; STAD; UCEC | miRanda | TCGA BLCA -0.101; TCGA BRCA -0.077; TCGA CESC -0.126; TCGA COAD -0.163; TCGA HNSC -0.066; TCGA KIRC -0.071; TCGA KIRP -0.062; TCGA LUAD -0.147; TCGA PAAD -0.198; TCGA PRAD -0.061; TCGA SARC -0.143; TCGA STAD -0.118; TCGA UCEC -0.092 |
hsa-miR-193a-3p | RALGPS1 | 15 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRanda | TCGA BLCA -0.199; TCGA BRCA -0.069; TCGA CESC -0.371; TCGA ESCA -0.388; TCGA HNSC -0.351; TCGA KIRC -0.923; TCGA KIRP -0.278; TCGA LGG -0.112; TCGA LUAD -0.148; TCGA LUSC -0.134; TCGA PAAD -0.323; TCGA PRAD -0.073; TCGA SARC -0.199; TCGA STAD -0.113; TCGA UCEC -0.068 |
hsa-miR-193a-3p | PTCHD1 | 9 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUAD; SARC; STAD; UCEC | miRanda | TCGA BLCA -0.509; TCGA BRCA -0.222; TCGA COAD -0.463; TCGA ESCA -0.561; TCGA HNSC -0.528; TCGA LUAD -0.23; TCGA SARC -0.65; TCGA STAD -0.548; TCGA UCEC -0.304 |
hsa-miR-193a-3p | GDF11 | 9 cancers: BLCA; CESC; COAD; HNSC; LIHC; LUSC; PAAD; PRAD; THCA | miRanda | TCGA BLCA -0.104; TCGA CESC -0.134; TCGA COAD -0.112; TCGA HNSC -0.261; TCGA LIHC -0.273; TCGA LUSC -0.1; TCGA PAAD -0.277; TCGA PRAD -0.297; TCGA THCA -0.067 |
hsa-miR-193a-3p | HERC1 | 14 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; LGG; LUAD; PAAD; PRAD; SARC; STAD; UCEC | miRanda | TCGA BLCA -0.1; TCGA BRCA -0.089; TCGA CESC -0.099; TCGA COAD -0.09; TCGA HNSC -0.146; TCGA KIRC -0.189; TCGA KIRP -0.088; TCGA LGG -0.07; TCGA LUAD -0.078; TCGA PAAD -0.257; TCGA PRAD -0.063; TCGA SARC -0.137; TCGA STAD -0.153; TCGA UCEC -0.088 |
hsa-miR-193a-3p | RGAG4 | 10 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; LGG; LIHC; PAAD; THCA | miRanda | TCGA BLCA -0.326; TCGA BRCA -0.175; TCGA CESC -0.478; TCGA COAD -0.225; TCGA HNSC -0.376; TCGA KIRC -0.326; TCGA LGG -0.058; TCGA LIHC -0.423; TCGA PAAD -0.669; TCGA THCA -0.202 |
hsa-miR-193a-3p | TNRC6B | 10 cancers: BLCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; LUSC; PRAD; SARC | miRanda; mirMAP | TCGA BLCA -0.09; TCGA CESC -0.124; TCGA ESCA -0.111; TCGA HNSC -0.202; TCGA KIRC -0.188; TCGA KIRP -0.15; TCGA LGG -0.088; TCGA LUSC -0.106; TCGA PRAD -0.09; TCGA SARC -0.1 |
hsa-miR-193a-3p | UBR3 | 11 cancers: BLCA; CESC; HNSC; KIRC; LGG; LUAD; PAAD; PRAD; SARC; THCA; STAD | miRanda | TCGA BLCA -0.093; TCGA CESC -0.101; TCGA HNSC -0.127; TCGA KIRC -0.233; TCGA LGG -0.059; TCGA LUAD -0.07; TCGA PAAD -0.122; TCGA PRAD -0.086; TCGA SARC -0.113; TCGA THCA -0.053; TCGA STAD -0.133 |
hsa-miR-193a-3p | TTC19 | 10 cancers: BLCA; CESC; HNSC; KIRC; KIRP; LGG; LUAD; PAAD; PRAD; UCEC | miRanda | TCGA BLCA -0.057; TCGA CESC -0.097; TCGA HNSC -0.104; TCGA KIRC -0.225; TCGA KIRP -0.125; TCGA LGG -0.078; TCGA LUAD -0.159; TCGA PAAD -0.129; TCGA PRAD -0.079; TCGA UCEC -0.066 |
hsa-miR-193a-3p | GPLD1 | 10 cancers: BLCA; BRCA; COAD; HNSC; KIRC; LGG; LUAD; PAAD; SARC; THCA | miRanda | TCGA BLCA -0.183; TCGA BRCA -0.202; TCGA COAD -0.168; TCGA HNSC -0.827; TCGA KIRC -0.476; TCGA LGG -0.164; TCGA LUAD -0.213; TCGA PAAD -0.486; TCGA SARC -0.26; TCGA THCA -0.111 |
hsa-miR-193a-3p | RPS6KA2 | 12 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LIHC; LUAD; PAAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.182; TCGA BRCA -0.173; TCGA CESC -0.243; TCGA ESCA -0.265; TCGA HNSC -0.193; TCGA KIRC -0.115; TCGA LIHC -0.302; TCGA LUAD -0.237; TCGA PAAD -0.124; TCGA THCA -0.114; TCGA STAD -0.131; TCGA UCEC -0.09 |
hsa-miR-193a-3p | IGSF9B | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LUAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.273; TCGA BRCA -0.217; TCGA CESC -0.294; TCGA COAD -0.296; TCGA ESCA -0.603; TCGA HNSC -0.377; TCGA KIRC -0.205; TCGA LGG -0.151; TCGA LUAD -0.349; TCGA SARC -0.783; TCGA STAD -0.434; TCGA UCEC -0.175 |
hsa-miR-193a-3p | TSPAN18 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUSC; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.166; TCGA BRCA -0.072; TCGA CESC -0.229; TCGA COAD -0.164; TCGA ESCA -0.415; TCGA HNSC -0.589; TCGA KIRP -0.146; TCGA LUSC -0.236; TCGA SARC -0.143; TCGA STAD -0.252; TCGA UCEC -0.132 |
hsa-miR-193a-3p | GRIK3 | 9 cancers: BLCA; BRCA; CESC; COAD; HNSC; LGG; PAAD; THCA; STAD | mirMAP | TCGA BLCA -0.234; TCGA BRCA -0.389; TCGA CESC -0.571; TCGA COAD -0.393; TCGA HNSC -0.331; TCGA LGG -0.095; TCGA PAAD -0.584; TCGA THCA -0.242; TCGA STAD -0.792 |
hsa-miR-193a-3p | CBX6 | 11 cancers: BLCA; BRCA; COAD; HNSC; KIRC; KIRP; LUSC; PAAD; PRAD; SARC; THCA | mirMAP | TCGA BLCA -0.115; TCGA BRCA -0.18; TCGA COAD -0.345; TCGA HNSC -0.483; TCGA KIRC -0.288; TCGA KIRP -0.14; TCGA LUSC -0.119; TCGA PAAD -0.543; TCGA PRAD -0.11; TCGA SARC -0.188; TCGA THCA -0.123 |
hsa-miR-193a-3p | MKLN1 | 10 cancers: BLCA; ESCA; HNSC; KIRC; LGG; LIHC; LUAD; LUSC; SARC; STAD | miRNATAP | TCGA BLCA -0.058; TCGA ESCA -0.136; TCGA HNSC -0.081; TCGA KIRC -0.151; TCGA LGG -0.071; TCGA LIHC -0.155; TCGA LUAD -0.088; TCGA LUSC -0.072; TCGA SARC -0.086; TCGA STAD -0.076 |
hsa-miR-193a-3p | TCF4 | 11 cancers: BRCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; LUSC; PAAD; STAD; UCEC | MirTarget; PITA; miRanda; miRNATAP | TCGA BRCA -0.194; TCGA CESC -0.447; TCGA COAD -0.279; TCGA ESCA -0.228; TCGA HNSC -0.323; TCGA LGG -0.129; TCGA LIHC -0.198; TCGA LUSC -0.166; TCGA PAAD -0.203; TCGA STAD -0.198; TCGA UCEC -0.174 |
hsa-miR-193a-3p | ETV1 | 10 cancers: BRCA; CESC; COAD; ESCA; HNSC; LGG; LUAD; PRAD; THCA; UCEC | MirTarget; PITA; miRanda; miRNATAP | TCGA BRCA -0.134; TCGA CESC -0.47; TCGA COAD -0.238; TCGA ESCA -0.281; TCGA HNSC -0.485; TCGA LGG -0.298; TCGA LUAD -0.195; TCGA PRAD -0.187; TCGA THCA -0.275; TCGA UCEC -0.187 |
hsa-miR-193a-3p | IL17RD | 9 cancers: BRCA; CESC; HNSC; KIRC; KIRP; LGG; SARC; THCA; UCEC | MirTarget | TCGA BRCA -0.215; TCGA CESC -0.221; TCGA HNSC -0.195; TCGA KIRC -0.524; TCGA KIRP -0.373; TCGA LGG -0.051; TCGA SARC -0.264; TCGA THCA -0.184; TCGA UCEC -0.248 |
hsa-miR-193a-3p | LUZP1 | 9 cancers: BRCA; COAD; KIRC; KIRP; LIHC; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BRCA -0.077; TCGA COAD -0.132; TCGA KIRC -0.157; TCGA KIRP -0.083; TCGA LIHC -0.208; TCGA PRAD -0.085; TCGA THCA -0.09; TCGA STAD -0.119; TCGA UCEC -0.069 |
hsa-miR-193a-3p | RGMA | 9 cancers: BRCA; COAD; HNSC; KIRC; LGG; LIHC; LUSC; SARC; STAD | MirTarget; miRanda | TCGA BRCA -0.269; TCGA COAD -0.359; TCGA HNSC -0.349; TCGA KIRC -0.23; TCGA LGG -0.069; TCGA LIHC -0.223; TCGA LUSC -0.478; TCGA SARC -0.356; TCGA STAD -0.579 |
hsa-miR-193a-3p | LRP4 | 9 cancers: BRCA; CESC; HNSC; LGG; LIHC; LUSC; THCA; STAD; UCEC | MirTarget; miRanda | TCGA BRCA -0.278; TCGA CESC -0.276; TCGA HNSC -0.245; TCGA LGG -0.244; TCGA LIHC -0.367; TCGA LUSC -0.239; TCGA THCA -0.768; TCGA STAD -0.269; TCGA UCEC -0.27 |
hsa-miR-193a-3p | RUNX1T1 | 10 cancers: BRCA; CESC; COAD; HNSC; KIRC; LGG; PAAD; SARC; STAD; UCEC | PITA; miRanda; miRNATAP | TCGA BRCA -0.342; TCGA CESC -0.681; TCGA COAD -0.426; TCGA HNSC -0.521; TCGA KIRC -0.312; TCGA LGG -0.178; TCGA PAAD -0.648; TCGA SARC -0.196; TCGA STAD -0.32; TCGA UCEC -0.38 |
hsa-miR-193a-3p | PDZRN3 | 10 cancers: BRCA; CESC; COAD; HNSC; KIRC; KIRP; LIHC; PAAD; SARC; STAD | miRanda | TCGA BRCA -0.23; TCGA CESC -0.343; TCGA COAD -0.171; TCGA HNSC -0.192; TCGA KIRC -0.674; TCGA KIRP -0.252; TCGA LIHC -0.32; TCGA PAAD -0.268; TCGA SARC -0.412; TCGA STAD -0.272 |
hsa-miR-193a-3p | ITPR1 | 11 cancers: BRCA; CESC; COAD; HNSC; KIRC; LIHC; LUAD; PAAD; SARC; STAD; UCEC | miRanda | TCGA BRCA -0.156; TCGA CESC -0.197; TCGA COAD -0.282; TCGA HNSC -0.251; TCGA KIRC -0.26; TCGA LIHC -0.185; TCGA LUAD -0.187; TCGA PAAD -0.306; TCGA SARC -0.271; TCGA STAD -0.269; TCGA UCEC -0.167 |
hsa-miR-193a-3p | OSBPL8 | 10 cancers: BRCA; CESC; COAD; KIRC; LGG; LIHC; PRAD; SARC; STAD; UCEC | miRanda | TCGA BRCA -0.084; TCGA CESC -0.123; TCGA COAD -0.114; TCGA KIRC -0.173; TCGA LGG -0.11; TCGA LIHC -0.146; TCGA PRAD -0.108; TCGA SARC -0.079; TCGA STAD -0.113; TCGA UCEC -0.112 |
hsa-miR-193a-3p | ZNF366 | 10 cancers: BRCA; CESC; COAD; HNSC; LIHC; LUAD; PAAD; SARC; STAD; UCEC | miRanda | TCGA BRCA -0.286; TCGA CESC -0.381; TCGA COAD -0.436; TCGA HNSC -0.377; TCGA LIHC -0.295; TCGA LUAD -0.187; TCGA PAAD -0.565; TCGA SARC -0.168; TCGA STAD -0.203; TCGA UCEC -0.266 |
hsa-miR-193a-3p | OGN | 9 cancers: BRCA; CESC; COAD; HNSC; KIRC; PAAD; SARC; STAD; UCEC | miRanda | TCGA BRCA -0.598; TCGA CESC -1.147; TCGA COAD -0.578; TCGA HNSC -0.899; TCGA KIRC -0.499; TCGA PAAD -0.619; TCGA SARC -0.536; TCGA STAD -0.743; TCGA UCEC -0.41 |
hsa-miR-193a-3p | PTPRG | 11 cancers: BRCA; CESC; COAD; HNSC; KIRC; KIRP; LIHC; LUAD; PAAD; THCA; STAD | miRanda | TCGA BRCA -0.111; TCGA CESC -0.318; TCGA COAD -0.15; TCGA HNSC -0.146; TCGA KIRC -0.26; TCGA KIRP -0.115; TCGA LIHC -0.477; TCGA LUAD -0.158; TCGA PAAD -0.231; TCGA THCA -0.122; TCGA STAD -0.137 |
hsa-miR-193a-3p | PDPR | 10 cancers: BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; SARC; STAD | miRanda | TCGA BRCA -0.089; TCGA CESC -0.171; TCGA ESCA -0.173; TCGA HNSC -0.175; TCGA KIRC -0.237; TCGA KIRP -0.161; TCGA LGG -0.088; TCGA LIHC -0.227; TCGA SARC -0.109; TCGA STAD -0.115 |
hsa-miR-193a-3p | ZSCAN18 | 9 cancers: BRCA; CESC; HNSC; KIRC; KIRP; LIHC; PAAD; SARC; STAD | miRanda | TCGA BRCA -0.116; TCGA CESC -0.524; TCGA HNSC -0.399; TCGA KIRC -0.299; TCGA KIRP -0.185; TCGA LIHC -0.255; TCGA PAAD -0.309; TCGA SARC -0.162; TCGA STAD -0.225 |
hsa-miR-193a-3p | NFIC | 9 cancers: BRCA; COAD; HNSC; KIRC; LUAD; PAAD; SARC; THCA; UCEC | miRanda; mirMAP | TCGA BRCA -0.088; TCGA COAD -0.181; TCGA HNSC -0.307; TCGA KIRC -0.211; TCGA LUAD -0.105; TCGA PAAD -0.233; TCGA SARC -0.271; TCGA THCA -0.106; TCGA UCEC -0.165 |
hsa-miR-193a-3p | PLEKHH2 | 11 cancers: BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; SARC; STAD; UCEC | miRanda | TCGA BRCA -0.231; TCGA CESC -0.381; TCGA ESCA -0.253; TCGA HNSC -0.258; TCGA KIRC -0.295; TCGA KIRP -0.421; TCGA LGG -0.115; TCGA LUAD -0.115; TCGA SARC -0.303; TCGA STAD -0.267; TCGA UCEC -0.149 |
hsa-miR-193a-3p | DAAM2 | 10 cancers: BRCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; LUAD; PAAD; STAD | miRanda | TCGA BRCA -0.166; TCGA CESC -0.493; TCGA COAD -0.337; TCGA ESCA -0.371; TCGA HNSC -0.322; TCGA LGG -0.131; TCGA LIHC -0.342; TCGA LUAD -0.318; TCGA PAAD -0.345; TCGA STAD -0.393 |
hsa-miR-193a-3p | SMG6 | 13 cancers: BRCA; CESC; COAD; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; SARC; STAD; UCEC | miRanda | TCGA BRCA -0.074; TCGA CESC -0.084; TCGA COAD -0.101; TCGA HNSC -0.071; TCGA KIRC -0.073; TCGA KIRP -0.108; TCGA LIHC -0.084; TCGA LUAD -0.059; TCGA LUSC -0.122; TCGA PAAD -0.166; TCGA SARC -0.079; TCGA STAD -0.057; TCGA UCEC -0.111 |
hsa-miR-193a-3p | PDGFRA | 9 cancers: BRCA; CESC; COAD; HNSC; KIRC; LGG; PAAD; STAD; UCEC | miRanda | TCGA BRCA -0.237; TCGA CESC -0.504; TCGA COAD -0.308; TCGA HNSC -0.385; TCGA KIRC -0.67; TCGA LGG -0.273; TCGA PAAD -0.293; TCGA STAD -0.167; TCGA UCEC -0.27 |
hsa-miR-193a-3p | THSD7A | 9 cancers: BRCA; CESC; COAD; HNSC; KIRC; LGG; PAAD; STAD; UCEC | miRanda | TCGA BRCA -0.277; TCGA CESC -0.784; TCGA COAD -0.428; TCGA HNSC -0.55; TCGA KIRC -0.876; TCGA LGG -0.129; TCGA PAAD -0.376; TCGA STAD -0.227; TCGA UCEC -0.143 |
hsa-miR-193a-3p | FARP1 | 9 cancers: BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LUAD; THCA; UCEC | miRanda | TCGA BRCA -0.112; TCGA CESC -0.221; TCGA ESCA -0.264; TCGA HNSC -0.415; TCGA KIRC -0.084; TCGA KIRP -0.223; TCGA LUAD -0.087; TCGA THCA -0.083; TCGA UCEC -0.056 |
hsa-miR-193a-3p | RAB30 | 10 cancers: BRCA; COAD; ESCA; HNSC; KIRC; LGG; LIHC; LUSC; SARC; STAD | miRanda | TCGA BRCA -0.116; TCGA COAD -0.235; TCGA ESCA -0.175; TCGA HNSC -0.178; TCGA KIRC -0.228; TCGA LGG -0.064; TCGA LIHC -0.354; TCGA LUSC -0.133; TCGA SARC -0.153; TCGA STAD -0.193 |
hsa-miR-193a-3p | UNC119B | 9 cancers: BRCA; HNSC; KIRC; KIRP; LUAD; PAAD; PRAD; STAD; UCEC | miRanda | TCGA BRCA -0.091; TCGA HNSC -0.194; TCGA KIRC -0.192; TCGA KIRP -0.088; TCGA LUAD -0.07; TCGA PAAD -0.3; TCGA PRAD -0.063; TCGA STAD -0.112; TCGA UCEC -0.075 |
hsa-miR-193a-3p | VAT1L | 9 cancers: BRCA; COAD; ESCA; HNSC; KIRC; LGG; PAAD; STAD; UCEC | miRanda | TCGA BRCA -0.129; TCGA COAD -0.34; TCGA ESCA -0.307; TCGA HNSC -0.243; TCGA KIRC -0.821; TCGA LGG -0.171; TCGA PAAD -0.365; TCGA STAD -0.321; TCGA UCEC -0.437 |
hsa-miR-193a-3p | LRRK2 | 10 cancers: BRCA; CESC; COAD; HNSC; KIRP; LUAD; SARC; THCA; STAD; UCEC | miRanda | TCGA BRCA -0.161; TCGA CESC -0.427; TCGA COAD -0.401; TCGA HNSC -0.245; TCGA KIRP -0.268; TCGA LUAD -0.582; TCGA SARC -0.334; TCGA THCA -0.645; TCGA STAD -0.43; TCGA UCEC -0.151 |
hsa-miR-193a-3p | ARHGAP19 | 9 cancers: BRCA; HNSC; KIRC; LUAD; LUSC; PRAD; THCA; STAD; UCEC | mirMAP; miRNATAP | TCGA BRCA -0.114; TCGA HNSC -0.124; TCGA KIRC -0.116; TCGA LUAD -0.118; TCGA LUSC -0.074; TCGA PRAD -0.057; TCGA THCA -0.24; TCGA STAD -0.097; TCGA UCEC -0.068 |
hsa-miR-193a-3p | PLXNC1 | 9 cancers: BRCA; CESC; COAD; HNSC; PAAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BRCA -0.218; TCGA CESC -0.508; TCGA COAD -0.598; TCGA HNSC -0.422; TCGA PAAD -0.584; TCGA SARC -0.339; TCGA THCA -0.315; TCGA STAD -0.18; TCGA UCEC -0.215 |
hsa-miR-193a-3p | RORA | 10 cancers: BRCA; CESC; COAD; HNSC; KIRP; LIHC; LUAD; LUSC; STAD; UCEC | miRNATAP | TCGA BRCA -0.126; TCGA CESC -0.347; TCGA COAD -0.315; TCGA HNSC -0.214; TCGA KIRP -0.168; TCGA LIHC -0.198; TCGA LUAD -0.095; TCGA LUSC -0.138; TCGA STAD -0.267; TCGA UCEC -0.149 |
hsa-miR-193a-3p | RGL1 | 10 cancers: BRCA; CESC; COAD; HNSC; LIHC; LUAD; PAAD; SARC; THCA; STAD | miRNATAP | TCGA BRCA -0.145; TCGA CESC -0.219; TCGA COAD -0.156; TCGA HNSC -0.266; TCGA LIHC -0.363; TCGA LUAD -0.138; TCGA PAAD -0.246; TCGA SARC -0.104; TCGA THCA -0.071; TCGA STAD -0.207 |
hsa-miR-193a-3p | DMXL2 | 9 cancers: CESC; COAD; HNSC; KIRC; LGG; PAAD; PRAD; SARC; THCA | MirTarget; PITA; miRanda; miRNATAP | TCGA CESC -0.154; TCGA COAD -0.134; TCGA HNSC -0.12; TCGA KIRC -0.084; TCGA LGG -0.073; TCGA PAAD -0.187; TCGA PRAD -0.08; TCGA SARC -0.094; TCGA THCA -0.058 |
hsa-miR-193a-3p | PIAS2 | 9 cancers: CESC; COAD; HNSC; KIRC; LIHC; LUSC; PAAD; PRAD; SARC | MirTarget | TCGA CESC -0.11; TCGA COAD -0.173; TCGA HNSC -0.182; TCGA KIRC -0.24; TCGA LIHC -0.105; TCGA LUSC -0.118; TCGA PAAD -0.183; TCGA PRAD -0.071; TCGA SARC -0.103 |
hsa-miR-193a-3p | KRAS | 9 cancers: CESC; COAD; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PRAD | MirTarget; miRanda; miRNATAP | TCGA CESC -0.085; TCGA COAD -0.071; TCGA HNSC -0.089; TCGA KIRC -0.128; TCGA KIRP -0.108; TCGA LIHC -0.128; TCGA LUAD -0.114; TCGA LUSC -0.087; TCGA PRAD -0.085 |
hsa-miR-193a-3p | OPHN1 | 11 cancers: CESC; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; PAAD; SARC; STAD; UCEC | miRanda | TCGA CESC -0.222; TCGA ESCA -0.131; TCGA HNSC -0.18; TCGA KIRC -0.491; TCGA KIRP -0.094; TCGA LGG -0.319; TCGA LUAD -0.135; TCGA PAAD -0.225; TCGA SARC -0.143; TCGA STAD -0.147; TCGA UCEC -0.103 |
hsa-miR-193a-3p | SLC2A13 | 12 cancers: CESC; COAD; KIRC; KIRP; LGG; LUAD; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRanda | TCGA CESC -0.389; TCGA COAD -0.172; TCGA KIRC -0.259; TCGA KIRP -0.157; TCGA LGG -0.2; TCGA LUAD -0.088; TCGA PAAD -0.315; TCGA PRAD -0.099; TCGA SARC -0.187; TCGA THCA -0.215; TCGA STAD -0.227; TCGA UCEC -0.107 |
hsa-miR-193a-3p | ARSB | 10 cancers: CESC; COAD; HNSC; LIHC; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRanda | TCGA CESC -0.169; TCGA COAD -0.155; TCGA HNSC -0.169; TCGA LIHC -0.21; TCGA PAAD -0.246; TCGA PRAD -0.109; TCGA SARC -0.078; TCGA THCA -0.094; TCGA STAD -0.164; TCGA UCEC -0.1 |
hsa-miR-193a-3p | GMCL1 | 9 cancers: CESC; COAD; HNSC; KIRC; LGG; LIHC; LUAD; LUSC; PRAD | miRanda | TCGA CESC -0.198; TCGA COAD -0.073; TCGA HNSC -0.22; TCGA KIRC -0.097; TCGA LGG -0.062; TCGA LIHC -0.146; TCGA LUAD -0.094; TCGA LUSC -0.118; TCGA PRAD -0.093 |
hsa-miR-193a-3p | ZNF510 | 10 cancers: CESC; ESCA; KIRC; KIRP; LGG; LUAD; PAAD; SARC; STAD; UCEC | miRanda | TCGA CESC -0.095; TCGA ESCA -0.128; TCGA KIRC -0.167; TCGA KIRP -0.093; TCGA LGG -0.11; TCGA LUAD -0.075; TCGA PAAD -0.212; TCGA SARC -0.112; TCGA STAD -0.108; TCGA UCEC -0.081 |
hsa-miR-193a-3p | TMEM150C | 9 cancers: CESC; HNSC; KIRC; KIRP; LIHC; PAAD; SARC; STAD; UCEC | miRanda | TCGA CESC -0.576; TCGA HNSC -0.479; TCGA KIRC -0.426; TCGA KIRP -0.211; TCGA LIHC -0.327; TCGA PAAD -0.294; TCGA SARC -0.236; TCGA STAD -0.173; TCGA UCEC -0.163 |
hsa-miR-193a-3p | UNC13B | 9 cancers: CESC; ESCA; KIRC; KIRP; LIHC; LUAD; SARC; THCA; STAD | miRanda | TCGA CESC -0.198; TCGA ESCA -0.217; TCGA KIRC -0.381; TCGA KIRP -0.144; TCGA LIHC -0.112; TCGA LUAD -0.347; TCGA SARC -0.151; TCGA THCA -0.094; TCGA STAD -0.119 |
hsa-miR-193a-3p | ZBTB6 | 10 cancers: COAD; ESCA; KIRC; LGG; LIHC; LUAD; PRAD; SARC; STAD; UCEC | MirTarget; miRanda | TCGA COAD -0.073; TCGA ESCA -0.095; TCGA KIRC -0.11; TCGA LGG -0.075; TCGA LIHC -0.081; TCGA LUAD -0.064; TCGA PRAD -0.094; TCGA SARC -0.054; TCGA STAD -0.086; TCGA UCEC -0.076 |
hsa-miR-193a-3p | ANKFY1 | 11 cancers: COAD; KIRC; KIRP; LGG; LIHC; LUAD; PAAD; PRAD; THCA; STAD; UCEC | PITA; miRanda | TCGA COAD -0.131; TCGA KIRC -0.174; TCGA KIRP -0.153; TCGA LGG -0.058; TCGA LIHC -0.169; TCGA LUAD -0.093; TCGA PAAD -0.158; TCGA PRAD -0.086; TCGA THCA -0.136; TCGA STAD -0.109; TCGA UCEC -0.051 |
hsa-miR-193a-3p | PAFAH1B1 | 10 cancers: COAD; KIRC; KIRP; LGG; LIHC; LUAD; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA COAD -0.129; TCGA KIRC -0.151; TCGA KIRP -0.122; TCGA LGG -0.059; TCGA LIHC -0.163; TCGA LUAD -0.075; TCGA PAAD -0.142; TCGA PRAD -0.065; TCGA STAD -0.087; TCGA UCEC -0.061 |
Enriched cancer pathways of putative targets