microRNA information: hsa-miR-20a-3p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-20a-3p | miRbase |
| Accession: | MIMAT0004493 | miRbase |
| Precursor name: | hsa-mir-20a | miRbase |
| Precursor accession: | MI0000076 | miRbase |
| Symbol: | MIR20A | HGNC |
| RefSeq ID: | NR_029492 | GenBank |
| Sequence: | ACUGCAUUAUGAGCACUUAAAG |
Reported expression in cancers: hsa-miR-20a-3p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-20a-3p | bladder cancer | deregulation | "miRNAs downregulated in bladder cancer such as miR ......" | 23712207 | |
| hsa-miR-20a-3p | bladder cancer | deregulation | "We screened 723 miRNAs by microarray and selected ......" | 23945108 | qPCR; Microarray |
| hsa-miR-20a-3p | bladder cancer | deregulation | "In GBC miRNAs with tumor suppressor activity miR-1 ......" | 26855538 | |
| hsa-miR-20a-3p | cervical and endocervical cancer | upregulation | "Our study demonstrated that miR-20a was upregulate ......" | 22449978 | |
| hsa-miR-20a-3p | cervical and endocervical cancer | upregulation | "Vote-counting analysis showed that up-regulation w ......" | 25920605 | |
| hsa-miR-20a-3p | cervical and endocervical cancer | upregulation | "Upregulation of miR 20a and miR 10a expression lev ......" | 26427662 | qPCR |
| hsa-miR-20a-3p | colon cancer | upregulation | "To present proof-of-principle application for empl ......" | 23741026 | qPCR; Microarray |
| hsa-miR-20a-3p | colorectal cancer | upregulation | "The aim of the present study was to investigate th ......" | 26622375 | qPCR |
| hsa-miR-20a-3p | colorectal cancer | upregulation | "Serum miRNAs were extracted from all subjects to a ......" | 27135244 | |
| hsa-miR-20a-3p | gastric cancer | upregulation | "The most highly expressed miRNAs in gastric cancer ......" | 19175831 | |
| hsa-miR-20a-3p | gastric cancer | upregulation | "The expression levels of miR-20a miR-21 miR-25 miR ......" | 22996433 | qPCR |
| hsa-miR-20a-3p | gastric cancer | upregulation | "We identified five miRNAs that were most consisten ......" | 24040025 | qPCR |
| hsa-miR-20a-3p | gastric cancer | upregulation | "We previously identified five miRNAs miR-1 miR-20a ......" | 24122958 | Reverse transcription PCR; qPCR |
| hsa-miR-20a-3p | gastric cancer | upregulation | "A total of 33 miRNAs were identified through the i ......" | 26059512 | Reverse transcription PCR; qPCR |
| hsa-miR-20a-3p | gastric cancer | upregulation | "Here we sought to investigate whether there was a ......" | 26286834 | |
| hsa-miR-20a-3p | head and neck cancer | deregulation | "A global miRNA profiling was done on 51 formalin-f ......" | 20145181 | Reverse transcription PCR |
| hsa-miR-20a-3p | liver cancer | upregulation | "MiR 20a Induces Cell Radioresistance by Activating ......" | 26031366 | |
| hsa-miR-20a-3p | liver cancer | upregulation | "Previous studies reported that miR-20a is among th ......" | 27748919 | |
| hsa-miR-20a-3p | lung cancer | upregulation | "miR 20a regulates expression of the iron exporter ......" | 26560875 | RNA-Seq |
| hsa-miR-20a-3p | pancreatic cancer | downregulation | "miRNAs that were differentially expressed in pancr ......" | 25485236 | Microarray; qPCR |
| hsa-miR-20a-3p | prostate cancer | upregulation | "The present study found miR-20a to be significantl ......" | 22785209 | |
| hsa-miR-20a-3p | prostate cancer | upregulation | "The present study found miR-20a was significantly ......" | 24464651 | |
| hsa-miR-20a-3p | thyroid cancer | upregulation | "There have been conflicting reports regarding the ......" | 24858712 | qPCR |
Reported cancer pathway affected by hsa-miR-20a-3p

| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-20a-3p | cervical and endocervical cancer | cell cycle pathway; Apoptosis pathway | "MiR 20a promotes cervical cancer proliferation and ......" | 25803820 | Western blot; Luciferase |
| hsa-miR-20a-3p | colon cancer | cell cycle pathway | "The Effects of miR 20a on p21: Two Mechanisms Bloc ......" | 26012475 | |
| hsa-miR-20a-3p | gastric cancer | Apoptosis pathway | "miR 20a enhances cisplatin resistance of human gas ......" | 26286834 | Luciferase |
| hsa-miR-20a-3p | gastric cancer | Apoptosis pathway | "miR 20a induces cisplatin resistance of a human ga ......" | 27357419 | Western blot; Flow cytometry; Luciferase |
| hsa-miR-20a-3p | head and neck cancer | cell cycle pathway | "A global miRNA profiling was done on 51 formalin-f ......" | 20145181 | Flow cytometry |
| hsa-miR-20a-3p | liver cancer | cell cycle pathway; Apoptosis pathway | "Decrease expression of microRNA 20a promotes cance ......" | 23594563 | Flow cytometry; Luciferase |
| hsa-miR-20a-3p | liver cancer | PI3K/Akt signaling pathway | "MiR 20a Induces Cell Radioresistance by Activating ......" | 26031366 | Colony formation; Luciferase |
| hsa-miR-20a-3p | lung cancer | Apoptosis pathway | "Apoptosis induction by antisense oligonucleotides ......" | 17384677 | |
| hsa-miR-20a-3p | sarcoma | Apoptosis pathway | "MiR 20a 5p represses multi drug resistance in oste ......" | 27499703 | Western blot; MTT assay; Flow cytometry |
Reported cancer prognosis affected by hsa-miR-20a-3p
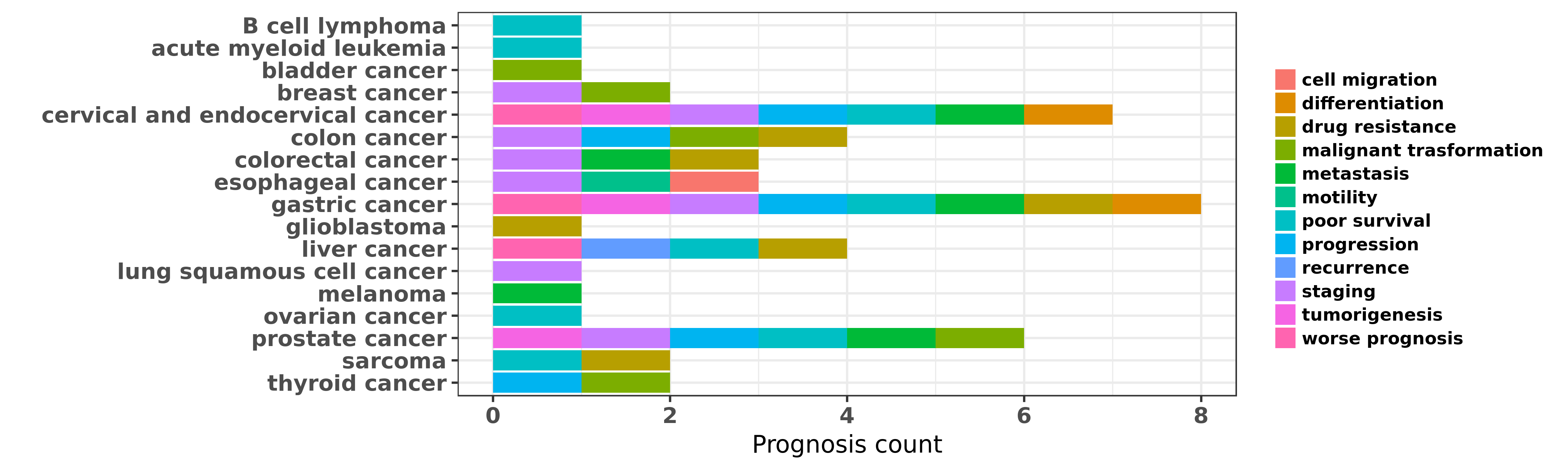
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-20a-3p | B cell lymphoma | poor survival | "In 22 primary cutaneous diffuse large B-cell lymph ......" | 25634356 | |
| hsa-miR-20a-3p | acute myeloid leukemia | poor survival | "The expression level of MN1 microRNA-20 miR-20a an ......" | 23515710 | |
| hsa-miR-20a-3p | bladder cancer | malignant trasformation | "We screened 723 miRNAs by microarray and selected ......" | 23945108 | |
| hsa-miR-20a-3p | breast cancer | malignant trasformation | "The relative concentrations of four circulating mi ......" | 22350790 | |
| hsa-miR-20a-3p | breast cancer | staging | "We employed qRT-PCR to determine expression level ......" | 27596294 | |
| hsa-miR-20a-3p | cervical and endocervical cancer | metastasis; tumorigenesis | "Aberrant expression of miR 20a and miR 203 in cerv ......" | 23725129 | |
| hsa-miR-20a-3p | cervical and endocervical cancer | staging; metastasis; differentiation | "We hypothesized that microRNA-20a miR-20a and micr ......" | 23819812 | |
| hsa-miR-20a-3p | cervical and endocervical cancer | metastasis; progression; tumorigenesis | "MiR 20a promotes cervical cancer proliferation and ......" | 25803820 | Western blot; Luciferase |
| hsa-miR-20a-3p | cervical and endocervical cancer | progression; worse prognosis; poor survival; staging; metastasis | "Upregulation of miR 20a and miR 10a expression lev ......" | 26427662 | |
| hsa-miR-20a-3p | colon cancer | staging | "To present proof-of-principle application for empl ......" | 23741026 | |
| hsa-miR-20a-3p | colon cancer | drug resistance; progression; malignant trasformation | "The Effects of miR 20a on p21: Two Mechanisms Bloc ......" | 26012475 | |
| hsa-miR-20a-3p | colorectal cancer | drug resistance | "miR 20a targets BNIP2 and contributes chemotherape ......" | 21242194 | Western blot |
| hsa-miR-20a-3p | colorectal cancer | staging | "Genome-wide microarray analysis of miRNA expressio ......" | 23673725 | |
| hsa-miR-20a-3p | colorectal cancer | staging | "A meta-analysis was performed to review three miRN ......" | 25027346 | |
| hsa-miR-20a-3p | colorectal cancer | metastasis | "microRNA 20a enhances the epithelial to mesenchyma ......" | 26622375 | Wound Healing Assay; Western blot |
| hsa-miR-20a-3p | colorectal cancer | metastasis | "MicroRNA 20a 5p promotes colorectal cancer invasio ......" | 27286257 | |
| hsa-miR-20a-3p | esophageal cancer | staging | "Expression of circulating microRNA 20a and let 7a ......" | 25914476 | |
| hsa-miR-20a-3p | esophageal cancer | cell migration; motility | "Here we evaluated microRNA expression in ESCC cell ......" | 27508097 | |
| hsa-miR-20a-3p | gastric cancer | worse prognosis | "Circulating miR-17-5p and miR-20a miR-17-5p/20a ha ......" | 22406928 | |
| hsa-miR-20a-3p | gastric cancer | metastasis; poor survival | "The expression levels of miR-20a miR-21 miR-25 miR ......" | 22996433 | |
| hsa-miR-20a-3p | gastric cancer | differentiation | "We found that the miR-17-92 cluster members miR-19 ......" | 23868977 | |
| hsa-miR-20a-3p | gastric cancer | progression; drug resistance; tumorigenesis | "Involvement of miR 20a in promoting gastric cancer ......" | 23924943 | Western blot; Luciferase |
| hsa-miR-20a-3p | gastric cancer | progression | "F box protein FBXO31 is down regulated in gastric ......" | 25115392 | Colony formation |
| hsa-miR-20a-3p | gastric cancer | staging | "A total of 33 miRNAs were identified through the i ......" | 26059512 | |
| hsa-miR-20a-3p | gastric cancer | drug resistance | "miR 20a enhances cisplatin resistance of human gas ......" | 26286834 | Luciferase |
| hsa-miR-20a-3p | gastric cancer | drug resistance | "miR 20a induces cisplatin resistance of a human ga ......" | 27357419 | Western blot; Flow cytometry; Luciferase |
| hsa-miR-20a-3p | glioblastoma | drug resistance | "miR 20a mediates temozolomide resistance in gliobl ......" | 25960225 | Western blot; Luciferase; RNAi |
| hsa-miR-20a-3p | liver cancer | poor survival; recurrence; worse prognosis | "Decrease expression of microRNA 20a promotes cance ......" | 23594563 | Flow cytometry; Luciferase |
| hsa-miR-20a-3p | liver cancer | drug resistance | "MiR 20a Induces Cell Radioresistance by Activating ......" | 26031366 | Colony formation; Luciferase |
| hsa-miR-20a-3p | lung squamous cell cancer | staging | "In order to find novel noninvasive biomarkers with ......" | 25421010 | |
| hsa-miR-20a-3p | melanoma | metastasis | "Circulating T cell natural killer NK natural kille ......" | 24370793 | Flow cytometry |
| hsa-miR-20a-3p | ovarian cancer | poor survival | "The Cox proportional hazards model and the log-ran ......" | 21345725 | |
| hsa-miR-20a-3p | prostate cancer | tumorigenesis | "Importance of miR 20a expression in prostate cance ......" | 20944140 | |
| hsa-miR-20a-3p | prostate cancer | staging | "Taqman based quantitative RT-PCR assays were perfo ......" | 22298119 | |
| hsa-miR-20a-3p | prostate cancer | progression | "Suppression of CX43 expression by miR 20a in the p ......" | 22785209 | Colony formation |
| hsa-miR-20a-3p | prostate cancer | staging; malignant trasformation | "Comparative microRNA profiling of prostate carcino ......" | 24337069 | |
| hsa-miR-20a-3p | prostate cancer | poor survival; progression | "miR 20a promotes prostate cancer invasion and migr ......" | 24464651 | Wound Healing Assay |
| hsa-miR-20a-3p | prostate cancer | metastasis; poor survival | "Multivariate Cox regression with bootstrapping val ......" | 24714754 | |
| hsa-miR-20a-3p | sarcoma | poor survival | "miR 20a encoded by the miR 17 92 cluster increases ......" | 22186140 | |
| hsa-miR-20a-3p | sarcoma | drug resistance | "MiR 20a 5p represses multi drug resistance in oste ......" | 27499703 | Western blot; MTT assay; Flow cytometry |
| hsa-miR-20a-3p | thyroid cancer | malignant trasformation; progression | "MiR 20a is upregulated in anaplastic thyroid cance ......" | 24858712 | Luciferase |
Reported gene related to hsa-miR-20a-3p

| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-20a-3p | cervical and endocervical cancer | ATG7 | "TIMP2 and ATG7 were proved to be direct targets of ......" | 25803820 |
| hsa-miR-20a-3p | prostate cancer | ATG7 | "Celastrol induced autophagy was inhibited by miR-2 ......" | 27501757 |
| hsa-miR-20a-3p | acute myeloid leukemia | FRTS1 | "In multivariable models high MN1 expression was as ......" | 23515710 |
| hsa-miR-20a-3p | liver cancer | FRTS1 | "Patients with lower miR-20a expression had signifi ......" | 23594563 |
| hsa-miR-20a-3p | lung cancer | SCLC1 | "We found earlier frequent overexpression of the mi ......" | 19597473 |
| hsa-miR-20a-3p | lung squamous cell cancer | SCLC1 | "Among them five downregulated miRNAs let-7 miR-20 ......" | 23117485 |
| hsa-miR-20a-3p | breast cancer | VEGFA | "Correlation analysis showed that the key miRNAs mi ......" | 22901144 |
| hsa-miR-20a-3p | endometrial cancer | VEGFA | "Moreover significant inverse correlations between ......" | 22904162 |
| hsa-miR-20a-3p | prostate cancer | ABL2 | "miR 20a promotes prostate cancer invasion and migr ......" | 24464651 |
| hsa-miR-20a-3p | melanoma | ADAR | "Luciferase assays showed that miR-17 but not miR-2 ......" | 24920276 |
| hsa-miR-20a-3p | ovarian cancer | APP | "miR 20a promotes proliferation and invasion by tar ......" | 20458444 |
| hsa-miR-20a-3p | prostate cancer | ARHGAP35 | "Furthermore we identified miR-20a directly targets ......" | 24464651 |
| hsa-miR-20a-3p | lymphoma | BCL2L11 | "We found that upregulated expression of miR-17 and ......" | 27044389 |
| hsa-miR-20a-3p | gastric cancer | BIRC7 | "And the upregulation of miR-20a was concurrent wit ......" | 26286834 |
| hsa-miR-20a-3p | colorectal cancer | BNIP2 | "miR 20a targets BNIP2 and contributes chemotherape ......" | 21242194 |
| hsa-miR-20a-3p | colon cancer | CDKN1A | "Using the poorly tumorigenic and TGF-β-sensitive ......" | 26012475 |
| hsa-miR-20a-3p | gastric cancer | CYLD | "miR 20a induces cisplatin resistance of a human ga ......" | 27357419 |
| hsa-miR-20a-3p | breast cancer | EGR1 | "Luciferase assays showed that whereas EGR1 stimula ......" | 23945289 |
| hsa-miR-20a-3p | gastric cancer | EGR2 | "Involvement of miR 20a in promoting gastric cancer ......" | 23924943 |
| hsa-miR-20a-3p | sarcoma | FAS | "We also found an inverse correlation between Fas a ......" | 22186140 |
| hsa-miR-20a-3p | sarcoma | FASLG | "Overexpression of miR-20a consistently resulted in ......" | 22186140 |
| hsa-miR-20a-3p | gastric cancer | FBXO31 | "F box protein FBXO31 is down regulated in gastric ......" | 25115392 |
| hsa-miR-20a-3p | prostate cancer | GJA1 | "Suppression of CX43 expression by miR 20a in the p ......" | 22785209 |
| hsa-miR-20a-3p | breast cancer | HIF1A | "Correlation analysis showed that the key miRNAs mi ......" | 22901144 |
| hsa-miR-20a-3p | sarcoma | KIF26B | "MiR 20a 5p represses multi drug resistance in oste ......" | 27499703 |
| hsa-miR-20a-3p | thyroid cancer | LIMK1 | "MiR 20a is upregulated in anaplastic thyroid cance ......" | 24858712 |
| hsa-miR-20a-3p | glioblastoma | LRIG1 | "miR 20a mediates temozolomide resistance in gliobl ......" | 25960225 |
| hsa-miR-20a-3p | liver cancer | MCL1 | "Subsequent investigations revealed that miR-20a di ......" | 23594563 |
| hsa-miR-20a-3p | acute myeloid leukemia | MN1 | "High MN1 expression was associated with spleen inv ......" | 23515710 |
| hsa-miR-20a-3p | colon cancer | MYC | "Finally miR-20a abrogated the TGF-β-mediated c-My ......" | 26012475 |
| hsa-miR-20a-3p | gastric cancer | NFKBIB | "miR 20a enhances cisplatin resistance of human gas ......" | 26286834 |
| hsa-miR-20a-3p | acute myeloid leukemia | NPM1 | "High MN1 expression was associated with spleen inv ......" | 23515710 |
| hsa-miR-20a-3p | liver cancer | PIK3CD | "The phosphatidylinositol 3-kinase inhibitor LY2940 ......" | 26031366 |
| hsa-miR-20a-3p | liver cancer | PTEN | "The expression of miR-20a and PTEN were detected i ......" | 26031366 |
| hsa-miR-20a-3p | liver cancer | RUNX3 | "MicroRNA 20a 5p targets RUNX3 to regulate prolifer ......" | 27748919 |
| hsa-miR-20a-3p | liver cancer | SLU7 | "Interestingly altered splicing of miR-17-92 and do ......" | 26804174 |
| hsa-miR-20a-3p | colorectal cancer | SMAD4 | "MicroRNA 20a 5p promotes colorectal cancer invasio ......" | 27286257 |
| hsa-miR-20a-3p | gastric cancer | TIMM8A | "Conversely ectopic expression of miR-20a dramatica ......" | 26286834 |
| hsa-miR-20a-3p | cervical and endocervical cancer | TIMP2 | "TIMP2 and ATG7 were proved to be direct targets of ......" | 25803820 |
| hsa-miR-20a-3p | cervical and endocervical cancer | TNKS2 | "miR 20a promotes migration and invasion by regulat ......" | 22449978 |
| hsa-miR-20a-3p | breast cancer | TNMD | "Inhibitory effect of miR-20a and 20b on basal and ......" | 26829385 |
| hsa-miR-20a-3p | gastric cancer | UBE2C | "We found that both miR-17 and miR-20a miR-17/20a t ......" | 25760688 |
Expression profile in cancer corhorts:

Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-20a-3p | RASSF3 | 12 cancers: BLCA; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD | miRNAWalker2 validate | TCGA BLCA -0.347; TCGA COAD -0.059; TCGA ESCA -0.246; TCGA HNSC -0.28; TCGA KIRP -0.247; TCGA LUAD -0.121; TCGA LUSC -0.31; TCGA PAAD -0.359; TCGA PRAD -0.265; TCGA SARC -0.523; TCGA THCA -0.204; TCGA STAD -0.161 |
hsa-miR-20a-3p | LCA5 | 13 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.154; TCGA CESC -0.227; TCGA COAD -0.214; TCGA ESCA -0.12; TCGA HNSC -0.288; TCGA KIRP -0.153; TCGA LUAD -0.188; TCGA LUSC -0.194; TCGA PAAD -0.185; TCGA PRAD -0.315; TCGA SARC -0.098; TCGA STAD -0.216; TCGA UCEC -0.205 |
hsa-miR-20a-3p | MMP16 | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUSC; PAAD; PRAD; THCA; STAD | MirTarget | TCGA BLCA -0.297; TCGA BRCA -0.12; TCGA COAD -0.399; TCGA ESCA -0.351; TCGA HNSC -0.269; TCGA LUSC -0.174; TCGA PAAD -0.42; TCGA PRAD -0.431; TCGA THCA -0.27; TCGA STAD -0.355 |
hsa-miR-20a-3p | CNTNAP1 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; LGG; LUSC; PAAD; PRAD; SARC; STAD | MirTarget | TCGA BLCA -0.342; TCGA BRCA -0.1; TCGA CESC -0.192; TCGA COAD -0.49; TCGA ESCA -0.418; TCGA LGG -0.216; TCGA LUSC -0.143; TCGA PAAD -0.341; TCGA PRAD -0.331; TCGA SARC -0.165; TCGA STAD -0.368 |
hsa-miR-20a-3p | KAT2B | 15 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.103; TCGA CESC -0.285; TCGA COAD -0.233; TCGA ESCA -0.417; TCGA HNSC -0.329; TCGA KIRP -0.172; TCGA LGG -0.082; TCGA LIHC -0.224; TCGA LUAD -0.227; TCGA LUSC -0.401; TCGA PRAD -0.153; TCGA SARC -0.271; TCGA THCA -0.083; TCGA STAD -0.267; TCGA UCEC -0.198 |
hsa-miR-20a-3p | GPR34 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.314; TCGA BRCA -0.162; TCGA CESC -0.29; TCGA COAD -0.36; TCGA ESCA -0.34; TCGA HNSC -0.382; TCGA LIHC -0.168; TCGA LUAD -0.315; TCGA LUSC -0.622; TCGA PAAD -0.227; TCGA PRAD -0.273; TCGA SARC -0.186; TCGA THCA -0.232; TCGA STAD -0.192; TCGA UCEC -0.286 |
hsa-miR-20a-3p | NDFIP1 | 9 cancers: BLCA; BRCA; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; UCEC | MirTarget | TCGA BLCA -0.074; TCGA BRCA -0.058; TCGA HNSC -0.094; TCGA KIRP -0.083; TCGA LGG -0.152; TCGA LIHC -0.103; TCGA LUAD -0.071; TCGA LUSC -0.158; TCGA UCEC -0.053 |
hsa-miR-20a-3p | FGD5 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.319; TCGA BRCA -0.193; TCGA CESC -0.319; TCGA COAD -0.252; TCGA ESCA -0.236; TCGA HNSC -0.299; TCGA LIHC -0.128; TCGA LUAD -0.255; TCGA LUSC -0.532; TCGA PAAD -0.334; TCGA PRAD -0.243; TCGA THCA -0.16; TCGA STAD -0.141; TCGA UCEC -0.272 |
hsa-miR-20a-3p | AKAP11 | 10 cancers: BLCA; CESC; HNSC; LGG; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.107; TCGA CESC -0.097; TCGA HNSC -0.123; TCGA LGG -0.098; TCGA LUAD -0.075; TCGA LUSC -0.142; TCGA PAAD -0.118; TCGA PRAD -0.186; TCGA STAD -0.069; TCGA UCEC -0.116 |
hsa-miR-20a-3p | SETD7 | 15 cancers: BLCA; COAD; ESCA; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.173; TCGA COAD -0.077; TCGA ESCA -0.12; TCGA HNSC -0.146; TCGA KIRP -0.095; TCGA LGG -0.117; TCGA LIHC -0.143; TCGA LUAD -0.123; TCGA LUSC -0.155; TCGA PAAD -0.163; TCGA PRAD -0.081; TCGA SARC -0.151; TCGA THCA -0.105; TCGA STAD -0.197; TCGA UCEC -0.249 |
hsa-miR-20a-3p | WDR47 | 9 cancers: BLCA; COAD; ESCA; LUAD; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.054; TCGA COAD -0.101; TCGA ESCA -0.165; TCGA LUAD -0.077; TCGA PAAD -0.189; TCGA PRAD -0.174; TCGA SARC -0.097; TCGA STAD -0.144; TCGA UCEC -0.088 |
hsa-miR-20a-3p | OSTM1 | 14 cancers: BLCA; COAD; ESCA; HNSC; KIRP; LGG; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.165; TCGA COAD -0.077; TCGA ESCA -0.176; TCGA HNSC -0.066; TCGA KIRP -0.132; TCGA LGG -0.088; TCGA LUAD -0.122; TCGA LUSC -0.237; TCGA PAAD -0.127; TCGA PRAD -0.156; TCGA SARC -0.112; TCGA THCA -0.064; TCGA STAD -0.195; TCGA UCEC -0.2 |
hsa-miR-20a-3p | KLHDC1 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.231; TCGA BRCA -0.178; TCGA CESC -0.35; TCGA COAD -0.227; TCGA ESCA -0.442; TCGA HNSC -0.225; TCGA LGG -0.101; TCGA LIHC -0.152; TCGA LUAD -0.101; TCGA LUSC -0.376; TCGA PRAD -0.103; TCGA SARC -0.183; TCGA STAD -0.316; TCGA UCEC -0.258 |
hsa-miR-20a-3p | EEA1 | 11 cancers: BLCA; CESC; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.131; TCGA CESC -0.163; TCGA ESCA -0.127; TCGA HNSC -0.173; TCGA LUAD -0.138; TCGA LUSC -0.125; TCGA PAAD -0.177; TCGA PRAD -0.143; TCGA SARC -0.141; TCGA STAD -0.103; TCGA UCEC -0.192 |
hsa-miR-20a-3p | CPEB3 | 13 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.19; TCGA BRCA -0.058; TCGA CESC -0.27; TCGA ESCA -0.318; TCGA HNSC -0.23; TCGA KIRP -0.137; TCGA LGG -0.15; TCGA LIHC -0.208; TCGA LUAD -0.118; TCGA LUSC -0.254; TCGA PRAD -0.059; TCGA STAD -0.129; TCGA UCEC -0.145 |
hsa-miR-20a-3p | CDK14 | 10 cancers: BLCA; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; PRAD; STAD | MirTarget | TCGA BLCA -0.168; TCGA COAD -0.561; TCGA ESCA -0.265; TCGA HNSC -0.168; TCGA LGG -0.134; TCGA LIHC -0.089; TCGA LUAD -0.171; TCGA LUSC -0.267; TCGA PRAD -0.075; TCGA STAD -0.256 |
hsa-miR-20a-3p | ANGPTL1 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.904; TCGA BRCA -0.258; TCGA CESC -0.594; TCGA COAD -1.116; TCGA ESCA -1.233; TCGA HNSC -0.725; TCGA LGG -0.181; TCGA LIHC -0.356; TCGA LUAD -0.324; TCGA LUSC -0.864; TCGA PAAD -0.36; TCGA PRAD -0.582; TCGA SARC -0.305; TCGA STAD -0.896; TCGA UCEC -0.401 |
hsa-miR-20a-3p | NFIA | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LIHC; LUAD; LUSC; PRAD; STAD | MirTarget; miRNATAP | TCGA BLCA -0.271; TCGA BRCA -0.141; TCGA CESC -0.203; TCGA ESCA -0.247; TCGA HNSC -0.118; TCGA LIHC -0.156; TCGA LUAD -0.144; TCGA LUSC -0.17; TCGA PRAD -0.094; TCGA STAD -0.323 |
hsa-miR-20a-3p | STAM2 | 11 cancers: BLCA; CESC; HNSC; KIRP; LGG; LIHC; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.074; TCGA CESC -0.105; TCGA HNSC -0.106; TCGA KIRP -0.087; TCGA LGG -0.073; TCGA LIHC -0.052; TCGA LUSC -0.122; TCGA PAAD -0.097; TCGA PRAD -0.082; TCGA STAD -0.053; TCGA UCEC -0.131 |
hsa-miR-20a-3p | RBMS3 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; STAD | MirTarget; mirMAP | TCGA BLCA -0.469; TCGA BRCA -0.122; TCGA CESC -0.208; TCGA COAD -0.492; TCGA ESCA -0.544; TCGA HNSC -0.413; TCGA LIHC -0.136; TCGA LUAD -0.381; TCGA LUSC -0.5; TCGA PAAD -0.475; TCGA PRAD -0.573; TCGA STAD -0.484 |
hsa-miR-20a-3p | CADPS2 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; PRAD; SARC; UCEC | MirTarget | TCGA BLCA -0.194; TCGA BRCA -0.138; TCGA CESC -0.171; TCGA COAD -0.134; TCGA ESCA -0.27; TCGA HNSC -0.269; TCGA KIRP -0.222; TCGA LGG -0.424; TCGA LIHC -0.151; TCGA LUAD -0.143; TCGA LUSC -0.407; TCGA PRAD -0.063; TCGA SARC -0.332; TCGA UCEC -0.228 |
hsa-miR-20a-3p | TCF21 | 10 cancers: BLCA; CESC; COAD; ESCA; LIHC; LUAD; LUSC; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.67; TCGA CESC -0.455; TCGA COAD -0.344; TCGA ESCA -0.663; TCGA LIHC -0.206; TCGA LUAD -0.411; TCGA LUSC -1.004; TCGA PRAD -0.266; TCGA STAD -0.258; TCGA UCEC -0.443 |
hsa-miR-20a-3p | SLC9A9 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.47; TCGA BRCA -0.06; TCGA CESC -0.461; TCGA COAD -0.525; TCGA ESCA -0.568; TCGA HNSC -0.231; TCGA LIHC -0.127; TCGA LUAD -0.195; TCGA LUSC -0.214; TCGA PAAD -0.218; TCGA PRAD -0.233; TCGA SARC -0.152; TCGA THCA -0.166; TCGA STAD -0.372; TCGA UCEC -0.31 |
hsa-miR-20a-3p | MAP1B | 11 cancers: BLCA; COAD; ESCA; HNSC; LGG; LIHC; PAAD; PRAD; SARC; THCA; STAD | MirTarget; miRNATAP | TCGA BLCA -0.505; TCGA COAD -0.442; TCGA ESCA -0.42; TCGA HNSC -0.278; TCGA LGG -0.17; TCGA LIHC -0.276; TCGA PAAD -0.36; TCGA PRAD -0.439; TCGA SARC -0.296; TCGA THCA -0.246; TCGA STAD -0.532 |
hsa-miR-20a-3p | CAPN2 | 14 cancers: BLCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.116; TCGA CESC -0.14; TCGA COAD -0.108; TCGA ESCA -0.114; TCGA HNSC -0.081; TCGA LGG -0.098; TCGA LIHC -0.13; TCGA LUAD -0.127; TCGA LUSC -0.245; TCGA PRAD -0.065; TCGA SARC -0.137; TCGA THCA -0.056; TCGA STAD -0.075; TCGA UCEC -0.151 |
hsa-miR-20a-3p | NPTN | 14 cancers: BLCA; COAD; ESCA; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.186; TCGA COAD -0.123; TCGA ESCA -0.097; TCGA HNSC -0.054; TCGA KIRP -0.091; TCGA LGG -0.167; TCGA LIHC -0.072; TCGA LUAD -0.067; TCGA LUSC -0.072; TCGA PAAD -0.149; TCGA PRAD -0.206; TCGA SARC -0.185; TCGA STAD -0.104; TCGA UCEC -0.114 |
hsa-miR-20a-3p | GPC6 | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; LGG; LUSC; PAAD; PRAD; STAD | MirTarget | TCGA BLCA -0.595; TCGA CESC -0.231; TCGA COAD -0.517; TCGA ESCA -0.463; TCGA HNSC -0.406; TCGA LGG -0.15; TCGA LUSC -0.314; TCGA PAAD -0.212; TCGA PRAD -0.169; TCGA STAD -0.215 |
hsa-miR-20a-3p | CPE | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LGG; LUSC; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.269; TCGA BRCA -0.211; TCGA COAD -0.281; TCGA ESCA -0.443; TCGA HNSC -0.203; TCGA LGG -0.332; TCGA LUSC -0.203; TCGA PRAD -0.107; TCGA SARC -0.179; TCGA STAD -0.478; TCGA UCEC -0.227 |
hsa-miR-20a-3p | SLC41A2 | 11 cancers: BLCA; COAD; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; SARC | MirTarget | TCGA BLCA -0.198; TCGA COAD -0.232; TCGA HNSC -0.291; TCGA KIRP -0.144; TCGA LGG -0.232; TCGA LIHC -0.186; TCGA LUAD -0.137; TCGA LUSC -0.362; TCGA PAAD -0.27; TCGA PRAD -0.298; TCGA SARC -0.106 |
hsa-miR-20a-3p | ANKRD50 | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.133; TCGA CESC -0.168; TCGA COAD -0.167; TCGA ESCA -0.115; TCGA HNSC -0.133; TCGA LIHC -0.142; TCGA LUAD -0.107; TCGA LUSC -0.127; TCGA PAAD -0.295; TCGA PRAD -0.09; TCGA STAD -0.158; TCGA UCEC -0.082 |
hsa-miR-20a-3p | FAT3 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LUAD; LUSC; PAAD; PRAD; SARC; STAD | MirTarget | TCGA BLCA -0.463; TCGA BRCA -0.206; TCGA CESC -0.352; TCGA COAD -0.446; TCGA ESCA -0.679; TCGA HNSC -0.324; TCGA LGG -0.279; TCGA LUAD -0.262; TCGA LUSC -0.416; TCGA PAAD -0.418; TCGA PRAD -0.715; TCGA SARC -0.314; TCGA STAD -0.783 |
hsa-miR-20a-3p | LPAR1 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUSC; PAAD; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.336; TCGA BRCA -0.13; TCGA CESC -0.285; TCGA COAD -0.255; TCGA ESCA -0.294; TCGA HNSC -0.254; TCGA LUSC -0.106; TCGA PAAD -0.179; TCGA PRAD -0.372; TCGA THCA -0.124; TCGA STAD -0.292; TCGA UCEC -0.118 |
hsa-miR-20a-3p | TLN1 | 12 cancers: BLCA; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.202; TCGA COAD -0.155; TCGA ESCA -0.272; TCGA HNSC -0.189; TCGA LIHC -0.061; TCGA LUAD -0.101; TCGA LUSC -0.21; TCGA PAAD -0.165; TCGA PRAD -0.238; TCGA SARC -0.211; TCGA STAD -0.253; TCGA UCEC -0.076 |
hsa-miR-20a-3p | SSPN | 13 cancers: BLCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.376; TCGA CESC -0.397; TCGA COAD -0.492; TCGA ESCA -0.491; TCGA HNSC -0.394; TCGA LGG -0.221; TCGA LIHC -0.284; TCGA LUAD -0.349; TCGA LUSC -0.364; TCGA PAAD -0.268; TCGA PRAD -0.369; TCGA STAD -0.454; TCGA UCEC -0.218 |
hsa-miR-20a-3p | NAV3 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; LGG; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.501; TCGA BRCA -0.215; TCGA CESC -0.318; TCGA COAD -0.571; TCGA ESCA -0.427; TCGA LGG -0.311; TCGA LUAD -0.257; TCGA LUSC -0.446; TCGA PAAD -0.224; TCGA PRAD -0.214; TCGA STAD -0.499; TCGA UCEC -0.338 |
hsa-miR-20a-3p | ITM2A | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.433; TCGA BRCA -0.156; TCGA CESC -0.483; TCGA COAD -0.483; TCGA ESCA -0.628; TCGA HNSC -0.453; TCGA LUAD -0.181; TCGA LUSC -0.598; TCGA PRAD -0.281; TCGA STAD -0.284; TCGA UCEC -0.52 |
hsa-miR-20a-3p | EBF1 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.48; TCGA BRCA -0.172; TCGA CESC -0.4; TCGA COAD -0.382; TCGA ESCA -0.469; TCGA HNSC -0.292; TCGA LIHC -0.205; TCGA LUAD -0.184; TCGA LUSC -0.474; TCGA PAAD -0.197; TCGA PRAD -0.239; TCGA THCA -0.165; TCGA STAD -0.369; TCGA UCEC -0.234 |
hsa-miR-20a-3p | FOXP2 | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; PAAD; PRAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.683; TCGA CESC -0.404; TCGA COAD -0.568; TCGA ESCA -0.747; TCGA HNSC -0.404; TCGA PAAD -0.403; TCGA PRAD -0.673; TCGA STAD -0.754; TCGA UCEC -0.442 |
hsa-miR-20a-3p | MPDZ | 11 cancers: BLCA; COAD; ESCA; HNSC; LIHC; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.192; TCGA COAD -0.536; TCGA ESCA -0.469; TCGA HNSC -0.27; TCGA LIHC -0.15; TCGA LUSC -0.204; TCGA PAAD -0.41; TCGA PRAD -0.191; TCGA SARC -0.096; TCGA STAD -0.494; TCGA UCEC -0.088 |
hsa-miR-20a-3p | SGTB | 12 cancers: BLCA; COAD; ESCA; KIRP; LGG; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD | MirTarget | TCGA BLCA -0.11; TCGA COAD -0.127; TCGA ESCA -0.149; TCGA KIRP -0.08; TCGA LGG -0.205; TCGA LUAD -0.072; TCGA LUSC -0.156; TCGA PAAD -0.127; TCGA PRAD -0.131; TCGA SARC -0.099; TCGA THCA -0.078; TCGA STAD -0.201 |
hsa-miR-20a-3p | F2R | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; THCA; STAD | MirTarget | TCGA BLCA -0.098; TCGA BRCA -0.175; TCGA CESC -0.137; TCGA COAD -0.288; TCGA ESCA -0.168; TCGA HNSC -0.18; TCGA LIHC -0.128; TCGA LUAD -0.118; TCGA LUSC -0.203; TCGA PAAD -0.276; TCGA THCA -0.305; TCGA STAD -0.099 |
hsa-miR-20a-3p | HIPK1 | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.069; TCGA CESC -0.09; TCGA COAD -0.055; TCGA ESCA -0.146; TCGA HNSC -0.161; TCGA LUAD -0.118; TCGA LUSC -0.146; TCGA PAAD -0.104; TCGA PRAD -0.144; TCGA SARC -0.063; TCGA STAD -0.068; TCGA UCEC -0.08 |
hsa-miR-20a-3p | CHRDL1 | 13 cancers: BLCA; CESC; COAD; ESCA; HNSC; LGG; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.915; TCGA CESC -0.656; TCGA COAD -1.064; TCGA ESCA -1.656; TCGA HNSC -0.902; TCGA LGG -0.429; TCGA LUAD -0.371; TCGA LUSC -0.915; TCGA PAAD -0.646; TCGA PRAD -0.636; TCGA SARC -0.603; TCGA STAD -0.995; TCGA UCEC -0.6 |
hsa-miR-20a-3p | KLF9 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.474; TCGA BRCA -0.111; TCGA CESC -0.256; TCGA COAD -0.334; TCGA ESCA -0.566; TCGA HNSC -0.248; TCGA LGG -0.238; TCGA LIHC -0.25; TCGA LUAD -0.194; TCGA LUSC -0.455; TCGA PAAD -0.181; TCGA PRAD -0.12; TCGA SARC -0.222; TCGA STAD -0.411; TCGA UCEC -0.297 |
hsa-miR-20a-3p | IGSF10 | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; STAD | MirTarget | TCGA BLCA -0.613; TCGA CESC -0.419; TCGA COAD -0.242; TCGA ESCA -0.452; TCGA HNSC -0.66; TCGA LIHC -0.285; TCGA LUAD -0.28; TCGA LUSC -0.418; TCGA PAAD -0.275; TCGA STAD -0.471 |
hsa-miR-20a-3p | AKAP6 | 10 cancers: BLCA; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PRAD; SARC; STAD | MirTarget | TCGA BLCA -0.592; TCGA COAD -0.303; TCGA ESCA -0.661; TCGA HNSC -0.409; TCGA LIHC -0.295; TCGA LUAD -0.132; TCGA LUSC -0.311; TCGA PRAD -0.285; TCGA SARC -0.398; TCGA STAD -0.676 |
hsa-miR-20a-3p | EPDR1 | 13 cancers: BLCA; CESC; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.432; TCGA CESC -0.27; TCGA HNSC -0.381; TCGA KIRP -0.114; TCGA LGG -0.238; TCGA LIHC -0.325; TCGA LUAD -0.196; TCGA LUSC -0.412; TCGA PAAD -0.245; TCGA PRAD -0.197; TCGA THCA -0.163; TCGA STAD -0.188; TCGA UCEC -0.229 |
hsa-miR-20a-3p | PPP1R12A | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.197; TCGA CESC -0.088; TCGA COAD -0.095; TCGA ESCA -0.177; TCGA HNSC -0.089; TCGA PAAD -0.14; TCGA PRAD -0.222; TCGA SARC -0.216; TCGA STAD -0.239; TCGA UCEC -0.129 |
hsa-miR-20a-3p | KCND2 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.173; TCGA BRCA -0.116; TCGA CESC -0.231; TCGA COAD -0.711; TCGA ESCA -0.321; TCGA HNSC -0.307; TCGA LIHC -0.24; TCGA PAAD -0.539; TCGA PRAD -0.222; TCGA STAD -0.205; TCGA UCEC -0.403 |
hsa-miR-20a-3p | ENDOD1 | 13 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LGG; LIHC; LUAD; PAAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.115; TCGA CESC -0.096; TCGA COAD -0.104; TCGA ESCA -0.237; TCGA HNSC -0.102; TCGA KIRP -0.122; TCGA LGG -0.13; TCGA LIHC -0.276; TCGA LUAD -0.259; TCGA PAAD -0.195; TCGA SARC -0.244; TCGA STAD -0.171; TCGA UCEC -0.138 |
hsa-miR-20a-3p | IGF1 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; THCA; STAD | mirMAP | TCGA BLCA -0.81; TCGA BRCA -0.257; TCGA CESC -0.766; TCGA COAD -0.828; TCGA ESCA -0.985; TCGA HNSC -0.574; TCGA LIHC -0.362; TCGA LUAD -0.272; TCGA LUSC -0.61; TCGA PAAD -0.554; TCGA PRAD -0.198; TCGA THCA -0.276; TCGA STAD -0.538 |
hsa-miR-20a-3p | SAMD4A | 12 cancers: BLCA; COAD; ESCA; HNSC; LGG; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.387; TCGA COAD -0.318; TCGA ESCA -0.357; TCGA HNSC -0.099; TCGA LGG -0.097; TCGA LUAD -0.204; TCGA LUSC -0.49; TCGA PAAD -0.15; TCGA PRAD -0.245; TCGA SARC -0.148; TCGA STAD -0.36; TCGA UCEC -0.245 |
hsa-miR-20a-3p | PREX2 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LUAD; LUSC; PAAD; PRAD; STAD | mirMAP | TCGA BLCA -0.462; TCGA BRCA -0.22; TCGA CESC -0.292; TCGA COAD -0.559; TCGA ESCA -0.689; TCGA HNSC -0.481; TCGA LGG -0.43; TCGA LUAD -0.33; TCGA LUSC -0.798; TCGA PAAD -0.543; TCGA PRAD -0.548; TCGA STAD -0.332 |
hsa-miR-20a-3p | MPPED2 | 9 cancers: BLCA; CESC; COAD; HNSC; LIHC; LUAD; PAAD; PRAD; STAD | mirMAP | TCGA BLCA -0.226; TCGA CESC -0.635; TCGA COAD -0.45; TCGA HNSC -0.384; TCGA LIHC -0.367; TCGA LUAD -0.128; TCGA PAAD -0.251; TCGA PRAD -0.246; TCGA STAD -0.478 |
hsa-miR-20a-3p | MYO9A | 12 cancers: BLCA; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD | mirMAP | TCGA BLCA -0.168; TCGA COAD -0.111; TCGA ESCA -0.255; TCGA HNSC -0.232; TCGA LGG -0.111; TCGA LIHC -0.225; TCGA LUAD -0.175; TCGA LUSC -0.161; TCGA PAAD -0.364; TCGA PRAD -0.333; TCGA SARC -0.183; TCGA STAD -0.254 |
hsa-miR-20a-3p | RORA | 13 cancers: BLCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.23; TCGA CESC -0.273; TCGA COAD -0.307; TCGA ESCA -0.493; TCGA HNSC -0.315; TCGA LGG -0.215; TCGA LIHC -0.166; TCGA LUAD -0.191; TCGA LUSC -0.239; TCGA PAAD -0.158; TCGA PRAD -0.189; TCGA STAD -0.298; TCGA UCEC -0.227 |
hsa-miR-20a-3p | FSTL3 | 9 cancers: BLCA; BRCA; COAD; ESCA; LIHC; LUAD; LUSC; STAD; UCEC | mirMAP | TCGA BLCA -0.218; TCGA BRCA -0.293; TCGA COAD -0.352; TCGA ESCA -0.318; TCGA LIHC -0.186; TCGA LUAD -0.154; TCGA LUSC -0.37; TCGA STAD -0.168; TCGA UCEC -0.126 |
hsa-miR-20a-3p | SPEG | 9 cancers: BLCA; COAD; ESCA; LGG; LIHC; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.652; TCGA COAD -0.646; TCGA ESCA -0.52; TCGA LGG -0.118; TCGA LIHC -0.32; TCGA PRAD -0.357; TCGA SARC -0.488; TCGA STAD -0.719; TCGA UCEC -0.276 |
hsa-miR-20a-3p | MRVI1 | 16 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.655; TCGA BRCA -0.155; TCGA CESC -0.427; TCGA COAD -0.487; TCGA ESCA -0.844; TCGA HNSC -0.26; TCGA LGG -0.342; TCGA LIHC -0.306; TCGA LUAD -0.298; TCGA LUSC -0.501; TCGA PAAD -0.462; TCGA PRAD -0.454; TCGA SARC -0.774; TCGA THCA -0.167; TCGA STAD -0.586; TCGA UCEC -0.46 |
hsa-miR-20a-3p | MGLL | 13 cancers: BLCA; BRCA; ESCA; HNSC; LGG; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.178; TCGA BRCA -0.152; TCGA ESCA -0.313; TCGA HNSC -0.308; TCGA LGG -0.22; TCGA LUAD -0.287; TCGA LUSC -0.537; TCGA PAAD -0.171; TCGA PRAD -0.169; TCGA SARC -0.233; TCGA THCA -0.187; TCGA STAD -0.177; TCGA UCEC -0.183 |
hsa-miR-20a-3p | RASAL2 | 9 cancers: BLCA; CESC; HNSC; KIRP; LUAD; LUSC; PAAD; PRAD; THCA | mirMAP | TCGA BLCA -0.069; TCGA CESC -0.112; TCGA HNSC -0.144; TCGA KIRP -0.084; TCGA LUAD -0.12; TCGA LUSC -0.144; TCGA PAAD -0.154; TCGA PRAD -0.128; TCGA THCA -0.105 |
hsa-miR-20a-3p | TNS1 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.757; TCGA BRCA -0.132; TCGA CESC -0.254; TCGA COAD -0.592; TCGA ESCA -0.779; TCGA HNSC -0.453; TCGA LGG -0.075; TCGA LUAD -0.265; TCGA LUSC -0.546; TCGA PAAD -0.304; TCGA PRAD -0.512; TCGA SARC -0.523; TCGA STAD -0.609; TCGA UCEC -0.269 |
hsa-miR-20a-3p | IRAK3 | 13 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.293; TCGA CESC -0.261; TCGA COAD -0.339; TCGA ESCA -0.411; TCGA HNSC -0.222; TCGA KIRP -0.205; TCGA LIHC -0.175; TCGA LUAD -0.187; TCGA LUSC -0.478; TCGA PRAD -0.38; TCGA THCA -0.172; TCGA STAD -0.116; TCGA UCEC -0.245 |
hsa-miR-20a-3p | PTGS1 | 11 cancers: BLCA; BRCA; COAD; ESCA; KIRP; LUAD; LUSC; PAAD; PRAD; THCA; STAD | mirMAP | TCGA BLCA -0.805; TCGA BRCA -0.092; TCGA COAD -0.372; TCGA ESCA -0.51; TCGA KIRP -0.169; TCGA LUAD -0.251; TCGA LUSC -0.387; TCGA PAAD -0.208; TCGA PRAD -0.498; TCGA THCA -0.215; TCGA STAD -0.26 |
hsa-miR-20a-3p | PCSK5 | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; PAAD; PRAD | mirMAP | TCGA BLCA -0.413; TCGA BRCA -0.187; TCGA COAD -0.395; TCGA ESCA -0.273; TCGA HNSC -0.284; TCGA LGG -0.33; TCGA LIHC -0.284; TCGA LUAD -0.296; TCGA LUSC -0.253; TCGA PAAD -0.413; TCGA PRAD -0.309 |
hsa-miR-20a-3p | ABLIM1 | 9 cancers: BLCA; CESC; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; PRAD | mirMAP | TCGA BLCA -0.121; TCGA CESC -0.22; TCGA ESCA -0.188; TCGA HNSC -0.14; TCGA LGG -0.101; TCGA LIHC -0.152; TCGA LUAD -0.085; TCGA LUSC -0.199; TCGA PRAD -0.124 |
hsa-miR-20a-3p | RASSF2 | 14 cancers: BLCA; CESC; COAD; ESCA; HNSC; LGG; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.18; TCGA CESC -0.322; TCGA COAD -0.428; TCGA ESCA -0.407; TCGA HNSC -0.279; TCGA LGG -0.155; TCGA LUAD -0.201; TCGA LUSC -0.511; TCGA PAAD -0.267; TCGA PRAD -0.156; TCGA SARC -0.13; TCGA THCA -0.211; TCGA STAD -0.27; TCGA UCEC -0.239 |
hsa-miR-20a-3p | STS | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; UCEC | mirMAP | TCGA BLCA -0.197; TCGA BRCA -0.104; TCGA CESC -0.238; TCGA COAD -0.2; TCGA ESCA -0.278; TCGA HNSC -0.246; TCGA LGG -0.252; TCGA LIHC -0.248; TCGA LUAD -0.242; TCGA LUSC -0.418; TCGA PAAD -0.219; TCGA PRAD -0.165; TCGA SARC -0.134; TCGA THCA -0.082; TCGA UCEC -0.206 |
hsa-miR-20a-3p | PRX | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUAD; LUSC; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.105; TCGA BRCA -0.235; TCGA COAD -0.206; TCGA ESCA -0.261; TCGA HNSC -0.094; TCGA LUAD -0.126; TCGA LUSC -0.531; TCGA PRAD -0.066; TCGA STAD -0.205; TCGA UCEC -0.198 |
hsa-miR-20a-3p | AGFG2 | 10 cancers: BLCA; BRCA; CESC; HNSC; LGG; LIHC; LUAD; LUSC; SARC; UCEC | mirMAP | TCGA BLCA -0.129; TCGA BRCA -0.123; TCGA CESC -0.175; TCGA HNSC -0.196; TCGA LGG -0.22; TCGA LIHC -0.123; TCGA LUAD -0.105; TCGA LUSC -0.16; TCGA SARC -0.085; TCGA UCEC -0.155 |
hsa-miR-20a-3p | ASPA | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.593; TCGA BRCA -0.243; TCGA CESC -0.553; TCGA COAD -0.603; TCGA ESCA -0.983; TCGA LUAD -0.327; TCGA LUSC -0.813; TCGA PAAD -0.303; TCGA PRAD -0.628; TCGA SARC -0.646; TCGA STAD -0.47; TCGA UCEC -0.452 |
hsa-miR-20a-3p | MDGA1 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; LGG; LIHC; LUAD; PAAD; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.311; TCGA BRCA -0.224; TCGA CESC -0.212; TCGA COAD -0.412; TCGA ESCA -0.43; TCGA LGG -0.199; TCGA LIHC -0.547; TCGA LUAD -0.263; TCGA PAAD -0.274; TCGA PRAD -0.196; TCGA THCA -0.146; TCGA STAD -0.262; TCGA UCEC -0.176 |
hsa-miR-20a-3p | RBM24 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.679; TCGA BRCA -0.282; TCGA CESC -0.351; TCGA COAD -0.256; TCGA ESCA -0.599; TCGA HNSC -0.308; TCGA LIHC -0.249; TCGA LUSC -0.313; TCGA PAAD -0.456; TCGA PRAD -0.261; TCGA SARC -0.419; TCGA STAD -0.628; TCGA UCEC -0.43 |
hsa-miR-20a-3p | BVES | 10 cancers: BLCA; COAD; ESCA; HNSC; LIHC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.346; TCGA COAD -0.519; TCGA ESCA -0.628; TCGA HNSC -0.212; TCGA LIHC -0.135; TCGA PAAD -0.324; TCGA PRAD -0.309; TCGA SARC -0.335; TCGA STAD -0.652; TCGA UCEC -0.151 |
hsa-miR-20a-3p | QKI | 11 cancers: BLCA; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.052; TCGA COAD -0.347; TCGA ESCA -0.24; TCGA HNSC -0.105; TCGA KIRP -0.071; TCGA LUAD -0.126; TCGA LUSC -0.181; TCGA PAAD -0.165; TCGA PRAD -0.147; TCGA STAD -0.267; TCGA UCEC -0.107 |
hsa-miR-20a-3p | CDH6 | 11 cancers: BLCA; BRCA; COAD; HNSC; LGG; LUAD; LUSC; PAAD; PRAD; THCA; STAD | mirMAP | TCGA BLCA -0.19; TCGA BRCA -0.121; TCGA COAD -0.211; TCGA HNSC -0.166; TCGA LGG -0.153; TCGA LUAD -0.295; TCGA LUSC -0.627; TCGA PAAD -0.28; TCGA PRAD -0.342; TCGA THCA -0.439; TCGA STAD -0.181 |
hsa-miR-20a-3p | ZBTB47 | 15 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.201; TCGA BRCA -0.131; TCGA COAD -0.28; TCGA ESCA -0.367; TCGA HNSC -0.204; TCGA KIRC -0.132; TCGA LGG -0.066; TCGA LIHC -0.131; TCGA LUAD -0.163; TCGA LUSC -0.26; TCGA PAAD -0.119; TCGA PRAD -0.104; TCGA SARC -0.22; TCGA STAD -0.294; TCGA UCEC -0.096 |
hsa-miR-20a-3p | FHL2 | 10 cancers: BLCA; BRCA; COAD; LGG; LIHC; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.143; TCGA BRCA -0.095; TCGA COAD -0.108; TCGA LGG -0.17; TCGA LIHC -0.428; TCGA PRAD -0.33; TCGA SARC -0.236; TCGA THCA -0.158; TCGA STAD -0.124; TCGA UCEC -0.12 |
hsa-miR-20a-3p | C1orf21 | 13 cancers: BLCA; BRCA; CESC; COAD; HNSC; LGG; LIHC; LUAD; LUSC; PAAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.217; TCGA BRCA -0.223; TCGA CESC -0.187; TCGA COAD -0.138; TCGA HNSC -0.063; TCGA LGG -0.104; TCGA LIHC -0.256; TCGA LUAD -0.166; TCGA LUSC -0.083; TCGA PAAD -0.188; TCGA SARC -0.207; TCGA STAD -0.194; TCGA UCEC -0.205 |
hsa-miR-20a-3p | SGIP1 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRP; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.269; TCGA BRCA -0.075; TCGA CESC -0.315; TCGA COAD -0.342; TCGA ESCA -0.469; TCGA KIRP -0.266; TCGA LUAD -0.198; TCGA LUSC -0.462; TCGA PAAD -0.38; TCGA PRAD -0.121; TCGA SARC -0.458; TCGA STAD -0.325; TCGA UCEC -0.278 |
hsa-miR-20a-3p | CHST3 | 11 cancers: BLCA; COAD; ESCA; HNSC; LIHC; LUAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.173; TCGA COAD -0.439; TCGA ESCA -0.222; TCGA HNSC -0.101; TCGA LIHC -0.143; TCGA LUAD -0.205; TCGA PRAD -0.362; TCGA SARC -0.094; TCGA THCA -0.121; TCGA STAD -0.265; TCGA UCEC -0.105 |
hsa-miR-20a-3p | KCNJ2 | 9 cancers: BLCA; CESC; HNSC; KIRP; LUAD; LUSC; PRAD; THCA; UCEC | mirMAP | TCGA BLCA -0.12; TCGA CESC -0.181; TCGA HNSC -0.321; TCGA KIRP -0.144; TCGA LUAD -0.208; TCGA LUSC -0.127; TCGA PRAD -0.164; TCGA THCA -0.256; TCGA UCEC -0.109 |
hsa-miR-20a-3p | ZNF831 | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; LGG; LUAD; LUSC; PRAD; UCEC | mirMAP | TCGA BLCA -0.362; TCGA CESC -0.56; TCGA COAD -0.48; TCGA ESCA -0.562; TCGA HNSC -0.404; TCGA LGG -0.277; TCGA LUAD -0.196; TCGA LUSC -0.529; TCGA PRAD -0.165; TCGA UCEC -0.219 |
hsa-miR-20a-3p | TMX4 | 10 cancers: BLCA; COAD; HNSC; KIRP; LGG; LUSC; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.137; TCGA COAD -0.106; TCGA HNSC -0.166; TCGA KIRP -0.207; TCGA LGG -0.124; TCGA LUSC -0.14; TCGA PRAD -0.13; TCGA SARC -0.132; TCGA STAD -0.101; TCGA UCEC -0.106 |
hsa-miR-20a-3p | TMOD2 | 14 cancers: BLCA; COAD; ESCA; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.118; TCGA COAD -0.126; TCGA ESCA -0.231; TCGA HNSC -0.193; TCGA KIRP -0.181; TCGA LGG -0.128; TCGA LIHC -0.205; TCGA LUAD -0.222; TCGA LUSC -0.152; TCGA PAAD -0.192; TCGA PRAD -0.238; TCGA THCA -0.176; TCGA STAD -0.275; TCGA UCEC -0.231 |
hsa-miR-20a-3p | PER2 | 9 cancers: BLCA; CESC; ESCA; HNSC; LGG; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.201; TCGA CESC -0.21; TCGA ESCA -0.196; TCGA HNSC -0.151; TCGA LGG -0.114; TCGA PRAD -0.168; TCGA SARC -0.122; TCGA STAD -0.059; TCGA UCEC -0.055 |
hsa-miR-20a-3p | SYT11 | 15 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.358; TCGA CESC -0.159; TCGA COAD -0.395; TCGA ESCA -0.383; TCGA HNSC -0.257; TCGA KIRP -0.218; TCGA LGG -0.113; TCGA LIHC -0.095; TCGA LUAD -0.095; TCGA LUSC -0.303; TCGA PAAD -0.244; TCGA PRAD -0.332; TCGA SARC -0.197; TCGA THCA -0.151; TCGA STAD -0.326 |
hsa-miR-20a-3p | DCLK1 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.451; TCGA BRCA -0.178; TCGA CESC -0.289; TCGA COAD -0.771; TCGA ESCA -0.568; TCGA HNSC -0.557; TCGA LGG -0.227; TCGA LIHC -0.246; TCGA PRAD -0.358; TCGA SARC -0.157; TCGA THCA -0.344; TCGA STAD -0.557; TCGA UCEC -0.25 |
hsa-miR-20a-3p | IL6ST | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LGG; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.43; TCGA BRCA -0.16; TCGA CESC -0.274; TCGA COAD -0.273; TCGA ESCA -0.399; TCGA HNSC -0.571; TCGA KIRP -0.274; TCGA LGG -0.164; TCGA LUAD -0.25; TCGA LUSC -0.511; TCGA PAAD -0.382; TCGA PRAD -0.638; TCGA SARC -0.286; TCGA STAD -0.264; TCGA UCEC -0.174 |
hsa-miR-20a-3p | KIAA1614 | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LGG; LUAD; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.293; TCGA BRCA -0.084; TCGA COAD -0.247; TCGA ESCA -0.226; TCGA HNSC -0.123; TCGA LGG -0.19; TCGA LUAD -0.099; TCGA PAAD -0.239; TCGA PRAD -0.421; TCGA SARC -0.155; TCGA STAD -0.293; TCGA UCEC -0.097 |
hsa-miR-20a-3p | FAM129A | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.58; TCGA BRCA -0.153; TCGA CESC -0.469; TCGA COAD -0.558; TCGA ESCA -0.66; TCGA HNSC -0.257; TCGA LIHC -0.237; TCGA LUAD -0.194; TCGA LUSC -0.191; TCGA PAAD -0.196; TCGA PRAD -0.244; TCGA SARC -0.495; TCGA THCA -0.166; TCGA STAD -0.608; TCGA UCEC -0.405 |
hsa-miR-20a-3p | TMEM127 | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LGG; LIHC; LUAD; PAAD; PRAD; SARC | mirMAP | TCGA BLCA -0.073; TCGA CESC -0.051; TCGA COAD -0.069; TCGA ESCA -0.076; TCGA HNSC -0.081; TCGA KIRP -0.074; TCGA LGG -0.062; TCGA LIHC -0.089; TCGA LUAD -0.075; TCGA PAAD -0.074; TCGA PRAD -0.098; TCGA SARC -0.052 |
hsa-miR-20a-3p | ENTPD1 | 14 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.261; TCGA CESC -0.152; TCGA COAD -0.217; TCGA ESCA -0.24; TCGA HNSC -0.072; TCGA KIRP -0.072; TCGA LIHC -0.08; TCGA LUAD -0.14; TCGA LUSC -0.221; TCGA PAAD -0.227; TCGA PRAD -0.119; TCGA SARC -0.26; TCGA STAD -0.203; TCGA UCEC -0.152 |
hsa-miR-20a-3p | FGF2 | 14 cancers: BLCA; COAD; ESCA; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.521; TCGA COAD -0.565; TCGA ESCA -0.636; TCGA HNSC -0.255; TCGA KIRP -0.186; TCGA LGG -0.145; TCGA LIHC -0.492; TCGA LUAD -0.234; TCGA LUSC -0.338; TCGA PAAD -0.428; TCGA PRAD -0.427; TCGA SARC -0.16; TCGA STAD -0.438; TCGA UCEC -0.455 |
hsa-miR-20a-3p | KIAA1644 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUSC; PAAD; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.79; TCGA BRCA -0.125; TCGA CESC -0.262; TCGA COAD -0.675; TCGA ESCA -0.558; TCGA HNSC -0.168; TCGA LIHC -0.329; TCGA LUSC -0.248; TCGA PAAD -0.46; TCGA PRAD -0.542; TCGA THCA -0.339; TCGA STAD -0.55; TCGA UCEC -0.494 |
hsa-miR-20a-3p | GLIPR1 | 15 cancers: BLCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.303; TCGA CESC -0.178; TCGA COAD -0.269; TCGA ESCA -0.237; TCGA HNSC -0.121; TCGA LGG -0.162; TCGA LIHC -0.248; TCGA LUAD -0.193; TCGA LUSC -0.362; TCGA PAAD -0.3; TCGA PRAD -0.254; TCGA SARC -0.177; TCGA THCA -0.131; TCGA STAD -0.203; TCGA UCEC -0.331 |
hsa-miR-20a-3p | JDP2 | 10 cancers: BLCA; BRCA; COAD; HNSC; LIHC; LUAD; LUSC; PRAD; THCA; STAD | mirMAP | TCGA BLCA -0.175; TCGA BRCA -0.166; TCGA COAD -0.099; TCGA HNSC -0.154; TCGA LIHC -0.223; TCGA LUAD -0.078; TCGA LUSC -0.255; TCGA PRAD -0.214; TCGA THCA -0.107; TCGA STAD -0.115 |
hsa-miR-20a-3p | NTRK3 | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; LGG; LUAD; LUSC; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.605; TCGA CESC -0.526; TCGA COAD -0.619; TCGA ESCA -0.977; TCGA HNSC -0.472; TCGA LGG -0.099; TCGA LUAD -0.399; TCGA LUSC -0.606; TCGA PRAD -0.399; TCGA SARC -0.349; TCGA STAD -0.618; TCGA UCEC -0.318 |
hsa-miR-20a-3p | MYOCD | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.952; TCGA BRCA -0.218; TCGA CESC -0.454; TCGA COAD -0.681; TCGA ESCA -1.288; TCGA LIHC -0.284; TCGA LUAD -0.594; TCGA LUSC -0.976; TCGA PAAD -0.741; TCGA PRAD -0.762; TCGA SARC -0.985; TCGA STAD -0.798; TCGA UCEC -0.741 |
hsa-miR-20a-3p | GULP1 | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUSC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.335; TCGA CESC -0.275; TCGA COAD -0.439; TCGA ESCA -0.225; TCGA HNSC -0.388; TCGA LUSC -0.268; TCGA PAAD -0.214; TCGA PRAD -0.089; TCGA STAD -0.228; TCGA UCEC -0.265 |
hsa-miR-20a-3p | ITGA9 | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.336; TCGA CESC -0.248; TCGA COAD -0.173; TCGA ESCA -0.648; TCGA HNSC -0.443; TCGA LUAD -0.256; TCGA LUSC -0.62; TCGA PAAD -0.404; TCGA PRAD -0.597; TCGA SARC -0.142; TCGA STAD -0.427; TCGA UCEC -0.19 |
hsa-miR-20a-3p | LPP | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUAD; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.401; TCGA CESC -0.201; TCGA COAD -0.246; TCGA ESCA -0.32; TCGA HNSC -0.211; TCGA LUAD -0.147; TCGA PAAD -0.364; TCGA PRAD -0.445; TCGA SARC -0.421; TCGA THCA -0.11; TCGA STAD -0.464; TCGA UCEC -0.172 |
hsa-miR-20a-3p | MARCH1 | 13 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.079; TCGA CESC -0.261; TCGA COAD -0.245; TCGA ESCA -0.132; TCGA HNSC -0.293; TCGA KIRP -0.201; TCGA LUAD -0.198; TCGA LUSC -0.316; TCGA PAAD -0.216; TCGA PRAD -0.135; TCGA SARC -0.165; TCGA THCA -0.191; TCGA STAD -0.117 |
hsa-miR-20a-3p | ARHGAP26 | 9 cancers: BLCA; CESC; ESCA; HNSC; LGG; LUAD; LUSC; PRAD; THCA | mirMAP | TCGA BLCA -0.083; TCGA CESC -0.308; TCGA ESCA -0.179; TCGA HNSC -0.234; TCGA LGG -0.284; TCGA LUAD -0.103; TCGA LUSC -0.187; TCGA PRAD -0.086; TCGA THCA -0.087 |
hsa-miR-20a-3p | NTRK2 | 11 cancers: BLCA; CESC; COAD; HNSC; LGG; LIHC; LUAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.639; TCGA CESC -0.543; TCGA COAD -0.6; TCGA HNSC -0.336; TCGA LGG -0.142; TCGA LIHC -0.253; TCGA LUAD -0.365; TCGA PRAD -0.509; TCGA SARC -0.432; TCGA STAD -0.463; TCGA UCEC -0.369 |
hsa-miR-20a-3p | SESN3 | 9 cancers: BLCA; COAD; ESCA; KIRP; LIHC; LUAD; PAAD; PRAD; STAD | mirMAP | TCGA BLCA -0.163; TCGA COAD -0.209; TCGA ESCA -0.308; TCGA KIRP -0.18; TCGA LIHC -0.295; TCGA LUAD -0.19; TCGA PAAD -0.273; TCGA PRAD -0.207; TCGA STAD -0.3 |
hsa-miR-20a-3p | SCN2B | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LGG; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.614; TCGA BRCA -0.375; TCGA COAD -0.812; TCGA ESCA -0.977; TCGA HNSC -0.381; TCGA LGG -0.371; TCGA LUAD -0.515; TCGA LUSC -0.683; TCGA PAAD -0.464; TCGA PRAD -0.54; TCGA STAD -0.813; TCGA UCEC -0.349 |
hsa-miR-20a-3p | GFRA1 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.67; TCGA BRCA -0.565; TCGA CESC -0.768; TCGA COAD -0.662; TCGA ESCA -1.042; TCGA HNSC -0.616; TCGA LIHC -0.296; TCGA LUAD -0.447; TCGA LUSC -0.736; TCGA PRAD -0.396; TCGA STAD -0.794; TCGA UCEC -0.205 |
hsa-miR-20a-3p | DST | 11 cancers: BLCA; COAD; HNSC; KIRP; LUAD; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.16; TCGA COAD -0.166; TCGA HNSC -0.11; TCGA KIRP -0.122; TCGA LUAD -0.191; TCGA PAAD -0.145; TCGA PRAD -0.336; TCGA SARC -0.189; TCGA THCA -0.17; TCGA STAD -0.295; TCGA UCEC -0.224 |
hsa-miR-20a-3p | SETBP1 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.405; TCGA BRCA -0.091; TCGA CESC -0.301; TCGA COAD -0.487; TCGA ESCA -0.524; TCGA HNSC -0.27; TCGA PAAD -0.165; TCGA PRAD -0.23; TCGA SARC -0.148; TCGA STAD -0.521; TCGA UCEC -0.197 |
hsa-miR-20a-3p | CCDC50 | 10 cancers: BLCA; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.091; TCGA COAD -0.069; TCGA ESCA -0.105; TCGA HNSC -0.093; TCGA LUAD -0.062; TCGA LUSC -0.104; TCGA PAAD -0.136; TCGA SARC -0.055; TCGA STAD -0.198; TCGA UCEC -0.111 |
hsa-miR-20a-3p | CAMK4 | 9 cancers: BLCA; COAD; ESCA; HNSC; LGG; LUAD; PRAD; THCA; STAD | mirMAP | TCGA BLCA -0.207; TCGA COAD -0.337; TCGA ESCA -0.32; TCGA HNSC -0.252; TCGA LGG -0.75; TCGA LUAD -0.142; TCGA PRAD -0.413; TCGA THCA -0.211; TCGA STAD -0.196 |
hsa-miR-20a-3p | GPD1L | 11 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LGG; LUAD; LUSC; PRAD; SARC; UCEC | mirMAP | TCGA BLCA -0.119; TCGA BRCA -0.133; TCGA CESC -0.137; TCGA ESCA -0.179; TCGA HNSC -0.242; TCGA LGG -0.15; TCGA LUAD -0.171; TCGA LUSC -0.356; TCGA PRAD -0.223; TCGA SARC -0.299; TCGA UCEC -0.109 |
hsa-miR-20a-3p | KCNMA1 | 16 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.674; TCGA BRCA -0.216; TCGA CESC -0.409; TCGA COAD -0.812; TCGA ESCA -1.255; TCGA HNSC -0.275; TCGA LGG -0.335; TCGA LIHC -0.199; TCGA LUAD -0.116; TCGA LUSC -0.4; TCGA PAAD -0.305; TCGA PRAD -0.453; TCGA SARC -0.426; TCGA THCA -0.29; TCGA STAD -0.865; TCGA UCEC -0.367 |
hsa-miR-20a-3p | ZNF208 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUSC; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.464; TCGA BRCA -0.109; TCGA CESC -0.554; TCGA COAD -0.457; TCGA ESCA -0.792; TCGA HNSC -0.28; TCGA LUSC -0.412; TCGA PRAD -0.349; TCGA SARC -0.214; TCGA STAD -0.448; TCGA UCEC -0.413 |
hsa-miR-20a-3p | DISC1 | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; THCA; STAD | mirMAP | TCGA BLCA -0.147; TCGA CESC -0.182; TCGA COAD -0.131; TCGA ESCA -0.159; TCGA HNSC -0.149; TCGA LGG -0.111; TCGA LIHC -0.225; TCGA THCA -0.121; TCGA STAD -0.057 |
hsa-miR-20a-3p | CTSS | 9 cancers: BLCA; CESC; HNSC; LUAD; LUSC; PRAD; SARC; THCA; UCEC | mirMAP | TCGA BLCA -0.134; TCGA CESC -0.377; TCGA HNSC -0.328; TCGA LUAD -0.206; TCGA LUSC -0.601; TCGA PRAD -0.2; TCGA SARC -0.152; TCGA THCA -0.278; TCGA UCEC -0.168 |
hsa-miR-20a-3p | FSTL1 | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.283; TCGA BRCA -0.072; TCGA COAD -0.392; TCGA ESCA -0.292; TCGA HNSC -0.215; TCGA LIHC -0.214; TCGA LUAD -0.125; TCGA LUSC -0.221; TCGA PAAD -0.36; TCGA PRAD -0.147; TCGA STAD -0.285; TCGA UCEC -0.099 |
hsa-miR-20a-3p | NFASC | 12 cancers: BLCA; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.405; TCGA COAD -0.209; TCGA ESCA -0.792; TCGA HNSC -0.184; TCGA LGG -0.196; TCGA LIHC -0.25; TCGA LUAD -0.226; TCGA LUSC -0.288; TCGA PRAD -0.312; TCGA SARC -0.655; TCGA STAD -0.602; TCGA UCEC -0.406 |
hsa-miR-20a-3p | PPM1L | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; LUAD; PAAD; PRAD; SARC; STAD | mirMAP | TCGA BLCA -0.198; TCGA CESC -0.334; TCGA COAD -0.188; TCGA ESCA -0.366; TCGA HNSC -0.425; TCGA LGG -0.116; TCGA LIHC -0.193; TCGA LUAD -0.09; TCGA PAAD -0.257; TCGA PRAD -0.42; TCGA SARC -0.273; TCGA STAD -0.228 |
hsa-miR-20a-3p | GALNT10 | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; THCA | mirMAP | TCGA BLCA -0.13; TCGA BRCA -0.219; TCGA COAD -0.068; TCGA ESCA -0.24; TCGA HNSC -0.156; TCGA KIRP -0.127; TCGA LIHC -0.201; TCGA LUAD -0.246; TCGA LUSC -0.295; TCGA PAAD -0.21; TCGA PRAD -0.094; TCGA THCA -0.14 |
hsa-miR-20a-3p | ZCCHC24 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.478; TCGA BRCA -0.274; TCGA CESC -0.191; TCGA COAD -0.235; TCGA ESCA -0.611; TCGA HNSC -0.282; TCGA LUAD -0.18; TCGA LUSC -0.52; TCGA PAAD -0.302; TCGA PRAD -0.161; TCGA STAD -0.457; TCGA UCEC -0.33 |
hsa-miR-20a-3p | GFRA2 | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PRAD; STAD | mirMAP | TCGA BLCA -0.42; TCGA BRCA -0.132; TCGA CESC -0.377; TCGA COAD -0.417; TCGA ESCA -0.724; TCGA HNSC -0.316; TCGA LUAD -0.228; TCGA LUSC -0.591; TCGA PRAD -0.303; TCGA STAD -0.45 |
hsa-miR-20a-3p | LONRF2 | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.511; TCGA BRCA -0.266; TCGA CESC -0.446; TCGA COAD -1.037; TCGA ESCA -0.936; TCGA HNSC -0.331; TCGA LGG -0.236; TCGA SARC -0.61; TCGA STAD -0.673; TCGA UCEC -0.163 |
hsa-miR-20a-3p | BCL2 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; LUAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.211; TCGA BRCA -0.139; TCGA CESC -0.157; TCGA COAD -0.19; TCGA ESCA -0.22; TCGA HNSC -0.21; TCGA LGG -0.069; TCGA LIHC -0.242; TCGA LUAD -0.102; TCGA PRAD -0.28; TCGA STAD -0.243; TCGA UCEC -0.108 |
hsa-miR-20a-3p | IL16 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.32; TCGA BRCA -0.133; TCGA CESC -0.409; TCGA COAD -0.305; TCGA ESCA -0.468; TCGA HNSC -0.247; TCGA LUAD -0.246; TCGA LUSC -0.504; TCGA PAAD -0.251; TCGA PRAD -0.153; TCGA SARC -0.198; TCGA THCA -0.216; TCGA STAD -0.155; TCGA UCEC -0.26 |
hsa-miR-20a-3p | AFF1 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.06; TCGA BRCA -0.066; TCGA CESC -0.151; TCGA COAD -0.068; TCGA ESCA -0.224; TCGA HNSC -0.133; TCGA LUAD -0.106; TCGA LUSC -0.175; TCGA PAAD -0.105; TCGA SARC -0.176; TCGA STAD -0.14; TCGA UCEC -0.118 |
hsa-miR-20a-3p | PPP1R3B | 10 cancers: BLCA; BRCA; ESCA; HNSC; KIRP; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.224; TCGA BRCA -0.07; TCGA ESCA -0.196; TCGA HNSC -0.186; TCGA KIRP -0.158; TCGA PAAD -0.127; TCGA PRAD -0.16; TCGA SARC -0.34; TCGA STAD -0.094; TCGA UCEC -0.253 |
hsa-miR-20a-3p | SNX18 | 14 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.072; TCGA CESC -0.122; TCGA COAD -0.205; TCGA ESCA -0.184; TCGA HNSC -0.121; TCGA KIRP -0.102; TCGA LIHC -0.058; TCGA LUAD -0.126; TCGA LUSC -0.146; TCGA PAAD -0.184; TCGA PRAD -0.115; TCGA SARC -0.18; TCGA STAD -0.138; TCGA UCEC -0.088 |
hsa-miR-20a-3p | HHIPL1 | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; PAAD; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.277; TCGA BRCA -0.121; TCGA COAD -0.387; TCGA ESCA -0.293; TCGA HNSC -0.142; TCGA PAAD -0.284; TCGA PRAD -0.182; TCGA THCA -0.149; TCGA STAD -0.171; TCGA UCEC -0.109 |
hsa-miR-20a-3p | PAPPA | 11 cancers: BLCA; COAD; ESCA; HNSC; LGG; LIHC; LUSC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.186; TCGA COAD -0.366; TCGA ESCA -0.371; TCGA HNSC -0.207; TCGA LGG -0.368; TCGA LIHC -0.265; TCGA LUSC -0.27; TCGA PAAD -0.314; TCGA PRAD -0.403; TCGA STAD -0.158; TCGA UCEC -0.518 |
hsa-miR-20a-3p | CAMK1D | 9 cancers: BLCA; CESC; ESCA; HNSC; LGG; LIHC; LUAD; PRAD; STAD | mirMAP | TCGA BLCA -0.188; TCGA CESC -0.223; TCGA ESCA -0.279; TCGA HNSC -0.131; TCGA LGG -0.084; TCGA LIHC -0.497; TCGA LUAD -0.13; TCGA PRAD -0.399; TCGA STAD -0.247 |
hsa-miR-20a-3p | FLRT2 | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.503; TCGA BRCA -0.121; TCGA COAD -0.548; TCGA ESCA -0.494; TCGA HNSC -0.185; TCGA LGG -0.252; TCGA LIHC -0.238; TCGA LUAD -0.148; TCGA LUSC -0.212; TCGA PRAD -0.423; TCGA STAD -0.414; TCGA UCEC -0.248 |
hsa-miR-20a-3p | THSD4 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD | mirMAP | TCGA BLCA -0.188; TCGA BRCA -0.41; TCGA CESC -0.248; TCGA COAD -0.131; TCGA ESCA -0.376; TCGA HNSC -0.183; TCGA LIHC -0.253; TCGA LUAD -0.178; TCGA LUSC -0.23; TCGA PAAD -0.248; TCGA PRAD -0.447; TCGA SARC -0.325; TCGA STAD -0.245 |
hsa-miR-20a-3p | LDLRAD2 | 9 cancers: BLCA; BRCA; COAD; ESCA; LIHC; LUAD; LUSC; SARC; STAD | mirMAP | TCGA BLCA -0.265; TCGA BRCA -0.205; TCGA COAD -0.164; TCGA ESCA -0.153; TCGA LIHC -0.12; TCGA LUAD -0.093; TCGA LUSC -0.153; TCGA SARC -0.167; TCGA STAD -0.177 |
hsa-miR-20a-3p | TLR7 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.245; TCGA BRCA -0.084; TCGA CESC -0.359; TCGA COAD -0.437; TCGA ESCA -0.45; TCGA HNSC -0.499; TCGA KIRP -0.223; TCGA LUAD -0.369; TCGA LUSC -0.668; TCGA PAAD -0.276; TCGA PRAD -0.223; TCGA SARC -0.311; TCGA THCA -0.304; TCGA STAD -0.165; TCGA UCEC -0.198 |
hsa-miR-20a-3p | ADARB1 | 10 cancers: BLCA; COAD; ESCA; LGG; LIHC; LUAD; LUSC; PAAD; STAD; UCEC | mirMAP | TCGA BLCA -0.288; TCGA COAD -0.218; TCGA ESCA -0.273; TCGA LGG -0.16; TCGA LIHC -0.152; TCGA LUAD -0.187; TCGA LUSC -0.355; TCGA PAAD -0.141; TCGA STAD -0.281; TCGA UCEC -0.145 |
hsa-miR-20a-3p | MYO5A | 13 cancers: BLCA; COAD; ESCA; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.145; TCGA COAD -0.425; TCGA ESCA -0.292; TCGA KIRP -0.187; TCGA LGG -0.128; TCGA LIHC -0.191; TCGA LUAD -0.158; TCGA LUSC -0.082; TCGA PAAD -0.216; TCGA PRAD -0.133; TCGA THCA -0.134; TCGA STAD -0.218; TCGA UCEC -0.134 |
hsa-miR-20a-3p | ZNF677 | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LUSC; PAAD; STAD | mirMAP | TCGA BLCA -0.226; TCGA CESC -0.292; TCGA COAD -0.422; TCGA ESCA -0.589; TCGA HNSC -0.333; TCGA KIRP -0.262; TCGA LUSC -0.476; TCGA PAAD -0.279; TCGA STAD -0.432 |
hsa-miR-20a-3p | RYR2 | 12 cancers: BLCA; CESC; COAD; ESCA; LGG; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP; miRNATAP | TCGA BLCA -0.698; TCGA CESC -0.365; TCGA COAD -0.474; TCGA ESCA -1.015; TCGA LGG -0.284; TCGA LUAD -0.337; TCGA LUSC -0.388; TCGA PAAD -0.225; TCGA PRAD -0.269; TCGA SARC -0.596; TCGA STAD -0.648; TCGA UCEC -0.249 |
hsa-miR-20a-3p | ITPRIPL2 | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; THCA; STAD | mirMAP | TCGA BLCA -0.07; TCGA BRCA -0.098; TCGA COAD -0.061; TCGA ESCA -0.107; TCGA HNSC -0.073; TCGA LUAD -0.077; TCGA LUSC -0.178; TCGA PAAD -0.099; TCGA PRAD -0.051; TCGA THCA -0.095; TCGA STAD -0.156 |
hsa-miR-20a-3p | DOK6 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.594; TCGA BRCA -0.137; TCGA CESC -0.494; TCGA COAD -0.444; TCGA ESCA -0.421; TCGA HNSC -0.362; TCGA LUAD -0.157; TCGA LUSC -0.308; TCGA PAAD -0.437; TCGA PRAD -0.229; TCGA STAD -0.37; TCGA UCEC -0.188 |
hsa-miR-20a-3p | PLXNA4 | 13 cancers: BLCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD | mirMAP; miRNATAP | TCGA BLCA -0.549; TCGA CESC -0.325; TCGA COAD -0.64; TCGA ESCA -0.717; TCGA HNSC -0.316; TCGA LGG -0.461; TCGA LIHC -0.285; TCGA LUAD -0.287; TCGA LUSC -0.446; TCGA PAAD -0.369; TCGA PRAD -0.34; TCGA SARC -0.7; TCGA STAD -0.467 |
hsa-miR-20a-3p | ZNF844 | 9 cancers: BLCA; CESC; ESCA; HNSC; LUAD; LUSC; PRAD; SARC; STAD | mirMAP | TCGA BLCA -0.099; TCGA CESC -0.254; TCGA ESCA -0.444; TCGA HNSC -0.336; TCGA LUAD -0.116; TCGA LUSC -0.282; TCGA PRAD -0.122; TCGA SARC -0.139; TCGA STAD -0.238 |
hsa-miR-20a-3p | C14orf132 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.317; TCGA BRCA -0.257; TCGA CESC -0.246; TCGA COAD -0.605; TCGA ESCA -0.559; TCGA HNSC -0.232; TCGA LGG -0.115; TCGA LUAD -0.265; TCGA LUSC -0.466; TCGA PAAD -0.291; TCGA PRAD -0.256; TCGA SARC -0.242; TCGA STAD -0.557; TCGA UCEC -0.361 |
hsa-miR-20a-3p | STON1 | 12 cancers: BLCA; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; UCEC | mirMAP | TCGA BLCA -0.492; TCGA COAD -0.58; TCGA ESCA -0.513; TCGA HNSC -0.106; TCGA LIHC -0.161; TCGA LUAD -0.224; TCGA LUSC -0.118; TCGA PAAD -0.399; TCGA PRAD -0.186; TCGA SARC -0.151; TCGA THCA -0.153; TCGA UCEC -0.283 |
hsa-miR-20a-3p | CPEB2 | 14 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.178; TCGA BRCA -0.183; TCGA COAD -0.058; TCGA ESCA -0.316; TCGA HNSC -0.129; TCGA KIRP -0.17; TCGA LGG -0.231; TCGA LIHC -0.189; TCGA LUAD -0.079; TCGA LUSC -0.28; TCGA PRAD -0.114; TCGA SARC -0.211; TCGA STAD -0.179; TCGA UCEC -0.25 |
hsa-miR-20a-3p | CACNB4 | 17 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.46; TCGA BRCA -0.092; TCGA CESC -0.243; TCGA COAD -0.226; TCGA ESCA -0.422; TCGA HNSC -0.272; TCGA KIRC -0.326; TCGA KIRP -0.445; TCGA LGG -0.254; TCGA LIHC -0.378; TCGA LUAD -0.215; TCGA LUSC -0.382; TCGA PAAD -0.188; TCGA PRAD -0.356; TCGA SARC -0.294; TCGA STAD -0.311; TCGA UCEC -0.158 |
hsa-miR-20a-3p | RUNX1T1 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.629; TCGA BRCA -0.212; TCGA CESC -0.634; TCGA COAD -0.551; TCGA ESCA -0.73; TCGA HNSC -0.548; TCGA LUAD -0.245; TCGA LUSC -0.677; TCGA PAAD -0.444; TCGA PRAD -0.577; TCGA STAD -0.516; TCGA UCEC -0.335 |
hsa-miR-20a-3p | VGLL3 | 12 cancers: BLCA; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.176; TCGA COAD -0.613; TCGA ESCA -0.464; TCGA HNSC -0.347; TCGA LIHC -0.316; TCGA LUAD -0.282; TCGA LUSC -0.63; TCGA PAAD -0.374; TCGA PRAD -0.481; TCGA SARC -0.522; TCGA STAD -0.363; TCGA UCEC -0.518 |
hsa-miR-20a-3p | MAF | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PRAD; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.308; TCGA BRCA -0.079; TCGA COAD -0.37; TCGA ESCA -0.304; TCGA HNSC -0.154; TCGA KIRC -0.146; TCGA LUAD -0.136; TCGA LUSC -0.104; TCGA PRAD -0.192; TCGA THCA -0.073; TCGA STAD -0.214; TCGA UCEC -0.398 |
hsa-miR-20a-3p | NCALD | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; THCA; STAD | miRNATAP | TCGA BLCA -0.349; TCGA CESC -0.272; TCGA COAD -0.145; TCGA ESCA -0.413; TCGA HNSC -0.175; TCGA KIRC -0.142; TCGA LUAD -0.103; TCGA LUSC -0.34; TCGA THCA -0.232; TCGA STAD -0.298 |
hsa-miR-20a-3p | SPRY1 | 11 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; PRAD; STAD | miRNATAP | TCGA BLCA -0.312; TCGA BRCA -0.144; TCGA CESC -0.271; TCGA ESCA -0.264; TCGA HNSC -0.236; TCGA LGG -0.165; TCGA LIHC -0.144; TCGA LUAD -0.146; TCGA LUSC -0.329; TCGA PRAD -0.261; TCGA STAD -0.133 |
hsa-miR-20a-3p | STAT5B | 10 cancers: BLCA; CESC; ESCA; HNSC; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.104; TCGA CESC -0.099; TCGA ESCA -0.137; TCGA HNSC -0.106; TCGA LUSC -0.127; TCGA PAAD -0.091; TCGA PRAD -0.134; TCGA SARC -0.2; TCGA STAD -0.151; TCGA UCEC -0.154 |
hsa-miR-20a-3p | PARD3B | 10 cancers: BLCA; CESC; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; SARC; STAD | miRNATAP | TCGA BLCA -0.43; TCGA CESC -0.359; TCGA ESCA -0.417; TCGA HNSC -0.494; TCGA LUAD -0.26; TCGA LUSC -0.459; TCGA PAAD -0.403; TCGA PRAD -0.286; TCGA SARC -0.226; TCGA STAD -0.225 |
hsa-miR-20a-3p | SLITRK3 | 10 cancers: BLCA; COAD; ESCA; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.674; TCGA COAD -0.951; TCGA ESCA -1.387; TCGA LUAD -0.256; TCGA LUSC -0.365; TCGA PAAD -0.555; TCGA PRAD -0.698; TCGA SARC -0.493; TCGA STAD -0.938; TCGA UCEC -0.575 |
hsa-miR-20a-3p | SOCS2 | 10 cancers: BLCA; BRCA; COAD; ESCA; LIHC; LUSC; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.098; TCGA BRCA -0.176; TCGA COAD -0.118; TCGA ESCA -0.336; TCGA LIHC -0.166; TCGA LUSC -0.344; TCGA SARC -0.349; TCGA THCA -0.154; TCGA STAD -0.175; TCGA UCEC -0.342 |
hsa-miR-20a-3p | RNF11 | 13 cancers: BLCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.103; TCGA CESC -0.12; TCGA COAD -0.117; TCGA ESCA -0.151; TCGA HNSC -0.116; TCGA LIHC -0.165; TCGA LUAD -0.099; TCGA LUSC -0.149; TCGA PAAD -0.097; TCGA PRAD -0.122; TCGA SARC -0.058; TCGA STAD -0.139; TCGA UCEC -0.182 |
hsa-miR-20a-3p | ERG | 13 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.264; TCGA BRCA -0.163; TCGA COAD -0.365; TCGA ESCA -0.339; TCGA HNSC -0.132; TCGA KIRP -0.164; TCGA LIHC -0.178; TCGA LUAD -0.281; TCGA LUSC -0.523; TCGA PAAD -0.196; TCGA THCA -0.135; TCGA STAD -0.24; TCGA UCEC -0.312 |
hsa-miR-20a-3p | NRXN2 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; LUAD; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.572; TCGA BRCA -0.295; TCGA CESC -0.469; TCGA COAD -0.211; TCGA ESCA -0.349; TCGA HNSC -0.196; TCGA LGG -0.142; TCGA LIHC -0.445; TCGA LUAD -0.186; TCGA THCA -0.151; TCGA STAD -0.515; TCGA UCEC -0.249 |
hsa-miR-20a-3p | SHC4 | 9 cancers: BLCA; COAD; ESCA; HNSC; LGG; LUAD; PAAD; PRAD; STAD | miRNATAP | TCGA BLCA -0.158; TCGA COAD -0.45; TCGA ESCA -0.325; TCGA HNSC -0.274; TCGA LGG -0.147; TCGA LUAD -0.166; TCGA PAAD -0.227; TCGA PRAD -0.215; TCGA STAD -0.389 |
hsa-miR-20a-3p | PPP3CB | 10 cancers: BLCA; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.114; TCGA COAD -0.061; TCGA ESCA -0.126; TCGA HNSC -0.067; TCGA KIRP -0.076; TCGA LUAD -0.054; TCGA LUSC -0.109; TCGA PRAD -0.102; TCGA STAD -0.135; TCGA UCEC -0.064 |
hsa-miR-20a-3p | RPS6KA5 | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; LGG; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.118; TCGA CESC -0.231; TCGA COAD -0.137; TCGA ESCA -0.207; TCGA HNSC -0.124; TCGA LGG -0.114; TCGA PRAD -0.266; TCGA SARC -0.26; TCGA STAD -0.17; TCGA UCEC -0.165 |
hsa-miR-20a-3p | COL14A1 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.596; TCGA BRCA -0.364; TCGA CESC -0.574; TCGA COAD -0.55; TCGA ESCA -0.709; TCGA HNSC -0.458; TCGA LIHC -0.276; TCGA LUAD -0.291; TCGA LUSC -0.577; TCGA PAAD -0.48; TCGA PRAD -0.336; TCGA SARC -0.486; TCGA THCA -0.186; TCGA STAD -0.52; TCGA UCEC -0.341 |
hsa-miR-20a-3p | ATRNL1 | 9 cancers: BRCA; COAD; ESCA; HNSC; LGG; LUAD; PRAD; STAD; UCEC | MirTarget; mirMAP | TCGA BRCA -0.208; TCGA COAD -0.648; TCGA ESCA -0.297; TCGA HNSC -0.246; TCGA LGG -0.238; TCGA LUAD -0.222; TCGA PRAD -0.437; TCGA STAD -0.449; TCGA UCEC -0.261 |
hsa-miR-20a-3p | TPRG1 | 12 cancers: BRCA; CESC; COAD; KIRC; LIHC; LUAD; PAAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BRCA -0.497; TCGA CESC -0.27; TCGA COAD -0.266; TCGA KIRC -0.175; TCGA LIHC -0.562; TCGA LUAD -0.267; TCGA PAAD -0.451; TCGA PRAD -0.288; TCGA SARC -0.259; TCGA THCA -0.172; TCGA STAD -0.153; TCGA UCEC -0.112 |
hsa-miR-20a-3p | SIAE | 9 cancers: BRCA; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; PRAD; THCA | MirTarget | TCGA BRCA -0.088; TCGA HNSC -0.143; TCGA KIRP -0.136; TCGA LGG -0.269; TCGA LIHC -0.337; TCGA LUAD -0.149; TCGA LUSC -0.32; TCGA PRAD -0.076; TCGA THCA -0.058 |
hsa-miR-20a-3p | PRLR | 9 cancers: BRCA; CESC; HNSC; KIRC; KIRP; LGG; PRAD; THCA; UCEC | mirMAP | TCGA BRCA -0.1; TCGA CESC -0.593; TCGA HNSC -0.437; TCGA KIRC -0.367; TCGA KIRP -0.346; TCGA LGG -0.15; TCGA PRAD -0.205; TCGA THCA -0.323; TCGA UCEC -0.289 |
hsa-miR-20a-3p | KLHL3 | 13 cancers: BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BRCA -0.159; TCGA CESC -0.251; TCGA ESCA -0.172; TCGA HNSC -0.195; TCGA KIRC -0.233; TCGA KIRP -0.298; TCGA LGG -0.175; TCGA LUAD -0.094; TCGA LUSC -0.172; TCGA PAAD -0.192; TCGA PRAD -0.172; TCGA STAD -0.252; TCGA UCEC -0.119 |
hsa-miR-20a-3p | THRB | 10 cancers: BRCA; COAD; ESCA; HNSC; LGG; LUAD; LUSC; SARC; STAD; UCEC | mirMAP | TCGA BRCA -0.119; TCGA COAD -0.266; TCGA ESCA -0.382; TCGA HNSC -0.268; TCGA LGG -0.296; TCGA LUAD -0.182; TCGA LUSC -0.283; TCGA SARC -0.209; TCGA STAD -0.304; TCGA UCEC -0.383 |
hsa-miR-20a-3p | ZFYVE1 | 10 cancers: BRCA; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PRAD; SARC; STAD | mirMAP | TCGA BRCA -0.077; TCGA COAD -0.142; TCGA ESCA -0.159; TCGA HNSC -0.096; TCGA LIHC -0.068; TCGA LUAD -0.054; TCGA LUSC -0.07; TCGA PRAD -0.061; TCGA SARC -0.071; TCGA STAD -0.092 |
hsa-miR-20a-3p | TANC2 | 9 cancers: BRCA; COAD; HNSC; LIHC; LUAD; LUSC; PAAD; THCA; STAD | mirMAP | TCGA BRCA -0.058; TCGA COAD -0.441; TCGA HNSC -0.145; TCGA LIHC -0.177; TCGA LUAD -0.1; TCGA LUSC -0.124; TCGA PAAD -0.184; TCGA THCA -0.1; TCGA STAD -0.133 |
hsa-miR-20a-3p | OPCML | 9 cancers: BRCA; COAD; ESCA; LGG; LUAD; PAAD; THCA; STAD; UCEC | mirMAP | TCGA BRCA -0.258; TCGA COAD -0.215; TCGA ESCA -0.719; TCGA LGG -0.147; TCGA LUAD -0.209; TCGA PAAD -0.298; TCGA THCA -0.681; TCGA STAD -0.218; TCGA UCEC -0.251 |
hsa-miR-20a-3p | NRIP1 | 9 cancers: BRCA; ESCA; HNSC; LIHC; LUAD; PAAD; SARC; STAD; UCEC | miRNATAP | TCGA BRCA -0.052; TCGA ESCA -0.135; TCGA HNSC -0.17; TCGA LIHC -0.073; TCGA LUAD -0.09; TCGA PAAD -0.094; TCGA SARC -0.103; TCGA STAD -0.057; TCGA UCEC -0.087 |
hsa-miR-20a-3p | PCMTD1 | 9 cancers: CESC; ESCA; HNSC; LIHC; LUAD; LUSC; SARC; STAD; UCEC | MirTarget | TCGA CESC -0.116; TCGA ESCA -0.162; TCGA HNSC -0.088; TCGA LIHC -0.084; TCGA LUAD -0.052; TCGA LUSC -0.145; TCGA SARC -0.164; TCGA STAD -0.197; TCGA UCEC -0.172 |
hsa-miR-20a-3p | FAM126B | 10 cancers: CESC; ESCA; HNSC; KIRP; LGG; LUAD; LUSC; PRAD; STAD; UCEC | MirTarget | TCGA CESC -0.107; TCGA ESCA -0.121; TCGA HNSC -0.073; TCGA KIRP -0.088; TCGA LGG -0.059; TCGA LUAD -0.08; TCGA LUSC -0.102; TCGA PRAD -0.069; TCGA STAD -0.087; TCGA UCEC -0.098 |
hsa-miR-20a-3p | SAP30L | 10 cancers: CESC; ESCA; HNSC; LGG; LUAD; LUSC; PAAD; SARC; STAD; UCEC | MirTarget | TCGA CESC -0.087; TCGA ESCA -0.102; TCGA HNSC -0.077; TCGA LGG -0.072; TCGA LUAD -0.097; TCGA LUSC -0.107; TCGA PAAD -0.097; TCGA SARC -0.096; TCGA STAD -0.082; TCGA UCEC -0.081 |
hsa-miR-20a-3p | FAM168A | 9 cancers: CESC; COAD; ESCA; HNSC; KIRP; LGG; PAAD; PRAD; STAD | mirMAP | TCGA CESC -0.136; TCGA COAD -0.129; TCGA ESCA -0.168; TCGA HNSC -0.146; TCGA KIRP -0.196; TCGA LGG -0.107; TCGA PAAD -0.273; TCGA PRAD -0.3; TCGA STAD -0.07 |
hsa-miR-20a-3p | NIPAL2 | 9 cancers: CESC; ESCA; HNSC; LGG; LIHC; LUSC; PRAD; SARC; UCEC | mirMAP | TCGA CESC -0.343; TCGA ESCA -0.172; TCGA HNSC -0.217; TCGA LGG -0.204; TCGA LIHC -0.176; TCGA LUSC -0.221; TCGA PRAD -0.302; TCGA SARC -0.212; TCGA UCEC -0.191 |
hsa-miR-20a-3p | MECP2 | 9 cancers: CESC; ESCA; KIRP; LIHC; LUAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA CESC -0.081; TCGA ESCA -0.089; TCGA KIRP -0.068; TCGA LIHC -0.051; TCGA LUAD -0.067; TCGA PRAD -0.095; TCGA SARC -0.09; TCGA STAD -0.118; TCGA UCEC -0.083 |
hsa-miR-20a-3p | CXorf23 | 9 cancers: CESC; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; STAD; UCEC | mirMAP | TCGA CESC -0.106; TCGA HNSC -0.092; TCGA KIRP -0.107; TCGA LGG -0.051; TCGA LIHC -0.127; TCGA LUAD -0.08; TCGA LUSC -0.051; TCGA STAD -0.083; TCGA UCEC -0.084 |
hsa-miR-20a-3p | CDH13 | 10 cancers: COAD; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA COAD -0.161; TCGA LIHC -0.249; TCGA LUAD -0.204; TCGA LUSC -0.216; TCGA PAAD -0.284; TCGA PRAD -0.122; TCGA SARC -0.174; TCGA THCA -0.253; TCGA STAD -0.142; TCGA UCEC -0.379 |
hsa-miR-20a-3p | NOS1 | 9 cancers: COAD; ESCA; HNSC; LGG; LUAD; LUSC; PAAD; PRAD; STAD | mirMAP | TCGA COAD -0.511; TCGA ESCA -0.644; TCGA HNSC -0.416; TCGA LGG -0.419; TCGA LUAD -0.216; TCGA LUSC -0.369; TCGA PAAD -0.426; TCGA PRAD -0.798; TCGA STAD -0.964 |
hsa-miR-20a-3p | EFNA5 | 9 cancers: COAD; ESCA; HNSC; LGG; LIHC; PAAD; PRAD; THCA; STAD | mirMAP | TCGA COAD -0.624; TCGA ESCA -0.283; TCGA HNSC -0.145; TCGA LGG -0.337; TCGA LIHC -0.246; TCGA PAAD -0.185; TCGA PRAD -0.41; TCGA THCA -0.196; TCGA STAD -0.281 |
hsa-miR-20a-3p | C17orf51 | 9 cancers: COAD; ESCA; KIRP; LIHC; PAAD; PRAD; THCA; STAD; UCEC | mirMAP | TCGA COAD -0.123; TCGA ESCA -0.164; TCGA KIRP -0.143; TCGA LIHC -0.215; TCGA PAAD -0.138; TCGA PRAD -0.189; TCGA THCA -0.073; TCGA STAD -0.207; TCGA UCEC -0.136 |
hsa-miR-20a-3p | MIER1 | 9 cancers: ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA ESCA -0.084; TCGA HNSC -0.086; TCGA LUAD -0.102; TCGA LUSC -0.12; TCGA PAAD -0.103; TCGA PRAD -0.152; TCGA SARC -0.069; TCGA STAD -0.055; TCGA UCEC -0.116 |
hsa-miR-20a-3p | MFAP3L | 9 cancers: HNSC; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget; mirMAP | TCGA HNSC -0.137; TCGA LGG -0.158; TCGA LIHC -0.554; TCGA LUAD -0.218; TCGA LUSC -0.191; TCGA PAAD -0.279; TCGA PRAD -0.252; TCGA STAD -0.218; TCGA UCEC -0.195 |
Enriched cancer pathways of putative targets