microRNA information: hsa-miR-195-5p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-195-5p | miRbase |
| Accession: | MIMAT0000461 | miRbase |
| Precursor name: | hsa-mir-195 | miRbase |
| Precursor accession: | MI0000489 | miRbase |
| Symbol: | MIR195 | HGNC |
| RefSeq ID: | NR_029712 | GenBank |
| Sequence: | UAGCAGCACAGAAAUAUUGGC |
Reported expression in cancers: hsa-miR-195-5p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-195-5p | bladder cancer | downregulation | "We identified a subset of 7 miRNAs miR-145 miR-30a ......" | 19378336 | qPCR |
| hsa-miR-195-5p | bladder cancer | downregulation | "microRNA 195 inhibits cell proliferation in bladde ......" | 26622447 | qPCR |
| hsa-miR-195-5p | breast cancer | downregulation | "Analysis of MiR 195 and MiR 497 expression regulat ......" | 21350001 | Northern blot; qPCR; RNA-Seq |
| hsa-miR-195-5p | breast cancer | downregulation | "Serum microRNA 195 is down regulated in breast can ......" | 25103018 | qPCR |
| hsa-miR-195-5p | cervical and endocervical cancer | downregulation | "The present study investigated the role of miR-195 ......" | 26622903 | |
| hsa-miR-195-5p | cervical and endocervical cancer | downregulation | "Here we found that miR-195 expression was down-reg ......" | 26631043 | |
| hsa-miR-195-5p | cervical and endocervical cancer | downregulation | "Quantitative real-time polymerase chain reaction w ......" | 27206216 | qPCR |
| hsa-miR-195-5p | colorectal cancer | downregulation | "In the present study we analyzed the roles of miR- ......" | 20727858 | |
| hsa-miR-195-5p | colorectal cancer | downregulation | "Downregulation of miR 195 correlates with lymph no ......" | 21390519 | qPCR |
| hsa-miR-195-5p | colorectal cancer | downregulation | "MiR-497 and miR-195 of the miR-15/16/195/424/497 f ......" | 22710713 | |
| hsa-miR-195-5p | endometrial cancer | deregulation | "A set of EEC-associated miRNAs in tissue and plasm ......" | 24491411 | RNA-Seq |
| hsa-miR-195-5p | esophageal cancer | downregulation | "In this study Agilent miRNA microarray was used to ......" | 24025765 | Microarray |
| hsa-miR-195-5p | gastric cancer | downregulation | "The BioMark™ 96.96 Dynamic Array Fluidigm Corpor ......" | 23212612 | qPCR |
| hsa-miR-195-5p | gastric cancer | downregulation | "The low expression of miR-195 and miR-378 was clos ......" | 23333942 | |
| hsa-miR-195-5p | gastric cancer | downregulation | "Expression level of microRNA 195 in the serum of p ......" | 27097947 | qPCR |
| hsa-miR-195-5p | gastric cancer | downregulation | "In our previous study we demonstrated that four mi ......" | 27529338 | Reverse transcription PCR; qPCR |
| hsa-miR-195-5p | glioblastoma | downregulation | "Previous studies have reported that microRNA-195 m ......" | 22217655 | |
| hsa-miR-195-5p | head and neck cancer | upregulation | "High expression of miR-142-3p miR-186-5p miR-195-5 ......" | 26057452 | |
| hsa-miR-195-5p | liver cancer | downregulation | "Down-regulation of miR-195 has been observed in va ......" | 19441017 | |
| hsa-miR-195-5p | liver cancer | downregulation | "There is frequent down-regulation of miR-195 expre ......" | 23468064 | |
| hsa-miR-195-5p | liver cancer | downregulation | "In this study we experimentally demonstrated throu ......" | 23487264 | |
| hsa-miR-195-5p | liver cancer | downregulation | "Among them two clustered miRNAs miR-195 and miR-49 ......" | 23544130 | RNA-Seq |
| hsa-miR-195-5p | liver cancer | downregulation | "The result of Real-time PCR reveals the expression ......" | 26823724 | qPCR |
| hsa-miR-195-5p | liver cancer | downregulation | "MiR-195 expression is frequently reduced in variou ......" | 27179445 | |
| hsa-miR-195-5p | lung squamous cell cancer | upregulation | "MiR-195 suppresses tumor growth and is associated ......" | 25840419 | |
| hsa-miR-195-5p | prostate cancer | downregulation | "miR-195 is downregulated in many human cancers. Ho ......" | 26650737 | |
| hsa-miR-195-5p | sarcoma | upregulation | "miR-195 one of the miR-16/15/195/424/497 family me ......" | 23162665 | |
| hsa-miR-195-5p | thyroid cancer | downregulation | "We also find that the expression of miR-195 is dow ......" | 26527888 |
Reported cancer pathway affected by hsa-miR-195-5p
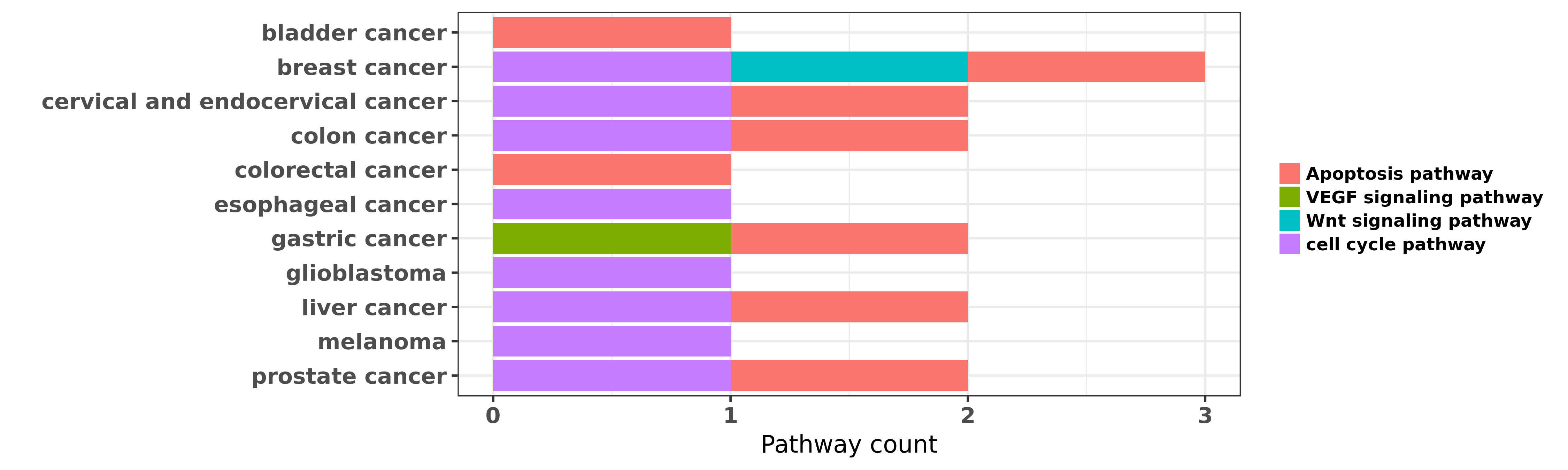
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-195-5p | bladder cancer | Apoptosis pathway | "MicroRNA 195 5p suppresses glucose uptake and prol ......" | 22265971 | |
| hsa-miR-195-5p | breast cancer | Apoptosis pathway | "Upregulation of miR 195 increases the sensitivity ......" | 23760062 | |
| hsa-miR-195-5p | breast cancer | cell cycle pathway | "MicroRNA 195 5p is a potential diagnostic and ther ......" | 24402230 | Colony formation; Western blot; Luciferase |
| hsa-miR-195-5p | breast cancer | Apoptosis pathway | "Upregulation of miR 195 enhances the radiosensitiv ......" | 26309570 | Colony formation; Western blot |
| hsa-miR-195-5p | breast cancer | Wnt signaling pathway | "Eribulin upregulates miR 195 expression and downre ......" | 26573286 | |
| hsa-miR-195-5p | breast cancer | Apoptosis pathway | "MicroRNA 195 inhibits proliferation invasion and m ......" | 26632252 | |
| hsa-miR-195-5p | cervical and endocervical cancer | cell cycle pathway | "MicroRNA 195 inhibits proliferation of cervical ca ......" | 26511972 | Western blot; Luciferase |
| hsa-miR-195-5p | cervical and endocervical cancer | Apoptosis pathway | "MiR 195 inhibits the proliferation of human cervic ......" | 26631043 | Luciferase |
| hsa-miR-195-5p | colon cancer | Apoptosis pathway | "MicroRNA 195 chemosensitizes colon cancer cells to ......" | 23526568 | |
| hsa-miR-195-5p | colon cancer | cell cycle pathway | "Tumor suppressive microRNA 195 5p regulates cell g ......" | 27347317 | MTT assay; Wound Healing Assay; Western blot; Luciferase |
| hsa-miR-195-5p | colorectal cancer | Apoptosis pathway | "microRNA 195 promotes apoptosis and suppresses tum ......" | 20727858 | |
| hsa-miR-195-5p | esophageal cancer | cell cycle pathway | "Differential expression of miR 195 in esophageal s ......" | 24025765 | |
| hsa-miR-195-5p | gastric cancer | VEGF signaling pathway | "MicroRNA 195 and microRNA 378 mediate tumor growth ......" | 23333942 | |
| hsa-miR-195-5p | gastric cancer | Apoptosis pathway | "In our previous study we demonstrated that four mi ......" | 27529338 | Colony formation |
| hsa-miR-195-5p | glioblastoma | cell cycle pathway | "MicroRNA 195 plays a tumor suppressor role in huma ......" | 22217655 | |
| hsa-miR-195-5p | liver cancer | cell cycle pathway | "MicroRNA 195 suppresses tumorigenicity and regulat ......" | 19441017 | |
| hsa-miR-195-5p | liver cancer | Apoptosis pathway | "The role of microRNA-195 in developing acquired dr ......" | 21947305 | Luciferase |
| hsa-miR-195-5p | liver cancer | Apoptosis pathway | "MiR 195 regulates cell apoptosis of human hepatoce ......" | 22888524 | Western blot; Flow cytometry; Luciferase |
| hsa-miR-195-5p | liver cancer | cell cycle pathway | "Among them two clustered miRNAs miR-195 and miR-49 ......" | 23544130 | |
| hsa-miR-195-5p | liver cancer | cell cycle pathway; Apoptosis pathway | "MicroRNA 195 acts as a tumor suppressor by directl ......" | 25174704 | Luciferase; Western blot |
| hsa-miR-195-5p | melanoma | cell cycle pathway | "Regulation of cell cycle checkpoint kinase WEE1 by ......" | 22847610 | |
| hsa-miR-195-5p | prostate cancer | Apoptosis pathway | "miR 195 Inhibits Tumor Progression by Targeting RP ......" | 26080838 | Luciferase |
| hsa-miR-195-5p | prostate cancer | cell cycle pathway | "Downregulation of miR 195 promotes prostate cancer ......" | 27175617 |
Reported cancer prognosis affected by hsa-miR-195-5p
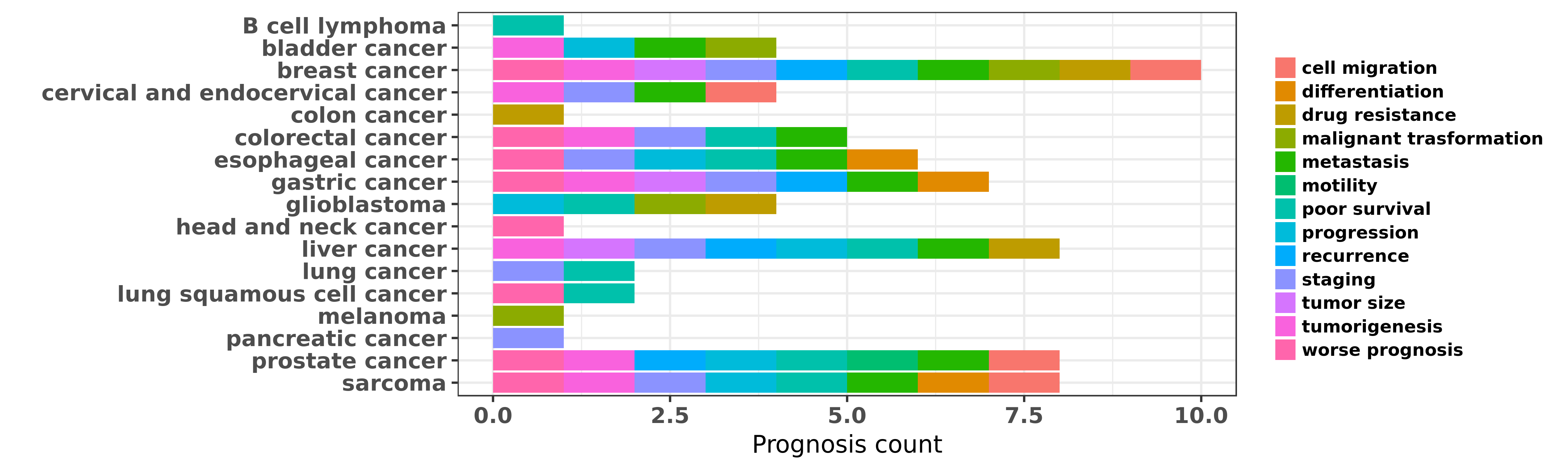
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-195-5p | B cell lymphoma | poor survival | "Finally eight miRNAs were found to correlate with ......" | 18537969 | |
| hsa-miR-195-5p | bladder cancer | tumorigenesis | "MicroRNA 195 5p suppresses glucose uptake and prol ......" | 22265971 | |
| hsa-miR-195-5p | bladder cancer | metastasis; progression | "The microRNA expression signature of bladder cance ......" | 24520312 | Western blot; Luciferase |
| hsa-miR-195-5p | bladder cancer | malignant trasformation | "microRNA 195 inhibits cell proliferation in bladde ......" | 26622447 | Western blot; Luciferase; MTT assay |
| hsa-miR-195-5p | breast cancer | worse prognosis; metastasis; tumor size; staging; recurrence; poor survival | "Correlation of miR 195 with invasiveness and progn ......" | 22800791 | |
| hsa-miR-195-5p | breast cancer | poor survival | "Upregulation of miR 195 increases the sensitivity ......" | 23760062 | |
| hsa-miR-195-5p | breast cancer | malignant trasformation | "In support of these findings in vitro functional s ......" | 24104550 | |
| hsa-miR-195-5p | breast cancer | tumorigenesis; cell migration | "MicroRNA 195 5p is a potential diagnostic and ther ......" | 24402230 | Colony formation; Western blot; Luciferase |
| hsa-miR-195-5p | breast cancer | tumor size | "The aim of the present study was to assess the eff ......" | 25009660 | |
| hsa-miR-195-5p | breast cancer | tumorigenesis; staging; metastasis; drug resistance | "Serum microRNA 195 is down regulated in breast can ......" | 25103018 | |
| hsa-miR-195-5p | breast cancer | metastasis | "Using real-time quantitative polymerase chain reac ......" | 26124926 | |
| hsa-miR-195-5p | breast cancer | drug resistance | "New miRNA-based drugs are also promising therapy f ......" | 26199650 | |
| hsa-miR-195-5p | breast cancer | staging | "Results showed differential pattern of expressions ......" | 26276721 | |
| hsa-miR-195-5p | breast cancer | poor survival; drug resistance | "Upregulation of miR 195 enhances the radiosensitiv ......" | 26309570 | Colony formation; Western blot |
| hsa-miR-195-5p | breast cancer | metastasis | "MicroRNA 195 inhibits proliferation invasion and m ......" | 26632252 | |
| hsa-miR-195-5p | breast cancer | staging | "We report for the first time an extremely high pre ......" | 27404381 | |
| hsa-miR-195-5p | cervical and endocervical cancer | tumorigenesis | "Serum miRNAs panel miR 16 2* miR 195 miR 2861 miR ......" | 26656154 | |
| hsa-miR-195-5p | cervical and endocervical cancer | staging; metastasis; cell migration | "MiR 195 Suppresses Cervical Cancer Migration and I ......" | 27206216 | Luciferase |
| hsa-miR-195-5p | colon cancer | drug resistance | "MicroRNA 195 chemosensitizes colon cancer cells to ......" | 23526568 | |
| hsa-miR-195-5p | colorectal cancer | metastasis; worse prognosis; tumorigenesis; poor survival; staging | "Downregulation of miR 195 correlates with lymph no ......" | 21390519 | |
| hsa-miR-195-5p | colorectal cancer | staging | "A meta-analysis was performed to review three miRN ......" | 25027346 | |
| hsa-miR-195-5p | colorectal cancer | metastasis | "Study on the molecular regulatory mechanism of Mic ......" | 26064276 | Western blot |
| hsa-miR-195-5p | colorectal cancer | staging | "In addition the qRT-PCR validation demonstrated th ......" | 26462034 | |
| hsa-miR-195-5p | colorectal cancer | poor survival | "In this study ten candidates were identified using ......" | 27126129 | |
| hsa-miR-195-5p | colorectal cancer | poor survival | "IGF2BP2 promotes colorectal cancer cell proliferat ......" | 27153315 | |
| hsa-miR-195-5p | esophageal cancer | worse prognosis; staging; metastasis; differentiation; progression; poor survival | "microRNA 195 Cdc42 axis acts as a prognostic facto ......" | 25400770 | |
| hsa-miR-195-5p | gastric cancer | metastasis; differentiation; tumor size | "The expressions of 60 candidate miRNAs in 30 gastr ......" | 21987613 | |
| hsa-miR-195-5p | gastric cancer | recurrence; worse prognosis | "Initial profiling using miR microarrays was perfor ......" | 22046085 | |
| hsa-miR-195-5p | gastric cancer | tumorigenesis | "The BioMark™ 96.96 Dynamic Array Fluidigm Corpor ......" | 23212612 | |
| hsa-miR-195-5p | gastric cancer | staging; metastasis; differentiation; worse prognosis | "Expression level of microRNA 195 in the serum of p ......" | 27097947 | |
| hsa-miR-195-5p | glioblastoma | drug resistance | "miR 195 miR 455 3p and miR 10a * are implicated in ......" | 20444541 | |
| hsa-miR-195-5p | glioblastoma | poor survival | "MiR 195 miR 196b miR 181c miR 21 expression levels ......" | 21895872 | |
| hsa-miR-195-5p | glioblastoma | progression; malignant trasformation | "MicroRNA 195 plays a tumor suppressor role in huma ......" | 22217655 | |
| hsa-miR-195-5p | head and neck cancer | worse prognosis | "High expression of miR-142-3p miR-186-5p miR-195-5 ......" | 26057452 | |
| hsa-miR-195-5p | liver cancer | progression | "MicroRNA 195 suppresses tumorigenicity and regulat ......" | 19441017 | |
| hsa-miR-195-5p | liver cancer | drug resistance | "The role of microRNA-195 in developing acquired dr ......" | 21947305 | Luciferase |
| hsa-miR-195-5p | liver cancer | staging | "MiR 195 regulates cell apoptosis of human hepatoce ......" | 22888524 | Western blot; Flow cytometry; Luciferase |
| hsa-miR-195-5p | liver cancer | metastasis; poor survival; recurrence | "MicroRNA 195 suppresses angiogenesis and metastasi ......" | 23468064 | Transwell assay |
| hsa-miR-195-5p | liver cancer | tumor size; metastasis; progression | "Genome wide screening reveals that miR 195 targets ......" | 23487264 | Luciferase |
| hsa-miR-195-5p | liver cancer | progression; tumorigenesis | "Among them two clustered miRNAs miR-195 and miR-49 ......" | 23544130 | |
| hsa-miR-195-5p | liver cancer | metastasis | "MicroRNA 195 functions as a tumor suppressor by in ......" | 25607636 | Western blot; Luciferase |
| hsa-miR-195-5p | liver cancer | metastasis; staging; poor survival; tumor size | "MiR 195 is a key negative regulator of hepatocellu ......" | 26823724 | Luciferase |
| hsa-miR-195-5p | lung cancer | staging; poor survival | "Using real-time RT-PCR we analyzed the expression ......" | 24282590 | |
| hsa-miR-195-5p | lung squamous cell cancer | poor survival; worse prognosis | "MiR 195 suppresses non small cell lung cancer by t ......" | 25840419 | |
| hsa-miR-195-5p | melanoma | malignant trasformation | "Regulation of cell cycle checkpoint kinase WEE1 by ......" | 22847610 | |
| hsa-miR-195-5p | pancreatic cancer | staging | "In addition from 10 to 50 weeks of age stage-speci ......" | 26516699 | |
| hsa-miR-195-5p | prostate cancer | progression; worse prognosis; metastasis; recurrence; poor survival | "miR 195 Inhibits Tumor Progression by Targeting RP ......" | 26080838 | Luciferase |
| hsa-miR-195-5p | prostate cancer | progression; motility | "MicroRNA 195 5p a new regulator of Fra 1 suppresse ......" | 26337460 | Transwell assay; Wound Healing Assay; Western blot; Luciferase |
| hsa-miR-195-5p | prostate cancer | metastasis; poor survival; recurrence | "MicroRNA 195 suppresses tumor cell proliferation a ......" | 26338045 | |
| hsa-miR-195-5p | prostate cancer | metastasis; poor survival; cell migration | "miR 195 Inhibits EMT by Targeting FGF2 in Prostate ......" | 26650737 | Western blot; Luciferase |
| hsa-miR-195-5p | prostate cancer | progression; tumorigenesis; worse prognosis | "Downregulation of miR 195 promotes prostate cancer ......" | 27175617 | |
| hsa-miR-195-5p | sarcoma | differentiation | "In order to investigate the involvement of miRNAs ......" | 23133552 | |
| hsa-miR-195-5p | sarcoma | tumorigenesis; cell migration | "microRNA 195 suppresses osteosarcoma cell invasion ......" | 23162665 | Western blot; Luciferase |
| hsa-miR-195-5p | sarcoma | progression; worse prognosis; poor survival; staging; metastasis | "Serum miR 195 is a diagnostic and prognostic marke ......" | 25498513 | |
| hsa-miR-195-5p | sarcoma | metastasis | "Second evaluation of miRNA concentration in indivi ......" | 25775010 | |
| hsa-miR-195-5p | sarcoma | metastasis; poor survival | "MicroRNA profiling identifies MiR 195 suppresses o ......" | 25823925 |
Reported gene related to hsa-miR-195-5p
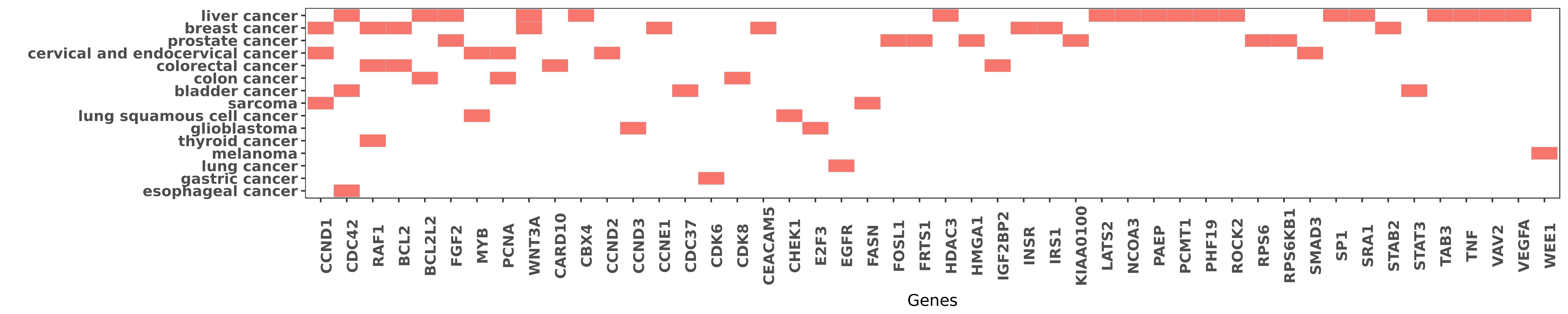
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-195-5p | breast cancer | BCL2 | "Previously we demonstrated that hsa-miR-195 target ......" | 26632252 |
| hsa-miR-195-5p | breast cancer | BCL2 | "Moreover knockdown of Raf-1 expression had similar ......" | 23760062 |
| hsa-miR-195-5p | breast cancer | BCL2 | "Upregulation of miR 195 enhances the radiosensitiv ......" | 26309570 |
| hsa-miR-195-5p | colorectal cancer | BCL2 | "Moreover important antiapoptotic Bcl-2 was identif ......" | 20727858 |
| hsa-miR-195-5p | bladder cancer | CDC42 | "A luciferase reporter assay was then conducted whi ......" | 26622447 |
| hsa-miR-195-5p | esophageal cancer | CDC42 | "microRNA 195 Cdc42 axis acts as a prognostic facto ......" | 25400770 |
| hsa-miR-195-5p | esophageal cancer | CDC42 | "Differential expression of miR 195 in esophageal s ......" | 24025765 |
| hsa-miR-195-5p | liver cancer | CDC42 | "Additionally miR-195 down-regulation led to increa ......" | 23468064 |
| hsa-miR-195-5p | breast cancer | RAF1 | "Raf-1 and Ccnd1 were identified as novel direct ta ......" | 21350001 |
| hsa-miR-195-5p | breast cancer | RAF1 | "Upregulation of miR 195 increases the sensitivity ......" | 23760062 |
| hsa-miR-195-5p | colorectal cancer | RAF1 | "IGF2BP2 promotes colorectal cancer cell proliferat ......" | 27153315 |
| hsa-miR-195-5p | thyroid cancer | RAF1 | "miR 195 is a key regulator of Raf1 in thyroid canc ......" | 26527888 |
| hsa-miR-195-5p | breast cancer | CCND1 | "Raf-1 and Ccnd1 were identified as novel direct ta ......" | 21350001 |
| hsa-miR-195-5p | cervical and endocervical cancer | CCND1 | "MiR 195 inhibits the proliferation of human cervic ......" | 26631043 |
| hsa-miR-195-5p | sarcoma | CCND1 | "MicroRNA profiling identifies MiR 195 suppresses o ......" | 25823925 |
| hsa-miR-195-5p | cervical and endocervical cancer | PCNA | "MicroRNA 195 inhibits proliferation of cervical ca ......" | 26511972 |
| hsa-miR-195-5p | cervical and endocervical cancer | PCNA | "MiR 195 inhibits the proliferation of human cervic ......" | 26631043 |
| hsa-miR-195-5p | colon cancer | PCNA | "Tumor suppressive microRNA 195 5p regulates cell g ......" | 27347317 |
| hsa-miR-195-5p | liver cancer | VEGFA | "In this study we used a new cationic peptide disul ......" | 27574422 |
| hsa-miR-195-5p | liver cancer | VEGFA | "Furthermore we revealed that miR-195 down-regulati ......" | 23468064 |
| hsa-miR-195-5p | liver cancer | VEGFA | "MiR 195 is a key negative regulator of hepatocellu ......" | 26823724 |
| hsa-miR-195-5p | colon cancer | BCL2L2 | "MicroRNA 195 chemosensitizes colon cancer cells to ......" | 23526568 |
| hsa-miR-195-5p | liver cancer | BCL2L2 | "These results indicate that miR-195 could improve ......" | 21947305 |
| hsa-miR-195-5p | liver cancer | FGF2 | "MiR 195 is a key negative regulator of hepatocellu ......" | 26823724 |
| hsa-miR-195-5p | prostate cancer | FGF2 | "miR 195 Inhibits EMT by Targeting FGF2 in Prostate ......" | 26650737 |
| hsa-miR-195-5p | cervical and endocervical cancer | MYB | "microRNA 195 inhibits the proliferation migration ......" | 26622903 |
| hsa-miR-195-5p | lung squamous cell cancer | MYB | "MicroRNA 195 inhibits non small cell lung cancer c ......" | 24486218 |
| hsa-miR-195-5p | breast cancer | WNT3A | "Eribulin upregulates miR 195 expression and downre ......" | 26573286 |
| hsa-miR-195-5p | liver cancer | WNT3A | "MicroRNA 195 acts as a tumor suppressor by directl ......" | 25174704 |
| hsa-miR-195-5p | colorectal cancer | CARD10 | "MicroRNA 195 inhibits colorectal cancer cell proli ......" | 24787958 |
| hsa-miR-195-5p | liver cancer | CBX4 | "MicroRNA 195 functions as a tumor suppressor by in ......" | 25607636 |
| hsa-miR-195-5p | cervical and endocervical cancer | CCND2 | "microRNA 195 inhibits the proliferation migration ......" | 26622903 |
| hsa-miR-195-5p | glioblastoma | CCND3 | "We identified E2F3 and CCND3 as functional downstr ......" | 22217655 |
| hsa-miR-195-5p | breast cancer | CCNE1 | "Furthermore through qPCR and western blot assays w ......" | 24402230 |
| hsa-miR-195-5p | bladder cancer | CDC37 | "The aim of the present study was to investigate th ......" | 26622447 |
| hsa-miR-195-5p | gastric cancer | CDK6 | "Expression of cyclin-dependent kinase 6 and vascul ......" | 23333942 |
| hsa-miR-195-5p | colon cancer | CDK8 | "Dual-luciferase reporter assays showed that miR-19 ......" | 27347317 |
| hsa-miR-195-5p | breast cancer | CEACAM5 | "The sensitivity and specificity of miR-195 in the ......" | 25103018 |
| hsa-miR-195-5p | lung squamous cell cancer | CHEK1 | "MiR 195 suppresses non small cell lung cancer by t ......" | 25840419 |
| hsa-miR-195-5p | glioblastoma | E2F3 | "We identified E2F3 and CCND3 as functional downstr ......" | 22217655 |
| hsa-miR-195-5p | lung cancer | EGFR | "Plasma levels of miR-195 and miR-122 expression we ......" | 24282590 |
| hsa-miR-195-5p | sarcoma | FASN | "microRNA 195 suppresses osteosarcoma cell invasion ......" | 23162665 |
| hsa-miR-195-5p | prostate cancer | FOSL1 | "MicroRNA 195 5p a new regulator of Fra 1 suppresse ......" | 26337460 |
| hsa-miR-195-5p | prostate cancer | FRTS1 | "Low miR-195 expression was characterised with a sh ......" | 26650737 |
| hsa-miR-195-5p | liver cancer | HDAC3 | "Expression of microRNA 195 is transactivated by Sp ......" | 27179445 |
| hsa-miR-195-5p | prostate cancer | HMGA1 | "Downregulation of miR 195 promotes prostate cancer ......" | 27175617 |
| hsa-miR-195-5p | colorectal cancer | IGF2BP2 | "IGF2BP2 promotes colorectal cancer cell proliferat ......" | 27153315 |
| hsa-miR-195-5p | breast cancer | INSR | "We show that miR-195 is inversely related with Ins ......" | 27133044 |
| hsa-miR-195-5p | breast cancer | IRS1 | "miR 195 inhibits tumor growth and angiogenesis thr ......" | 27133044 |
| hsa-miR-195-5p | prostate cancer | KIAA0100 | "MicroRNA 195 suppresses tumor cell proliferation a ......" | 26338045 |
| hsa-miR-195-5p | liver cancer | LATS2 | "MiR 195 regulates cell apoptosis of human hepatoce ......" | 22888524 |
| hsa-miR-195-5p | liver cancer | NCOA3 | "In the current study by way of informatics softwar ......" | 24740565 |
| hsa-miR-195-5p | liver cancer | PAEP | "ST21-H3R5-polyethylene glycol PEG exhibited excell ......" | 27574422 |
| hsa-miR-195-5p | liver cancer | PCMT1 | "Hsa miR 195 targets PCMT1 in hepatocellular carcin ......" | 25119594 |
| hsa-miR-195-5p | liver cancer | PHF19 | "MicroRNA 195 5p acts as an anti oncogene by target ......" | 25955388 |
| hsa-miR-195-5p | liver cancer | ROCK2 | "In this study we used a new cationic peptide disul ......" | 27574422 |
| hsa-miR-195-5p | prostate cancer | RPS6 | "The roles of miR-195 and its candidate target gene ......" | 26080838 |
| hsa-miR-195-5p | prostate cancer | RPS6KB1 | "miR 195 Inhibits Tumor Progression by Targeting RP ......" | 26080838 |
| hsa-miR-195-5p | cervical and endocervical cancer | SMAD3 | "MiR 195 Suppresses Cervical Cancer Migration and I ......" | 27206216 |
| hsa-miR-195-5p | liver cancer | SP1 | "Expression of microRNA 195 is transactivated by Sp ......" | 27179445 |
| hsa-miR-195-5p | liver cancer | SRA1 | "MicroRNA 195 regulates steroid receptor coactivato ......" | 24740565 |
| hsa-miR-195-5p | breast cancer | STAB2 | "We found that persistently elevated postoperative ......" | 25503185 |
| hsa-miR-195-5p | bladder cancer | STAT3 | "The aim of the present study was to investigate th ......" | 26622447 |
| hsa-miR-195-5p | liver cancer | TAB3 | "Genome wide screening reveals that miR 195 targets ......" | 23487264 |
| hsa-miR-195-5p | liver cancer | TNF | "Genome wide screening reveals that miR 195 targets ......" | 23487264 |
| hsa-miR-195-5p | liver cancer | VAV2 | "Additionally miR-195 down-regulation led to increa ......" | 23468064 |
| hsa-miR-195-5p | melanoma | WEE1 | "Regulation of cell cycle checkpoint kinase WEE1 by ......" | 22847610 |
Expression profile in cancer corhorts:
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-195-5p | CDK4 | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; SARC; THCA; STAD; UCEC | miRNAWalker2 validate; miRTarBase | TCGA BLCA -0.058; TCGA BRCA -0.169; TCGA COAD -0.179; TCGA ESCA -0.16; TCGA HNSC -0.099; TCGA LIHC -0.182; TCGA LUAD -0.113; TCGA LUSC -0.314; TCGA SARC -0.473; TCGA THCA -0.117; TCGA STAD -0.082; TCGA UCEC -0.119 |
hsa-miR-195-5p | CHUK | 9 cancers: BLCA; BRCA; CESC; ESCA; KIRC; LUAD; PRAD; STAD; UCEC | miRNAWalker2 validate; MirTarget | TCGA BLCA -0.052; TCGA BRCA -0.096; TCGA CESC -0.051; TCGA ESCA -0.133; TCGA KIRC -0.157; TCGA LUAD -0.064; TCGA PRAD -0.095; TCGA STAD -0.209; TCGA UCEC -0.077 |
hsa-miR-195-5p | NOLC1 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; LIHC; LUAD; LUSC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.095; TCGA BRCA -0.151; TCGA CESC -0.069; TCGA COAD -0.146; TCGA ESCA -0.229; TCGA KIRC -0.078; TCGA LIHC -0.065; TCGA LUAD -0.099; TCGA LUSC -0.197; TCGA STAD -0.297; TCGA UCEC -0.139 |
hsa-miR-195-5p | TMC6 | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LUAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.146; TCGA BRCA -0.233; TCGA CESC -0.076; TCGA ESCA -0.123; TCGA HNSC -0.116; TCGA LUAD -0.118; TCGA SARC -0.137; TCGA THCA -0.581; TCGA STAD -0.159; TCGA UCEC -0.223 |
hsa-miR-195-5p | TPI1 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; UCEC | miRNAWalker2 validate | TCGA BLCA -0.186; TCGA BRCA -0.284; TCGA CESC -0.132; TCGA COAD -0.181; TCGA ESCA -0.146; TCGA HNSC -0.174; TCGA KIRP -0.076; TCGA LIHC -0.086; TCGA LUAD -0.19; TCGA LUSC -0.374; TCGA PAAD -0.327; TCGA PRAD -0.162; TCGA SARC -0.072; TCGA UCEC -0.108 |
hsa-miR-195-5p | VEGFA | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LUAD; STAD; UCEC | miRNAWalker2 validate; miRTarBase; MirTarget | TCGA BLCA -0.219; TCGA BRCA -0.219; TCGA CESC -0.182; TCGA COAD -0.171; TCGA ESCA -0.182; TCGA HNSC -0.266; TCGA LGG -0.126; TCGA LUAD -0.216; TCGA STAD -0.293; TCGA UCEC -0.2 |
hsa-miR-195-5p | ATP5G3 | 11 cancers: BLCA; BRCA; CESC; COAD; KIRC; LIHC; LUAD; LUSC; PAAD; PRAD; UCEC | MirTarget | TCGA BLCA -0.137; TCGA BRCA -0.116; TCGA CESC -0.056; TCGA COAD -0.112; TCGA KIRC -0.147; TCGA LIHC -0.053; TCGA LUAD -0.117; TCGA LUSC -0.265; TCGA PAAD -0.133; TCGA PRAD -0.161; TCGA UCEC -0.145 |
hsa-miR-195-5p | SMARCD2 | 12 cancers: BLCA; BRCA; COAD; KIRP; LIHC; LUAD; LUSC; PRAD; SARC; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.085; TCGA BRCA -0.18; TCGA COAD -0.066; TCGA KIRP -0.092; TCGA LIHC -0.062; TCGA LUAD -0.096; TCGA LUSC -0.216; TCGA PRAD -0.08; TCGA SARC -0.057; TCGA THCA -0.052; TCGA STAD -0.098; TCGA UCEC -0.132 |
hsa-miR-195-5p | ZNF622 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.087; TCGA BRCA -0.155; TCGA CESC -0.122; TCGA COAD -0.091; TCGA ESCA -0.086; TCGA HNSC -0.113; TCGA KIRP -0.055; TCGA LIHC -0.087; TCGA LUAD -0.062; TCGA LUSC -0.102; TCGA PAAD -0.163; TCGA PRAD -0.195; TCGA SARC -0.054; TCGA THCA -0.139; TCGA UCEC -0.121 |
hsa-miR-195-5p | PRR13 | 9 cancers: BLCA; BRCA; CESC; HNSC; LIHC; PRAD; SARC; THCA; UCEC | MirTarget | TCGA BLCA -0.059; TCGA BRCA -0.143; TCGA CESC -0.057; TCGA HNSC -0.071; TCGA LIHC -0.094; TCGA PRAD -0.094; TCGA SARC -0.078; TCGA THCA -0.074; TCGA UCEC -0.071 |
hsa-miR-195-5p | CCDC18 | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LGG; LUAD; LUSC; STAD; UCEC | MirTarget | TCGA BLCA -0.124; TCGA BRCA -0.138; TCGA COAD -0.127; TCGA ESCA -0.23; TCGA HNSC -0.078; TCGA KIRP -0.114; TCGA LGG -0.134; TCGA LUAD -0.22; TCGA LUSC -0.282; TCGA STAD -0.285; TCGA UCEC -0.144 |
hsa-miR-195-5p | RAB10 | 11 cancers: BLCA; BRCA; CESC; ESCA; LIHC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.083; TCGA BRCA -0.128; TCGA CESC -0.062; TCGA ESCA -0.119; TCGA LIHC -0.056; TCGA LUAD -0.124; TCGA LUSC -0.234; TCGA PRAD -0.066; TCGA SARC -0.053; TCGA STAD -0.134; TCGA UCEC -0.117 |
hsa-miR-195-5p | PDIA6 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.057; TCGA BRCA -0.231; TCGA CESC -0.119; TCGA COAD -0.14; TCGA ESCA -0.086; TCGA HNSC -0.091; TCGA LIHC -0.109; TCGA LUAD -0.138; TCGA LUSC -0.238; TCGA PAAD -0.163; TCGA PRAD -0.225; TCGA SARC -0.114; TCGA STAD -0.086; TCGA UCEC -0.211 |
hsa-miR-195-5p | ANKRD13D | 9 cancers: BLCA; BRCA; HNSC; KIRP; LIHC; LUAD; LUSC; THCA; STAD | MirTarget | TCGA BLCA -0.121; TCGA BRCA -0.138; TCGA HNSC -0.174; TCGA KIRP -0.059; TCGA LIHC -0.109; TCGA LUAD -0.13; TCGA LUSC -0.16; TCGA THCA -0.18; TCGA STAD -0.074 |
hsa-miR-195-5p | PFKFB4 | 14 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.292; TCGA BRCA -0.284; TCGA CESC -0.274; TCGA ESCA -0.399; TCGA HNSC -0.201; TCGA LIHC -0.357; TCGA LUAD -0.283; TCGA LUSC -0.336; TCGA PAAD -0.267; TCGA PRAD -0.181; TCGA SARC -0.094; TCGA THCA -0.088; TCGA STAD -0.268; TCGA UCEC -0.148 |
hsa-miR-195-5p | GNAI3 | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUSC; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.057; TCGA BRCA -0.115; TCGA COAD -0.056; TCGA ESCA -0.127; TCGA HNSC -0.064; TCGA LUSC -0.098; TCGA PRAD -0.052; TCGA SARC -0.06; TCGA STAD -0.133; TCGA UCEC -0.103 |
hsa-miR-195-5p | EIF2B2 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; UCEC | MirTarget | TCGA BLCA -0.085; TCGA BRCA -0.089; TCGA CESC -0.089; TCGA COAD -0.121; TCGA ESCA -0.147; TCGA HNSC -0.143; TCGA LUAD -0.079; TCGA LUSC -0.12; TCGA PAAD -0.086; TCGA PRAD -0.086; TCGA UCEC -0.07 |
hsa-miR-195-5p | ATG9A | 11 cancers: BLCA; BRCA; CESC; LIHC; LUAD; LUSC; PAAD; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.107; TCGA BRCA -0.057; TCGA CESC -0.075; TCGA LIHC -0.096; TCGA LUAD -0.084; TCGA LUSC -0.077; TCGA PAAD -0.116; TCGA PRAD -0.075; TCGA THCA -0.117; TCGA STAD -0.056; TCGA UCEC -0.102 |
hsa-miR-195-5p | BZW1 | 12 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LGG; LUAD; LUSC; PAAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.079; TCGA BRCA -0.145; TCGA CESC -0.147; TCGA ESCA -0.089; TCGA HNSC -0.077; TCGA LGG -0.071; TCGA LUAD -0.135; TCGA LUSC -0.142; TCGA PAAD -0.123; TCGA SARC -0.135; TCGA STAD -0.176; TCGA UCEC -0.146 |
hsa-miR-195-5p | TBL1XR1 | 9 cancers: BLCA; BRCA; ESCA; HNSC; KIRP; LUAD; LUSC; STAD; UCEC | MirTarget | TCGA BLCA -0.085; TCGA BRCA -0.095; TCGA ESCA -0.207; TCGA HNSC -0.077; TCGA KIRP -0.078; TCGA LUAD -0.053; TCGA LUSC -0.387; TCGA STAD -0.269; TCGA UCEC -0.168 |
hsa-miR-195-5p | JARID2 | 9 cancers: BLCA; BRCA; CESC; HNSC; LIHC; LUAD; LUSC; SARC; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.104; TCGA BRCA -0.152; TCGA CESC -0.087; TCGA HNSC -0.075; TCGA LIHC -0.156; TCGA LUAD -0.107; TCGA LUSC -0.274; TCGA SARC -0.106; TCGA UCEC -0.071 |
hsa-miR-195-5p | WWC1 | 9 cancers: BLCA; CESC; COAD; KIRC; KIRP; PAAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.154; TCGA CESC -0.311; TCGA COAD -0.089; TCGA KIRC -0.142; TCGA KIRP -0.089; TCGA PAAD -0.16; TCGA SARC -0.378; TCGA STAD -0.201; TCGA UCEC -0.187 |
hsa-miR-195-5p | TTPAL | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LUAD; LUSC; STAD; UCEC | MirTarget | TCGA BLCA -0.076; TCGA BRCA -0.063; TCGA CESC -0.067; TCGA ESCA -0.208; TCGA HNSC -0.201; TCGA KIRC -0.097; TCGA LUAD -0.08; TCGA LUSC -0.286; TCGA STAD -0.187; TCGA UCEC -0.118 |
hsa-miR-195-5p | TIMM22 | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; PAAD; PRAD; SARC | MirTarget | TCGA BLCA -0.062; TCGA CESC -0.054; TCGA COAD -0.116; TCGA ESCA -0.087; TCGA HNSC -0.07; TCGA KIRP -0.073; TCGA PAAD -0.096; TCGA PRAD -0.074; TCGA SARC -0.072 |
hsa-miR-195-5p | SPTLC1 | 10 cancers: BLCA; COAD; ESCA; KIRC; LGG; LUSC; PAAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.084; TCGA COAD -0.056; TCGA ESCA -0.143; TCGA KIRC -0.116; TCGA LGG -0.05; TCGA LUSC -0.099; TCGA PAAD -0.164; TCGA SARC -0.067; TCGA STAD -0.162; TCGA UCEC -0.058 |
hsa-miR-195-5p | IPPK | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LGG; LUAD; LUSC; PRAD; STAD | MirTarget | TCGA BLCA -0.096; TCGA BRCA -0.141; TCGA COAD -0.095; TCGA ESCA -0.185; TCGA HNSC -0.115; TCGA KIRC -0.143; TCGA LGG -0.069; TCGA LUAD -0.118; TCGA LUSC -0.287; TCGA PRAD -0.177; TCGA STAD -0.087 |
hsa-miR-195-5p | FASN | 10 cancers: BLCA; BRCA; CESC; ESCA; LIHC; LUAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.138; TCGA BRCA -0.133; TCGA CESC -0.226; TCGA ESCA -0.155; TCGA LIHC -0.238; TCGA LUAD -0.175; TCGA PRAD -0.279; TCGA SARC -0.098; TCGA STAD -0.318; TCGA UCEC -0.216 |
hsa-miR-195-5p | PSME3 | 13 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.089; TCGA BRCA -0.103; TCGA CESC -0.079; TCGA ESCA -0.166; TCGA HNSC -0.061; TCGA KIRC -0.14; TCGA LIHC -0.12; TCGA LUAD -0.115; TCGA LUSC -0.205; TCGA PAAD -0.066; TCGA PRAD -0.076; TCGA STAD -0.093; TCGA UCEC -0.092 |
hsa-miR-195-5p | HN1L | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; STAD | MirTarget | TCGA BLCA -0.101; TCGA BRCA -0.254; TCGA CESC -0.072; TCGA COAD -0.105; TCGA ESCA -0.186; TCGA HNSC -0.137; TCGA KIRP -0.152; TCGA LUAD -0.073; TCGA LUSC -0.261; TCGA PAAD -0.211; TCGA STAD -0.232 |
hsa-miR-195-5p | ERCC6 | 9 cancers: BLCA; COAD; ESCA; KIRC; LUAD; LUSC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.159; TCGA COAD -0.082; TCGA ESCA -0.171; TCGA KIRC -0.113; TCGA LUAD -0.06; TCGA LUSC -0.175; TCGA THCA -0.09; TCGA STAD -0.16; TCGA UCEC -0.055 |
hsa-miR-195-5p | TFDP2 | 9 cancers: BLCA; BRCA; HNSC; KIRC; LUAD; LUSC; PRAD; THCA; UCEC | MirTarget; mirMAP | TCGA BLCA -0.121; TCGA BRCA -0.076; TCGA HNSC -0.069; TCGA KIRC -0.153; TCGA LUAD -0.132; TCGA LUSC -0.444; TCGA PRAD -0.1; TCGA THCA -0.102; TCGA UCEC -0.215 |
hsa-miR-195-5p | YIPF6 | 11 cancers: BLCA; BRCA; COAD; ESCA; KIRC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.068; TCGA BRCA -0.161; TCGA COAD -0.134; TCGA ESCA -0.086; TCGA KIRC -0.162; TCGA LUAD -0.134; TCGA LUSC -0.168; TCGA PAAD -0.141; TCGA PRAD -0.123; TCGA STAD -0.093; TCGA UCEC -0.158 |
hsa-miR-195-5p | APLN | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; PAAD; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.226; TCGA BRCA -0.151; TCGA CESC -0.468; TCGA COAD -0.29; TCGA ESCA -0.521; TCGA HNSC -0.406; TCGA LIHC -0.631; TCGA PAAD -0.886; TCGA THCA -0.287; TCGA STAD -0.483; TCGA UCEC -0.224 |
hsa-miR-195-5p | WBP11 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.078; TCGA BRCA -0.143; TCGA CESC -0.055; TCGA COAD -0.089; TCGA ESCA -0.137; TCGA KIRC -0.103; TCGA LUAD -0.113; TCGA LUSC -0.141; TCGA PRAD -0.071; TCGA SARC -0.064; TCGA STAD -0.13; TCGA UCEC -0.115 |
hsa-miR-195-5p | CASK | 11 cancers: BLCA; BRCA; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.089; TCGA BRCA -0.083; TCGA ESCA -0.171; TCGA HNSC -0.11; TCGA KIRP -0.123; TCGA LIHC -0.061; TCGA LUAD -0.068; TCGA LUSC -0.266; TCGA PAAD -0.172; TCGA STAD -0.244; TCGA UCEC -0.094 |
hsa-miR-195-5p | CLSPN | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.156; TCGA BRCA -0.749; TCGA COAD -0.343; TCGA ESCA -0.413; TCGA HNSC -0.164; TCGA KIRP -0.175; TCGA LIHC -0.333; TCGA LUAD -0.557; TCGA LUSC -0.79; TCGA THCA -0.217; TCGA STAD -0.606; TCGA UCEC -0.425 |
hsa-miR-195-5p | TMEM33 | 12 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.125; TCGA BRCA -0.086; TCGA CESC -0.079; TCGA ESCA -0.123; TCGA HNSC -0.062; TCGA KIRC -0.194; TCGA LUAD -0.078; TCGA LUSC -0.092; TCGA PAAD -0.106; TCGA SARC -0.07; TCGA STAD -0.348; TCGA UCEC -0.093 |
hsa-miR-195-5p | PSMA5 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.153; TCGA BRCA -0.244; TCGA CESC -0.095; TCGA COAD -0.121; TCGA ESCA -0.125; TCGA HNSC -0.112; TCGA LIHC -0.094; TCGA LUAD -0.09; TCGA LUSC -0.149; TCGA PAAD -0.113; TCGA PRAD -0.136; TCGA THCA -0.1; TCGA STAD -0.125; TCGA UCEC -0.129 |
hsa-miR-195-5p | MAP7 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; KIRP; LUAD; LUSC; PAAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.287; TCGA BRCA -0.199; TCGA CESC -0.139; TCGA COAD -0.1; TCGA ESCA -0.253; TCGA KIRC -0.155; TCGA KIRP -0.074; TCGA LUAD -0.145; TCGA LUSC -0.3; TCGA PAAD -0.193; TCGA STAD -0.377; TCGA UCEC -0.224 |
hsa-miR-195-5p | CLDN12 | 11 cancers: BLCA; ESCA; KIRC; KIRP; LUAD; LUSC; PAAD; SARC; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.122; TCGA ESCA -0.212; TCGA KIRC -0.143; TCGA KIRP -0.156; TCGA LUAD -0.125; TCGA LUSC -0.221; TCGA PAAD -0.305; TCGA SARC -0.051; TCGA THCA -0.11; TCGA STAD -0.358; TCGA UCEC -0.064 |
hsa-miR-195-5p | STX1A | 11 cancers: BLCA; BRCA; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; SARC; THCA; UCEC | MirTarget | TCGA BLCA -0.232; TCGA BRCA -0.252; TCGA HNSC -0.333; TCGA KIRP -0.171; TCGA LGG -0.192; TCGA LIHC -0.235; TCGA LUAD -0.357; TCGA LUSC -0.419; TCGA SARC -0.157; TCGA THCA -0.055; TCGA UCEC -0.121 |
hsa-miR-195-5p | CAPRIN1 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; LIHC; LUAD; LUSC; PAAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.084; TCGA BRCA -0.12; TCGA CESC -0.068; TCGA COAD -0.083; TCGA ESCA -0.138; TCGA KIRC -0.121; TCGA LIHC -0.081; TCGA LUAD -0.07; TCGA LUSC -0.116; TCGA PAAD -0.087; TCGA THCA -0.058; TCGA STAD -0.228; TCGA UCEC -0.071 |
hsa-miR-195-5p | SH3BP2 | 10 cancers: BLCA; CESC; HNSC; KIRP; LGG; LUAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.167; TCGA CESC -0.069; TCGA HNSC -0.066; TCGA KIRP -0.065; TCGA LGG -0.114; TCGA LUAD -0.08; TCGA SARC -0.076; TCGA THCA -0.146; TCGA STAD -0.072; TCGA UCEC -0.083 |
hsa-miR-195-5p | COPS7B | 13 cancers: BLCA; BRCA; COAD; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.078; TCGA BRCA -0.074; TCGA COAD -0.067; TCGA HNSC -0.066; TCGA KIRC -0.054; TCGA KIRP -0.085; TCGA LGG -0.051; TCGA LIHC -0.161; TCGA LUAD -0.122; TCGA LUSC -0.145; TCGA THCA -0.089; TCGA STAD -0.061; TCGA UCEC -0.058 |
hsa-miR-195-5p | SSRP1 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; STAD; UCEC | MirTarget | TCGA BLCA -0.077; TCGA BRCA -0.122; TCGA CESC -0.073; TCGA COAD -0.136; TCGA ESCA -0.171; TCGA HNSC -0.087; TCGA LIHC -0.086; TCGA LUAD -0.106; TCGA LUSC -0.237; TCGA STAD -0.119; TCGA UCEC -0.085 |
hsa-miR-195-5p | TMEM41A | 13 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LUSC; PAAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.093; TCGA BRCA -0.158; TCGA COAD -0.098; TCGA ESCA -0.198; TCGA HNSC -0.123; TCGA KIRC -0.063; TCGA KIRP -0.092; TCGA LUSC -0.325; TCGA PAAD -0.197; TCGA SARC -0.147; TCGA THCA -0.295; TCGA STAD -0.217; TCGA UCEC -0.211 |
hsa-miR-195-5p | SEH1L | 9 cancers: BLCA; BRCA; COAD; ESCA; LUAD; LUSC; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.069; TCGA BRCA -0.071; TCGA COAD -0.112; TCGA ESCA -0.158; TCGA LUAD -0.069; TCGA LUSC -0.208; TCGA THCA -0.077; TCGA STAD -0.236; TCGA UCEC -0.136 |
hsa-miR-195-5p | ZBTB33 | 10 cancers: BLCA; COAD; ESCA; KIRC; LIHC; LUSC; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.131; TCGA COAD -0.078; TCGA ESCA -0.09; TCGA KIRC -0.101; TCGA LIHC -0.101; TCGA LUSC -0.214; TCGA SARC -0.064; TCGA THCA -0.161; TCGA STAD -0.25; TCGA UCEC -0.111 |
hsa-miR-195-5p | SPTBN2 | 9 cancers: BLCA; BRCA; KIRC; LUAD; LUSC; PAAD; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.218; TCGA BRCA -0.238; TCGA KIRC -0.895; TCGA LUAD -0.34; TCGA LUSC -0.679; TCGA PAAD -0.226; TCGA THCA -0.793; TCGA STAD -0.449; TCGA UCEC -0.249 |
hsa-miR-195-5p | ATXN7L3 | 13 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; PRAD; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.137; TCGA BRCA -0.086; TCGA CESC -0.051; TCGA ESCA -0.122; TCGA HNSC -0.068; TCGA LGG -0.062; TCGA LIHC -0.159; TCGA LUAD -0.167; TCGA LUSC -0.196; TCGA PRAD -0.063; TCGA THCA -0.051; TCGA STAD -0.079; TCGA UCEC -0.063 |
hsa-miR-195-5p | PWWP2B | 11 cancers: BLCA; BRCA; CESC; KIRC; LGG; LIHC; LUAD; PRAD; SARC; THCA; UCEC | MirTarget | TCGA BLCA -0.149; TCGA BRCA -0.197; TCGA CESC -0.087; TCGA KIRC -0.212; TCGA LGG -0.102; TCGA LIHC -0.133; TCGA LUAD -0.087; TCGA PRAD -0.164; TCGA SARC -0.079; TCGA THCA -0.16; TCGA UCEC -0.066 |
hsa-miR-195-5p | PLRG1 | 9 cancers: BLCA; COAD; ESCA; KIRC; LUSC; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.078; TCGA COAD -0.05; TCGA ESCA -0.077; TCGA KIRC -0.067; TCGA LUSC -0.058; TCGA PRAD -0.056; TCGA THCA -0.158; TCGA STAD -0.054; TCGA UCEC -0.055 |
hsa-miR-195-5p | ZBTB9 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; PAAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.136; TCGA BRCA -0.159; TCGA CESC -0.094; TCGA COAD -0.072; TCGA ESCA -0.136; TCGA HNSC -0.074; TCGA KIRC -0.109; TCGA LIHC -0.207; TCGA LUAD -0.117; TCGA LUSC -0.18; TCGA PAAD -0.112; TCGA STAD -0.134; TCGA UCEC -0.14 |
hsa-miR-195-5p | SLC25A15 | 9 cancers: BLCA; BRCA; CESC; ESCA; KIRC; LUAD; LUSC; STAD; UCEC | MirTarget | TCGA BLCA -0.135; TCGA BRCA -0.134; TCGA CESC -0.099; TCGA ESCA -0.204; TCGA KIRC -0.214; TCGA LUAD -0.168; TCGA LUSC -0.287; TCGA STAD -0.209; TCGA UCEC -0.116 |
hsa-miR-195-5p | CDC25A | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LIHC; LUAD; LUSC; PAAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.121; TCGA BRCA -0.526; TCGA CESC -0.132; TCGA COAD -0.195; TCGA ESCA -0.375; TCGA HNSC -0.139; TCGA KIRC -0.226; TCGA LGG -0.106; TCGA LIHC -0.345; TCGA LUAD -0.467; TCGA LUSC -0.794; TCGA PAAD -0.314; TCGA SARC -0.115; TCGA STAD -0.432; TCGA UCEC -0.34 |
hsa-miR-195-5p | DOLPP1 | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LGG; LUAD; LUSC; PAAD; SARC; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.127; TCGA BRCA -0.152; TCGA COAD -0.103; TCGA ESCA -0.225; TCGA HNSC -0.076; TCGA KIRC -0.087; TCGA LGG -0.055; TCGA LUAD -0.141; TCGA LUSC -0.325; TCGA PAAD -0.16; TCGA SARC -0.068; TCGA UCEC -0.058 |
hsa-miR-195-5p | USP14 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.057; TCGA BRCA -0.166; TCGA CESC -0.09; TCGA COAD -0.17; TCGA ESCA -0.167; TCGA HNSC -0.072; TCGA KIRC -0.104; TCGA LGG -0.077; TCGA LIHC -0.113; TCGA LUAD -0.139; TCGA LUSC -0.224; TCGA PAAD -0.092; TCGA PRAD -0.106; TCGA STAD -0.226; TCGA UCEC -0.167 |
hsa-miR-195-5p | TRIM59 | 12 cancers: BLCA; BRCA; COAD; ESCA; KIRP; LIHC; LUAD; LUSC; PAAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.094; TCGA BRCA -0.368; TCGA COAD -0.125; TCGA ESCA -0.322; TCGA KIRP -0.073; TCGA LIHC -0.232; TCGA LUAD -0.171; TCGA LUSC -0.553; TCGA PAAD -0.192; TCGA THCA -0.166; TCGA STAD -0.418; TCGA UCEC -0.101 |
hsa-miR-195-5p | VPS33B | 13 cancers: BLCA; BRCA; COAD; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PRAD; THCA; STAD | MirTarget | TCGA BLCA -0.057; TCGA BRCA -0.062; TCGA COAD -0.067; TCGA HNSC -0.068; TCGA KIRC -0.054; TCGA KIRP -0.066; TCGA LGG -0.069; TCGA LIHC -0.098; TCGA LUAD -0.078; TCGA LUSC -0.09; TCGA PRAD -0.133; TCGA THCA -0.057; TCGA STAD -0.06 |
hsa-miR-195-5p | CBX4 | 10 cancers: BLCA; BRCA; CESC; HNSC; LIHC; LUAD; LUSC; PAAD; THCA; STAD | MirTarget; miRNATAP | TCGA BLCA -0.057; TCGA BRCA -0.225; TCGA CESC -0.063; TCGA HNSC -0.091; TCGA LIHC -0.175; TCGA LUAD -0.154; TCGA LUSC -0.308; TCGA PAAD -0.132; TCGA THCA -0.092; TCGA STAD -0.1 |
hsa-miR-195-5p | CEP55 | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.183; TCGA BRCA -0.739; TCGA COAD -0.196; TCGA ESCA -0.529; TCGA HNSC -0.244; TCGA KIRP -0.265; TCGA LIHC -0.415; TCGA LUAD -0.454; TCGA LUSC -0.945; TCGA THCA -0.11; TCGA STAD -0.512; TCGA UCEC -0.37 |
hsa-miR-195-5p | UBE2K | 12 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LGG; LIHC; LUAD; LUSC; PAAD; STAD; UCEC | MirTarget | TCGA BLCA -0.069; TCGA BRCA -0.069; TCGA ESCA -0.092; TCGA HNSC -0.053; TCGA KIRC -0.056; TCGA LGG -0.068; TCGA LIHC -0.05; TCGA LUAD -0.087; TCGA LUSC -0.115; TCGA PAAD -0.094; TCGA STAD -0.152; TCGA UCEC -0.063 |
hsa-miR-195-5p | GPN1 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; SARC; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.057; TCGA BRCA -0.124; TCGA CESC -0.064; TCGA COAD -0.075; TCGA ESCA -0.106; TCGA HNSC -0.076; TCGA KIRP -0.066; TCGA LIHC -0.081; TCGA LUAD -0.111; TCGA LUSC -0.225; TCGA PAAD -0.086; TCGA SARC -0.095; TCGA THCA -0.092; TCGA STAD -0.056; TCGA UCEC -0.182 |
hsa-miR-195-5p | FBXO22 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.111; TCGA BRCA -0.124; TCGA CESC -0.097; TCGA COAD -0.154; TCGA ESCA -0.193; TCGA HNSC -0.063; TCGA KIRC -0.084; TCGA LUAD -0.073; TCGA LUSC -0.243; TCGA PRAD -0.06; TCGA THCA -0.095; TCGA STAD -0.182; TCGA UCEC -0.067 |
hsa-miR-195-5p | CHEK1 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.096; TCGA BRCA -0.386; TCGA CESC -0.094; TCGA COAD -0.157; TCGA ESCA -0.337; TCGA HNSC -0.174; TCGA KIRP -0.07; TCGA LIHC -0.272; TCGA LUAD -0.346; TCGA LUSC -0.623; TCGA PAAD -0.187; TCGA PRAD -0.185; TCGA THCA -0.145; TCGA STAD -0.398; TCGA UCEC -0.227 |
hsa-miR-195-5p | NUFIP2 | 9 cancers: BLCA; BRCA; ESCA; KIRC; KIRP; LUAD; LUSC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.065; TCGA BRCA -0.108; TCGA ESCA -0.089; TCGA KIRC -0.066; TCGA KIRP -0.069; TCGA LUAD -0.093; TCGA LUSC -0.136; TCGA STAD -0.214; TCGA UCEC -0.09 |
hsa-miR-195-5p | GATAD2A | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LUAD; LUSC; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.084; TCGA BRCA -0.12; TCGA CESC -0.082; TCGA ESCA -0.179; TCGA HNSC -0.059; TCGA LUAD -0.058; TCGA LUSC -0.14; TCGA THCA -0.125; TCGA STAD -0.153; TCGA UCEC -0.117 |
hsa-miR-195-5p | GALNT7 | 9 cancers: BLCA; BRCA; KIRC; KIRP; LUAD; LUSC; SARC; THCA; STAD | MirTarget | TCGA BLCA -0.123; TCGA BRCA -0.191; TCGA KIRC -0.133; TCGA KIRP -0.136; TCGA LUAD -0.14; TCGA LUSC -0.262; TCGA SARC -0.134; TCGA THCA -0.528; TCGA STAD -0.144 |
hsa-miR-195-5p | UBE2Q1 | 9 cancers: BLCA; BRCA; ESCA; HNSC; LIHC; LUAD; LUSC; THCA; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.07; TCGA BRCA -0.133; TCGA ESCA -0.092; TCGA HNSC -0.054; TCGA LIHC -0.129; TCGA LUAD -0.085; TCGA LUSC -0.165; TCGA THCA -0.059; TCGA UCEC -0.094 |
hsa-miR-195-5p | NOB1 | 10 cancers: BLCA; COAD; ESCA; HNSC; KIRP; LIHC; LUSC; SARC; THCA; STAD | MirTarget | TCGA BLCA -0.123; TCGA COAD -0.106; TCGA ESCA -0.117; TCGA HNSC -0.065; TCGA KIRP -0.069; TCGA LIHC -0.071; TCGA LUSC -0.24; TCGA SARC -0.089; TCGA THCA -0.089; TCGA STAD -0.114 |
hsa-miR-195-5p | GTF3C5 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; STAD | MirTarget | TCGA BLCA -0.116; TCGA BRCA -0.105; TCGA CESC -0.081; TCGA COAD -0.088; TCGA ESCA -0.229; TCGA HNSC -0.117; TCGA KIRP -0.066; TCGA LIHC -0.107; TCGA LUAD -0.105; TCGA LUSC -0.266; TCGA PAAD -0.116; TCGA STAD -0.132 |
hsa-miR-195-5p | NAPG | 12 cancers: BLCA; CESC; ESCA; HNSC; KIRC; LGG; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.072; TCGA CESC -0.054; TCGA ESCA -0.154; TCGA HNSC -0.097; TCGA KIRC -0.127; TCGA LGG -0.065; TCGA LUAD -0.057; TCGA LUSC -0.089; TCGA PAAD -0.172; TCGA PRAD -0.097; TCGA STAD -0.166; TCGA UCEC -0.079 |
hsa-miR-195-5p | CUL2 | 9 cancers: BLCA; BRCA; ESCA; LIHC; LUAD; LUSC; PRAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.087; TCGA BRCA -0.112; TCGA ESCA -0.155; TCGA LIHC -0.051; TCGA LUAD -0.08; TCGA LUSC -0.106; TCGA PRAD -0.075; TCGA STAD -0.13; TCGA UCEC -0.116 |
hsa-miR-195-5p | SAMD10 | 9 cancers: BLCA; BRCA; KIRP; LIHC; LUAD; LUSC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.147; TCGA BRCA -0.174; TCGA KIRP -0.116; TCGA LIHC -0.239; TCGA LUAD -0.159; TCGA LUSC -0.109; TCGA THCA -0.205; TCGA STAD -0.18; TCGA UCEC -0.101 |
hsa-miR-195-5p | LASP1 | 10 cancers: BLCA; BRCA; CESC; KIRP; LIHC; LUAD; PAAD; SARC; THCA; UCEC | mirMAP | TCGA BLCA -0.101; TCGA BRCA -0.147; TCGA CESC -0.11; TCGA KIRP -0.116; TCGA LIHC -0.156; TCGA LUAD -0.114; TCGA PAAD -0.098; TCGA SARC -0.064; TCGA THCA -0.173; TCGA UCEC -0.145 |
hsa-miR-195-5p | RPUSD1 | 12 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; THCA; UCEC | mirMAP | TCGA BLCA -0.092; TCGA BRCA -0.251; TCGA CESC -0.07; TCGA ESCA -0.118; TCGA HNSC -0.123; TCGA KIRP -0.064; TCGA LIHC -0.109; TCGA LUAD -0.115; TCGA LUSC -0.28; TCGA PAAD -0.13; TCGA THCA -0.068; TCGA UCEC -0.073 |
hsa-miR-195-5p | HMGB3 | 15 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; PAAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.184; TCGA BRCA -0.558; TCGA COAD -0.145; TCGA ESCA -0.282; TCGA HNSC -0.22; TCGA KIRC -0.252; TCGA KIRP -0.261; TCGA LGG -0.086; TCGA LUAD -0.319; TCGA LUSC -0.586; TCGA PAAD -0.17; TCGA SARC -0.09; TCGA THCA -0.057; TCGA STAD -0.444; TCGA UCEC -0.152 |
hsa-miR-195-5p | MTA3 | 9 cancers: BLCA; BRCA; CESC; HNSC; LIHC; LUAD; LUSC; PRAD; UCEC | mirMAP | TCGA BLCA -0.061; TCGA BRCA -0.118; TCGA CESC -0.065; TCGA HNSC -0.063; TCGA LIHC -0.17; TCGA LUAD -0.113; TCGA LUSC -0.211; TCGA PRAD -0.088; TCGA UCEC -0.05 |
hsa-miR-195-5p | MOCOS | 9 cancers: BLCA; BRCA; COAD; ESCA; LUAD; LUSC; PRAD; THCA; STAD | mirMAP | TCGA BLCA -0.195; TCGA BRCA -0.126; TCGA COAD -0.242; TCGA ESCA -0.368; TCGA LUAD -0.117; TCGA LUSC -0.197; TCGA PRAD -0.194; TCGA THCA -0.126; TCGA STAD -0.32 |
hsa-miR-195-5p | STK24 | 9 cancers: BLCA; CESC; HNSC; LUAD; LUSC; PAAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.078; TCGA CESC -0.086; TCGA HNSC -0.051; TCGA LUAD -0.08; TCGA LUSC -0.122; TCGA PAAD -0.1; TCGA THCA -0.07; TCGA STAD -0.084; TCGA UCEC -0.088 |
hsa-miR-195-5p | NUTF2 | 11 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRP; LIHC; LUSC; PAAD; SARC; STAD | mirMAP | TCGA BLCA -0.093; TCGA BRCA -0.192; TCGA CESC -0.097; TCGA ESCA -0.133; TCGA HNSC -0.177; TCGA KIRP -0.111; TCGA LIHC -0.125; TCGA LUSC -0.133; TCGA PAAD -0.144; TCGA SARC -0.06; TCGA STAD -0.112 |
hsa-miR-195-5p | PYCRL | 12 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; UCEC | mirMAP | TCGA BLCA -0.107; TCGA BRCA -0.318; TCGA CESC -0.077; TCGA ESCA -0.288; TCGA HNSC -0.171; TCGA LIHC -0.149; TCGA LUAD -0.146; TCGA LUSC -0.368; TCGA PAAD -0.275; TCGA PRAD -0.139; TCGA SARC -0.082; TCGA UCEC -0.065 |
hsa-miR-195-5p | TMEM104 | 14 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; UCEC | mirMAP | TCGA BLCA -0.068; TCGA BRCA -0.212; TCGA CESC -0.081; TCGA ESCA -0.117; TCGA HNSC -0.095; TCGA KIRC -0.129; TCGA KIRP -0.097; TCGA LIHC -0.186; TCGA LUAD -0.119; TCGA LUSC -0.067; TCGA PAAD -0.151; TCGA PRAD -0.163; TCGA SARC -0.088; TCGA UCEC -0.119 |
hsa-miR-195-5p | STK35 | 10 cancers: BLCA; BRCA; CESC; KIRP; LIHC; LUAD; LUSC; PAAD; STAD; UCEC | mirMAP | TCGA BLCA -0.094; TCGA BRCA -0.147; TCGA CESC -0.069; TCGA KIRP -0.07; TCGA LIHC -0.164; TCGA LUAD -0.118; TCGA LUSC -0.246; TCGA PAAD -0.14; TCGA STAD -0.14; TCGA UCEC -0.139 |
hsa-miR-195-5p | DDA1 | 11 cancers: BLCA; BRCA; CESC; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; THCA; UCEC | mirMAP | TCGA BLCA -0.103; TCGA BRCA -0.099; TCGA CESC -0.055; TCGA HNSC -0.09; TCGA KIRP -0.063; TCGA LIHC -0.097; TCGA LUAD -0.055; TCGA LUSC -0.064; TCGA PAAD -0.118; TCGA THCA -0.11; TCGA UCEC -0.101 |
hsa-miR-195-5p | GRSF1 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; KIRP; LUAD; LUSC; PAAD; STAD; UCEC | mirMAP | TCGA BLCA -0.093; TCGA BRCA -0.11; TCGA CESC -0.053; TCGA COAD -0.095; TCGA ESCA -0.105; TCGA KIRC -0.147; TCGA KIRP -0.058; TCGA LUAD -0.075; TCGA LUSC -0.077; TCGA PAAD -0.141; TCGA STAD -0.137; TCGA UCEC -0.104 |
hsa-miR-195-5p | TTYH3 | 13 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; PAAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.113; TCGA BRCA -0.293; TCGA CESC -0.144; TCGA ESCA -0.273; TCGA HNSC -0.191; TCGA LGG -0.121; TCGA LIHC -0.135; TCGA LUAD -0.207; TCGA LUSC -0.303; TCGA PAAD -0.173; TCGA SARC -0.225; TCGA STAD -0.272; TCGA UCEC -0.161 |
hsa-miR-195-5p | TYSND1 | 9 cancers: BLCA; BRCA; ESCA; HNSC; LIHC; LUAD; LUSC; PRAD; STAD | mirMAP | TCGA BLCA -0.159; TCGA BRCA -0.053; TCGA ESCA -0.196; TCGA HNSC -0.065; TCGA LIHC -0.207; TCGA LUAD -0.053; TCGA LUSC -0.317; TCGA PRAD -0.073; TCGA STAD -0.09 |
hsa-miR-195-5p | FBXO41 | 9 cancers: BLCA; BRCA; HNSC; KIRP; LUAD; LUSC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.134; TCGA BRCA -0.108; TCGA HNSC -0.157; TCGA KIRP -0.155; TCGA LUAD -0.279; TCGA LUSC -0.473; TCGA THCA -0.302; TCGA STAD -0.223; TCGA UCEC -0.152 |
hsa-miR-195-5p | LRPAP1 | 10 cancers: BLCA; BRCA; COAD; KIRC; LIHC; PAAD; PRAD; SARC; THCA; UCEC | mirMAP | TCGA BLCA -0.097; TCGA BRCA -0.102; TCGA COAD -0.12; TCGA KIRC -0.092; TCGA LIHC -0.082; TCGA PAAD -0.133; TCGA PRAD -0.266; TCGA SARC -0.063; TCGA THCA -0.134; TCGA UCEC -0.091 |
hsa-miR-195-5p | LETM1 | 9 cancers: BLCA; BRCA; CESC; KIRC; LIHC; LUAD; LUSC; STAD; UCEC | mirMAP | TCGA BLCA -0.116; TCGA BRCA -0.128; TCGA CESC -0.092; TCGA KIRC -0.132; TCGA LIHC -0.087; TCGA LUAD -0.127; TCGA LUSC -0.167; TCGA STAD -0.197; TCGA UCEC -0.067 |
hsa-miR-195-5p | LMNB2 | 14 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.106; TCGA BRCA -0.289; TCGA COAD -0.13; TCGA ESCA -0.331; TCGA HNSC -0.17; TCGA KIRP -0.072; TCGA LIHC -0.18; TCGA LUAD -0.299; TCGA LUSC -0.461; TCGA PAAD -0.164; TCGA SARC -0.111; TCGA THCA -0.104; TCGA STAD -0.294; TCGA UCEC -0.158 |
hsa-miR-195-5p | GNB1L | 13 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.088; TCGA BRCA -0.166; TCGA COAD -0.125; TCGA ESCA -0.121; TCGA HNSC -0.113; TCGA LGG -0.063; TCGA LIHC -0.231; TCGA LUAD -0.155; TCGA LUSC -0.31; TCGA PRAD -0.117; TCGA THCA -0.137; TCGA STAD -0.112; TCGA UCEC -0.051 |
hsa-miR-195-5p | TMEM201 | 10 cancers: BLCA; BRCA; COAD; ESCA; KIRC; LIHC; LUAD; LUSC; STAD; UCEC | mirMAP | TCGA BLCA -0.117; TCGA BRCA -0.081; TCGA COAD -0.093; TCGA ESCA -0.203; TCGA KIRC -0.131; TCGA LIHC -0.211; TCGA LUAD -0.109; TCGA LUSC -0.306; TCGA STAD -0.311; TCGA UCEC -0.056 |
hsa-miR-195-5p | E2F7 | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; THCA; STAD | miRNATAP | TCGA BLCA -0.259; TCGA BRCA -0.576; TCGA COAD -0.257; TCGA ESCA -0.45; TCGA HNSC -0.236; TCGA KIRP -0.217; TCGA LIHC -0.366; TCGA LUAD -0.428; TCGA LUSC -1.071; TCGA THCA -0.337; TCGA STAD -0.719 |
hsa-miR-195-5p | AMMECR1 | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.117; TCGA BRCA -0.185; TCGA COAD -0.084; TCGA ESCA -0.21; TCGA HNSC -0.137; TCGA KIRC -0.093; TCGA KIRP -0.094; TCGA LUAD -0.082; TCGA LUSC -0.304; TCGA THCA -0.083; TCGA STAD -0.218; TCGA UCEC -0.107 |
hsa-miR-195-5p | PCMT1 | 11 cancers: BLCA; BRCA; CESC; ESCA; KIRC; LGG; LUSC; PRAD; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.06; TCGA BRCA -0.146; TCGA CESC -0.067; TCGA ESCA -0.114; TCGA KIRC -0.068; TCGA LGG -0.052; TCGA LUSC -0.055; TCGA PRAD -0.133; TCGA THCA -0.066; TCGA STAD -0.081; TCGA UCEC -0.118 |
hsa-miR-195-5p | IPO9 | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.08; TCGA BRCA -0.167; TCGA COAD -0.083; TCGA ESCA -0.157; TCGA HNSC -0.059; TCGA LIHC -0.127; TCGA LUAD -0.093; TCGA LUSC -0.232; TCGA PAAD -0.065; TCGA STAD -0.151; TCGA UCEC -0.088 |
hsa-miR-195-5p | ZNF697 | 9 cancers: BLCA; CESC; ESCA; HNSC; LGG; LUAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.097; TCGA CESC -0.132; TCGA ESCA -0.212; TCGA HNSC -0.109; TCGA LGG -0.131; TCGA LUAD -0.112; TCGA SARC -0.164; TCGA STAD -0.227; TCGA UCEC -0.116 |
hsa-miR-195-5p | DCAF7 | 10 cancers: BLCA; BRCA; ESCA; KIRC; KIRP; LIHC; LUAD; LUSC; STAD; UCEC | miRNATAP | TCGA BLCA -0.071; TCGA BRCA -0.171; TCGA ESCA -0.078; TCGA KIRC -0.095; TCGA KIRP -0.062; TCGA LIHC -0.101; TCGA LUAD -0.101; TCGA LUSC -0.098; TCGA STAD -0.107; TCGA UCEC -0.1 |
hsa-miR-195-5p | CDCA4 | 13 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.166; TCGA BRCA -0.238; TCGA COAD -0.211; TCGA ESCA -0.328; TCGA HNSC -0.23; TCGA KIRP -0.057; TCGA LGG -0.086; TCGA LIHC -0.179; TCGA LUAD -0.248; TCGA LUSC -0.63; TCGA THCA -0.186; TCGA STAD -0.234; TCGA UCEC -0.123 |
hsa-miR-195-5p | SUMO3 | 9 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LIHC; LUSC; SARC; UCEC | miRNATAP | TCGA BLCA -0.062; TCGA BRCA -0.158; TCGA COAD -0.07; TCGA ESCA -0.106; TCGA HNSC -0.055; TCGA LIHC -0.059; TCGA LUSC -0.085; TCGA SARC -0.12; TCGA UCEC -0.089 |
hsa-miR-195-5p | HNRNPA2B1 | 11 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; STAD; UCEC | miRNATAP | TCGA BLCA -0.066; TCGA BRCA -0.141; TCGA ESCA -0.168; TCGA HNSC -0.066; TCGA KIRC -0.06; TCGA KIRP -0.056; TCGA LIHC -0.075; TCGA LUAD -0.086; TCGA LUSC -0.201; TCGA STAD -0.238; TCGA UCEC -0.062 |
hsa-miR-195-5p | BAZ2A | 9 cancers: BLCA; CESC; COAD; ESCA; LIHC; LUAD; LUSC; STAD; UCEC | miRNATAP | TCGA BLCA -0.059; TCGA CESC -0.082; TCGA COAD -0.055; TCGA ESCA -0.07; TCGA LIHC -0.065; TCGA LUAD -0.152; TCGA LUSC -0.086; TCGA STAD -0.105; TCGA UCEC -0.096 |
hsa-miR-195-5p | HDGF | 13 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.117; TCGA BRCA -0.276; TCGA CESC -0.111; TCGA ESCA -0.137; TCGA HNSC -0.116; TCGA LIHC -0.062; TCGA LUAD -0.152; TCGA LUSC -0.314; TCGA PAAD -0.17; TCGA PRAD -0.058; TCGA THCA -0.077; TCGA STAD -0.099; TCGA UCEC -0.217 |
hsa-miR-195-5p | CCNJ | 9 cancers: BLCA; BRCA; ESCA; KIRP; LGG; LUAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.115; TCGA BRCA -0.129; TCGA ESCA -0.128; TCGA KIRP -0.058; TCGA LGG -0.07; TCGA LUAD -0.138; TCGA SARC -0.111; TCGA STAD -0.303; TCGA UCEC -0.052 |
hsa-miR-195-5p | HMGA1 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.253; TCGA BRCA -0.439; TCGA CESC -0.131; TCGA COAD -0.141; TCGA ESCA -0.235; TCGA HNSC -0.177; TCGA KIRC -0.297; TCGA KIRP -0.211; TCGA LIHC -0.424; TCGA LUAD -0.381; TCGA LUSC -0.597; TCGA SARC -0.249; TCGA THCA -0.368; TCGA STAD -0.232; TCGA UCEC -0.484 |
hsa-miR-195-5p | KLC2 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; THCA; STAD | miRNATAP | TCGA BLCA -0.103; TCGA BRCA -0.231; TCGA CESC -0.061; TCGA COAD -0.094; TCGA ESCA -0.2; TCGA HNSC -0.065; TCGA KIRC -0.108; TCGA LGG -0.068; TCGA LIHC -0.127; TCGA LUAD -0.201; TCGA LUSC -0.29; TCGA PAAD -0.107; TCGA PRAD -0.119; TCGA THCA -0.088; TCGA STAD -0.082 |
hsa-miR-195-5p | SRPK1 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; LIHC; LUAD; LUSC; STAD; UCEC | miRNATAP | TCGA BLCA -0.132; TCGA BRCA -0.251; TCGA CESC -0.068; TCGA COAD -0.097; TCGA ESCA -0.214; TCGA KIRC -0.144; TCGA LIHC -0.205; TCGA LUAD -0.212; TCGA LUSC -0.328; TCGA STAD -0.277; TCGA UCEC -0.192 |
hsa-miR-195-5p | SMYD5 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; SARC; THCA; STAD | miRNATAP | TCGA BLCA -0.054; TCGA BRCA -0.084; TCGA CESC -0.07; TCGA COAD -0.069; TCGA ESCA -0.109; TCGA HNSC -0.068; TCGA LIHC -0.185; TCGA LUAD -0.109; TCGA LUSC -0.273; TCGA PAAD -0.091; TCGA SARC -0.079; TCGA THCA -0.079; TCGA STAD -0.064 |
hsa-miR-195-5p | MOV10 | 13 cancers: BLCA; BRCA; COAD; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.181; TCGA BRCA -0.179; TCGA COAD -0.086; TCGA HNSC -0.067; TCGA LIHC -0.145; TCGA LUAD -0.146; TCGA LUSC -0.118; TCGA PAAD -0.081; TCGA PRAD -0.079; TCGA SARC -0.053; TCGA THCA -0.144; TCGA STAD -0.113; TCGA UCEC -0.069 |
hsa-miR-195-5p | SIX4 | 9 cancers: BLCA; BRCA; ESCA; KIRC; LIHC; LUAD; LUSC; SARC; STAD | miRNATAP | TCGA BLCA -0.142; TCGA BRCA -0.215; TCGA ESCA -0.258; TCGA KIRC -0.487; TCGA LIHC -0.465; TCGA LUAD -0.202; TCGA LUSC -0.514; TCGA SARC -0.247; TCGA STAD -0.311 |
hsa-miR-195-5p | FAM60A | 10 cancers: BLCA; ESCA; KIRC; KIRP; LIHC; LUAD; LUSC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.162; TCGA ESCA -0.181; TCGA KIRC -0.215; TCGA KIRP -0.097; TCGA LIHC -0.119; TCGA LUAD -0.193; TCGA LUSC -0.352; TCGA THCA -0.122; TCGA STAD -0.373; TCGA UCEC -0.167 |
hsa-miR-195-5p | TXNDC17 | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LIHC; LUSC; PAAD; PRAD; THCA; UCEC | miRNATAP | TCGA BLCA -0.157; TCGA BRCA -0.229; TCGA COAD -0.09; TCGA ESCA -0.141; TCGA HNSC -0.174; TCGA KIRP -0.066; TCGA LIHC -0.076; TCGA LUSC -0.241; TCGA PAAD -0.146; TCGA PRAD -0.152; TCGA THCA -0.098; TCGA UCEC -0.058 |
hsa-miR-195-5p | AP2A1 | 11 cancers: BLCA; BRCA; CESC; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA | miRNATAP | TCGA BLCA -0.061; TCGA BRCA -0.171; TCGA CESC -0.088; TCGA LGG -0.051; TCGA LIHC -0.09; TCGA LUAD -0.134; TCGA LUSC -0.114; TCGA PAAD -0.088; TCGA PRAD -0.084; TCGA SARC -0.053; TCGA THCA -0.116 |
hsa-miR-195-5p | FAM189B | 11 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; STAD; UCEC | miRNATAP | TCGA BLCA -0.09; TCGA BRCA -0.209; TCGA CESC -0.084; TCGA ESCA -0.132; TCGA HNSC -0.075; TCGA KIRP -0.064; TCGA LIHC -0.288; TCGA LUAD -0.176; TCGA LUSC -0.378; TCGA STAD -0.118; TCGA UCEC -0.144 |
hsa-miR-195-5p | G2E3 | 10 cancers: BLCA; BRCA; COAD; ESCA; KIRC; LGG; LUAD; LUSC; STAD; UCEC | miRNATAP | TCGA BLCA -0.075; TCGA BRCA -0.06; TCGA COAD -0.127; TCGA ESCA -0.207; TCGA KIRC -0.085; TCGA LGG -0.068; TCGA LUAD -0.116; TCGA LUSC -0.284; TCGA STAD -0.281; TCGA UCEC -0.094 |
hsa-miR-195-5p | OTX1 | 12 cancers: BLCA; BRCA; COAD; ESCA; LGG; LIHC; LUAD; LUSC; PAAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.17; TCGA BRCA -0.449; TCGA COAD -0.352; TCGA ESCA -0.429; TCGA LGG -0.189; TCGA LIHC -0.339; TCGA LUAD -0.549; TCGA LUSC -1.068; TCGA PAAD -0.399; TCGA SARC -0.219; TCGA STAD -0.684; TCGA UCEC -0.448 |
hsa-miR-195-5p | LYPLA2 | 11 cancers: BLCA; BRCA; CESC; HNSC; LGG; LIHC; LUSC; PAAD; SARC; THCA; UCEC | miRNATAP | TCGA BLCA -0.165; TCGA BRCA -0.18; TCGA CESC -0.063; TCGA HNSC -0.122; TCGA LGG -0.123; TCGA LIHC -0.091; TCGA LUSC -0.082; TCGA PAAD -0.13; TCGA SARC -0.159; TCGA THCA -0.235; TCGA UCEC -0.118 |
hsa-miR-195-5p | TFAP4 | 10 cancers: BLCA; BRCA; ESCA; KIRP; LIHC; LUAD; LUSC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.155; TCGA BRCA -0.081; TCGA ESCA -0.166; TCGA KIRP -0.106; TCGA LIHC -0.122; TCGA LUAD -0.092; TCGA LUSC -0.493; TCGA THCA -0.141; TCGA STAD -0.204; TCGA UCEC -0.09 |
hsa-miR-195-5p | OTUB1 | 11 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LIHC; LUSC; PRAD; SARC; THCA; UCEC | miRNATAP | TCGA BLCA -0.126; TCGA BRCA -0.173; TCGA CESC -0.101; TCGA ESCA -0.113; TCGA HNSC -0.116; TCGA LIHC -0.101; TCGA LUSC -0.128; TCGA PRAD -0.09; TCGA SARC -0.057; TCGA THCA -0.078; TCGA UCEC -0.065 |
hsa-miR-195-5p | EIF4A3 | 12 cancers: BLCA; BRCA; CESC; COAD; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | RAID | TCGA BLCA -0.073; TCGA BRCA -0.19; TCGA CESC -0.073; TCGA COAD -0.117; TCGA KIRP -0.05; TCGA LIHC -0.105; TCGA LUAD -0.157; TCGA LUSC -0.258; TCGA PAAD -0.125; TCGA PRAD -0.066; TCGA STAD -0.151; TCGA UCEC -0.194 |
hsa-miR-195-5p | PTBP1 | 9 cancers: BLCA; BRCA; COAD; LIHC; LUAD; LUSC; PAAD; SARC; STAD | RAID | TCGA BLCA -0.114; TCGA BRCA -0.141; TCGA COAD -0.051; TCGA LIHC -0.069; TCGA LUAD -0.126; TCGA LUSC -0.182; TCGA PAAD -0.088; TCGA SARC -0.066; TCGA STAD -0.19 |
hsa-miR-195-5p | CAND1 | 10 cancers: BRCA; CESC; COAD; KIRC; LUAD; LUSC; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BRCA -0.146; TCGA CESC -0.085; TCGA COAD -0.102; TCGA KIRC -0.098; TCGA LUAD -0.119; TCGA LUSC -0.226; TCGA SARC -0.059; TCGA THCA -0.094; TCGA STAD -0.236; TCGA UCEC -0.121 |
hsa-miR-195-5p | CCNE1 | 15 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PRAD; THCA; STAD; UCEC | miRNAWalker2 validate; MirTarget; miRNATAP | TCGA BRCA -0.617; TCGA CESC -0.115; TCGA COAD -0.175; TCGA ESCA -0.431; TCGA HNSC -0.31; TCGA KIRC -0.127; TCGA KIRP -0.097; TCGA LGG -0.188; TCGA LIHC -0.297; TCGA LUAD -0.518; TCGA LUSC -0.919; TCGA PRAD -0.141; TCGA THCA -0.127; TCGA STAD -0.508; TCGA UCEC -0.504 |
hsa-miR-195-5p | COPB1 | 10 cancers: BRCA; CESC; COAD; ESCA; KIRC; LUAD; LUSC; PAAD; PRAD; STAD | miRNAWalker2 validate | TCGA BRCA -0.12; TCGA CESC -0.1; TCGA COAD -0.069; TCGA ESCA -0.119; TCGA KIRC -0.066; TCGA LUAD -0.086; TCGA LUSC -0.084; TCGA PAAD -0.101; TCGA PRAD -0.082; TCGA STAD -0.159 |
hsa-miR-195-5p | KPNA1 | 9 cancers: BRCA; ESCA; KIRC; LUAD; LUSC; PRAD; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BRCA -0.122; TCGA ESCA -0.163; TCGA KIRC -0.07; TCGA LUAD -0.078; TCGA LUSC -0.23; TCGA PRAD -0.08; TCGA THCA -0.062; TCGA STAD -0.107; TCGA UCEC -0.064 |
hsa-miR-195-5p | GOLT1B | 15 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BRCA -0.166; TCGA CESC -0.074; TCGA COAD -0.099; TCGA ESCA -0.161; TCGA HNSC -0.116; TCGA KIRP -0.055; TCGA LGG -0.078; TCGA LIHC -0.103; TCGA LUAD -0.171; TCGA LUSC -0.2; TCGA PAAD -0.13; TCGA PRAD -0.072; TCGA SARC -0.089; TCGA STAD -0.237; TCGA UCEC -0.133 |
hsa-miR-195-5p | ENTPD7 | 11 cancers: BRCA; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; SARC; STAD; UCEC | MirTarget | TCGA BRCA -0.25; TCGA ESCA -0.177; TCGA HNSC -0.094; TCGA KIRC -0.08; TCGA KIRP -0.121; TCGA LGG -0.065; TCGA LUAD -0.192; TCGA LUSC -0.312; TCGA SARC -0.22; TCGA STAD -0.096; TCGA UCEC -0.08 |
hsa-miR-195-5p | SEC61A1 | 11 cancers: BRCA; COAD; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD | MirTarget | TCGA BRCA -0.186; TCGA COAD -0.079; TCGA HNSC -0.097; TCGA LIHC -0.076; TCGA LUAD -0.104; TCGA LUSC -0.156; TCGA PAAD -0.226; TCGA PRAD -0.087; TCGA SARC -0.102; TCGA THCA -0.06; TCGA STAD -0.068 |
hsa-miR-195-5p | SGPL1 | 9 cancers: BRCA; ESCA; KIRC; LUAD; LUSC; SARC; THCA; STAD; UCEC | MirTarget | TCGA BRCA -0.145; TCGA ESCA -0.172; TCGA KIRC -0.125; TCGA LUAD -0.118; TCGA LUSC -0.226; TCGA SARC -0.077; TCGA THCA -0.108; TCGA STAD -0.274; TCGA UCEC -0.072 |
hsa-miR-195-5p | SUZ12 | 9 cancers: BRCA; COAD; ESCA; KIRC; LIHC; LUAD; LUSC; STAD; UCEC | MirTarget; miRNATAP | TCGA BRCA -0.07; TCGA COAD -0.091; TCGA ESCA -0.182; TCGA KIRC -0.098; TCGA LIHC -0.051; TCGA LUAD -0.147; TCGA LUSC -0.254; TCGA STAD -0.272; TCGA UCEC -0.106 |
hsa-miR-195-5p | CALU | 10 cancers: BRCA; ESCA; KIRP; LIHC; LUAD; LUSC; PAAD; SARC; STAD; UCEC | MirTarget | TCGA BRCA -0.228; TCGA ESCA -0.126; TCGA KIRP -0.092; TCGA LIHC -0.053; TCGA LUAD -0.167; TCGA LUSC -0.177; TCGA PAAD -0.218; TCGA SARC -0.059; TCGA STAD -0.068; TCGA UCEC -0.112 |
hsa-miR-195-5p | AVL9 | 11 cancers: BRCA; CESC; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; PAAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BRCA -0.156; TCGA CESC -0.113; TCGA ESCA -0.17; TCGA HNSC -0.089; TCGA KIRC -0.197; TCGA LIHC -0.135; TCGA LUAD -0.216; TCGA LUSC -0.305; TCGA PAAD -0.176; TCGA STAD -0.267; TCGA UCEC -0.169 |
hsa-miR-195-5p | IARS | 12 cancers: BRCA; CESC; COAD; ESCA; KIRC; LGG; LIHC; LUAD; LUSC; PRAD; STAD; UCEC | MirTarget | TCGA BRCA -0.158; TCGA CESC -0.086; TCGA COAD -0.175; TCGA ESCA -0.179; TCGA KIRC -0.128; TCGA LGG -0.081; TCGA LIHC -0.12; TCGA LUAD -0.125; TCGA LUSC -0.259; TCGA PRAD -0.102; TCGA STAD -0.246; TCGA UCEC -0.095 |
hsa-miR-195-5p | RAD23B | 11 cancers: BRCA; CESC; COAD; ESCA; KIRC; LUAD; LUSC; PAAD; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BRCA -0.113; TCGA CESC -0.056; TCGA COAD -0.071; TCGA ESCA -0.222; TCGA KIRC -0.072; TCGA LUAD -0.113; TCGA LUSC -0.211; TCGA PAAD -0.081; TCGA THCA -0.246; TCGA STAD -0.179; TCGA UCEC -0.058 |
hsa-miR-195-5p | CCDC85C | 11 cancers: BRCA; CESC; HNSC; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; THCA; UCEC | MirTarget | TCGA BRCA -0.092; TCGA CESC -0.117; TCGA HNSC -0.107; TCGA LGG -0.103; TCGA LIHC -0.165; TCGA LUAD -0.087; TCGA LUSC -0.226; TCGA PAAD -0.178; TCGA PRAD -0.074; TCGA THCA -0.062; TCGA UCEC -0.121 |
hsa-miR-195-5p | SUPT16H | 9 cancers: BRCA; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; STAD; UCEC | MirTarget; miRNATAP | TCGA BRCA -0.074; TCGA COAD -0.116; TCGA ESCA -0.181; TCGA HNSC -0.052; TCGA KIRC -0.09; TCGA LUAD -0.102; TCGA LUSC -0.196; TCGA STAD -0.194; TCGA UCEC -0.086 |
hsa-miR-195-5p | YWHAQ | 12 cancers: BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BRCA -0.131; TCGA CESC -0.063; TCGA COAD -0.076; TCGA ESCA -0.172; TCGA HNSC -0.091; TCGA LIHC -0.059; TCGA LUAD -0.084; TCGA LUSC -0.179; TCGA PAAD -0.073; TCGA PRAD -0.071; TCGA STAD -0.111; TCGA UCEC -0.104 |
hsa-miR-195-5p | RTN3 | 9 cancers: BRCA; CESC; ESCA; KIRC; LIHC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BRCA -0.056; TCGA CESC -0.083; TCGA ESCA -0.152; TCGA KIRC -0.104; TCGA LIHC -0.076; TCGA PAAD -0.138; TCGA PRAD -0.086; TCGA STAD -0.149; TCGA UCEC -0.108 |
hsa-miR-195-5p | CABLES2 | 11 cancers: BRCA; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; THCA; STAD; UCEC | MirTarget | TCGA BRCA -0.277; TCGA HNSC -0.058; TCGA KIRP -0.063; TCGA LGG -0.081; TCGA LIHC -0.11; TCGA LUAD -0.155; TCGA LUSC -0.361; TCGA PAAD -0.216; TCGA THCA -0.056; TCGA STAD -0.122; TCGA UCEC -0.182 |
hsa-miR-195-5p | NRBP1 | 10 cancers: BRCA; KIRP; LGG; LIHC; LUAD; LUSC; PRAD; SARC; THCA; UCEC | MirTarget; miRNATAP | TCGA BRCA -0.154; TCGA KIRP -0.05; TCGA LGG -0.072; TCGA LIHC -0.141; TCGA LUAD -0.094; TCGA LUSC -0.177; TCGA PRAD -0.088; TCGA SARC -0.108; TCGA THCA -0.11; TCGA UCEC -0.083 |
hsa-miR-195-5p | G6PD | 9 cancers: BRCA; HNSC; KIRP; LIHC; LUAD; LUSC; PRAD; SARC; UCEC | MirTarget | TCGA BRCA -0.259; TCGA HNSC -0.106; TCGA KIRP -0.137; TCGA LIHC -0.433; TCGA LUAD -0.222; TCGA LUSC -0.53; TCGA PRAD -0.11; TCGA SARC -0.082; TCGA UCEC -0.09 |
hsa-miR-195-5p | SLC35F5 | 11 cancers: BRCA; ESCA; KIRC; KIRP; LGG; LUAD; LUSC; PAAD; SARC; STAD; UCEC | MirTarget | TCGA BRCA -0.077; TCGA ESCA -0.095; TCGA KIRC -0.204; TCGA KIRP -0.082; TCGA LGG -0.062; TCGA LUAD -0.092; TCGA LUSC -0.263; TCGA PAAD -0.114; TCGA SARC -0.104; TCGA STAD -0.117; TCGA UCEC -0.22 |
hsa-miR-195-5p | UBFD1 | 12 cancers: BRCA; CESC; ESCA; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BRCA -0.157; TCGA CESC -0.073; TCGA ESCA -0.097; TCGA KIRC -0.08; TCGA KIRP -0.058; TCGA LIHC -0.087; TCGA LUAD -0.122; TCGA LUSC -0.208; TCGA PAAD -0.158; TCGA THCA -0.112; TCGA STAD -0.187; TCGA UCEC -0.136 |
hsa-miR-195-5p | FAM91A1 | 10 cancers: BRCA; CESC; ESCA; HNSC; KIRC; LUAD; LUSC; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BRCA -0.136; TCGA CESC -0.062; TCGA ESCA -0.261; TCGA HNSC -0.108; TCGA KIRC -0.054; TCGA LUAD -0.101; TCGA LUSC -0.23; TCGA SARC -0.1; TCGA STAD -0.342; TCGA UCEC -0.141 |
hsa-miR-195-5p | SNRPB2 | 10 cancers: BRCA; CESC; COAD; HNSC; LIHC; LUAD; LUSC; PAAD; STAD; UCEC | MirTarget | TCGA BRCA -0.158; TCGA CESC -0.066; TCGA COAD -0.12; TCGA HNSC -0.089; TCGA LIHC -0.089; TCGA LUAD -0.059; TCGA LUSC -0.129; TCGA PAAD -0.106; TCGA STAD -0.108; TCGA UCEC -0.146 |
hsa-miR-195-5p | WIPI2 | 10 cancers: BRCA; CESC; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; UCEC | MirTarget; miRNATAP | TCGA BRCA -0.06; TCGA CESC -0.063; TCGA HNSC -0.091; TCGA KIRC -0.124; TCGA KIRP -0.103; TCGA LIHC -0.075; TCGA LUAD -0.06; TCGA LUSC -0.133; TCGA PAAD -0.159; TCGA UCEC -0.097 |
hsa-miR-195-5p | MAPRE1 | 11 cancers: BRCA; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BRCA -0.155; TCGA ESCA -0.117; TCGA HNSC -0.056; TCGA KIRC -0.063; TCGA LIHC -0.077; TCGA LUAD -0.07; TCGA LUSC -0.166; TCGA PAAD -0.101; TCGA PRAD -0.053; TCGA STAD -0.096; TCGA UCEC -0.154 |
hsa-miR-195-5p | ZDHHC23 | 10 cancers: BRCA; KIRC; KIRP; LGG; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BRCA -0.144; TCGA KIRC -0.361; TCGA KIRP -0.165; TCGA LGG -0.195; TCGA LUAD -0.161; TCGA LUSC -0.237; TCGA PAAD -0.198; TCGA PRAD -0.135; TCGA STAD -0.358; TCGA UCEC -0.148 |
hsa-miR-195-5p | CARM1 | 9 cancers: BRCA; CESC; ESCA; LGG; LIHC; LUAD; LUSC; PAAD; THCA | MirTarget | TCGA BRCA -0.18; TCGA CESC -0.054; TCGA ESCA -0.15; TCGA LGG -0.077; TCGA LIHC -0.103; TCGA LUAD -0.119; TCGA LUSC -0.251; TCGA PAAD -0.111; TCGA THCA -0.1 |
hsa-miR-195-5p | CHORDC1 | 9 cancers: BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; STAD | MirTarget | TCGA BRCA -0.158; TCGA CESC -0.147; TCGA COAD -0.136; TCGA ESCA -0.268; TCGA HNSC -0.178; TCGA LIHC -0.092; TCGA LUAD -0.163; TCGA LUSC -0.307; TCGA STAD -0.33 |
hsa-miR-195-5p | CAMSAP1 | 9 cancers: BRCA; COAD; ESCA; KIRP; LUAD; LUSC; PAAD; THCA; STAD | MirTarget; miRNATAP | TCGA BRCA -0.051; TCGA COAD -0.078; TCGA ESCA -0.103; TCGA KIRP -0.142; TCGA LUAD -0.075; TCGA LUSC -0.182; TCGA PAAD -0.081; TCGA THCA -0.105; TCGA STAD -0.183 |
hsa-miR-195-5p | RDH13 | 11 cancers: BRCA; CESC; COAD; HNSC; KIRP; LIHC; LUAD; LUSC; PRAD; STAD; UCEC | MirTarget | TCGA BRCA -0.144; TCGA CESC -0.122; TCGA COAD -0.103; TCGA HNSC -0.128; TCGA KIRP -0.114; TCGA LIHC -0.09; TCGA LUAD -0.131; TCGA LUSC -0.153; TCGA PRAD -0.097; TCGA STAD -0.135; TCGA UCEC -0.124 |
hsa-miR-195-5p | HMGA2 | 12 cancers: BRCA; ESCA; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; SARC; THCA; STAD; UCEC | MirTarget | TCGA BRCA -0.403; TCGA ESCA -0.575; TCGA HNSC -0.819; TCGA KIRP -0.662; TCGA LGG -0.379; TCGA LIHC -0.674; TCGA LUAD -0.455; TCGA LUSC -1.214; TCGA SARC -1.383; TCGA THCA -1.522; TCGA STAD -0.589; TCGA UCEC -0.656 |
hsa-miR-195-5p | ARFGAP1 | 11 cancers: BRCA; CESC; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; SARC; THCA; STAD | mirMAP | TCGA BRCA -0.215; TCGA CESC -0.063; TCGA HNSC -0.172; TCGA KIRP -0.06; TCGA LIHC -0.147; TCGA LUAD -0.162; TCGA LUSC -0.105; TCGA PAAD -0.169; TCGA SARC -0.084; TCGA THCA -0.067; TCGA STAD -0.071 |
hsa-miR-195-5p | CD276 | 11 cancers: BRCA; ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BRCA -0.223; TCGA ESCA -0.234; TCGA HNSC -0.202; TCGA KIRP -0.119; TCGA LUAD -0.122; TCGA LUSC -0.23; TCGA PAAD -0.185; TCGA SARC -0.131; TCGA THCA -0.246; TCGA STAD -0.103; TCGA UCEC -0.06 |
hsa-miR-195-5p | SNX8 | 9 cancers: BRCA; CESC; LIHC; LUAD; LUSC; SARC; THCA; STAD; UCEC | mirMAP | TCGA BRCA -0.119; TCGA CESC -0.07; TCGA LIHC -0.132; TCGA LUAD -0.109; TCGA LUSC -0.098; TCGA SARC -0.101; TCGA THCA -0.167; TCGA STAD -0.093; TCGA UCEC -0.079 |
hsa-miR-195-5p | SURF4 | 11 cancers: BRCA; CESC; COAD; ESCA; HNSC; LGG; LUAD; LUSC; PAAD; PRAD; SARC | mirMAP | TCGA BRCA -0.146; TCGA CESC -0.106; TCGA COAD -0.079; TCGA ESCA -0.132; TCGA HNSC -0.052; TCGA LGG -0.055; TCGA LUAD -0.063; TCGA LUSC -0.065; TCGA PAAD -0.175; TCGA PRAD -0.053; TCGA SARC -0.114 |
hsa-miR-195-5p | IGF2BP1 | 9 cancers: BRCA; CESC; HNSC; LIHC; LUAD; LUSC; SARC; STAD; UCEC | mirMAP | TCGA BRCA -0.659; TCGA CESC -0.638; TCGA HNSC -0.755; TCGA LIHC -1.287; TCGA LUAD -0.753; TCGA LUSC -1.243; TCGA SARC -0.562; TCGA STAD -0.507; TCGA UCEC -0.799 |
hsa-miR-195-5p | STX3 | 9 cancers: BRCA; CESC; HNSC; KIRP; LIHC; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BRCA -0.196; TCGA CESC -0.119; TCGA HNSC -0.13; TCGA KIRP -0.084; TCGA LIHC -0.084; TCGA PRAD -0.087; TCGA THCA -0.228; TCGA STAD -0.146; TCGA UCEC -0.144 |
hsa-miR-195-5p | DAG1 | 9 cancers: BRCA; CESC; KIRC; LUAD; LUSC; PAAD; THCA; STAD; UCEC | mirMAP | TCGA BRCA -0.056; TCGA CESC -0.068; TCGA KIRC -0.136; TCGA LUAD -0.058; TCGA LUSC -0.066; TCGA PAAD -0.2; TCGA THCA -0.093; TCGA STAD -0.146; TCGA UCEC -0.135 |
hsa-miR-195-5p | ATP2A2 | 9 cancers: BRCA; ESCA; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; STAD | miRNATAP | TCGA BRCA -0.096; TCGA ESCA -0.102; TCGA KIRC -0.055; TCGA KIRP -0.079; TCGA LIHC -0.051; TCGA LUAD -0.17; TCGA LUSC -0.268; TCGA PAAD -0.11; TCGA STAD -0.197 |
hsa-miR-195-5p | ONECUT2 | 9 cancers: BRCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; UCEC | miRNATAP | TCGA BRCA -0.46; TCGA HNSC -0.265; TCGA KIRC -0.324; TCGA KIRP -0.553; TCGA LGG -0.172; TCGA LIHC -0.192; TCGA LUAD -0.243; TCGA LUSC -0.798; TCGA UCEC -0.426 |
hsa-miR-195-5p | ZNF367 | 10 cancers: BRCA; COAD; ESCA; KIRC; LUAD; LUSC; PAAD; THCA; STAD; UCEC | miRNATAP | TCGA BRCA -0.342; TCGA COAD -0.151; TCGA ESCA -0.335; TCGA KIRC -0.101; TCGA LUAD -0.258; TCGA LUSC -0.556; TCGA PAAD -0.185; TCGA THCA -0.148; TCGA STAD -0.474; TCGA UCEC -0.246 |
hsa-miR-195-5p | AKIRIN1 | 9 cancers: BRCA; CESC; COAD; HNSC; KIRC; LGG; LUAD; PRAD; UCEC | miRNATAP | TCGA BRCA -0.075; TCGA CESC -0.071; TCGA COAD -0.061; TCGA HNSC -0.054; TCGA KIRC -0.189; TCGA LGG -0.092; TCGA LUAD -0.089; TCGA PRAD -0.1; TCGA UCEC -0.236 |
hsa-miR-195-5p | NAA25 | 10 cancers: BRCA; COAD; ESCA; KIRC; KIRP; LIHC; LUAD; LUSC; STAD; UCEC | miRNATAP | TCGA BRCA -0.081; TCGA COAD -0.136; TCGA ESCA -0.165; TCGA KIRC -0.053; TCGA KIRP -0.108; TCGA LIHC -0.096; TCGA LUAD -0.169; TCGA LUSC -0.28; TCGA STAD -0.348; TCGA UCEC -0.114 |
hsa-miR-195-5p | ZBTB34 | 9 cancers: CESC; HNSC; KIRC; KIRP; LGG; SARC; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA CESC -0.055; TCGA HNSC -0.063; TCGA KIRC -0.075; TCGA KIRP -0.069; TCGA LGG -0.059; TCGA SARC -0.096; TCGA THCA -0.053; TCGA STAD -0.115; TCGA UCEC -0.071 |
hsa-miR-195-5p | SPRYD3 | 9 cancers: CESC; COAD; KIRC; KIRP; LIHC; PAAD; PRAD; SARC; THCA | MirTarget; miRNATAP | TCGA CESC -0.07; TCGA COAD -0.057; TCGA KIRC -0.088; TCGA KIRP -0.067; TCGA LIHC -0.093; TCGA PAAD -0.104; TCGA PRAD -0.147; TCGA SARC -0.064; TCGA THCA -0.065 |
hsa-miR-195-5p | PEX5 | 9 cancers: CESC; ESCA; LIHC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget; miRNATAP | TCGA CESC -0.083; TCGA ESCA -0.131; TCGA LIHC -0.073; TCGA LUAD -0.063; TCGA LUSC -0.093; TCGA PAAD -0.121; TCGA PRAD -0.127; TCGA STAD -0.137; TCGA UCEC -0.054 |
hsa-miR-195-5p | PSAT1 | 10 cancers: COAD; ESCA; LGG; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA COAD -0.26; TCGA ESCA -0.339; TCGA LGG -0.145; TCGA LUAD -0.313; TCGA LUSC -1.056; TCGA PAAD -0.281; TCGA PRAD -0.132; TCGA SARC -0.17; TCGA STAD -0.409; TCGA UCEC -0.473 |
hsa-miR-195-5p | CPD | 10 cancers: KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; SARC; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA KIRC -0.117; TCGA KIRP -0.125; TCGA LIHC -0.129; TCGA LUAD -0.262; TCGA LUSC -0.11; TCGA PAAD -0.141; TCGA SARC -0.1; TCGA THCA -0.18; TCGA STAD -0.22; TCGA UCEC -0.126 |
hsa-miR-195-5p | ACVR2B | 9 cancers: KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; THCA; STAD; UCEC | miRNATAP | TCGA KIRC -0.146; TCGA KIRP -0.102; TCGA LIHC -0.076; TCGA LUAD -0.14; TCGA LUSC -0.194; TCGA PAAD -0.187; TCGA THCA -0.081; TCGA STAD -0.252; TCGA UCEC -0.069 |
Enriched cancer pathways of putative targets