microRNA information: hsa-miR-193b-3p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-193b-3p | miRbase |
| Accession: | MIMAT0002819 | miRbase |
| Precursor name: | hsa-mir-193b | miRbase |
| Precursor accession: | MI0003137 | miRbase |
| Symbol: | MIR193B | HGNC |
| RefSeq ID: | NR_030177 | GenBank |
| Sequence: | AACUGGCCCUCAAAGUCCCGCU |
Reported expression in cancers: hsa-miR-193b-3p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-193b-3p | breast cancer | upregulation | "In this study expression profiles of microRNAs miR ......" | 23183268 | Microarray; qPCR |
| hsa-miR-193b-3p | gastric cancer | deregulation | "Association of miR 193b down regulation and miR 19 ......" | 25374225 | qPCR; Microarray |
| hsa-miR-193b-3p | liver cancer | downregulation | "The expression profiles of miR-193b were compared ......" | 20655737 | qPCR |
| hsa-miR-193b-3p | lung squamous cell cancer | downregulation | "Here we investigated the role of microRNA-193b miR ......" | 22491710 | |
| hsa-miR-193b-3p | lymphoma | deregulation | "We found miR-34a miR-146a and miR-193b to be up-re ......" | 25862860 | |
| hsa-miR-193b-3p | melanoma | downregulation | "Notably miR-193b was significantly down-regulated ......" | 20304954 | |
| hsa-miR-193b-3p | melanoma | downregulation | "Among the miRNAs that were commonly and significan ......" | 27149382 | |
| hsa-miR-193b-3p | ovarian cancer | downregulation | "Tissue miR 193b as a Novel Biomarker for Patients ......" | 26675282 | |
| hsa-miR-193b-3p | pancreatic cancer | downregulation | "Here we identified miR-193b as a tumor-suppressive ......" | 25905463 | |
| hsa-miR-193b-3p | prostate cancer | deregulation | "Upregulation of miR-122 miR-335 miR-184 miR-193 mi ......" | 23781281 |
Reported cancer pathway affected by hsa-miR-193b-3p
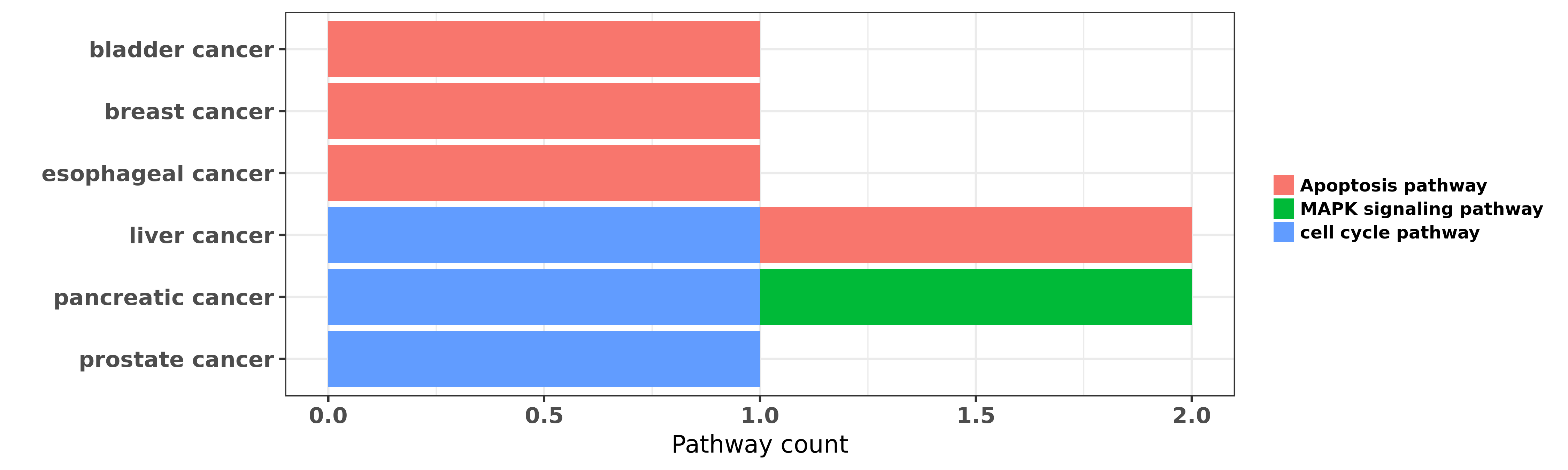
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-193b-3p | bladder cancer | Apoptosis pathway | "Bladder cancer cell lines were exposed to normoxic ......" | 26196183 | Flow cytometry |
| hsa-miR-193b-3p | breast cancer | Apoptosis pathway | "Profiling studies also exposed a rapid metformin-i ......" | 25213330 | |
| hsa-miR-193b-3p | esophageal cancer | Apoptosis pathway | "MiR 193b promotes autophagy and non apoptotic cell ......" | 26878873 | Colony formation |
| hsa-miR-193b-3p | liver cancer | cell cycle pathway | "MicroRNA 193b regulates proliferation migration an ......" | 20655737 | Luciferase |
| hsa-miR-193b-3p | liver cancer | Apoptosis pathway | "Restoration of miR 193b sensitizes Hepatitis B vir ......" | 25034398 | Western blot; Luciferase; Flow cytometry |
| hsa-miR-193b-3p | pancreatic cancer | cell cycle pathway | "Deregulation of the MiR 193b KRAS Axis Contributes ......" | 25905463 | |
| hsa-miR-193b-3p | pancreatic cancer | MAPK signaling pathway | "In our previous studies we identified four microRN ......" | 27380024 | |
| hsa-miR-193b-3p | prostate cancer | cell cycle pathway | "miR 193b is an epigenetically regulated putative t ......" | 20073067 |
Reported cancer prognosis affected by hsa-miR-193b-3p
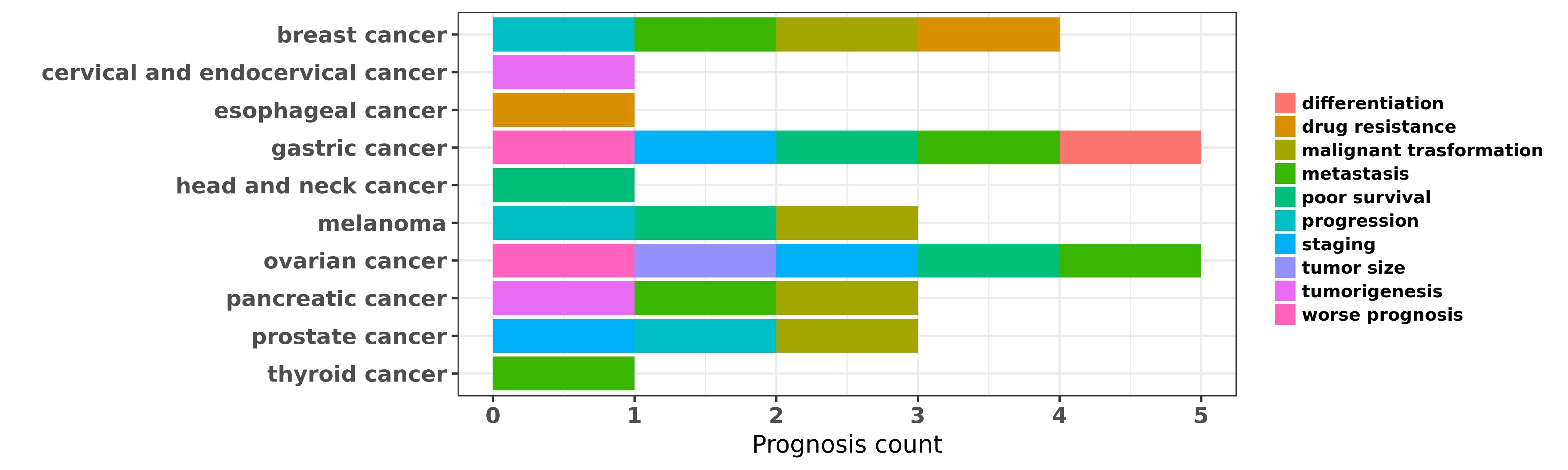
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-193b-3p | breast cancer | progression; metastasis | "Downregulation of miR 193b contributes to enhance ......" | 19701247 | |
| hsa-miR-193b-3p | breast cancer | malignant trasformation | "In support of these findings in vitro functional s ......" | 24104550 | |
| hsa-miR-193b-3p | breast cancer | progression | "Tumor suppressive microRNA 193b promotes breast ca ......" | 25550792 | Luciferase |
| hsa-miR-193b-3p | breast cancer | drug resistance | "miR 193b Modulates Resistance to Doxorubicin in Hu ......" | 26526790 | |
| hsa-miR-193b-3p | cervical and endocervical cancer | tumorigenesis | "The methylation status was determined using Human ......" | 26797462 | |
| hsa-miR-193b-3p | esophageal cancer | drug resistance | "An in-vitro model of acquired chemotherapy resista ......" | 25356050 | |
| hsa-miR-193b-3p | esophageal cancer | drug resistance | "Differentially expressed miRNAs between ESCC patho ......" | 26445467 | |
| hsa-miR-193b-3p | gastric cancer | metastasis | "We detected the expression profiles of miRNA by mi ......" | 23372348 | Western blot; Wound Healing Assay |
| hsa-miR-193b-3p | gastric cancer | worse prognosis; staging; metastasis; differentiation; poor survival | "Association of miR 193b down regulation and miR 19 ......" | 25374225 | |
| hsa-miR-193b-3p | head and neck cancer | poor survival | "Alteration in miR-193b expression level does not s ......" | 25677760 | |
| hsa-miR-193b-3p | head and neck cancer | poor survival | "In this study we analyzed miRNA-seq data obtained ......" | 27109697 | |
| hsa-miR-193b-3p | melanoma | poor survival | "Furthermore low expression of miR-191 and high exp ......" | 20357817 | |
| hsa-miR-193b-3p | melanoma | malignant trasformation; progression | "miR 193b Regulates Mcl 1 in Melanoma; In a previou ......" | 21893020 | |
| hsa-miR-193b-3p | ovarian cancer | metastasis | "Microenvironment induced downregulation of miR 193 ......" | 25798837 | |
| hsa-miR-193b-3p | ovarian cancer | poor survival; staging; metastasis; tumor size; worse prognosis | "Tissue miR 193b as a Novel Biomarker for Patients ......" | 26675282 | |
| hsa-miR-193b-3p | pancreatic cancer | metastasis | "miR 193b directly targets STMN1 and uPA genes and ......" | 25215905 | |
| hsa-miR-193b-3p | pancreatic cancer | malignant trasformation; tumorigenesis | "Deregulation of the MiR 193b KRAS Axis Contributes ......" | 25905463 | |
| hsa-miR-193b-3p | prostate cancer | progression; malignant trasformation | "CFTR suppresses tumor progression through miR 193b ......" | 22797075 | |
| hsa-miR-193b-3p | prostate cancer | staging | "Epigenetically altered miR 193b targets cyclin D1 ......" | 26129688 | |
| hsa-miR-193b-3p | thyroid cancer | metastasis | "To identify the miRNA signature in PTC with LN met ......" | 23609190 |
Reported gene related to hsa-miR-193b-3p
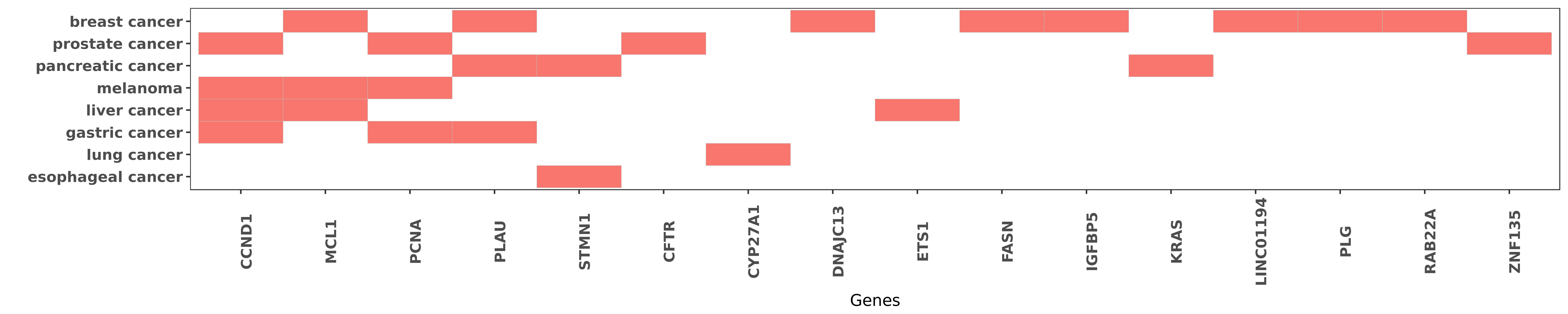
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-193b-3p | gastric cancer | CCND1 | "MicroRNA 193b inhibits the proliferation migration ......" | 27071318 |
| hsa-miR-193b-3p | liver cancer | CCND1 | "CCND1 and ETS1 were revealed to be regulated by mi ......" | 20655737 |
| hsa-miR-193b-3p | melanoma | CCND1 | "MicroRNA 193b represses cell proliferation and reg ......" | 20304954 |
| hsa-miR-193b-3p | melanoma | CCND1 | "In a previous study we reported that miR-193b repr ......" | 21893020 |
| hsa-miR-193b-3p | prostate cancer | CCND1 | "Epigenetically altered miR 193b targets cyclin D1 ......" | 26129688 |
| hsa-miR-193b-3p | gastric cancer | PCNA | "MicroRNA 193b inhibits the proliferation migration ......" | 27071318 |
| hsa-miR-193b-3p | melanoma | PCNA | "MicroRNA 193b represses cell proliferation and reg ......" | 20304954 |
| hsa-miR-193b-3p | melanoma | PCNA | "In a previous study we reported that miR-193b repr ......" | 21893020 |
| hsa-miR-193b-3p | prostate cancer | PCNA | "Epigenetically altered miR 193b targets cyclin D1 ......" | 26129688 |
| hsa-miR-193b-3p | breast cancer | MCL1 | "miR 193b Modulates Resistance to Doxorubicin in Hu ......" | 26526790 |
| hsa-miR-193b-3p | liver cancer | MCL1 | "Expression of miR-193b and myeloid cell leukemia-1 ......" | 25034398 |
| hsa-miR-193b-3p | melanoma | MCL1 | "miR 193b Regulates Mcl 1 in Melanoma; Herein we de ......" | 21893020 |
| hsa-miR-193b-3p | breast cancer | PLAU | "Downregulation of miR 193b contributes to enhance ......" | 19701247 |
| hsa-miR-193b-3p | gastric cancer | PLAU | "Among the altered miRNAs TGF-β1 induced the down- ......" | 23372348 |
| hsa-miR-193b-3p | pancreatic cancer | PLAU | "miR 193b directly targets STMN1 and uPA genes and ......" | 25215905 |
| hsa-miR-193b-3p | esophageal cancer | STMN1 | "The critical mRNA targets of miR-193b are unknown ......" | 26878873 |
| hsa-miR-193b-3p | pancreatic cancer | STMN1 | "miR 193b directly targets STMN1 and uPA genes and ......" | 25215905 |
| hsa-miR-193b-3p | prostate cancer | CFTR | "Taken together the present study has demonstrated ......" | 22797075 |
| hsa-miR-193b-3p | lung cancer | CYP27A1 | "Only miR-193 was associated with biochemical marke ......" | 23142363 |
| hsa-miR-193b-3p | breast cancer | DNAJC13 | "Tumor suppressive microRNA 193b promotes breast ca ......" | 25550792 |
| hsa-miR-193b-3p | liver cancer | ETS1 | "CCND1 and ETS1 were revealed to be regulated by mi ......" | 20655737 |
| hsa-miR-193b-3p | breast cancer | FASN | "Profiling studies also exposed a rapid metformin-i ......" | 25213330 |
| hsa-miR-193b-3p | breast cancer | IGFBP5 | "Experimental and bioinformatics studies have shown ......" | 25983620 |
| hsa-miR-193b-3p | pancreatic cancer | KRAS | "Deregulation of the MiR 193b KRAS Axis Contributes ......" | 25905463 |
| hsa-miR-193b-3p | breast cancer | LINC01194 | "Here we applied a high-throughput strategy using q ......" | 21512034 |
| hsa-miR-193b-3p | breast cancer | PLG | "Downregulation of miR 193b contributes to enhance ......" | 19701247 |
| hsa-miR-193b-3p | breast cancer | RAB22A | "Tumor suppressive microRNA 193b promotes breast ca ......" | 25550792 |
| hsa-miR-193b-3p | prostate cancer | ZNF135 | "We found reduced miR-193b expression P < 0.05 in s ......" | 26129688 |
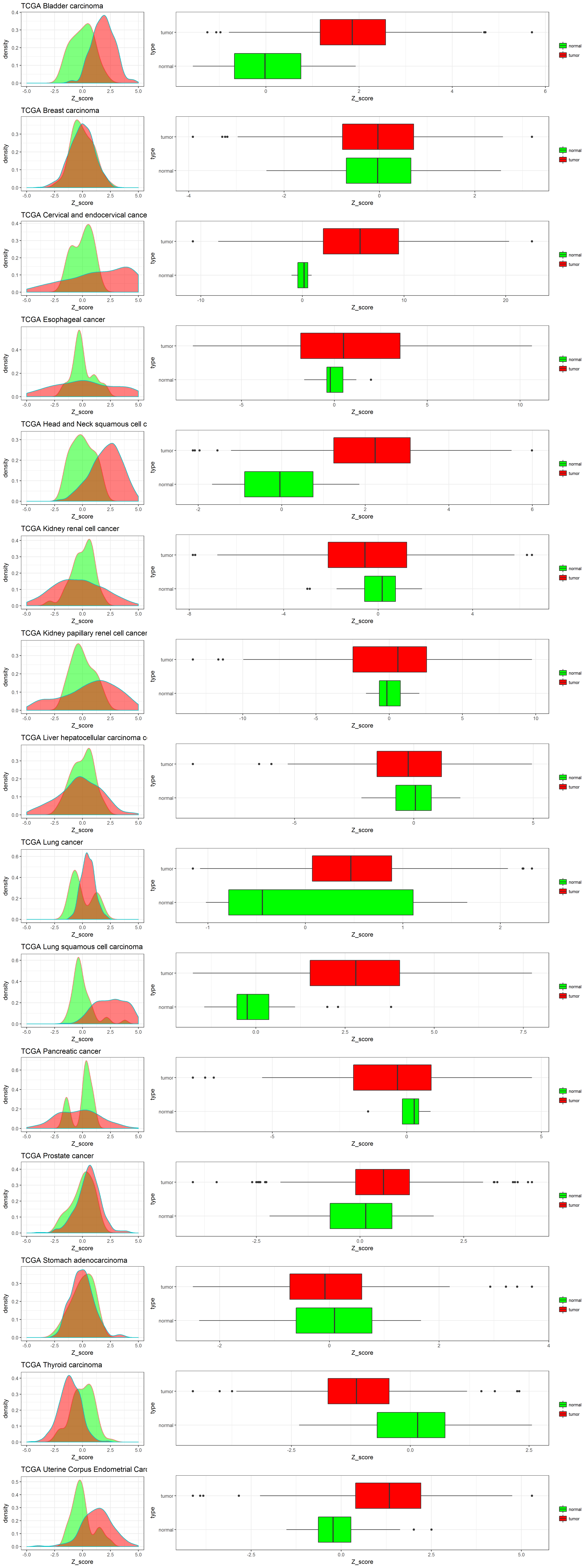
Expression profile in cancer corhorts:
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-193b-3p | ABI2 | 16 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate; MirTarget | TCGA BLCA -0.107; TCGA BRCA -0.063; TCGA CESC -0.153; TCGA ESCA -0.092; TCGA HNSC -0.153; TCGA KIRC -0.065; TCGA KIRP -0.097; TCGA LIHC -0.243; TCGA LUAD -0.066; TCGA LUSC -0.092; TCGA PAAD -0.057; TCGA PRAD -0.152; TCGA SARC -0.087; TCGA THCA -0.074; TCGA STAD -0.095; TCGA UCEC -0.085 |
hsa-miR-193b-3p | ADARB1 | 9 cancers: BLCA; BRCA; KIRC; KIRP; LIHC; LUAD; LUSC; STAD; UCEC | miRNAWalker2 validate; MirTarget | TCGA BLCA -0.151; TCGA BRCA -0.055; TCGA KIRC -0.158; TCGA KIRP -0.251; TCGA LIHC -0.115; TCGA LUAD -0.06; TCGA LUSC -0.303; TCGA STAD -0.199; TCGA UCEC -0.097 |
hsa-miR-193b-3p | AKAP13 | 13 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; PRAD; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.19; TCGA BRCA -0.06; TCGA ESCA -0.211; TCGA HNSC -0.131; TCGA KIRC -0.114; TCGA KIRP -0.061; TCGA LGG -0.106; TCGA LUAD -0.114; TCGA LUSC -0.209; TCGA PRAD -0.169; TCGA THCA -0.091; TCGA STAD -0.069; TCGA UCEC -0.078 |
hsa-miR-193b-3p | ALDH1A2 | 9 cancers: BLCA; BRCA; CESC; HNSC; KIRP; LUAD; LUSC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.433; TCGA BRCA -0.274; TCGA CESC -0.63; TCGA HNSC -0.647; TCGA KIRP -0.671; TCGA LUAD -0.148; TCGA LUSC -0.617; TCGA STAD -0.418; TCGA UCEC -0.384 |
hsa-miR-193b-3p | APOLD1 | 12 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.341; TCGA BRCA -0.08; TCGA CESC -0.174; TCGA ESCA -0.378; TCGA HNSC -0.177; TCGA KIRC -0.483; TCGA LUAD -0.097; TCGA LUSC -0.319; TCGA PAAD -0.169; TCGA SARC -0.174; TCGA STAD -0.26; TCGA UCEC -0.274 |
hsa-miR-193b-3p | ASH1L | 10 cancers: BLCA; COAD; ESCA; HNSC; KIRC; KIRP; OV; PRAD; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.117; TCGA COAD -0.068; TCGA ESCA -0.107; TCGA HNSC -0.19; TCGA KIRC -0.113; TCGA KIRP -0.065; TCGA OV -0.09; TCGA PRAD -0.174; TCGA STAD -0.109; TCGA UCEC -0.088 |
hsa-miR-193b-3p | ATOH8 | 15 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.813; TCGA BRCA -0.438; TCGA CESC -0.189; TCGA ESCA -0.593; TCGA HNSC -0.451; TCGA KIRC -0.346; TCGA KIRP -0.236; TCGA LGG -0.305; TCGA LUAD -0.254; TCGA LUSC -0.635; TCGA PAAD -0.411; TCGA PRAD -0.163; TCGA SARC -0.19; TCGA STAD -0.297; TCGA UCEC -0.377 |
hsa-miR-193b-3p | C5 | 9 cancers: BLCA; CESC; ESCA; HNSC; LUAD; LUSC; PAAD; THCA; UCEC | miRNAWalker2 validate | TCGA BLCA -0.13; TCGA CESC -0.112; TCGA ESCA -0.193; TCGA HNSC -0.372; TCGA LUAD -0.244; TCGA LUSC -0.303; TCGA PAAD -0.245; TCGA THCA -0.075; TCGA UCEC -0.218 |
hsa-miR-193b-3p | CTDSP2 | 9 cancers: BLCA; COAD; HNSC; KIRC; KIRP; LUSC; PRAD; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.102; TCGA COAD -0.057; TCGA HNSC -0.105; TCGA KIRC -0.095; TCGA KIRP -0.104; TCGA LUSC -0.079; TCGA PRAD -0.113; TCGA STAD -0.064; TCGA UCEC -0.059 |
hsa-miR-193b-3p | DACT2 | 11 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LIHC; LUSC; PAAD; PRAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.302; TCGA BRCA -0.65; TCGA CESC -0.296; TCGA HNSC -0.607; TCGA KIRC -0.298; TCGA KIRP -0.662; TCGA LIHC -0.733; TCGA LUSC -0.225; TCGA PAAD -0.491; TCGA PRAD -0.792; TCGA UCEC -0.183 |
hsa-miR-193b-3p | DCAF7 | 13 cancers: BLCA; CESC; ESCA; HNSC; KIRP; LGG; LIHC; LUAD; OV; PAAD; PRAD; SARC; THCA | miRNAWalker2 validate; MirTarget; miRNATAP | TCGA BLCA -0.074; TCGA CESC -0.09; TCGA ESCA -0.149; TCGA HNSC -0.149; TCGA KIRP -0.086; TCGA LGG -0.106; TCGA LIHC -0.172; TCGA LUAD -0.065; TCGA OV -0.148; TCGA PAAD -0.135; TCGA PRAD -0.171; TCGA SARC -0.062; TCGA THCA -0.057 |
hsa-miR-193b-3p | EEF2 | 9 cancers: BLCA; BRCA; HNSC; LGG; LIHC; LUAD; PRAD; THCA; STAD | miRNAWalker2 validate | TCGA BLCA -0.088; TCGA BRCA -0.11; TCGA HNSC -0.099; TCGA LGG -0.231; TCGA LIHC -0.071; TCGA LUAD -0.072; TCGA PRAD -0.091; TCGA THCA -0.152; TCGA STAD -0.069 |
hsa-miR-193b-3p | EFHD1 | 10 cancers: BLCA; CESC; ESCA; HNSC; KIRP; LUSC; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.197; TCGA CESC -0.307; TCGA ESCA -0.369; TCGA HNSC -0.351; TCGA KIRP -0.252; TCGA LUSC -0.265; TCGA SARC -0.431; TCGA THCA -0.085; TCGA STAD -0.197; TCGA UCEC -0.325 |
hsa-miR-193b-3p | EP300 | 9 cancers: BLCA; COAD; HNSC; KIRC; KIRP; OV; PRAD; SARC; THCA | miRNAWalker2 validate | TCGA BLCA -0.106; TCGA COAD -0.073; TCGA HNSC -0.144; TCGA KIRC -0.094; TCGA KIRP -0.085; TCGA OV -0.068; TCGA PRAD -0.147; TCGA SARC -0.072; TCGA THCA -0.058 |
hsa-miR-193b-3p | ESR1 | 9 cancers: BLCA; CESC; HNSC; LUAD; LUSC; PAAD; PRAD; THCA; STAD | miRNAWalker2 validate; miRTarBase | TCGA BLCA -0.251; TCGA CESC -0.284; TCGA HNSC -0.171; TCGA LUAD -0.174; TCGA LUSC -0.183; TCGA PAAD -0.271; TCGA PRAD -0.136; TCGA THCA -0.209; TCGA STAD -0.291 |
hsa-miR-193b-3p | ETS1 | 10 cancers: BLCA; BRCA; COAD; ESCA; KIRC; LUAD; LUSC; PRAD; STAD; UCEC | miRNAWalker2 validate; miRTarBase | TCGA BLCA -0.116; TCGA BRCA -0.093; TCGA COAD -0.171; TCGA ESCA -0.226; TCGA KIRC -0.271; TCGA LUAD -0.087; TCGA LUSC -0.339; TCGA PRAD -0.081; TCGA STAD -0.099; TCGA UCEC -0.086 |
hsa-miR-193b-3p | GCC2 | 13 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PRAD; SARC; STAD | miRNAWalker2 validate | TCGA BLCA -0.083; TCGA CESC -0.102; TCGA COAD -0.131; TCGA ESCA -0.497; TCGA HNSC -0.181; TCGA KIRC -0.06; TCGA KIRP -0.065; TCGA LIHC -0.137; TCGA LUAD -0.096; TCGA LUSC -0.101; TCGA PRAD -0.206; TCGA SARC -0.083; TCGA STAD -0.081 |
hsa-miR-193b-3p | GIMAP2 | 9 cancers: BLCA; CESC; ESCA; HNSC; LUAD; LUSC; PRAD; SARC; STAD | miRNAWalker2 validate | TCGA BLCA -0.099; TCGA CESC -0.135; TCGA ESCA -0.314; TCGA HNSC -0.158; TCGA LUAD -0.188; TCGA LUSC -0.351; TCGA PRAD -0.08; TCGA SARC -0.132; TCGA STAD -0.182 |
hsa-miR-193b-3p | HACE1 | 13 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; LIHC; LUSC; OV; PAAD; PRAD; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.062; TCGA BRCA -0.051; TCGA CESC -0.086; TCGA COAD -0.079; TCGA HNSC -0.145; TCGA KIRC -0.129; TCGA LIHC -0.118; TCGA LUSC -0.078; TCGA OV -0.155; TCGA PAAD -0.109; TCGA PRAD -0.177; TCGA STAD -0.14; TCGA UCEC -0.114 |
hsa-miR-193b-3p | HERC1 | 11 cancers: BLCA; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.164; TCGA ESCA -0.13; TCGA HNSC -0.196; TCGA KIRC -0.073; TCGA LUAD -0.056; TCGA LUSC -0.11; TCGA PAAD -0.171; TCGA PRAD -0.128; TCGA SARC -0.086; TCGA STAD -0.164; TCGA UCEC -0.054 |
hsa-miR-193b-3p | KANK2 | 11 cancers: BLCA; CESC; ESCA; HNSC; KIRC; LUAD; LUSC; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.299; TCGA CESC -0.114; TCGA ESCA -0.202; TCGA HNSC -0.075; TCGA KIRC -0.117; TCGA LUAD -0.09; TCGA LUSC -0.371; TCGA SARC -0.228; TCGA THCA -0.19; TCGA STAD -0.318; TCGA UCEC -0.219 |
hsa-miR-193b-3p | MAP3K3 | 11 cancers: BLCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; SARC; STAD; UCEC | miRNAWalker2 validate; miRNATAP | TCGA BLCA -0.12; TCGA HNSC -0.125; TCGA KIRC -0.068; TCGA KIRP -0.136; TCGA LIHC -0.05; TCGA LUAD -0.117; TCGA LUSC -0.224; TCGA PAAD -0.086; TCGA SARC -0.096; TCGA STAD -0.142; TCGA UCEC -0.125 |
hsa-miR-193b-3p | MTR | 10 cancers: BLCA; CESC; HNSC; KIRC; LIHC; LUAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.105; TCGA CESC -0.069; TCGA HNSC -0.14; TCGA KIRC -0.053; TCGA LIHC -0.122; TCGA LUAD -0.062; TCGA PRAD -0.057; TCGA SARC -0.145; TCGA STAD -0.173; TCGA UCEC -0.136 |
hsa-miR-193b-3p | NCOA2 | 9 cancers: BLCA; COAD; ESCA; HNSC; LUAD; LUSC; PRAD; SARC; STAD | miRNAWalker2 validate | TCGA BLCA -0.146; TCGA COAD -0.256; TCGA ESCA -0.204; TCGA HNSC -0.248; TCGA LUAD -0.173; TCGA LUSC -0.12; TCGA PRAD -0.241; TCGA SARC -0.255; TCGA STAD -0.11 |
hsa-miR-193b-3p | PLXNC1 | 10 cancers: BLCA; CESC; COAD; HNSC; LUAD; LUSC; PRAD; THCA; STAD; UCEC | miRNAWalker2 validate; miRNATAP | TCGA BLCA -0.185; TCGA CESC -0.165; TCGA COAD -0.462; TCGA HNSC -0.428; TCGA LUAD -0.199; TCGA LUSC -0.371; TCGA PRAD -0.255; TCGA THCA -0.271; TCGA STAD -0.163; TCGA UCEC -0.231 |
hsa-miR-193b-3p | PM20D2 | 10 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; PAAD; PRAD | miRNAWalker2 validate | TCGA BLCA -0.152; TCGA BRCA -0.332; TCGA ESCA -0.134; TCGA HNSC -0.277; TCGA KIRC -0.139; TCGA KIRP -0.059; TCGA LGG -0.165; TCGA LIHC -0.268; TCGA PAAD -0.244; TCGA PRAD -0.176 |
hsa-miR-193b-3p | PRICKLE4 | 10 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; KIRP; LGG; LUSC; SARC; UCEC | miRNAWalker2 validate | TCGA BLCA -0.158; TCGA BRCA -0.224; TCGA ESCA -0.575; TCGA HNSC -0.325; TCGA KIRC -0.323; TCGA KIRP -0.262; TCGA LGG -0.111; TCGA LUSC -0.194; TCGA SARC -0.223; TCGA UCEC -0.111 |
hsa-miR-193b-3p | PTPRG | 11 cancers: BLCA; CESC; COAD; ESCA; KIRC; LUAD; LUSC; PRAD; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.213; TCGA CESC -0.185; TCGA COAD -0.123; TCGA ESCA -0.378; TCGA KIRC -0.2; TCGA LUAD -0.253; TCGA LUSC -0.296; TCGA PRAD -0.184; TCGA THCA -0.087; TCGA STAD -0.161; TCGA UCEC -0.099 |
hsa-miR-193b-3p | RASSF3 | 9 cancers: BLCA; COAD; ESCA; HNSC; LUAD; LUSC; PRAD; SARC; STAD | miRNAWalker2 validate | TCGA BLCA -0.195; TCGA COAD -0.158; TCGA ESCA -0.373; TCGA HNSC -0.141; TCGA LUAD -0.116; TCGA LUSC -0.252; TCGA PRAD -0.13; TCGA SARC -0.36; TCGA STAD -0.138 |
hsa-miR-193b-3p | SNRK | 13 cancers: BLCA; COAD; ESCA; KIRC; KIRP; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.157; TCGA COAD -0.057; TCGA ESCA -0.16; TCGA KIRC -0.203; TCGA KIRP -0.064; TCGA LUAD -0.103; TCGA LUSC -0.235; TCGA PAAD -0.092; TCGA PRAD -0.113; TCGA SARC -0.094; TCGA THCA -0.061; TCGA STAD -0.194; TCGA UCEC -0.103 |
hsa-miR-193b-3p | SNX18 | 10 cancers: BLCA; CESC; HNSC; KIRP; LGG; LUAD; PRAD; SARC; THCA; STAD | miRNAWalker2 validate | TCGA BLCA -0.108; TCGA CESC -0.077; TCGA HNSC -0.054; TCGA KIRP -0.076; TCGA LGG -0.127; TCGA LUAD -0.139; TCGA PRAD -0.099; TCGA SARC -0.09; TCGA THCA -0.11; TCGA STAD -0.1 |
hsa-miR-193b-3p | TACC1 | 14 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.291; TCGA CESC -0.112; TCGA COAD -0.265; TCGA ESCA -0.343; TCGA HNSC -0.369; TCGA KIRC -0.168; TCGA LIHC -0.143; TCGA LUAD -0.175; TCGA LUSC -0.385; TCGA PAAD -0.172; TCGA PRAD -0.115; TCGA SARC -0.191; TCGA STAD -0.23; TCGA UCEC -0.211 |
hsa-miR-193b-3p | TAF9B | 12 cancers: BLCA; CESC; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.098; TCGA CESC -0.104; TCGA HNSC -0.149; TCGA KIRC -0.08; TCGA KIRP -0.05; TCGA LUAD -0.108; TCGA LUSC -0.056; TCGA PAAD -0.163; TCGA PRAD -0.119; TCGA SARC -0.079; TCGA STAD -0.162; TCGA UCEC -0.146 |
hsa-miR-193b-3p | TGFBR3 | 12 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate; MirTarget; miRNATAP | TCGA BLCA -0.543; TCGA BRCA -0.187; TCGA CESC -0.156; TCGA ESCA -0.499; TCGA HNSC -0.625; TCGA LUAD -0.23; TCGA LUSC -0.44; TCGA PAAD -0.267; TCGA PRAD -0.125; TCGA SARC -0.341; TCGA STAD -0.483; TCGA UCEC -0.176 |
hsa-miR-193b-3p | TLE4 | 10 cancers: BLCA; BRCA; CESC; COAD; KIRP; LIHC; LUAD; PAAD; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.249; TCGA BRCA -0.2; TCGA CESC -0.186; TCGA COAD -0.169; TCGA KIRP -0.177; TCGA LIHC -0.12; TCGA LUAD -0.219; TCGA PAAD -0.099; TCGA STAD -0.241; TCGA UCEC -0.198 |
hsa-miR-193b-3p | TMEM204 | 11 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; SARC; UCEC | miRNAWalker2 validate | TCGA BLCA -0.206; TCGA BRCA -0.111; TCGA CESC -0.109; TCGA ESCA -0.186; TCGA HNSC -0.092; TCGA KIRC -0.361; TCGA LIHC -0.111; TCGA LUAD -0.06; TCGA LUSC -0.327; TCGA SARC -0.135; TCGA UCEC -0.164 |
hsa-miR-193b-3p | TRAK2 | 13 cancers: BLCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD | miRNAWalker2 validate | TCGA BLCA -0.147; TCGA CESC -0.18; TCGA COAD -0.083; TCGA ESCA -0.261; TCGA HNSC -0.192; TCGA LIHC -0.082; TCGA LUAD -0.109; TCGA LUSC -0.191; TCGA PAAD -0.074; TCGA PRAD -0.087; TCGA SARC -0.107; TCGA THCA -0.15; TCGA STAD -0.149 |
hsa-miR-193b-3p | UBR1 | 10 cancers: BLCA; COAD; ESCA; KIRC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.125; TCGA COAD -0.164; TCGA ESCA -0.118; TCGA KIRC -0.079; TCGA LUAD -0.075; TCGA LUSC -0.066; TCGA PRAD -0.135; TCGA SARC -0.127; TCGA STAD -0.127; TCGA UCEC -0.079 |
hsa-miR-193b-3p | SON | 9 cancers: BLCA; COAD; HNSC; KIRC; KIRP; LUAD; PAAD; PRAD; STAD | MirTarget | TCGA BLCA -0.074; TCGA COAD -0.107; TCGA HNSC -0.09; TCGA KIRC -0.07; TCGA KIRP -0.066; TCGA LUAD -0.058; TCGA PAAD -0.063; TCGA PRAD -0.143; TCGA STAD -0.063 |
hsa-miR-193b-3p | TCF4 | 10 cancers: BLCA; CESC; HNSC; KIRC; LGG; LUAD; LUSC; PRAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.263; TCGA CESC -0.161; TCGA HNSC -0.245; TCGA KIRC -0.301; TCGA LGG -0.141; TCGA LUAD -0.091; TCGA LUSC -0.097; TCGA PRAD -0.069; TCGA STAD -0.209; TCGA UCEC -0.225 |
hsa-miR-193b-3p | FAM84A | 12 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LIHC; OV; PAAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.369; TCGA BRCA -0.248; TCGA CESC -0.24; TCGA HNSC -0.387; TCGA KIRC -0.25; TCGA KIRP -0.244; TCGA LIHC -0.295; TCGA OV -0.284; TCGA PAAD -0.214; TCGA THCA -0.204; TCGA STAD -0.319; TCGA UCEC -0.139 |
hsa-miR-193b-3p | VASH1 | 10 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; KIRP; LIHC; LUSC; PAAD; UCEC | MirTarget | TCGA BLCA -0.165; TCGA BRCA -0.139; TCGA ESCA -0.225; TCGA HNSC -0.184; TCGA KIRC -0.221; TCGA KIRP -0.063; TCGA LIHC -0.198; TCGA LUSC -0.307; TCGA PAAD -0.166; TCGA UCEC -0.076 |
hsa-miR-193b-3p | ETV1 | 10 cancers: BLCA; CESC; HNSC; KIRP; LGG; LUAD; LUSC; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.173; TCGA CESC -0.334; TCGA HNSC -0.539; TCGA KIRP -0.35; TCGA LGG -0.379; TCGA LUAD -0.33; TCGA LUSC -0.352; TCGA THCA -0.238; TCGA STAD -0.204; TCGA UCEC -0.245 |
hsa-miR-193b-3p | ARHGEF12 | 13 cancers: BLCA; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PRAD; SARC; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.118; TCGA COAD -0.091; TCGA ESCA -0.147; TCGA HNSC -0.066; TCGA KIRC -0.066; TCGA KIRP -0.061; TCGA LUAD -0.093; TCGA LUSC -0.069; TCGA PRAD -0.155; TCGA SARC -0.116; TCGA THCA -0.114; TCGA STAD -0.101; TCGA UCEC -0.054 |
hsa-miR-193b-3p | KIAA0430 | 11 cancers: BLCA; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.161; TCGA COAD -0.059; TCGA ESCA -0.074; TCGA HNSC -0.069; TCGA KIRC -0.051; TCGA LUAD -0.056; TCGA LUSC -0.05; TCGA PRAD -0.121; TCGA SARC -0.088; TCGA STAD -0.188; TCGA UCEC -0.06 |
hsa-miR-193b-3p | INO80D | 9 cancers: BLCA; CESC; ESCA; HNSC; KIRP; LIHC; PRAD; SARC; STAD | MirTarget; miRNATAP | TCGA BLCA -0.077; TCGA CESC -0.099; TCGA ESCA -0.159; TCGA HNSC -0.063; TCGA KIRP -0.067; TCGA LIHC -0.076; TCGA PRAD -0.116; TCGA SARC -0.091; TCGA STAD -0.068 |
hsa-miR-193b-3p | SPATA13 | 9 cancers: BLCA; CESC; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; SARC | MirTarget | TCGA BLCA -0.237; TCGA CESC -0.29; TCGA ESCA -0.783; TCGA HNSC -0.601; TCGA KIRC -0.095; TCGA KIRP -0.13; TCGA LUAD -0.112; TCGA LUSC -0.3; TCGA SARC -0.198 |
hsa-miR-193b-3p | ING5 | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; OV; PAAD | MirTarget; mirMAP; miRNATAP | TCGA BLCA -0.053; TCGA BRCA -0.127; TCGA CESC -0.073; TCGA ESCA -0.103; TCGA HNSC -0.077; TCGA KIRC -0.064; TCGA KIRP -0.099; TCGA LGG -0.07; TCGA OV -0.096; TCGA PAAD -0.117 |
hsa-miR-193b-3p | JMY | 12 cancers: BLCA; CESC; COAD; HNSC; LGG; LUAD; OV; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.2; TCGA CESC -0.102; TCGA COAD -0.089; TCGA HNSC -0.126; TCGA LGG -0.064; TCGA LUAD -0.084; TCGA OV -0.095; TCGA PAAD -0.132; TCGA PRAD -0.11; TCGA SARC -0.17; TCGA STAD -0.235; TCGA UCEC -0.053 |
hsa-miR-193b-3p | CSRNP1 | 9 cancers: BLCA; BRCA; ESCA; KIRC; KIRP; LGG; LUAD; LUSC; UCEC | MirTarget | TCGA BLCA -0.239; TCGA BRCA -0.117; TCGA ESCA -0.255; TCGA KIRC -0.178; TCGA KIRP -0.091; TCGA LGG -0.09; TCGA LUAD -0.153; TCGA LUSC -0.373; TCGA UCEC -0.085 |
hsa-miR-193b-3p | KIT | 12 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRP; LIHC; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.69; TCGA BRCA -0.456; TCGA CESC -0.536; TCGA ESCA -0.42; TCGA HNSC -0.693; TCGA KIRP -0.412; TCGA LIHC -0.324; TCGA LUSC -0.548; TCGA PAAD -0.217; TCGA PRAD -0.176; TCGA STAD -0.495; TCGA UCEC -0.346 |
hsa-miR-193b-3p | BEND4 | 9 cancers: BLCA; COAD; ESCA; HNSC; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.455; TCGA COAD -0.407; TCGA ESCA -0.449; TCGA HNSC -0.607; TCGA LUSC -0.403; TCGA PAAD -0.41; TCGA PRAD -0.445; TCGA STAD -0.445; TCGA UCEC -0.197 |
hsa-miR-193b-3p | ZBTB40 | 12 cancers: BLCA; BRCA; ESCA; KIRC; KIRP; LGG; LUSC; OV; PAAD; PRAD; SARC; STAD | MirTarget | TCGA BLCA -0.098; TCGA BRCA -0.07; TCGA ESCA -0.079; TCGA KIRC -0.088; TCGA KIRP -0.072; TCGA LGG -0.094; TCGA LUSC -0.073; TCGA OV -0.082; TCGA PAAD -0.16; TCGA PRAD -0.116; TCGA SARC -0.136; TCGA STAD -0.105 |
hsa-miR-193b-3p | ADAMTSL3 | 13 cancers: BLCA; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.747; TCGA COAD -0.28; TCGA ESCA -0.448; TCGA HNSC -0.852; TCGA KIRC -0.263; TCGA KIRP -0.444; TCGA LUAD -0.197; TCGA LUSC -0.636; TCGA PAAD -0.309; TCGA PRAD -0.367; TCGA SARC -0.472; TCGA STAD -0.583; TCGA UCEC -0.48 |
hsa-miR-193b-3p | ZMAT3 | 9 cancers: BLCA; COAD; HNSC; LUAD; PAAD; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.186; TCGA COAD -0.174; TCGA HNSC -0.319; TCGA LUAD -0.088; TCGA PAAD -0.115; TCGA PRAD -0.11; TCGA THCA -0.31; TCGA STAD -0.203; TCGA UCEC -0.09 |
hsa-miR-193b-3p | RGMA | 10 cancers: BLCA; BRCA; CESC; HNSC; KIRP; LGG; LIHC; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.347; TCGA BRCA -0.382; TCGA CESC -0.236; TCGA HNSC -0.553; TCGA KIRP -0.255; TCGA LGG -0.18; TCGA LIHC -0.227; TCGA SARC -0.24; TCGA STAD -0.676; TCGA UCEC -0.276 |
hsa-miR-193b-3p | PER3 | 13 cancers: BLCA; CESC; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.19; TCGA CESC -0.149; TCGA HNSC -0.163; TCGA KIRC -0.236; TCGA KIRP -0.076; TCGA LGG -0.147; TCGA LUAD -0.18; TCGA LUSC -0.197; TCGA PRAD -0.159; TCGA SARC -0.111; TCGA THCA -0.128; TCGA STAD -0.225; TCGA UCEC -0.127 |
hsa-miR-193b-3p | CBX7 | 13 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.451; TCGA BRCA -0.314; TCGA CESC -0.253; TCGA ESCA -0.316; TCGA HNSC -0.504; TCGA KIRC -0.189; TCGA KIRP -0.13; TCGA LUAD -0.154; TCGA LUSC -0.265; TCGA PAAD -0.345; TCGA SARC -0.453; TCGA STAD -0.388; TCGA UCEC -0.255 |
hsa-miR-193b-3p | CTNND2 | 10 cancers: BLCA; CESC; COAD; ESCA; LGG; LIHC; LUAD; LUSC; PAAD; STAD | MirTarget | TCGA BLCA -0.471; TCGA CESC -0.263; TCGA COAD -0.258; TCGA ESCA -0.417; TCGA LGG -0.106; TCGA LIHC -1.327; TCGA LUAD -0.242; TCGA LUSC -0.617; TCGA PAAD -0.502; TCGA STAD -0.575 |
hsa-miR-193b-3p | FLI1 | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; STAD; UCEC | MirTarget | TCGA BLCA -0.144; TCGA BRCA -0.117; TCGA COAD -0.251; TCGA ESCA -0.172; TCGA HNSC -0.204; TCGA KIRC -0.209; TCGA LUAD -0.174; TCGA LUSC -0.446; TCGA STAD -0.214; TCGA UCEC -0.155 |
hsa-miR-193b-3p | TAPT1 | 14 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.151; TCGA CESC -0.116; TCGA COAD -0.063; TCGA ESCA -0.155; TCGA HNSC -0.15; TCGA KIRC -0.076; TCGA LGG -0.109; TCGA LUAD -0.143; TCGA LUSC -0.172; TCGA PAAD -0.128; TCGA PRAD -0.128; TCGA SARC -0.11; TCGA STAD -0.066; TCGA UCEC -0.095 |
hsa-miR-193b-3p | FAT4 | 12 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PRAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.5; TCGA BRCA -0.109; TCGA CESC -0.206; TCGA ESCA -0.59; TCGA HNSC -0.469; TCGA KIRC -0.215; TCGA KIRP -0.29; TCGA LUAD -0.29; TCGA LUSC -0.569; TCGA PRAD -0.154; TCGA STAD -0.383; TCGA UCEC -0.181 |
hsa-miR-193b-3p | PIKFYVE | 11 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LUSC; OV; PRAD; SARC; STAD | MirTarget | TCGA BLCA -0.116; TCGA CESC -0.106; TCGA COAD -0.069; TCGA ESCA -0.116; TCGA HNSC -0.164; TCGA KIRP -0.064; TCGA LUSC -0.084; TCGA OV -0.07; TCGA PRAD -0.217; TCGA SARC -0.102; TCGA STAD -0.129 |
hsa-miR-193b-3p | BHLHE41 | 12 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; PRAD; THCA; STAD | MirTarget | TCGA BLCA -0.181; TCGA BRCA -0.188; TCGA CESC -0.255; TCGA ESCA -0.261; TCGA HNSC -0.295; TCGA KIRC -0.161; TCGA LIHC -0.397; TCGA LUAD -0.139; TCGA LUSC -0.214; TCGA PRAD -0.375; TCGA THCA -0.304; TCGA STAD -0.198 |
hsa-miR-193b-3p | RPS6KA2 | 10 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LUAD; LUSC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.312; TCGA BRCA -0.091; TCGA ESCA -0.384; TCGA HNSC -0.202; TCGA KIRC -0.254; TCGA LUAD -0.127; TCGA LUSC -0.36; TCGA THCA -0.142; TCGA STAD -0.104; TCGA UCEC -0.086 |
hsa-miR-193b-3p | IGSF9B | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.717; TCGA BRCA -0.155; TCGA CESC -0.346; TCGA COAD -0.256; TCGA ESCA -0.848; TCGA HNSC -0.352; TCGA KIRC -0.333; TCGA KIRP -0.176; TCGA LGG -0.16; TCGA LUAD -0.243; TCGA PRAD -0.201; TCGA SARC -0.896; TCGA STAD -0.272; TCGA UCEC -0.388 |
hsa-miR-193b-3p | SEC14L1 | 9 cancers: BLCA; CESC; ESCA; HNSC; KIRC; KIRP; LUSC; PRAD; THCA | mirMAP | TCGA BLCA -0.058; TCGA CESC -0.07; TCGA ESCA -0.142; TCGA HNSC -0.095; TCGA KIRC -0.216; TCGA KIRP -0.068; TCGA LUSC -0.141; TCGA PRAD -0.112; TCGA THCA -0.063 |
hsa-miR-193b-3p | CABLES1 | 9 cancers: BLCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC | mirMAP | TCGA BLCA -0.136; TCGA CESC -0.213; TCGA ESCA -0.685; TCGA HNSC -0.215; TCGA KIRC -0.096; TCGA KIRP -0.085; TCGA LGG -0.211; TCGA LUAD -0.169; TCGA LUSC -0.456 |
hsa-miR-193b-3p | RHBDL3 | 10 cancers: BLCA; BRCA; CESC; HNSC; LGG; LUAD; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.389; TCGA BRCA -0.155; TCGA CESC -0.353; TCGA HNSC -0.571; TCGA LGG -0.252; TCGA LUAD -0.141; TCGA PAAD -0.305; TCGA PRAD -0.233; TCGA STAD -0.164; TCGA UCEC -0.259 |
hsa-miR-193b-3p | PRDM16 | 11 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LIHC; LUAD; LUSC; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.172; TCGA BRCA -0.133; TCGA CESC -0.293; TCGA ESCA -1.074; TCGA HNSC -0.225; TCGA LIHC -0.677; TCGA LUAD -0.381; TCGA LUSC -0.237; TCGA SARC -0.667; TCGA STAD -0.18; TCGA UCEC -0.36 |
hsa-miR-193b-3p | GRIK3 | 9 cancers: BLCA; CESC; COAD; HNSC; KIRC; LGG; LUAD; PAAD; STAD | mirMAP | TCGA BLCA -0.711; TCGA CESC -0.281; TCGA COAD -0.414; TCGA HNSC -0.621; TCGA KIRC -0.701; TCGA LGG -0.238; TCGA LUAD -0.158; TCGA PAAD -0.529; TCGA STAD -0.741 |
hsa-miR-193b-3p | MTHFR | 9 cancers: BLCA; BRCA; ESCA; KIRC; LGG; LUSC; PAAD; SARC; THCA | mirMAP | TCGA BLCA -0.118; TCGA BRCA -0.106; TCGA ESCA -0.334; TCGA KIRC -0.087; TCGA LGG -0.056; TCGA LUSC -0.151; TCGA PAAD -0.099; TCGA SARC -0.179; TCGA THCA -0.139 |
hsa-miR-193b-3p | CBX6 | 12 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; LIHC; PAAD; PRAD; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.246; TCGA BRCA -0.237; TCGA CESC -0.118; TCGA COAD -0.224; TCGA HNSC -0.378; TCGA KIRC -0.115; TCGA LIHC -0.379; TCGA PAAD -0.223; TCGA PRAD -0.246; TCGA SARC -0.175; TCGA THCA -0.142; TCGA STAD -0.185 |
hsa-miR-193b-3p | SLCO2A1 | 12 cancers: BLCA; BRCA; COAD; ESCA; KIRC; KIRP; LUAD; LUSC; PAAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.198; TCGA BRCA -0.079; TCGA COAD -0.191; TCGA ESCA -0.47; TCGA KIRC -0.414; TCGA KIRP -0.2; TCGA LUAD -0.193; TCGA LUSC -0.45; TCGA PAAD -0.232; TCGA SARC -0.205; TCGA STAD -0.265; TCGA UCEC -0.313 |
hsa-miR-193b-3p | RORA | 10 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; LUAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.168; TCGA BRCA -0.062; TCGA CESC -0.144; TCGA COAD -0.211; TCGA HNSC -0.262; TCGA KIRC -0.175; TCGA KIRP -0.183; TCGA LUAD -0.135; TCGA STAD -0.256; TCGA UCEC -0.161 |
hsa-miR-193b-3p | HELZ | 9 cancers: BLCA; COAD; HNSC; KIRP; LUAD; OV; PAAD; PRAD; STAD | miRNATAP | TCGA BLCA -0.075; TCGA COAD -0.053; TCGA HNSC -0.065; TCGA KIRP -0.107; TCGA LUAD -0.063; TCGA OV -0.079; TCGA PAAD -0.131; TCGA PRAD -0.142; TCGA STAD -0.108 |
hsa-miR-193b-3p | NOVA1 | 10 cancers: BLCA; CESC; HNSC; KIRP; LGG; OV; PAAD; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.814; TCGA CESC -0.538; TCGA HNSC -0.64; TCGA KIRP -0.095; TCGA LGG -0.21; TCGA OV -0.267; TCGA PAAD -0.68; TCGA PRAD -0.192; TCGA STAD -0.582; TCGA UCEC -0.498 |
hsa-miR-193b-3p | MKLN1 | 9 cancers: BLCA; ESCA; HNSC; KIRP; LGG; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.062; TCGA ESCA -0.145; TCGA HNSC -0.081; TCGA KIRP -0.07; TCGA LGG -0.115; TCGA PRAD -0.055; TCGA SARC -0.065; TCGA STAD -0.089; TCGA UCEC -0.06 |
hsa-miR-193b-3p | SOS2 | 11 cancers: BLCA; COAD; KIRC; LGG; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.087; TCGA COAD -0.077; TCGA KIRC -0.063; TCGA LGG -0.064; TCGA LUAD -0.055; TCGA LUSC -0.052; TCGA PAAD -0.1; TCGA PRAD -0.138; TCGA SARC -0.118; TCGA STAD -0.139; TCGA UCEC -0.068 |
hsa-miR-193b-3p | RUNX1T1 | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LUAD; LUSC; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.524; TCGA CESC -0.35; TCGA COAD -0.22; TCGA ESCA -0.242; TCGA HNSC -0.552; TCGA KIRC -0.293; TCGA LGG -0.201; TCGA LUAD -0.156; TCGA LUSC -0.521; TCGA SARC -0.178; TCGA STAD -0.481; TCGA UCEC -0.316 |
hsa-miR-193b-3p | KCNH8 | 12 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.399; TCGA BRCA -0.682; TCGA CESC -0.583; TCGA ESCA -0.796; TCGA HNSC -0.764; TCGA KIRC -0.273; TCGA KIRP -0.622; TCGA PAAD -0.298; TCGA PRAD -0.587; TCGA SARC -0.313; TCGA STAD -0.531; TCGA UCEC -0.481 |
hsa-miR-193b-3p | ARHGAP19 | 12 cancers: BRCA; COAD; HNSC; KIRC; KIRP; LGG; LIHC; LUSC; PAAD; PRAD; THCA; UCEC | miRNAWalker2 validate; mirMAP | TCGA BRCA -0.116; TCGA COAD -0.11; TCGA HNSC -0.164; TCGA KIRC -0.064; TCGA KIRP -0.104; TCGA LGG -0.061; TCGA LIHC -0.143; TCGA LUSC -0.06; TCGA PAAD -0.105; TCGA PRAD -0.084; TCGA THCA -0.221; TCGA UCEC -0.056 |
hsa-miR-193b-3p | IL17RD | 11 cancers: BRCA; HNSC; KIRC; KIRP; LGG; LIHC; OV; SARC; THCA; STAD; UCEC | miRNAWalker2 validate; MirTarget | TCGA BRCA -0.269; TCGA HNSC -0.369; TCGA KIRC -0.214; TCGA KIRP -0.152; TCGA LGG -0.15; TCGA LIHC -0.514; TCGA OV -0.154; TCGA SARC -0.186; TCGA THCA -0.318; TCGA STAD -0.297; TCGA UCEC -0.274 |
hsa-miR-193b-3p | ST6GALNAC5 | 9 cancers: BRCA; HNSC; KIRP; LIHC; LUAD; LUSC; PRAD; THCA; STAD | MirTarget | TCGA BRCA -0.284; TCGA HNSC -0.122; TCGA KIRP -0.469; TCGA LIHC -0.47; TCGA LUAD -0.157; TCGA LUSC -0.462; TCGA PRAD -0.366; TCGA THCA -0.892; TCGA STAD -0.245 |
hsa-miR-193b-3p | ING1 | 9 cancers: BRCA; CESC; HNSC; KIRC; KIRP; LGG; OV; PRAD; UCEC | MirTarget | TCGA BRCA -0.054; TCGA CESC -0.053; TCGA HNSC -0.053; TCGA KIRC -0.069; TCGA KIRP -0.079; TCGA LGG -0.099; TCGA OV -0.078; TCGA PRAD -0.132; TCGA UCEC -0.065 |
hsa-miR-193b-3p | NEDD9 | 13 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PRAD; SARC; UCEC | MirTarget | TCGA BRCA -0.124; TCGA CESC -0.168; TCGA COAD -0.146; TCGA ESCA -0.388; TCGA HNSC -0.221; TCGA KIRC -0.307; TCGA KIRP -0.108; TCGA LIHC -0.186; TCGA LUAD -0.199; TCGA LUSC -0.31; TCGA PRAD -0.152; TCGA SARC -0.177; TCGA UCEC -0.17 |
hsa-miR-193b-3p | MBP | 9 cancers: BRCA; COAD; ESCA; LUAD; PAAD; PRAD; SARC; THCA; STAD | mirMAP | TCGA BRCA -0.162; TCGA COAD -0.157; TCGA ESCA -0.105; TCGA LUAD -0.102; TCGA PAAD -0.233; TCGA PRAD -0.059; TCGA SARC -0.328; TCGA THCA -0.088; TCGA STAD -0.109 |
hsa-miR-193b-3p | PSIP1 | 10 cancers: CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LUSC; PAAD; PRAD | miRNAWalker2 validate | TCGA CESC -0.088; TCGA COAD -0.084; TCGA ESCA -0.1; TCGA HNSC -0.229; TCGA KIRC -0.107; TCGA KIRP -0.057; TCGA LGG -0.071; TCGA LUSC -0.089; TCGA PAAD -0.189; TCGA PRAD -0.069 |
hsa-miR-193b-3p | SP4 | 13 cancers: CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA CESC -0.096; TCGA COAD -0.19; TCGA ESCA -0.095; TCGA HNSC -0.248; TCGA KIRC -0.108; TCGA KIRP -0.14; TCGA LUAD -0.107; TCGA LUSC -0.073; TCGA OV -0.121; TCGA PAAD -0.152; TCGA PRAD -0.189; TCGA STAD -0.178; TCGA UCEC -0.103 |
hsa-miR-193b-3p | KCNJ2 | 10 cancers: CESC; COAD; HNSC; KIRC; KIRP; LIHC; LUAD; PAAD; PRAD; THCA | miRNATAP | TCGA CESC -0.241; TCGA COAD -0.144; TCGA HNSC -0.43; TCGA KIRC -0.179; TCGA KIRP -0.2; TCGA LIHC -0.15; TCGA LUAD -0.163; TCGA PAAD -0.292; TCGA PRAD -0.115; TCGA THCA -0.3 |
hsa-miR-193b-3p | ELK3 | 9 cancers: COAD; KIRC; LIHC; LUAD; LUSC; PRAD; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA COAD -0.129; TCGA KIRC -0.24; TCGA LIHC -0.151; TCGA LUAD -0.06; TCGA LUSC -0.094; TCGA PRAD -0.161; TCGA THCA -0.111; TCGA STAD -0.12; TCGA UCEC -0.128 |
hsa-miR-193b-3p | MIB1 | 10 cancers: COAD; HNSC; KIRP; LGG; LIHC; OV; PRAD; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA COAD -0.125; TCGA HNSC -0.07; TCGA KIRP -0.06; TCGA LGG -0.121; TCGA LIHC -0.123; TCGA OV -0.092; TCGA PRAD -0.099; TCGA THCA -0.12; TCGA STAD -0.089; TCGA UCEC -0.065 |
hsa-miR-193b-3p | TMEM30A | 9 cancers: COAD; HNSC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD | MirTarget; miRNATAP | TCGA COAD -0.084; TCGA HNSC -0.072; TCGA LUAD -0.094; TCGA LUSC -0.073; TCGA PAAD -0.057; TCGA PRAD -0.156; TCGA SARC -0.08; TCGA THCA -0.078; TCGA STAD -0.065 |
hsa-miR-193b-3p | GATAD2B | 9 cancers: HNSC; KIRC; LGG; LIHC; OV; PRAD; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA HNSC -0.099; TCGA KIRC -0.054; TCGA LGG -0.098; TCGA LIHC -0.082; TCGA OV -0.097; TCGA PRAD -0.06; TCGA THCA -0.052; TCGA STAD -0.062; TCGA UCEC -0.065 |
Enriched cancer pathways of putative targets