microRNA information: hsa-miR-193a-5p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-193a-5p | miRbase |
| Accession: | MIMAT0004614 | miRbase |
| Precursor name: | hsa-mir-193a | miRbase |
| Precursor accession: | MI0000487 | miRbase |
| Symbol: | MIR193A | HGNC |
| RefSeq ID: | NR_029710 | GenBank |
| Sequence: | UGGGUCUUUGCGGGCGAGAUGA |
Reported expression in cancers: hsa-miR-193a-5p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-193a-5p | bladder cancer | upregulation | "Identification and validation of reference genes f ......" | 25738263 | qPCR; RNA-Seq |
| hsa-miR-193a-5p | colorectal cancer | downregulation | "Downregulation of miR 193a 5p correlates with lymp ......" | 25232258 | |
| hsa-miR-193a-5p | lung squamous cell cancer | downregulation | "MicroRNA 193a 3p and 5p suppress the metastasis of ......" | 24469061 | |
| hsa-miR-193a-5p | prostate cancer | deregulation | "Upregulation of miR-122 miR-335 miR-184 miR-193 mi ......" | 23781281 |
Reported cancer pathway affected by hsa-miR-193a-5p
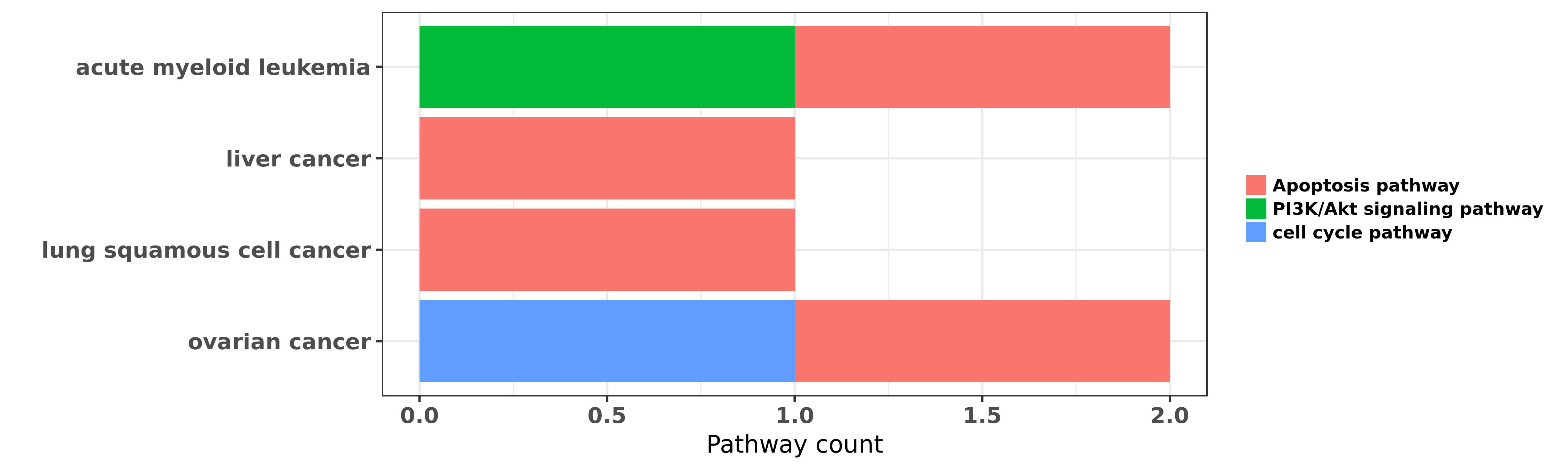
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-193a-5p | acute myeloid leukemia | Apoptosis pathway | "MicroRNA 193a represses c kit expression and funct ......" | 21399664 | Luciferase |
| hsa-miR-193a-5p | acute myeloid leukemia | Apoptosis pathway; PI3K/Akt signaling pathway | "Epigenetic silencing of microRNA 193a contributes ......" | 23223432 | |
| hsa-miR-193a-5p | acute myeloid leukemia | Apoptosis pathway | "Long non coding RNA HOTAIR modulates c KIT express ......" | 25979172 | Colony formation |
| hsa-miR-193a-5p | liver cancer | Apoptosis pathway | "Effects of miR 193a and sorafenib on hepatocellula ......" | 24330766 | Luciferase; Western blot |
| hsa-miR-193a-5p | lung squamous cell cancer | Apoptosis pathway | "Demethylation of miR 9 3 and miR 193a genes suppre ......" | 24356455 | Flow cytometry; Western blot |
| hsa-miR-193a-5p | ovarian cancer | Apoptosis pathway; cell cycle pathway | "Gain of function microRNA screens identify miR 193 ......" | 23588298 |
Reported cancer prognosis affected by hsa-miR-193a-5p
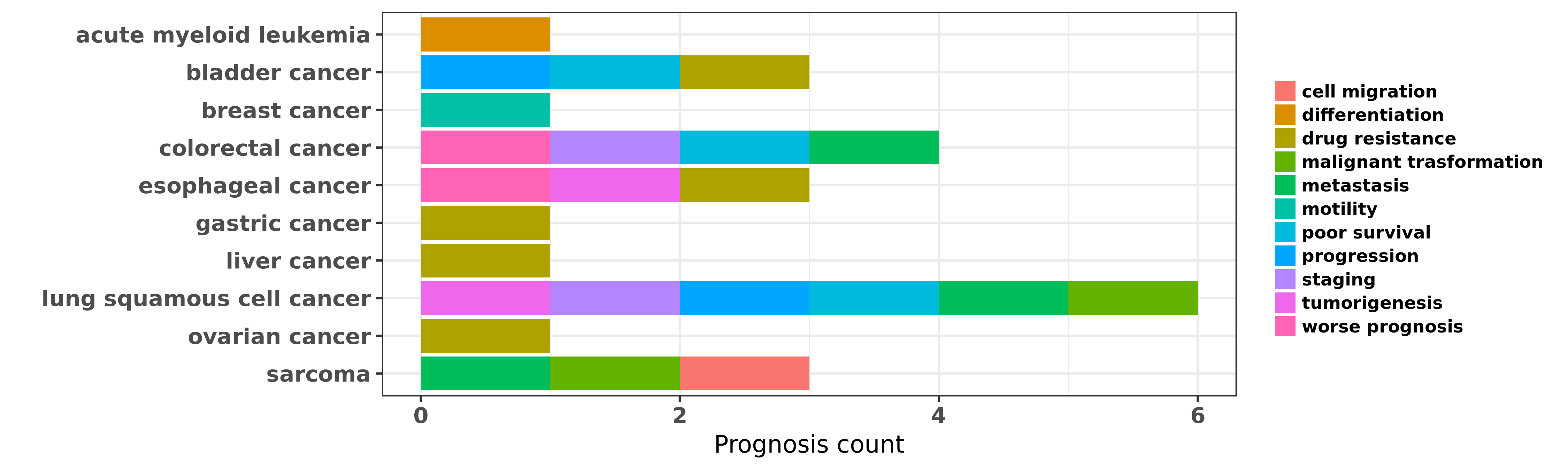
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-193a-5p | acute myeloid leukemia | differentiation | "MicroRNA 193a represses c kit expression and funct ......" | 21399664 | Luciferase |
| hsa-miR-193a-5p | acute myeloid leukemia | differentiation | "Epigenetic silencing of microRNA 193a contributes ......" | 23223432 | |
| hsa-miR-193a-5p | bladder cancer | drug resistance | "The DNA methylation regulated miR 193a 3p dictates ......" | 25188512 | |
| hsa-miR-193a-5p | bladder cancer | drug resistance | "miR 193a 3p regulates the multi drug resistance of ......" | 25311867 | |
| hsa-miR-193a-5p | bladder cancer | drug resistance | "MiR 193a 3p promotes the multi chemoresistance of ......" | 25444900 | |
| hsa-miR-193a-5p | bladder cancer | drug resistance | "The miR 193a 3p regulated PSEN1 gene suppresses th ......" | 25542424 | |
| hsa-miR-193a-5p | bladder cancer | progression; poor survival | "Of the eight most important progression-related mi ......" | 25990459 | |
| hsa-miR-193a-5p | bladder cancer | drug resistance | "The miR 193a 3p regulated ING5 gene activates the ......" | 25991669 | |
| hsa-miR-193a-5p | bladder cancer | drug resistance | "MiR 193a 5p Targets the Coding Region of AP 2α mR ......" | 27698912 | Western blot; Luciferase |
| hsa-miR-193a-5p | breast cancer | motility | "Arm Selection Preference of MicroRNA 193a Varies i ......" | 27307030 | |
| hsa-miR-193a-5p | colorectal cancer | metastasis; worse prognosis; poor survival; staging | "Downregulation of miR 193a 5p correlates with lymp ......" | 25232258 | |
| hsa-miR-193a-5p | esophageal cancer | drug resistance | "miR 193a 3p regulation of chemoradiation resistanc ......" | 26743123 | |
| hsa-miR-193a-5p | esophageal cancer | drug resistance; tumorigenesis; worse prognosis | "MiR 193a 5p/ERBB2 act as concurrent chemoradiation ......" | 27203740 | |
| hsa-miR-193a-5p | gastric cancer | drug resistance | "Downregulation of microRNA 193 3p inhibits tumor p ......" | 26753960 | Western blot; Luciferase |
| hsa-miR-193a-5p | liver cancer | drug resistance | "DNA methylation regulated miR 193a 3p dictates res ......" | 22117060 | |
| hsa-miR-193a-5p | lung squamous cell cancer | staging; malignant trasformation | "Genome wide miRNA expression profiling identifies ......" | 22282464 | |
| hsa-miR-193a-5p | lung squamous cell cancer | progression; tumorigenesis; poor survival | "Demethylation of miR 9 3 and miR 193a genes suppre ......" | 24356455 | Flow cytometry; Western blot |
| hsa-miR-193a-5p | lung squamous cell cancer | metastasis | "MicroRNA 193a 3p and 5p suppress the metastasis of ......" | 24469061 | |
| hsa-miR-193a-5p | lung squamous cell cancer | metastasis | "Quantitative proteomic analysis of the metastasis ......" | 25833338 | Western blot |
| hsa-miR-193a-5p | ovarian cancer | drug resistance | "Of these 10 miRNAs miR-193a-5p miR-375 miR-339-3p ......" | 26485143 | |
| hsa-miR-193a-5p | sarcoma | malignant trasformation | "Differential microRNA expression profiles between ......" | 24287458 | |
| hsa-miR-193a-5p | sarcoma | metastasis; cell migration | "MiR 193a 3p and miR 193a 5p suppress the metastasi ......" | 26913720 |
Reported gene related to hsa-miR-193a-5p

| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-193a-5p | lung squamous cell cancer | MCL1 | "Treatment with 5-AzaC concomitantly upregulated ex ......" | 24356455 |
| hsa-miR-193a-5p | ovarian cancer | MCL1 | "We demonstrated that miR-193a decreased the amount ......" | 23588298 |
| hsa-miR-193a-5p | bladder cancer | PSEN1 | "The miR 193a 3p regulated PSEN1 gene suppresses th ......" | 25542424 |
| hsa-miR-193a-5p | esophageal cancer | PSEN1 | "miR 193a 3p regulation of chemoradiation resistanc ......" | 26743123 |
| hsa-miR-193a-5p | acute myeloid leukemia | PTEN | "Our study identifies miR-193a and PTEN as targets ......" | 23223432 |
| hsa-miR-193a-5p | gastric cancer | PTEN | "Downregulation of microRNA 193 3p inhibits tumor p ......" | 26753960 |
| hsa-miR-193a-5p | liver cancer | CASP2 | "This newly identified miR-193a-3p-SRSF2 axis highl ......" | 22117060 |
| hsa-miR-193a-5p | lung cancer | CYP27A1 | "Only miR-193 was associated with biochemical marke ......" | 23142363 |
| hsa-miR-193a-5p | esophageal cancer | ERBB2 | "Furthermore miR-193a-5p reduced ERBB2 expression b ......" | 27203740 |
| hsa-miR-193a-5p | lung cancer | ERBB4 | "miR 193a 3p functions as a tumor suppressor in lun ......" | 25391651 |
| hsa-miR-193a-5p | liver cancer | F2 | "We found miR-193a down-regulation in HCC respect t ......" | 24330766 |
| hsa-miR-193a-5p | breast cancer | FASN | "Profiling studies also exposed a rapid metformin-i ......" | 25213330 |
| hsa-miR-193a-5p | acute myeloid leukemia | HOTAIR | "Finally HOTAIR modulated c-KIT expression by compe ......" | 25979172 |
| hsa-miR-193a-5p | bladder cancer | HOXC9 | "MiR 193a 3p promotes the multi chemoresistance of ......" | 25444900 |
| hsa-miR-193a-5p | bladder cancer | ING5 | "The miR 193a 3p regulated ING5 gene activates the ......" | 25991669 |
| hsa-miR-193a-5p | bladder cancer | LOXL4 | "miR 193a 3p regulates the multi drug resistance of ......" | 25311867 |
| hsa-miR-193a-5p | bladder cancer | OA5 | "MiR 193a 5p Targets the Coding Region of AP 2α mR ......" | 27698912 |
| hsa-miR-193a-5p | liver cancer | PLAU | "The molecular interaction between miR-193a and uPA ......" | 24330766 |
| hsa-miR-193a-5p | sarcoma | SMARCB1 | "Differential microRNA expression profiles between ......" | 24287458 |
| hsa-miR-193a-5p | liver cancer | SRSF2 | "DNA methylation regulated miR 193a 3p dictates res ......" | 22117060 |
| hsa-miR-193a-5p | lung squamous cell cancer | UCA1 | "LncRNA UCA1 exerts oncogenic functions in non smal ......" | 26655272 |
Expression profile in cancer corhorts:
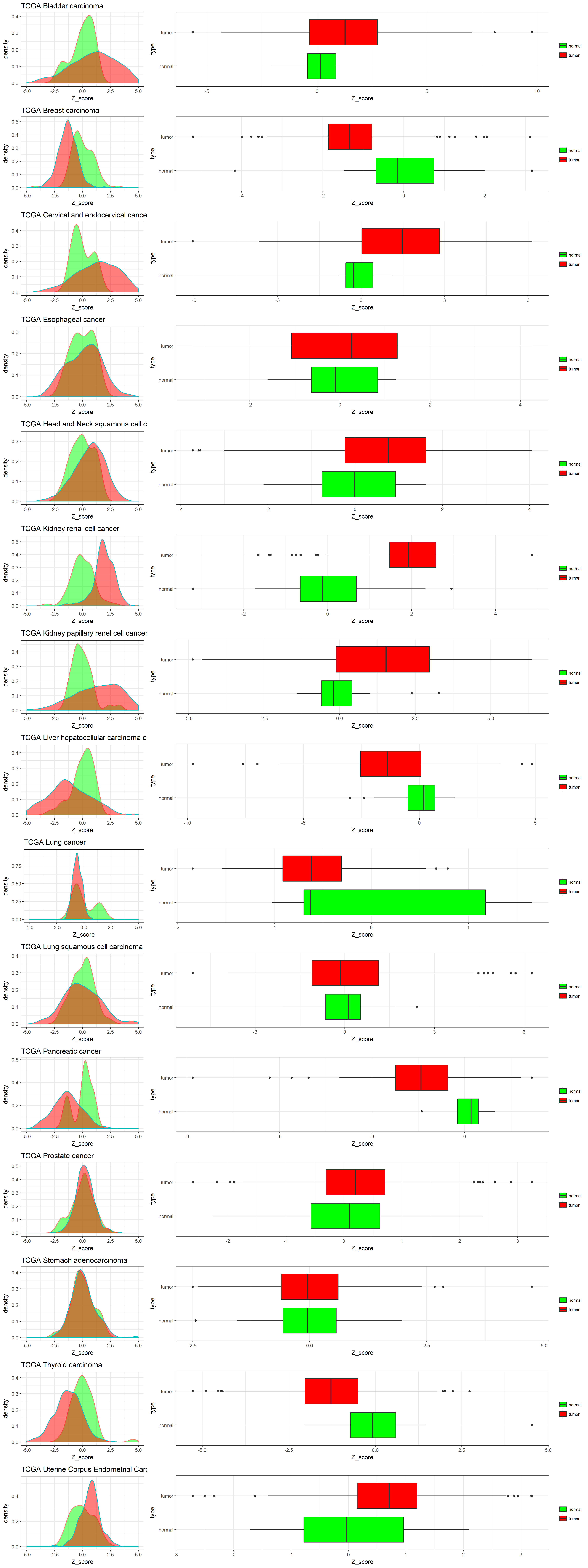
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-193a-5p | GBA2 | 11 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LUAD; PAAD; SARC; STAD | MirTarget; PITA; TargetScan; miRNATAP | TCGA BLCA -0.071; TCGA BRCA -0.113; TCGA CESC -0.148; TCGA ESCA -0.165; TCGA HNSC -0.135; TCGA KIRC -0.374; TCGA KIRP -0.259; TCGA LUAD -0.057; TCGA PAAD -0.186; TCGA SARC -0.129; TCGA STAD -0.147 |
hsa-miR-193a-5p | ACBD4 | 12 cancers: BLCA; CESC; ESCA; HNSC; KIRP; LGG; LUAD; PAAD; PRAD; SARC; THCA; UCEC | MirTarget | TCGA BLCA -0.245; TCGA CESC -0.167; TCGA ESCA -0.308; TCGA HNSC -0.344; TCGA KIRP -0.111; TCGA LGG -0.061; TCGA LUAD -0.134; TCGA PAAD -0.348; TCGA PRAD -0.078; TCGA SARC -0.269; TCGA THCA -0.067; TCGA UCEC -0.087 |
hsa-miR-193a-5p | NOVA1 | 12 cancers: BLCA; CESC; ESCA; HNSC; LGG; LIHC; LUSC; PAAD; PRAD; SARC; THCA; UCEC | PITA; TargetScan; miRNATAP | TCGA BLCA -0.427; TCGA CESC -0.713; TCGA ESCA -0.591; TCGA HNSC -0.248; TCGA LGG -0.333; TCGA LIHC -0.357; TCGA LUSC -0.445; TCGA PAAD -0.413; TCGA PRAD -0.24; TCGA SARC -0.349; TCGA THCA -0.486; TCGA UCEC -0.376 |
hsa-miR-193a-5p | DLX5 | 9 cancers: BLCA; BRCA; CESC; HNSC; LIHC; PRAD; SARC; THCA; UCEC | PITA | TCGA BLCA -0.23; TCGA BRCA -0.299; TCGA CESC -0.696; TCGA HNSC -0.184; TCGA LIHC -0.486; TCGA PRAD -0.158; TCGA SARC -0.454; TCGA THCA -0.449; TCGA UCEC -0.39 |
hsa-miR-193a-5p | MBD6 | 9 cancers: BLCA; BRCA; CESC; HNSC; LUAD; LUSC; PAAD; SARC; THCA | PITA; TargetScan | TCGA BLCA -0.118; TCGA BRCA -0.14; TCGA CESC -0.104; TCGA HNSC -0.058; TCGA LUAD -0.069; TCGA LUSC -0.129; TCGA PAAD -0.187; TCGA SARC -0.13; TCGA THCA -0.11 |
hsa-miR-193a-5p | KBTBD3 | 11 cancers: BLCA; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; PRAD; SARC; STAD | PITA | TCGA BLCA -0.151; TCGA ESCA -0.301; TCGA HNSC -0.255; TCGA KIRC -0.265; TCGA KIRP -0.078; TCGA LUAD -0.163; TCGA LUSC -0.201; TCGA PAAD -0.114; TCGA PRAD -0.067; TCGA SARC -0.1; TCGA STAD -0.119 |
hsa-miR-193a-5p | CNTD1 | 11 cancers: BLCA; ESCA; HNSC; KIRC; LGG; LIHC; LUAD; LUSC; PAAD; SARC; THCA | TargetScan | TCGA BLCA -0.095; TCGA ESCA -0.226; TCGA HNSC -0.328; TCGA KIRC -0.351; TCGA LGG -0.154; TCGA LIHC -0.17; TCGA LUAD -0.245; TCGA LUSC -0.202; TCGA PAAD -0.367; TCGA SARC -0.09; TCGA THCA -0.148 |
hsa-miR-193a-5p | GOLGA6L10 | 9 cancers: BLCA; CESC; HNSC; LGG; LUAD; LUSC; PAAD; SARC; UCEC | TargetScan | TCGA BLCA -0.339; TCGA CESC -0.298; TCGA HNSC -0.211; TCGA LGG -0.111; TCGA LUAD -0.3; TCGA LUSC -0.23; TCGA PAAD -0.225; TCGA SARC -0.182; TCGA UCEC -0.143 |
hsa-miR-193a-5p | GOLGA6L9 | 9 cancers: BLCA; BRCA; CESC; HNSC; LGG; LUAD; LUSC; PAAD; THCA | TargetScan | TCGA BLCA -0.363; TCGA BRCA -0.152; TCGA CESC -0.314; TCGA HNSC -0.373; TCGA LGG -0.137; TCGA LUAD -0.254; TCGA LUSC -0.311; TCGA PAAD -0.355; TCGA THCA -0.077 |
hsa-miR-193a-5p | KIAA0895L | 10 cancers: BLCA; CESC; HNSC; LGG; LIHC; LUAD; LUSC; PAAD; SARC; THCA | TargetScan; miRNATAP | TCGA BLCA -0.273; TCGA CESC -0.149; TCGA HNSC -0.239; TCGA LGG -0.324; TCGA LIHC -0.213; TCGA LUAD -0.261; TCGA LUSC -0.237; TCGA PAAD -0.286; TCGA SARC -0.124; TCGA THCA -0.197 |
hsa-miR-193a-5p | MARCH3 | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUSC; PAAD; PRAD; STAD; UCEC | TargetScan | TCGA BLCA -0.124; TCGA CESC -0.271; TCGA COAD -0.365; TCGA ESCA -0.534; TCGA HNSC -0.426; TCGA LUSC -0.417; TCGA PAAD -0.215; TCGA PRAD -0.123; TCGA STAD -0.336; TCGA UCEC -0.148 |
hsa-miR-193a-5p | SYNGR1 | 9 cancers: BLCA; CESC; HNSC; KIRC; KIRP; LGG; LIHC; PAAD; UCEC | TargetScan | TCGA BLCA -0.373; TCGA CESC -0.279; TCGA HNSC -0.589; TCGA KIRC -0.541; TCGA KIRP -0.141; TCGA LGG -0.103; TCGA LIHC -0.741; TCGA PAAD -0.245; TCGA UCEC -0.167 |
hsa-miR-193a-5p | ALDH5A1 | 10 cancers: BLCA; CESC; HNSC; KIRC; KIRP; LGG; LUAD; OV; PAAD; THCA | mirMAP | TCGA BLCA -0.211; TCGA CESC -0.164; TCGA HNSC -0.513; TCGA KIRC -0.571; TCGA KIRP -0.253; TCGA LGG -0.231; TCGA LUAD -0.16; TCGA OV -0.172; TCGA PAAD -0.292; TCGA THCA -0.07 |
hsa-miR-193a-5p | LONRF2 | 9 cancers: BLCA; CESC; HNSC; KIRC; LUAD; LUSC; PAAD; SARC; THCA | mirMAP | TCGA BLCA -0.534; TCGA CESC -0.751; TCGA HNSC -1.001; TCGA KIRC -1.294; TCGA LUAD -0.443; TCGA LUSC -0.607; TCGA PAAD -0.514; TCGA SARC -1.417; TCGA THCA -0.282 |
hsa-miR-193a-5p | SAMD12 | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LUAD; LUSC; SARC; STAD | mirMAP | TCGA BLCA -0.327; TCGA BRCA -0.102; TCGA CESC -0.369; TCGA ESCA -0.688; TCGA HNSC -0.841; TCGA KIRC -0.88; TCGA LUAD -0.129; TCGA LUSC -0.746; TCGA SARC -0.581; TCGA STAD -0.228 |
hsa-miR-193a-5p | ZFP41 | 10 cancers: BLCA; BRCA; CESC; KIRP; LGG; LIHC; LUAD; OV; PAAD; SARC | mirMAP | TCGA BLCA -0.116; TCGA BRCA -0.114; TCGA CESC -0.109; TCGA KIRP -0.073; TCGA LGG -0.165; TCGA LIHC -0.186; TCGA LUAD -0.071; TCGA OV -0.123; TCGA PAAD -0.277; TCGA SARC -0.119 |
hsa-miR-193a-5p | PLEKHH1 | 9 cancers: BLCA; CESC; ESCA; HNSC; KIRC; KIRP; LIHC; PAAD; STAD | miRNATAP | TCGA BLCA -0.437; TCGA CESC -0.317; TCGA ESCA -0.357; TCGA HNSC -0.17; TCGA KIRC -0.52; TCGA KIRP -0.157; TCGA LIHC -0.188; TCGA PAAD -0.601; TCGA STAD -0.178 |
hsa-miR-193a-5p | CDC14B | 9 cancers: BLCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; LUSC; STAD | miRNATAP | TCGA BLCA -0.14; TCGA CESC -0.313; TCGA ESCA -0.467; TCGA HNSC -0.107; TCGA KIRC -0.399; TCGA KIRP -0.155; TCGA LGG -0.065; TCGA LUSC -0.235; TCGA STAD -0.103 |
hsa-miR-193a-5p | WNK2 | 9 cancers: BRCA; CESC; HNSC; KIRC; KIRP; LUSC; PAAD; PRAD; SARC | mirMAP | TCGA BRCA -0.204; TCGA CESC -0.529; TCGA HNSC -1.368; TCGA KIRC -0.865; TCGA KIRP -0.427; TCGA LUSC -0.613; TCGA PAAD -0.701; TCGA PRAD -0.269; TCGA SARC -0.44 |
hsa-miR-193a-5p | SPEG | 9 cancers: BRCA; HNSC; KIRC; LIHC; LUAD; PRAD; SARC; THCA; UCEC | miRNATAP | TCGA BRCA -0.191; TCGA HNSC -0.244; TCGA KIRC -0.331; TCGA LIHC -0.473; TCGA LUAD -0.237; TCGA PRAD -0.177; TCGA SARC -0.652; TCGA THCA -0.116; TCGA UCEC -0.24 |
hsa-miR-193a-5p | GATS | 10 cancers: CESC; ESCA; HNSC; LGG; LIHC; LUSC; PAAD; PRAD; THCA; UCEC | MirTarget | TCGA CESC -0.331; TCGA ESCA -0.213; TCGA HNSC -0.512; TCGA LGG -0.143; TCGA LIHC -0.458; TCGA LUSC -0.297; TCGA PAAD -0.27; TCGA PRAD -0.085; TCGA THCA -0.14; TCGA UCEC -0.106 |
hsa-miR-193a-5p | KCNH8 | 10 cancers: CESC; HNSC; LGG; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | PITA; TargetScan | TCGA CESC -0.629; TCGA HNSC -0.941; TCGA LGG -0.394; TCGA LUAD -0.294; TCGA LUSC -1.098; TCGA PAAD -0.655; TCGA PRAD -0.38; TCGA SARC -0.588; TCGA STAD -0.508; TCGA UCEC -0.435 |
hsa-miR-193a-5p | HDDC2 | 9 cancers: HNSC; KIRC; KIRP; LGG; LIHC; LUSC; OV; PAAD; THCA | TargetScan | TCGA HNSC -0.199; TCGA KIRC -0.21; TCGA KIRP -0.087; TCGA LGG -0.094; TCGA LIHC -0.199; TCGA LUSC -0.131; TCGA OV -0.141; TCGA PAAD -0.136; TCGA THCA -0.095 |
Enriched cancer pathways of putative targets