microRNA information: hsa-miR-194-3p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-194-3p | miRbase |
| Accession: | MIMAT0004671 | miRbase |
| Precursor name: | hsa-mir-194-1 | miRbase |
| Precursor accession: | MI0000488 | miRbase |
| Symbol: | MIR194-1 | HGNC |
| RefSeq ID: | NR_029711 | GenBank |
| Sequence: | CCAGUGGGGCUGCUGUUAUCUG |
Reported expression in cancers: hsa-miR-194-3p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-194-3p | bladder cancer | downregulation | "In the present study we investigated the roles and ......" | 27133066 | |
| hsa-miR-194-3p | colorectal cancer | downregulation | "MiR 194 commonly repressed in colorectal cancer su ......" | 25602366 | Microarray |
| hsa-miR-194-3p | gastric cancer | upregulation | "We performed miRNA microarray and quantitative rev ......" | 23385731 | Reverse transcription PCR; Microarray; qPCR |
| hsa-miR-194-3p | kidney renal cell cancer | downregulation | "miR 192 miR 194 and miR 215: a convergent microRNA ......" | 23715501 | |
| hsa-miR-194-3p | kidney renal cell cancer | downregulation | "miR-194 was reported to be downregulated in severa ......" | 26860079 | |
| hsa-miR-194-3p | liver cancer | upregulation | "Up-regulation of miR-15a miR-16 miR-27b miR-30b mi ......" | 24314246 | qPCR |
| hsa-miR-194-3p | lung squamous cell cancer | downregulation | "Earlier studies have demonstrated that miR-194 was ......" | 26909612 | |
| hsa-miR-194-3p | melanoma | upregulation | "miR-194 is a tumor-suppressor gene in multiple tum ......" | 27573550 | |
| hsa-miR-194-3p | ovarian cancer | upregulation | "In this study we explored the role of miR-194 in o ......" | 27486333 | qPCR |
| hsa-miR-194-3p | prostate cancer | upregulation | "A trend of increased expression >40% of miR-135b a ......" | 18949015 | qPCR |
Reported cancer pathway affected by hsa-miR-194-3p
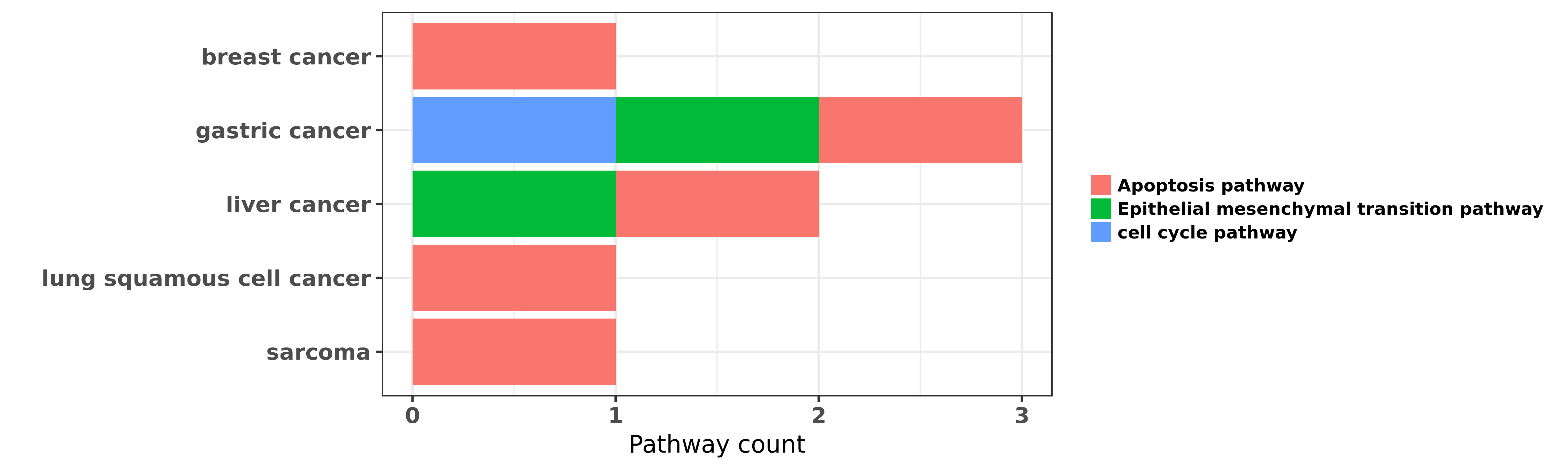
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-194-3p | breast cancer | Apoptosis pathway | "We have performed miRNA microarray profiling befor ......" | 22829924 | Luciferase |
| hsa-miR-194-3p | gastric cancer | cell cycle pathway | "Inverse association between miR 194 expression and ......" | 21845495 | Western blot |
| hsa-miR-194-3p | gastric cancer | Epithelial mesenchymal transition pathway | "We hypothesized that miR-194 may control Forkhead ......" | 24748184 | |
| hsa-miR-194-3p | gastric cancer | cell cycle pathway; Apoptosis pathway | "miR 194 targets RBX1 gene to modulate proliferatio ......" | 25412959 | Flow cytometry; Transwell assay; Wound Healing Assay; Western blot; Luciferase |
| hsa-miR-194-3p | liver cancer | Epithelial mesenchymal transition pathway | "miR 194 is a marker of hepatic epithelial cells an ......" | 20979124 | |
| hsa-miR-194-3p | liver cancer | Apoptosis pathway | "In this study we aimed to analyze the role of micr ......" | 26722431 | |
| hsa-miR-194-3p | lung squamous cell cancer | Apoptosis pathway | "miR 194 inhibits the proliferation invasion migrat ......" | 26909612 | |
| hsa-miR-194-3p | sarcoma | Apoptosis pathway | "Studies have shown that miR-194 functions as a tum ......" | 25096247 | Western blot; Luciferase |
Reported cancer prognosis affected by hsa-miR-194-3p
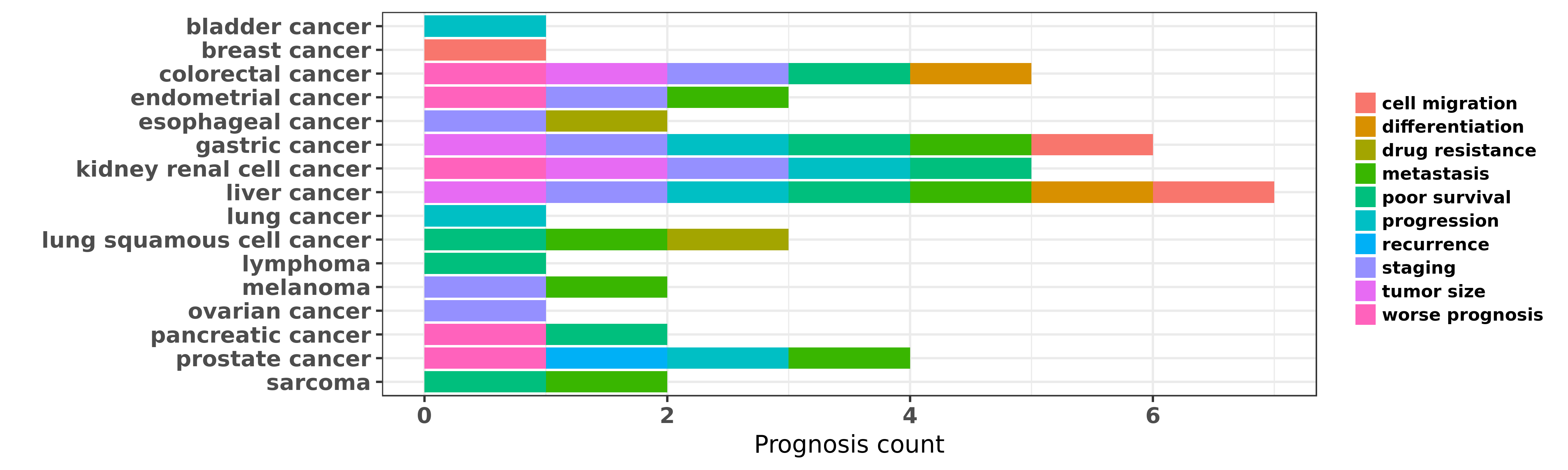
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-194-3p | bladder cancer | progression | "MiR 194 inhibits cell proliferation and invasion v ......" | 27133066 | |
| hsa-miR-194-3p | breast cancer | cell migration | "We have performed miRNA microarray profiling befor ......" | 22829924 | Luciferase |
| hsa-miR-194-3p | colorectal cancer | staging; differentiation; tumor size; poor survival | "MiR 194 commonly repressed in colorectal cancer su ......" | 25602366 | Luciferase |
| hsa-miR-194-3p | colorectal cancer | worse prognosis | "Circulating levels of the miRNAs miR 194 and miR 2 ......" | 26318304 | |
| hsa-miR-194-3p | colorectal cancer | poor survival | "The expression levels of miR-206 miR-219 miR-192 m ......" | 27666868 | |
| hsa-miR-194-3p | colorectal cancer | staging; poor survival | "Significant inverse correlation between the expres ......" | 27747302 | |
| hsa-miR-194-3p | endometrial cancer | metastasis | "Furthermore we discovered that the expression of B ......" | 21851624 | |
| hsa-miR-194-3p | endometrial cancer | staging; worse prognosis | "Prognostic significance of miR 194 in endometrial ......" | 24040515 | |
| hsa-miR-194-3p | esophageal cancer | drug resistance; staging | "Initially a microarray-based approach was performe ......" | 23649428 | |
| hsa-miR-194-3p | gastric cancer | staging; tumor size; progression | "Inverse association between miR 194 expression and ......" | 21845495 | Western blot |
| hsa-miR-194-3p | gastric cancer | staging | "We performed miRNA microarray and quantitative rev ......" | 23385731 | |
| hsa-miR-194-3p | gastric cancer | cell migration; progression | "We hypothesized that miR-194 may control Forkhead ......" | 24748184 | |
| hsa-miR-194-3p | gastric cancer | poor survival; metastasis; tumor size | "miR 194 targets RBX1 gene to modulate proliferatio ......" | 25412959 | Flow cytometry; Transwell assay; Wound Healing Assay; Western blot; Luciferase |
| hsa-miR-194-3p | kidney renal cell cancer | progression | "miR 192 miR 194 and miR 215: a convergent microRNA ......" | 23715501 | Luciferase |
| hsa-miR-194-3p | kidney renal cell cancer | staging; poor survival; tumor size; worse prognosis; progression | "miR-194 was reported to be downregulated in severa ......" | 26860079 | |
| hsa-miR-194-3p | liver cancer | metastasis; differentiation | "miR 194 is a marker of hepatic epithelial cells an ......" | 20979124 | |
| hsa-miR-194-3p | liver cancer | staging | "Gα12gep oncogene inhibits FOXO1 in hepatocellular ......" | 24631529 | |
| hsa-miR-194-3p | liver cancer | cell migration; metastasis | "NF κB signaling relieves negative regulation by m ......" | 26221053 | Luciferase |
| hsa-miR-194-3p | liver cancer | poor survival; staging; metastasis; tumor size; progression | "In this study we aimed to analyze the role of micr ......" | 26722431 | |
| hsa-miR-194-3p | lung cancer | progression | "We succeeded in establishing the MTA1 knockdown NS ......" | 22576802 | RNAi |
| hsa-miR-194-3p | lung squamous cell cancer | metastasis | "miR 194 suppresses metastasis of non small cell lu ......" | 23584484 | |
| hsa-miR-194-3p | lung squamous cell cancer | metastasis; drug resistance; poor survival | "miR 194 inhibits the proliferation invasion migrat ......" | 26909612 | |
| hsa-miR-194-3p | lymphoma | poor survival | "Functional studies indicated that CDKN1A/p21 and S ......" | 22102710 | |
| hsa-miR-194-3p | melanoma | metastasis; staging | "miR 194 is a negative regulator of GEF H1 pathway ......" | 27573550 | |
| hsa-miR-194-3p | ovarian cancer | staging | "The reactivation of p53 was detected by Western bl ......" | 22340095 | Western blot |
| hsa-miR-194-3p | pancreatic cancer | worse prognosis; poor survival | "The aim of this study was to investigate microRNAs ......" | 25906450 | |
| hsa-miR-194-3p | prostate cancer | recurrence; progression; metastasis; worse prognosis | "MicroRNAs in the serum of patients who had experie ......" | 23846169 | |
| hsa-miR-194-3p | sarcoma | metastasis; poor survival | "Studies have shown that miR-194 functions as a tum ......" | 25096247 | Western blot; Luciferase |
Reported gene related to hsa-miR-194-3p

| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-194-3p | lung squamous cell cancer | BMP1 | "miR 194 suppresses metastasis of non small cell lu ......" | 23584484 |
| hsa-miR-194-3p | prostate cancer | BMP1 | "Bone morphogenetic protein 1 BMP1 was shown to be ......" | 26820911 |
| hsa-miR-194-3p | gastric cancer | CDH2 | "The results of real-time PCR and western blot high ......" | 21845495 |
| hsa-miR-194-3p | liver cancer | CDH2 | "The overexpression of miR-194 in liver mesenchymal ......" | 20979124 |
| hsa-miR-194-3p | colorectal cancer | MAP4K4 | "In addition using dual-luciferase reporter gene as ......" | 25602366 |
| hsa-miR-194-3p | liver cancer | MAP4K4 | "Mitogen-activated protein kinase 4 MAP4K4 a potent ......" | 26722431 |
| hsa-miR-194-3p | bladder cancer | AKT2 | "Long noncoding RNA H19 contributes to gallbladder ......" | 26803515 |
| hsa-miR-194-3p | melanoma | ARHGEF2 | "miR 194 is a negative regulator of GEF H1 pathway ......" | 27573550 |
| hsa-miR-194-3p | liver cancer | C21ORF91 | "Transcripts encoding tripartite motif containing 2 ......" | 26221053 |
| hsa-miR-194-3p | endometrial cancer | CDH1 | "Ectopic expression of miR-194 in EC cells induced ......" | 21851624 |
| hsa-miR-194-3p | lymphoma | CDKN1A | "miR-20a/b and miR-194 target CDKN1A and SOCS2 in f ......" | 22102710 |
| hsa-miR-194-3p | lung squamous cell cancer | CDKN1B | "miR 194 suppresses metastasis of non small cell lu ......" | 23584484 |
| hsa-miR-194-3p | breast cancer | DNM2 | "Increased miR-194 expression markedly reduced leve ......" | 22829924 |
| hsa-miR-194-3p | breast cancer | ERBB2 | "Forced expression of miR-194 in breast cancer cell ......" | 22829924 |
| hsa-miR-194-3p | gastric cancer | F2 | "Though there was no significant difference between ......" | 21845495 |
| hsa-miR-194-3p | lung squamous cell cancer | FOXA1 | "miR 194 inhibits the proliferation invasion migrat ......" | 26909612 |
| hsa-miR-194-3p | gastric cancer | FOXM1 | "We hypothesized that miR-194 may control Forkhead ......" | 24748184 |
| hsa-miR-194-3p | liver cancer | FOXO1 | "Gα12gep oncogene inhibits FOXO1 in hepatocellular ......" | 24631529 |
| hsa-miR-194-3p | bladder cancer | H19 | "Long noncoding RNA H19 contributes to gallbladder ......" | 26803515 |
| hsa-miR-194-3p | liver cancer | MAP2K6 | "Mitogen-activated protein kinase 4 MAP4K4 a potent ......" | 26722431 |
| hsa-miR-194-3p | liver cancer | NFASC | "NF κB signaling relieves negative regulation by m ......" | 26221053 |
| hsa-miR-194-3p | lung squamous cell cancer | NUDC | "Human nuclear distribution C hNUDC was predicted t ......" | 27035759 |
| hsa-miR-194-3p | colorectal cancer | PER1 | "Significant inverse correlation between the expres ......" | 27747302 |
| hsa-miR-194-3p | ovarian cancer | PTPN12 | "Meanwhile bioinformatics tools were used to identi ......" | 27486333 |
| hsa-miR-194-3p | bladder cancer | RAP2B | "MiR 194 inhibits cell proliferation and invasion v ......" | 27133066 |
| hsa-miR-194-3p | gastric cancer | RBX1 | "miR 194 targets RBX1 gene to modulate proliferatio ......" | 25412959 |
| hsa-miR-194-3p | lymphoma | SOCS2 | "Functional studies indicated that CDKN1A/p21 and S ......" | 22102710 |
| hsa-miR-194-3p | colon cancer | THBS1 | "p53 responsive miR 194 inhibits thrombospondin 1 a ......" | 22028325 |
| hsa-miR-194-3p | colon cancer | TP53 | "p53 responsive miR 194 inhibits thrombospondin 1 a ......" | 22028325 |
| hsa-miR-194-3p | liver cancer | TRIM23 | "Transcripts encoding tripartite motif containing 2 ......" | 26221053 |
| hsa-miR-194-3p | endometrial cancer | VIM | "Ectopic expression of miR-194 in EC cells induced ......" | 21851624 |
Expression profile in cancer corhorts:
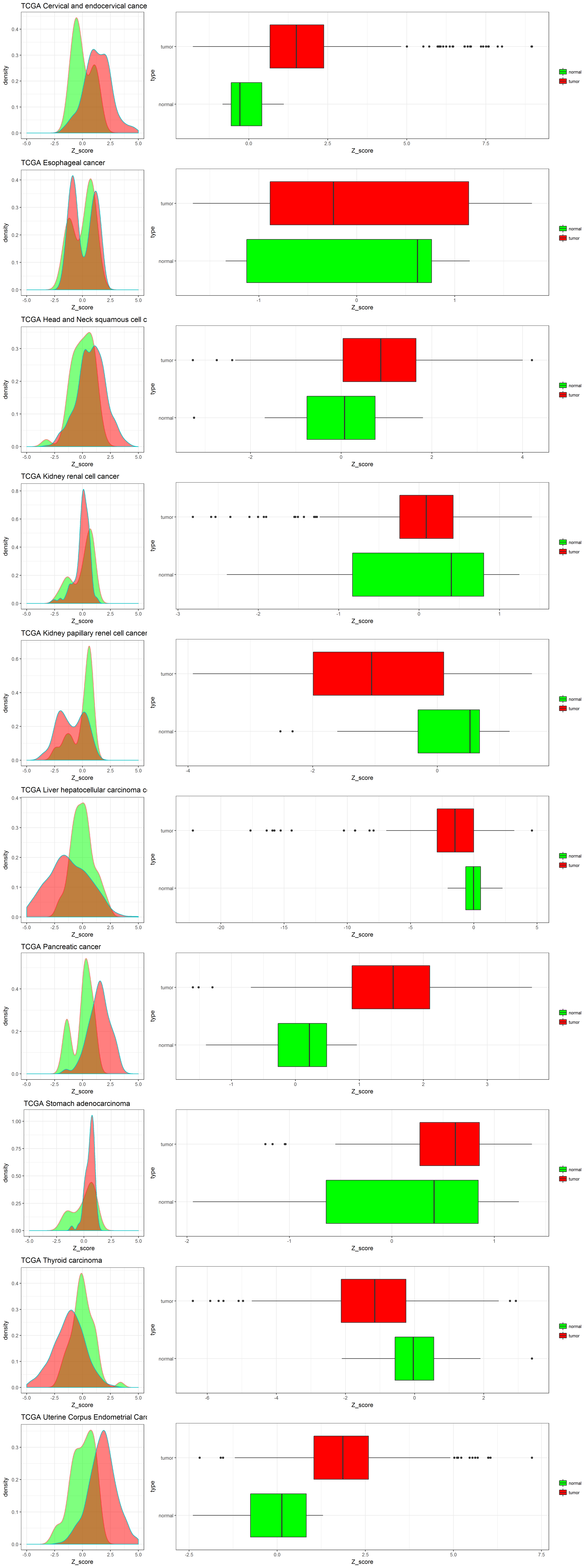
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-194-3p | NUMBL | 10 cancers: CESC; COAD; ESCA; HNSC; KIRP; LIHC; PAAD; THCA; STAD; UCEC | MirTarget | TCGA CESC -0.141; TCGA COAD -0.337; TCGA ESCA -0.129; TCGA HNSC -0.089; TCGA KIRP -0.108; TCGA LIHC -0.288; TCGA PAAD -0.2; TCGA THCA -0.071; TCGA STAD -0.187; TCGA UCEC -0.139 |
hsa-miR-194-3p | MXRA8 | 10 cancers: CESC; COAD; ESCA; HNSC; LIHC; OV; PAAD; THCA; STAD; UCEC | MirTarget | TCGA CESC -0.109; TCGA COAD -0.62; TCGA ESCA -0.106; TCGA HNSC -0.245; TCGA LIHC -0.561; TCGA OV -0.384; TCGA PAAD -0.336; TCGA THCA -0.341; TCGA STAD -0.203; TCGA UCEC -0.261 |
hsa-miR-194-3p | PLXDC1 | 10 cancers: CESC; COAD; HNSC; KIRP; LIHC; OV; PAAD; THCA; STAD; UCEC | MirTarget | TCGA CESC -0.1; TCGA COAD -0.312; TCGA HNSC -0.111; TCGA KIRP -0.111; TCGA LIHC -0.67; TCGA OV -0.173; TCGA PAAD -0.239; TCGA THCA -0.356; TCGA STAD -0.059; TCGA UCEC -0.202 |
hsa-miR-194-3p | ANTXR1 | 10 cancers: CESC; COAD; ESCA; HNSC; KIRP; LIHC; OV; PAAD; STAD; UCEC | MirTarget | TCGA CESC -0.121; TCGA COAD -0.64; TCGA ESCA -0.176; TCGA HNSC -0.145; TCGA KIRP -0.209; TCGA LIHC -0.475; TCGA OV -0.181; TCGA PAAD -0.225; TCGA STAD -0.188; TCGA UCEC -0.229 |
hsa-miR-194-3p | CAV1 | 9 cancers: CESC; COAD; ESCA; HNSC; KIRP; LIHC; PAAD; STAD; UCEC | MirTarget | TCGA CESC -0.237; TCGA COAD -0.487; TCGA ESCA -0.267; TCGA HNSC -0.138; TCGA KIRP -0.068; TCGA LIHC -0.285; TCGA PAAD -0.459; TCGA STAD -0.354; TCGA UCEC -0.288 |
hsa-miR-194-3p | COL8A2 | 12 cancers: CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; OV; PAAD; THCA; STAD; UCEC | MirTarget | TCGA CESC -0.239; TCGA COAD -0.726; TCGA ESCA -0.309; TCGA HNSC -0.133; TCGA KIRC -0.196; TCGA KIRP -0.115; TCGA LIHC -0.539; TCGA OV -0.228; TCGA PAAD -0.287; TCGA THCA -0.548; TCGA STAD -0.336; TCGA UCEC -0.18 |
hsa-miR-194-3p | NLGN4X | 10 cancers: CESC; COAD; ESCA; HNSC; KIRP; OV; PAAD; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA CESC -0.195; TCGA COAD -0.532; TCGA ESCA -0.25; TCGA HNSC -0.176; TCGA KIRP -0.204; TCGA OV -0.179; TCGA PAAD -0.338; TCGA THCA -0.213; TCGA STAD -0.228; TCGA UCEC -0.159 |
hsa-miR-194-3p | TNFAIP6 | 10 cancers: CESC; COAD; ESCA; HNSC; KIRP; LIHC; OV; PAAD; THCA; STAD | MirTarget | TCGA CESC -0.221; TCGA COAD -0.618; TCGA ESCA -0.207; TCGA HNSC -0.152; TCGA KIRP -0.346; TCGA LIHC -0.861; TCGA OV -0.194; TCGA PAAD -0.448; TCGA THCA -0.691; TCGA STAD -0.106 |
hsa-miR-194-3p | GAS7 | 10 cancers: CESC; COAD; ESCA; HNSC; LIHC; OV; PAAD; THCA; STAD; UCEC | mirMAP | TCGA CESC -0.138; TCGA COAD -0.546; TCGA ESCA -0.13; TCGA HNSC -0.183; TCGA LIHC -0.414; TCGA OV -0.177; TCGA PAAD -0.292; TCGA THCA -0.322; TCGA STAD -0.288; TCGA UCEC -0.237 |
hsa-miR-194-3p | GPC1 | 9 cancers: CESC; COAD; ESCA; KIRC; KIRP; LIHC; PAAD; THCA; STAD | mirMAP | TCGA CESC -0.274; TCGA COAD -0.35; TCGA ESCA -0.361; TCGA KIRC -0.13; TCGA KIRP -0.202; TCGA LIHC -0.238; TCGA PAAD -0.174; TCGA THCA -0.165; TCGA STAD -0.171 |
hsa-miR-194-3p | EVC | 9 cancers: CESC; COAD; ESCA; HNSC; LIHC; OV; PAAD; STAD; UCEC | mirMAP | TCGA CESC -0.197; TCGA COAD -0.651; TCGA ESCA -0.295; TCGA HNSC -0.138; TCGA LIHC -0.378; TCGA OV -0.097; TCGA PAAD -0.238; TCGA STAD -0.325; TCGA UCEC -0.128 |
hsa-miR-194-3p | KIAA1644 | 9 cancers: CESC; COAD; ESCA; HNSC; LIHC; OV; THCA; STAD; UCEC | mirMAP | TCGA CESC -0.126; TCGA COAD -0.723; TCGA ESCA -0.254; TCGA HNSC -0.223; TCGA LIHC -0.444; TCGA OV -0.314; TCGA THCA -0.317; TCGA STAD -0.408; TCGA UCEC -0.4 |
hsa-miR-194-3p | C1QTNF1 | 10 cancers: CESC; COAD; KIRC; KIRP; LIHC; OV; PAAD; THCA; STAD; UCEC | mirMAP | TCGA CESC -0.108; TCGA COAD -0.208; TCGA KIRC -0.085; TCGA KIRP -0.144; TCGA LIHC -0.357; TCGA OV -0.147; TCGA PAAD -0.135; TCGA THCA -0.288; TCGA STAD -0.117; TCGA UCEC -0.245 |
hsa-miR-194-3p | PELI3 | 9 cancers: CESC; COAD; ESCA; KIRP; LIHC; PAAD; THCA; STAD; UCEC | mirMAP | TCGA CESC -0.085; TCGA COAD -0.117; TCGA ESCA -0.125; TCGA KIRP -0.054; TCGA LIHC -0.093; TCGA PAAD -0.096; TCGA THCA -0.096; TCGA STAD -0.16; TCGA UCEC -0.072 |
hsa-miR-194-3p | SH3PXD2B | 10 cancers: CESC; COAD; ESCA; HNSC; LIHC; OV; PAAD; THCA; STAD; UCEC | mirMAP | TCGA CESC -0.109; TCGA COAD -0.227; TCGA ESCA -0.125; TCGA HNSC -0.11; TCGA LIHC -0.35; TCGA OV -0.125; TCGA PAAD -0.232; TCGA THCA -0.228; TCGA STAD -0.169; TCGA UCEC -0.105 |
hsa-miR-194-3p | DIRAS1 | 10 cancers: CESC; COAD; ESCA; HNSC; LIHC; OV; PAAD; THCA; STAD; UCEC | mirMAP | TCGA CESC -0.291; TCGA COAD -0.76; TCGA ESCA -0.353; TCGA HNSC -0.441; TCGA LIHC -0.665; TCGA OV -0.284; TCGA PAAD -0.249; TCGA THCA -0.821; TCGA STAD -0.461; TCGA UCEC -0.419 |
hsa-miR-194-3p | SRRM3 | 11 cancers: CESC; COAD; ESCA; KIRC; KIRP; LGG; LIHC; PAAD; THCA; STAD; UCEC | mirMAP | TCGA CESC -0.34; TCGA COAD -0.522; TCGA ESCA -0.331; TCGA KIRC -0.244; TCGA KIRP -0.167; TCGA LGG -0.169; TCGA LIHC -0.858; TCGA PAAD -0.205; TCGA THCA -0.163; TCGA STAD -0.274; TCGA UCEC -0.173 |
hsa-miR-194-3p | MXRA7 | 9 cancers: CESC; COAD; ESCA; KIRC; KIRP; LIHC; PAAD; STAD; UCEC | mirMAP | TCGA CESC -0.14; TCGA COAD -0.461; TCGA ESCA -0.196; TCGA KIRC -0.106; TCGA KIRP -0.09; TCGA LIHC -0.218; TCGA PAAD -0.193; TCGA STAD -0.371; TCGA UCEC -0.166 |
hsa-miR-194-3p | GAS2L1 | 9 cancers: CESC; COAD; ESCA; KIRC; KIRP; LGG; LIHC; PAAD; THCA | mirMAP | TCGA CESC -0.094; TCGA COAD -0.1; TCGA ESCA -0.149; TCGA KIRC -0.064; TCGA KIRP -0.061; TCGA LGG -0.096; TCGA LIHC -0.178; TCGA PAAD -0.15; TCGA THCA -0.129 |
hsa-miR-194-3p | ITGA5 | 9 cancers: CESC; COAD; ESCA; HNSC; LIHC; PAAD; THCA; STAD; UCEC | miRNATAP | TCGA CESC -0.168; TCGA COAD -0.566; TCGA ESCA -0.122; TCGA HNSC -0.108; TCGA LIHC -0.071; TCGA PAAD -0.326; TCGA THCA -0.173; TCGA STAD -0.212; TCGA UCEC -0.068 |
hsa-miR-194-3p | CSDC2 | 9 cancers: COAD; ESCA; HNSC; LGG; LIHC; OV; PAAD; STAD; UCEC | MirTarget | TCGA COAD -0.971; TCGA ESCA -0.093; TCGA HNSC -0.554; TCGA LGG -0.192; TCGA LIHC -0.186; TCGA OV -0.261; TCGA PAAD -0.279; TCGA STAD -0.334; TCGA UCEC -0.529 |
hsa-miR-194-3p | ZBTB47 | 9 cancers: COAD; HNSC; KIRC; KIRP; LGG; LIHC; PAAD; STAD; UCEC | MirTarget | TCGA COAD -0.316; TCGA HNSC -0.106; TCGA KIRC -0.147; TCGA KIRP -0.059; TCGA LGG -0.074; TCGA LIHC -0.233; TCGA PAAD -0.118; TCGA STAD -0.234; TCGA UCEC -0.103 |
hsa-miR-194-3p | NPTXR | 10 cancers: COAD; ESCA; KIRC; KIRP; LIHC; OV; PAAD; THCA; STAD; UCEC | mirMAP | TCGA COAD -0.771; TCGA ESCA -0.219; TCGA KIRC -0.2; TCGA KIRP -0.1; TCGA LIHC -0.424; TCGA OV -0.13; TCGA PAAD -0.298; TCGA THCA -0.363; TCGA STAD -0.419; TCGA UCEC -0.16 |
Enriched cancer pathways of putative targets