microRNA information: hsa-miR-203a-3p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-203a-3p | miRbase |
| Accession: | MIMAT0000264 | miRbase |
| Precursor name: | hsa-mir-203a | miRbase |
| Precursor accession: | MI0000283 | miRbase |
| Symbol: | NA | HGNC |
| RefSeq ID: | NA | GenBank |
| Sequence: | GUGAAAUGUUUAGGACCACUAG |
Reported expression in cancers: hsa-miR-203a-3p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-203a-3p | acute myeloid leukemia | upregulation | "By multivariate analysis we identified that high e ......" | 25428263 | |
| hsa-miR-203a-3p | bladder cancer | deregulation | "In this article we define for the first time that ......" | 21836020 | |
| hsa-miR-203a-3p | breast cancer | upregulation | "In this study the authors observed a significant u ......" | 22207897 | |
| hsa-miR-203a-3p | breast cancer | upregulation | "Western blot and QPCR were used to determine the e ......" | 22260523 | qPCR |
| hsa-miR-203a-3p | breast cancer | upregulation | "We previously observed that miR-203 was highly upr ......" | 27517632 | |
| hsa-miR-203a-3p | cervical and endocervical cancer | downregulation | "We analyzed serum levels of miR-20a and miR-203 in ......" | 23819812 | qPCR |
| hsa-miR-203a-3p | cervical and endocervical cancer | downregulation | "In this study we investigated the roles and mechan ......" | 23867971 | qPCR |
| hsa-miR-203a-3p | cervical and endocervical cancer | downregulation | "Down-regulation was reported most consistently for ......" | 25920605 | |
| hsa-miR-203a-3p | cervical and endocervical cancer | downregulation | "In the current study the expressions of miR-203 an ......" | 26490989 | qPCR |
| hsa-miR-203a-3p | colorectal cancer | downregulation | "Aberrant expression of miR 203 and its clinical si ......" | 21063914 | qPCR |
| hsa-miR-203a-3p | colorectal cancer | downregulation | "MicroRNAs miRNAs are short non-coding RNAs that re ......" | 27236538 | |
| hsa-miR-203a-3p | colorectal cancer | downregulation | "In this study miR-203 was found to be downregulate ......" | 27253631 | |
| hsa-miR-203a-3p | colorectal cancer | downregulation | "While it is known that miR-203 is frequently downr ......" | 27376958 | |
| hsa-miR-203a-3p | colorectal cancer | downregulation | "The expressions of miR-203 in the tissues of 122 C ......" | 27714672 | Microarray |
| hsa-miR-203a-3p | esophageal cancer | downregulation | "The expression of miR-203 has been reported to be ......" | 24001611 | |
| hsa-miR-203a-3p | esophageal cancer | downregulation | "Finally we identified miR-203 as a direct target o ......" | 24994936 | |
| hsa-miR-203a-3p | esophageal cancer | downregulation | "Methylation mediated repression of potential tumor ......" | 26577858 | |
| hsa-miR-203a-3p | gastric cancer | downregulation | "In this study we reported that miR-203 is signific ......" | 25373785 | |
| hsa-miR-203a-3p | head and neck cancer | downregulation | "Conversely decreased expressions of miR-153 miR-20 ......" | 25677760 | |
| hsa-miR-203a-3p | head and neck cancer | downregulation | "The miR-200 family and miR-203 were downregulated ......" | 26882562 | |
| hsa-miR-203a-3p | kidney renal cell cancer | upregulation | "miR 203a regulates proliferation migration and apo ......" | 25123268 | |
| hsa-miR-203a-3p | liver cancer | upregulation | "Although recent studies have shown the utility of ......" | 26109910 | Reverse transcription PCR; qPCR |
| hsa-miR-203a-3p | lung cancer | deregulation | "MiRNA profiles were obtained using microarray and ......" | 25359683 | qPCR; Microarray |
| hsa-miR-203a-3p | lung squamous cell cancer | downregulation | "Association between downexpression of MiR 203 and ......" | 26307752 | Reverse transcription PCR; qPCR |
| hsa-miR-203a-3p | lung squamous cell cancer | downregulation | "Moreover analysis of NSCLC patient samples reveale ......" | 27177222 | |
| hsa-miR-203a-3p | melanoma | downregulation | "To identify improved molecular markers to support ......" | 22223089 | Microarray |
| hsa-miR-203a-3p | melanoma | downregulation | "We recently reported that miR-203 is down-regulate ......" | 22354972 | |
| hsa-miR-203a-3p | melanoma | downregulation | "Presently we examined the expression profile of mi ......" | 23638671 | qPCR; Microarray |
| hsa-miR-203a-3p | melanoma | downregulation | "We have elucidated the antitumor mechanisms of ant ......" | 26225581 | |
| hsa-miR-203a-3p | melanoma | downregulation | "Expression of miR 203 is decreased and associated ......" | 26722525 | qPCR |
| hsa-miR-203a-3p | ovarian cancer | upregulation | "The purpose of this study was to investigate wheth ......" | 23918241 | qPCR |
| hsa-miR-203a-3p | ovarian cancer | upregulation | "The role and expression of miR 100 and miR 203 pro ......" | 27347348 | qPCR |
| hsa-miR-203a-3p | prostate cancer | downregulation | "Here we show that miR-203 is downregulated in clin ......" | 21368580 | |
| hsa-miR-203a-3p | prostate cancer | downregulation | "MiR 130a miR 203 and miR 205 jointly repress key o ......" | 22391564 | |
| hsa-miR-203a-3p | prostate cancer | downregulation | "miR-203 levels are down-regulated in clinical samp ......" | 25004126 | |
| hsa-miR-203a-3p | prostate cancer | downregulation | "However the exact mechanisms of miR-203 in PCa are ......" | 25636908 | qPCR |
Reported cancer pathway affected by hsa-miR-203a-3p

| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-203a-3p | bladder cancer | Apoptosis pathway | "Here we evaluated the expression of microRNA-203 m ......" | 21205209 | Western blot; Flow cytometry; Luciferase |
| hsa-miR-203a-3p | bladder cancer | Apoptosis pathway | "In this article we define for the first time that ......" | 21836020 | |
| hsa-miR-203a-3p | bladder cancer | Apoptosis pathway | "To address this issue we employed RT-qPCR to evalu ......" | 26599571 | Western blot; Luciferase |
| hsa-miR-203a-3p | breast cancer | cell cycle pathway; Apoptosis pathway | "Epigenetic Silencing of miR 203 Upregulates SNAI2 ......" | 22393463 | Luciferase |
| hsa-miR-203a-3p | breast cancer | Apoptosis pathway | "Kallistatin via PKC-ERK activation reduced miR-203 ......" | 26790955 | |
| hsa-miR-203a-3p | cervical and endocervical cancer | cell cycle pathway | "In cervical cancer miR-203 levels are decreased al ......" | 25658920 | Colony formation |
| hsa-miR-203a-3p | colon cancer | Apoptosis pathway | "miR 203 reverses chemoresistance in p53 mutated co ......" | 21354697 | |
| hsa-miR-203a-3p | colorectal cancer | Apoptosis pathway | "In addition we indicated that CPEB4 is a novel tar ......" | 26361147 | |
| hsa-miR-203a-3p | esophageal cancer | Apoptosis pathway | "MiR 203 suppresses tumor growth and invasion and d ......" | 24001611 | |
| hsa-miR-203a-3p | esophageal cancer | cell cycle pathway; Apoptosis pathway | "miR 203 is a direct transcriptional target of E2F1 ......" | 25216463 | Luciferase |
| hsa-miR-203a-3p | gastric cancer | PI3K/Akt signaling pathway | "Overexpression of miR-203 or knockdown of its targ ......" | 26980572 | |
| hsa-miR-203a-3p | gastric cancer | Apoptosis pathway | "Berberine modulates cisplatin sensitivity of human ......" | 27142767 | |
| hsa-miR-203a-3p | glioblastoma | Epithelial mesenchymal transition pathway | "MiR 203 downregulation is responsible for chemores ......" | 25871397 | Western blot; Luciferase; Wound Healing Assay |
| hsa-miR-203a-3p | head and neck cancer | Epithelial mesenchymal transition pathway | "The miR-200 family and miR-203 were downregulated ......" | 26882562 | |
| hsa-miR-203a-3p | kidney renal cell cancer | Apoptosis pathway | "miR 203a regulates proliferation migration and apo ......" | 25123268 | Luciferase |
| hsa-miR-203a-3p | liver cancer | Apoptosis pathway | "miR 203 suppresses the proliferation and metastasi ......" | 26179263 | |
| hsa-miR-203a-3p | liver cancer | Epithelial mesenchymal transition pathway; Apoptosis pathway | "Downregulation of miRNA 30c and miR 203a is associ ......" | 26210453 | Western blot |
| hsa-miR-203a-3p | liver cancer | Apoptosis pathway | "EGR1 mediates miR 203a suppress the hepatocellular ......" | 27244890 | Luciferase |
| hsa-miR-203a-3p | lung cancer | Apoptosis pathway | "We aimed to determine the expression of microRNA-2 ......" | 23073851 | Western blot; MTT assay; Flow cytometry; Luciferase |
| hsa-miR-203a-3p | lung cancer | Apoptosis pathway | "miR 203 inhibits cell proliferation and migration ......" | 24040137 | |
| hsa-miR-203a-3p | lung cancer | Apoptosis pathway | "miR 203 suppresses the proliferation and migration ......" | 25140799 | Luciferase |
| hsa-miR-203a-3p | lung cancer | Wnt signaling pathway | "miR 203 Inhibits Frizzled 2 Expression via CD82/KA ......" | 26132195 | |
| hsa-miR-203a-3p | lung cancer | Apoptosis pathway | "The present study investigated the relationship be ......" | 26397233 | |
| hsa-miR-203a-3p | lung cancer | Epithelial mesenchymal transition pathway | "Direct interaction between miR 203 and ZEB2 suppre ......" | 27733346 | |
| hsa-miR-203a-3p | lung squamous cell cancer | Epithelial mesenchymal transition pathway | "MiR 145 and miR 203 represses TGF β induced epith ......" | 27237033 | Western blot; RNAi; Luciferase |
| hsa-miR-203a-3p | melanoma | cell cycle pathway | "We recently reported that miR-203 is down-regulate ......" | 22354972 | Luciferase |
| hsa-miR-203a-3p | prostate cancer | Epithelial mesenchymal transition pathway | "Regulatory Role of mir 203 in Prostate Cancer Prog ......" | 21159887 | |
| hsa-miR-203a-3p | sarcoma | cell cycle pathway; Apoptosis pathway | "MiR 34a and miR 203 Inhibit Survivin Expression to ......" | 27326248 | Colony formation |
Reported cancer prognosis affected by hsa-miR-203a-3p

| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-203a-3p | bladder cancer | progression; poor survival; worse prognosis | "To address this issue we employed RT-qPCR to evalu ......" | 26599571 | Western blot; Luciferase |
| hsa-miR-203a-3p | breast cancer | drug resistance | "Anti miR 203 Upregulates SOCS3 Expression in Breas ......" | 22207897 | |
| hsa-miR-203a-3p | breast cancer | malignant trasformation; tumorigenesis; metastasis | "Epigenetic Silencing of miR 203 Upregulates SNAI2 ......" | 22393463 | Luciferase |
| hsa-miR-203a-3p | breast cancer | cell migration | "This study was performed to investigate the effect ......" | 22713668 | Luciferase |
| hsa-miR-203a-3p | breast cancer | drug resistance | "In addition our analysis showed inverse expression ......" | 24039897 | Luciferase |
| hsa-miR-203a-3p | breast cancer | progression; metastasis | "Targeting of Runx2 by miR 135 and miR 203 Impairs ......" | 25634212 | |
| hsa-miR-203a-3p | breast cancer | metastasis | "These included miR-141 miR-144 miR-193b miR-200a m ......" | 26785733 | |
| hsa-miR-203a-3p | breast cancer | progression | "Kallistatin via PKC-ERK activation reduced miR-203 ......" | 26790955 | |
| hsa-miR-203a-3p | breast cancer | staging | "The level of miR-203a in particular was shown to b ......" | 27431784 | |
| hsa-miR-203a-3p | cervical and endocervical cancer | metastasis; progression; tumorigenesis | "Aberrant expression of miR 20a and miR 203 in cerv ......" | 23725129 | |
| hsa-miR-203a-3p | cervical and endocervical cancer | staging; metastasis | "We hypothesized that microRNA-20a miR-20a and micr ......" | 23819812 | |
| hsa-miR-203a-3p | cervical and endocervical cancer | metastasis; poor survival; progression | "In the current study the expressions of miR-203 an ......" | 26490989 | |
| hsa-miR-203a-3p | cervical and endocervical cancer | differentiation; tumorigenesis | "The miR-203 and one of its targets ΔNp63 are know ......" | 27466497 | |
| hsa-miR-203a-3p | colon cancer | drug resistance; poor survival | "miR 203 reverses chemoresistance in p53 mutated co ......" | 21354697 | |
| hsa-miR-203a-3p | colon cancer | metastasis; differentiation | "Maintenance of the stemness in CD44+ HCT 15 and HC ......" | 24145190 | Colony formation |
| hsa-miR-203a-3p | colorectal cancer | staging; tumor size | "Aberrant expression of miR 203 and its clinical si ......" | 21063914 | |
| hsa-miR-203a-3p | colorectal cancer | staging; poor survival | "TaqMan quantitative real-time PCR was used to quan ......" | 23719259 | |
| hsa-miR-203a-3p | colorectal cancer | drug resistance | "miR 203 induces oxaliplatin resistance in colorect ......" | 24145123 | |
| hsa-miR-203a-3p | colorectal cancer | drug resistance | "miR 203 enhances chemosensitivity to 5 fluorouraci ......" | 25482885 | |
| hsa-miR-203a-3p | colorectal cancer | staging | "We performed miRNA profiling of 1547 human miRNAs ......" | 26045793 | |
| hsa-miR-203a-3p | colorectal cancer | metastasis; staging; poor survival; worse prognosis | "In spite of the important function of microRNA miR ......" | 26701878 | |
| hsa-miR-203a-3p | colorectal cancer | staging; worse prognosis | "Comprehensive analyses showed that plasma miR-96 d ......" | 26863633 | |
| hsa-miR-203a-3p | colorectal cancer | staging; metastasis; poor survival | "While it is known that miR-203 is frequently downr ......" | 27376958 | |
| hsa-miR-203a-3p | colorectal cancer | staging; metastasis; tumor size; progression | "miR 203 is a predictive biomarker for colorectal c ......" | 27714672 | |
| hsa-miR-203a-3p | esophageal cancer | progression; poor survival | "miR 203 inhibits the migration and invasion of eso ......" | 22940702 | |
| hsa-miR-203a-3p | esophageal cancer | differentiation; worse prognosis | "MicroRNA miR-203 has been shown to induce squamous ......" | 24692008 | Flow cytometry |
| hsa-miR-203a-3p | esophageal cancer | metastasis | "EGF induced C/EBPβ participates in EMT by decreas ......" | 24994936 | |
| hsa-miR-203a-3p | esophageal cancer | progression | "miR 203 is a direct transcriptional target of E2F1 ......" | 25216463 | Luciferase |
| hsa-miR-203a-3p | esophageal cancer | worse prognosis; staging; metastasis; differentiation; poor survival | "Methylation mediated repression of potential tumor ......" | 26577858 | |
| hsa-miR-203a-3p | esophageal cancer | progression | "Regulatory Role of miR 203 in Occurrence and Progr ......" | 27029934 | |
| hsa-miR-203a-3p | gastric cancer | metastasis; tumorigenesis; cell migration | "miR 203 suppression in gastric carcinoma promotes ......" | 26194864 | Luciferase; Cell migration assay |
| hsa-miR-203a-3p | gastric cancer | staging; metastasis; poor survival; worse prognosis | "In the first step of this study preliminary experi ......" | 26233325 | |
| hsa-miR-203a-3p | gastric cancer | metastasis; progression | "MiR 203 inhibits tumor invasion and metastasis in ......" | 27542403 | Colony formation |
| hsa-miR-203a-3p | glioblastoma | drug resistance | "MiR 203 downregulation is responsible for chemores ......" | 25871397 | Western blot; Luciferase; Wound Healing Assay |
| hsa-miR-203a-3p | head and neck cancer | worse prognosis | "Conversely decreased expressions of miR-153 miR-20 ......" | 25677760 | |
| hsa-miR-203a-3p | head and neck cancer | metastasis | "The most differentially expressed microRNAs were v ......" | 25956054 | |
| hsa-miR-203a-3p | head and neck cancer | recurrence; drug resistance | "Moreover low expression of the most important miR ......" | 26265694 | |
| hsa-miR-203a-3p | kidney renal cell cancer | malignant trasformation | "Specifically we documented the overexpression of m ......" | 19513557 | |
| hsa-miR-203a-3p | kidney renal cell cancer | staging; poor survival; progression; worse prognosis | "miR 203a regulates proliferation migration and apo ......" | 25123268 | Luciferase |
| hsa-miR-203a-3p | liver cancer | tumorigenesis | "miR 124 and miR 203 are epigenetically silenced tu ......" | 19843643 | |
| hsa-miR-203a-3p | liver cancer | worse prognosis; recurrence; poor survival | "miR 203 expression predicts outcome after liver tr ......" | 21786180 | |
| hsa-miR-203a-3p | liver cancer | recurrence; staging; metastasis; progression; tumorigenesis | "Although recent studies have shown the utility of ......" | 26109910 | |
| hsa-miR-203a-3p | liver cancer | metastasis; staging; poor survival | "miR 203 suppresses the proliferation and metastasi ......" | 26179263 | |
| hsa-miR-203a-3p | liver cancer | cell migration; poor survival; worse prognosis | "Downregulation of miRNA 30c and miR 203a is associ ......" | 26210453 | Western blot |
| hsa-miR-203a-3p | liver cancer | progression; tumorigenesis | "EGR1 mediates miR 203a suppress the hepatocellular ......" | 27244890 | Luciferase |
| hsa-miR-203a-3p | lung cancer | metastasis | "Moreover expression levels of 84 tumor metastasis- ......" | 23296057 | |
| hsa-miR-203a-3p | lung cancer | cell migration; metastasis | "miR 203 Inhibits Frizzled 2 Expression via CD82/KA ......" | 26132195 | |
| hsa-miR-203a-3p | lung cancer | drug resistance; cell migration | "Direct interaction between miR 203 and ZEB2 suppre ......" | 27733346 | |
| hsa-miR-203a-3p | lung squamous cell cancer | tumorigenesis; drug resistance | "It has been recently reported that epidermal growt ......" | 24286402 | |
| hsa-miR-203a-3p | lung squamous cell cancer | worse prognosis; poor survival; staging; metastasis; tumor size | "Association between downexpression of MiR 203 and ......" | 26307752 | |
| hsa-miR-203a-3p | lung squamous cell cancer | cell migration | "MiR 145 and miR 203 represses TGF β induced epith ......" | 27237033 | Western blot; RNAi; Luciferase |
| hsa-miR-203a-3p | lung squamous cell cancer | cell migration | "Therapeutic intervention of silymarin on the migra ......" | 27429844 | Cell migration assay |
| hsa-miR-203a-3p | lymphoma | staging | "Herein we used a global quantitative real-time pol ......" | 25503151 | |
| hsa-miR-203a-3p | melanoma | staging | "To identify improved molecular markers to support ......" | 22223089 | |
| hsa-miR-203a-3p | melanoma | malignant trasformation | "We recently reported that miR-203 is down-regulate ......" | 22354972 | Luciferase |
| hsa-miR-203a-3p | melanoma | metastasis | "Here we demonstrate a significant correlation betw ......" | 22956368 | |
| hsa-miR-203a-3p | melanoma | poor survival | "Presently we examined the expression profile of mi ......" | 23638671 | |
| hsa-miR-203a-3p | melanoma | metastasis | "Six microRNAs miR-9 miR-145 miR-150 miR-155 miR-20 ......" | 23863473 | |
| hsa-miR-203a-3p | melanoma | drug resistance | "MicroRNA miR-203 is known to be downregulated and ......" | 23884313 | Luciferase |
| hsa-miR-203a-3p | melanoma | metastasis | "MiR 203 inhibits melanoma invasive and proliferati ......" | 25475727 | |
| hsa-miR-203a-3p | melanoma | worse prognosis; staging; poor survival | "Expression of miR 203 is decreased and associated ......" | 26722525 | |
| hsa-miR-203a-3p | ovarian cancer | staging | "Levels of 8 microRNAs miR-21 miR-141 miR-200a miR- ......" | 18589210 | |
| hsa-miR-203a-3p | ovarian cancer | staging; recurrence; progression; poor survival; worse prognosis | "MicroRNA-203 miR-203 possessing tumor suppressive ......" | 23918241 | |
| hsa-miR-203a-3p | ovarian cancer | poor survival | "miR 203 Functions as a Tumor Suppressor by Inhibit ......" | 26819680 | |
| hsa-miR-203a-3p | ovarian cancer | staging; poor survival; progression; worse prognosis | "The role and expression of miR 100 and miR 203 pro ......" | 27347348 | |
| hsa-miR-203a-3p | ovarian cancer | metastasis; poor survival | "MiR 203 promotes the growth and migration of ovari ......" | 27655286 | |
| hsa-miR-203a-3p | pancreatic cancer | poor survival | "We measured the levels of miR-155 miR-203 miR-210 ......" | 19551852 | |
| hsa-miR-203a-3p | pancreatic cancer | worse prognosis; poor survival | "The aim of this study is to evaluate the correlati ......" | 20652642 | |
| hsa-miR-203a-3p | pancreatic cancer | cell migration | "miR 203 inhibits tumor cell migration and invasion ......" | 24520289 | |
| hsa-miR-203a-3p | prostate cancer | metastasis; progression; staging; motility | "Regulatory Role of mir 203 in Prostate Cancer Prog ......" | 21159887 | |
| hsa-miR-203a-3p | prostate cancer | drug resistance | "The altered expression of miR-221/-222 as previous ......" | 22127852 | |
| hsa-miR-203a-3p | prostate cancer | metastasis; progression | "Loss of tumor suppressor mir 203 mediates overexpr ......" | 24980827 | |
| hsa-miR-203a-3p | prostate cancer | metastasis; drug resistance; progression | "Loss of EGFR signaling regulated miR 203 promotes ......" | 25004126 | |
| hsa-miR-203a-3p | prostate cancer | metastasis | "MiR 203 down regulates Rap1A and suppresses cell p ......" | 25636908 | Luciferase |
| hsa-miR-203a-3p | prostate cancer | worse prognosis | "Diagnostic and prognostic values of tissue hsa miR ......" | 26499781 | |
| hsa-miR-203a-3p | sarcoma | differentiation | "miR 203 a tumor suppressor frequently down regulat ......" | 24247238 | |
| hsa-miR-203a-3p | sarcoma | progression; differentiation | "Therefore in this review we focus on the regulatio ......" | 24831881 | |
| hsa-miR-203a-3p | sarcoma | progression; tumorigenesis | "miR 203 Acts as a Tumor Suppressor Gene in Osteosa ......" | 26382657 | |
| hsa-miR-203a-3p | sarcoma | progression; poor survival | "MiR 203 Determines Poor Outcome and Suppresses Tum ......" | 26584294 | Luciferase |
| hsa-miR-203a-3p | sarcoma | poor survival | "MiR 34a and miR 203 Inhibit Survivin Expression to ......" | 27326248 | Colony formation |
| hsa-miR-203a-3p | thyroid cancer | recurrence; poor survival; worse prognosis | "To investigate the potential role of serum miR-203 ......" | 27689614 |
Reported gene related to hsa-miR-203a-3p

| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-203a-3p | breast cancer | LASP1 | "Luciferase assays were also performed to validate ......" | 22713668 |
| hsa-miR-203a-3p | esophageal cancer | LASP1 | "miR 203 inhibits the migration and invasion of eso ......" | 22940702 |
| hsa-miR-203a-3p | lung squamous cell cancer | LASP1 | "LASP 1 regulated by miR 203 promotes tumor prolife ......" | 26683818 |
| hsa-miR-203a-3p | prostate cancer | LASP1 | "Loss of tumor suppressor mir 203 mediates overexpr ......" | 24980827 |
| hsa-miR-203a-3p | breast cancer | SNAI2 | "Epigenetic Silencing of miR 203 Upregulates SNAI2 ......" | 22393463 |
| hsa-miR-203a-3p | gastric cancer | SNAI2 | "miR 203 suppression in gastric carcinoma promotes ......" | 26194864 |
| hsa-miR-203a-3p | glioblastoma | SNAI2 | "MiR 203 downregulation is responsible for chemores ......" | 25871397 |
| hsa-miR-203a-3p | ovarian cancer | SNAI2 | "miR-203 expression was downregulated whereas expre ......" | 26819680 |
| hsa-miR-203a-3p | breast cancer | TP53 | "Knockdown of miR-203 following cisplatin treatment ......" | 22207897 |
| hsa-miR-203a-3p | colon cancer | TP53 | "miR 203 reverses chemoresistance in p53 mutated co ......" | 21354697 |
| hsa-miR-203a-3p | lung cancer | TP53 | "In the present study adriamycin and nutlin-3 were ......" | 26397233 |
| hsa-miR-203a-3p | ovarian cancer | TP53 | "Here we showed that the expression of miR-203 was ......" | 27655286 |
| hsa-miR-203a-3p | cervical and endocervical cancer | TP63 | "As the role of p63 and miR-203 in cervical carcino ......" | 27466497 |
| hsa-miR-203a-3p | esophageal cancer | TP63 | "Quantitative real-time polymerase chain reaction q ......" | 27029934 |
| hsa-miR-203a-3p | esophageal cancer | TP63 | "MicroRNA miR-203 has been shown to induce squamous ......" | 24692008 |
| hsa-miR-203a-3p | sarcoma | TP63 | "Mechanistically miR-203 exerts its tumor-suppressi ......" | 24247238 |
| hsa-miR-203a-3p | bladder cancer | BCL2L2 | "Western blotting and luciferase reporter assay sho ......" | 26599571 |
| hsa-miR-203a-3p | bladder cancer | BCL2L2 | "We further verified that miR-203 directly targeted ......" | 21205209 |
| hsa-miR-203a-3p | gastric cancer | BCL2L2 | "Importantly we showed that miR-203 was able to tar ......" | 27142767 |
| hsa-miR-203a-3p | breast cancer | BMI1 | "MiR-200c and miR-203 overexpression in breast canc ......" | 24039897 |
| hsa-miR-203a-3p | lung squamous cell cancer | BMI1 | "In the present study the association between B-cel ......" | 26137120 |
| hsa-miR-203a-3p | melanoma | BMI1 | "MiR 203 inhibits melanoma invasive and proliferati ......" | 25475727 |
| hsa-miR-203a-3p | breast cancer | SOCS3 | "Kallistatin via PKC-ERK activation reduced miR-203 ......" | 26790955 |
| hsa-miR-203a-3p | breast cancer | SOCS3 | "Anti miR 203 suppresses ER positive breast cancer ......" | 27517632 |
| hsa-miR-203a-3p | breast cancer | SOCS3 | "Anti miR 203 Upregulates SOCS3 Expression in Breas ......" | 22207897 |
| hsa-miR-203a-3p | bladder cancer | AKT2 | "Also we define Akt2 and Src as novel miR-203 targe ......" | 21836020 |
| hsa-miR-203a-3p | colon cancer | AKT2 | "miR 203 reverses chemoresistance in p53 mutated co ......" | 21354697 |
| hsa-miR-203a-3p | colorectal cancer | ATM | "miR 203 induces oxaliplatin resistance in colorect ......" | 24145123 |
| hsa-miR-203a-3p | gastric cancer | ATM | "MiR 203 inhibits tumor invasion and metastasis in ......" | 27542403 |
| hsa-miR-203a-3p | breast cancer | BIRC5 | "Luciferase assays were also performed to validate ......" | 22713668 |
| hsa-miR-203a-3p | colorectal cancer | BIRC5 | "miR 203 is a predictive biomarker for colorectal c ......" | 27714672 |
| hsa-miR-203a-3p | gastric cancer | CDH1 | "In addition miR-203 overexpression significantly s ......" | 27542403 |
| hsa-miR-203a-3p | lung squamous cell cancer | CDH1 | "Therapeutic intervention of silymarin on the migra ......" | 27429844 |
| hsa-miR-203a-3p | liver cancer | FRTS1 | "Patients with higher miR-203 expression had signif ......" | 21786180 |
| hsa-miR-203a-3p | thyroid cancer | FRTS1 | "The association between serum miR-203 expressions ......" | 27689614 |
| hsa-miR-203a-3p | esophageal cancer | PDLIM5 | "Genome-wide gene expression data and target site i ......" | 22940702 |
| hsa-miR-203a-3p | prostate cancer | PDLIM5 | "Loss of tumor suppressor mir 203 mediates overexpr ......" | 24980827 |
| hsa-miR-203a-3p | esophageal cancer | RAN | "MiR 203 suppresses tumor growth and invasion and d ......" | 24001611 |
| hsa-miR-203a-3p | esophageal cancer | RAN | "The efficient delivery of miR-203 significantly su ......" | 24154605 |
| hsa-miR-203a-3p | breast cancer | RUNX2 | "Targeting of Runx2 by miR 135 and miR 203 Impairs ......" | 25634212 |
| hsa-miR-203a-3p | prostate cancer | RUNX2 | "Importantly miR-203 regulates a cohort of pro-meta ......" | 21159887 |
| hsa-miR-203a-3p | head and neck cancer | SIK1 | "Interestingly we identified NUAK family SNF1-like ......" | 26882562 |
| hsa-miR-203a-3p | pancreatic cancer | SIK1 | "miR 203 promotes proliferation migration and invas ......" | 26719072 |
| hsa-miR-203a-3p | colon cancer | SNAI1 | "In addition the significant upregulation of the pr ......" | 24145190 |
| hsa-miR-203a-3p | gastric cancer | SNAI1 | "In addition miR-203 overexpression significantly s ......" | 27542403 |
| hsa-miR-203a-3p | bladder cancer | SRC | "Also we define Akt2 and Src as novel miR-203 targe ......" | 21836020 |
| hsa-miR-203a-3p | lung cancer | SRC | "miR 203 suppresses the proliferation and migration ......" | 25140799 |
| hsa-miR-203a-3p | lung cancer | ZEB2 | "Direct interaction between miR 203 and ZEB2 suppre ......" | 27733346 |
| hsa-miR-203a-3p | prostate cancer | ZEB2 | "Importantly miR-203 regulates a cohort of pro-meta ......" | 21159887 |
| hsa-miR-203a-3p | colorectal cancer | ABCG2 | "A novel miR 203 DNMT3b ABCG2 regulatory pathway pr ......" | 27253631 |
| hsa-miR-203a-3p | lymphoma | ABL1 | "Treatment of lymphoma B cells with demethylating a ......" | 21454413 |
| hsa-miR-203a-3p | liver cancer | ADAM9 | "miR 203 suppresses the proliferation and metastasi ......" | 26179263 |
| hsa-miR-203a-3p | cervical and endocervical cancer | BANF1 | "We also demonstrate that miR-203 targets the 3' un ......" | 25658920 |
| hsa-miR-203a-3p | breast cancer | BAX | "Knockdown of miR-203 following cisplatin treatment ......" | 22207897 |
| hsa-miR-203a-3p | lung cancer | BBC3 | "In the present study adriamycin and nutlin-3 were ......" | 26397233 |
| hsa-miR-203a-3p | esophageal cancer | BMP4 | "Our cDNA microarray analysis demonstrated the upre ......" | 24692008 |
| hsa-miR-203a-3p | melanoma | CAMP | "In addition the data indicated that exogenous miR- ......" | 23884313 |
| hsa-miR-203a-3p | gastric cancer | CASK | "Down regulation of miR 203 induced by Helicobacter ......" | 25373785 |
| hsa-miR-203a-3p | pancreatic cancer | CAV1 | "miR 203 inhibits tumor cell migration and invasion ......" | 24520289 |
| hsa-miR-203a-3p | colon cancer | CD44 | "By manipulating the expression of CD44 in HCT-15 a ......" | 24145190 |
| hsa-miR-203a-3p | lung cancer | CD82 | "Among these miRNAs CD82 caused upregulation of miR ......" | 26132195 |
| hsa-miR-203a-3p | colorectal cancer | CPEB4 | "In addition we indicated that CPEB4 is a novel tar ......" | 26361147 |
| hsa-miR-203a-3p | lung cancer | DICER1 | "A modest decrease in Drosha and Dicer mRNA levels ......" | 23296057 |
| hsa-miR-203a-3p | colorectal cancer | DMPK | "Furthermore low expression level of miR-203 was co ......" | 27714672 |
| hsa-miR-203a-3p | lung squamous cell cancer | DNMT1 | "In contrast increased expression of both DNA methy ......" | 27177222 |
| hsa-miR-203a-3p | colorectal cancer | DNMT3B | "A novel miR 203 DNMT3b ABCG2 regulatory pathway pr ......" | 27253631 |
| hsa-miR-203a-3p | lung cancer | DROSHA | "A modest decrease in Drosha and Dicer mRNA levels ......" | 23296057 |
| hsa-miR-203a-3p | esophageal cancer | E2F1 | "miR 203 is a direct transcriptional target of E2F1 ......" | 25216463 |
| hsa-miR-203a-3p | melanoma | E2F3 | "miR-203 significantly suppressed the luciferase ac ......" | 22354972 |
| hsa-miR-203a-3p | esophageal cancer | EGF | "EGF induced C/EBPβ participates in EMT by decreas ......" | 24994936 |
| hsa-miR-203a-3p | prostate cancer | EGFR | "Loss of EGFR signaling regulated miR 203 promotes ......" | 25004126 |
| hsa-miR-203a-3p | liver cancer | EGR1 | "In this study we found that the overexpression of ......" | 27244890 |
| hsa-miR-203a-3p | colorectal cancer | EIF5A2 | "Mechanistically we identified EIF5A2 as a direct a ......" | 27376958 |
| hsa-miR-203a-3p | melanoma | ERBB3 | "Our data suggest that miR-203 is a new prognostic ......" | 23638671 |
| hsa-miR-203a-3p | lung squamous cell cancer | ERGIC3 | "ERGIC3 which is regulated by miR 203a is a potenti ......" | 26177443 |
| hsa-miR-203a-3p | colorectal cancer | F2 | "Moreover low expression of miR-203 was correlated ......" | 21063914 |
| hsa-miR-203a-3p | lung cancer | FZD1 | "miR 203 Inhibits Frizzled 2 Expression via CD82/KA ......" | 26132195 |
| hsa-miR-203a-3p | lung cancer | FZD2 | "miR 203 Inhibits Frizzled 2 Expression via CD82/KA ......" | 26132195 |
| hsa-miR-203a-3p | liver cancer | HOXD3 | "Through bioinformatic analysis and luciferase assa ......" | 27244890 |
| hsa-miR-203a-3p | liver cancer | HULC | "miR 203 suppresses the proliferation and metastasi ......" | 26179263 |
| hsa-miR-203a-3p | pancreatic cancer | IL12A | "Tumor necrosis factor-α TNF-α and interleukin-12 ......" | 25290620 |
| hsa-miR-203a-3p | cervical and endocervical cancer | IRF1 | "Promoter analysis revealed that IRF1 a transcripti ......" | 25658920 |
| hsa-miR-203a-3p | melanoma | KIF5B | "The target gene of miR-203 involved in the mechani ......" | 23884313 |
| hsa-miR-203a-3p | sarcoma | LIF | "Mechanistically miR-203 exerts its tumor-suppressi ......" | 24247238 |
| hsa-miR-203a-3p | endometrial cancer | MLH1 | "Methylation status of miR-203 was significantly as ......" | 24530564 |
| hsa-miR-203a-3p | melanoma | MMS | "Our data suggest that miR-203 is a new prognostic ......" | 23638671 |
| hsa-miR-203a-3p | liver cancer | NME1 | "Expression of miR-203 was negatively correlated to ......" | 26109910 |
| hsa-miR-203a-3p | head and neck cancer | NUAK1 | "Interestingly we identified NUAK family SNF1-like ......" | 26882562 |
| hsa-miR-203a-3p | lung squamous cell cancer | PCSK9 | "Functionally in vitro effects of miR-203 on prolif ......" | 26307752 |
| hsa-miR-203a-3p | ovarian cancer | PDHB | "Mechanically miR-203 targeted the 3'-UTR of pyruva ......" | 27655286 |
| hsa-miR-203a-3p | gastric cancer | PIBF1 | "Overexpression of miR-203 or knockdown of its targ ......" | 26980572 |
| hsa-miR-203a-3p | glioblastoma | PLD2 | "The transfection of the miR-203 mimic into U251 ce ......" | 24270883 |
| hsa-miR-203a-3p | sarcoma | RAB22A | "miR 203 Acts as a Tumor Suppressor Gene in Osteosa ......" | 26382657 |
| hsa-miR-203a-3p | prostate cancer | RAC1 | "Mechanistic dissection revealed that miR-203 media ......" | 25636908 |
| hsa-miR-203a-3p | prostate cancer | RAP1A | "MiR 203 down regulates Rap1A and suppresses cell p ......" | 25636908 |
| hsa-miR-203a-3p | breast cancer | SERPINA4 | "Kallistatin via PKC-ERK activation reduced miR-203 ......" | 26790955 |
| hsa-miR-203a-3p | colorectal cancer | SIK2 | "Overexpression of miR 203 sensitizes paclitaxel Ta ......" | 27236538 |
| hsa-miR-203a-3p | lung squamous cell cancer | SMAD3 | "MiR 145 and miR 203 represses TGF β induced epith ......" | 27237033 |
| hsa-miR-203a-3p | endometrial cancer | SOX4 | "Transfection of a miR-203 mimic decreased SOX4 gen ......" | 24530564 |
| hsa-miR-203a-3p | sarcoma | TBK1 | "MiR 203 Determines Poor Outcome and Suppresses Tum ......" | 26584294 |
| hsa-miR-203a-3p | lung cancer | TIMM8A | "However the role of miR-203 in suppressing chemoth ......" | 27733346 |
| hsa-miR-203a-3p | esophageal cancer | TJP1 | "Our cDNA microarray analysis demonstrated the upre ......" | 24692008 |
| hsa-miR-203a-3p | pancreatic cancer | TLR4 | "Pancreatic cancer derived exosomes regulate the ex ......" | 25290620 |
| hsa-miR-203a-3p | prostate cancer | TXK | "Loss of EGFR signaling regulated miR 203 promotes ......" | 25004126 |
| hsa-miR-203a-3p | colorectal cancer | TYMS | "miR 203 enhances chemosensitivity to 5 fluorouraci ......" | 25482885 |
| hsa-miR-203a-3p | melanoma | TYR | "At present we found that exogenous miR-203 increas ......" | 23884313 |
| hsa-miR-203a-3p | cervical and endocervical cancer | VEGFA | "miR 203 suppresses tumor growth and angiogenesis b ......" | 23867971 |
| hsa-miR-203a-3p | lung squamous cell cancer | ZEB1 | "Therapeutic intervention of silymarin on the migra ......" | 27429844 |
| hsa-miR-203a-3p | melanoma | ZNF148 | "miR-203 significantly suppressed the luciferase ac ......" | 22354972 |
| hsa-miR-203a-3p | colorectal cancer | ZNF217 | "MiR 203 suppresses ZNF217 upregulation in colorect ......" | 25621839 |
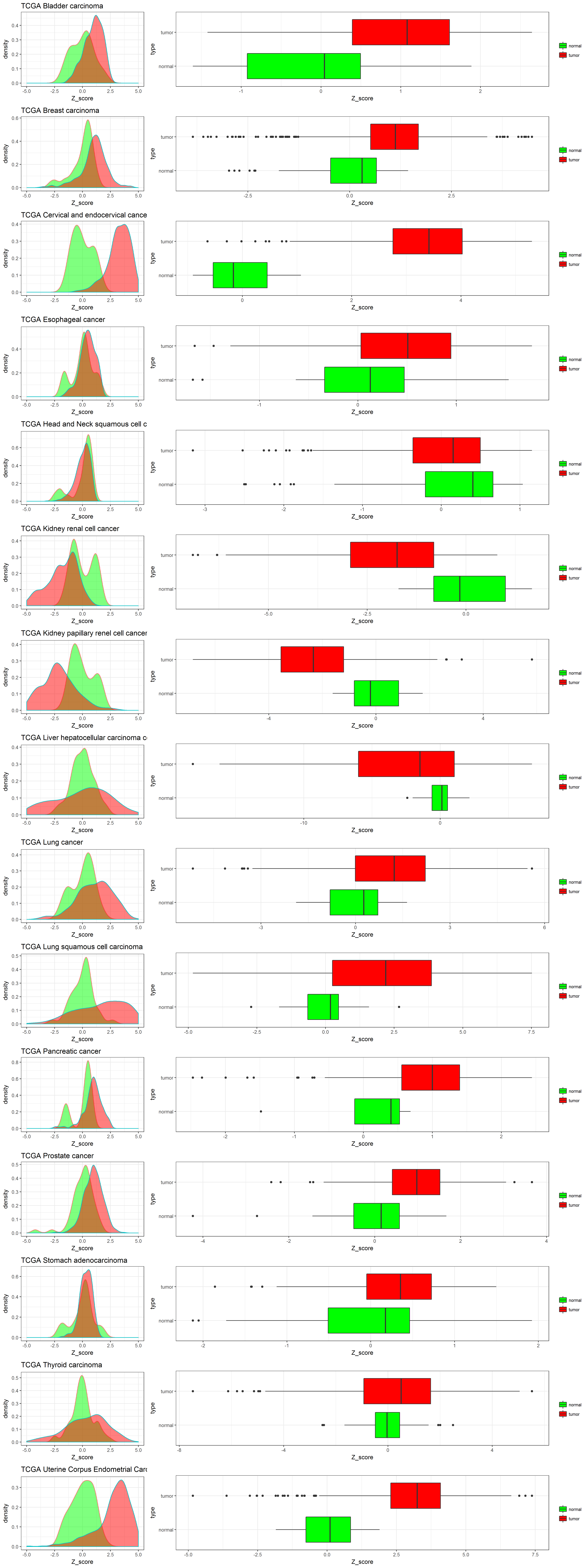
Expression profile in cancer corhorts:
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-203a-3p | ABL1 | 9 cancers: BLCA; CESC; ESCA; HNSC; LGG; PAAD; PRAD; STAD; UCEC | miRTarBase | TCGA BLCA -0.076; TCGA CESC -0.06; TCGA ESCA -0.072; TCGA HNSC -0.113; TCGA LGG -0.083; TCGA PAAD -0.075; TCGA PRAD -0.085; TCGA STAD -0.147; TCGA UCEC -0.108 |
hsa-miR-203a-3p | EDNRA | 11 cancers: BLCA; CESC; COAD; ESCA; HNSC; LGG; LUAD; LUSC; PRAD; STAD; UCEC | miRTarBase | TCGA BLCA -0.157; TCGA CESC -0.186; TCGA COAD -0.236; TCGA ESCA -0.076; TCGA HNSC -0.081; TCGA LGG -0.217; TCGA LUAD -0.078; TCGA LUSC -0.197; TCGA PRAD -0.424; TCGA STAD -0.272; TCGA UCEC -0.274 |
hsa-miR-203a-3p | ZEB2 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRTarBase | TCGA BLCA -0.208; TCGA BRCA -0.108; TCGA CESC -0.075; TCGA COAD -0.347; TCGA ESCA -0.192; TCGA HNSC -0.224; TCGA KIRC -0.077; TCGA LUAD -0.074; TCGA LUSC -0.223; TCGA PAAD -0.094; TCGA PRAD -0.377; TCGA SARC -0.128; TCGA THCA -0.196; TCGA STAD -0.227; TCGA UCEC -0.233 |
hsa-miR-203a-3p | ABCD2 | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.307; TCGA BRCA -0.19; TCGA COAD -0.358; TCGA ESCA -0.3; TCGA HNSC -0.203; TCGA KIRC -0.183; TCGA LUSC -0.229; TCGA PAAD -0.231; TCGA PRAD -0.545; TCGA STAD -0.301; TCGA UCEC -0.242 |
hsa-miR-203a-3p | RGS5 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUSC; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.123; TCGA BRCA -0.203; TCGA CESC -0.18; TCGA COAD -0.349; TCGA ESCA -0.357; TCGA HNSC -0.168; TCGA KIRC -0.111; TCGA LUSC -0.168; TCGA PRAD -0.242; TCGA THCA -0.443; TCGA STAD -0.34; TCGA UCEC -0.159 |
hsa-miR-203a-3p | CCR1 | 12 cancers: BLCA; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; PRAD; SARC; STAD | MirTarget | TCGA BLCA -0.178; TCGA COAD -0.264; TCGA ESCA -0.173; TCGA HNSC -0.196; TCGA KIRC -0.175; TCGA KIRP -0.082; TCGA LGG -0.175; TCGA LUAD -0.115; TCGA LUSC -0.237; TCGA PRAD -0.159; TCGA SARC -0.159; TCGA STAD -0.065 |
hsa-miR-203a-3p | NBEA | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUSC; PAAD; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.195; TCGA BRCA -0.181; TCGA CESC -0.494; TCGA COAD -0.621; TCGA ESCA -0.391; TCGA HNSC -0.331; TCGA LUSC -0.241; TCGA PAAD -0.352; TCGA PRAD -0.234; TCGA THCA -0.288; TCGA STAD -0.617; TCGA UCEC -0.262 |
hsa-miR-203a-3p | CLIC4 | 13 cancers: BLCA; COAD; HNSC; KIRP; LGG; LUAD; LUSC; OV; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.141; TCGA COAD -0.278; TCGA HNSC -0.075; TCGA KIRP -0.149; TCGA LGG -0.152; TCGA LUAD -0.106; TCGA LUSC -0.098; TCGA OV -0.055; TCGA PRAD -0.34; TCGA SARC -0.126; TCGA THCA -0.09; TCGA STAD -0.285; TCGA UCEC -0.147 |
hsa-miR-203a-3p | HDX | 11 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.129; TCGA CESC -0.227; TCGA COAD -0.189; TCGA ESCA -0.127; TCGA HNSC -0.203; TCGA LUAD -0.19; TCGA LUSC -0.303; TCGA PAAD -0.112; TCGA PRAD -0.132; TCGA STAD -0.312; TCGA UCEC -0.14 |
hsa-miR-203a-3p | SEMA3A | 10 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; LUAD; LUSC; PRAD; STAD | MirTarget | TCGA BLCA -0.157; TCGA BRCA -0.079; TCGA CESC -0.276; TCGA COAD -0.301; TCGA HNSC -0.113; TCGA KIRC -0.094; TCGA LUAD -0.23; TCGA LUSC -0.128; TCGA PRAD -0.51; TCGA STAD -0.351 |
hsa-miR-203a-3p | PKD2 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.07; TCGA BRCA -0.085; TCGA CESC -0.118; TCGA COAD -0.31; TCGA ESCA -0.08; TCGA HNSC -0.127; TCGA KIRP -0.051; TCGA LUAD -0.112; TCGA PAAD -0.11; TCGA PRAD -0.225; TCGA STAD -0.323; TCGA UCEC -0.195 |
hsa-miR-203a-3p | COL4A4 | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUSC; OV; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.239; TCGA CESC -0.18; TCGA COAD -0.417; TCGA ESCA -0.483; TCGA HNSC -0.302; TCGA LUSC -0.465; TCGA OV -0.177; TCGA PRAD -0.341; TCGA STAD -0.313; TCGA UCEC -0.1 |
hsa-miR-203a-3p | TGFB2 | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.138; TCGA CESC -0.268; TCGA COAD -0.347; TCGA ESCA -0.166; TCGA HNSC -0.213; TCGA LUAD -0.127; TCGA LUSC -0.31; TCGA PRAD -0.324; TCGA STAD -0.194; TCGA UCEC -0.177 |
hsa-miR-203a-3p | VCAN | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LUAD; LUSC; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.132; TCGA CESC -0.159; TCGA COAD -0.382; TCGA ESCA -0.17; TCGA HNSC -0.318; TCGA KIRC -0.147; TCGA LGG -0.183; TCGA LUAD -0.178; TCGA LUSC -0.122; TCGA PRAD -0.426; TCGA STAD -0.167; TCGA UCEC -0.099 |
hsa-miR-203a-3p | FSTL1 | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LUAD; LUSC; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.143; TCGA CESC -0.237; TCGA COAD -0.328; TCGA ESCA -0.198; TCGA HNSC -0.248; TCGA KIRC -0.166; TCGA LGG -0.109; TCGA LUAD -0.107; TCGA LUSC -0.127; TCGA PRAD -0.168; TCGA STAD -0.269; TCGA UCEC -0.1 |
hsa-miR-203a-3p | SV2B | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; STAD; UCEC | MirTarget | TCGA BLCA -0.272; TCGA BRCA -0.094; TCGA COAD -0.712; TCGA ESCA -0.203; TCGA HNSC -0.41; TCGA LUAD -0.152; TCGA LUSC -0.184; TCGA PAAD -0.215; TCGA STAD -0.366; TCGA UCEC -0.223 |
hsa-miR-203a-3p | B3GALT2 | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.187; TCGA BRCA -0.099; TCGA COAD -0.354; TCGA ESCA -0.171; TCGA HNSC -0.21; TCGA LUSC -0.308; TCGA PAAD -0.253; TCGA PRAD -0.444; TCGA STAD -0.141; TCGA UCEC -0.254 |
hsa-miR-203a-3p | ARHGAP42 | 11 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LGG; LUSC; PRAD; SARC; UCEC | MirTarget | TCGA BLCA -0.082; TCGA BRCA -0.087; TCGA CESC -0.244; TCGA ESCA -0.334; TCGA HNSC -0.141; TCGA KIRC -0.133; TCGA LGG -0.083; TCGA LUSC -0.217; TCGA PRAD -0.116; TCGA SARC -0.119; TCGA UCEC -0.076 |
hsa-miR-203a-3p | PRICKLE2 | 11 cancers: BLCA; BRCA; CESC; COAD; LUAD; LUSC; PAAD; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.099; TCGA BRCA -0.09; TCGA CESC -0.143; TCGA COAD -0.317; TCGA LUAD -0.084; TCGA LUSC -0.124; TCGA PAAD -0.153; TCGA PRAD -0.401; TCGA THCA -0.184; TCGA STAD -0.408; TCGA UCEC -0.161 |
hsa-miR-203a-3p | UNC5C | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.177; TCGA BRCA -0.113; TCGA CESC -0.361; TCGA COAD -0.374; TCGA ESCA -0.364; TCGA HNSC -0.423; TCGA LUAD -0.104; TCGA LUSC -0.357; TCGA PRAD -0.52; TCGA STAD -0.133; TCGA UCEC -0.219 |
hsa-miR-203a-3p | ISL1 | 10 cancers: BLCA; CESC; COAD; ESCA; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.117; TCGA CESC -0.196; TCGA COAD -0.386; TCGA ESCA -0.371; TCGA LUAD -0.191; TCGA LUSC -0.226; TCGA PAAD -0.317; TCGA PRAD -0.286; TCGA STAD -0.492; TCGA UCEC -0.171 |
hsa-miR-203a-3p | AP1S2 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.157; TCGA BRCA -0.072; TCGA CESC -0.094; TCGA COAD -0.283; TCGA ESCA -0.121; TCGA HNSC -0.137; TCGA KIRC -0.092; TCGA LUAD -0.137; TCGA LUSC -0.134; TCGA PAAD -0.2; TCGA PRAD -0.247; TCGA SARC -0.108; TCGA STAD -0.321; TCGA UCEC -0.117 |
hsa-miR-203a-3p | ZEB1 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LIHC; LUAD; LUSC; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.215; TCGA BRCA -0.096; TCGA CESC -0.234; TCGA COAD -0.444; TCGA ESCA -0.241; TCGA HNSC -0.228; TCGA KIRC -0.066; TCGA LGG -0.09; TCGA LIHC -0.052; TCGA LUAD -0.072; TCGA LUSC -0.149; TCGA PRAD -0.412; TCGA THCA -0.29; TCGA STAD -0.395; TCGA UCEC -0.31 |
hsa-miR-203a-3p | RERG | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUSC; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.181; TCGA BRCA -0.181; TCGA CESC -0.26; TCGA COAD -0.581; TCGA ESCA -0.208; TCGA HNSC -0.238; TCGA KIRP -0.107; TCGA LUSC -0.257; TCGA PRAD -0.33; TCGA SARC -0.102; TCGA THCA -0.446; TCGA STAD -0.562; TCGA UCEC -0.44 |
hsa-miR-203a-3p | PCDHB7 | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUSC; OV; PAAD; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.087; TCGA CESC -0.13; TCGA COAD -0.478; TCGA ESCA -0.141; TCGA HNSC -0.151; TCGA LUSC -0.078; TCGA OV -0.213; TCGA PAAD -0.087; TCGA PRAD -0.259; TCGA THCA -0.172; TCGA STAD -0.248; TCGA UCEC -0.106 |
hsa-miR-203a-3p | ROR1 | 13 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; OV; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.223; TCGA CESC -0.339; TCGA COAD -0.53; TCGA ESCA -0.279; TCGA HNSC -0.134; TCGA KIRC -0.084; TCGA LUAD -0.19; TCGA LUSC -0.427; TCGA OV -0.105; TCGA PRAD -0.586; TCGA SARC -0.112; TCGA STAD -0.217; TCGA UCEC -0.214 |
hsa-miR-203a-3p | SOX5 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; OV; PAAD; PRAD; STAD | MirTarget | TCGA BLCA -0.157; TCGA BRCA -0.166; TCGA CESC -0.401; TCGA COAD -0.43; TCGA ESCA -0.218; TCGA HNSC -0.271; TCGA LIHC -0.101; TCGA OV -0.168; TCGA PAAD -0.195; TCGA PRAD -0.319; TCGA STAD -0.317 |
hsa-miR-203a-3p | GNE | 9 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LUSC; OV; PAAD; UCEC | MirTarget | TCGA BLCA -0.07; TCGA BRCA -0.087; TCGA CESC -0.186; TCGA ESCA -0.112; TCGA HNSC -0.128; TCGA LUSC -0.062; TCGA OV -0.072; TCGA PAAD -0.064; TCGA UCEC -0.082 |
hsa-miR-203a-3p | RAB39B | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; THCA; STAD | MirTarget | TCGA BLCA -0.155; TCGA BRCA -0.136; TCGA CESC -0.098; TCGA COAD -0.266; TCGA ESCA -0.188; TCGA HNSC -0.239; TCGA LUAD -0.133; TCGA LUSC -0.242; TCGA PAAD -0.578; TCGA THCA -0.229; TCGA STAD -0.275 |
hsa-miR-203a-3p | BEND4 | 9 cancers: BLCA; COAD; ESCA; HNSC; LUSC; OV; PAAD; STAD; UCEC | MirTarget | TCGA BLCA -0.196; TCGA COAD -0.265; TCGA ESCA -0.337; TCGA HNSC -0.238; TCGA LUSC -0.25; TCGA OV -0.275; TCGA PAAD -0.275; TCGA STAD -0.253; TCGA UCEC -0.182 |
hsa-miR-203a-3p | TSC22D1 | 9 cancers: BLCA; CESC; ESCA; HNSC; LIHC; LUSC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.072; TCGA CESC -0.153; TCGA ESCA -0.081; TCGA HNSC -0.076; TCGA LIHC -0.118; TCGA LUSC -0.085; TCGA THCA -0.137; TCGA STAD -0.088; TCGA UCEC -0.059 |
hsa-miR-203a-3p | SCN9A | 9 cancers: BLCA; BRCA; COAD; HNSC; KIRC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.191; TCGA BRCA -0.136; TCGA COAD -0.623; TCGA HNSC -0.203; TCGA KIRC -0.169; TCGA PAAD -0.226; TCGA PRAD -0.298; TCGA STAD -0.458; TCGA UCEC -0.249 |
hsa-miR-203a-3p | SLC4A4 | 9 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRP; LUSC; THCA; UCEC | MirTarget | TCGA BLCA -0.19; TCGA BRCA -0.266; TCGA CESC -0.575; TCGA ESCA -0.747; TCGA HNSC -0.286; TCGA KIRP -0.151; TCGA LUSC -0.423; TCGA THCA -0.461; TCGA UCEC -0.162 |
hsa-miR-203a-3p | RRAGD | 9 cancers: BLCA; COAD; HNSC; KIRP; LIHC; LUAD; PAAD; SARC; STAD | MirTarget | TCGA BLCA -0.094; TCGA COAD -0.275; TCGA HNSC -0.1; TCGA KIRP -0.086; TCGA LIHC -0.076; TCGA LUAD -0.211; TCGA PAAD -0.179; TCGA SARC -0.086; TCGA STAD -0.133 |
hsa-miR-203a-3p | IGFBP5 | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUAD; OV; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.162; TCGA CESC -0.444; TCGA COAD -0.528; TCGA ESCA -0.168; TCGA HNSC -0.344; TCGA LUAD -0.092; TCGA OV -0.156; TCGA PRAD -0.297; TCGA STAD -0.299; TCGA UCEC -0.219 |
hsa-miR-203a-3p | KITLG | 11 cancers: BLCA; CESC; ESCA; HNSC; LIHC; LUSC; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.091; TCGA CESC -0.084; TCGA ESCA -0.09; TCGA HNSC -0.099; TCGA LIHC -0.094; TCGA LUSC -0.072; TCGA PRAD -0.261; TCGA SARC -0.071; TCGA THCA -0.323; TCGA STAD -0.085; TCGA UCEC -0.175 |
hsa-miR-203a-3p | AKAP7 | 9 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRP; LUSC; OV; STAD | MirTarget | TCGA BLCA -0.096; TCGA BRCA -0.069; TCGA CESC -0.305; TCGA ESCA -0.155; TCGA HNSC -0.196; TCGA KIRP -0.118; TCGA LUSC -0.088; TCGA OV -0.083; TCGA STAD -0.071 |
hsa-miR-203a-3p | TFPI | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LGG; LUSC; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.125; TCGA BRCA -0.105; TCGA CESC -0.226; TCGA COAD -0.188; TCGA ESCA -0.337; TCGA HNSC -0.209; TCGA KIRP -0.095; TCGA LGG -0.346; TCGA LUSC -0.226; TCGA PRAD -0.44; TCGA THCA -0.263; TCGA STAD -0.147; TCGA UCEC -0.261 |
hsa-miR-203a-3p | PLXDC2 | 10 cancers: BLCA; COAD; KIRC; KIRP; LGG; LUAD; LUSC; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.062; TCGA COAD -0.436; TCGA KIRC -0.124; TCGA KIRP -0.096; TCGA LGG -0.108; TCGA LUAD -0.125; TCGA LUSC -0.086; TCGA PRAD -0.138; TCGA STAD -0.165; TCGA UCEC -0.129 |
hsa-miR-203a-3p | PKHD1L1 | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUSC; PAAD; PRAD; STAD | MirTarget | TCGA BLCA -0.222; TCGA BRCA -0.149; TCGA CESC -0.503; TCGA COAD -0.48; TCGA ESCA -0.559; TCGA HNSC -0.204; TCGA LUSC -0.488; TCGA PAAD -0.308; TCGA PRAD -0.268; TCGA STAD -0.401 |
hsa-miR-203a-3p | DGKB | 9 cancers: BLCA; COAD; ESCA; HNSC; LUAD; OV; PAAD; STAD; UCEC | MirTarget | TCGA BLCA -0.287; TCGA COAD -0.521; TCGA ESCA -0.269; TCGA HNSC -0.145; TCGA LUAD -0.206; TCGA OV -0.185; TCGA PAAD -0.151; TCGA STAD -0.562; TCGA UCEC -0.355 |
hsa-miR-203a-3p | CYTIP | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LGG; LUAD; LUSC; PRAD; SARC | MirTarget | TCGA BLCA -0.198; TCGA BRCA -0.061; TCGA COAD -0.208; TCGA ESCA -0.216; TCGA HNSC -0.116; TCGA KIRC -0.258; TCGA LGG -0.141; TCGA LUAD -0.072; TCGA LUSC -0.267; TCGA PRAD -0.194; TCGA SARC -0.127 |
hsa-miR-203a-3p | RNF38 | 10 cancers: BLCA; LIHC; LUSC; OV; PAAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.052; TCGA LIHC -0.061; TCGA LUSC -0.057; TCGA OV -0.063; TCGA PAAD -0.069; TCGA PRAD -0.091; TCGA SARC -0.068; TCGA THCA -0.078; TCGA STAD -0.098; TCGA UCEC -0.1 |
hsa-miR-203a-3p | EBF3 | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.165; TCGA BRCA -0.2; TCGA COAD -0.546; TCGA ESCA -0.261; TCGA HNSC -0.213; TCGA LUAD -0.201; TCGA LUSC -0.142; TCGA PAAD -0.132; TCGA PRAD -0.164; TCGA THCA -0.424; TCGA STAD -0.305; TCGA UCEC -0.191 |
hsa-miR-203a-3p | BACH2 | 10 cancers: BLCA; CESC; COAD; HNSC; LUAD; OV; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.223; TCGA CESC -0.129; TCGA COAD -0.438; TCGA HNSC -0.289; TCGA LUAD -0.088; TCGA OV -0.155; TCGA PAAD -0.27; TCGA PRAD -0.117; TCGA STAD -0.259; TCGA UCEC -0.217 |
hsa-miR-203a-3p | SPARC | 13 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LUAD; LUSC; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.121; TCGA CESC -0.145; TCGA COAD -0.357; TCGA ESCA -0.091; TCGA HNSC -0.218; TCGA KIRC -0.224; TCGA LGG -0.116; TCGA LUAD -0.064; TCGA LUSC -0.141; TCGA PRAD -0.308; TCGA THCA -0.094; TCGA STAD -0.134; TCGA UCEC -0.135 |
hsa-miR-203a-3p | CSGALNACT1 | 9 cancers: BLCA; COAD; ESCA; HNSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.094; TCGA COAD -0.209; TCGA ESCA -0.148; TCGA HNSC -0.114; TCGA PAAD -0.182; TCGA PRAD -0.14; TCGA SARC -0.133; TCGA STAD -0.101; TCGA UCEC -0.066 |
hsa-miR-203a-3p | CORO1C | 9 cancers: BLCA; KIRC; KIRP; LGG; LUAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.081; TCGA KIRC -0.146; TCGA KIRP -0.116; TCGA LGG -0.096; TCGA LUAD -0.099; TCGA PRAD -0.284; TCGA SARC -0.108; TCGA STAD -0.11; TCGA UCEC -0.112 |
hsa-miR-203a-3p | RUNDC3B | 16 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; OV; PAAD; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.079; TCGA BRCA -0.121; TCGA CESC -0.193; TCGA COAD -0.175; TCGA ESCA -0.154; TCGA HNSC -0.07; TCGA KIRP -0.213; TCGA LIHC -0.076; TCGA LUAD -0.163; TCGA LUSC -0.09; TCGA OV -0.094; TCGA PAAD -0.335; TCGA PRAD -0.213; TCGA THCA -0.147; TCGA STAD -0.242; TCGA UCEC -0.17 |
hsa-miR-203a-3p | FAM180A | 9 cancers: BLCA; COAD; LGG; LUAD; LUSC; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.217; TCGA COAD -0.605; TCGA LGG -0.103; TCGA LUAD -0.099; TCGA LUSC -0.139; TCGA PRAD -0.168; TCGA SARC -0.359; TCGA STAD -0.388; TCGA UCEC -0.275 |
hsa-miR-203a-3p | CCDC50 | 13 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LUSC; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.082; TCGA BRCA -0.07; TCGA COAD -0.103; TCGA ESCA -0.119; TCGA HNSC -0.117; TCGA KIRC -0.051; TCGA KIRP -0.055; TCGA LGG -0.108; TCGA LUSC -0.122; TCGA PRAD -0.05; TCGA THCA -0.097; TCGA STAD -0.165; TCGA UCEC -0.076 |
hsa-miR-203a-3p | ZFHX4 | 10 cancers: BLCA; BRCA; COAD; HNSC; LGG; LIHC; LUAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.204; TCGA BRCA -0.066; TCGA COAD -0.76; TCGA HNSC -0.098; TCGA LGG -0.056; TCGA LIHC -0.099; TCGA LUAD -0.11; TCGA PRAD -0.205; TCGA STAD -0.434; TCGA UCEC -0.216 |
hsa-miR-203a-3p | FAT3 | 9 cancers: BLCA; CESC; COAD; HNSC; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.164; TCGA CESC -0.22; TCGA COAD -0.429; TCGA HNSC -0.351; TCGA LUSC -0.531; TCGA PAAD -0.124; TCGA PRAD -0.582; TCGA STAD -0.666; TCGA UCEC -0.2 |
hsa-miR-203a-3p | TMEM56 | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LUSC; OV; PAAD; PRAD; STAD | MirTarget | TCGA BLCA -0.112; TCGA BRCA -0.147; TCGA CESC -0.346; TCGA ESCA -0.366; TCGA HNSC -0.306; TCGA LUSC -0.179; TCGA OV -0.096; TCGA PAAD -0.104; TCGA PRAD -0.215; TCGA STAD -0.171 |
hsa-miR-203a-3p | SCN2A | 10 cancers: BLCA; COAD; ESCA; HNSC; LIHC; LUAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.093; TCGA COAD -0.473; TCGA ESCA -0.217; TCGA HNSC -0.226; TCGA LIHC -0.211; TCGA LUAD -0.22; TCGA PRAD -0.236; TCGA SARC -0.134; TCGA STAD -0.389; TCGA UCEC -0.15 |
hsa-miR-203a-3p | DAB2 | 14 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.155; TCGA BRCA -0.062; TCGA CESC -0.107; TCGA ESCA -0.209; TCGA HNSC -0.291; TCGA KIRC -0.15; TCGA KIRP -0.093; TCGA LGG -0.189; TCGA LUAD -0.077; TCGA LUSC -0.189; TCGA PRAD -0.132; TCGA THCA -0.093; TCGA STAD -0.192; TCGA UCEC -0.189 |
hsa-miR-203a-3p | PDE3B | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; OV; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.131; TCGA BRCA -0.203; TCGA COAD -0.109; TCGA ESCA -0.214; TCGA HNSC -0.191; TCGA OV -0.121; TCGA PAAD -0.313; TCGA PRAD -0.122; TCGA STAD -0.121; TCGA UCEC -0.124 |
hsa-miR-203a-3p | SFRP1 | 10 cancers: BLCA; CESC; COAD; LUAD; LUSC; PAAD; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.206; TCGA CESC -0.346; TCGA COAD -0.818; TCGA LUAD -0.153; TCGA LUSC -0.216; TCGA PAAD -0.386; TCGA PRAD -0.287; TCGA THCA -0.601; TCGA STAD -0.705; TCGA UCEC -0.184 |
hsa-miR-203a-3p | DNM3 | 9 cancers: BLCA; CESC; ESCA; HNSC; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.083; TCGA CESC -0.159; TCGA ESCA -0.197; TCGA HNSC -0.108; TCGA LUSC -0.288; TCGA PAAD -0.087; TCGA PRAD -0.185; TCGA STAD -0.166; TCGA UCEC -0.068 |
hsa-miR-203a-3p | MYEF2 | 9 cancers: BLCA; CESC; ESCA; HNSC; LGG; LIHC; LUSC; PAAD; STAD | MirTarget | TCGA BLCA -0.182; TCGA CESC -0.526; TCGA ESCA -0.326; TCGA HNSC -0.296; TCGA LGG -0.052; TCGA LIHC -0.127; TCGA LUSC -0.28; TCGA PAAD -0.213; TCGA STAD -0.561 |
hsa-miR-203a-3p | ADAMTS6 | 11 cancers: BLCA; CESC; COAD; ESCA; HNSC; LGG; LUAD; LUSC; OV; PRAD; STAD | MirTarget | TCGA BLCA -0.173; TCGA CESC -0.252; TCGA COAD -0.343; TCGA ESCA -0.224; TCGA HNSC -0.253; TCGA LGG -0.147; TCGA LUAD -0.164; TCGA LUSC -0.164; TCGA OV -0.105; TCGA PRAD -0.225; TCGA STAD -0.15 |
hsa-miR-203a-3p | RAPGEF1 | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.088; TCGA BRCA -0.05; TCGA COAD -0.08; TCGA ESCA -0.06; TCGA HNSC -0.127; TCGA KIRC -0.073; TCGA LUSC -0.077; TCGA PAAD -0.095; TCGA PRAD -0.103; TCGA STAD -0.096; TCGA UCEC -0.067 |
hsa-miR-203a-3p | KCNQ5 | 9 cancers: BLCA; COAD; HNSC; KIRC; LUAD; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.284; TCGA COAD -0.505; TCGA HNSC -0.14; TCGA KIRC -0.343; TCGA LUAD -0.274; TCGA PAAD -0.191; TCGA PRAD -0.308; TCGA STAD -0.397; TCGA UCEC -0.163 |
hsa-miR-203a-3p | CTSS | 9 cancers: BLCA; ESCA; HNSC; KIRC; KIRP; LGG; LUSC; PRAD; SARC | MirTarget | TCGA BLCA -0.08; TCGA ESCA -0.26; TCGA HNSC -0.161; TCGA KIRC -0.219; TCGA KIRP -0.125; TCGA LGG -0.191; TCGA LUSC -0.241; TCGA PRAD -0.16; TCGA SARC -0.107 |
hsa-miR-203a-3p | CDON | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; OV; PAAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.145; TCGA BRCA -0.181; TCGA CESC -0.363; TCGA COAD -0.388; TCGA ESCA -0.205; TCGA HNSC -0.144; TCGA KIRC -0.296; TCGA LIHC -0.094; TCGA LUAD -0.185; TCGA LUSC -0.268; TCGA OV -0.09; TCGA PAAD -0.222; TCGA THCA -0.432; TCGA STAD -0.403; TCGA UCEC -0.183 |
hsa-miR-203a-3p | TMEM100 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LUSC; PAAD; STAD; UCEC | MirTarget | TCGA BLCA -0.284; TCGA BRCA -0.183; TCGA CESC -0.495; TCGA COAD -0.587; TCGA ESCA -0.36; TCGA HNSC -0.238; TCGA LGG -0.117; TCGA LUSC -0.448; TCGA PAAD -0.123; TCGA STAD -0.62; TCGA UCEC -0.201 |
hsa-miR-203a-3p | GPRIN3 | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; SARC | MirTarget | TCGA BLCA -0.141; TCGA BRCA -0.162; TCGA CESC -0.103; TCGA ESCA -0.355; TCGA HNSC -0.158; TCGA LUAD -0.081; TCGA LUSC -0.266; TCGA PAAD -0.237; TCGA PRAD -0.241; TCGA SARC -0.097 |
hsa-miR-203a-3p | MEF2C | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.186; TCGA BRCA -0.128; TCGA CESC -0.149; TCGA COAD -0.333; TCGA ESCA -0.237; TCGA HNSC -0.266; TCGA KIRC -0.107; TCGA LUAD -0.117; TCGA LUSC -0.216; TCGA PAAD -0.118; TCGA PRAD -0.319; TCGA THCA -0.208; TCGA STAD -0.299; TCGA UCEC -0.241 |
hsa-miR-203a-3p | PXDN | 14 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.145; TCGA CESC -0.336; TCGA COAD -0.325; TCGA ESCA -0.101; TCGA HNSC -0.245; TCGA KIRC -0.17; TCGA KIRP -0.167; TCGA LGG -0.163; TCGA LUAD -0.142; TCGA LUSC -0.19; TCGA PRAD -0.17; TCGA THCA -0.111; TCGA STAD -0.152; TCGA UCEC -0.123 |
hsa-miR-203a-3p | MPDZ | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.134; TCGA BRCA -0.106; TCGA CESC -0.342; TCGA COAD -0.524; TCGA ESCA -0.211; TCGA HNSC -0.309; TCGA LIHC -0.112; TCGA LUAD -0.094; TCGA LUSC -0.249; TCGA OV -0.155; TCGA PAAD -0.22; TCGA PRAD -0.196; TCGA STAD -0.41; TCGA UCEC -0.197 |
hsa-miR-203a-3p | PRSS35 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; LUAD; LUSC; PAAD; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.226; TCGA BRCA -0.114; TCGA CESC -0.26; TCGA COAD -0.218; TCGA ESCA -0.327; TCGA LUAD -0.11; TCGA LUSC -0.172; TCGA PAAD -0.166; TCGA PRAD -0.458; TCGA THCA -0.39; TCGA STAD -0.256; TCGA UCEC -0.412 |
hsa-miR-203a-3p | SGTB | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.121; TCGA BRCA -0.067; TCGA COAD -0.197; TCGA ESCA -0.062; TCGA HNSC -0.108; TCGA KIRC -0.053; TCGA LUAD -0.118; TCGA LUSC -0.084; TCGA PAAD -0.186; TCGA SARC -0.054; TCGA STAD -0.168; TCGA UCEC -0.138 |
hsa-miR-203a-3p | ANTXR2 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LUAD; LUSC; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.182; TCGA BRCA -0.094; TCGA CESC -0.221; TCGA COAD -0.13; TCGA ESCA -0.223; TCGA HNSC -0.132; TCGA LGG -0.135; TCGA LUAD -0.079; TCGA LUSC -0.183; TCGA SARC -0.129; TCGA THCA -0.142; TCGA STAD -0.315; TCGA UCEC -0.299 |
hsa-miR-203a-3p | CPS1 | 10 cancers: BLCA; BRCA; COAD; ESCA; LGG; OV; PRAD; SARC; THCA; UCEC | MirTarget | TCGA BLCA -0.11; TCGA BRCA -0.118; TCGA COAD -0.354; TCGA ESCA -0.266; TCGA LGG -0.104; TCGA OV -0.192; TCGA PRAD -0.247; TCGA SARC -0.132; TCGA THCA -0.282; TCGA UCEC -0.141 |
hsa-miR-203a-3p | LIFR | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.194; TCGA BRCA -0.229; TCGA CESC -0.372; TCGA COAD -0.513; TCGA ESCA -0.343; TCGA HNSC -0.362; TCGA LUAD -0.137; TCGA LUSC -0.407; TCGA PAAD -0.239; TCGA THCA -0.36; TCGA STAD -0.322; TCGA UCEC -0.199 |
hsa-miR-203a-3p | IKZF1 | 12 cancers: BLCA; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; PRAD; SARC; STAD | MirTarget | TCGA BLCA -0.193; TCGA COAD -0.284; TCGA ESCA -0.196; TCGA HNSC -0.126; TCGA KIRC -0.303; TCGA KIRP -0.064; TCGA LGG -0.124; TCGA LUAD -0.069; TCGA LUSC -0.282; TCGA PRAD -0.253; TCGA SARC -0.111; TCGA STAD -0.109 |
hsa-miR-203a-3p | CNTN3 | 9 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LUSC; PAAD; THCA; STAD | MirTarget | TCGA BLCA -0.17; TCGA BRCA -0.07; TCGA CESC -0.268; TCGA ESCA -0.332; TCGA HNSC -0.109; TCGA LUSC -0.234; TCGA PAAD -0.192; TCGA THCA -0.331; TCGA STAD -0.373 |
hsa-miR-203a-3p | S1PR1 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUSC; PAAD; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.143; TCGA BRCA -0.161; TCGA CESC -0.151; TCGA COAD -0.381; TCGA ESCA -0.311; TCGA HNSC -0.153; TCGA LUSC -0.242; TCGA PAAD -0.175; TCGA PRAD -0.223; TCGA THCA -0.084; TCGA STAD -0.246; TCGA UCEC -0.168 |
hsa-miR-203a-3p | SLC2A3 | 10 cancers: BLCA; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.154; TCGA COAD -0.26; TCGA ESCA -0.219; TCGA HNSC -0.221; TCGA KIRC -0.192; TCGA LUAD -0.081; TCGA LUSC -0.217; TCGA PRAD -0.208; TCGA STAD -0.122; TCGA UCEC -0.104 |
hsa-miR-203a-3p | HGF | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.307; TCGA BRCA -0.136; TCGA CESC -0.205; TCGA COAD -0.461; TCGA ESCA -0.519; TCGA HNSC -0.321; TCGA LUAD -0.154; TCGA LUSC -0.433; TCGA PRAD -0.503; TCGA SARC -0.123; TCGA THCA -0.121; TCGA STAD -0.201; TCGA UCEC -0.22 |
hsa-miR-203a-3p | PDGFD | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.099; TCGA BRCA -0.179; TCGA CESC -0.211; TCGA COAD -0.19; TCGA ESCA -0.317; TCGA HNSC -0.232; TCGA KIRC -0.157; TCGA LUAD -0.153; TCGA LUSC -0.204; TCGA PRAD -0.22; TCGA THCA -0.385; TCGA STAD -0.241; TCGA UCEC -0.1 |
hsa-miR-203a-3p | GLRB | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; PRAD; STAD | MirTarget | TCGA BLCA -0.161; TCGA BRCA -0.239; TCGA CESC -0.574; TCGA COAD -0.551; TCGA ESCA -0.251; TCGA HNSC -0.428; TCGA KIRP -0.132; TCGA LUAD -0.159; TCGA LUSC -0.324; TCGA PAAD -0.164; TCGA PRAD -0.235; TCGA STAD -0.432 |
hsa-miR-203a-3p | RGAG4 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.184; TCGA BRCA -0.147; TCGA CESC -0.229; TCGA COAD -0.415; TCGA ESCA -0.248; TCGA HNSC -0.316; TCGA LUAD -0.1; TCGA LUSC -0.24; TCGA OV -0.109; TCGA PAAD -0.371; TCGA PRAD -0.102; TCGA STAD -0.435; TCGA UCEC -0.188 |
hsa-miR-203a-3p | GNB4 | 10 cancers: BLCA; COAD; HNSC; KIRC; LGG; LUAD; LUSC; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.111; TCGA COAD -0.372; TCGA HNSC -0.078; TCGA KIRC -0.083; TCGA LGG -0.153; TCGA LUAD -0.079; TCGA LUSC -0.072; TCGA PRAD -0.107; TCGA STAD -0.243; TCGA UCEC -0.095 |
hsa-miR-203a-3p | PCSK2 | 9 cancers: BLCA; BRCA; CESC; COAD; ESCA; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.159; TCGA BRCA -0.16; TCGA CESC -0.147; TCGA COAD -0.674; TCGA ESCA -0.261; TCGA PAAD -0.55; TCGA PRAD -0.413; TCGA STAD -0.656; TCGA UCEC -0.13 |
hsa-miR-203a-3p | DCBLD2 | 10 cancers: BLCA; CESC; COAD; HNSC; LIHC; LUAD; LUSC; PRAD; SARC; STAD | MirTarget | TCGA BLCA -0.112; TCGA CESC -0.127; TCGA COAD -0.215; TCGA HNSC -0.157; TCGA LIHC -0.064; TCGA LUAD -0.099; TCGA LUSC -0.116; TCGA PRAD -0.21; TCGA SARC -0.085; TCGA STAD -0.229 |
hsa-miR-203a-3p | LIMCH1 | 11 cancers: BRCA; CESC; COAD; ESCA; HNSC; LUSC; PAAD; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BRCA -0.106; TCGA CESC -0.281; TCGA COAD -0.404; TCGA ESCA -0.137; TCGA HNSC -0.119; TCGA LUSC -0.244; TCGA PAAD -0.233; TCGA PRAD -0.159; TCGA THCA -0.141; TCGA STAD -0.172; TCGA UCEC -0.097 |
hsa-miR-203a-3p | ATM | 10 cancers: BRCA; CESC; ESCA; HNSC; KIRC; LGG; LUSC; PAAD; STAD; UCEC | MirTarget | TCGA BRCA -0.07; TCGA CESC -0.107; TCGA ESCA -0.139; TCGA HNSC -0.105; TCGA KIRC -0.084; TCGA LGG -0.058; TCGA LUSC -0.099; TCGA PAAD -0.08; TCGA STAD -0.115; TCGA UCEC -0.091 |
hsa-miR-203a-3p | DGKE | 10 cancers: BRCA; CESC; ESCA; HNSC; LUAD; PAAD; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BRCA -0.11; TCGA CESC -0.116; TCGA ESCA -0.119; TCGA HNSC -0.072; TCGA LUAD -0.083; TCGA PAAD -0.166; TCGA PRAD -0.116; TCGA THCA -0.143; TCGA STAD -0.143; TCGA UCEC -0.096 |
hsa-miR-203a-3p | GLCCI1 | 11 cancers: BRCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; LUSC; OV; PAAD; STAD | MirTarget | TCGA BRCA -0.086; TCGA CESC -0.084; TCGA COAD -0.123; TCGA ESCA -0.181; TCGA HNSC -0.068; TCGA LGG -0.053; TCGA LIHC -0.058; TCGA LUSC -0.214; TCGA OV -0.098; TCGA PAAD -0.175; TCGA STAD -0.167 |
hsa-miR-203a-3p | TMEM47 | 12 cancers: BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BRCA -0.095; TCGA CESC -0.308; TCGA COAD -0.347; TCGA ESCA -0.249; TCGA HNSC -0.256; TCGA LUAD -0.064; TCGA LUSC -0.199; TCGA PRAD -0.284; TCGA SARC -0.132; TCGA THCA -0.351; TCGA STAD -0.335; TCGA UCEC -0.115 |
hsa-miR-203a-3p | ZNF441 | 9 cancers: BRCA; CESC; ESCA; HNSC; LUSC; PAAD; THCA; STAD; UCEC | MirTarget | TCGA BRCA -0.065; TCGA CESC -0.142; TCGA ESCA -0.159; TCGA HNSC -0.085; TCGA LUSC -0.086; TCGA PAAD -0.187; TCGA THCA -0.06; TCGA STAD -0.119; TCGA UCEC -0.073 |
hsa-miR-203a-3p | PPM1E | 10 cancers: BRCA; CESC; COAD; ESCA; LUAD; LUSC; PAAD; THCA; STAD; UCEC | MirTarget | TCGA BRCA -0.094; TCGA CESC -0.176; TCGA COAD -0.203; TCGA ESCA -0.176; TCGA LUAD -0.282; TCGA LUSC -0.256; TCGA PAAD -0.439; TCGA THCA -0.325; TCGA STAD -0.286; TCGA UCEC -0.151 |
hsa-miR-203a-3p | SESTD1 | 10 cancers: BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUSC; PRAD; SARC; STAD | MirTarget | TCGA BRCA -0.094; TCGA CESC -0.158; TCGA COAD -0.114; TCGA ESCA -0.166; TCGA HNSC -0.167; TCGA LIHC -0.056; TCGA LUSC -0.103; TCGA PRAD -0.179; TCGA SARC -0.07; TCGA STAD -0.148 |
hsa-miR-203a-3p | KCNIP4 | 11 cancers: BRCA; CESC; COAD; ESCA; HNSC; LUSC; OV; PAAD; THCA; STAD; UCEC | MirTarget | TCGA BRCA -0.06; TCGA CESC -0.131; TCGA COAD -0.117; TCGA ESCA -0.084; TCGA HNSC -0.062; TCGA LUSC -0.057; TCGA OV -0.143; TCGA PAAD -0.085; TCGA THCA -0.179; TCGA STAD -0.151; TCGA UCEC -0.249 |
hsa-miR-203a-3p | DPY19L3 | 9 cancers: BRCA; CESC; ESCA; HNSC; LGG; LUSC; OV; PAAD; UCEC | MirTarget | TCGA BRCA -0.055; TCGA CESC -0.12; TCGA ESCA -0.115; TCGA HNSC -0.075; TCGA LGG -0.074; TCGA LUSC -0.09; TCGA OV -0.091; TCGA PAAD -0.099; TCGA UCEC -0.087 |
hsa-miR-203a-3p | TTN | 9 cancers: BRCA; COAD; HNSC; KIRC; LUSC; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BRCA -0.097; TCGA COAD -0.159; TCGA HNSC -0.214; TCGA KIRC -0.157; TCGA LUSC -0.129; TCGA PRAD -0.184; TCGA THCA -0.234; TCGA STAD -0.137; TCGA UCEC -0.066 |
hsa-miR-203a-3p | EYA4 | 11 cancers: CESC; COAD; HNSC; LIHC; LUSC; OV; PAAD; PRAD; THCA; STAD; UCEC | miRTarBase | TCGA CESC -0.432; TCGA COAD -0.376; TCGA HNSC -0.239; TCGA LIHC -0.153; TCGA LUSC -0.276; TCGA OV -0.258; TCGA PAAD -0.168; TCGA PRAD -0.488; TCGA THCA -0.378; TCGA STAD -0.346; TCGA UCEC -0.171 |
hsa-miR-203a-3p | GUCY1A2 | 9 cancers: CESC; COAD; ESCA; HNSC; LUSC; PRAD; THCA; STAD; UCEC | MirTarget | TCGA CESC -0.233; TCGA COAD -0.302; TCGA ESCA -0.118; TCGA HNSC -0.189; TCGA LUSC -0.178; TCGA PRAD -0.28; TCGA THCA -0.209; TCGA STAD -0.134; TCGA UCEC -0.224 |
hsa-miR-203a-3p | SP4 | 12 cancers: CESC; COAD; ESCA; HNSC; LUAD; LUSC; OV; PAAD; PRAD; THCA; STAD; UCEC | MirTarget | TCGA CESC -0.071; TCGA COAD -0.113; TCGA ESCA -0.127; TCGA HNSC -0.125; TCGA LUAD -0.136; TCGA LUSC -0.084; TCGA OV -0.15; TCGA PAAD -0.188; TCGA PRAD -0.097; TCGA THCA -0.053; TCGA STAD -0.107; TCGA UCEC -0.103 |
hsa-miR-203a-3p | CLSTN3 | 11 cancers: CESC; COAD; ESCA; HNSC; KIRC; LUSC; OV; PAAD; PRAD; THCA; STAD | MirTarget | TCGA CESC -0.078; TCGA COAD -0.193; TCGA ESCA -0.071; TCGA HNSC -0.153; TCGA KIRC -0.086; TCGA LUSC -0.117; TCGA OV -0.16; TCGA PAAD -0.224; TCGA PRAD -0.074; TCGA THCA -0.126; TCGA STAD -0.147 |
Enriched cancer pathways of putative targets