microRNA information: hsa-miR-200a-3p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-200a-3p | miRbase |
| Accession: | MIMAT0000682 | miRbase |
| Precursor name: | hsa-mir-200a | miRbase |
| Precursor accession: | MI0000737 | miRbase |
| Symbol: | MIR200A | HGNC |
| RefSeq ID: | NR_029834 | GenBank |
| Sequence: | UAACACUGUCUGGUAACGAUGU |
Reported expression in cancers: hsa-miR-200a-3p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-200a-3p | bladder cancer | deregulation | "In other recruited 17 patients with BUC who were d ......" | 22886973 | Reverse transcription PCR; Microarray |
| hsa-miR-200a-3p | bladder cancer | deregulation | "We screened 723 miRNAs by microarray and selected ......" | 23945108 | qPCR; Microarray |
| hsa-miR-200a-3p | breast cancer | downregulation | "Expression levels of miR-200a miR-200b miR-200c mi ......" | 26201425 | qPCR |
| hsa-miR-200a-3p | breast cancer | downregulation | "Loss of expression of miR-200 family members has b ......" | 27717206 | |
| hsa-miR-200a-3p | cervical and endocervical cancer | upregulation | "Vote-counting analysis showed that up-regulation w ......" | 25920605 | |
| hsa-miR-200a-3p | colorectal cancer | downregulation | "Here we used in situ hybridization and immunohisto ......" | 23441132 | in situ hybridization; qPCR |
| hsa-miR-200a-3p | colorectal cancer | upregulation | "The processes of EMT and metastasis are highly reg ......" | 25422078 | |
| hsa-miR-200a-3p | endometrial cancer | upregulation | "We evaluated the differential expressions of miRNA ......" | 21035172 | Microarray |
| hsa-miR-200a-3p | endometrial cancer | upregulation | "Using integrated statistical analyses we identifie ......" | 25750291 | |
| hsa-miR-200a-3p | endometrial cancer | upregulation | "The expression level of miR-200a was significantly ......" | 26045795 | |
| hsa-miR-200a-3p | esophageal cancer | deregulation | "The expression profiles of miRNAs in paired EC and ......" | 23761828 | Microarray; qPCR |
| hsa-miR-200a-3p | gastric cancer | downregulation | "Although microRNA-200 miR-200 family members are t ......" | 25502084 | |
| hsa-miR-200a-3p | gastric cancer | downregulation | "To find a potentially useful prognostic predictor ......" | 26137064 | |
| hsa-miR-200a-3p | head and neck cancer | downregulation | "The miR-200 family and miR-203 were downregulated ......" | 26882562 | |
| hsa-miR-200a-3p | kidney renal cell cancer | downregulation | "The set of miRNAs with significantly decreased exp ......" | 23074016 | |
| hsa-miR-200a-3p | kidney renal cell cancer | downregulation | "Here we report that the expression of miR-200a was ......" | 25813153 | |
| hsa-miR-200a-3p | liver cancer | downregulation | "We investigated the diagnostic impact of microRNA- ......" | 24895326 | qPCR |
| hsa-miR-200a-3p | liver cancer | downregulation | "Although microRNA-200a miR-200a is frequently down ......" | 25797260 | |
| hsa-miR-200a-3p | liver cancer | downregulation | "MiR-200 family is an important regulator of epithe ......" | 25909223 | |
| hsa-miR-200a-3p | liver cancer | downregulation | "Expression of the microRNA-200 family was investig ......" | 26447841 | qPCR |
| hsa-miR-200a-3p | lung cancer | upregulation | "Among these 28 miRNAs were up-regulated in both th ......" | 27189341 | qPCR |
| hsa-miR-200a-3p | lymphoma | downregulation | "Intriguingly both hsa-miR-363 and hsa-miR-200a bel ......" | 26893685 | |
| hsa-miR-200a-3p | melanoma | deregulation | "A functional role of microRNAs miRNAs or miRs in n ......" | 20957176 | |
| hsa-miR-200a-3p | melanoma | downregulation | "Here we demonstrate a significant correlation betw ......" | 22956368 | |
| hsa-miR-200a-3p | melanoma | upregulation | "Arsenic exposed Keratinocytes Exhibit Differential ......" | 27054085 | qPCR |
| hsa-miR-200a-3p | ovarian cancer | upregulation | "To identify the micro-ribonucleic acids miRNAs exp ......" | 24816756 | qPCR |
| hsa-miR-200a-3p | ovarian cancer | upregulation | "miR 200a overexpression in advanced ovarian carcin ......" | 25374174 | qPCR; Microarray |
| hsa-miR-200a-3p | ovarian cancer | upregulation | "The prognostic value of the miR 200 family in ovar ......" | 26910180 | |
| hsa-miR-200a-3p | prostate cancer | upregulation | "Here we demonstrate the involvement of miR-200 b i ......" | 24391862 | |
| hsa-miR-200a-3p | sarcoma | deregulation | "The most significantly downregulated miRNAs were m ......" | 24027049 | |
| hsa-miR-200a-3p | thyroid cancer | upregulation | "The miR-200 family was recently identified as a su ......" | 22797360 | |
| hsa-miR-200a-3p | thyroid cancer | downregulation | "MiR 200 Regulates Epithelial Mesenchymal Transitio ......" | 25542369 |
Reported cancer pathway affected by hsa-miR-200a-3p
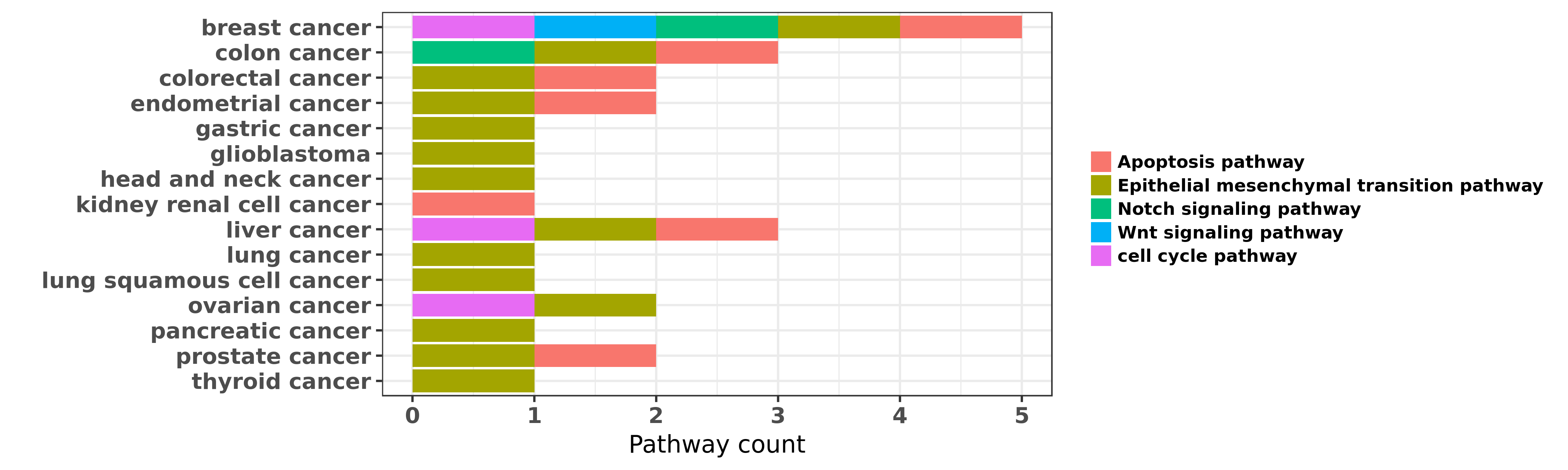
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-200a-3p | breast cancer | cell cycle pathway; Apoptosis pathway | "The genes encoding microRNAs of the human miR-200 ......" | 20514023 | Luciferase |
| hsa-miR-200a-3p | breast cancer | Epithelial mesenchymal transition pathway | "Phosphoglucose isomerase/autocrine motility factor ......" | 21389093 | |
| hsa-miR-200a-3p | breast cancer | Epithelial mesenchymal transition pathway | "Global miRNA expression profiling was performed on ......" | 22294488 | |
| hsa-miR-200a-3p | breast cancer | Epithelial mesenchymal transition pathway | "Epigenetic modulation of the miR 200 family is ass ......" | 23525011 | |
| hsa-miR-200a-3p | breast cancer | Epithelial mesenchymal transition pathway | "MiR 200 can repress breast cancer metastasis throu ......" | 24037528 | |
| hsa-miR-200a-3p | breast cancer | Epithelial mesenchymal transition pathway | "Some miRNAs especially the miR-200 family miR-9 an ......" | 25086633 | |
| hsa-miR-200a-3p | breast cancer | Wnt signaling pathway; Apoptosis pathway; Notch signaling pathway | "The miRNAs and their clusters such as the miR-200 ......" | 26712794 | |
| hsa-miR-200a-3p | colon cancer | Epithelial mesenchymal transition pathway | "MicroRNA 200 miR 200 cluster regulation by achaete ......" | 25371200 | |
| hsa-miR-200a-3p | colon cancer | Apoptosis pathway; Notch signaling pathway | "Niclosamide inhibits colon cancer progression thro ......" | 27460529 | Flow cytometry; Cell migration assay |
| hsa-miR-200a-3p | colorectal cancer | Apoptosis pathway; Epithelial mesenchymal transition pathway | "MiR 200a regulates epithelial to mesenchymal trans ......" | 24504363 | |
| hsa-miR-200a-3p | colorectal cancer | Epithelial mesenchymal transition pathway | "Epithelial-mesenchymal transition EMT and changes ......" | 25757925 | |
| hsa-miR-200a-3p | colorectal cancer | Apoptosis pathway | "Expression of miR 200a in colorectal carcinoma cel ......" | 25818799 | Cell migration assay |
| hsa-miR-200a-3p | colorectal cancer | Epithelial mesenchymal transition pathway | "The miR-200 family presented as the most powerful ......" | 27157610 | |
| hsa-miR-200a-3p | endometrial cancer | Epithelial mesenchymal transition pathway | "Tamoxifen represses miR 200 microRNAs and promotes ......" | 23295740 | |
| hsa-miR-200a-3p | endometrial cancer | Apoptosis pathway | "Moreover the study explored the involvement of mic ......" | 27497669 | Western blot; Wound Healing Assay |
| hsa-miR-200a-3p | gastric cancer | Epithelial mesenchymal transition pathway | "The microRNA-200 miR-200 family has been reported ......" | 22311119 | |
| hsa-miR-200a-3p | gastric cancer | Epithelial mesenchymal transition pathway | "In addition the microRNA miR-200 family plays a ce ......" | 25411357 | |
| hsa-miR-200a-3p | glioblastoma | Epithelial mesenchymal transition pathway | "NPV LDE 225 Erismodegib inhibits epithelial mesenc ......" | 23482671 | Luciferase; Western blot |
| hsa-miR-200a-3p | head and neck cancer | Epithelial mesenchymal transition pathway | "Epithelial-mesenchymal-transition EMT is a critica ......" | 24424572 | |
| hsa-miR-200a-3p | kidney renal cell cancer | Apoptosis pathway | "MicroRNA 200a 3p suppresses tumor proliferation an ......" | 26797273 | |
| hsa-miR-200a-3p | liver cancer | cell cycle pathway | "Expression of the microRNA-200 miR-200 family has ......" | 23760980 | Flow cytometry; MTT assay |
| hsa-miR-200a-3p | liver cancer | cell cycle pathway | "microRNA 200a is an independent prognostic factor ......" | 24009066 | Western blot |
| hsa-miR-200a-3p | liver cancer | Epithelial mesenchymal transition pathway | "We investigated the diagnostic impact of microRNA- ......" | 24895326 | |
| hsa-miR-200a-3p | liver cancer | Epithelial mesenchymal transition pathway | "Overexpression of miR 200a suppresses epithelial m ......" | 25412960 | |
| hsa-miR-200a-3p | liver cancer | Epithelial mesenchymal transition pathway | "MiR-200 family is an important regulator of epithe ......" | 25909223 | |
| hsa-miR-200a-3p | liver cancer | Epithelial mesenchymal transition pathway; Apoptosis pathway | "MicroRNA 200a inhibits epithelial mesenchymal tran ......" | 26617701 | Western blot; Wound Healing Assay; MTT assay |
| hsa-miR-200a-3p | liver cancer | Epithelial mesenchymal transition pathway | "LncRNA HULC enhances epithelial mesenchymal transi ......" | 27285757 | |
| hsa-miR-200a-3p | lung cancer | Epithelial mesenchymal transition pathway | "BMP4 depletion by miR 200 inhibits tumorigenesis a ......" | 26395571 | Western blot; Luciferase |
| hsa-miR-200a-3p | lung cancer | Epithelial mesenchymal transition pathway | "Microarray analysis using 2000 probes revealed 87 ......" | 27189341 | |
| hsa-miR-200a-3p | lung squamous cell cancer | Epithelial mesenchymal transition pathway | "The microRNA‑200 miR-200 family is a powerful re ......" | 23708087 | Western blot |
| hsa-miR-200a-3p | lung squamous cell cancer | Epithelial mesenchymal transition pathway | "The difference in miRNA expression profiles betwee ......" | 26783187 | |
| hsa-miR-200a-3p | ovarian cancer | Epithelial mesenchymal transition pathway | "Differences in miRNA expression between HGSC CCC a ......" | 24512620 | |
| hsa-miR-200a-3p | ovarian cancer | Epithelial mesenchymal transition pathway | "Epithelial mesenchymal transition associated miRNA ......" | 24952258 | |
| hsa-miR-200a-3p | ovarian cancer | cell cycle pathway | "Upregulation of microRNA 200a associates with tumo ......" | 25997962 | Colony formation |
| hsa-miR-200a-3p | ovarian cancer | Epithelial mesenchymal transition pathway | "The miR 200 family differentially regulates sensit ......" | 26025631 | |
| hsa-miR-200a-3p | pancreatic cancer | Epithelial mesenchymal transition pathway | "MiR 200a inhibits epithelial mesenchymal transitio ......" | 24521357 | |
| hsa-miR-200a-3p | prostate cancer | Epithelial mesenchymal transition pathway | "miR 200 regulates PDGF D mediated epithelial mesen ......" | 19544444 | |
| hsa-miR-200a-3p | prostate cancer | Apoptosis pathway | "Nevertheless decreased expressions of tumor suppre ......" | 26843836 | |
| hsa-miR-200a-3p | thyroid cancer | Epithelial mesenchymal transition pathway | "We identified two significantly decreased microRNA ......" | 20498632 | |
| hsa-miR-200a-3p | thyroid cancer | Epithelial mesenchymal transition pathway | "The miR 200 family regulates the epithelial mesenc ......" | 22797360 | Western blot |
| hsa-miR-200a-3p | thyroid cancer | Epithelial mesenchymal transition pathway | "MiR 200 Regulates Epithelial Mesenchymal Transitio ......" | 25542369 | Western blot |
Reported cancer prognosis affected by hsa-miR-200a-3p
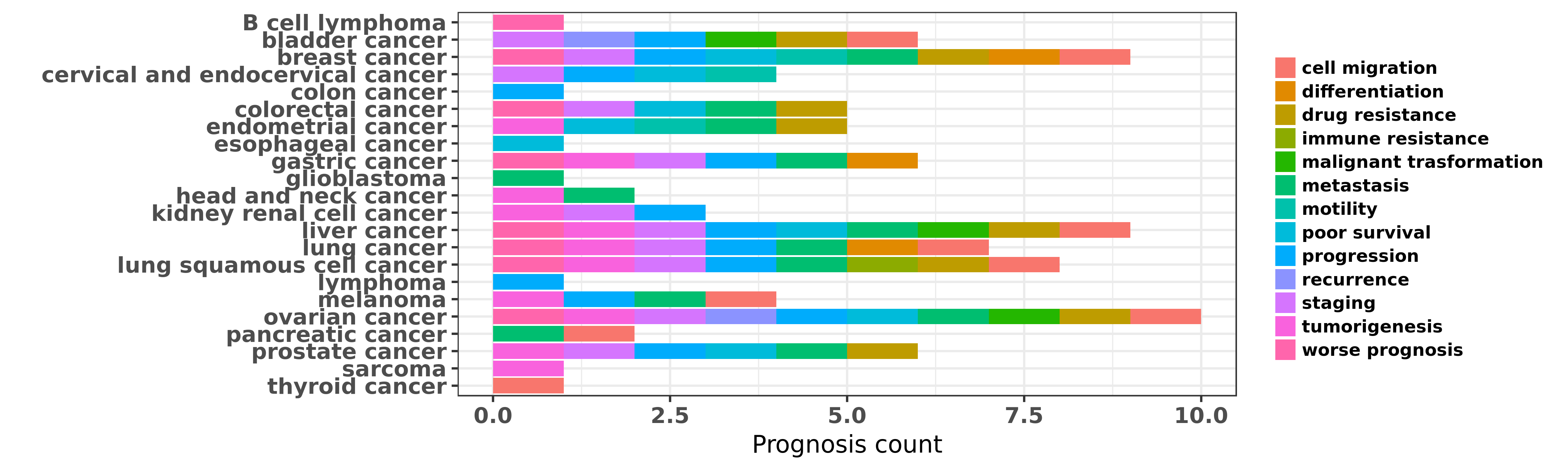
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-200a-3p | B cell lymphoma | worse prognosis | "Inhibition of ZEB1 by miR 200 characterizes Helico ......" | 24390222 | |
| hsa-miR-200a-3p | bladder cancer | drug resistance; cell migration | "miR 200 expression regulates epithelial to mesench ......" | 19671845 | Western blot; Luciferase |
| hsa-miR-200a-3p | bladder cancer | staging; progression | "Coordinated epigenetic repression of the miR 200 f ......" | 20473948 | |
| hsa-miR-200a-3p | bladder cancer | recurrence | "Datasets of GSE20418 and GSE19717 were used for an ......" | 22961325 | |
| hsa-miR-200a-3p | bladder cancer | malignant trasformation | "We screened 723 miRNAs by microarray and selected ......" | 23945108 | |
| hsa-miR-200a-3p | breast cancer | drug resistance | "The drug resistance of MCF-7/ADR cells was evaluat ......" | 18971180 | Flow cytometry; MTT assay |
| hsa-miR-200a-3p | breast cancer | metastasis | "miR 200 enhances mouse breast cancer cell coloniza ......" | 19787069 | |
| hsa-miR-200a-3p | breast cancer | progression | "The genes encoding microRNAs of the human miR-200 ......" | 20514023 | Luciferase |
| hsa-miR-200a-3p | breast cancer | motility; metastasis | "Phosphoglucose isomerase/autocrine motility factor ......" | 21389093 | |
| hsa-miR-200a-3p | breast cancer | metastasis | "Decreased expression of miRNAs of the miR-200 fami ......" | 21682933 | |
| hsa-miR-200a-3p | breast cancer | metastasis; staging; progression | "Global miRNA expression profiling was performed on ......" | 22294488 | |
| hsa-miR-200a-3p | breast cancer | differentiation | "In order to identify which miRNAs are involved in ......" | 22562546 | |
| hsa-miR-200a-3p | breast cancer | differentiation | "As the family of miR-200 microRNAs has been shown ......" | 23112837 | |
| hsa-miR-200a-3p | breast cancer | metastasis; drug resistance | "MicroRNA 200a promotes anoikis resistance and meta ......" | 23340296 | Luciferase; Colony formation |
| hsa-miR-200a-3p | breast cancer | progression | "Epigenetic modulation of the miR 200 family is ass ......" | 23525011 | |
| hsa-miR-200a-3p | breast cancer | drug resistance; progression | "Reduced expression of miR 200 family members contr ......" | 23626803 | |
| hsa-miR-200a-3p | breast cancer | metastasis | "Whole genome miR array analysis using PELP1-overex ......" | 23975430 | |
| hsa-miR-200a-3p | breast cancer | metastasis; staging; poor survival | "MiR 200 can repress breast cancer metastasis throu ......" | 24037528 | |
| hsa-miR-200a-3p | breast cancer | progression | "Loss of microRNA 200a expression correlates with t ......" | 24280074 | |
| hsa-miR-200a-3p | breast cancer | metastasis | "The expression of miRNAs in patients with primary ......" | 24846313 | |
| hsa-miR-200a-3p | breast cancer | metastasis; cell migration | "The microRNA-200 miR-200 family is found to inhibi ......" | 24925028 | |
| hsa-miR-200a-3p | breast cancer | metastasis; progression | "Members of three miRNA families i.e miR-17 miR-200 ......" | 25001613 | |
| hsa-miR-200a-3p | breast cancer | worse prognosis | "Some miRNAs especially the miR-200 family miR-9 an ......" | 25086633 | |
| hsa-miR-200a-3p | breast cancer | drug resistance | "We created a doxorubicin-resistant MCF-7 MCF-/Adr ......" | 25451164 | MTT assay |
| hsa-miR-200a-3p | breast cancer | drug resistance | "Therefore we investigated the role of known EMT re ......" | 25746005 | |
| hsa-miR-200a-3p | breast cancer | progression | "ADAM12 L is a direct target of the miR 29 and miR ......" | 25886595 | Western blot; Luciferase |
| hsa-miR-200a-3p | breast cancer | metastasis | "The upregulation of fibronectin and lysyl oxidase ......" | 26068592 | |
| hsa-miR-200a-3p | breast cancer | poor survival; cell migration; metastasis | "miR 200a inhibits migration of triple negative bre ......" | 26088362 | |
| hsa-miR-200a-3p | breast cancer | metastasis | "These included miR-141 miR-144 miR-193b miR-200a m ......" | 26785733 | |
| hsa-miR-200a-3p | breast cancer | differentiation | "miR 9 and miR 200 Regulate PDGFRβ Mediated Endoth ......" | 27402080 | |
| hsa-miR-200a-3p | breast cancer | progression; poor survival | "We identified eight microRNAs miR-10a miR-10b miR- ......" | 27433802 | |
| hsa-miR-200a-3p | breast cancer | poor survival | "In this multicenter study a 10-miRNA classifier in ......" | 27566954 | |
| hsa-miR-200a-3p | breast cancer | drug resistance | "Loss of expression of miR-200 family members has b ......" | 27717206 | |
| hsa-miR-200a-3p | breast cancer | drug resistance | "Specifically we will discuss key miRNAs involved i ......" | 27721895 | |
| hsa-miR-200a-3p | cervical and endocervical cancer | poor survival; motility | "Using an established PCR-based miRNA assay to anal ......" | 20124485 | |
| hsa-miR-200a-3p | cervical and endocervical cancer | staging; progression | "An initial screening of miRNA expression was perfo ......" | 26171195 | |
| hsa-miR-200a-3p | colon cancer | progression | "Niclosamide inhibits colon cancer progression thro ......" | 27460529 | Flow cytometry; Cell migration assay |
| hsa-miR-200a-3p | colorectal cancer | metastasis | "Applying real-time PCR we detected the expression ......" | 21827717 | |
| hsa-miR-200a-3p | colorectal cancer | metastasis | "Although the microRNA-200 miR-200 family is a cruc ......" | 22735571 | |
| hsa-miR-200a-3p | colorectal cancer | metastasis; staging | "Here we used in situ hybridization and immunohisto ......" | 23441132 | |
| hsa-miR-200a-3p | colorectal cancer | staging; metastasis | "In the first phase we selected candidate miRNAs as ......" | 23982750 | |
| hsa-miR-200a-3p | colorectal cancer | worse prognosis; poor survival | "MiR 200a regulates epithelial to mesenchymal trans ......" | 24504363 | |
| hsa-miR-200a-3p | colorectal cancer | poor survival; staging | "Role of miR 200 family members in survival of colo ......" | 24510588 | |
| hsa-miR-200a-3p | colorectal cancer | metastasis | "The processes of EMT and metastasis are highly reg ......" | 25422078 | |
| hsa-miR-200a-3p | colorectal cancer | drug resistance | "Epithelial-mesenchymal transition EMT and changes ......" | 25757925 | |
| hsa-miR-200a-3p | colorectal cancer | metastasis | "Expression of miR 200a in colorectal carcinoma cel ......" | 25818799 | Cell migration assay |
| hsa-miR-200a-3p | colorectal cancer | metastasis | "MiR 200 suppresses metastases of colorectal cancer ......" | 26242262 | Luciferase |
| hsa-miR-200a-3p | colorectal cancer | worse prognosis; poor survival | "The miR-200 family presented as the most powerful ......" | 27157610 | |
| hsa-miR-200a-3p | colorectal cancer | worse prognosis | "Plasma miR 122 and miR 200 family are prognostic m ......" | 27632639 | |
| hsa-miR-200a-3p | endometrial cancer | tumorigenesis | "We compared the expression profiles of 723 human m ......" | 21897839 | |
| hsa-miR-200a-3p | endometrial cancer | motility; drug resistance | "Tamoxifen represses miR 200 microRNAs and promotes ......" | 23295740 | |
| hsa-miR-200a-3p | endometrial cancer | tumorigenesis | "The miRNA profiles were analyzed by miRNA microarr ......" | 24742567 | |
| hsa-miR-200a-3p | endometrial cancer | metastasis; poor survival | "The expression level of miR-200a was significantly ......" | 26045795 | |
| hsa-miR-200a-3p | endometrial cancer | metastasis | "The abnormal expression of the long noncoding RNA ......" | 27693631 | |
| hsa-miR-200a-3p | esophageal cancer | poor survival | "In this study we selected 10 miRNAs and analyzed t ......" | 20309880 | |
| hsa-miR-200a-3p | gastric cancer | tumorigenesis | "The effect of microRNA abnormalities in carcinogen ......" | 20484038 | |
| hsa-miR-200a-3p | gastric cancer | differentiation | "The microRNA-200 miR-200 family has been reported ......" | 22311119 | |
| hsa-miR-200a-3p | gastric cancer | worse prognosis | "Integrated microRNA network analyses identify a po ......" | 24352645 | |
| hsa-miR-200a-3p | gastric cancer | metastasis | "In addition the microRNA miR-200 family plays a ce ......" | 25411357 | |
| hsa-miR-200a-3p | gastric cancer | tumorigenesis; progression | "Although microRNA-200 miR-200 family members are t ......" | 25502084 | Wound Healing Assay |
| hsa-miR-200a-3p | gastric cancer | staging; metastasis | "In the first step of this study preliminary experi ......" | 26233325 | |
| hsa-miR-200a-3p | glioblastoma | metastasis | "We determined CSF levels of several cancer-associa ......" | 22492962 | |
| hsa-miR-200a-3p | head and neck cancer | metastasis; tumorigenesis | "Epithelial-mesenchymal-transition EMT is a critica ......" | 24424572 | |
| hsa-miR-200a-3p | kidney renal cell cancer | progression; tumorigenesis | "MicroRNA-429 miR-429 a short noncoding RNA belongi ......" | 25953723 | |
| hsa-miR-200a-3p | kidney renal cell cancer | staging | "Comparisons of RCC and BC expression signatures re ......" | 27357429 | |
| hsa-miR-200a-3p | liver cancer | malignant trasformation | "Our study identified and validated miR-224 overexp ......" | 18433021 | |
| hsa-miR-200a-3p | liver cancer | cell migration | "MicroRNA 200a and 200b mediated hepatocellular car ......" | 22868917 | Cell migration assay |
| hsa-miR-200a-3p | liver cancer | metastasis | "Epigenetic activation of the MiR 200 family contri ......" | 23222811 | |
| hsa-miR-200a-3p | liver cancer | drug resistance | "Expression of the microRNA-200 miR-200 family has ......" | 23760980 | Flow cytometry; MTT assay |
| hsa-miR-200a-3p | liver cancer | metastasis; progression | "The aim of this study was to assess the role of th ......" | 24135722 | |
| hsa-miR-200a-3p | liver cancer | staging; progression | "An expression analysis of miR 200a in serum and li ......" | 25203708 | |
| hsa-miR-200a-3p | liver cancer | poor survival; worse prognosis | "Eleven miRNAs miR- miR-19a miR-101-3p miR-199a-5p ......" | 25275448 | |
| hsa-miR-200a-3p | liver cancer | worse prognosis; metastasis; progression; poor survival | "miR 200a suppresses cell growth and migration by t ......" | 25482402 | |
| hsa-miR-200a-3p | liver cancer | metastasis | "MicroRNA 200a suppresses metastatic potential of s ......" | 25797260 | |
| hsa-miR-200a-3p | liver cancer | tumorigenesis; cell migration; metastasis | "MiR-200 family is an important regulator of epithe ......" | 25909223 | |
| hsa-miR-200a-3p | liver cancer | metastasis; tumorigenesis | "LncRNA HULC enhances epithelial mesenchymal transi ......" | 27285757 | |
| hsa-miR-200a-3p | lung cancer | metastasis; worse prognosis; tumorigenesis | "miR 200 Inhibits lung adenocarcinoma cell invasion ......" | 21115742 | |
| hsa-miR-200a-3p | lung cancer | metastasis | "The Notch ligand Jagged2 promotes lung adenocarcin ......" | 21403400 | |
| hsa-miR-200a-3p | lung cancer | differentiation | "The rBM 3-D culture-specific miRNA profile was hig ......" | 23036707 | |
| hsa-miR-200a-3p | lung cancer | metastasis | "Loss of miR-200s has been shown to enhance cancer ......" | 24791940 | |
| hsa-miR-200a-3p | lung cancer | metastasis; cell migration | "The miR 200 family and the miR 183~96~182 cluster ......" | 25798833 | |
| hsa-miR-200a-3p | lung cancer | staging; tumorigenesis | "MicroRNA-200 miR-200 has emerged as a regulator of ......" | 26314828 | |
| hsa-miR-200a-3p | lung cancer | metastasis; tumorigenesis | "BMP4 depletion by miR 200 inhibits tumorigenesis a ......" | 26395571 | Western blot; Luciferase |
| hsa-miR-200a-3p | lung cancer | staging | "Microarray analysis using 2000 probes revealed 87 ......" | 27189341 | |
| hsa-miR-200a-3p | lung cancer | progression | "Interestingly mir-200 family mir-200a mir-200b and ......" | 27695346 | |
| hsa-miR-200a-3p | lung squamous cell cancer | metastasis; immune resistance | "The microRNA‑200 miR-200 family is a powerful re ......" | 23708087 | Western blot |
| hsa-miR-200a-3p | lung squamous cell cancer | worse prognosis; staging | "The miRNA family miR-200 has been associated with ......" | 25003366 | |
| hsa-miR-200a-3p | lung squamous cell cancer | drug resistance; cell migration | "MicroRNA 200a Targets EGFR and c Met to Inhibit Mi ......" | 26184032 | |
| hsa-miR-200a-3p | lung squamous cell cancer | progression; tumorigenesis | "The microRNA miR-200 family has been demonstrated ......" | 27602157 | |
| hsa-miR-200a-3p | lymphoma | progression | "The profiles of miRNAs in conjunctival MALT lympho ......" | 22183793 | Luciferase |
| hsa-miR-200a-3p | melanoma | metastasis; progression; cell migration | "A functional role of microRNAs miRNAs or miRs in n ......" | 20957176 | |
| hsa-miR-200a-3p | melanoma | progression; metastasis | "Loss of microRNA 200a and c and microRNA 203 expre ......" | 22956368 | |
| hsa-miR-200a-3p | melanoma | tumorigenesis | "Arsenic exposed Keratinocytes Exhibit Differential ......" | 27054085 | |
| hsa-miR-200a-3p | ovarian cancer | worse prognosis | "The microRNA expression profiles were examined usi ......" | 18451233 | |
| hsa-miR-200a-3p | ovarian cancer | staging | "Levels of 8 microRNAs miR-21 miR-141 miR-200a miR- ......" | 18589210 | |
| hsa-miR-200a-3p | ovarian cancer | staging; poor survival; recurrence; cell migration | "A miR 200 microRNA cluster as prognostic marker in ......" | 19501389 | |
| hsa-miR-200a-3p | ovarian cancer | tumorigenesis | "Regulation of miR 200 family microRNAs and ZEB tra ......" | 19854497 | Luciferase |
| hsa-miR-200a-3p | ovarian cancer | worse prognosis | "We highlight the role of the let-7 and miR-200 fam ......" | 20083225 | |
| hsa-miR-200a-3p | ovarian cancer | progression; poor survival; drug resistance | "The miR 200 family controls beta tubulin III expre ......" | 21051560 | |
| hsa-miR-200a-3p | ovarian cancer | progression | "MicroRNA 200a inhibits CD133/1+ ovarian cancer ste ......" | 21529905 | Western blot; Luciferase; Wound Healing Assay |
| hsa-miR-200a-3p | ovarian cancer | poor survival; recurrence | "In this study miR-187 and miR-200a were found to b ......" | 21725366 | |
| hsa-miR-200a-3p | ovarian cancer | worse prognosis; staging | "Two families of miRNAs miR-200 and let-7 are frequ ......" | 23237306 | |
| hsa-miR-200a-3p | ovarian cancer | staging | "The biphasic expression pattern of miR 200a and E ......" | 23888941 | |
| hsa-miR-200a-3p | ovarian cancer | drug resistance | "Sequence variation among members of the miR 200 mi ......" | 24447705 | |
| hsa-miR-200a-3p | ovarian cancer | poor survival; progression; worse prognosis | "The role of miR 200a in vasculogenic mimicry and i ......" | 24503464 | Western blot; Luciferase |
| hsa-miR-200a-3p | ovarian cancer | progression; poor survival | "Differences in miRNA expression between HGSC CCC a ......" | 24512620 | |
| hsa-miR-200a-3p | ovarian cancer | metastasis; recurrence | "We recently determined that the ectopic over-expre ......" | 24802724 | |
| hsa-miR-200a-3p | ovarian cancer | drug resistance | "Hybridization was carried out on miRNA microarray ......" | 24805828 | |
| hsa-miR-200a-3p | ovarian cancer | metastasis; drug resistance | "For example deficiencies of enzymes including Dice ......" | 24822185 | |
| hsa-miR-200a-3p | ovarian cancer | progression; metastasis | "Epithelial mesenchymal transition associated miRNA ......" | 24952258 | |
| hsa-miR-200a-3p | ovarian cancer | poor survival; staging; progression | "Clinicopathological and prognostic implications of ......" | 24966949 | |
| hsa-miR-200a-3p | ovarian cancer | drug resistance | "Involvement of miR 200a in chemosensitivity regula ......" | 25327865 | Western blot; Luciferase |
| hsa-miR-200a-3p | ovarian cancer | staging; progression | "miR 200a overexpression in advanced ovarian carcin ......" | 25374174 | |
| hsa-miR-200a-3p | ovarian cancer | poor survival | "The authors used quantitative polymerase chain rea ......" | 25556270 | Western blot |
| hsa-miR-200a-3p | ovarian cancer | drug resistance | "Upregulation of microRNA 200a associates with tumo ......" | 25997962 | Colony formation |
| hsa-miR-200a-3p | ovarian cancer | progression; staging; worse prognosis; poor survival | "Expression of serum miR 200a miR 200b and miR 200c ......" | 26063644 | |
| hsa-miR-200a-3p | ovarian cancer | progression; poor survival | "The prognostic value of the miR 200 family in ovar ......" | 26910180 | |
| hsa-miR-200a-3p | ovarian cancer | malignant trasformation; staging; metastasis | "Diagnostic and prognostic relevance of circulating ......" | 26943577 | |
| hsa-miR-200a-3p | ovarian cancer | metastasis | "The miR-200 family especially miR-200c has been sh ......" | 27601996 | |
| hsa-miR-200a-3p | ovarian cancer | poor survival | "Furthermore some of the identified DNA-methylated ......" | 27746113 | |
| hsa-miR-200a-3p | ovarian cancer | malignant trasformation | "Circulating Cell Free miR 373 miR 200a miR 200b an ......" | 27753009 | |
| hsa-miR-200a-3p | pancreatic cancer | metastasis | "Additionally the miR-200 family regulates several ......" | 24040120 | Luciferase |
| hsa-miR-200a-3p | pancreatic cancer | cell migration | "MiR 200a inhibits epithelial mesenchymal transitio ......" | 24521357 | |
| hsa-miR-200a-3p | prostate cancer | staging | "Biochemical relapse following radical prostatectom ......" | 22161972 | |
| hsa-miR-200a-3p | prostate cancer | metastasis | "Regulation of epithelial plasticity by miR 424 and ......" | 24193225 | |
| hsa-miR-200a-3p | prostate cancer | metastasis; progression | "Here we demonstrate the involvement of miR-200 b i ......" | 24391862 | |
| hsa-miR-200a-3p | prostate cancer | poor survival | "Non-responders to docetaxel and patients with shor ......" | 24714754 | |
| hsa-miR-200a-3p | prostate cancer | progression; tumorigenesis | "Expression of 1205 human miRNAs and miRNA*s were e ......" | 25768283 | |
| hsa-miR-200a-3p | prostate cancer | drug resistance | "Nevertheless decreased expressions of tumor suppre ......" | 26843836 | |
| hsa-miR-200a-3p | sarcoma | tumorigenesis | "MiR-141 which belong to miR-200 family take a part ......" | 24307282 | |
| hsa-miR-200a-3p | thyroid cancer | cell migration | "However specific miRNAs are downregulated in ATC s ......" | 25202329 |
Reported gene related to hsa-miR-200a-3p
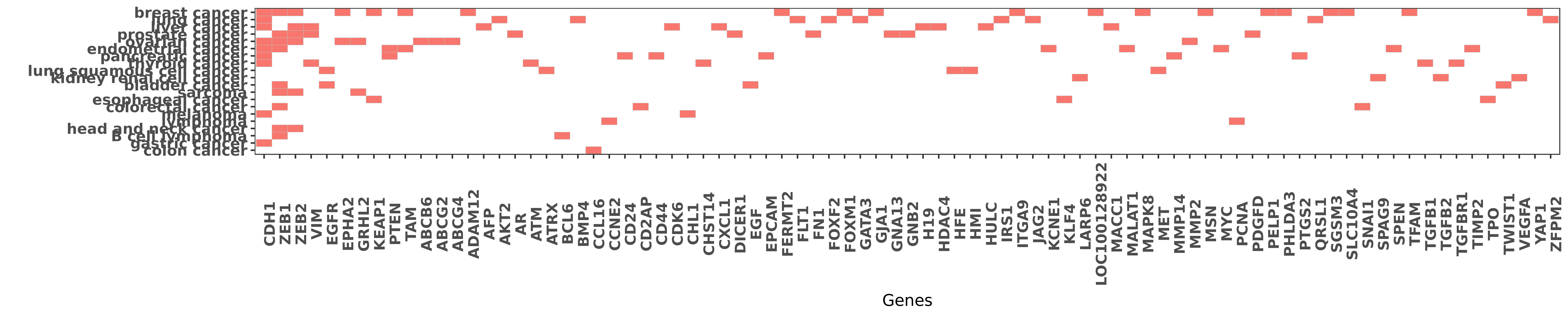
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-200a-3p | breast cancer | CDH1 | "β1 integrin inhibition elicits a prometastatic sw ......" | 24518294 |
| hsa-miR-200a-3p | breast cancer | CDH1 | "These findings are surprising since the miR-200 fa ......" | 19787069 |
| hsa-miR-200a-3p | breast cancer | CDH1 | "Here we demonstrate that the tumor suppressive miR ......" | 26062653 |
| hsa-miR-200a-3p | breast cancer | CDH1 | "We establish that miR-200a directs cell migration ......" | 26088362 |
| hsa-miR-200a-3p | endometrial cancer | CDH1 | "Moreover the study explored the involvement of mic ......" | 27497669 |
| hsa-miR-200a-3p | gastric cancer | CDH1 | "E-cadherin expression was partially restored by tr ......" | 20484038 |
| hsa-miR-200a-3p | liver cancer | CDH1 | "Spearman's Rho analysis revealed a significant neg ......" | 24895326 |
| hsa-miR-200a-3p | liver cancer | CDH1 | "We found that overexpression of miR-200 family mem ......" | 22868917 |
| hsa-miR-200a-3p | lung cancer | CDH1 | "However the expression of miR-200a was not signifi ......" | 27189341 |
| hsa-miR-200a-3p | melanoma | CDH1 | "MiR-200 in situ hybridization and E-cadherin immun ......" | 22956368 |
| hsa-miR-200a-3p | ovarian cancer | CDH1 | "The biphasic expression pattern of miR 200a and E ......" | 23888941 |
| hsa-miR-200a-3p | ovarian cancer | CDH1 | "Our findings showed that miR-101 represents a redu ......" | 24677166 |
| hsa-miR-200a-3p | ovarian cancer | CDH1 | "MicroRNA 200a inhibits CD133/1+ ovarian cancer ste ......" | 21529905 |
| hsa-miR-200a-3p | pancreatic cancer | CDH1 | "In a panel of 23 pancreatic cell lines we observed ......" | 20551052 |
| hsa-miR-200a-3p | thyroid cancer | CDH1 | "Restoration of miR-200 expression by pre-miR-200a/ ......" | 25542369 |
| hsa-miR-200a-3p | B cell lymphoma | ZEB1 | "Inhibition of ZEB1 by miR 200 characterizes Helico ......" | 24390222 |
| hsa-miR-200a-3p | bladder cancer | ZEB1 | "ZEB1 is also known to repress miR-200c-141 transcr ......" | 20473948 |
| hsa-miR-200a-3p | breast cancer | ZEB1 | "Conversely ectopic expression of miR-200 inhibited ......" | 27402080 |
| hsa-miR-200a-3p | breast cancer | ZEB1 | "Concomitant with miR-200 decrease there was an inc ......" | 23626803 |
| hsa-miR-200a-3p | breast cancer | ZEB1 | "MiR 200 can repress breast cancer metastasis throu ......" | 24037528 |
| hsa-miR-200a-3p | colorectal cancer | ZEB1 | "MiR 200 suppresses metastases of colorectal cancer ......" | 26242262 |
| hsa-miR-200a-3p | endometrial cancer | ZEB1 | "This finding was accompanied by a sharp downregula ......" | 23743934 |
| hsa-miR-200a-3p | head and neck cancer | ZEB1 | "Furthermore our data suggest that the promoter hyp ......" | 24424572 |
| hsa-miR-200a-3p | ovarian cancer | ZEB1 | "We used qRT-PCR to examine expression of the miR-2 ......" | 19854497 |
| hsa-miR-200a-3p | ovarian cancer | ZEB1 | "GRHL2 miR 200 ZEB1 maintains the epithelial status ......" | 26887977 |
| hsa-miR-200a-3p | prostate cancer | ZEB1 | "Recent studies have shown that the miR-200 family ......" | 19544444 |
| hsa-miR-200a-3p | sarcoma | ZEB1 | "This circuit comprises the microRNA 200 miR-200 fa ......" | 27402864 |
| hsa-miR-200a-3p | sarcoma | ZEB1 | "Moreover a series of loss-of-function and gain-of- ......" | 24815002 |
| hsa-miR-200a-3p | breast cancer | ZEB2 | "This switch involved activation of the transformin ......" | 24518294 |
| hsa-miR-200a-3p | breast cancer | ZEB2 | "These findings are surprising since the miR-200 fa ......" | 19787069 |
| hsa-miR-200a-3p | head and neck cancer | ZEB2 | "Furthermore our data suggest that the promoter hyp ......" | 24424572 |
| hsa-miR-200a-3p | liver cancer | ZEB2 | "MicroRNA 200a suppresses metastatic potential of s ......" | 25797260 |
| hsa-miR-200a-3p | ovarian cancer | ZEB2 | "We used qRT-PCR to examine expression of the miR-2 ......" | 19854497 |
| hsa-miR-200a-3p | ovarian cancer | ZEB2 | "MicroRNA 200a inhibits CD133/1+ ovarian cancer ste ......" | 21529905 |
| hsa-miR-200a-3p | prostate cancer | ZEB2 | "Recent studies have shown that the miR-200 family ......" | 19544444 |
| hsa-miR-200a-3p | sarcoma | ZEB2 | "Moreover a series of loss-of-function and gain-of- ......" | 24815002 |
| hsa-miR-200a-3p | breast cancer | EPHA2 | "Our studies propose EphA2 as a novel and important ......" | 22562546 |
| hsa-miR-200a-3p | breast cancer | EPHA2 | "miR 200a inhibits migration of triple negative bre ......" | 26088362 |
| hsa-miR-200a-3p | ovarian cancer | EPHA2 | "In addition our results suggested that miR-200a in ......" | 24503464 |
| hsa-miR-200a-3p | endometrial cancer | PTEN | "PTEN might be a potential target of miR-141 and mi ......" | 24742567 |
| hsa-miR-200a-3p | pancreatic cancer | PTEN | "Re expression of miR 200 by novel approaches regul ......" | 22637745 |
| hsa-miR-200a-3p | pancreatic cancer | PTEN | "Anti tumor activity of a novel compound CDF is med ......" | 21408027 |
| hsa-miR-200a-3p | liver cancer | VIM | "Spearman's Rho analysis revealed a significant neg ......" | 24895326 |
| hsa-miR-200a-3p | prostate cancer | VIM | "In contrast mesenchymal markers fibronectin and vi ......" | 24391862 |
| hsa-miR-200a-3p | thyroid cancer | VIM | "Restoration of miR-200 expression by pre-miR-200a/ ......" | 25542369 |
| hsa-miR-200a-3p | liver cancer | AFP | "Real-time quantitative PCR and enzyme-linked immun ......" | 25203708 |
| hsa-miR-200a-3p | liver cancer | AFP | "Multivariate analysis demonstrated that AFP satell ......" | 25275448 |
| hsa-miR-200a-3p | thyroid cancer | ATM | "However specific miRNAs are downregulated in ATC s ......" | 25202329 |
| hsa-miR-200a-3p | thyroid cancer | ATM | "This study was set to study the molecular mechanis ......" | 25542369 |
| hsa-miR-200a-3p | pancreatic cancer | CD44 | "Pancreatic cancer cells with EMT phenotype display ......" | 24521357 |
| hsa-miR-200a-3p | pancreatic cancer | CD44 | "In the current study we showed for the first time ......" | 21408027 |
| hsa-miR-200a-3p | bladder cancer | EGFR | "Protein expression and signaling pathway modulatio ......" | 19671845 |
| hsa-miR-200a-3p | lung squamous cell cancer | EGFR | "MicroRNA 200a Targets EGFR and c Met to Inhibit Mi ......" | 26184032 |
| hsa-miR-200a-3p | ovarian cancer | GRHL2 | "GRHL2 miR 200 ZEB1 maintains the epithelial status ......" | 26887977 |
| hsa-miR-200a-3p | sarcoma | GRHL2 | "This circuit comprises the microRNA 200 miR-200 fa ......" | 27402864 |
| hsa-miR-200a-3p | breast cancer | KEAP1 | "miR 200a regulates Nrf2 activation by targeting Ke ......" | 21926171 |
| hsa-miR-200a-3p | esophageal cancer | KEAP1 | "MSA could also significantly induce miR-200a expre ......" | 26341629 |
| hsa-miR-200a-3p | lung cancer | QRSL1 | "It was reported that miR-200 can activate PI3K/AKT ......" | 26314828 |
| hsa-miR-200a-3p | lung cancer | QRSL1 | "Mechanistically Jagged2 was found to promote metas ......" | 21403400 |
| hsa-miR-200a-3p | breast cancer | TAM | "miR-200 family expression was progressively reduce ......" | 23626803 |
| hsa-miR-200a-3p | endometrial cancer | TAM | "When treated with TAM ECC-1 and Ishikawa cells wer ......" | 23295740 |
| hsa-miR-200a-3p | ovarian cancer | ABCB6 | "Furthermore miR-200a regulation of chemoresistance ......" | 25327865 |
| hsa-miR-200a-3p | ovarian cancer | ABCG2 | "Finally the interaction between miR-200a and ABCG2 ......" | 25327865 |
| hsa-miR-200a-3p | ovarian cancer | ABCG4 | "Furthermore miR-200a regulation of chemoresistance ......" | 25327865 |
| hsa-miR-200a-3p | breast cancer | ADAM12 | "ADAM12 L is a direct target of the miR 29 and miR ......" | 25886595 |
| hsa-miR-200a-3p | lung cancer | AKT2 | "These results suggest that AKT2 can regulate miR-2 ......" | 27189341 |
| hsa-miR-200a-3p | prostate cancer | AR | "We identified miR-200 b as a downstream target of ......" | 24391862 |
| hsa-miR-200a-3p | lung squamous cell cancer | ATRX | "Additionally miR-200 family downregulates HNRNPR3 ......" | 23708087 |
| hsa-miR-200a-3p | B cell lymphoma | BCL6 | "In 30 H pylori-positive and 30 H pylori-negative g ......" | 24390222 |
| hsa-miR-200a-3p | lung cancer | BMP4 | "BMP4 depletion by miR 200 inhibits tumorigenesis a ......" | 26395571 |
| hsa-miR-200a-3p | colon cancer | CCL16 | "This observation indicates a common molecular axis ......" | 25832648 |
| hsa-miR-200a-3p | lymphoma | CCNE2 | "Targetscan analysis suggested cyclin E2 as potenti ......" | 22183793 |
| hsa-miR-200a-3p | pancreatic cancer | CD24 | "Pancreatic cancer cells with EMT phenotype display ......" | 24521357 |
| hsa-miR-200a-3p | colorectal cancer | CD2AP | "Importantly epigenetic silencing of the miR-200 fa ......" | 27157610 |
| hsa-miR-200a-3p | liver cancer | CDK6 | "microRNA 200a is an independent prognostic factor ......" | 24009066 |
| hsa-miR-200a-3p | melanoma | CHL1 | "Overall our findings call into question the genera ......" | 20957176 |
| hsa-miR-200a-3p | thyroid cancer | CHST14 | "We identified two significantly decreased microRNA ......" | 20498632 |
| hsa-miR-200a-3p | liver cancer | CXCL1 | "Using computational analysis we identified the mic ......" | 27542259 |
| hsa-miR-200a-3p | prostate cancer | DICER1 | "These findings suggest possibilities that miR-200a ......" | 25768283 |
| hsa-miR-200a-3p | bladder cancer | EGF | "miR 200 expression regulates epithelial to mesench ......" | 19671845 |
| hsa-miR-200a-3p | pancreatic cancer | EPCAM | "In the current study we showed for the first time ......" | 21408027 |
| hsa-miR-200a-3p | breast cancer | FERMT2 | "Here we report that Kindlin 2 markedly downregulat ......" | 23483548 |
| hsa-miR-200a-3p | lung cancer | FLT1 | "Forced miR-200 expression suppressed Flt1 levels i ......" | 21115742 |
| hsa-miR-200a-3p | prostate cancer | FN1 | "In contrast mesenchymal markers fibronectin and vi ......" | 24391862 |
| hsa-miR-200a-3p | lung cancer | FOXF2 | "The miR 200 family and the miR 183~96~182 cluster ......" | 25798833 |
| hsa-miR-200a-3p | breast cancer | FOXM1 | "SUMOylation of FOXM1B alters its transcriptional a ......" | 24918286 |
| hsa-miR-200a-3p | lung cancer | GATA3 | "Reciprocally miR-200 inhibited expression of Gata3 ......" | 21403400 |
| hsa-miR-200a-3p | breast cancer | GJA1 | "Identification of miR 200a as a novel suppressor o ......" | 26283635 |
| hsa-miR-200a-3p | prostate cancer | GNA13 | "MicroRNA 182 and microRNA 200a control G protein s ......" | 23329838 |
| hsa-miR-200a-3p | prostate cancer | GNB2 | "MicroRNA 182 and microRNA 200a control G protein s ......" | 23329838 |
| hsa-miR-200a-3p | liver cancer | H19 | "Epigenetic activation of the MiR 200 family contri ......" | 23222811 |
| hsa-miR-200a-3p | liver cancer | HDAC4 | "Furthermore our results suggested that the histone ......" | 21837748 |
| hsa-miR-200a-3p | lung squamous cell cancer | HFE | "Additionally miR-200 family downregulates HNRNPR3 ......" | 23708087 |
| hsa-miR-200a-3p | lung squamous cell cancer | HMI | "The miR-200 family and these potential targets are ......" | 23708087 |
| hsa-miR-200a-3p | liver cancer | HULC | "LncRNA HULC enhances epithelial mesenchymal transi ......" | 27285757 |
| hsa-miR-200a-3p | lung cancer | IRS1 | "Taken together our results suggest that miR-200 ma ......" | 26314828 |
| hsa-miR-200a-3p | breast cancer | ITGA9 | "β1 integrin inhibition elicits a prometastatic sw ......" | 24518294 |
| hsa-miR-200a-3p | lung cancer | JAG2 | "The Notch ligand Jagged2 promotes lung adenocarcin ......" | 21403400 |
| hsa-miR-200a-3p | endometrial cancer | KCNE1 | "Furthermore it was observed that the expression pa ......" | 26788150 |
| hsa-miR-200a-3p | esophageal cancer | KLF4 | "Moreover MSA-induced miR-200a expression was depen ......" | 26341629 |
| hsa-miR-200a-3p | kidney renal cell cancer | LARP6 | "In addition our study uncovered that miR-200a-3p d ......" | 26797273 |
| hsa-miR-200a-3p | breast cancer | LOC100128922 | "Identification of miR 200a as a novel suppressor o ......" | 26283635 |
| hsa-miR-200a-3p | liver cancer | MACC1 | "miR 200a suppresses cell growth and migration by t ......" | 25482402 |
| hsa-miR-200a-3p | endometrial cancer | MALAT1 | "The abnormal expression of the long noncoding RNA ......" | 27693631 |
| hsa-miR-200a-3p | breast cancer | MAPK8 | "SUMOylation of FOXM1B alters its transcriptional a ......" | 24918286 |
| hsa-miR-200a-3p | lung squamous cell cancer | MET | "MicroRNA 200a Targets EGFR and c Met to Inhibit Mi ......" | 26184032 |
| hsa-miR-200a-3p | pancreatic cancer | MMP14 | "Re expression of miR 200 by novel approaches regul ......" | 22637745 |
| hsa-miR-200a-3p | ovarian cancer | MMP2 | "Interestingly our findings show that catalpol trea ......" | 25347277 |
| hsa-miR-200a-3p | breast cancer | MSN | "MiR 200 can repress breast cancer metastasis throu ......" | 24037528 |
| hsa-miR-200a-3p | endometrial cancer | MYC | "Tamoxifen represses miR 200 microRNAs and promotes ......" | 23295740 |
| hsa-miR-200a-3p | lymphoma | PCNA | "Targetscan analysis suggested cyclin E2 as potenti ......" | 22183793 |
| hsa-miR-200a-3p | prostate cancer | PDGFD | "miR 200 regulates PDGF D mediated epithelial mesen ......" | 19544444 |
| hsa-miR-200a-3p | breast cancer | PELP1 | "Whole genome miR array analysis using PELP1-overex ......" | 23975430 |
| hsa-miR-200a-3p | breast cancer | PHLDA3 | "Our data indicate that Kindlin 2 plays a novel rol ......" | 23483548 |
| hsa-miR-200a-3p | pancreatic cancer | PTGS2 | "In a xenograft mouse model of human PC CDF treatme ......" | 21408027 |
| hsa-miR-200a-3p | breast cancer | SGSM3 | "The genes encoding microRNAs of the human miR-200 ......" | 20514023 |
| hsa-miR-200a-3p | breast cancer | SLC10A4 | "Here we investigated P4 downregulation of miR-141 ......" | 25241899 |
| hsa-miR-200a-3p | colorectal cancer | SNAI1 | "EMT-TFs and microRNAs such as ZEB1/2 and miR-200 o ......" | 27573895 |
| hsa-miR-200a-3p | kidney renal cell cancer | SPAG9 | "MicroRNA 200a 3p suppresses tumor proliferation an ......" | 26797273 |
| hsa-miR-200a-3p | endometrial cancer | SPEN | "This finding was accompanied by a sharp downregula ......" | 23743934 |
| hsa-miR-200a-3p | breast cancer | TFAM | "In this study we showed that miR-200a expression l ......" | 24684598 |
| hsa-miR-200a-3p | thyroid cancer | TGFB1 | "Inhibition of TGFbeta receptor 1 TGFBR1 in these c ......" | 20498632 |
| hsa-miR-200a-3p | kidney renal cell cancer | TGFB2 | "Tumor suppressive microRNA 200a inhibits renal cel ......" | 25813153 |
| hsa-miR-200a-3p | thyroid cancer | TGFBR1 | "Inhibition of TGFbeta receptor 1 TGFBR1 in these c ......" | 20498632 |
| hsa-miR-200a-3p | endometrial cancer | TIMP2 | "We found that miR-200b repressed TIMP2 expression ......" | 23205572 |
| hsa-miR-200a-3p | esophageal cancer | TPO | "MSA could also significantly induce miR-200a expre ......" | 26341629 |
| hsa-miR-200a-3p | bladder cancer | TWIST1 | "In addition we observe that the mesoderm transcrip ......" | 20473948 |
| hsa-miR-200a-3p | kidney renal cell cancer | VEGFA | "We also found strong anti-correlation between VEGF ......" | 20420713 |
| hsa-miR-200a-3p | breast cancer | YAP1 | "MicroRNA 200a promotes anoikis resistance and meta ......" | 23340296 |
| hsa-miR-200a-3p | lung cancer | ZFPM2 | "It was reported that miR-200 can activate PI3K/AKT ......" | 26314828 |
Expression profile in cancer corhorts:
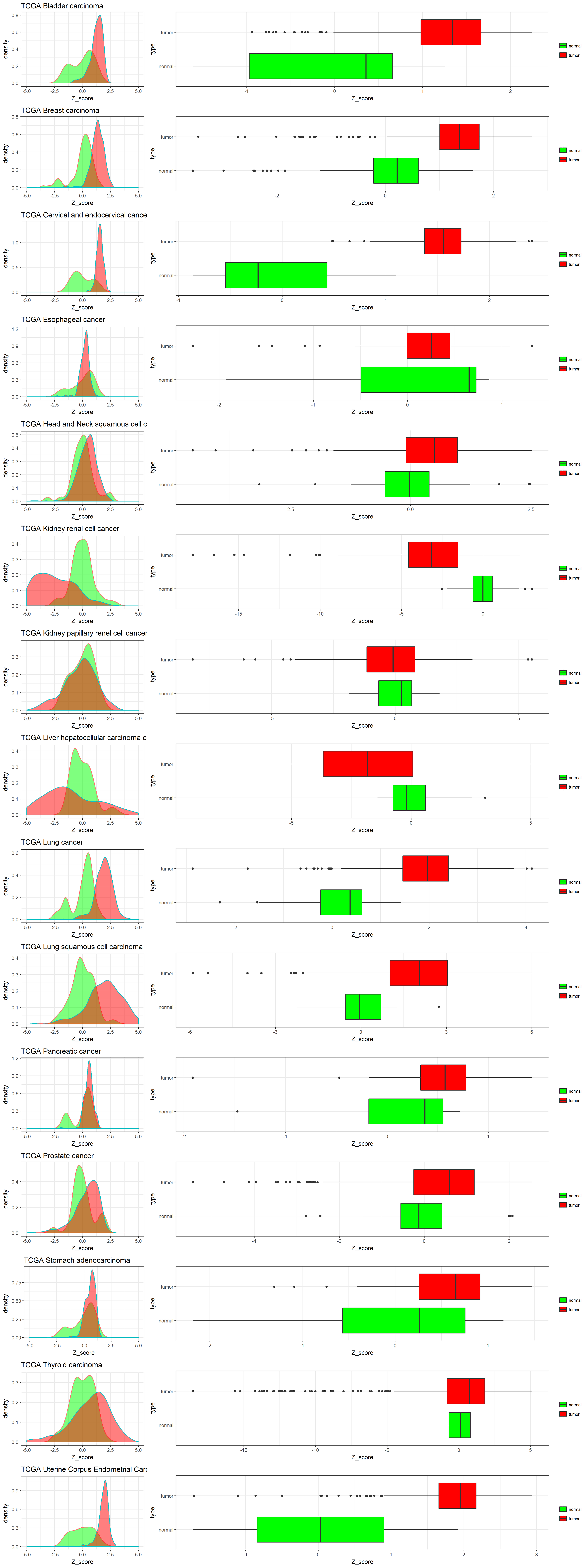
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-200a-3p | KLHL20 | 11 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; OV; PAAD; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.071; TCGA BRCA -0.126; TCGA CESC -0.084; TCGA COAD -0.089; TCGA HNSC -0.076; TCGA KIRC -0.071; TCGA KIRP -0.157; TCGA OV -0.055; TCGA PAAD -0.056; TCGA STAD -0.082; TCGA UCEC -0.126 |
hsa-miR-200a-3p | PTPRD | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LUSC; OV; PAAD; STAD; UCEC | miRNAWalker2 validate; miRNATAP | TCGA BLCA -0.485; TCGA BRCA -0.264; TCGA CESC -0.294; TCGA ESCA -0.382; TCGA HNSC -0.517; TCGA LUSC -0.37; TCGA OV -0.458; TCGA PAAD -0.225; TCGA STAD -0.272; TCGA UCEC -0.347 |
hsa-miR-200a-3p | RAB30 | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LIHC; LUAD; OV; PAAD; STAD; UCEC | miRNAWalker2 validate; miRNATAP | TCGA BLCA -0.198; TCGA CESC -0.166; TCGA COAD -0.153; TCGA ESCA -0.092; TCGA HNSC -0.219; TCGA KIRP -0.369; TCGA LIHC -0.066; TCGA LUAD -0.101; TCGA OV -0.135; TCGA PAAD -0.166; TCGA STAD -0.136; TCGA UCEC -0.166 |
hsa-miR-200a-3p | RASSF2 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.095; TCGA BRCA -0.122; TCGA CESC -0.147; TCGA COAD -0.455; TCGA ESCA -0.136; TCGA HNSC -0.286; TCGA KIRC -0.413; TCGA LUAD -0.141; TCGA LUSC -0.476; TCGA OV -0.084; TCGA PAAD -0.485; TCGA PRAD -0.07; TCGA STAD -0.244; TCGA UCEC -0.254 |
hsa-miR-200a-3p | RIN2 | 9 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LUAD; PAAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.133; TCGA BRCA -0.072; TCGA CESC -0.101; TCGA ESCA -0.069; TCGA HNSC -0.111; TCGA KIRC -0.055; TCGA LUAD -0.063; TCGA PAAD -0.063; TCGA UCEC -0.08 |
hsa-miR-200a-3p | SEPT7 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; OV; PAAD; STAD; UCEC | miRNAWalker2 validate; MirTarget; miRNATAP | TCGA BLCA -0.093; TCGA BRCA -0.073; TCGA CESC -0.094; TCGA COAD -0.103; TCGA ESCA -0.087; TCGA HNSC -0.083; TCGA KIRC -0.114; TCGA LUAD -0.104; TCGA LUSC -0.054; TCGA OV -0.086; TCGA PAAD -0.121; TCGA STAD -0.124; TCGA UCEC -0.12 |
hsa-miR-200a-3p | SHC1 | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; OV; PAAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.063; TCGA CESC -0.159; TCGA COAD -0.054; TCGA ESCA -0.089; TCGA HNSC -0.1; TCGA KIRC -0.247; TCGA OV -0.075; TCGA PAAD -0.073; TCGA UCEC -0.051 |
hsa-miR-200a-3p | TCF7L1 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; KIRP; OV; PAAD; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.317; TCGA BRCA -0.151; TCGA CESC -0.203; TCGA COAD -0.326; TCGA ESCA -0.382; TCGA KIRC -0.07; TCGA KIRP -0.195; TCGA OV -0.145; TCGA PAAD -0.18; TCGA STAD -0.405; TCGA UCEC -0.079 |
hsa-miR-200a-3p | VCAM1 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.343; TCGA BRCA -0.192; TCGA CESC -0.238; TCGA COAD -0.505; TCGA ESCA -0.118; TCGA HNSC -0.366; TCGA LUAD -0.225; TCGA LUSC -0.436; TCGA OV -0.213; TCGA PAAD -0.394; TCGA PRAD -0.097; TCGA STAD -0.117; TCGA UCEC -0.295 |
hsa-miR-200a-3p | WASF3 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUSC; OV; PAAD; THCA; STAD; UCEC | miRNAWalker2 validate; miRTarBase | TCGA BLCA -0.101; TCGA BRCA -0.406; TCGA CESC -0.371; TCGA COAD -0.603; TCGA ESCA -0.258; TCGA HNSC -0.142; TCGA LUSC -0.331; TCGA OV -0.186; TCGA PAAD -0.218; TCGA THCA -0.08; TCGA STAD -0.376; TCGA UCEC -0.242 |
hsa-miR-200a-3p | YAP1 | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; PAAD; STAD; UCEC | miRNAWalker2 validate; MirTarget | TCGA BLCA -0.116; TCGA BRCA -0.179; TCGA CESC -0.205; TCGA COAD -0.085; TCGA ESCA -0.071; TCGA HNSC -0.128; TCGA LUAD -0.07; TCGA PAAD -0.161; TCGA STAD -0.118; TCGA UCEC -0.108 |
hsa-miR-200a-3p | ZEB1 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | TCGA BLCA -0.369; TCGA BRCA -0.32; TCGA CESC -0.321; TCGA COAD -0.575; TCGA ESCA -0.213; TCGA HNSC -0.468; TCGA KIRC -0.205; TCGA KIRP -0.159; TCGA LUAD -0.186; TCGA LUSC -0.387; TCGA OV -0.259; TCGA PAAD -0.279; TCGA PRAD -0.089; TCGA STAD -0.452; TCGA UCEC -0.446 |
hsa-miR-200a-3p | ZEB2 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | TCGA BLCA -0.407; TCGA BRCA -0.35; TCGA CESC -0.338; TCGA COAD -0.505; TCGA ESCA -0.209; TCGA HNSC -0.505; TCGA KIRC -0.187; TCGA LUAD -0.247; TCGA LUSC -0.531; TCGA PAAD -0.376; TCGA PRAD -0.103; TCGA STAD -0.259; TCGA UCEC -0.337 |
hsa-miR-200a-3p | ZFPM2 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | miRNAWalker2 validate; miRTarBase | TCGA BLCA -0.516; TCGA BRCA -0.372; TCGA CESC -0.29; TCGA COAD -0.715; TCGA ESCA -0.38; TCGA HNSC -0.526; TCGA KIRC -0.326; TCGA KIRP -0.27; TCGA LUAD -0.257; TCGA LUSC -0.54; TCGA OV -0.179; TCGA PAAD -0.42; TCGA PRAD -0.201; TCGA STAD -0.525; TCGA UCEC -0.624 |
hsa-miR-200a-3p | WIPF1 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; OV; PAAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.354; TCGA BRCA -0.168; TCGA CESC -0.291; TCGA COAD -0.415; TCGA ESCA -0.224; TCGA HNSC -0.309; TCGA KIRC -0.301; TCGA KIRP -0.234; TCGA LUAD -0.186; TCGA LUSC -0.349; TCGA OV -0.106; TCGA PAAD -0.367; TCGA STAD -0.193; TCGA UCEC -0.198 |
hsa-miR-200a-3p | TMEFF1 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; OV; PAAD; UCEC | MirTarget | TCGA BLCA -0.121; TCGA BRCA -0.061; TCGA CESC -0.12; TCGA COAD -0.318; TCGA ESCA -0.188; TCGA HNSC -0.165; TCGA KIRC -0.222; TCGA KIRP -0.532; TCGA LUAD -0.167; TCGA OV -0.177; TCGA PAAD -0.143; TCGA UCEC -0.121 |
hsa-miR-200a-3p | ABL2 | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD | MirTarget; miRNATAP | TCGA BLCA -0.137; TCGA CESC -0.135; TCGA COAD -0.131; TCGA ESCA -0.1; TCGA HNSC -0.195; TCGA KIRC -0.216; TCGA KIRP -0.102; TCGA LUAD -0.069; TCGA LUSC -0.156; TCGA PAAD -0.193 |
hsa-miR-200a-3p | CBL | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget; mirMAP | TCGA BLCA -0.082; TCGA BRCA -0.107; TCGA CESC -0.068; TCGA COAD -0.059; TCGA ESCA -0.05; TCGA HNSC -0.129; TCGA KIRC -0.115; TCGA KIRP -0.104; TCGA LUAD -0.13; TCGA LUSC -0.059; TCGA PAAD -0.137; TCGA PRAD -0.073; TCGA STAD -0.088; TCGA UCEC -0.109 |
hsa-miR-200a-3p | KPNA3 | 9 cancers: BLCA; BRCA; HNSC; KIRC; KIRP; LUAD; OV; PAAD; PRAD | MirTarget; miRNATAP | TCGA BLCA -0.106; TCGA BRCA -0.083; TCGA HNSC -0.134; TCGA KIRC -0.056; TCGA KIRP -0.103; TCGA LUAD -0.137; TCGA OV -0.079; TCGA PAAD -0.077; TCGA PRAD -0.057 |
hsa-miR-200a-3p | TCF4 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.243; TCGA BRCA -0.295; TCGA CESC -0.15; TCGA COAD -0.465; TCGA ESCA -0.168; TCGA HNSC -0.236; TCGA KIRC -0.281; TCGA KIRP -0.159; TCGA LUAD -0.167; TCGA LUSC -0.123; TCGA OV -0.142; TCGA PAAD -0.33; TCGA PRAD -0.059; TCGA STAD -0.215; TCGA UCEC -0.224 |
hsa-miR-200a-3p | CLIC4 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.381; TCGA BRCA -0.115; TCGA CESC -0.215; TCGA COAD -0.373; TCGA ESCA -0.246; TCGA HNSC -0.164; TCGA LUAD -0.193; TCGA LUSC -0.175; TCGA PAAD -0.247; TCGA PRAD -0.113; TCGA STAD -0.351; TCGA UCEC -0.218 |
hsa-miR-200a-3p | KIF17 | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; LUAD; LUSC; PAAD; STAD; UCEC | MirTarget | TCGA BLCA -0.255; TCGA BRCA -0.19; TCGA CESC -0.276; TCGA COAD -0.532; TCGA ESCA -0.281; TCGA LUAD -0.228; TCGA LUSC -0.344; TCGA PAAD -0.366; TCGA STAD -0.187; TCGA UCEC -0.152 |
hsa-miR-200a-3p | KLF12 | 13 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.277; TCGA BRCA -0.129; TCGA COAD -0.264; TCGA ESCA -0.211; TCGA HNSC -0.178; TCGA LIHC -0.089; TCGA LUAD -0.1; TCGA LUSC -0.199; TCGA OV -0.084; TCGA PAAD -0.252; TCGA PRAD -0.106; TCGA STAD -0.265; TCGA UCEC -0.178 |
hsa-miR-200a-3p | RAB11FIP2 | 11 cancers: BLCA; BRCA; CESC; COAD; HNSC; LIHC; LUAD; LUSC; PAAD; STAD; UCEC | MirTarget | TCGA BLCA -0.05; TCGA BRCA -0.145; TCGA CESC -0.129; TCGA COAD -0.132; TCGA HNSC -0.138; TCGA LIHC -0.055; TCGA LUAD -0.083; TCGA LUSC -0.199; TCGA PAAD -0.091; TCGA STAD -0.142; TCGA UCEC -0.186 |
hsa-miR-200a-3p | WWTR1 | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.296; TCGA BRCA -0.183; TCGA COAD -0.657; TCGA ESCA -0.249; TCGA HNSC -0.249; TCGA KIRP -0.22; TCGA LUAD -0.168; TCGA LUSC -0.103; TCGA PAAD -0.225; TCGA PRAD -0.109; TCGA STAD -0.367; TCGA UCEC -0.091 |
hsa-miR-200a-3p | GEM | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUSC; OV; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.489; TCGA BRCA -0.273; TCGA CESC -0.195; TCGA COAD -0.495; TCGA ESCA -0.139; TCGA HNSC -0.337; TCGA KIRC -0.179; TCGA KIRP -0.16; TCGA LUSC -0.402; TCGA OV -0.293; TCGA PAAD -0.3; TCGA PRAD -0.098; TCGA STAD -0.391; TCGA UCEC -0.338 |
hsa-miR-200a-3p | CCND2 | 9 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LUAD; STAD; UCEC | MirTarget | TCGA BLCA -0.323; TCGA BRCA -0.261; TCGA CESC -0.415; TCGA ESCA -0.158; TCGA HNSC -0.1; TCGA KIRC -0.213; TCGA LUAD -0.174; TCGA STAD -0.219; TCGA UCEC -0.349 |
hsa-miR-200a-3p | TGFB2 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.35; TCGA BRCA -0.157; TCGA CESC -0.351; TCGA COAD -0.393; TCGA ESCA -0.252; TCGA HNSC -0.246; TCGA LUAD -0.154; TCGA LUSC -0.269; TCGA PAAD -0.165; TCGA PRAD -0.082; TCGA STAD -0.19; TCGA UCEC -0.173 |
hsa-miR-200a-3p | STAT4 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; UCEC | MirTarget | TCGA BLCA -0.282; TCGA BRCA -0.214; TCGA CESC -0.283; TCGA COAD -0.266; TCGA ESCA -0.143; TCGA HNSC -0.149; TCGA KIRC -0.34; TCGA KIRP -0.311; TCGA LUAD -0.126; TCGA LUSC -0.369; TCGA PAAD -0.437; TCGA UCEC -0.063 |
hsa-miR-200a-3p | RFTN1 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.227; TCGA BRCA -0.225; TCGA CESC -0.351; TCGA COAD -0.395; TCGA ESCA -0.243; TCGA HNSC -0.288; TCGA KIRC -0.198; TCGA KIRP -0.388; TCGA LIHC -0.071; TCGA LUSC -0.467; TCGA PAAD -0.316; TCGA PRAD -0.108; TCGA STAD -0.169; TCGA UCEC -0.264 |
hsa-miR-200a-3p | VCAN | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; OV; PAAD; PRAD; STAD | MirTarget; miRNATAP | TCGA BLCA -0.33; TCGA BRCA -0.115; TCGA CESC -0.339; TCGA COAD -0.554; TCGA ESCA -0.173; TCGA HNSC -0.623; TCGA KIRC -0.397; TCGA LUAD -0.181; TCGA LUSC -0.249; TCGA OV -0.275; TCGA PAAD -0.284; TCGA PRAD -0.111; TCGA STAD -0.086 |
hsa-miR-200a-3p | CDC42EP3 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.214; TCGA BRCA -0.131; TCGA CESC -0.241; TCGA COAD -0.269; TCGA ESCA -0.27; TCGA HNSC -0.287; TCGA LUAD -0.111; TCGA LUSC -0.228; TCGA OV -0.08; TCGA PAAD -0.108; TCGA PRAD -0.094; TCGA STAD -0.344; TCGA UCEC -0.245 |
hsa-miR-200a-3p | ERG | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; OV; PAAD; STAD; UCEC | MirTarget | TCGA BLCA -0.209; TCGA BRCA -0.329; TCGA CESC -0.358; TCGA COAD -0.473; TCGA ESCA -0.151; TCGA HNSC -0.268; TCGA KIRC -0.141; TCGA KIRP -0.196; TCGA LUAD -0.165; TCGA LUSC -0.319; TCGA OV -0.119; TCGA PAAD -0.36; TCGA STAD -0.213; TCGA UCEC -0.209 |
hsa-miR-200a-3p | SV2B | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; OV; PAAD; STAD; UCEC | MirTarget | TCGA BLCA -0.458; TCGA BRCA -0.395; TCGA CESC -0.394; TCGA COAD -0.929; TCGA ESCA -0.425; TCGA HNSC -0.355; TCGA KIRP -0.63; TCGA LUAD -0.244; TCGA OV -0.303; TCGA PAAD -0.289; TCGA STAD -0.391; TCGA UCEC -0.361 |
hsa-miR-200a-3p | DUSP3 | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.161; TCGA BRCA -0.097; TCGA COAD -0.096; TCGA ESCA -0.069; TCGA HNSC -0.179; TCGA KIRP -0.193; TCGA LUAD -0.085; TCGA LUSC -0.16; TCGA PAAD -0.069; TCGA PRAD -0.057; TCGA STAD -0.215; TCGA UCEC -0.109 |
hsa-miR-200a-3p | PAPPA | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUSC; OV; PAAD; PRAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.344; TCGA BRCA -0.225; TCGA CESC -0.497; TCGA COAD -0.47; TCGA ESCA -0.196; TCGA HNSC -0.454; TCGA LUSC -0.293; TCGA OV -0.274; TCGA PAAD -0.256; TCGA PRAD -0.07; TCGA STAD -0.157; TCGA UCEC -0.449 |
hsa-miR-200a-3p | UNC5C | 12 cancers: BLCA; BRCA; COAD; HNSC; KIRP; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.413; TCGA BRCA -0.247; TCGA COAD -0.38; TCGA HNSC -0.566; TCGA KIRP -0.34; TCGA LUAD -0.22; TCGA LUSC -0.705; TCGA OV -0.425; TCGA PAAD -0.327; TCGA PRAD -0.139; TCGA STAD -0.074; TCGA UCEC -0.168 |
hsa-miR-200a-3p | MAP7D1 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; OV; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.152; TCGA BRCA -0.087; TCGA CESC -0.163; TCGA COAD -0.17; TCGA ESCA -0.216; TCGA HNSC -0.158; TCGA KIRC -0.114; TCGA LUAD -0.104; TCGA LUSC -0.137; TCGA OV -0.065; TCGA STAD -0.242; TCGA UCEC -0.082 |
hsa-miR-200a-3p | ADARB1 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; STAD; UCEC | MirTarget | TCGA BLCA -0.287; TCGA BRCA -0.114; TCGA CESC -0.228; TCGA COAD -0.228; TCGA ESCA -0.268; TCGA HNSC -0.179; TCGA KIRC -0.115; TCGA LUAD -0.136; TCGA LUSC -0.419; TCGA PAAD -0.276; TCGA STAD -0.315; TCGA UCEC -0.223 |
hsa-miR-200a-3p | GNG7 | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUSC; PAAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.218; TCGA BRCA -0.085; TCGA CESC -0.308; TCGA COAD -0.271; TCGA ESCA -0.201; TCGA HNSC -0.131; TCGA LUSC -0.383; TCGA PAAD -0.23; TCGA STAD -0.422; TCGA UCEC -0.41 |
hsa-miR-200a-3p | IRS2 | 10 cancers: BLCA; BRCA; HNSC; KIRC; KIRP; LIHC; LUSC; OV; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.154; TCGA BRCA -0.176; TCGA HNSC -0.142; TCGA KIRC -0.113; TCGA KIRP -0.279; TCGA LIHC -0.099; TCGA LUSC -0.309; TCGA OV -0.114; TCGA STAD -0.116; TCGA UCEC -0.329 |
hsa-miR-200a-3p | RORA | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; PAAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.16; TCGA BRCA -0.151; TCGA CESC -0.125; TCGA COAD -0.424; TCGA ESCA -0.176; TCGA HNSC -0.138; TCGA LIHC -0.086; TCGA PAAD -0.221; TCGA STAD -0.265; TCGA UCEC -0.168 |
hsa-miR-200a-3p | PFKM | 9 cancers: BLCA; CESC; ESCA; HNSC; KIRP; LUAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.106; TCGA CESC -0.206; TCGA ESCA -0.096; TCGA HNSC -0.203; TCGA KIRP -0.231; TCGA LUAD -0.067; TCGA THCA -0.066; TCGA STAD -0.188; TCGA UCEC -0.235 |
hsa-miR-200a-3p | ZCCHC24 | 16 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.349; TCGA BRCA -0.347; TCGA CESC -0.29; TCGA COAD -0.471; TCGA ESCA -0.312; TCGA HNSC -0.457; TCGA KIRC -0.167; TCGA KIRP -0.343; TCGA LIHC -0.061; TCGA LUAD -0.112; TCGA LUSC -0.424; TCGA OV -0.273; TCGA PAAD -0.325; TCGA PRAD -0.089; TCGA STAD -0.47; TCGA UCEC -0.451 |
hsa-miR-200a-3p | ANGPTL1 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.638; TCGA BRCA -0.662; TCGA CESC -0.465; TCGA COAD -1.439; TCGA ESCA -0.428; TCGA HNSC -0.801; TCGA KIRP -0.325; TCGA LUAD -0.252; TCGA LUSC -0.727; TCGA OV -0.363; TCGA PAAD -0.547; TCGA PRAD -0.162; TCGA STAD -0.906; TCGA UCEC -0.596 |
hsa-miR-200a-3p | TTBK2 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.117; TCGA BRCA -0.238; TCGA CESC -0.123; TCGA COAD -0.216; TCGA ESCA -0.173; TCGA HNSC -0.209; TCGA KIRP -0.173; TCGA LUAD -0.086; TCGA PAAD -0.149; TCGA PRAD -0.079; TCGA STAD -0.225; TCGA UCEC -0.163 |
hsa-miR-200a-3p | PLCL1 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.332; TCGA BRCA -0.18; TCGA CESC -0.425; TCGA COAD -0.393; TCGA ESCA -0.172; TCGA HNSC -0.415; TCGA LUAD -0.117; TCGA LUSC -0.374; TCGA OV -0.103; TCGA PAAD -0.316; TCGA PRAD -0.233; TCGA STAD -0.388; TCGA UCEC -0.444 |
hsa-miR-200a-3p | DYNC1I1 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; KIRP; LIHC; OV; PRAD; STAD | MirTarget | TCGA BLCA -0.127; TCGA BRCA -0.246; TCGA CESC -0.236; TCGA COAD -0.611; TCGA ESCA -0.314; TCGA KIRC -0.134; TCGA KIRP -0.966; TCGA LIHC -0.274; TCGA OV -0.384; TCGA PRAD -0.069; TCGA STAD -0.564 |
hsa-miR-200a-3p | PRKACB | 12 cancers: BLCA; BRCA; CESC; HNSC; LIHC; LUAD; LUSC; OV; PRAD; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.09; TCGA BRCA -0.1; TCGA CESC -0.104; TCGA HNSC -0.241; TCGA LIHC -0.076; TCGA LUAD -0.079; TCGA LUSC -0.167; TCGA OV -0.147; TCGA PRAD -0.07; TCGA THCA -0.054; TCGA STAD -0.23; TCGA UCEC -0.199 |
hsa-miR-200a-3p | CCDC80 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.633; TCGA BRCA -0.438; TCGA CESC -0.389; TCGA COAD -0.829; TCGA ESCA -0.521; TCGA HNSC -0.475; TCGA KIRC -0.278; TCGA KIRP -0.637; TCGA LUAD -0.297; TCGA LUSC -0.315; TCGA OV -0.361; TCGA PAAD -0.51; TCGA PRAD -0.171; TCGA STAD -0.563; TCGA UCEC -0.409 |
hsa-miR-200a-3p | LHFP | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; OV; PAAD; STAD; UCEC | MirTarget | TCGA BLCA -0.282; TCGA BRCA -0.382; TCGA CESC -0.3; TCGA COAD -0.545; TCGA ESCA -0.283; TCGA HNSC -0.35; TCGA KIRC -0.25; TCGA KIRP -0.364; TCGA LUAD -0.136; TCGA LUSC -0.4; TCGA OV -0.16; TCGA PAAD -0.3; TCGA STAD -0.312; TCGA UCEC -0.331 |
hsa-miR-200a-3p | TMEM110 | 9 cancers: BLCA; CESC; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD | MirTarget; miRNATAP | TCGA BLCA -0.05; TCGA CESC -0.053; TCGA ESCA -0.058; TCGA HNSC -0.081; TCGA KIRP -0.115; TCGA LIHC -0.098; TCGA LUAD -0.062; TCGA LUSC -0.165; TCGA PAAD -0.107 |
hsa-miR-200a-3p | ACO1 | 10 cancers: BLCA; BRCA; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; UCEC | MirTarget | TCGA BLCA -0.118; TCGA BRCA -0.256; TCGA HNSC -0.118; TCGA KIRP -0.298; TCGA LIHC -0.09; TCGA LUAD -0.089; TCGA LUSC -0.175; TCGA PAAD -0.075; TCGA PRAD -0.05; TCGA UCEC -0.093 |
hsa-miR-200a-3p | ZDHHC15 | 9 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUSC; PAAD; STAD; UCEC | MirTarget | TCGA BLCA -0.262; TCGA BRCA -0.165; TCGA COAD -0.712; TCGA ESCA -0.263; TCGA HNSC -0.18; TCGA LUSC -0.281; TCGA PAAD -0.139; TCGA STAD -0.5; TCGA UCEC -0.295 |
hsa-miR-200a-3p | PHYHIPL | 9 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUSC; OV; PRAD; STAD | MirTarget; miRNATAP | TCGA BLCA -0.162; TCGA BRCA -0.263; TCGA COAD -0.487; TCGA ESCA -0.188; TCGA HNSC -0.128; TCGA LUSC -0.228; TCGA OV -0.228; TCGA PRAD -0.083; TCGA STAD -0.357 |
hsa-miR-200a-3p | TTLL7 | 11 cancers: BLCA; BRCA; COAD; HNSC; KIRC; LIHC; LUAD; LUSC; OV; THCA; STAD | MirTarget | TCGA BLCA -0.287; TCGA BRCA -0.22; TCGA COAD -0.517; TCGA HNSC -0.241; TCGA KIRC -0.223; TCGA LIHC -0.176; TCGA LUAD -0.137; TCGA LUSC -0.217; TCGA OV -0.205; TCGA THCA -0.075; TCGA STAD -0.446 |
hsa-miR-200a-3p | NECAB1 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; OV; PAAD; STAD; UCEC | MirTarget | TCGA BLCA -0.389; TCGA BRCA -0.362; TCGA CESC -0.438; TCGA COAD -0.791; TCGA ESCA -0.416; TCGA HNSC -0.213; TCGA LUAD -0.208; TCGA LUSC -0.461; TCGA OV -0.281; TCGA PAAD -0.205; TCGA STAD -0.699; TCGA UCEC -0.602 |
hsa-miR-200a-3p | ATXN1 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; PAAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.235; TCGA BRCA -0.084; TCGA CESC -0.081; TCGA COAD -0.134; TCGA ESCA -0.095; TCGA HNSC -0.183; TCGA KIRC -0.115; TCGA LUAD -0.078; TCGA PAAD -0.143; TCGA STAD -0.21; TCGA UCEC -0.104 |
hsa-miR-200a-3p | GSN | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LUSC; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.243; TCGA BRCA -0.311; TCGA COAD -0.162; TCGA ESCA -0.176; TCGA HNSC -0.082; TCGA KIRC -0.122; TCGA LUSC -0.154; TCGA PRAD -0.074; TCGA STAD -0.327; TCGA UCEC -0.202 |
hsa-miR-200a-3p | ST3GAL3 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; OV; PAAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.138; TCGA BRCA -0.107; TCGA CESC -0.079; TCGA COAD -0.162; TCGA ESCA -0.241; TCGA HNSC -0.059; TCGA LIHC -0.113; TCGA OV -0.061; TCGA PAAD -0.127; TCGA STAD -0.323; TCGA UCEC -0.165 |
hsa-miR-200a-3p | GIT2 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; STAD; UCEC | MirTarget | TCGA BLCA -0.063; TCGA BRCA -0.081; TCGA CESC -0.072; TCGA COAD -0.084; TCGA ESCA -0.08; TCGA HNSC -0.092; TCGA KIRC -0.216; TCGA KIRP -0.065; TCGA LUAD -0.07; TCGA LUSC -0.123; TCGA PAAD -0.148; TCGA STAD -0.126; TCGA UCEC -0.089 |
hsa-miR-200a-3p | DCUN1D3 | 12 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; STAD; UCEC | MirTarget | TCGA BLCA -0.074; TCGA BRCA -0.163; TCGA CESC -0.085; TCGA COAD -0.078; TCGA HNSC -0.09; TCGA KIRC -0.113; TCGA KIRP -0.067; TCGA LUAD -0.12; TCGA LUSC -0.197; TCGA PAAD -0.099; TCGA STAD -0.053; TCGA UCEC -0.051 |
hsa-miR-200a-3p | TMEM170B | 12 cancers: BLCA; BRCA; COAD; HNSC; KIRP; LIHC; LUSC; OV; PAAD; PRAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.084; TCGA BRCA -0.241; TCGA COAD -0.199; TCGA HNSC -0.202; TCGA KIRP -0.185; TCGA LIHC -0.126; TCGA LUSC -0.125; TCGA OV -0.117; TCGA PAAD -0.177; TCGA PRAD -0.055; TCGA STAD -0.285; TCGA UCEC -0.174 |
hsa-miR-200a-3p | SPAG9 | 12 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.053; TCGA BRCA -0.133; TCGA CESC -0.082; TCGA COAD -0.086; TCGA HNSC -0.122; TCGA KIRC -0.055; TCGA KIRP -0.116; TCGA LUAD -0.085; TCGA LUSC -0.086; TCGA PAAD -0.105; TCGA STAD -0.089; TCGA UCEC -0.128 |
hsa-miR-200a-3p | PXK | 13 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.159; TCGA BRCA -0.133; TCGA COAD -0.107; TCGA ESCA -0.053; TCGA HNSC -0.215; TCGA KIRP -0.209; TCGA LIHC -0.069; TCGA LUAD -0.159; TCGA LUSC -0.228; TCGA PAAD -0.244; TCGA PRAD -0.071; TCGA STAD -0.084; TCGA UCEC -0.172 |
hsa-miR-200a-3p | SDC2 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.268; TCGA BRCA -0.211; TCGA CESC -0.29; TCGA COAD -0.473; TCGA ESCA -0.208; TCGA HNSC -0.419; TCGA LIHC -0.06; TCGA LUAD -0.124; TCGA LUSC -0.311; TCGA OV -0.23; TCGA PAAD -0.157; TCGA PRAD -0.095; TCGA STAD -0.267; TCGA UCEC -0.195 |
hsa-miR-200a-3p | APBB2 | 10 cancers: BLCA; BRCA; CESC; HNSC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.157; TCGA BRCA -0.076; TCGA CESC -0.125; TCGA HNSC -0.221; TCGA LUAD -0.114; TCGA LUSC -0.282; TCGA PAAD -0.11; TCGA PRAD -0.063; TCGA STAD -0.128; TCGA UCEC -0.083 |
hsa-miR-200a-3p | EGFR | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LUAD; PRAD; STAD | MirTarget | TCGA BLCA -0.167; TCGA BRCA -0.53; TCGA CESC -0.314; TCGA ESCA -0.169; TCGA HNSC -0.186; TCGA KIRC -0.128; TCGA KIRP -0.202; TCGA LUAD -0.136; TCGA PRAD -0.071; TCGA STAD -0.15 |
hsa-miR-200a-3p | TSHZ3 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; OV; PAAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.359; TCGA BRCA -0.242; TCGA CESC -0.252; TCGA COAD -0.515; TCGA ESCA -0.347; TCGA HNSC -0.343; TCGA KIRC -0.205; TCGA LUAD -0.233; TCGA LUSC -0.243; TCGA OV -0.132; TCGA PAAD -0.234; TCGA STAD -0.407; TCGA UCEC -0.422 |
hsa-miR-200a-3p | TRIM23 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; OV; PAAD; STAD; UCEC | MirTarget | TCGA BLCA -0.075; TCGA BRCA -0.107; TCGA CESC -0.065; TCGA COAD -0.105; TCGA ESCA -0.117; TCGA HNSC -0.137; TCGA KIRC -0.054; TCGA KIRP -0.244; TCGA LIHC -0.08; TCGA LUAD -0.076; TCGA LUSC -0.127; TCGA OV -0.108; TCGA PAAD -0.096; TCGA STAD -0.16; TCGA UCEC -0.161 |
hsa-miR-200a-3p | TRAM2 | 11 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; UCEC | MirTarget | TCGA BLCA -0.2; TCGA BRCA -0.136; TCGA CESC -0.089; TCGA COAD -0.078; TCGA HNSC -0.112; TCGA KIRC -0.139; TCGA KIRP -0.201; TCGA LUAD -0.083; TCGA LUSC -0.059; TCGA PAAD -0.184; TCGA UCEC -0.128 |
hsa-miR-200a-3p | IL6R | 12 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRP; LIHC; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.28; TCGA BRCA -0.235; TCGA CESC -0.19; TCGA COAD -0.203; TCGA HNSC -0.212; TCGA KIRP -0.368; TCGA LIHC -0.16; TCGA LUSC -0.313; TCGA PAAD -0.177; TCGA PRAD -0.092; TCGA STAD -0.181; TCGA UCEC -0.148 |
hsa-miR-200a-3p | RNF38 | 9 cancers: BLCA; BRCA; CESC; HNSC; KIRP; LUSC; PAAD; STAD; UCEC | MirTarget; mirMAP | TCGA BLCA -0.056; TCGA BRCA -0.057; TCGA CESC -0.059; TCGA HNSC -0.055; TCGA KIRP -0.065; TCGA LUSC -0.116; TCGA PAAD -0.065; TCGA STAD -0.159; TCGA UCEC -0.149 |
hsa-miR-200a-3p | LMO3 | 10 cancers: BLCA; BRCA; COAD; ESCA; KIRP; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.317; TCGA BRCA -0.268; TCGA COAD -1.216; TCGA ESCA -0.302; TCGA KIRP -0.394; TCGA LUSC -0.369; TCGA PAAD -0.216; TCGA PRAD -0.122; TCGA STAD -0.682; TCGA UCEC -0.411 |
hsa-miR-200a-3p | TNS1 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.437; TCGA BRCA -0.446; TCGA CESC -0.206; TCGA COAD -0.787; TCGA ESCA -0.336; TCGA HNSC -0.417; TCGA KIRP -0.178; TCGA LUSC -0.439; TCGA PAAD -0.25; TCGA PRAD -0.068; TCGA STAD -0.622; TCGA UCEC -0.388 |
hsa-miR-200a-3p | ZNF385D | 13 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.367; TCGA BRCA -0.246; TCGA COAD -1.009; TCGA ESCA -0.431; TCGA HNSC -0.534; TCGA LIHC -0.421; TCGA LUAD -0.231; TCGA LUSC -0.599; TCGA OV -0.326; TCGA PAAD -0.527; TCGA PRAD -0.093; TCGA STAD -0.501; TCGA UCEC -0.312 |
hsa-miR-200a-3p | GAB1 | 11 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LUAD; LUSC; OV; PAAD; STAD; UCEC | MirTarget | TCGA BLCA -0.077; TCGA BRCA -0.098; TCGA CESC -0.082; TCGA ESCA -0.113; TCGA HNSC -0.147; TCGA LUAD -0.12; TCGA LUSC -0.138; TCGA OV -0.067; TCGA PAAD -0.075; TCGA STAD -0.212; TCGA UCEC -0.168 |
hsa-miR-200a-3p | CRYBG3 | 9 cancers: BLCA; BRCA; COAD; HNSC; LIHC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.066; TCGA BRCA -0.377; TCGA COAD -0.16; TCGA HNSC -0.098; TCGA LIHC -0.088; TCGA PAAD -0.125; TCGA PRAD -0.054; TCGA STAD -0.141; TCGA UCEC -0.235 |
hsa-miR-200a-3p | SH2B3 | 11 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; UCEC | MirTarget | TCGA BLCA -0.229; TCGA BRCA -0.153; TCGA CESC -0.166; TCGA COAD -0.158; TCGA HNSC -0.269; TCGA KIRC -0.313; TCGA KIRP -0.124; TCGA LUAD -0.199; TCGA LUSC -0.383; TCGA PAAD -0.251; TCGA UCEC -0.148 |
hsa-miR-200a-3p | HAS2 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.354; TCGA BRCA -0.354; TCGA CESC -0.526; TCGA COAD -0.313; TCGA ESCA -0.304; TCGA HNSC -0.461; TCGA LUAD -0.267; TCGA LUSC -0.441; TCGA OV -0.348; TCGA PAAD -0.392; TCGA PRAD -0.087; TCGA STAD -0.096; TCGA UCEC -0.175 |
hsa-miR-200a-3p | CDK17 | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.092; TCGA CESC -0.155; TCGA COAD -0.137; TCGA ESCA -0.147; TCGA HNSC -0.076; TCGA KIRC -0.168; TCGA KIRP -0.06; TCGA LUAD -0.12; TCGA LUSC -0.063; TCGA PAAD -0.121; TCGA STAD -0.12; TCGA UCEC -0.142 |
hsa-miR-200a-3p | LHX6 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; KIRP; LUAD; PAAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.163; TCGA BRCA -0.282; TCGA CESC -0.254; TCGA COAD -0.455; TCGA ESCA -0.128; TCGA KIRC -0.266; TCGA KIRP -0.203; TCGA LUAD -0.161; TCGA PAAD -0.24; TCGA STAD -0.221; TCGA UCEC -0.148 |
hsa-miR-200a-3p | ABL1 | 11 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LUSC; PAAD; STAD; UCEC | MirTarget | TCGA BLCA -0.148; TCGA BRCA -0.07; TCGA CESC -0.081; TCGA ESCA -0.124; TCGA HNSC -0.168; TCGA KIRC -0.087; TCGA KIRP -0.096; TCGA LUSC -0.089; TCGA PAAD -0.098; TCGA STAD -0.203; TCGA UCEC -0.131 |
hsa-miR-200a-3p | NRIP3 | 10 cancers: BLCA; CESC; COAD; ESCA; KIRC; KIRP; LUAD; PAAD; STAD; UCEC | MirTarget | TCGA BLCA -0.053; TCGA CESC -0.135; TCGA COAD -0.317; TCGA ESCA -0.174; TCGA KIRC -0.113; TCGA KIRP -0.224; TCGA LUAD -0.12; TCGA PAAD -0.222; TCGA STAD -0.229; TCGA UCEC -0.108 |
hsa-miR-200a-3p | ITGAV | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; PAAD; STAD; UCEC | MirTarget | TCGA BLCA -0.132; TCGA BRCA -0.1; TCGA CESC -0.135; TCGA COAD -0.215; TCGA ESCA -0.096; TCGA HNSC -0.175; TCGA KIRP -0.355; TCGA LUAD -0.127; TCGA PAAD -0.122; TCGA STAD -0.093; TCGA UCEC -0.114 |
hsa-miR-200a-3p | FAT3 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.134; TCGA BRCA -0.457; TCGA CESC -0.227; TCGA COAD -0.534; TCGA ESCA -0.514; TCGA HNSC -0.417; TCGA KIRP -0.607; TCGA LUAD -0.191; TCGA LUSC -0.342; TCGA OV -0.314; TCGA PAAD -0.414; TCGA PRAD -0.15; TCGA STAD -0.814; TCGA UCEC -0.361 |
hsa-miR-200a-3p | KIAA0408 | 9 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUSC; PAAD; PRAD; STAD | MirTarget | TCGA BLCA -0.48; TCGA BRCA -0.901; TCGA COAD -0.797; TCGA ESCA -0.607; TCGA HNSC -0.609; TCGA LUSC -0.775; TCGA PAAD -0.593; TCGA PRAD -0.278; TCGA STAD -0.657 |
hsa-miR-200a-3p | PRKD1 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; OV; PAAD; STAD; UCEC | MirTarget | TCGA BLCA -0.136; TCGA BRCA -0.349; TCGA CESC -0.197; TCGA COAD -0.724; TCGA ESCA -0.322; TCGA HNSC -0.245; TCGA LUAD -0.086; TCGA LUSC -0.266; TCGA OV -0.235; TCGA PAAD -0.218; TCGA STAD -0.358; TCGA UCEC -0.266 |
hsa-miR-200a-3p | CD274 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; PRAD; THCA | MirTarget | TCGA BLCA -0.32; TCGA BRCA -0.105; TCGA CESC -0.349; TCGA COAD -0.253; TCGA ESCA -0.197; TCGA HNSC -0.28; TCGA KIRP -0.667; TCGA LUAD -0.46; TCGA LUSC -0.375; TCGA PAAD -0.253; TCGA PRAD -0.069; TCGA THCA -0.063 |
hsa-miR-200a-3p | STX2 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.364; TCGA BRCA -0.127; TCGA CESC -0.089; TCGA COAD -0.216; TCGA ESCA -0.137; TCGA HNSC -0.19; TCGA KIRC -0.13; TCGA LUAD -0.085; TCGA LUSC -0.298; TCGA OV -0.087; TCGA PAAD -0.12; TCGA PRAD -0.085; TCGA STAD -0.25; TCGA UCEC -0.232 |
hsa-miR-200a-3p | PLEK | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; PRAD | MirTarget | TCGA BLCA -0.442; TCGA BRCA -0.098; TCGA CESC -0.271; TCGA COAD -0.325; TCGA ESCA -0.182; TCGA HNSC -0.319; TCGA KIRC -0.368; TCGA KIRP -0.389; TCGA LUAD -0.255; TCGA LUSC -0.439; TCGA PAAD -0.529; TCGA PRAD -0.063 |
hsa-miR-200a-3p | SLC16A7 | 10 cancers: BLCA; BRCA; COAD; HNSC; KIRP; OV; PRAD; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.19; TCGA BRCA -0.577; TCGA COAD -0.451; TCGA HNSC -0.231; TCGA KIRP -0.587; TCGA OV -0.241; TCGA PRAD -0.108; TCGA THCA -0.072; TCGA STAD -0.278; TCGA UCEC -0.353 |
hsa-miR-200a-3p | STAT5B | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.066; TCGA BRCA -0.166; TCGA CESC -0.14; TCGA COAD -0.108; TCGA ESCA -0.063; TCGA HNSC -0.147; TCGA LUAD -0.057; TCGA LUSC -0.138; TCGA PAAD -0.104; TCGA STAD -0.194; TCGA UCEC -0.215 |
hsa-miR-200a-3p | PPM1K | 13 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.217; TCGA BRCA -0.064; TCGA CESC -0.149; TCGA COAD -0.161; TCGA HNSC -0.15; TCGA KIRP -0.3; TCGA LIHC -0.058; TCGA LUAD -0.113; TCGA LUSC -0.305; TCGA PAAD -0.278; TCGA PRAD -0.062; TCGA STAD -0.254; TCGA UCEC -0.249 |
hsa-miR-200a-3p | GJC1 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.223; TCGA BRCA -0.145; TCGA CESC -0.241; TCGA COAD -0.45; TCGA ESCA -0.244; TCGA HNSC -0.275; TCGA KIRC -0.556; TCGA KIRP -0.344; TCGA LUAD -0.163; TCGA LUSC -0.141; TCGA OV -0.224; TCGA PAAD -0.233; TCGA PRAD -0.118; TCGA STAD -0.438; TCGA UCEC -0.399 |
hsa-miR-200a-3p | SCHIP1 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; OV; PAAD; UCEC | MirTarget | TCGA BLCA -0.203; TCGA BRCA -0.134; TCGA CESC -0.105; TCGA COAD -0.459; TCGA ESCA -0.287; TCGA HNSC -0.1; TCGA LIHC -0.068; TCGA LUAD -0.168; TCGA OV -0.123; TCGA PAAD -0.216; TCGA UCEC -0.178 |
hsa-miR-200a-3p | FRMD4A | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; STAD | MirTarget; miRNATAP | TCGA BLCA -0.098; TCGA BRCA -0.152; TCGA CESC -0.1; TCGA COAD -0.206; TCGA ESCA -0.148; TCGA HNSC -0.134; TCGA LUAD -0.168; TCGA LUSC -0.22; TCGA PAAD -0.303; TCGA STAD -0.227 |
hsa-miR-200a-3p | FAM20C | 9 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUSC; STAD; UCEC | MirTarget | TCGA BLCA -0.436; TCGA BRCA -0.09; TCGA CESC -0.117; TCGA COAD -0.439; TCGA ESCA -0.235; TCGA HNSC -0.191; TCGA LUSC -0.126; TCGA STAD -0.219; TCGA UCEC -0.079 |
hsa-miR-200a-3p | MBNL1 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; PAAD; PRAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.14; TCGA BRCA -0.153; TCGA CESC -0.096; TCGA COAD -0.084; TCGA ESCA -0.128; TCGA HNSC -0.075; TCGA KIRC -0.114; TCGA KIRP -0.09; TCGA LUAD -0.082; TCGA PAAD -0.179; TCGA PRAD -0.056; TCGA STAD -0.175; TCGA UCEC -0.131 |
hsa-miR-200a-3p | TNS3 | 9 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRP; LUAD; LUSC; PAAD | MirTarget; miRNATAP | TCGA BLCA -0.114; TCGA BRCA -0.146; TCGA CESC -0.095; TCGA COAD -0.132; TCGA HNSC -0.407; TCGA KIRP -0.124; TCGA LUAD -0.193; TCGA LUSC -0.476; TCGA PAAD -0.149 |
hsa-miR-200a-3p | SULF2 | 11 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; OV; PAAD; PRAD; UCEC | MirTarget | TCGA BLCA -0.409; TCGA CESC -0.356; TCGA COAD -0.227; TCGA ESCA -0.204; TCGA HNSC -0.302; TCGA KIRC -0.213; TCGA LUAD -0.144; TCGA OV -0.184; TCGA PAAD -0.175; TCGA PRAD -0.062; TCGA UCEC -0.216 |
hsa-miR-200a-3p | OSBPL8 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; OV; PAAD; STAD; UCEC | MirTarget | TCGA BLCA -0.083; TCGA BRCA -0.097; TCGA CESC -0.098; TCGA COAD -0.091; TCGA ESCA -0.068; TCGA HNSC -0.136; TCGA KIRP -0.19; TCGA LUAD -0.131; TCGA OV -0.088; TCGA PAAD -0.172; TCGA STAD -0.061; TCGA UCEC -0.134 |
hsa-miR-200a-3p | JAZF1 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.36; TCGA BRCA -0.187; TCGA CESC -0.256; TCGA COAD -0.444; TCGA ESCA -0.179; TCGA HNSC -0.265; TCGA KIRC -0.248; TCGA KIRP -0.227; TCGA LIHC -0.08; TCGA LUAD -0.083; TCGA LUSC -0.27; TCGA PAAD -0.122; TCGA PRAD -0.091; TCGA STAD -0.271; TCGA UCEC -0.213 |
hsa-miR-200a-3p | BICD2 | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; LUAD; OV; PAAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.084; TCGA BRCA -0.081; TCGA CESC -0.237; TCGA COAD -0.092; TCGA ESCA -0.297; TCGA LUAD -0.092; TCGA OV -0.052; TCGA PAAD -0.21; TCGA STAD -0.175; TCGA UCEC -0.094 |
hsa-miR-200a-3p | SAMD8 | 9 cancers: BLCA; BRCA; COAD; HNSC; KIRP; LUAD; PRAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.089; TCGA BRCA -0.171; TCGA COAD -0.125; TCGA HNSC -0.106; TCGA KIRP -0.21; TCGA LUAD -0.159; TCGA PRAD -0.06; TCGA STAD -0.085; TCGA UCEC -0.051 |
hsa-miR-200a-3p | GLI2 | 9 cancers: BLCA; BRCA; COAD; ESCA; KIRC; KIRP; PAAD; STAD; UCEC | MirTarget | TCGA BLCA -0.402; TCGA BRCA -0.277; TCGA COAD -0.555; TCGA ESCA -0.231; TCGA KIRC -0.317; TCGA KIRP -0.274; TCGA PAAD -0.258; TCGA STAD -0.266; TCGA UCEC -0.434 |
hsa-miR-200a-3p | PDGFRA | 12 cancers: BLCA; BRCA; CESC; COAD; HNSC; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.3; TCGA BRCA -0.404; TCGA CESC -0.228; TCGA COAD -0.414; TCGA HNSC -0.419; TCGA LUAD -0.223; TCGA LUSC -0.389; TCGA OV -0.176; TCGA PAAD -0.496; TCGA PRAD -0.105; TCGA STAD -0.132; TCGA UCEC -0.347 |
hsa-miR-200a-3p | PPFIBP1 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.153; TCGA BRCA -0.148; TCGA CESC -0.12; TCGA COAD -0.121; TCGA ESCA -0.11; TCGA HNSC -0.204; TCGA LUAD -0.212; TCGA LUSC -0.157; TCGA PAAD -0.092; TCGA PRAD -0.059; TCGA STAD -0.105; TCGA UCEC -0.092 |
hsa-miR-200a-3p | KCTD20 | 12 cancers: BLCA; BRCA; COAD; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.102; TCGA BRCA -0.137; TCGA COAD -0.062; TCGA HNSC -0.131; TCGA KIRC -0.124; TCGA KIRP -0.212; TCGA LUAD -0.096; TCGA LUSC -0.113; TCGA PAAD -0.189; TCGA PRAD -0.056; TCGA STAD -0.114; TCGA UCEC -0.137 |
hsa-miR-200a-3p | CHRDL1 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.745; TCGA BRCA -0.746; TCGA CESC -0.591; TCGA COAD -1.515; TCGA ESCA -0.744; TCGA HNSC -0.672; TCGA KIRP -0.727; TCGA LUAD -0.202; TCGA LUSC -0.778; TCGA OV -0.431; TCGA PAAD -0.814; TCGA PRAD -0.158; TCGA STAD -0.946; TCGA UCEC -0.751 |
hsa-miR-200a-3p | SCN2B | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.599; TCGA BRCA -0.627; TCGA CESC -0.497; TCGA COAD -1.043; TCGA ESCA -0.496; TCGA HNSC -0.531; TCGA LUAD -0.195; TCGA LUSC -0.542; TCGA PAAD -0.374; TCGA STAD -0.794; TCGA UCEC -0.649 |
hsa-miR-200a-3p | SLC25A46 | 11 cancers: BLCA; BRCA; CESC; HNSC; KIRP; LIHC; LUAD; LUSC; OV; PRAD; UCEC | MirTarget | TCGA BLCA -0.09; TCGA BRCA -0.109; TCGA CESC -0.051; TCGA HNSC -0.162; TCGA KIRP -0.239; TCGA LIHC -0.081; TCGA LUAD -0.091; TCGA LUSC -0.101; TCGA OV -0.057; TCGA PRAD -0.055; TCGA UCEC -0.052 |
hsa-miR-200a-3p | NFASC | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUSC; PRAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.237; TCGA BRCA -0.074; TCGA CESC -0.367; TCGA COAD -0.379; TCGA ESCA -0.507; TCGA HNSC -0.138; TCGA LUSC -0.277; TCGA PRAD -0.073; TCGA STAD -0.639; TCGA UCEC -0.349 |
hsa-miR-200a-3p | COL15A1 | 14 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.308; TCGA BRCA -0.24; TCGA CESC -0.262; TCGA COAD -0.464; TCGA HNSC -0.441; TCGA KIRC -0.439; TCGA KIRP -0.315; TCGA LUAD -0.254; TCGA LUSC -0.447; TCGA OV -0.127; TCGA PAAD -0.266; TCGA PRAD -0.113; TCGA STAD -0.219; TCGA UCEC -0.287 |
hsa-miR-200a-3p | WNK1 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.086; TCGA BRCA -0.062; TCGA CESC -0.105; TCGA COAD -0.082; TCGA ESCA -0.081; TCGA HNSC -0.167; TCGA KIRP -0.225; TCGA LUAD -0.071; TCGA PRAD -0.075; TCGA STAD -0.115; TCGA UCEC -0.073 |
hsa-miR-200a-3p | HGF | 13 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRP; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.566; TCGA BRCA -0.396; TCGA CESC -0.281; TCGA COAD -0.596; TCGA HNSC -0.464; TCGA KIRP -0.352; TCGA LUAD -0.253; TCGA LUSC -0.653; TCGA OV -0.21; TCGA PAAD -0.52; TCGA PRAD -0.107; TCGA STAD -0.181; TCGA UCEC -0.3 |
hsa-miR-200a-3p | CYP26B1 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; OV; PAAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.365; TCGA BRCA -0.396; TCGA CESC -0.299; TCGA COAD -0.282; TCGA ESCA -0.219; TCGA HNSC -0.33; TCGA KIRP -0.182; TCGA LUAD -0.201; TCGA LUSC -0.243; TCGA OV -0.345; TCGA PAAD -0.329; TCGA UCEC -0.29 |
hsa-miR-200a-3p | CNTN1 | 9 cancers: BLCA; BRCA; CESC; COAD; ESCA; LUAD; PRAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.634; TCGA BRCA -0.448; TCGA CESC -0.288; TCGA COAD -1.123; TCGA ESCA -0.863; TCGA LUAD -0.338; TCGA PRAD -0.121; TCGA STAD -0.9; TCGA UCEC -0.226 |
hsa-miR-200a-3p | DOCK4 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.153; TCGA BRCA -0.186; TCGA CESC -0.292; TCGA COAD -0.296; TCGA ESCA -0.086; TCGA HNSC -0.244; TCGA KIRC -0.177; TCGA KIRP -0.177; TCGA LUAD -0.167; TCGA LUSC -0.339; TCGA PAAD -0.194; TCGA PRAD -0.06; TCGA STAD -0.13; TCGA UCEC -0.24 |
hsa-miR-200a-3p | OSBPL6 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUSC; OV; PRAD; THCA; STAD | MirTarget; miRNATAP | TCGA BLCA -0.557; TCGA BRCA -0.118; TCGA CESC -0.258; TCGA COAD -0.373; TCGA ESCA -0.625; TCGA HNSC -0.081; TCGA KIRP -0.7; TCGA LUSC -0.177; TCGA OV -0.406; TCGA PRAD -0.05; TCGA THCA -0.09; TCGA STAD -0.335 |
hsa-miR-200a-3p | ZC3H12C | 10 cancers: BLCA; BRCA; CESC; HNSC; KIRP; LUAD; LUSC; PAAD; STAD; UCEC | MirTarget | TCGA BLCA -0.078; TCGA BRCA -0.25; TCGA CESC -0.109; TCGA HNSC -0.151; TCGA KIRP -0.165; TCGA LUAD -0.101; TCGA LUSC -0.112; TCGA PAAD -0.11; TCGA STAD -0.168; TCGA UCEC -0.182 |
hsa-miR-200a-3p | FRMD6 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; OV; PAAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.134; TCGA BRCA -0.164; TCGA CESC -0.466; TCGA COAD -0.614; TCGA ESCA -0.497; TCGA HNSC -0.112; TCGA KIRC -0.211; TCGA LUAD -0.234; TCGA OV -0.176; TCGA PAAD -0.333; TCGA STAD -0.389; TCGA UCEC -0.38 |
hsa-miR-200a-3p | TMEM130 | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUSC; OV; STAD; UCEC | MirTarget | TCGA BLCA -0.246; TCGA BRCA -0.141; TCGA CESC -0.277; TCGA COAD -0.904; TCGA ESCA -0.362; TCGA HNSC -0.35; TCGA LUSC -0.535; TCGA OV -0.165; TCGA STAD -0.549; TCGA UCEC -0.196 |
hsa-miR-200a-3p | LTBP1 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.191; TCGA BRCA -0.156; TCGA CESC -0.109; TCGA COAD -0.439; TCGA ESCA -0.329; TCGA HNSC -0.327; TCGA LUAD -0.154; TCGA LUSC -0.108; TCGA PAAD -0.239; TCGA PRAD -0.063; TCGA STAD -0.364; TCGA UCEC -0.099 |
hsa-miR-200a-3p | OLFM1 | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; LUSC; PRAD; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.361; TCGA BRCA -0.161; TCGA CESC -0.257; TCGA COAD -0.566; TCGA ESCA -0.345; TCGA LUSC -0.246; TCGA PRAD -0.084; TCGA THCA -0.088; TCGA STAD -0.306; TCGA UCEC -0.496 |
hsa-miR-200a-3p | LRRC8A | 9 cancers: BLCA; CESC; ESCA; KIRC; KIRP; LUAD; OV; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.095; TCGA CESC -0.096; TCGA ESCA -0.114; TCGA KIRC -0.15; TCGA KIRP -0.108; TCGA LUAD -0.056; TCGA OV -0.071; TCGA STAD -0.05; TCGA UCEC -0.068 |
hsa-miR-200a-3p | HIPK3 | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.205; TCGA BRCA -0.242; TCGA COAD -0.17; TCGA ESCA -0.074; TCGA HNSC -0.372; TCGA KIRP -0.271; TCGA LUAD -0.132; TCGA LUSC -0.161; TCGA PRAD -0.143; TCGA STAD -0.202; TCGA UCEC -0.164 |
hsa-miR-200a-3p | PCDH9 | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; OV; PAAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.521; TCGA BRCA -0.554; TCGA COAD -0.721; TCGA ESCA -0.504; TCGA HNSC -0.495; TCGA KIRP -0.288; TCGA LUAD -0.237; TCGA LUSC -0.575; TCGA OV -0.458; TCGA PAAD -0.445; TCGA STAD -0.721; TCGA UCEC -0.246 |
hsa-miR-200a-3p | FOXN3 | 14 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; OV; PAAD; PRAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.111; TCGA BRCA -0.309; TCGA COAD -0.126; TCGA ESCA -0.115; TCGA HNSC -0.144; TCGA KIRC -0.107; TCGA KIRP -0.111; TCGA LIHC -0.074; TCGA LUAD -0.066; TCGA OV -0.067; TCGA PAAD -0.194; TCGA PRAD -0.075; TCGA STAD -0.218; TCGA UCEC -0.211 |
hsa-miR-200a-3p | SCN4A | 13 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LIHC; LUSC; OV; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.184; TCGA BRCA -0.496; TCGA COAD -0.497; TCGA ESCA -0.165; TCGA HNSC -0.93; TCGA KIRP -0.318; TCGA LIHC -0.178; TCGA LUSC -0.182; TCGA OV -0.348; TCGA PAAD -0.494; TCGA PRAD -0.214; TCGA STAD -0.243; TCGA UCEC -0.187 |
hsa-miR-200a-3p | AGPAT4 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; STAD; UCEC | mirMAP | TCGA BLCA -0.202; TCGA BRCA -0.179; TCGA CESC -0.17; TCGA COAD -0.316; TCGA ESCA -0.156; TCGA HNSC -0.167; TCGA LUAD -0.116; TCGA LUSC -0.193; TCGA PAAD -0.134; TCGA STAD -0.212; TCGA UCEC -0.076 |
hsa-miR-200a-3p | GAB2 | 9 cancers: BLCA; BRCA; HNSC; KIRC; KIRP; LUSC; PAAD; STAD; UCEC | mirMAP | TCGA BLCA -0.069; TCGA BRCA -0.078; TCGA HNSC -0.218; TCGA KIRC -0.153; TCGA KIRP -0.182; TCGA LUSC -0.38; TCGA PAAD -0.07; TCGA STAD -0.084; TCGA UCEC -0.208 |
hsa-miR-200a-3p | CELF2 | 11 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LUSC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.42; TCGA BRCA -0.351; TCGA CESC -0.233; TCGA ESCA -0.323; TCGA HNSC -0.337; TCGA KIRC -0.138; TCGA LUSC -0.493; TCGA PAAD -0.517; TCGA PRAD -0.107; TCGA STAD -0.402; TCGA UCEC -0.26 |
hsa-miR-200a-3p | NAV3 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; OV; PAAD; STAD; UCEC | mirMAP | TCGA BLCA -0.528; TCGA BRCA -0.286; TCGA CESC -0.336; TCGA COAD -0.743; TCGA ESCA -0.476; TCGA HNSC -0.497; TCGA KIRC -0.331; TCGA LUAD -0.314; TCGA LUSC -0.495; TCGA OV -0.129; TCGA PAAD -0.424; TCGA STAD -0.418; TCGA UCEC -0.365 |
hsa-miR-200a-3p | CYLD | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; STAD; UCEC | mirMAP | TCGA BLCA -0.156; TCGA BRCA -0.055; TCGA CESC -0.094; TCGA COAD -0.104; TCGA ESCA -0.145; TCGA HNSC -0.149; TCGA KIRP -0.106; TCGA LUAD -0.058; TCGA LUSC -0.219; TCGA PAAD -0.124; TCGA STAD -0.113; TCGA UCEC -0.076 |
hsa-miR-200a-3p | CD59 | 9 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; STAD | mirMAP | TCGA BLCA -0.136; TCGA BRCA -0.088; TCGA CESC -0.177; TCGA COAD -0.1; TCGA ESCA -0.105; TCGA HNSC -0.144; TCGA LUAD -0.073; TCGA LUSC -0.185; TCGA STAD -0.13 |
hsa-miR-200a-3p | SIGLEC1 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; STAD | mirMAP | TCGA BLCA -0.459; TCGA BRCA -0.101; TCGA CESC -0.356; TCGA COAD -0.564; TCGA ESCA -0.333; TCGA HNSC -0.366; TCGA KIRC -0.522; TCGA KIRP -0.513; TCGA LUAD -0.268; TCGA LUSC -0.555; TCGA PAAD -0.425; TCGA STAD -0.162 |
hsa-miR-200a-3p | IRAK3 | 9 cancers: BLCA; BRCA; COAD; HNSC; KIRC; LUSC; PAAD; PRAD; UCEC | mirMAP | TCGA BLCA -0.235; TCGA BRCA -0.318; TCGA COAD -0.386; TCGA HNSC -0.135; TCGA KIRC -0.195; TCGA LUSC -0.284; TCGA PAAD -0.175; TCGA PRAD -0.071; TCGA UCEC -0.238 |
hsa-miR-200a-3p | GRAP2 | 9 cancers: BLCA; BRCA; CESC; HNSC; KIRC; LUAD; LUSC; PAAD; UCEC | mirMAP | TCGA BLCA -0.349; TCGA BRCA -0.123; TCGA CESC -0.239; TCGA HNSC -0.151; TCGA KIRC -0.355; TCGA LUAD -0.104; TCGA LUSC -0.395; TCGA PAAD -0.381; TCGA UCEC -0.12 |
hsa-miR-200a-3p | RASA4 | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUSC; STAD; UCEC | mirMAP | TCGA BLCA -0.107; TCGA BRCA -0.114; TCGA CESC -0.2; TCGA COAD -0.223; TCGA ESCA -0.15; TCGA HNSC -0.201; TCGA KIRC -0.135; TCGA LUSC -0.19; TCGA STAD -0.192; TCGA UCEC -0.24 |
hsa-miR-200a-3p | PPARGC1A | 10 cancers: BLCA; BRCA; CESC; HNSC; KIRP; LUSC; OV; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.453; TCGA BRCA -0.314; TCGA CESC -0.242; TCGA HNSC -0.367; TCGA KIRP -0.277; TCGA LUSC -0.24; TCGA OV -0.549; TCGA THCA -0.152; TCGA STAD -0.132; TCGA UCEC -0.259 |
hsa-miR-200a-3p | BACH2 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; OV; PAAD; STAD; UCEC | mirMAP | TCGA BLCA -0.458; TCGA BRCA -0.288; TCGA CESC -0.216; TCGA COAD -0.587; TCGA ESCA -0.366; TCGA HNSC -0.293; TCGA KIRP -0.55; TCGA LUAD -0.175; TCGA OV -0.231; TCGA PAAD -0.477; TCGA STAD -0.269; TCGA UCEC -0.334 |
hsa-miR-200a-3p | BVES | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUAD; LUSC; OV; PAAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.416; TCGA BRCA -0.296; TCGA COAD -0.711; TCGA ESCA -0.419; TCGA HNSC -0.483; TCGA LUAD -0.209; TCGA LUSC -0.269; TCGA OV -0.171; TCGA PAAD -0.235; TCGA THCA -0.061; TCGA STAD -0.607; TCGA UCEC -0.321 |
hsa-miR-200a-3p | HPCAL4 | 10 cancers: BLCA; CESC; ESCA; HNSC; KIRP; LUSC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.18; TCGA CESC -0.249; TCGA ESCA -0.311; TCGA HNSC -0.257; TCGA KIRP -0.42; TCGA LUSC -0.269; TCGA PAAD -0.329; TCGA PRAD -0.134; TCGA STAD -0.489; TCGA UCEC -0.169 |
hsa-miR-200a-3p | RIMS3 | 9 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LUSC; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.209; TCGA BRCA -0.147; TCGA CESC -0.309; TCGA ESCA -0.188; TCGA HNSC -0.22; TCGA LUSC -0.299; TCGA PRAD -0.072; TCGA STAD -0.38; TCGA UCEC -0.339 |
hsa-miR-200a-3p | SEPT6 | 9 cancers: BLCA; COAD; ESCA; HNSC; LUAD; LUSC; OV; PAAD; STAD | mirMAP | TCGA BLCA -0.236; TCGA COAD -0.182; TCGA ESCA -0.155; TCGA HNSC -0.166; TCGA LUAD -0.08; TCGA LUSC -0.277; TCGA OV -0.08; TCGA PAAD -0.328; TCGA STAD -0.256 |
hsa-miR-200a-3p | HS3ST3B1 | 9 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRP; LUAD; LUSC; PRAD | mirMAP | TCGA BLCA -0.362; TCGA BRCA -0.115; TCGA CESC -0.226; TCGA ESCA -0.182; TCGA HNSC -0.295; TCGA KIRP -0.34; TCGA LUAD -0.26; TCGA LUSC -0.162; TCGA PRAD -0.082 |
hsa-miR-200a-3p | HIVEP3 | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; STAD | mirMAP | TCGA BLCA -0.215; TCGA CESC -0.292; TCGA COAD -0.16; TCGA ESCA -0.161; TCGA HNSC -0.223; TCGA KIRC -0.08; TCGA KIRP -0.11; TCGA LUAD -0.141; TCGA LUSC -0.257; TCGA STAD -0.074 |
hsa-miR-200a-3p | WDFY4 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUSC; PAAD; STAD | mirMAP | TCGA BLCA -0.431; TCGA BRCA -0.079; TCGA CESC -0.221; TCGA COAD -0.416; TCGA ESCA -0.229; TCGA HNSC -0.357; TCGA KIRC -0.33; TCGA KIRP -0.3; TCGA LUSC -0.623; TCGA PAAD -0.707; TCGA STAD -0.096 |
hsa-miR-200a-3p | MAP1B | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; OV; PAAD; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.45; TCGA BRCA -0.287; TCGA CESC -0.353; TCGA COAD -0.586; TCGA ESCA -0.357; TCGA HNSC -0.369; TCGA KIRC -0.333; TCGA KIRP -0.609; TCGA LUAD -0.23; TCGA OV -0.177; TCGA PAAD -0.171; TCGA PRAD -0.121; TCGA THCA -0.063; TCGA STAD -0.509; TCGA UCEC -0.206 |
hsa-miR-200a-3p | ETS1 | 12 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; LUAD; LUSC; OV; PAAD; STAD; UCEC | mirMAP | TCGA BLCA -0.131; TCGA BRCA -0.227; TCGA CESC -0.217; TCGA COAD -0.296; TCGA HNSC -0.364; TCGA KIRC -0.239; TCGA LUAD -0.224; TCGA LUSC -0.481; TCGA OV -0.088; TCGA PAAD -0.318; TCGA STAD -0.051; TCGA UCEC -0.157 |
hsa-miR-200a-3p | KIAA1644 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUSC; OV; PAAD; STAD; UCEC | mirMAP | TCGA BLCA -0.619; TCGA BRCA -0.127; TCGA CESC -0.582; TCGA COAD -0.846; TCGA ESCA -0.452; TCGA HNSC -0.488; TCGA KIRC -0.44; TCGA KIRP -0.447; TCGA LUSC -0.47; TCGA OV -0.295; TCGA PAAD -0.281; TCGA STAD -0.633; TCGA UCEC -0.547 |
hsa-miR-200a-3p | NTRK3 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUSC; OV; PAAD; STAD; UCEC | mirMAP | TCGA BLCA -0.332; TCGA BRCA -0.338; TCGA CESC -0.368; TCGA COAD -0.856; TCGA ESCA -0.266; TCGA HNSC -0.297; TCGA LUSC -0.48; TCGA OV -0.303; TCGA PAAD -0.334; TCGA STAD -0.585; TCGA UCEC -0.391 |
hsa-miR-200a-3p | FTO | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; PAAD; STAD; UCEC | mirMAP | TCGA BLCA -0.086; TCGA BRCA -0.086; TCGA CESC -0.078; TCGA COAD -0.125; TCGA ESCA -0.13; TCGA HNSC -0.152; TCGA KIRC -0.061; TCGA LIHC -0.05; TCGA LUAD -0.056; TCGA LUSC -0.135; TCGA PAAD -0.087; TCGA STAD -0.162; TCGA UCEC -0.117 |
hsa-miR-200a-3p | WTIP | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUSC; OV; PAAD; STAD; UCEC | mirMAP | TCGA BLCA -0.089; TCGA BRCA -0.154; TCGA CESC -0.156; TCGA COAD -0.365; TCGA ESCA -0.279; TCGA HNSC -0.136; TCGA LUSC -0.294; TCGA OV -0.092; TCGA PAAD -0.187; TCGA STAD -0.264; TCGA UCEC -0.191 |
hsa-miR-200a-3p | COL8A1 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; OV; PAAD; PRAD; STAD | mirMAP | TCGA BLCA -0.302; TCGA BRCA -0.162; TCGA CESC -0.31; TCGA COAD -0.765; TCGA ESCA -0.231; TCGA HNSC -0.597; TCGA KIRC -0.528; TCGA KIRP -0.406; TCGA LUAD -0.225; TCGA LUSC -0.548; TCGA OV -0.246; TCGA PAAD -0.371; TCGA PRAD -0.084; TCGA STAD -0.358 |
hsa-miR-200a-3p | TMEM108 | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LUSC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.364; TCGA BRCA -0.131; TCGA COAD -0.474; TCGA ESCA -0.334; TCGA HNSC -0.308; TCGA KIRC -0.205; TCGA KIRP -0.247; TCGA LUSC -0.192; TCGA PAAD -0.25; TCGA PRAD -0.082; TCGA STAD -0.353; TCGA UCEC -0.189 |
hsa-miR-200a-3p | RNF217 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; PAAD; STAD | mirMAP | TCGA BLCA -0.324; TCGA BRCA -0.238; TCGA CESC -0.148; TCGA COAD -0.375; TCGA ESCA -0.485; TCGA HNSC -0.087; TCGA KIRC -0.154; TCGA KIRP -0.202; TCGA LIHC -0.164; TCGA LUAD -0.138; TCGA PAAD -0.226; TCGA STAD -0.378 |
hsa-miR-200a-3p | GNA12 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; STAD; UCEC | mirMAP | TCGA BLCA -0.168; TCGA BRCA -0.061; TCGA CESC -0.179; TCGA COAD -0.115; TCGA ESCA -0.088; TCGA HNSC -0.161; TCGA KIRC -0.138; TCGA KIRP -0.148; TCGA LUAD -0.129; TCGA LUSC -0.097; TCGA PAAD -0.147; TCGA STAD -0.094; TCGA UCEC -0.116 |
hsa-miR-200a-3p | CREB5 | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; STAD | mirMAP | TCGA BLCA -0.404; TCGA BRCA -0.33; TCGA COAD -0.311; TCGA ESCA -0.152; TCGA HNSC -0.282; TCGA KIRC -0.14; TCGA LUAD -0.111; TCGA LUSC -0.132; TCGA PAAD -0.198; TCGA STAD -0.264 |
hsa-miR-200a-3p | FOXO1 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; STAD; UCEC | mirMAP | TCGA BLCA -0.081; TCGA BRCA -0.355; TCGA CESC -0.136; TCGA COAD -0.139; TCGA ESCA -0.08; TCGA HNSC -0.225; TCGA LUAD -0.058; TCGA LUSC -0.152; TCGA PAAD -0.254; TCGA STAD -0.095; TCGA UCEC -0.161 |
hsa-miR-200a-3p | FER | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.192; TCGA BRCA -0.237; TCGA CESC -0.189; TCGA COAD -0.16; TCGA ESCA -0.157; TCGA HNSC -0.139; TCGA KIRP -0.17; TCGA LIHC -0.055; TCGA LUAD -0.148; TCGA LUSC -0.193; TCGA PAAD -0.112; TCGA PRAD -0.057; TCGA STAD -0.188; TCGA UCEC -0.083 |
hsa-miR-200a-3p | WWC2 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; STAD; UCEC | mirMAP | TCGA BLCA -0.179; TCGA BRCA -0.196; TCGA CESC -0.191; TCGA COAD -0.378; TCGA ESCA -0.081; TCGA HNSC -0.259; TCGA LUAD -0.171; TCGA LUSC -0.491; TCGA PAAD -0.135; TCGA STAD -0.157; TCGA UCEC -0.083 |
hsa-miR-200a-3p | GFRA1 | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; LIHC; LUSC; OV; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.282; TCGA CESC -0.45; TCGA COAD -0.683; TCGA ESCA -0.228; TCGA HNSC -0.388; TCGA LIHC -0.241; TCGA LUSC -0.379; TCGA OV -0.441; TCGA PAAD -0.384; TCGA PRAD -0.112; TCGA STAD -0.648; TCGA UCEC -0.436 |
hsa-miR-200a-3p | DST | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; STAD; UCEC | mirMAP | TCGA BLCA -0.145; TCGA BRCA -0.29; TCGA CESC -0.318; TCGA COAD -0.161; TCGA ESCA -0.189; TCGA HNSC -0.135; TCGA KIRP -0.118; TCGA LUAD -0.212; TCGA STAD -0.294; TCGA UCEC -0.222 |
hsa-miR-200a-3p | CAMK4 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.285; TCGA BRCA -0.093; TCGA CESC -0.267; TCGA COAD -0.238; TCGA ESCA -0.129; TCGA HNSC -0.205; TCGA KIRC -0.202; TCGA KIRP -0.208; TCGA LUAD -0.159; TCGA LUSC -0.221; TCGA OV -0.216; TCGA PAAD -0.249; TCGA PRAD -0.107; TCGA STAD -0.163; TCGA UCEC -0.201 |
hsa-miR-200a-3p | PDE1C | 11 cancers: BLCA; BRCA; COAD; HNSC; KIRP; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.38; TCGA BRCA -0.391; TCGA COAD -0.21; TCGA HNSC -0.339; TCGA KIRP -0.515; TCGA LUAD -0.149; TCGA LUSC -0.48; TCGA PAAD -0.198; TCGA PRAD -0.202; TCGA STAD -0.4; TCGA UCEC -0.332 |
hsa-miR-200a-3p | PI4K2A | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; STAD | mirMAP | TCGA BLCA -0.054; TCGA CESC -0.109; TCGA COAD -0.101; TCGA ESCA -0.139; TCGA HNSC -0.133; TCGA KIRP -0.211; TCGA LUAD -0.132; TCGA LUSC -0.1; TCGA PAAD -0.134; TCGA STAD -0.077 |
hsa-miR-200a-3p | EIF4E3 | 9 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUSC; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.057; TCGA BRCA -0.148; TCGA COAD -0.11; TCGA ESCA -0.115; TCGA HNSC -0.166; TCGA LUSC -0.362; TCGA PRAD -0.072; TCGA STAD -0.221; TCGA UCEC -0.151 |
hsa-miR-200a-3p | HEYL | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.174; TCGA BRCA -0.054; TCGA CESC -0.27; TCGA COAD -0.321; TCGA ESCA -0.092; TCGA HNSC -0.342; TCGA KIRC -0.398; TCGA KIRP -0.212; TCGA LUAD -0.064; TCGA LUSC -0.373; TCGA PAAD -0.215; TCGA PRAD -0.05; TCGA STAD -0.206; TCGA UCEC -0.234 |
hsa-miR-200a-3p | CTSB | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD | mirMAP | TCGA BLCA -0.213; TCGA CESC -0.098; TCGA COAD -0.152; TCGA ESCA -0.107; TCGA HNSC -0.138; TCGA KIRC -0.134; TCGA KIRP -0.309; TCGA LUAD -0.186; TCGA LUSC -0.174; TCGA PAAD -0.177 |
hsa-miR-200a-3p | ABCA1 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; STAD | mirMAP | TCGA BLCA -0.102; TCGA BRCA -0.253; TCGA CESC -0.223; TCGA COAD -0.261; TCGA ESCA -0.237; TCGA HNSC -0.254; TCGA KIRC -0.297; TCGA KIRP -0.192; TCGA LUAD -0.179; TCGA LUSC -0.125; TCGA PAAD -0.189; TCGA STAD -0.163 |
hsa-miR-200a-3p | GFRA2 | 13 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; OV; PAAD; STAD; UCEC | mirMAP | TCGA BLCA -0.366; TCGA BRCA -0.439; TCGA COAD -0.519; TCGA ESCA -0.407; TCGA HNSC -0.364; TCGA KIRC -0.174; TCGA LIHC -0.114; TCGA LUAD -0.258; TCGA LUSC -0.497; TCGA OV -0.239; TCGA PAAD -0.432; TCGA STAD -0.505; TCGA UCEC -0.339 |
hsa-miR-200a-3p | SHE | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUSC; OV; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.253; TCGA BRCA -0.398; TCGA CESC -0.231; TCGA COAD -0.519; TCGA ESCA -0.126; TCGA HNSC -0.341; TCGA KIRP -0.145; TCGA LUSC -0.556; TCGA OV -0.152; TCGA PAAD -0.347; TCGA PRAD -0.082; TCGA STAD -0.281; TCGA UCEC -0.38 |
hsa-miR-200a-3p | KCNK3 | 10 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; LUSC; STAD; UCEC | mirMAP | TCGA BLCA -0.436; TCGA BRCA -0.246; TCGA CESC -0.231; TCGA COAD -0.934; TCGA HNSC -0.441; TCGA KIRC -0.231; TCGA KIRP -0.719; TCGA LUSC -0.488; TCGA STAD -0.714; TCGA UCEC -0.378 |
hsa-miR-200a-3p | ANO5 | 10 cancers: BLCA; BRCA; COAD; HNSC; LIHC; LUSC; OV; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.189; TCGA BRCA -0.311; TCGA COAD -0.567; TCGA HNSC -0.651; TCGA LIHC -0.118; TCGA LUSC -0.405; TCGA OV -0.29; TCGA THCA -0.105; TCGA STAD -0.424; TCGA UCEC -0.335 |
hsa-miR-200a-3p | LRRC15 | 11 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; UCEC | mirMAP | TCGA BLCA -0.479; TCGA CESC -0.326; TCGA COAD -0.546; TCGA ESCA -0.241; TCGA HNSC -0.68; TCGA KIRC -0.556; TCGA KIRP -0.324; TCGA LUAD -0.269; TCGA LUSC -0.386; TCGA PAAD -0.51; TCGA UCEC -0.195 |
hsa-miR-200a-3p | SH3PXD2B | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.148; TCGA BRCA -0.102; TCGA CESC -0.091; TCGA COAD -0.164; TCGA ESCA -0.202; TCGA HNSC -0.19; TCGA KIRC -0.423; TCGA KIRP -0.227; TCGA LUAD -0.076; TCGA LUSC -0.132; TCGA PAAD -0.237; TCGA PRAD -0.079; TCGA STAD -0.208; TCGA UCEC -0.091 |
hsa-miR-200a-3p | AMZ1 | 10 cancers: BLCA; CESC; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; UCEC | mirMAP | TCGA BLCA -0.212; TCGA CESC -0.285; TCGA ESCA -0.125; TCGA HNSC -0.166; TCGA KIRC -0.506; TCGA KIRP -0.379; TCGA LUAD -0.122; TCGA LUSC -0.452; TCGA PAAD -0.194; TCGA UCEC -0.185 |
hsa-miR-200a-3p | ARL10 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; PAAD; STAD; UCEC | mirMAP | TCGA BLCA -0.211; TCGA BRCA -0.225; TCGA CESC -0.268; TCGA COAD -0.414; TCGA ESCA -0.314; TCGA HNSC -0.194; TCGA KIRC -0.111; TCGA LUAD -0.182; TCGA PAAD -0.193; TCGA STAD -0.307; TCGA UCEC -0.237 |
hsa-miR-200a-3p | FAM26E | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.378; TCGA BRCA -0.299; TCGA CESC -0.327; TCGA COAD -0.593; TCGA ESCA -0.222; TCGA HNSC -0.475; TCGA KIRC -0.12; TCGA KIRP -0.448; TCGA LUAD -0.272; TCGA LUSC -0.435; TCGA OV -0.112; TCGA PAAD -0.326; TCGA PRAD -0.098; TCGA STAD -0.189; TCGA UCEC -0.284 |
hsa-miR-200a-3p | FAM133A | 9 cancers: BLCA; BRCA; ESCA; LIHC; LUAD; OV; PAAD; STAD; UCEC | mirMAP | TCGA BLCA -0.297; TCGA BRCA -0.209; TCGA ESCA -0.352; TCGA LIHC -0.441; TCGA LUAD -0.288; TCGA OV -0.267; TCGA PAAD -0.183; TCGA STAD -0.491; TCGA UCEC -0.274 |
hsa-miR-200a-3p | ST6GALNAC3 | 13 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRP; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.208; TCGA BRCA -0.326; TCGA CESC -0.297; TCGA COAD -0.15; TCGA HNSC -0.286; TCGA KIRP -0.319; TCGA LUAD -0.174; TCGA LUSC -0.38; TCGA OV -0.259; TCGA PAAD -0.198; TCGA PRAD -0.129; TCGA STAD -0.199; TCGA UCEC -0.351 |
hsa-miR-200a-3p | OLFML2A | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; LUAD; OV; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.25; TCGA BRCA -0.202; TCGA CESC -0.439; TCGA COAD -0.317; TCGA ESCA -0.218; TCGA KIRC -0.534; TCGA LUAD -0.151; TCGA OV -0.16; TCGA PAAD -0.162; TCGA PRAD -0.069; TCGA STAD -0.239; TCGA UCEC -0.362 |
hsa-miR-200a-3p | BACE1 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; STAD; UCEC | mirMAP; miRNATAP | TCGA BLCA -0.188; TCGA BRCA -0.151; TCGA CESC -0.063; TCGA COAD -0.203; TCGA ESCA -0.149; TCGA HNSC -0.196; TCGA LUAD -0.141; TCGA LUSC -0.135; TCGA PAAD -0.138; TCGA STAD -0.209; TCGA UCEC -0.11 |
hsa-miR-200a-3p | TEAD1 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.152; TCGA BRCA -0.223; TCGA CESC -0.063; TCGA COAD -0.149; TCGA ESCA -0.055; TCGA HNSC -0.214; TCGA LUAD -0.102; TCGA LUSC -0.1; TCGA PAAD -0.073; TCGA PRAD -0.073; TCGA STAD -0.316; TCGA UCEC -0.144 |
hsa-miR-200a-3p | PRELP | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; STAD; UCEC | mirMAP | TCGA BLCA -0.444; TCGA BRCA -0.401; TCGA CESC -0.311; TCGA COAD -1.083; TCGA ESCA -0.499; TCGA HNSC -0.451; TCGA KIRP -0.305; TCGA LUAD -0.134; TCGA LUSC -0.539; TCGA PAAD -0.243; TCGA STAD -0.719; TCGA UCEC -0.533 |
hsa-miR-200a-3p | AFAP1 | 11 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LUAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.226; TCGA BRCA -0.072; TCGA CESC -0.077; TCGA ESCA -0.065; TCGA HNSC -0.172; TCGA KIRC -0.169; TCGA KIRP -0.122; TCGA LUAD -0.071; TCGA PRAD -0.053; TCGA STAD -0.179; TCGA UCEC -0.116 |
hsa-miR-200a-3p | SPN | 10 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD | mirMAP | TCGA BLCA -0.287; TCGA BRCA -0.064; TCGA CESC -0.18; TCGA COAD -0.235; TCGA HNSC -0.271; TCGA KIRC -0.312; TCGA KIRP -0.177; TCGA LUAD -0.215; TCGA LUSC -0.65; TCGA PAAD -0.394 |
hsa-miR-200a-3p | ITSN1 | 11 cancers: BLCA; BRCA; COAD; HNSC; KIRP; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.073; TCGA BRCA -0.282; TCGA COAD -0.095; TCGA HNSC -0.121; TCGA KIRP -0.114; TCGA LUAD -0.085; TCGA LUSC -0.147; TCGA PAAD -0.113; TCGA PRAD -0.063; TCGA STAD -0.1; TCGA UCEC -0.118 |
hsa-miR-200a-3p | GIMAP1 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; UCEC | mirMAP | TCGA BLCA -0.274; TCGA BRCA -0.196; TCGA CESC -0.333; TCGA COAD -0.35; TCGA ESCA -0.141; TCGA HNSC -0.275; TCGA KIRC -0.159; TCGA KIRP -0.203; TCGA LUAD -0.164; TCGA LUSC -0.5; TCGA PAAD -0.379; TCGA UCEC -0.272 |
hsa-miR-200a-3p | NEURL1B | 9 cancers: BLCA; BRCA; ESCA; KIRC; KIRP; LUSC; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.217; TCGA BRCA -0.083; TCGA ESCA -0.14; TCGA KIRC -0.216; TCGA KIRP -0.187; TCGA LUSC -0.119; TCGA PRAD -0.069; TCGA STAD -0.203; TCGA UCEC -0.18 |
hsa-miR-200a-3p | PLXNA4 | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUAD; LUSC; OV; PAAD; STAD | mirMAP; miRNATAP | TCGA BLCA -0.52; TCGA BRCA -0.52; TCGA COAD -0.843; TCGA ESCA -0.33; TCGA HNSC -0.489; TCGA LUAD -0.149; TCGA LUSC -0.542; TCGA OV -0.227; TCGA PAAD -0.465; TCGA STAD -0.468 |
hsa-miR-200a-3p | MYH10 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.138; TCGA BRCA -0.148; TCGA CESC -0.204; TCGA COAD -0.422; TCGA ESCA -0.406; TCGA HNSC -0.246; TCGA KIRP -0.106; TCGA LUAD -0.15; TCGA LUSC -0.323; TCGA OV -0.102; TCGA PAAD -0.21; TCGA PRAD -0.081; TCGA STAD -0.287; TCGA UCEC -0.149 |
hsa-miR-200a-3p | CXCL12 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.497; TCGA BRCA -0.361; TCGA CESC -0.45; TCGA COAD -0.468; TCGA ESCA -0.309; TCGA HNSC -0.578; TCGA KIRP -0.395; TCGA LUAD -0.293; TCGA LUSC -0.564; TCGA OV -0.302; TCGA PAAD -0.649; TCGA PRAD -0.122; TCGA STAD -0.44; TCGA UCEC -0.543 |
hsa-miR-200a-3p | NAMPT | 9 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LUAD; PAAD; PRAD | miRNATAP | TCGA BLCA -0.164; TCGA BRCA -0.089; TCGA CESC -0.133; TCGA HNSC -0.08; TCGA KIRC -0.106; TCGA KIRP -0.163; TCGA LUAD -0.214; TCGA PAAD -0.114; TCGA PRAD -0.104 |
hsa-miR-200a-3p | BRMS1L | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRP; OV; PAAD; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.106; TCGA BRCA -0.06; TCGA CESC -0.066; TCGA COAD -0.109; TCGA ESCA -0.101; TCGA KIRP -0.118; TCGA OV -0.07; TCGA PAAD -0.08; TCGA PRAD -0.068; TCGA STAD -0.093; TCGA UCEC -0.118 |
hsa-miR-200a-3p | DLC1 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUSC; OV; PAAD; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.158; TCGA BRCA -0.334; TCGA CESC -0.244; TCGA COAD -0.372; TCGA ESCA -0.096; TCGA HNSC -0.425; TCGA KIRC -0.103; TCGA LUSC -0.624; TCGA OV -0.126; TCGA PAAD -0.296; TCGA PRAD -0.123; TCGA STAD -0.298; TCGA UCEC -0.207 |
hsa-miR-200a-3p | SYNPO2 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; PRAD; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.489; TCGA BRCA -0.445; TCGA CESC -0.327; TCGA COAD -1.004; TCGA ESCA -0.438; TCGA HNSC -0.678; TCGA KIRP -0.332; TCGA LUAD -0.224; TCGA LUSC -0.309; TCGA PAAD -0.382; TCGA PRAD -0.096; TCGA THCA -0.05; TCGA STAD -0.913; TCGA UCEC -0.507 |
hsa-miR-200a-3p | PALM2-AKAP2 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; OV; PAAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.371; TCGA BRCA -0.335; TCGA CESC -0.323; TCGA COAD -0.385; TCGA ESCA -0.12; TCGA HNSC -0.38; TCGA KIRC -0.147; TCGA KIRP -0.217; TCGA LUAD -0.249; TCGA LUSC -0.546; TCGA OV -0.069; TCGA PAAD -0.306; TCGA STAD -0.258; TCGA UCEC -0.29 |
hsa-miR-200a-3p | NRP1 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.246; TCGA BRCA -0.273; TCGA CESC -0.237; TCGA COAD -0.407; TCGA ESCA -0.129; TCGA HNSC -0.371; TCGA KIRC -0.234; TCGA LUAD -0.177; TCGA LUSC -0.395; TCGA PAAD -0.202; TCGA STAD -0.158; TCGA UCEC -0.164 |
hsa-miR-200a-3p | MAP3K3 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.161; TCGA BRCA -0.135; TCGA CESC -0.12; TCGA COAD -0.239; TCGA ESCA -0.148; TCGA HNSC -0.216; TCGA LUAD -0.082; TCGA LUSC -0.251; TCGA PAAD -0.215; TCGA STAD -0.231; TCGA UCEC -0.189 |
hsa-miR-200a-3p | IGDCC4 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.282; TCGA BRCA -0.244; TCGA CESC -0.279; TCGA COAD -0.26; TCGA ESCA -0.123; TCGA HNSC -0.58; TCGA KIRC -0.258; TCGA LUAD -0.177; TCGA LUSC -0.294; TCGA OV -0.121; TCGA PAAD -0.522; TCGA PRAD -0.137; TCGA STAD -0.158; TCGA UCEC -0.293 |
hsa-miR-200a-3p | PTEN | 10 cancers: BLCA; BRCA; CESC; HNSC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.061; TCGA BRCA -0.109; TCGA CESC -0.073; TCGA HNSC -0.148; TCGA LUAD -0.093; TCGA LUSC -0.204; TCGA PAAD -0.126; TCGA PRAD -0.122; TCGA STAD -0.075; TCGA UCEC -0.092 |
hsa-miR-200a-3p | ARID5B | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; PAAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.137; TCGA BRCA -0.244; TCGA CESC -0.138; TCGA COAD -0.128; TCGA ESCA -0.135; TCGA HNSC -0.131; TCGA LUAD -0.065; TCGA PAAD -0.279; TCGA STAD -0.098; TCGA UCEC -0.123 |
hsa-miR-200a-3p | MARCH1 | 11 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; STAD | miRNATAP | TCGA BLCA -0.178; TCGA CESC -0.13; TCGA COAD -0.264; TCGA ESCA -0.12; TCGA HNSC -0.291; TCGA KIRC -0.191; TCGA KIRP -0.267; TCGA LUAD -0.244; TCGA LUSC -0.376; TCGA PAAD -0.453; TCGA STAD -0.057 |
hsa-miR-200a-3p | SOX5 | 10 cancers: BLCA; BRCA; COAD; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; STAD | miRNATAP | TCGA BLCA -0.227; TCGA BRCA -0.351; TCGA COAD -0.512; TCGA HNSC -0.276; TCGA LIHC -0.087; TCGA LUAD -0.177; TCGA LUSC -0.238; TCGA PAAD -0.141; TCGA PRAD -0.098; TCGA STAD -0.35 |
hsa-miR-200a-3p | ANK2 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.579; TCGA BRCA -0.461; TCGA CESC -0.34; TCGA COAD -0.892; TCGA ESCA -0.386; TCGA HNSC -0.596; TCGA KIRP -0.957; TCGA LUAD -0.173; TCGA LUSC -0.583; TCGA PAAD -0.454; TCGA THCA -0.092; TCGA STAD -0.628; TCGA UCEC -0.28 |
hsa-miR-200a-3p | PCDH17 | 12 cancers: BLCA; CESC; COAD; HNSC; KIRC; KIRP; LUAD; LUSC; OV; PAAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.111; TCGA CESC -0.141; TCGA COAD -0.334; TCGA HNSC -0.344; TCGA KIRC -0.386; TCGA KIRP -0.412; TCGA LUAD -0.135; TCGA LUSC -0.315; TCGA OV -0.114; TCGA PAAD -0.227; TCGA STAD -0.101; TCGA UCEC -0.145 |
hsa-miR-200a-3p | LPP | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; PRAD; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.194; TCGA BRCA -0.161; TCGA COAD -0.181; TCGA ESCA -0.124; TCGA HNSC -0.136; TCGA KIRP -0.17; TCGA PRAD -0.141; TCGA THCA -0.067; TCGA STAD -0.37; TCGA UCEC -0.134 |
hsa-miR-200a-3p | GRIN3A | 9 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LUAD; LUSC; PAAD; UCEC | miRNATAP | TCGA BLCA -0.189; TCGA BRCA -0.096; TCGA CESC -0.11; TCGA ESCA -0.097; TCGA HNSC -0.138; TCGA LUAD -0.159; TCGA LUSC -0.231; TCGA PAAD -0.21; TCGA UCEC -0.136 |
hsa-miR-200a-3p | CDON | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; PAAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.341; TCGA BRCA -0.09; TCGA COAD -0.352; TCGA ESCA -0.133; TCGA HNSC -0.101; TCGA KIRC -0.363; TCGA KIRP -0.346; TCGA PAAD -0.253; TCGA STAD -0.496; TCGA UCEC -0.316 |
hsa-miR-200a-3p | SPRED1 | 9 cancers: BLCA; BRCA; CESC; HNSC; KIRC; LUAD; LUSC; PRAD; UCEC | miRNATAP | TCGA BLCA -0.18; TCGA BRCA -0.132; TCGA CESC -0.093; TCGA HNSC -0.164; TCGA KIRC -0.171; TCGA LUAD -0.148; TCGA LUSC -0.195; TCGA PRAD -0.06; TCGA UCEC -0.129 |
hsa-miR-200a-3p | RHOBTB1 | 9 cancers: BLCA; BRCA; COAD; HNSC; KIRC; KIRP; LUSC; PRAD; UCEC | miRNATAP | TCGA BLCA -0.071; TCGA BRCA -0.1; TCGA COAD -0.107; TCGA HNSC -0.342; TCGA KIRC -0.155; TCGA KIRP -0.197; TCGA LUSC -0.242; TCGA PRAD -0.058; TCGA UCEC -0.081 |
hsa-miR-200a-3p | MAP2 | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; OV; PRAD; STAD | miRNATAP | TCGA BLCA -0.22; TCGA BRCA -0.117; TCGA COAD -0.5; TCGA ESCA -0.174; TCGA HNSC -0.24; TCGA KIRP -0.22; TCGA LIHC -0.106; TCGA LUAD -0.109; TCGA LUSC -0.243; TCGA OV -0.2; TCGA PRAD -0.22; TCGA STAD -0.327 |
hsa-miR-200a-3p | DZIP1 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; OV; PAAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.268; TCGA BRCA -0.223; TCGA CESC -0.258; TCGA COAD -0.61; TCGA ESCA -0.468; TCGA HNSC -0.112; TCGA LUAD -0.139; TCGA OV -0.187; TCGA PAAD -0.246; TCGA STAD -0.427; TCGA UCEC -0.323 |
hsa-miR-200a-3p | TTYH2 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.317; TCGA BRCA -0.165; TCGA CESC -0.404; TCGA COAD -0.36; TCGA ESCA -0.256; TCGA HNSC -0.217; TCGA KIRC -0.276; TCGA KIRP -0.34; TCGA LUAD -0.092; TCGA LUSC -0.189; TCGA OV -0.093; TCGA PAAD -0.35; TCGA PRAD -0.052; TCGA STAD -0.246; TCGA UCEC -0.355 |
hsa-miR-200a-3p | MAF | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; OV; PAAD; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.314; TCGA BRCA -0.239; TCGA CESC -0.38; TCGA COAD -0.402; TCGA ESCA -0.346; TCGA HNSC -0.215; TCGA KIRC -0.283; TCGA KIRP -0.32; TCGA LIHC -0.051; TCGA LUAD -0.126; TCGA OV -0.132; TCGA PAAD -0.251; TCGA PRAD -0.103; TCGA STAD -0.21; TCGA UCEC -0.366 |
hsa-miR-200a-3p | SVIL | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; PAAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.227; TCGA BRCA -0.15; TCGA CESC -0.144; TCGA COAD -0.284; TCGA ESCA -0.283; TCGA HNSC -0.144; TCGA LUAD -0.093; TCGA PAAD -0.175; TCGA STAD -0.554; TCGA UCEC -0.283 |
hsa-miR-200a-3p | NPTX1 | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRP; LUSC; OV; STAD; UCEC | miRNATAP | TCGA BLCA -0.23; TCGA BRCA -0.154; TCGA CESC -0.389; TCGA COAD -1.098; TCGA ESCA -0.723; TCGA KIRP -0.395; TCGA LUSC -0.281; TCGA OV -0.19; TCGA STAD -0.958; TCGA UCEC -0.206 |
hsa-miR-200a-3p | RARB | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; OV; PAAD; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.237; TCGA BRCA -0.106; TCGA COAD -0.406; TCGA ESCA -0.178; TCGA HNSC -0.145; TCGA KIRP -0.129; TCGA OV -0.127; TCGA PAAD -0.176; TCGA PRAD -0.063; TCGA STAD -0.231; TCGA UCEC -0.158 |
hsa-miR-200a-3p | NRP2 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.45; TCGA BRCA -0.188; TCGA CESC -0.2; TCGA COAD -0.555; TCGA ESCA -0.3; TCGA HNSC -0.172; TCGA KIRC -0.354; TCGA KIRP -0.412; TCGA LUAD -0.211; TCGA LUSC -0.346; TCGA PAAD -0.201; TCGA PRAD -0.118; TCGA STAD -0.45; TCGA UCEC -0.238 |
hsa-miR-200a-3p | SLC6A17 | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; THCA; STAD | miRNATAP | TCGA BLCA -0.296; TCGA CESC -0.314; TCGA COAD -0.549; TCGA ESCA -0.232; TCGA HNSC -0.452; TCGA KIRP -0.628; TCGA LUAD -0.281; TCGA LUSC -0.304; TCGA THCA -0.196; TCGA STAD -0.344 |
hsa-miR-200a-3p | PLXNC1 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.397; TCGA BRCA -0.192; TCGA CESC -0.266; TCGA COAD -0.562; TCGA ESCA -0.153; TCGA HNSC -0.636; TCGA KIRC -0.291; TCGA KIRP -0.58; TCGA LUAD -0.234; TCGA LUSC -0.682; TCGA PAAD -0.317; TCGA PRAD -0.114; TCGA STAD -0.083; TCGA UCEC -0.265 |
hsa-miR-200a-3p | MDGA1 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; OV; PAAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.328; TCGA BRCA -0.145; TCGA CESC -0.172; TCGA COAD -0.491; TCGA ESCA -0.372; TCGA HNSC -0.156; TCGA KIRP -0.215; TCGA LUAD -0.262; TCGA OV -0.14; TCGA PAAD -0.39; TCGA STAD -0.349; TCGA UCEC -0.315 |
hsa-miR-200a-3p | SETBP1 | 9 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; PAAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.317; TCGA BRCA -0.138; TCGA CESC -0.272; TCGA COAD -0.503; TCGA ESCA -0.404; TCGA HNSC -0.275; TCGA PAAD -0.232; TCGA STAD -0.496; TCGA UCEC -0.273 |
hsa-miR-200a-3p | EGR2 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.408; TCGA BRCA -0.269; TCGA CESC -0.189; TCGA COAD -0.534; TCGA ESCA -0.31; TCGA HNSC -0.253; TCGA LUAD -0.174; TCGA LUSC -0.374; TCGA PAAD -0.306; TCGA PRAD -0.09; TCGA STAD -0.162; TCGA UCEC -0.226 |
hsa-miR-200a-3p | EPB41L3 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.245; TCGA BRCA -0.176; TCGA CESC -0.18; TCGA COAD -0.498; TCGA ESCA -0.309; TCGA HNSC -0.241; TCGA KIRP -0.225; TCGA LUAD -0.26; TCGA LUSC -0.33; TCGA PAAD -0.246; TCGA THCA -0.064; TCGA STAD -0.268; TCGA UCEC -0.272 |
hsa-miR-200a-3p | NMNAT2 | 9 cancers: BLCA; BRCA; COAD; HNSC; KIRC; LUSC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.251; TCGA BRCA -0.382; TCGA COAD -0.45; TCGA HNSC -0.211; TCGA KIRC -0.34; TCGA LUSC -0.424; TCGA THCA -0.105; TCGA STAD -0.333; TCGA UCEC -0.104 |
hsa-miR-200a-3p | KLF6 | 10 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; LUAD; LUSC; STAD; UCEC | miRNATAP | TCGA BLCA -0.117; TCGA BRCA -0.181; TCGA CESC -0.213; TCGA COAD -0.076; TCGA HNSC -0.171; TCGA KIRC -0.101; TCGA LUAD -0.179; TCGA LUSC -0.392; TCGA STAD -0.051; TCGA UCEC -0.166 |
hsa-miR-200a-3p | COL11A1 | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; OV; PAAD | miRNATAP | TCGA BLCA -0.27; TCGA CESC -0.473; TCGA COAD -0.772; TCGA ESCA -0.368; TCGA HNSC -0.617; TCGA KIRC -0.828; TCGA KIRP -0.569; TCGA LUAD -0.287; TCGA OV -0.484; TCGA PAAD -0.378 |
hsa-miR-200a-3p | SAMD4A | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.321; TCGA BRCA -0.3; TCGA CESC -0.337; TCGA COAD -0.397; TCGA ESCA -0.161; TCGA HNSC -0.315; TCGA KIRC -0.077; TCGA KIRP -0.079; TCGA LUAD -0.207; TCGA LUSC -0.319; TCGA PAAD -0.238; TCGA STAD -0.369; TCGA UCEC -0.322 |
hsa-miR-200a-3p | ASTN1 | 11 cancers: BRCA; COAD; ESCA; KIRP; LUAD; OV; PAAD; PRAD; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BRCA -0.36; TCGA COAD -0.903; TCGA ESCA -0.619; TCGA KIRP -1.238; TCGA LUAD -0.227; TCGA OV -0.296; TCGA PAAD -0.268; TCGA PRAD -0.22; TCGA THCA -0.148; TCGA STAD -0.863; TCGA UCEC -0.314 |
hsa-miR-200a-3p | CREB3L2 | 9 cancers: BRCA; COAD; ESCA; HNSC; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BRCA -0.067; TCGA COAD -0.107; TCGA ESCA -0.077; TCGA HNSC -0.215; TCGA LUSC -0.172; TCGA PAAD -0.11; TCGA PRAD -0.063; TCGA STAD -0.135; TCGA UCEC -0.143 |
hsa-miR-200a-3p | PRTG | 11 cancers: BRCA; CESC; COAD; HNSC; KIRP; LUSC; OV; PAAD; PRAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BRCA -0.283; TCGA CESC -0.382; TCGA COAD -0.68; TCGA HNSC -0.412; TCGA KIRP -0.395; TCGA LUSC -0.617; TCGA OV -0.198; TCGA PAAD -0.3; TCGA PRAD -0.117; TCGA STAD -0.368; TCGA UCEC -0.381 |
hsa-miR-200a-3p | ANO6 | 14 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BRCA -0.258; TCGA CESC -0.163; TCGA COAD -0.149; TCGA ESCA -0.108; TCGA HNSC -0.242; TCGA KIRC -0.065; TCGA KIRP -0.212; TCGA LIHC -0.057; TCGA LUAD -0.193; TCGA LUSC -0.238; TCGA PAAD -0.144; TCGA PRAD -0.072; TCGA STAD -0.162; TCGA UCEC -0.23 |
hsa-miR-200a-3p | YWHAG | 9 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; PRAD | MirTarget; miRNATAP | TCGA BRCA -0.093; TCGA CESC -0.102; TCGA COAD -0.063; TCGA ESCA -0.073; TCGA HNSC -0.11; TCGA KIRC -0.108; TCGA KIRP -0.133; TCGA LUAD -0.112; TCGA PRAD -0.063 |
hsa-miR-200a-3p | ST3GAL5 | 12 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUSC; OV; PAAD; STAD; UCEC | MirTarget | TCGA BRCA -0.151; TCGA CESC -0.286; TCGA COAD -0.246; TCGA ESCA -0.108; TCGA HNSC -0.157; TCGA KIRC -0.143; TCGA KIRP -0.159; TCGA LUSC -0.341; TCGA OV -0.083; TCGA PAAD -0.136; TCGA STAD -0.146; TCGA UCEC -0.219 |
hsa-miR-200a-3p | RAB8B | 12 cancers: BRCA; CESC; COAD; HNSC; KIRC; LUAD; LUSC; OV; PAAD; THCA; STAD; UCEC | MirTarget | TCGA BRCA -0.15; TCGA CESC -0.167; TCGA COAD -0.213; TCGA HNSC -0.192; TCGA KIRC -0.09; TCGA LUAD -0.199; TCGA LUSC -0.307; TCGA OV -0.077; TCGA PAAD -0.164; TCGA THCA -0.06; TCGA STAD -0.074; TCGA UCEC -0.233 |
hsa-miR-200a-3p | ADAM22 | 12 cancers: BRCA; COAD; HNSC; KIRP; LUAD; LUSC; OV; PAAD; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BRCA -0.323; TCGA COAD -0.276; TCGA HNSC -0.165; TCGA KIRP -0.715; TCGA LUAD -0.211; TCGA LUSC -0.218; TCGA OV -0.33; TCGA PAAD -0.237; TCGA PRAD -0.148; TCGA THCA -0.14; TCGA STAD -0.176; TCGA UCEC -0.274 |
hsa-miR-200a-3p | GLDN | 9 cancers: BRCA; CESC; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; STAD | MirTarget | TCGA BRCA -0.425; TCGA CESC -0.165; TCGA ESCA -0.166; TCGA HNSC -0.172; TCGA LUAD -0.275; TCGA LUSC -0.212; TCGA PAAD -0.195; TCGA PRAD -0.169; TCGA STAD -0.313 |
hsa-miR-200a-3p | IDS | 9 cancers: BRCA; COAD; HNSC; KIRC; KIRP; LUAD; LUSC; STAD; UCEC | mirMAP | TCGA BRCA -0.074; TCGA COAD -0.182; TCGA HNSC -0.114; TCGA KIRC -0.083; TCGA KIRP -0.136; TCGA LUAD -0.131; TCGA LUSC -0.119; TCGA STAD -0.157; TCGA UCEC -0.095 |
hsa-miR-200a-3p | EHD3 | 10 cancers: BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; STAD; UCEC | mirMAP | TCGA BRCA -0.071; TCGA CESC -0.204; TCGA COAD -0.38; TCGA ESCA -0.286; TCGA HNSC -0.2; TCGA LUAD -0.132; TCGA LUSC -0.085; TCGA PAAD -0.222; TCGA STAD -0.238; TCGA UCEC -0.097 |
hsa-miR-200a-3p | C20orf194 | 9 cancers: BRCA; CESC; COAD; ESCA; HNSC; LUSC; PAAD; STAD; UCEC | mirMAP | TCGA BRCA -0.174; TCGA CESC -0.107; TCGA COAD -0.634; TCGA ESCA -0.263; TCGA HNSC -0.105; TCGA LUSC -0.091; TCGA PAAD -0.119; TCGA STAD -0.358; TCGA UCEC -0.161 |
hsa-miR-200a-3p | ZNF423 | 12 cancers: BRCA; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BRCA -0.373; TCGA COAD -0.367; TCGA ESCA -0.274; TCGA HNSC -0.497; TCGA KIRC -0.201; TCGA LUAD -0.176; TCGA LUSC -0.49; TCGA OV -0.155; TCGA PAAD -0.275; TCGA PRAD -0.076; TCGA STAD -0.414; TCGA UCEC -0.213 |
hsa-miR-200a-3p | ZFP37 | 9 cancers: BRCA; CESC; COAD; ESCA; LUAD; OV; PAAD; STAD; UCEC | mirMAP | TCGA BRCA -0.141; TCGA CESC -0.175; TCGA COAD -0.205; TCGA ESCA -0.137; TCGA LUAD -0.119; TCGA OV -0.152; TCGA PAAD -0.161; TCGA STAD -0.174; TCGA UCEC -0.135 |
hsa-miR-200a-3p | SHROOM4 | 9 cancers: BRCA; CESC; HNSC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BRCA -0.267; TCGA CESC -0.119; TCGA HNSC -0.391; TCGA LUAD -0.092; TCGA LUSC -0.406; TCGA PAAD -0.254; TCGA PRAD -0.08; TCGA STAD -0.152; TCGA UCEC -0.121 |
hsa-miR-200a-3p | AR | 11 cancers: BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUSC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BRCA -0.294; TCGA CESC -0.229; TCGA COAD -0.661; TCGA ESCA -0.208; TCGA HNSC -0.598; TCGA LIHC -0.322; TCGA LUSC -0.508; TCGA PAAD -0.369; TCGA PRAD -0.094; TCGA STAD -0.615; TCGA UCEC -0.155 |
hsa-miR-200a-3p | PKIA | 10 cancers: BRCA; CESC; COAD; ESCA; HNSC; LUAD; OV; PRAD; STAD; UCEC | mirMAP | TCGA BRCA -0.362; TCGA CESC -0.279; TCGA COAD -0.532; TCGA ESCA -0.251; TCGA HNSC -0.453; TCGA LUAD -0.136; TCGA OV -0.515; TCGA PRAD -0.106; TCGA STAD -0.422; TCGA UCEC -0.331 |
hsa-miR-200a-3p | SNAP91 | 9 cancers: BRCA; COAD; ESCA; KIRP; OV; PAAD; THCA; STAD; UCEC | miRNATAP | TCGA BRCA -0.127; TCGA COAD -0.82; TCGA ESCA -0.554; TCGA KIRP -0.49; TCGA OV -0.31; TCGA PAAD -0.327; TCGA THCA -0.107; TCGA STAD -0.793; TCGA UCEC -0.367 |
hsa-miR-200a-3p | RBMS1 | 10 cancers: BRCA; CESC; COAD; ESCA; HNSC; LUAD; OV; PAAD; STAD; UCEC | miRNATAP | TCGA BRCA -0.203; TCGA CESC -0.201; TCGA COAD -0.517; TCGA ESCA -0.236; TCGA HNSC -0.074; TCGA LUAD -0.12; TCGA OV -0.054; TCGA PAAD -0.215; TCGA STAD -0.23; TCGA UCEC -0.185 |
hsa-miR-200a-3p | SLC23A2 | 9 cancers: BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUSC; STAD; UCEC | miRNATAP | TCGA BRCA -0.124; TCGA CESC -0.139; TCGA COAD -0.133; TCGA ESCA -0.088; TCGA HNSC -0.103; TCGA LIHC -0.067; TCGA LUSC -0.089; TCGA STAD -0.121; TCGA UCEC -0.228 |
hsa-miR-200a-3p | DSTYK | 9 cancers: CESC; COAD; ESCA; HNSC; KIRP; LUSC; PAAD; STAD; UCEC | MirTarget | TCGA CESC -0.097; TCGA COAD -0.092; TCGA ESCA -0.102; TCGA HNSC -0.085; TCGA KIRP -0.101; TCGA LUSC -0.063; TCGA PAAD -0.119; TCGA STAD -0.131; TCGA UCEC -0.113 |
hsa-miR-200a-3p | KIF3C | 11 cancers: CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; OV; PAAD; STAD; UCEC | mirMAP | TCGA CESC -0.256; TCGA COAD -0.264; TCGA ESCA -0.289; TCGA HNSC -0.112; TCGA KIRC -0.197; TCGA KIRP -0.263; TCGA LUAD -0.172; TCGA OV -0.099; TCGA PAAD -0.131; TCGA STAD -0.292; TCGA UCEC -0.137 |
hsa-miR-200a-3p | GNA13 | 9 cancers: COAD; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; STAD; UCEC | MirTarget | TCGA COAD -0.072; TCGA HNSC -0.094; TCGA KIRC -0.117; TCGA KIRP -0.175; TCGA LUAD -0.145; TCGA LUSC -0.056; TCGA PAAD -0.149; TCGA STAD -0.053; TCGA UCEC -0.074 |
Enriched cancer pathways of putative targets