microRNA information: hsa-miR-218-5p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-218-5p | miRbase |
| Accession: | MIMAT0000275 | miRbase |
| Precursor name: | hsa-mir-218-1 | miRbase |
| Precursor accession: | MI0000294 | miRbase |
| Symbol: | MIR218-1 | HGNC |
| RefSeq ID: | NR_029631 | GenBank |
| Sequence: | UUGUGCUUGAUCUAACCAUGU |
Reported expression in cancers: hsa-miR-218-5p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-218-5p | bladder cancer | deregulation | "In GBC miRNAs with tumor suppressor activity miR-1 ......" | 26855538 | |
| hsa-miR-218-5p | breast cancer | upregulation | "Here we report that miR-218-5p is highly expressed ......" | 27738322 | |
| hsa-miR-218-5p | cervical and endocervical cancer | downregulation | "Here we reported that miR-218 microRNA-218 was dow ......" | 27285984 | |
| hsa-miR-218-5p | colon cancer | downregulation | "In this study miR-218 was found to be downregulate ......" | 23255074 | |
| hsa-miR-218-5p | esophageal cancer | downregulation | "The expression of microRNA-218 miR-218 is downregu ......" | 25812647 | |
| hsa-miR-218-5p | esophageal cancer | downregulation | "In the present study we found that the expression ......" | 25999024 | |
| hsa-miR-218-5p | gastric cancer | downregulation | "Large-scale microarray assays have indicated that ......" | 19890957 | Microarray |
| hsa-miR-218-5p | gastric cancer | downregulation | "The results demonstrated that expression of miR-21 ......" | 25694126 | |
| hsa-miR-218-5p | gastric cancer | downregulation | "The microRNA miR-218 influences various pathobiolo ......" | 26261515 | |
| hsa-miR-218-5p | gastric cancer | downregulation | "miR 218 tissue expression level is associated with ......" | 27323107 | Reverse transcription PCR |
| hsa-miR-218-5p | head and neck cancer | downregulation | "Conversely decreased expressions of miR-153 miR-20 ......" | 25677760 | |
| hsa-miR-218-5p | kidney renal cell cancer | downregulation | "We analyzed 70 matched pairs of clear cell renal c ......" | 21784468 | qPCR; Microarray |
| hsa-miR-218-5p | liver cancer | downregulation | "To investigate the expression of microRNA-218 miR- ......" | 23996750 | qPCR |
| hsa-miR-218-5p | liver cancer | downregulation | "Prognostic significance of miR 218 in human hepato ......" | 25110121 | |
| hsa-miR-218-5p | liver cancer | downregulation | "Here we investigated the role of microRNA 218 miR- ......" | 25374061 | qPCR |
| hsa-miR-218-5p | lung cancer | deregulation | "Therefore we conduct to systematically identify mi ......" | 26870998 | Microarray |
| hsa-miR-218-5p | lung squamous cell cancer | downregulation | "We confirmed decreased expression of mature miR-21 ......" | 20838434 | qPCR |
| hsa-miR-218-5p | melanoma | downregulation | "In this study we analyzed the miR-218 expression l ......" | 24839010 | |
| hsa-miR-218-5p | pancreatic cancer | downregulation | "microRNA-218 miR-218 is a vertebrate-specific miRN ......" | 26662432 | |
| hsa-miR-218-5p | prostate cancer | downregulation | "Our recent studies of the microRNA miRNA expressio ......" | 24815849 | |
| hsa-miR-218-5p | sarcoma | deregulation | "We utilized quantitative real-time PCR to evaluate ......" | 26446495 | qPCR |
Reported cancer pathway affected by hsa-miR-218-5p
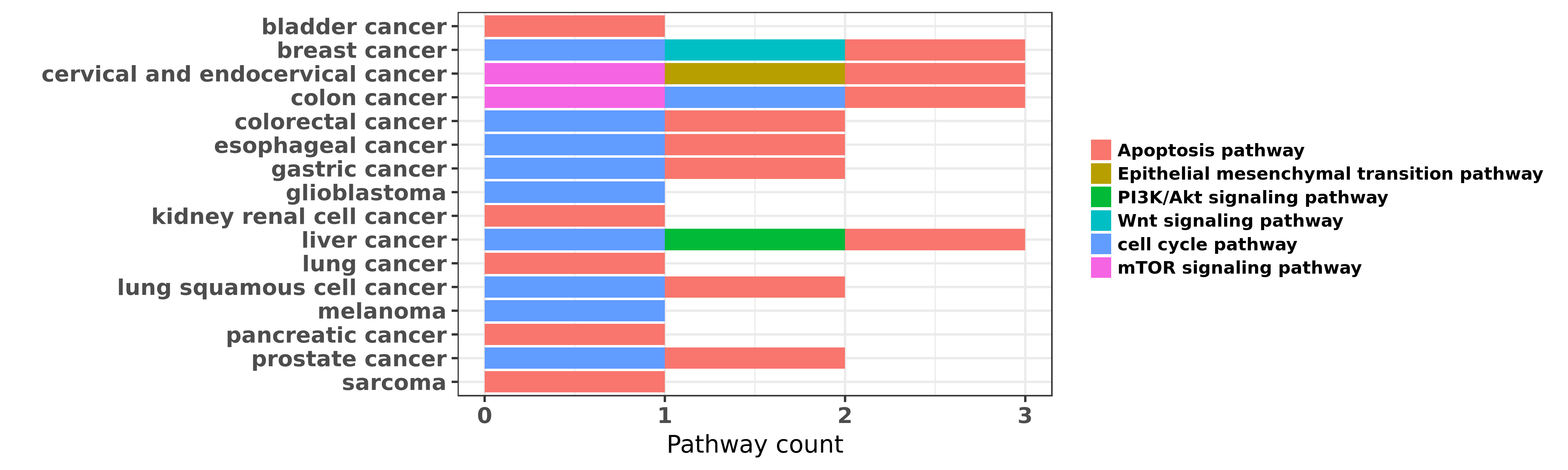
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-218-5p | bladder cancer | Apoptosis pathway | "miR 218 on the genomic loss region of chromosome 4 ......" | 21519788 | Flow cytometry |
| hsa-miR-218-5p | breast cancer | cell cycle pathway | "miR 7 and miR 218 epigenetically control tumor sup ......" | 22705304 | |
| hsa-miR-218-5p | breast cancer | Apoptosis pathway | "MiR 218 regulates cisplatin chemosensitivity in br ......" | 25394901 | |
| hsa-miR-218-5p | breast cancer | Apoptosis pathway | "miR 218 targets survivin and regulates resistance ......" | 25900794 | Western blot; Flow cytometry; Luciferase; RNAi |
| hsa-miR-218-5p | breast cancer | cell cycle pathway; Apoptosis pathway | "Tumor suppressing roles of miR 214 and miR 218 in ......" | 27109339 | |
| hsa-miR-218-5p | breast cancer | Wnt signaling pathway | "Antagonizing miR 218 5p attenuates Wnt signaling a ......" | 27738322 | |
| hsa-miR-218-5p | cervical and endocervical cancer | Apoptosis pathway; mTOR signaling pathway | "MiR 218 impairs tumor growth and increases chemo s ......" | 23443110 | |
| hsa-miR-218-5p | cervical and endocervical cancer | Apoptosis pathway | "We previously reported frequent loss of microRNA-2 ......" | 24843318 | |
| hsa-miR-218-5p | cervical and endocervical cancer | Epithelial mesenchymal transition pathway | "Here we reported that miR-218 microRNA-218 was dow ......" | 27285984 | |
| hsa-miR-218-5p | colon cancer | cell cycle pathway; Apoptosis pathway | "In this study miR-218 was found to be downregulate ......" | 23255074 | Luciferase |
| hsa-miR-218-5p | colon cancer | mTOR signaling pathway | "miR 218 inhibits the invasion and migration of col ......" | 25760926 | |
| hsa-miR-218-5p | colorectal cancer | Apoptosis pathway; cell cycle pathway | "To further explore the possible mechanisms we eval ......" | 26442524 | |
| hsa-miR-218-5p | esophageal cancer | cell cycle pathway; Apoptosis pathway | "miR 218 suppresses tumor growth and enhances the c ......" | 25482044 | Colony formation |
| hsa-miR-218-5p | esophageal cancer | Apoptosis pathway | "In the present study we found that the expression ......" | 25999024 | Western blot; Luciferase |
| hsa-miR-218-5p | gastric cancer | Apoptosis pathway | "Large-scale microarray assays have indicated that ......" | 19890957 | Luciferase |
| hsa-miR-218-5p | gastric cancer | cell cycle pathway; Apoptosis pathway | "Reduced expression of miR 218 and its significance ......" | 20510072 | |
| hsa-miR-218-5p | gastric cancer | Apoptosis pathway | "MiR 218 inhibits multidrug resistance MDR of gastr ......" | 26261515 | |
| hsa-miR-218-5p | gastric cancer | Apoptosis pathway | "The present study aims to investigate the effects ......" | 27696291 | Flow cytometry; Western blot |
| hsa-miR-218-5p | glioblastoma | cell cycle pathway | "MiR 218 Inhibited Growth and Metabolism of Human G ......" | 26012781 | Flow cytometry; Western blot; Luciferase |
| hsa-miR-218-5p | kidney renal cell cancer | Apoptosis pathway | "The expression of miR-218 in renal cell carcinoma ......" | 27133059 | Flow cytometry; MTT assay; Transwell assay; Western blot; Luciferase |
| hsa-miR-218-5p | liver cancer | Apoptosis pathway | "To investigate the expression of microRNA-218 miR- ......" | 23996750 | Western blot; Flow cytometry; MTT assay |
| hsa-miR-218-5p | liver cancer | Apoptosis pathway | "Prognostic significance of miR 218 in human hepato ......" | 25110121 | |
| hsa-miR-218-5p | liver cancer | PI3K/Akt signaling pathway | "miR 218 modulate hepatocellular carcinoma cell pro ......" | 25120782 | Western blot |
| hsa-miR-218-5p | liver cancer | cell cycle pathway | "In the present study miR-218 and miR-520a were obs ......" | 25816091 | Luciferase |
| hsa-miR-218-5p | lung cancer | Apoptosis pathway | "MiR 205 and miR 218 expression is associated with ......" | 25917317 | |
| hsa-miR-218-5p | lung squamous cell cancer | cell cycle pathway; Apoptosis pathway | "Downregulation of microRNA-218 miR-218 is found in ......" | 26282001 | MTT assay; Flow cytometry; Western blot; Luciferase |
| hsa-miR-218-5p | melanoma | cell cycle pathway | "In this study we analyzed the miR-218 expression l ......" | 24839010 | Western blot; Luciferase |
| hsa-miR-218-5p | pancreatic cancer | Apoptosis pathway | "To detect the expression profiles of microRNA-218 ......" | 26157321 | Flow cytometry; Cell migration assay; Western blot; Luciferase |
| hsa-miR-218-5p | prostate cancer | Apoptosis pathway | "This study tried to explore the regulative role of ......" | 25511701 | |
| hsa-miR-218-5p | prostate cancer | cell cycle pathway | "MiR 218 impedes IL 6 induced prostate cancer cell ......" | 26986507 | |
| hsa-miR-218-5p | sarcoma | Apoptosis pathway | "miR 218 expression in osteosarcoma tissues and its ......" | 25479631 | Flow cytometry; Cell proliferation assay; Cell Proliferation Assay |
Reported cancer prognosis affected by hsa-miR-218-5p
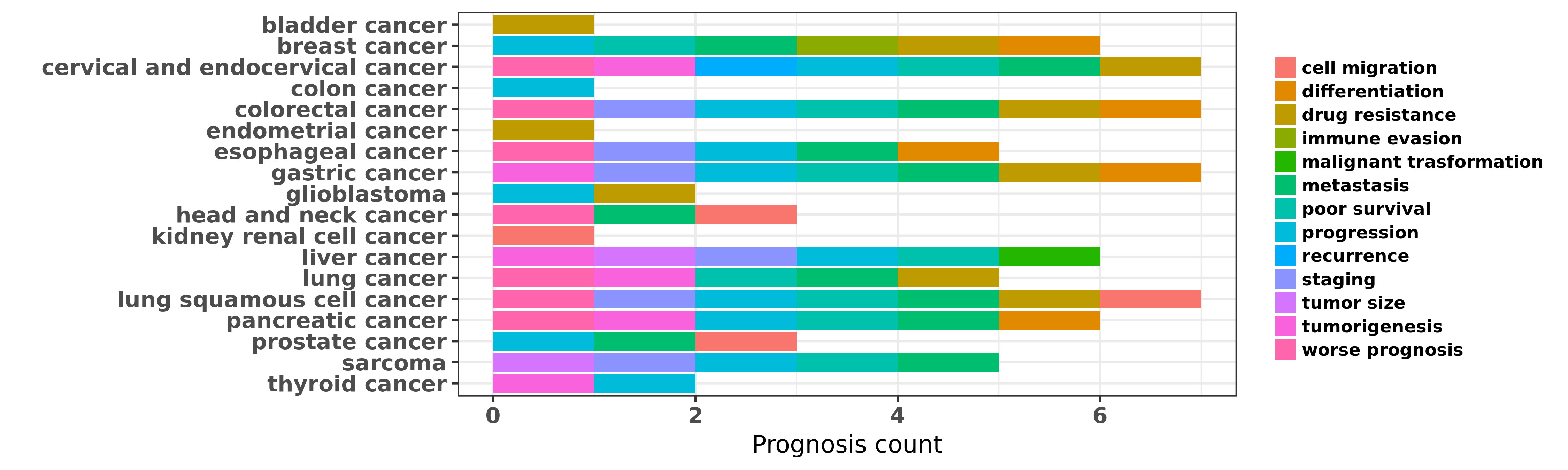
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-218-5p | bladder cancer | drug resistance | "To solve the problem we applied miRNA response ele ......" | 23442927 | Luciferase |
| hsa-miR-218-5p | breast cancer | drug resistance; poor survival | "MiR 218 regulates cisplatin chemosensitivity in br ......" | 25394901 | |
| hsa-miR-218-5p | breast cancer | drug resistance; immune evasion | "miR 218 targets survivin and regulates resistance ......" | 25900794 | Western blot; Flow cytometry; Luciferase; RNAi |
| hsa-miR-218-5p | breast cancer | metastasis; differentiation; progression | "Antagonizing miR 218 5p attenuates Wnt signaling a ......" | 27738322 | |
| hsa-miR-218-5p | cervical and endocervical cancer | metastasis | "MiR 218 impairs tumor growth and increases chemo s ......" | 23443110 | |
| hsa-miR-218-5p | cervical and endocervical cancer | progression; worse prognosis; poor survival; drug resistance | "We previously reported frequent loss of microRNA-2 ......" | 24843318 | |
| hsa-miR-218-5p | cervical and endocervical cancer | poor survival; recurrence | "In the present study global microRNA profiling for ......" | 25473903 | |
| hsa-miR-218-5p | cervical and endocervical cancer | tumorigenesis | "The methylation status was determined using Human ......" | 26797462 | |
| hsa-miR-218-5p | colon cancer | progression | "In this study miR-218 was found to be downregulate ......" | 23255074 | Luciferase |
| hsa-miR-218-5p | colorectal cancer | worse prognosis; staging; metastasis; differentiation; poor survival; progression | "Decreased expression of miR 218 is associated with ......" | 24294377 | |
| hsa-miR-218-5p | colorectal cancer | drug resistance | "To further explore the possible mechanisms we eval ......" | 26442524 | |
| hsa-miR-218-5p | colorectal cancer | metastasis | "MACC1 is post transcriptionally regulated by miR 2 ......" | 27462788 | Luciferase; Colony formation |
| hsa-miR-218-5p | endometrial cancer | drug resistance | "MiR 218 inhibits HMGB1 mediated autophagy in endom ......" | 26261543 | |
| hsa-miR-218-5p | esophageal cancer | worse prognosis | "The impact of pri miR 218 rs11134527 on the risk a ......" | 25337271 | |
| hsa-miR-218-5p | esophageal cancer | metastasis; progression; staging | "miR 218 suppresses tumor growth and enhances the c ......" | 25482044 | Colony formation |
| hsa-miR-218-5p | esophageal cancer | staging; metastasis; progression; differentiation | "The expression of microRNA-218 miR-218 is downregu ......" | 25812647 | |
| hsa-miR-218-5p | esophageal cancer | staging; metastasis | "In the present study we found that the expression ......" | 25999024 | Western blot; Luciferase |
| hsa-miR-218-5p | gastric cancer | drug resistance; tumorigenesis | "Large-scale microarray assays have indicated that ......" | 19890957 | Luciferase |
| hsa-miR-218-5p | gastric cancer | metastasis | "MiR 218 inhibits invasion and metastasis of gastri ......" | 20300657 | |
| hsa-miR-218-5p | gastric cancer | metastasis; tumorigenesis | "Plasma microRNAs miR 223 miR 21 and miR 218 as nov ......" | 22860003 | |
| hsa-miR-218-5p | gastric cancer | metastasis; staging; poor survival | "To investigate the expression of microRNA-218 miR- ......" | 24944481 | |
| hsa-miR-218-5p | gastric cancer | drug resistance | "A miRNA microarray containing human mature and pre ......" | 25170221 | |
| hsa-miR-218-5p | gastric cancer | metastasis | "Thermo chemotherapy Induced miR 218 upregulation i ......" | 25694126 | |
| hsa-miR-218-5p | gastric cancer | drug resistance | "MiR 218 inhibits multidrug resistance MDR of gastr ......" | 26261515 | |
| hsa-miR-218-5p | gastric cancer | progression; staging; metastasis; differentiation; poor survival | "miR 218 tissue expression level is associated with ......" | 27323107 | |
| hsa-miR-218-5p | gastric cancer | progression; tumorigenesis | "miR 218 inhibits proliferation migration and EMT o ......" | 27642088 | Western blot; Luciferase |
| hsa-miR-218-5p | glioblastoma | drug resistance | "miR 218 opposes a critical RTK HIF pathway in mese ......" | 24368849 | |
| hsa-miR-218-5p | glioblastoma | progression | "MiR 218 Inhibited Growth and Metabolism of Human G ......" | 26012781 | Flow cytometry; Western blot; Luciferase |
| hsa-miR-218-5p | head and neck cancer | cell migration | "Recent our microRNA miRNA expression signature rev ......" | 23159910 | Luciferase |
| hsa-miR-218-5p | head and neck cancer | worse prognosis | "Conversely decreased expressions of miR-153 miR-20 ......" | 25677760 | |
| hsa-miR-218-5p | head and neck cancer | metastasis; cell migration | "Tumor suppressive microRNAs miR 26a/b miR 29a/b/c ......" | 26490187 | Luciferase |
| hsa-miR-218-5p | kidney renal cell cancer | cell migration | "Our microRNA expression signature of renal cell ca ......" | 23454155 | Luciferase |
| hsa-miR-218-5p | liver cancer | staging; tumor size; malignant trasformation | "To investigate the expression of microRNA-218 miR- ......" | 23996750 | Western blot; Flow cytometry; MTT assay |
| hsa-miR-218-5p | liver cancer | poor survival | "Prognostic significance of miR 218 in human hepato ......" | 25110121 | |
| hsa-miR-218-5p | liver cancer | tumorigenesis | "miR 218 modulate hepatocellular carcinoma cell pro ......" | 25120782 | Western blot |
| hsa-miR-218-5p | liver cancer | progression | "In the present study miR-218 and miR-520a were obs ......" | 25816091 | Luciferase |
| hsa-miR-218-5p | lung cancer | drug resistance; tumorigenesis | "MiR 205 and miR 218 expression is associated with ......" | 25917317 | |
| hsa-miR-218-5p | lung cancer | drug resistance | "miR 218 suppressed the growth of lung carcinoma by ......" | 26409449 | Luciferase |
| hsa-miR-218-5p | lung cancer | metastasis; poor survival; tumorigenesis | "ADAM9 enhances CDCP1 protein expression by suppres ......" | 26553452 | Luciferase |
| hsa-miR-218-5p | lung cancer | worse prognosis | "In this study the miRNAs were detected in normal a ......" | 27247934 | |
| hsa-miR-218-5p | lung squamous cell cancer | progression; metastasis; poor survival | "Paxillin PXN gene mutations are associated with lu ......" | 21159652 | Colony formation |
| hsa-miR-218-5p | lung squamous cell cancer | cell migration | "The aim of this study is to investigate the potent ......" | 24247270 | Western blot |
| hsa-miR-218-5p | lung squamous cell cancer | progression; staging | "With the use of next-generation deep sequencing te ......" | 25813410 | |
| hsa-miR-218-5p | lung squamous cell cancer | drug resistance | "Downregulation of microRNA-218 miR-218 is found in ......" | 26282001 | MTT assay; Flow cytometry; Western blot; Luciferase |
| hsa-miR-218-5p | lung squamous cell cancer | staging; drug resistance | "Testing group: Quantitative reverse transcriptase ......" | 26563758 | |
| hsa-miR-218-5p | lung squamous cell cancer | worse prognosis | "In the present study we investigated whether singl ......" | 26632718 | |
| hsa-miR-218-5p | lung squamous cell cancer | cell migration | "Downregulation of microRNA-218 miR-218 was frequen ......" | 27633630 | Luciferase |
| hsa-miR-218-5p | pancreatic cancer | progression; tumorigenesis; metastasis; differentiation; poor survival; worse prognosis | "microRNA-218 miR-218 is a vertebrate-specific miRN ......" | 26662432 | |
| hsa-miR-218-5p | prostate cancer | metastasis | "Change in expression of miR let7c miR 100 and miR ......" | 19372056 | |
| hsa-miR-218-5p | prostate cancer | metastasis; cell migration | "Our recent studies of the microRNA miRNA expressio ......" | 24815849 | Luciferase |
| hsa-miR-218-5p | prostate cancer | progression | "MiR 218 impedes IL 6 induced prostate cancer cell ......" | 26986507 | |
| hsa-miR-218-5p | sarcoma | metastasis | "Upon transfection with a miR-218 expression vector ......" | 23886165 | Western blot |
| hsa-miR-218-5p | sarcoma | staging; metastasis; tumor size; progression | "miR 218 expression in osteosarcoma tissues and its ......" | 25479631 | Flow cytometry; Cell proliferation assay; Cell Proliferation Assay |
| hsa-miR-218-5p | sarcoma | poor survival; staging; metastasis | "Up regulation of miR 130b expression level and dow ......" | 26446495 | |
| hsa-miR-218-5p | thyroid cancer | progression; tumorigenesis | "Down regulation of miR 218 2 and its host gene SLI ......" | 23720784 |
Reported gene related to hsa-miR-218-5p
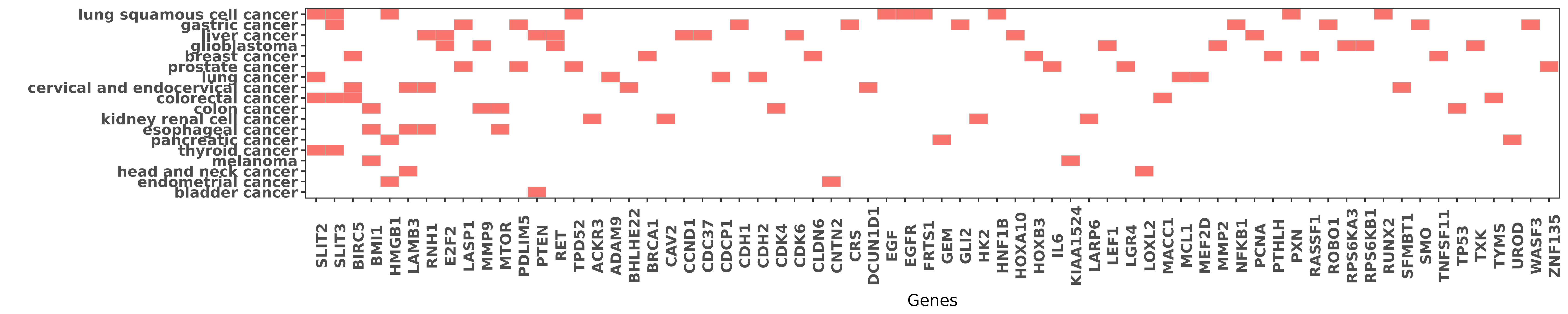
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-218-5p | cervical and endocervical cancer | LAMB3 | "Polymorphisms involved in the miR 218 LAMB3 pathwa ......" | 20163849 |
| hsa-miR-218-5p | cervical and endocervical cancer | LAMB3 | "However this association was not observed for the ......" | 23320911 |
| hsa-miR-218-5p | cervical and endocervical cancer | LAMB3 | "We also demonstrate that the epithelial cell-speci ......" | 17998940 |
| hsa-miR-218-5p | esophageal cancer | LAMB3 | "The aim of this study was to investigate whether p ......" | 25337271 |
| hsa-miR-218-5p | head and neck cancer | LAMB3 | "Furthermore we focused on LAMB3 which has a miR-21 ......" | 23159910 |
| hsa-miR-218-5p | colorectal cancer | SLIT2 | "The expression levels of miR-218 SLIT2 and SLIT3 w ......" | 24294377 |
| hsa-miR-218-5p | colorectal cancer | SLIT2 | "Further we performed methylation-specific PCR of t ......" | 27462788 |
| hsa-miR-218-5p | lung cancer | SLIT2 | "MiR-218 was generated from pri-mir-218-1 which is ......" | 24705471 |
| hsa-miR-218-5p | lung squamous cell cancer | SLIT2 | "MiR-218 encoded on 4p15.31 and 5q35.1 within two h ......" | 20838434 |
| hsa-miR-218-5p | thyroid cancer | SLIT2 | "The aim of our work was to investigate the express ......" | 23720784 |
| hsa-miR-218-5p | colorectal cancer | SLIT3 | "The expression levels of miR-218 SLIT2 and SLIT3 w ......" | 24294377 |
| hsa-miR-218-5p | colorectal cancer | SLIT3 | "Further we performed methylation-specific PCR of t ......" | 27462788 |
| hsa-miR-218-5p | gastric cancer | SLIT3 | "miR-218 expression is decreased along with the exp ......" | 20300657 |
| hsa-miR-218-5p | lung squamous cell cancer | SLIT3 | "MiR-218 encoded on 4p15.31 and 5q35.1 within two h ......" | 20838434 |
| hsa-miR-218-5p | thyroid cancer | SLIT3 | "Down regulation of miR 218 2 and its host gene SLI ......" | 23720784 |
| hsa-miR-218-5p | endometrial cancer | HMGB1 | "MiR 218 inhibits HMGB1 mediated autophagy in endom ......" | 26261543 |
| hsa-miR-218-5p | lung squamous cell cancer | HMGB1 | "Enhanced green fluorescent protein reporter assay ......" | 24247270 |
| hsa-miR-218-5p | pancreatic cancer | HMGB1 | "miR-218 promoted the sensitivity of PANC-1 cells t ......" | 26157323 |
| hsa-miR-218-5p | pancreatic cancer | HMGB1 | "To detect the expression profiles of microRNA-218 ......" | 26157321 |
| hsa-miR-218-5p | breast cancer | BIRC5 | "miR-218 binds survivin BIRC5 mRNA 3'-UTR and down- ......" | 25900794 |
| hsa-miR-218-5p | cervical and endocervical cancer | BIRC5 | "Survivin BIRC5 was subsequently identified as an i ......" | 25473903 |
| hsa-miR-218-5p | colorectal cancer | BIRC5 | "MiR-218 promoted apoptosis inhibited cell prolifer ......" | 26442524 |
| hsa-miR-218-5p | colon cancer | BMI1 | "Potential target genes of miR-218 were predicted a ......" | 23255074 |
| hsa-miR-218-5p | esophageal cancer | BMI1 | "Western blot analysis and luciferase reporter assa ......" | 25999024 |
| hsa-miR-218-5p | melanoma | BMI1 | "We then used the Dual-Luciferase assay system quan ......" | 24839010 |
| hsa-miR-218-5p | cervical and endocervical cancer | RNH1 | "A pri miR 218 variant and risk of cervical carcino ......" | 23320911 |
| hsa-miR-218-5p | esophageal cancer | RNH1 | "The impact of pri miR 218 rs11134527 on the risk a ......" | 25337271 |
| hsa-miR-218-5p | liver cancer | RNH1 | "Association between pri miR 218 polymorphism and r ......" | 22011248 |
| hsa-miR-218-5p | lung cancer | ADAM9 | "ADAM9 up regulates N cadherin via miR 218 suppress ......" | 24705471 |
| hsa-miR-218-5p | lung cancer | ADAM9 | "ADAM9 enhances CDCP1 protein expression by suppres ......" | 26553452 |
| hsa-miR-218-5p | glioblastoma | E2F2 | "MiR 218 Inhibited Growth and Metabolism of Human G ......" | 26012781 |
| hsa-miR-218-5p | liver cancer | E2F2 | "Furthermore a dual-luciferase reporter assay ident ......" | 25816091 |
| hsa-miR-218-5p | lung squamous cell cancer | EGFR | "Higher expression levels of miR-21 AmiR-27a and mi ......" | 26563758 |
| hsa-miR-218-5p | lung squamous cell cancer | EGFR | "Tumor suppressive miR 218 5p inhibits cancer cell ......" | 27057632 |
| hsa-miR-218-5p | gastric cancer | LASP1 | "The present study aims to investigate the effects ......" | 27696291 |
| hsa-miR-218-5p | prostate cancer | LASP1 | "Gene expression data and in silico analysis demon ......" | 24815849 |
| hsa-miR-218-5p | colon cancer | MMP9 | "The upregulation of miR-218 inhibited the activati ......" | 25760926 |
| hsa-miR-218-5p | glioblastoma | MMP9 | "Importantly western blot assays demonstrated that ......" | 22766851 |
| hsa-miR-218-5p | colon cancer | MTOR | "In the present study the expression status of miR- ......" | 25760926 |
| hsa-miR-218-5p | esophageal cancer | MTOR | "Importantly this study also showed that miR-218 re ......" | 25482044 |
| hsa-miR-218-5p | gastric cancer | PDLIM5 | "The present study aims to investigate the effects ......" | 27696291 |
| hsa-miR-218-5p | prostate cancer | PDLIM5 | "Gene expression data and in silico analysis demon ......" | 24815849 |
| hsa-miR-218-5p | bladder cancer | PTEN | "In addition we found that miR-218 regulated the ex ......" | 25967457 |
| hsa-miR-218-5p | liver cancer | PTEN | "Our results indicate that miR-218 modulates HCC de ......" | 25374061 |
| hsa-miR-218-5p | glioblastoma | RET | "Importantly miR-218 targets multiple components of ......" | 24368849 |
| hsa-miR-218-5p | liver cancer | RET | "Overexpression of miR 218 inhibits hepatocellular ......" | 25374061 |
| hsa-miR-218-5p | lung squamous cell cancer | TPD52 | "Moreover direct binding of miR-218 to the 3'-UTR o ......" | 27633630 |
| hsa-miR-218-5p | prostate cancer | TPD52 | "This study tried to explore the regulative role of ......" | 25511701 |
| hsa-miR-218-5p | kidney renal cell cancer | ACKR3 | "Overexpression of miR-218 mimic resulted in signif ......" | 27133059 |
| hsa-miR-218-5p | cervical and endocervical cancer | BHLHE22 | "MicroRNA-218 miR-218 can target laminin-5 beta3 LA ......" | 20163849 |
| hsa-miR-218-5p | breast cancer | BRCA1 | "MiR 218 regulates cisplatin chemosensitivity in br ......" | 25394901 |
| hsa-miR-218-5p | kidney renal cell cancer | CAV2 | "Gene expression studies and luciferase reporter as ......" | 23454155 |
| hsa-miR-218-5p | liver cancer | CCND1 | "Western blotting analysis demonstrated that tumori ......" | 25120782 |
| hsa-miR-218-5p | liver cancer | CDC37 | "Restoring miR-218 expression resulted in downregul ......" | 25110121 |
| hsa-miR-218-5p | lung cancer | CDCP1 | "ADAM9 enhances CDCP1 protein expression by suppres ......" | 26553452 |
| hsa-miR-218-5p | gastric cancer | CDH1 | "Thermo chemotherapy Induced miR 218 upregulation i ......" | 25694126 |
| hsa-miR-218-5p | lung cancer | CDH2 | "ADAM9 up regulates N cadherin via miR 218 suppress ......" | 24705471 |
| hsa-miR-218-5p | colon cancer | CDK4 | "In addition miR-218 suppressed protein expression ......" | 23255074 |
| hsa-miR-218-5p | liver cancer | CDK6 | "Ectopic expression of miR-218 in HepG2 cells resul ......" | 23996750 |
| hsa-miR-218-5p | breast cancer | CLDN6 | "Stable overexpression of miR-7 and miR-218 was acc ......" | 22705304 |
| hsa-miR-218-5p | endometrial cancer | CNTN2 | "HMGB1-mediated autophagy could be suppressed by mi ......" | 26261543 |
| hsa-miR-218-5p | gastric cancer | CRS | "miRNA microarray chip analysis found that the leve ......" | 25170221 |
| hsa-miR-218-5p | cervical and endocervical cancer | DCUN1D1 | "An inverse correlation was observed between the ex ......" | 27285984 |
| hsa-miR-218-5p | lung squamous cell cancer | EGF | "In this study we identified a specific binding sit ......" | 27057632 |
| hsa-miR-218-5p | lung squamous cell cancer | FRTS1 | "The prognostic value of miR-218 and PXN expression ......" | 21159652 |
| hsa-miR-218-5p | pancreatic cancer | GEM | "The purpose of this study was to examine the effec ......" | 26157323 |
| hsa-miR-218-5p | gastric cancer | GLI2 | "Thermo chemotherapy Induced miR 218 upregulation i ......" | 25694126 |
| hsa-miR-218-5p | kidney renal cell cancer | HK2 | "The expression of miR-218 in renal cell carcinoma ......" | 27133059 |
| hsa-miR-218-5p | lung squamous cell cancer | HNF1B | "Luciferase reporter assays and Western blots were ......" | 26282001 |
| hsa-miR-218-5p | liver cancer | HOXA10 | "miR 218 modulate hepatocellular carcinoma cell pro ......" | 25120782 |
| hsa-miR-218-5p | breast cancer | HOXB3 | "In this study we found that the expression levels ......" | 22705304 |
| hsa-miR-218-5p | prostate cancer | IL6 | "MiR 218 impedes IL 6 induced prostate cancer cell ......" | 26986507 |
| hsa-miR-218-5p | melanoma | KIAA1524 | "We then used the Dual-Luciferase assay system quan ......" | 24839010 |
| hsa-miR-218-5p | kidney renal cell cancer | LARP6 | "MiR-218 mimic and NC were transfected into renal c ......" | 27133059 |
| hsa-miR-218-5p | glioblastoma | LEF1 | "MiR 218 reverses high invasiveness of glioblastoma ......" | 22766851 |
| hsa-miR-218-5p | prostate cancer | LGR4 | "MiR 218 impedes IL 6 induced prostate cancer cell ......" | 26986507 |
| hsa-miR-218-5p | head and neck cancer | LOXL2 | "Tumor suppressive microRNAs miR 26a/b miR 29a/b/c ......" | 26490187 |
| hsa-miR-218-5p | colorectal cancer | MACC1 | "MACC1 is post transcriptionally regulated by miR 2 ......" | 27462788 |
| hsa-miR-218-5p | lung cancer | MCL1 | "MiR 205 and miR 218 expression is associated with ......" | 25917317 |
| hsa-miR-218-5p | lung cancer | MEF2D | "miR 218 suppressed the growth of lung carcinoma by ......" | 26409449 |
| hsa-miR-218-5p | glioblastoma | MMP2 | "The data showed an inverse correlation in 60 GBM t ......" | 22766851 |
| hsa-miR-218-5p | gastric cancer | NFKB1 | "Overexpression of miR-218 also inhibited NF-kappaB ......" | 19890957 |
| hsa-miR-218-5p | liver cancer | PCNA | "Western blotting analysis demonstrated that tumori ......" | 25120782 |
| hsa-miR-218-5p | breast cancer | PTHLH | "Furthermore miR-218-5p also mediates the Wnt-depen ......" | 27738322 |
| hsa-miR-218-5p | lung squamous cell cancer | PXN | "On this basis we hypothesized that PXN overexpress ......" | 21159652 |
| hsa-miR-218-5p | breast cancer | RASSF1 | "Stable overexpression of miR-7 and miR-218 was acc ......" | 22705304 |
| hsa-miR-218-5p | gastric cancer | ROBO1 | "MiR 218 inhibits invasion and metastasis of gastri ......" | 20300657 |
| hsa-miR-218-5p | glioblastoma | RPS6KA3 | "We found that decreased miR-218 expression in GBM ......" | 25943352 |
| hsa-miR-218-5p | glioblastoma | RPS6KB1 | "We found that decreased miR-218 expression in GBM ......" | 25943352 |
| hsa-miR-218-5p | lung squamous cell cancer | RUNX2 | "Luciferase reporter assays and Western blots were ......" | 26282001 |
| hsa-miR-218-5p | cervical and endocervical cancer | SFMBT1 | "Importantly HPV16 E6 downregulated the expression ......" | 27285984 |
| hsa-miR-218-5p | gastric cancer | SMO | "We identified the smoothened SMO gene as a functio ......" | 26261515 |
| hsa-miR-218-5p | breast cancer | TNFSF11 | "Antagonizing miR-218-5p reduced the expression of ......" | 27738322 |
| hsa-miR-218-5p | colon cancer | TP53 | "In addition miR-218 suppressed protein expression ......" | 23255074 |
| hsa-miR-218-5p | glioblastoma | TXK | "Importantly miR-218 targets multiple components of ......" | 24368849 |
| hsa-miR-218-5p | colorectal cancer | TYMS | "Furthermore miR-218 enhanced 5-FU cytotoxicity in ......" | 26442524 |
| hsa-miR-218-5p | pancreatic cancer | UROD | "To detect the expression profiles of microRNA-218 ......" | 26157321 |
| hsa-miR-218-5p | gastric cancer | WASF3 | "miR 218 inhibits proliferation migration and EMT o ......" | 27642088 |
| hsa-miR-218-5p | prostate cancer | ZNF135 | "Expression analysis of 14 miRNAs was carried out u ......" | 19372056 |
Expression profile in cancer corhorts:
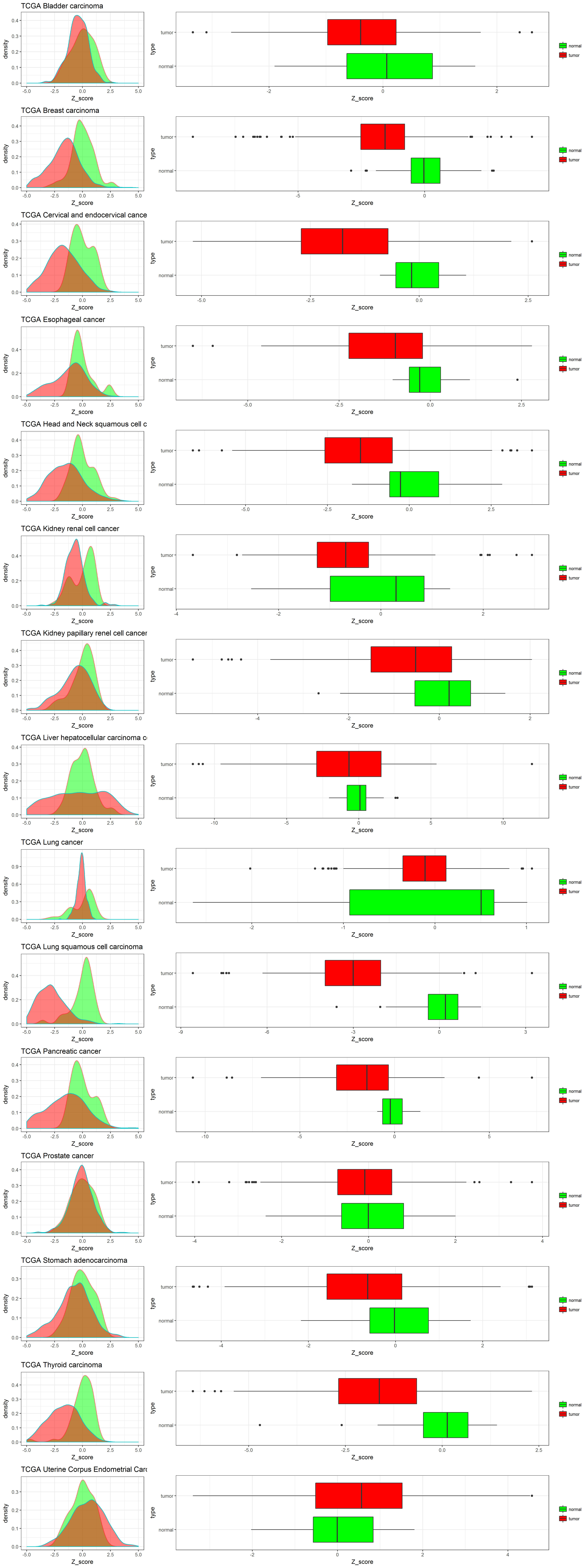
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-218-5p | EBP | 10 cancers: BLCA; BRCA; COAD; ESCA; LIHC; LUAD; LUSC; PAAD; THCA; STAD | miRNAWalker2 validate | TCGA BLCA -0.06; TCGA BRCA -0.194; TCGA COAD -0.152; TCGA ESCA -0.152; TCGA LIHC -0.22; TCGA LUAD -0.104; TCGA LUSC -0.156; TCGA PAAD -0.16; TCGA THCA -0.06; TCGA STAD -0.164 |
hsa-miR-218-5p | EFNA1 | 11 cancers: BLCA; BRCA; CESC; ESCA; KIRC; LIHC; LUAD; OV; PAAD; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.21; TCGA BRCA -0.223; TCGA CESC -0.262; TCGA ESCA -0.099; TCGA KIRC -0.168; TCGA LIHC -0.089; TCGA LUAD -0.134; TCGA OV -0.12; TCGA PAAD -0.291; TCGA STAD -0.175; TCGA UCEC -0.176 |
hsa-miR-218-5p | FOXN2 | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; LGG; LUAD; PRAD; STAD | miRNAWalker2 validate | TCGA BLCA -0.122; TCGA CESC -0.057; TCGA COAD -0.085; TCGA ESCA -0.093; TCGA HNSC -0.18; TCGA LGG -0.057; TCGA LUAD -0.063; TCGA PRAD -0.181; TCGA STAD -0.191 |
hsa-miR-218-5p | SBNO1 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; PRAD; SARC; STAD | miRNAWalker2 validate | TCGA BLCA -0.198; TCGA BRCA -0.134; TCGA CESC -0.132; TCGA COAD -0.142; TCGA ESCA -0.069; TCGA HNSC -0.142; TCGA KIRP -0.141; TCGA LUAD -0.152; TCGA LUSC -0.106; TCGA PRAD -0.284; TCGA SARC -0.155; TCGA STAD -0.139 |
hsa-miR-218-5p | SERINC3 | 9 cancers: BLCA; ESCA; HNSC; KIRP; LIHC; PAAD; PRAD; THCA; STAD | miRNAWalker2 validate | TCGA BLCA -0.12; TCGA ESCA -0.057; TCGA HNSC -0.144; TCGA KIRP -0.074; TCGA LIHC -0.051; TCGA PAAD -0.104; TCGA PRAD -0.144; TCGA THCA -0.114; TCGA STAD -0.063 |
hsa-miR-218-5p | SLC11A2 | 9 cancers: BLCA; BRCA; COAD; ESCA; LIHC; LUAD; PRAD; THCA; STAD | miRNAWalker2 validate | TCGA BLCA -0.075; TCGA BRCA -0.11; TCGA COAD -0.191; TCGA ESCA -0.121; TCGA LIHC -0.057; TCGA LUAD -0.057; TCGA PRAD -0.22; TCGA THCA -0.118; TCGA STAD -0.19 |
hsa-miR-218-5p | SLC38A1 | 10 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; LUAD; LUSC; SARC; THCA | miRNAWalker2 validate | TCGA BLCA -0.089; TCGA BRCA -0.205; TCGA CESC -0.079; TCGA COAD -0.116; TCGA HNSC -0.139; TCGA KIRC -0.196; TCGA LUAD -0.108; TCGA LUSC -0.217; TCGA SARC -0.412; TCGA THCA -0.096 |
hsa-miR-218-5p | SP1 | 10 cancers: BLCA; CESC; COAD; KIRC; LGG; LUAD; PAAD; PRAD; STAD; UCEC | miRNAWalker2 validate; miRTarBase | TCGA BLCA -0.08; TCGA CESC -0.059; TCGA COAD -0.099; TCGA KIRC -0.073; TCGA LGG -0.084; TCGA LUAD -0.055; TCGA PAAD -0.082; TCGA PRAD -0.116; TCGA STAD -0.102; TCGA UCEC -0.055 |
hsa-miR-218-5p | SSR1 | 12 cancers: BLCA; BRCA; COAD; HNSC; LGG; LIHC; LUAD; LUSC; OV; PRAD; THCA; STAD | miRNAWalker2 validate; miRNATAP | TCGA BLCA -0.112; TCGA BRCA -0.103; TCGA COAD -0.146; TCGA HNSC -0.105; TCGA LGG -0.067; TCGA LIHC -0.052; TCGA LUAD -0.097; TCGA LUSC -0.148; TCGA OV -0.057; TCGA PRAD -0.125; TCGA THCA -0.076; TCGA STAD -0.129 |
hsa-miR-218-5p | THOC2 | 9 cancers: BLCA; BRCA; COAD; ESCA; LGG; LUAD; PRAD; SARC; STAD | miRNAWalker2 validate | TCGA BLCA -0.095; TCGA BRCA -0.096; TCGA COAD -0.108; TCGA ESCA -0.132; TCGA LGG -0.064; TCGA LUAD -0.064; TCGA PRAD -0.145; TCGA SARC -0.063; TCGA STAD -0.144 |
hsa-miR-218-5p | TPD52 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRP; LUAD; LUSC; OV; PAAD; PRAD; SARC; THCA; STAD | miRNAWalker2 validate; MirTarget | TCGA BLCA -0.068; TCGA BRCA -0.507; TCGA CESC -0.081; TCGA COAD -0.237; TCGA ESCA -0.377; TCGA KIRP -0.161; TCGA LUAD -0.237; TCGA LUSC -0.222; TCGA OV -0.109; TCGA PAAD -0.164; TCGA PRAD -0.283; TCGA SARC -0.128; TCGA THCA -0.062; TCGA STAD -0.203 |
hsa-miR-218-5p | WIPF2 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; PAAD; PRAD; SARC; STAD | MirTarget | TCGA BLCA -0.126; TCGA BRCA -0.088; TCGA CESC -0.099; TCGA COAD -0.085; TCGA ESCA -0.219; TCGA HNSC -0.135; TCGA LUAD -0.096; TCGA PAAD -0.096; TCGA PRAD -0.144; TCGA SARC -0.059; TCGA STAD -0.1 |
hsa-miR-218-5p | PPP2R5A | 9 cancers: BLCA; BRCA; CESC; COAD; LIHC; OV; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.071; TCGA BRCA -0.094; TCGA CESC -0.051; TCGA COAD -0.07; TCGA LIHC -0.099; TCGA OV -0.074; TCGA SARC -0.187; TCGA STAD -0.07; TCGA UCEC -0.113 |
hsa-miR-218-5p | CCNA2 | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LGG; LUAD; LUSC; PAAD; PRAD; STAD | MirTarget | TCGA BLCA -0.145; TCGA BRCA -0.488; TCGA COAD -0.289; TCGA ESCA -0.159; TCGA HNSC -0.141; TCGA KIRC -0.148; TCGA LGG -0.139; TCGA LUAD -0.386; TCGA LUSC -0.691; TCGA PAAD -0.264; TCGA PRAD -0.297; TCGA STAD -0.416 |
hsa-miR-218-5p | PLEKHF2 | 10 cancers: BLCA; BRCA; CESC; COAD; HNSC; LGG; LUSC; OV; PRAD; STAD | MirTarget | TCGA BLCA -0.071; TCGA BRCA -0.289; TCGA CESC -0.093; TCGA COAD -0.095; TCGA HNSC -0.152; TCGA LGG -0.066; TCGA LUSC -0.062; TCGA OV -0.151; TCGA PRAD -0.22; TCGA STAD -0.09 |
hsa-miR-218-5p | GALNT3 | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUSC; PAAD; PRAD; THCA; STAD | MirTarget; miRNATAP | TCGA BLCA -0.274; TCGA CESC -0.162; TCGA COAD -0.181; TCGA ESCA -0.44; TCGA HNSC -0.125; TCGA LUSC -0.176; TCGA PAAD -0.269; TCGA PRAD -0.209; TCGA THCA -0.197; TCGA STAD -0.405 |
hsa-miR-218-5p | MRPL19 | 10 cancers: BLCA; BRCA; COAD; ESCA; LUAD; LUSC; PAAD; PRAD; THCA; STAD | MirTarget | TCGA BLCA -0.068; TCGA BRCA -0.157; TCGA COAD -0.11; TCGA ESCA -0.084; TCGA LUAD -0.143; TCGA LUSC -0.151; TCGA PAAD -0.088; TCGA PRAD -0.157; TCGA THCA -0.113; TCGA STAD -0.159 |
hsa-miR-218-5p | ZWILCH | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PRAD; THCA; STAD | MirTarget | TCGA BLCA -0.08; TCGA BRCA -0.233; TCGA CESC -0.091; TCGA COAD -0.215; TCGA ESCA -0.186; TCGA HNSC -0.062; TCGA LUAD -0.174; TCGA LUSC -0.295; TCGA PRAD -0.238; TCGA THCA -0.077; TCGA STAD -0.295 |
hsa-miR-218-5p | RNF139 | 9 cancers: BLCA; BRCA; CESC; COAD; HNSC; LUAD; LUSC; OV; PRAD | MirTarget | TCGA BLCA -0.054; TCGA BRCA -0.143; TCGA CESC -0.087; TCGA COAD -0.063; TCGA HNSC -0.072; TCGA LUAD -0.051; TCGA LUSC -0.082; TCGA OV -0.12; TCGA PRAD -0.145 |
hsa-miR-218-5p | FANCI | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; KIRP; LGG; LUAD; LUSC; PAAD; PRAD; THCA; STAD | MirTarget | TCGA BLCA -0.072; TCGA BRCA -0.347; TCGA CESC -0.075; TCGA COAD -0.226; TCGA ESCA -0.12; TCGA KIRC -0.205; TCGA KIRP -0.087; TCGA LGG -0.068; TCGA LUAD -0.283; TCGA LUSC -0.432; TCGA PAAD -0.151; TCGA PRAD -0.214; TCGA THCA -0.3; TCGA STAD -0.262 |
hsa-miR-218-5p | UEVLD | 13 cancers: BLCA; BRCA; ESCA; HNSC; KIRP; LUAD; OV; PAAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.148; TCGA BRCA -0.123; TCGA ESCA -0.07; TCGA HNSC -0.159; TCGA KIRP -0.076; TCGA LUAD -0.067; TCGA OV -0.071; TCGA PAAD -0.157; TCGA PRAD -0.118; TCGA SARC -0.065; TCGA THCA -0.068; TCGA STAD -0.139; TCGA UCEC -0.056 |
hsa-miR-218-5p | FBXO28 | 13 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.075; TCGA BRCA -0.162; TCGA COAD -0.081; TCGA ESCA -0.071; TCGA HNSC -0.064; TCGA LUAD -0.051; TCGA LUSC -0.062; TCGA PAAD -0.133; TCGA PRAD -0.152; TCGA SARC -0.109; TCGA THCA -0.055; TCGA STAD -0.149; TCGA UCEC -0.076 |
hsa-miR-218-5p | ARF6 | 10 cancers: BLCA; BRCA; CESC; COAD; HNSC; LUSC; OV; PRAD; THCA; STAD | MirTarget | TCGA BLCA -0.07; TCGA BRCA -0.105; TCGA CESC -0.064; TCGA COAD -0.108; TCGA HNSC -0.152; TCGA LUSC -0.163; TCGA OV -0.058; TCGA PRAD -0.195; TCGA THCA -0.073; TCGA STAD -0.115 |
hsa-miR-218-5p | EDEM1 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LIHC; OV; PRAD; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.105; TCGA BRCA -0.068; TCGA CESC -0.142; TCGA COAD -0.065; TCGA ESCA -0.086; TCGA HNSC -0.071; TCGA KIRP -0.056; TCGA LIHC -0.062; TCGA OV -0.072; TCGA PRAD -0.19; TCGA THCA -0.122; TCGA STAD -0.145; TCGA UCEC -0.1 |
hsa-miR-218-5p | POLA1 | 10 cancers: BLCA; BRCA; COAD; KIRP; LGG; LUAD; LUSC; PRAD; SARC; STAD | MirTarget | TCGA BLCA -0.122; TCGA BRCA -0.134; TCGA COAD -0.108; TCGA KIRP -0.084; TCGA LGG -0.089; TCGA LUAD -0.085; TCGA LUSC -0.134; TCGA PRAD -0.08; TCGA SARC -0.13; TCGA STAD -0.158 |
hsa-miR-218-5p | SERBP1 | 10 cancers: BLCA; COAD; HNSC; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; STAD | MirTarget | TCGA BLCA -0.074; TCGA COAD -0.127; TCGA HNSC -0.11; TCGA LGG -0.058; TCGA LIHC -0.071; TCGA LUAD -0.066; TCGA LUSC -0.164; TCGA PAAD -0.054; TCGA PRAD -0.092; TCGA STAD -0.133 |
hsa-miR-218-5p | ZNF468 | 9 cancers: BLCA; BRCA; COAD; ESCA; LUAD; OV; PRAD; THCA; STAD | MirTarget | TCGA BLCA -0.084; TCGA BRCA -0.168; TCGA COAD -0.169; TCGA ESCA -0.255; TCGA LUAD -0.09; TCGA OV -0.071; TCGA PRAD -0.182; TCGA THCA -0.141; TCGA STAD -0.277 |
hsa-miR-218-5p | RBM47 | 9 cancers: BLCA; BRCA; COAD; ESCA; LGG; LUAD; PAAD; PRAD; STAD | MirTarget | TCGA BLCA -0.083; TCGA BRCA -0.186; TCGA COAD -0.195; TCGA ESCA -0.51; TCGA LGG -0.135; TCGA LUAD -0.113; TCGA PAAD -0.282; TCGA PRAD -0.188; TCGA STAD -0.344 |
hsa-miR-218-5p | BRCC3 | 13 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; OV; PAAD; PRAD; THCA; STAD | MirTarget; miRNATAP | TCGA BLCA -0.119; TCGA BRCA -0.177; TCGA COAD -0.095; TCGA ESCA -0.197; TCGA HNSC -0.062; TCGA KIRP -0.059; TCGA LUAD -0.078; TCGA LUSC -0.113; TCGA OV -0.085; TCGA PAAD -0.074; TCGA PRAD -0.126; TCGA THCA -0.066; TCGA STAD -0.214 |
hsa-miR-218-5p | DCAF10 | 11 cancers: BLCA; BRCA; CESC; HNSC; LIHC; LUAD; PAAD; PRAD; SARC; THCA; STAD | MirTarget | TCGA BLCA -0.084; TCGA BRCA -0.18; TCGA CESC -0.071; TCGA HNSC -0.076; TCGA LIHC -0.065; TCGA LUAD -0.099; TCGA PAAD -0.065; TCGA PRAD -0.179; TCGA SARC -0.053; TCGA THCA -0.063; TCGA STAD -0.073 |
hsa-miR-218-5p | GHITM | 11 cancers: BLCA; BRCA; COAD; ESCA; LIHC; LUAD; PAAD; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.086; TCGA BRCA -0.135; TCGA COAD -0.14; TCGA ESCA -0.07; TCGA LIHC -0.055; TCGA LUAD -0.051; TCGA PAAD -0.067; TCGA PRAD -0.06; TCGA THCA -0.161; TCGA STAD -0.065; TCGA UCEC -0.059 |
hsa-miR-218-5p | TMX1 | 10 cancers: BLCA; BRCA; COAD; HNSC; LGG; LUAD; LUSC; OV; PRAD; STAD | MirTarget; miRNATAP | TCGA BLCA -0.09; TCGA BRCA -0.066; TCGA COAD -0.126; TCGA HNSC -0.107; TCGA LGG -0.128; TCGA LUAD -0.061; TCGA LUSC -0.171; TCGA OV -0.057; TCGA PRAD -0.135; TCGA STAD -0.161 |
hsa-miR-218-5p | ARPP19 | 11 cancers: BLCA; BRCA; CESC; COAD; HNSC; LUAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.117; TCGA BRCA -0.066; TCGA CESC -0.057; TCGA COAD -0.124; TCGA HNSC -0.057; TCGA LUAD -0.056; TCGA PRAD -0.083; TCGA SARC -0.149; TCGA THCA -0.087; TCGA STAD -0.067; TCGA UCEC -0.053 |
hsa-miR-218-5p | CCDC6 | 9 cancers: BLCA; BRCA; COAD; ESCA; PAAD; PRAD; SARC; THCA; STAD | MirTarget; miRNATAP | TCGA BLCA -0.089; TCGA BRCA -0.127; TCGA COAD -0.076; TCGA ESCA -0.094; TCGA PAAD -0.083; TCGA PRAD -0.336; TCGA SARC -0.214; TCGA THCA -0.118; TCGA STAD -0.211 |
hsa-miR-218-5p | THAP6 | 10 cancers: BLCA; BRCA; COAD; KIRC; LGG; LIHC; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.059; TCGA BRCA -0.084; TCGA COAD -0.096; TCGA KIRC -0.084; TCGA LGG -0.059; TCGA LIHC -0.062; TCGA PRAD -0.118; TCGA SARC -0.081; TCGA STAD -0.058; TCGA UCEC -0.058 |
hsa-miR-218-5p | DCAF12 | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; LGG; LUAD; PRAD; THCA; STAD | MirTarget | TCGA BLCA -0.052; TCGA BRCA -0.132; TCGA CESC -0.053; TCGA COAD -0.069; TCGA ESCA -0.069; TCGA LGG -0.109; TCGA LUAD -0.107; TCGA PRAD -0.16; TCGA THCA -0.099; TCGA STAD -0.179 |
hsa-miR-218-5p | PARP12 | 9 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LGG; LUSC; STAD | MirTarget; miRNATAP | TCGA BLCA -0.14; TCGA BRCA -0.207; TCGA CESC -0.154; TCGA ESCA -0.15; TCGA HNSC -0.208; TCGA KIRC -0.19; TCGA LGG -0.053; TCGA LUSC -0.068; TCGA STAD -0.134 |
hsa-miR-218-5p | UBE2D1 | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRP; LUAD; LUSC; PRAD; STAD | MirTarget | TCGA BLCA -0.075; TCGA BRCA -0.082; TCGA CESC -0.062; TCGA ESCA -0.065; TCGA HNSC -0.056; TCGA KIRP -0.055; TCGA LUAD -0.096; TCGA LUSC -0.133; TCGA PRAD -0.136; TCGA STAD -0.084 |
hsa-miR-218-5p | YY1 | 10 cancers: BLCA; BRCA; COAD; HNSC; LUAD; LUSC; PAAD; PRAD; THCA; STAD | MirTarget | TCGA BLCA -0.099; TCGA BRCA -0.145; TCGA COAD -0.09; TCGA HNSC -0.089; TCGA LUAD -0.13; TCGA LUSC -0.209; TCGA PAAD -0.099; TCGA PRAD -0.163; TCGA THCA -0.065; TCGA STAD -0.097 |
hsa-miR-218-5p | SLC39A1 | 11 cancers: BLCA; BRCA; ESCA; KIRC; LGG; LUAD; LUSC; PAAD; PRAD; THCA; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.055; TCGA BRCA -0.122; TCGA ESCA -0.095; TCGA KIRC -0.142; TCGA LGG -0.095; TCGA LUAD -0.124; TCGA LUSC -0.101; TCGA PAAD -0.097; TCGA PRAD -0.061; TCGA THCA -0.052; TCGA UCEC -0.083 |
hsa-miR-218-5p | DNAJC13 | 10 cancers: BLCA; BRCA; CESC; HNSC; KIRP; LUSC; PRAD; SARC; THCA; STAD | MirTarget; miRNATAP | TCGA BLCA -0.081; TCGA BRCA -0.061; TCGA CESC -0.068; TCGA HNSC -0.113; TCGA KIRP -0.056; TCGA LUSC -0.118; TCGA PRAD -0.089; TCGA SARC -0.071; TCGA THCA -0.108; TCGA STAD -0.082 |
hsa-miR-218-5p | RRP1B | 11 cancers: BLCA; BRCA; CESC; COAD; HNSC; LGG; LIHC; LUAD; LUSC; PRAD; STAD | MirTarget | TCGA BLCA -0.051; TCGA BRCA -0.075; TCGA CESC -0.104; TCGA COAD -0.087; TCGA HNSC -0.062; TCGA LGG -0.052; TCGA LIHC -0.062; TCGA LUAD -0.076; TCGA LUSC -0.154; TCGA PRAD -0.102; TCGA STAD -0.157 |
hsa-miR-218-5p | XPO5 | 13 cancers: BLCA; BRCA; ESCA; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; THCA; STAD | MirTarget | TCGA BLCA -0.055; TCGA BRCA -0.178; TCGA ESCA -0.142; TCGA HNSC -0.075; TCGA KIRP -0.063; TCGA LGG -0.075; TCGA LIHC -0.087; TCGA LUAD -0.133; TCGA LUSC -0.243; TCGA PAAD -0.109; TCGA PRAD -0.187; TCGA THCA -0.102; TCGA STAD -0.225 |
hsa-miR-218-5p | DOLPP1 | 10 cancers: BLCA; BRCA; COAD; ESCA; LIHC; LUAD; LUSC; PAAD; PRAD; STAD | MirTarget; miRNATAP | TCGA BLCA -0.085; TCGA BRCA -0.146; TCGA COAD -0.152; TCGA ESCA -0.195; TCGA LIHC -0.127; TCGA LUAD -0.089; TCGA LUSC -0.177; TCGA PAAD -0.103; TCGA PRAD -0.132; TCGA STAD -0.128 |
hsa-miR-218-5p | PKN2 | 11 cancers: BLCA; COAD; ESCA; HNSC; LGG; LUAD; LUSC; PAAD; PRAD; SARC; STAD | MirTarget | TCGA BLCA -0.126; TCGA COAD -0.133; TCGA ESCA -0.13; TCGA HNSC -0.095; TCGA LGG -0.076; TCGA LUAD -0.074; TCGA LUSC -0.067; TCGA PAAD -0.102; TCGA PRAD -0.128; TCGA SARC -0.06; TCGA STAD -0.209 |
hsa-miR-218-5p | PANX1 | 10 cancers: BLCA; BRCA; CESC; HNSC; KIRP; LIHC; LUAD; LUSC; SARC; THCA | MirTarget | TCGA BLCA -0.113; TCGA BRCA -0.098; TCGA CESC -0.091; TCGA HNSC -0.252; TCGA KIRP -0.069; TCGA LIHC -0.097; TCGA LUAD -0.102; TCGA LUSC -0.191; TCGA SARC -0.056; TCGA THCA -0.137 |
hsa-miR-218-5p | DHTKD1 | 11 cancers: BLCA; BRCA; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.088; TCGA BRCA -0.277; TCGA LGG -0.057; TCGA LIHC -0.156; TCGA LUAD -0.127; TCGA LUSC -0.111; TCGA PAAD -0.072; TCGA PRAD -0.116; TCGA THCA -0.156; TCGA STAD -0.093; TCGA UCEC -0.05 |
hsa-miR-218-5p | KCTD9 | 10 cancers: BLCA; CESC; COAD; HNSC; KIRC; KIRP; SARC; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.151; TCGA CESC -0.108; TCGA COAD -0.184; TCGA HNSC -0.115; TCGA KIRC -0.095; TCGA KIRP -0.058; TCGA SARC -0.14; TCGA THCA -0.126; TCGA STAD -0.141; TCGA UCEC -0.076 |
hsa-miR-218-5p | ADIPOR2 | 9 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRP; LIHC; LUAD; PAAD | MirTarget; miRNATAP | TCGA BLCA -0.107; TCGA BRCA -0.15; TCGA CESC -0.102; TCGA COAD -0.155; TCGA ESCA -0.068; TCGA KIRP -0.059; TCGA LIHC -0.085; TCGA LUAD -0.118; TCGA PAAD -0.154 |
hsa-miR-218-5p | TJP2 | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; LGG; PAAD; SARC; STAD | MirTarget | TCGA BLCA -0.105; TCGA CESC -0.098; TCGA COAD -0.097; TCGA ESCA -0.313; TCGA HNSC -0.188; TCGA LGG -0.13; TCGA PAAD -0.115; TCGA SARC -0.327; TCGA STAD -0.142 |
hsa-miR-218-5p | UBE2H | 9 cancers: BLCA; BRCA; HNSC; KIRP; LUAD; LUSC; PAAD; PRAD; THCA | MirTarget; miRNATAP | TCGA BLCA -0.134; TCGA BRCA -0.156; TCGA HNSC -0.073; TCGA KIRP -0.054; TCGA LUAD -0.132; TCGA LUSC -0.069; TCGA PAAD -0.162; TCGA PRAD -0.127; TCGA THCA -0.061 |
hsa-miR-218-5p | ARL5B | 9 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUAD; PAAD; PRAD; STAD | MirTarget | TCGA BLCA -0.102; TCGA BRCA -0.116; TCGA COAD -0.17; TCGA ESCA -0.194; TCGA HNSC -0.126; TCGA LUAD -0.141; TCGA PAAD -0.177; TCGA PRAD -0.282; TCGA STAD -0.271 |
hsa-miR-218-5p | USP32 | 9 cancers: BLCA; BRCA; CESC; COAD; HNSC; LUAD; PRAD; THCA; STAD | MirTarget | TCGA BLCA -0.109; TCGA BRCA -0.194; TCGA CESC -0.062; TCGA COAD -0.062; TCGA HNSC -0.139; TCGA LUAD -0.151; TCGA PRAD -0.204; TCGA THCA -0.125; TCGA STAD -0.103 |
hsa-miR-218-5p | DCUN1D1 | 11 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRP; LUAD; LUSC; PRAD; SARC; STAD | MirTarget | TCGA BLCA -0.136; TCGA BRCA -0.155; TCGA CESC -0.098; TCGA COAD -0.098; TCGA HNSC -0.104; TCGA KIRP -0.083; TCGA LUAD -0.1; TCGA LUSC -0.196; TCGA PRAD -0.256; TCGA SARC -0.052; TCGA STAD -0.183 |
hsa-miR-218-5p | PHF6 | 10 cancers: BLCA; BRCA; COAD; KIRP; LGG; LUAD; LUSC; PAAD; PRAD; STAD | MirTarget; miRNATAP | TCGA BLCA -0.109; TCGA BRCA -0.149; TCGA COAD -0.109; TCGA KIRP -0.076; TCGA LGG -0.067; TCGA LUAD -0.079; TCGA LUSC -0.145; TCGA PAAD -0.077; TCGA PRAD -0.192; TCGA STAD -0.177 |
hsa-miR-218-5p | ZBTB11 | 9 cancers: BLCA; BRCA; CESC; COAD; HNSC; LUAD; LUSC; PRAD; STAD | MirTarget; miRNATAP | TCGA BLCA -0.103; TCGA BRCA -0.079; TCGA CESC -0.077; TCGA COAD -0.074; TCGA HNSC -0.098; TCGA LUAD -0.065; TCGA LUSC -0.096; TCGA PRAD -0.199; TCGA STAD -0.112 |
hsa-miR-218-5p | KPNA4 | 12 cancers: BLCA; BRCA; CESC; COAD; HNSC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD | MirTarget; miRNATAP | TCGA BLCA -0.087; TCGA BRCA -0.127; TCGA CESC -0.085; TCGA COAD -0.091; TCGA HNSC -0.09; TCGA LUAD -0.104; TCGA LUSC -0.195; TCGA PAAD -0.118; TCGA PRAD -0.056; TCGA SARC -0.054; TCGA THCA -0.097; TCGA STAD -0.083 |
hsa-miR-218-5p | SFMBT1 | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; LIHC; PAAD; PRAD; SARC; STAD | MirTarget; miRNATAP | TCGA BLCA -0.062; TCGA BRCA -0.108; TCGA CESC -0.055; TCGA COAD -0.107; TCGA ESCA -0.273; TCGA LIHC -0.082; TCGA PAAD -0.085; TCGA PRAD -0.106; TCGA SARC -0.093; TCGA STAD -0.231 |
hsa-miR-218-5p | GPRC5A | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; PAAD; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.172; TCGA BRCA -0.527; TCGA CESC -0.229; TCGA COAD -0.173; TCGA ESCA -1.013; TCGA HNSC -0.333; TCGA PAAD -0.414; TCGA PRAD -0.249; TCGA THCA -0.193; TCGA STAD -0.592; TCGA UCEC -0.245 |
hsa-miR-218-5p | KPNA6 | 10 cancers: BLCA; BRCA; CESC; COAD; HNSC; LUSC; PRAD; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.096; TCGA BRCA -0.088; TCGA CESC -0.056; TCGA COAD -0.063; TCGA HNSC -0.078; TCGA LUSC -0.073; TCGA PRAD -0.101; TCGA SARC -0.068; TCGA THCA -0.109; TCGA STAD -0.056 |
hsa-miR-218-5p | EIF2AK2 | 11 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRP; LUAD; LUSC; PRAD; THCA; STAD | mirMAP | TCGA BLCA -0.209; TCGA BRCA -0.259; TCGA CESC -0.103; TCGA COAD -0.07; TCGA HNSC -0.243; TCGA KIRP -0.13; TCGA LUAD -0.183; TCGA LUSC -0.182; TCGA PRAD -0.267; TCGA THCA -0.13; TCGA STAD -0.136 |
hsa-miR-218-5p | POLR1A | 13 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; STAD | mirMAP | TCGA BLCA -0.072; TCGA BRCA -0.124; TCGA CESC -0.106; TCGA ESCA -0.101; TCGA HNSC -0.118; TCGA KIRP -0.079; TCGA LGG -0.056; TCGA LIHC -0.068; TCGA LUAD -0.126; TCGA LUSC -0.171; TCGA PAAD -0.13; TCGA PRAD -0.168; TCGA STAD -0.177 |
hsa-miR-218-5p | SMC1A | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUAD; LUSC; PRAD; SARC; STAD | mirMAP | TCGA BLCA -0.105; TCGA BRCA -0.128; TCGA COAD -0.121; TCGA ESCA -0.062; TCGA HNSC -0.078; TCGA LUAD -0.124; TCGA LUSC -0.1; TCGA PRAD -0.116; TCGA SARC -0.067; TCGA STAD -0.183 |
hsa-miR-218-5p | PVR | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.09; TCGA BRCA -0.105; TCGA CESC -0.122; TCGA COAD -0.117; TCGA ESCA -0.225; TCGA HNSC -0.12; TCGA LIHC -0.053; TCGA LUAD -0.133; TCGA THCA -0.1; TCGA STAD -0.121; TCGA UCEC -0.111 |
hsa-miR-218-5p | SENP1 | 9 cancers: BLCA; BRCA; COAD; KIRP; LGG; LUAD; LUSC; PRAD; STAD | mirMAP; miRNATAP | TCGA BLCA -0.092; TCGA BRCA -0.099; TCGA COAD -0.058; TCGA KIRP -0.057; TCGA LGG -0.067; TCGA LUAD -0.15; TCGA LUSC -0.11; TCGA PRAD -0.158; TCGA STAD -0.078 |
hsa-miR-218-5p | AVL9 | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; THCA; STAD | mirMAP | TCGA BLCA -0.105; TCGA BRCA -0.152; TCGA COAD -0.088; TCGA ESCA -0.211; TCGA HNSC -0.105; TCGA LIHC -0.053; TCGA LUAD -0.165; TCGA LUSC -0.193; TCGA PAAD -0.243; TCGA PRAD -0.316; TCGA THCA -0.119; TCGA STAD -0.249 |
hsa-miR-218-5p | PSMA2 | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; THCA | mirMAP | TCGA BLCA -0.056; TCGA BRCA -0.147; TCGA COAD -0.091; TCGA ESCA -0.063; TCGA HNSC -0.055; TCGA LGG -0.054; TCGA LIHC -0.053; TCGA LUAD -0.071; TCGA LUSC -0.144; TCGA PAAD -0.097; TCGA PRAD -0.085; TCGA THCA -0.062 |
hsa-miR-218-5p | CEP72 | 10 cancers: BLCA; BRCA; COAD; ESCA; KIRC; LUAD; LUSC; PAAD; PRAD; STAD | mirMAP | TCGA BLCA -0.094; TCGA BRCA -0.179; TCGA COAD -0.231; TCGA ESCA -0.144; TCGA KIRC -0.141; TCGA LUAD -0.178; TCGA LUSC -0.291; TCGA PAAD -0.284; TCGA PRAD -0.117; TCGA STAD -0.245 |
hsa-miR-218-5p | ICMT | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUSC; PAAD; THCA; STAD | mirMAP | TCGA BLCA -0.085; TCGA CESC -0.057; TCGA COAD -0.073; TCGA ESCA -0.075; TCGA HNSC -0.066; TCGA LUSC -0.116; TCGA PAAD -0.066; TCGA THCA -0.075; TCGA STAD -0.101 |
hsa-miR-218-5p | LGALS8 | 11 cancers: BLCA; BRCA; COAD; KIRC; LIHC; LUAD; PAAD; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.112; TCGA BRCA -0.121; TCGA COAD -0.075; TCGA KIRC -0.053; TCGA LIHC -0.08; TCGA LUAD -0.059; TCGA PAAD -0.089; TCGA PRAD -0.206; TCGA THCA -0.093; TCGA STAD -0.07; TCGA UCEC -0.08 |
hsa-miR-218-5p | LPGAT1 | 11 cancers: BLCA; BRCA; HNSC; KIRC; LUAD; LUSC; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.068; TCGA BRCA -0.165; TCGA HNSC -0.066; TCGA KIRC -0.114; TCGA LUAD -0.167; TCGA LUSC -0.13; TCGA PRAD -0.174; TCGA SARC -0.057; TCGA THCA -0.054; TCGA STAD -0.099; TCGA UCEC -0.096 |
hsa-miR-218-5p | IPPK | 10 cancers: BLCA; BRCA; CESC; COAD; HNSC; LUAD; LUSC; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.19; TCGA BRCA -0.163; TCGA CESC -0.288; TCGA COAD -0.098; TCGA HNSC -0.214; TCGA LUAD -0.056; TCGA LUSC -0.219; TCGA PRAD -0.109; TCGA STAD -0.123; TCGA UCEC -0.073 |
hsa-miR-218-5p | ARHGAP32 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUSC; PAAD; PRAD; THCA; STAD | mirMAP | TCGA BLCA -0.107; TCGA BRCA -0.131; TCGA CESC -0.158; TCGA COAD -0.206; TCGA ESCA -0.274; TCGA HNSC -0.176; TCGA KIRC -0.075; TCGA LUSC -0.193; TCGA PAAD -0.312; TCGA PRAD -0.212; TCGA THCA -0.094; TCGA STAD -0.195 |
hsa-miR-218-5p | SLC31A1 | 9 cancers: BLCA; BRCA; COAD; ESCA; KIRP; LIHC; PRAD; THCA; STAD | mirMAP | TCGA BLCA -0.077; TCGA BRCA -0.088; TCGA COAD -0.151; TCGA ESCA -0.195; TCGA KIRP -0.072; TCGA LIHC -0.082; TCGA PRAD -0.115; TCGA THCA -0.158; TCGA STAD -0.244 |
hsa-miR-218-5p | ZBTB7B | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; OV; PAAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.126; TCGA BRCA -0.19; TCGA CESC -0.221; TCGA COAD -0.091; TCGA ESCA -0.141; TCGA HNSC -0.139; TCGA KIRC -0.125; TCGA LUAD -0.127; TCGA LUSC -0.112; TCGA OV -0.063; TCGA PAAD -0.137; TCGA SARC -0.13; TCGA THCA -0.078; TCGA STAD -0.115; TCGA UCEC -0.066 |
hsa-miR-218-5p | KIAA1958 | 9 cancers: BLCA; BRCA; COAD; ESCA; LGG; LUAD; PRAD; SARC; STAD | mirMAP | TCGA BLCA -0.122; TCGA BRCA -0.152; TCGA COAD -0.155; TCGA ESCA -0.145; TCGA LGG -0.06; TCGA LUAD -0.097; TCGA PRAD -0.282; TCGA SARC -0.269; TCGA STAD -0.225 |
hsa-miR-218-5p | STARD5 | 10 cancers: BLCA; CESC; COAD; HNSC; KIRC; LIHC; LUSC; OV; SARC; UCEC | mirMAP | TCGA BLCA -0.146; TCGA CESC -0.341; TCGA COAD -0.19; TCGA HNSC -0.284; TCGA KIRC -0.069; TCGA LIHC -0.081; TCGA LUSC -0.401; TCGA OV -0.067; TCGA SARC -0.14; TCGA UCEC -0.099 |
hsa-miR-218-5p | VANGL1 | 10 cancers: BLCA; BRCA; COAD; HNSC; LGG; LUAD; LUSC; PAAD; PRAD; STAD | mirMAP | TCGA BLCA -0.149; TCGA BRCA -0.228; TCGA COAD -0.095; TCGA HNSC -0.177; TCGA LGG -0.092; TCGA LUAD -0.098; TCGA LUSC -0.262; TCGA PAAD -0.18; TCGA PRAD -0.175; TCGA STAD -0.167 |
hsa-miR-218-5p | MYO1D | 10 cancers: BLCA; CESC; ESCA; HNSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.089; TCGA CESC -0.118; TCGA ESCA -0.227; TCGA HNSC -0.088; TCGA PAAD -0.094; TCGA PRAD -0.11; TCGA SARC -0.155; TCGA THCA -0.148; TCGA STAD -0.11; TCGA UCEC -0.057 |
hsa-miR-218-5p | ASB7 | 9 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; PRAD; STAD | mirMAP | TCGA BLCA -0.074; TCGA BRCA -0.084; TCGA CESC -0.065; TCGA COAD -0.094; TCGA ESCA -0.105; TCGA HNSC -0.061; TCGA LIHC -0.057; TCGA PRAD -0.144; TCGA STAD -0.121 |
hsa-miR-218-5p | PPARA | 9 cancers: BLCA; COAD; ESCA; LIHC; LUSC; PAAD; PRAD; SARC; UCEC | mirMAP | TCGA BLCA -0.107; TCGA COAD -0.114; TCGA ESCA -0.11; TCGA LIHC -0.185; TCGA LUSC -0.071; TCGA PAAD -0.111; TCGA PRAD -0.099; TCGA SARC -0.061; TCGA UCEC -0.094 |
hsa-miR-218-5p | LRP10 | 11 cancers: BLCA; CESC; ESCA; HNSC; LGG; LUAD; OV; PAAD; SARC; THCA; UCEC | mirMAP | TCGA BLCA -0.07; TCGA CESC -0.052; TCGA ESCA -0.144; TCGA HNSC -0.105; TCGA LGG -0.073; TCGA LUAD -0.052; TCGA OV -0.08; TCGA PAAD -0.124; TCGA SARC -0.064; TCGA THCA -0.052; TCGA UCEC -0.089 |
hsa-miR-218-5p | CHML | 11 cancers: BLCA; BRCA; COAD; KIRC; KIRP; LUAD; PAAD; PRAD; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.082; TCGA BRCA -0.237; TCGA COAD -0.127; TCGA KIRC -0.071; TCGA KIRP -0.07; TCGA LUAD -0.183; TCGA PAAD -0.176; TCGA PRAD -0.272; TCGA SARC -0.161; TCGA THCA -0.121; TCGA STAD -0.213 |
hsa-miR-218-5p | INTS6 | 9 cancers: BLCA; CESC; HNSC; KIRP; LUAD; LUSC; PRAD; THCA; STAD | miRNATAP | TCGA BLCA -0.105; TCGA CESC -0.055; TCGA HNSC -0.093; TCGA KIRP -0.06; TCGA LUAD -0.071; TCGA LUSC -0.083; TCGA PRAD -0.088; TCGA THCA -0.072; TCGA STAD -0.062 |
hsa-miR-218-5p | ANKRD52 | 9 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUAD; LUSC; PRAD; STAD | miRNATAP | TCGA BLCA -0.067; TCGA BRCA -0.084; TCGA COAD -0.088; TCGA ESCA -0.078; TCGA HNSC -0.061; TCGA LUAD -0.11; TCGA LUSC -0.076; TCGA PRAD -0.165; TCGA STAD -0.108 |
hsa-miR-218-5p | PTP4A1 | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.099; TCGA BRCA -0.114; TCGA COAD -0.152; TCGA ESCA -0.191; TCGA HNSC -0.135; TCGA LUAD -0.152; TCGA PRAD -0.171; TCGA SARC -0.147; TCGA STAD -0.134; TCGA UCEC -0.067 |
hsa-miR-218-5p | RPS6KB1 | 10 cancers: BLCA; BRCA; CESC; COAD; HNSC; LIHC; LUAD; LUSC; PRAD; STAD | miRNATAP | TCGA BLCA -0.085; TCGA BRCA -0.154; TCGA CESC -0.051; TCGA COAD -0.1; TCGA HNSC -0.068; TCGA LIHC -0.082; TCGA LUAD -0.163; TCGA LUSC -0.113; TCGA PRAD -0.175; TCGA STAD -0.113 |
hsa-miR-218-5p | USP9X | 9 cancers: BLCA; BRCA; ESCA; HNSC; KIRP; LUAD; PRAD; SARC; STAD | miRNATAP | TCGA BLCA -0.108; TCGA BRCA -0.107; TCGA ESCA -0.069; TCGA HNSC -0.088; TCGA KIRP -0.072; TCGA LUAD -0.081; TCGA PRAD -0.121; TCGA SARC -0.063; TCGA STAD -0.109 |
hsa-miR-218-5p | GFPT1 | 11 cancers: BLCA; BRCA; COAD; ESCA; KIRP; LUAD; PAAD; PRAD; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.072; TCGA BRCA -0.235; TCGA COAD -0.139; TCGA ESCA -0.395; TCGA KIRP -0.109; TCGA LUAD -0.188; TCGA PAAD -0.244; TCGA PRAD -0.18; TCGA THCA -0.184; TCGA STAD -0.32; TCGA UCEC -0.077 |
hsa-miR-218-5p | KIAA1522 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; LUAD; LUSC; PAAD; PRAD; SARC; STAD | miRNATAP | TCGA BLCA -0.132; TCGA BRCA -0.218; TCGA CESC -0.121; TCGA COAD -0.14; TCGA ESCA -0.185; TCGA LUAD -0.076; TCGA LUSC -0.289; TCGA PAAD -0.288; TCGA PRAD -0.135; TCGA SARC -0.216; TCGA STAD -0.27 |
hsa-miR-218-5p | FCHO2 | 9 cancers: BLCA; COAD; HNSC; KIRC; LGG; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.104; TCGA COAD -0.069; TCGA HNSC -0.084; TCGA KIRC -0.078; TCGA LGG -0.062; TCGA SARC -0.071; TCGA THCA -0.191; TCGA STAD -0.152; TCGA UCEC -0.085 |
hsa-miR-218-5p | RCOR1 | 10 cancers: BLCA; BRCA; CESC; COAD; HNSC; LGG; PAAD; PRAD; THCA; STAD | miRNATAP | TCGA BLCA -0.181; TCGA BRCA -0.103; TCGA CESC -0.137; TCGA COAD -0.147; TCGA HNSC -0.229; TCGA LGG -0.053; TCGA PAAD -0.173; TCGA PRAD -0.142; TCGA THCA -0.074; TCGA STAD -0.206 |
hsa-miR-218-5p | KLF3 | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUSC; PAAD; PRAD; STAD | miRNATAP | TCGA BLCA -0.115; TCGA CESC -0.071; TCGA COAD -0.103; TCGA ESCA -0.128; TCGA HNSC -0.115; TCGA LUSC -0.062; TCGA PAAD -0.148; TCGA PRAD -0.147; TCGA STAD -0.188 |
hsa-miR-218-5p | RAB6A | 12 cancers: BLCA; BRCA; COAD; HNSC; KIRP; LUAD; LUSC; OV; PAAD; PRAD; THCA; STAD | miRNATAP | TCGA BLCA -0.1; TCGA BRCA -0.112; TCGA COAD -0.111; TCGA HNSC -0.096; TCGA KIRP -0.058; TCGA LUAD -0.109; TCGA LUSC -0.054; TCGA OV -0.085; TCGA PAAD -0.09; TCGA PRAD -0.107; TCGA THCA -0.061; TCGA STAD -0.063 |
hsa-miR-218-5p | TMPO | 9 cancers: BLCA; BRCA; COAD; ESCA; LGG; LUAD; LUSC; PRAD; STAD | miRNATAP | TCGA BLCA -0.072; TCGA BRCA -0.155; TCGA COAD -0.183; TCGA ESCA -0.166; TCGA LGG -0.065; TCGA LUAD -0.209; TCGA LUSC -0.139; TCGA PRAD -0.134; TCGA STAD -0.279 |
hsa-miR-218-5p | HOXD10 | 10 cancers: BLCA; CESC; HNSC; LGG; LIHC; LUAD; LUSC; PAAD; THCA; UCEC | miRNATAP | TCGA BLCA -0.266; TCGA CESC -0.322; TCGA HNSC -0.528; TCGA LGG -0.128; TCGA LIHC -0.427; TCGA LUAD -0.397; TCGA LUSC -1.053; TCGA PAAD -0.561; TCGA THCA -0.34; TCGA UCEC -0.124 |
hsa-miR-218-5p | STYX | 12 cancers: BLCA; BRCA; CESC; HNSC; LGG; LIHC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.1; TCGA BRCA -0.076; TCGA CESC -0.061; TCGA HNSC -0.112; TCGA LGG -0.064; TCGA LIHC -0.065; TCGA LUAD -0.094; TCGA LUSC -0.163; TCGA PRAD -0.114; TCGA SARC -0.059; TCGA STAD -0.058; TCGA UCEC -0.052 |
hsa-miR-218-5p | SPOPL | 10 cancers: BLCA; BRCA; COAD; HNSC; LUAD; OV; PAAD; PRAD; SARC; STAD | miRNATAP | TCGA BLCA -0.052; TCGA BRCA -0.151; TCGA COAD -0.057; TCGA HNSC -0.125; TCGA LUAD -0.083; TCGA OV -0.055; TCGA PAAD -0.089; TCGA PRAD -0.196; TCGA SARC -0.12; TCGA STAD -0.089 |
hsa-miR-218-5p | SETD7 | 9 cancers: BLCA; CESC; LGG; LIHC; LUAD; PRAD; SARC; THCA; UCEC | miRNATAP | TCGA BLCA -0.12; TCGA CESC -0.13; TCGA LGG -0.052; TCGA LIHC -0.068; TCGA LUAD -0.106; TCGA PRAD -0.149; TCGA SARC -0.084; TCGA THCA -0.232; TCGA UCEC -0.271 |
hsa-miR-218-5p | RNF19B | 9 cancers: BLCA; BRCA; CESC; COAD; HNSC; LUSC; OV; THCA; STAD | miRNATAP | TCGA BLCA -0.142; TCGA BRCA -0.051; TCGA CESC -0.201; TCGA COAD -0.087; TCGA HNSC -0.14; TCGA LUSC -0.134; TCGA OV -0.092; TCGA THCA -0.1; TCGA STAD -0.143 |
hsa-miR-218-5p | NXT2 | 9 cancers: BLCA; BRCA; COAD; LGG; LIHC; LUSC; OV; PRAD; STAD | miRNATAP | TCGA BLCA -0.112; TCGA BRCA -0.098; TCGA COAD -0.15; TCGA LGG -0.051; TCGA LIHC -0.075; TCGA LUSC -0.101; TCGA OV -0.073; TCGA PRAD -0.177; TCGA STAD -0.222 |
hsa-miR-218-5p | LMNB1 | 14 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; PAAD; PRAD; THCA; STAD | miRNATAP | TCGA BLCA -0.086; TCGA BRCA -0.386; TCGA COAD -0.186; TCGA ESCA -0.147; TCGA HNSC -0.067; TCGA KIRC -0.199; TCGA KIRP -0.137; TCGA LGG -0.126; TCGA LUAD -0.243; TCGA LUSC -0.307; TCGA PAAD -0.141; TCGA PRAD -0.232; TCGA THCA -0.132; TCGA STAD -0.354 |
hsa-miR-218-5p | ZFP91 | 9 cancers: BLCA; BRCA; COAD; HNSC; LUSC; PRAD; SARC; THCA; STAD | miRNATAP | TCGA BLCA -0.123; TCGA BRCA -0.063; TCGA COAD -0.101; TCGA HNSC -0.107; TCGA LUSC -0.067; TCGA PRAD -0.167; TCGA SARC -0.064; TCGA THCA -0.081; TCGA STAD -0.076 |
hsa-miR-218-5p | EIF3J | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; LUAD; LUSC; PRAD; THCA; STAD | miRNATAP | TCGA BLCA -0.054; TCGA BRCA -0.091; TCGA CESC -0.086; TCGA COAD -0.131; TCGA ESCA -0.118; TCGA LUAD -0.092; TCGA LUSC -0.089; TCGA PRAD -0.121; TCGA THCA -0.058; TCGA STAD -0.095 |
hsa-miR-218-5p | CDK12 | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; STAD | miRNATAP | TCGA BLCA -0.094; TCGA BRCA -0.145; TCGA COAD -0.116; TCGA ESCA -0.177; TCGA HNSC -0.114; TCGA LUAD -0.101; TCGA LUSC -0.067; TCGA PAAD -0.1; TCGA PRAD -0.192; TCGA STAD -0.212 |
hsa-miR-218-5p | FAM126B | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUAD; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.122; TCGA CESC -0.089; TCGA COAD -0.07; TCGA ESCA -0.133; TCGA HNSC -0.162; TCGA LUAD -0.051; TCGA PRAD -0.187; TCGA STAD -0.106; TCGA UCEC -0.06 |
hsa-miR-218-5p | NAA15 | 11 cancers: BLCA; BRCA; COAD; HNSC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD | miRNATAP | TCGA BLCA -0.088; TCGA BRCA -0.132; TCGA COAD -0.148; TCGA HNSC -0.099; TCGA LUAD -0.134; TCGA LUSC -0.128; TCGA PAAD -0.095; TCGA PRAD -0.17; TCGA SARC -0.058; TCGA THCA -0.052; TCGA STAD -0.18 |
hsa-miR-218-5p | AZIN1 | 12 cancers: BLCA; BRCA; COAD; ESCA; KIRP; LUAD; LUSC; OV; PAAD; PRAD; THCA; STAD | miRNATAP | TCGA BLCA -0.062; TCGA BRCA -0.18; TCGA COAD -0.092; TCGA ESCA -0.112; TCGA KIRP -0.098; TCGA LUAD -0.092; TCGA LUSC -0.135; TCGA OV -0.096; TCGA PAAD -0.172; TCGA PRAD -0.16; TCGA THCA -0.078; TCGA STAD -0.202 |
hsa-miR-218-5p | RAB1A | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; PRAD; STAD | miRNATAP | TCGA BLCA -0.056; TCGA BRCA -0.136; TCGA COAD -0.08; TCGA ESCA -0.132; TCGA HNSC -0.072; TCGA KIRP -0.054; TCGA LUAD -0.113; TCGA LUSC -0.118; TCGA PAAD -0.096; TCGA PRAD -0.071; TCGA STAD -0.102 |
hsa-miR-218-5p | PSMC2 | 9 cancers: BLCA; BRCA; CESC; LIHC; LUAD; LUSC; PAAD; PRAD; THCA | miRNATAP | TCGA BLCA -0.06; TCGA BRCA -0.167; TCGA CESC -0.055; TCGA LIHC -0.088; TCGA LUAD -0.091; TCGA LUSC -0.118; TCGA PAAD -0.053; TCGA PRAD -0.064; TCGA THCA -0.065 |
hsa-miR-218-5p | ARL8B | 12 cancers: BLCA; BRCA; CESC; HNSC; LUSC; OV; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.097; TCGA BRCA -0.161; TCGA CESC -0.165; TCGA HNSC -0.108; TCGA LUSC -0.051; TCGA OV -0.077; TCGA PAAD -0.053; TCGA PRAD -0.084; TCGA SARC -0.065; TCGA THCA -0.096; TCGA STAD -0.051; TCGA UCEC -0.076 |
hsa-miR-218-5p | KPNA1 | 12 cancers: BLCA; BRCA; CESC; COAD; HNSC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD | miRNATAP | TCGA BLCA -0.078; TCGA BRCA -0.161; TCGA CESC -0.083; TCGA COAD -0.064; TCGA HNSC -0.103; TCGA LUAD -0.106; TCGA LUSC -0.163; TCGA PAAD -0.077; TCGA PRAD -0.121; TCGA SARC -0.077; TCGA THCA -0.098; TCGA STAD -0.082 |
hsa-miR-218-5p | UBE2G1 | 9 cancers: BLCA; BRCA; CESC; COAD; HNSC; LUSC; PRAD; THCA; STAD | miRNATAP | TCGA BLCA -0.1; TCGA BRCA -0.08; TCGA CESC -0.09; TCGA COAD -0.075; TCGA HNSC -0.086; TCGA LUSC -0.101; TCGA PRAD -0.154; TCGA THCA -0.15; TCGA STAD -0.062 |
hsa-miR-218-5p | SIAH2 | 10 cancers: BLCA; BRCA; HNSC; KIRC; LIHC; LUAD; LUSC; OV; PAAD; SARC | miRNATAP | TCGA BLCA -0.056; TCGA BRCA -0.338; TCGA HNSC -0.092; TCGA KIRC -0.084; TCGA LIHC -0.174; TCGA LUAD -0.108; TCGA LUSC -0.304; TCGA OV -0.089; TCGA PAAD -0.113; TCGA SARC -0.093 |
hsa-miR-218-5p | NET1 | 13 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.212; TCGA BRCA -0.19; TCGA COAD -0.072; TCGA ESCA -0.086; TCGA HNSC -0.078; TCGA LIHC -0.067; TCGA LUAD -0.102; TCGA LUSC -0.297; TCGA OV -0.104; TCGA PAAD -0.327; TCGA PRAD -0.183; TCGA STAD -0.094; TCGA UCEC -0.057 |
hsa-miR-218-5p | CENPO | 9 cancers: BRCA; COAD; ESCA; KIRP; LUAD; LUSC; PAAD; PRAD; STAD | miRNAWalker2 validate | TCGA BRCA -0.246; TCGA COAD -0.127; TCGA ESCA -0.152; TCGA KIRP -0.094; TCGA LUAD -0.205; TCGA LUSC -0.273; TCGA PAAD -0.15; TCGA PRAD -0.248; TCGA STAD -0.144 |
hsa-miR-218-5p | EARS2 | 10 cancers: BRCA; COAD; ESCA; LIHC; LUAD; LUSC; PAAD; PRAD; THCA; STAD | miRNAWalker2 validate | TCGA BRCA -0.203; TCGA COAD -0.096; TCGA ESCA -0.097; TCGA LIHC -0.067; TCGA LUAD -0.063; TCGA LUSC -0.132; TCGA PAAD -0.141; TCGA PRAD -0.147; TCGA THCA -0.113; TCGA STAD -0.129 |
hsa-miR-218-5p | LASP1 | 9 cancers: BRCA; COAD; ESCA; HNSC; KIRC; LUAD; PAAD; STAD; UCEC | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | TCGA BRCA -0.126; TCGA COAD -0.09; TCGA ESCA -0.258; TCGA HNSC -0.077; TCGA KIRC -0.067; TCGA LUAD -0.06; TCGA PAAD -0.072; TCGA STAD -0.133; TCGA UCEC -0.051 |
hsa-miR-218-5p | MLX | 11 cancers: BRCA; CESC; COAD; ESCA; LIHC; LUAD; PAAD; PRAD; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BRCA -0.068; TCGA CESC -0.052; TCGA COAD -0.146; TCGA ESCA -0.193; TCGA LIHC -0.133; TCGA LUAD -0.087; TCGA PAAD -0.085; TCGA PRAD -0.101; TCGA THCA -0.129; TCGA STAD -0.12; TCGA UCEC -0.074 |
hsa-miR-218-5p | NACC1 | 10 cancers: BRCA; COAD; HNSC; KIRP; LUAD; LUSC; PAAD; PRAD; THCA; STAD | miRNAWalker2 validate; MirTarget; miRNATAP | TCGA BRCA -0.262; TCGA COAD -0.095; TCGA HNSC -0.067; TCGA KIRP -0.063; TCGA LUAD -0.09; TCGA LUSC -0.207; TCGA PAAD -0.107; TCGA PRAD -0.074; TCGA THCA -0.066; TCGA STAD -0.093 |
hsa-miR-218-5p | PPIA | 12 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; OV; PAAD; THCA | miRNAWalker2 validate | TCGA BRCA -0.156; TCGA CESC -0.062; TCGA COAD -0.113; TCGA ESCA -0.083; TCGA HNSC -0.076; TCGA KIRC -0.073; TCGA LIHC -0.079; TCGA LUAD -0.091; TCGA LUSC -0.174; TCGA OV -0.072; TCGA PAAD -0.124; TCGA THCA -0.063 |
hsa-miR-218-5p | TOB1 | 9 cancers: BRCA; COAD; ESCA; LIHC; PAAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate; miRTarBase; miRNATAP | TCGA BRCA -0.191; TCGA COAD -0.162; TCGA ESCA -0.312; TCGA LIHC -0.118; TCGA PAAD -0.175; TCGA PRAD -0.171; TCGA SARC -0.196; TCGA STAD -0.182; TCGA UCEC -0.07 |
hsa-miR-218-5p | BIRC5 | 12 cancers: BRCA; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; THCA; STAD | miRTarBase; MirTarget | TCGA BRCA -0.683; TCGA COAD -0.215; TCGA ESCA -0.163; TCGA HNSC -0.111; TCGA LGG -0.183; TCGA LIHC -0.142; TCGA LUAD -0.523; TCGA LUSC -0.889; TCGA PAAD -0.311; TCGA PRAD -0.31; TCGA THCA -0.199; TCGA STAD -0.418 |
hsa-miR-218-5p | RCC1 | 11 cancers: BRCA; CESC; COAD; ESCA; KIRC; LGG; LUAD; LUSC; PRAD; THCA; STAD | MirTarget; miRNATAP | TCGA BRCA -0.296; TCGA CESC -0.056; TCGA COAD -0.2; TCGA ESCA -0.319; TCGA KIRC -0.19; TCGA LGG -0.211; TCGA LUAD -0.161; TCGA LUSC -0.428; TCGA PRAD -0.232; TCGA THCA -0.202; TCGA STAD -0.306 |
hsa-miR-218-5p | ICK | 9 cancers: BRCA; KIRC; LGG; LIHC; OV; PAAD; PRAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BRCA -0.064; TCGA KIRC -0.075; TCGA LGG -0.051; TCGA LIHC -0.218; TCGA OV -0.067; TCGA PAAD -0.149; TCGA PRAD -0.186; TCGA STAD -0.112; TCGA UCEC -0.052 |
hsa-miR-218-5p | SEC61A1 | 10 cancers: BRCA; COAD; KIRP; LGG; LUAD; LUSC; PAAD; PRAD; THCA; STAD | MirTarget; miRNATAP | TCGA BRCA -0.155; TCGA COAD -0.078; TCGA KIRP -0.08; TCGA LGG -0.091; TCGA LUAD -0.12; TCGA LUSC -0.156; TCGA PAAD -0.145; TCGA PRAD -0.07; TCGA THCA -0.129; TCGA STAD -0.131 |
hsa-miR-218-5p | FAF2 | 9 cancers: BRCA; COAD; ESCA; HNSC; KIRC; LUSC; PAAD; PRAD; STAD | MirTarget | TCGA BRCA -0.107; TCGA COAD -0.064; TCGA ESCA -0.068; TCGA HNSC -0.095; TCGA KIRC -0.125; TCGA LUSC -0.058; TCGA PAAD -0.074; TCGA PRAD -0.109; TCGA STAD -0.148 |
hsa-miR-218-5p | LARP4B | 9 cancers: BRCA; CESC; COAD; ESCA; HNSC; PAAD; PRAD; THCA; STAD | MirTarget; miRNATAP | TCGA BRCA -0.161; TCGA CESC -0.072; TCGA COAD -0.097; TCGA ESCA -0.199; TCGA HNSC -0.053; TCGA PAAD -0.112; TCGA PRAD -0.091; TCGA THCA -0.086; TCGA STAD -0.201 |
hsa-miR-218-5p | MTPAP | 9 cancers: BRCA; COAD; ESCA; LUAD; LUSC; PAAD; PRAD; THCA; STAD | MirTarget | TCGA BRCA -0.122; TCGA COAD -0.109; TCGA ESCA -0.142; TCGA LUAD -0.138; TCGA LUSC -0.141; TCGA PAAD -0.052; TCGA PRAD -0.158; TCGA THCA -0.067; TCGA STAD -0.158 |
hsa-miR-218-5p | HCN3 | 9 cancers: BRCA; CESC; ESCA; KIRC; LIHC; LUAD; LUSC; STAD; UCEC | MirTarget; miRNATAP | TCGA BRCA -0.163; TCGA CESC -0.083; TCGA ESCA -0.136; TCGA KIRC -0.199; TCGA LIHC -0.159; TCGA LUAD -0.198; TCGA LUSC -0.289; TCGA STAD -0.108; TCGA UCEC -0.072 |
hsa-miR-218-5p | SERP1 | 11 cancers: BRCA; CESC; COAD; HNSC; KIRP; LIHC; LUSC; OV; PAAD; PRAD; STAD | MirTarget; miRNATAP | TCGA BRCA -0.185; TCGA CESC -0.081; TCGA COAD -0.124; TCGA HNSC -0.104; TCGA KIRP -0.098; TCGA LIHC -0.063; TCGA LUSC -0.063; TCGA OV -0.083; TCGA PAAD -0.15; TCGA PRAD -0.172; TCGA STAD -0.093 |
hsa-miR-218-5p | IMPAD1 | 9 cancers: BRCA; COAD; KIRP; LIHC; LUAD; PAAD; PRAD; THCA; STAD | MirTarget; miRNATAP | TCGA BRCA -0.096; TCGA COAD -0.095; TCGA KIRP -0.063; TCGA LIHC -0.074; TCGA LUAD -0.104; TCGA PAAD -0.091; TCGA PRAD -0.119; TCGA THCA -0.054; TCGA STAD -0.057 |
hsa-miR-218-5p | GTPBP10 | 9 cancers: BRCA; COAD; ESCA; LIHC; LUAD; LUSC; PAAD; PRAD; STAD | MirTarget | TCGA BRCA -0.065; TCGA COAD -0.105; TCGA ESCA -0.173; TCGA LIHC -0.091; TCGA LUAD -0.076; TCGA LUSC -0.104; TCGA PAAD -0.124; TCGA PRAD -0.187; TCGA STAD -0.126 |
hsa-miR-218-5p | TMCO1 | 11 cancers: BRCA; CESC; ESCA; KIRC; LIHC; LUAD; LUSC; OV; PAAD; PRAD; STAD | MirTarget | TCGA BRCA -0.208; TCGA CESC -0.069; TCGA ESCA -0.147; TCGA KIRC -0.06; TCGA LIHC -0.079; TCGA LUAD -0.094; TCGA LUSC -0.16; TCGA OV -0.065; TCGA PAAD -0.097; TCGA PRAD -0.063; TCGA STAD -0.064 |
hsa-miR-218-5p | HOOK1 | 9 cancers: BRCA; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PRAD; STAD | MirTarget; miRNATAP | TCGA BRCA -0.318; TCGA COAD -0.206; TCGA ESCA -0.319; TCGA HNSC -0.261; TCGA KIRC -0.159; TCGA LUAD -0.19; TCGA LUSC -0.123; TCGA PRAD -0.242; TCGA STAD -0.382 |
hsa-miR-218-5p | RNF103 | 9 cancers: BRCA; ESCA; LIHC; LUAD; OV; PAAD; PRAD; SARC; UCEC | MirTarget | TCGA BRCA -0.136; TCGA ESCA -0.245; TCGA LIHC -0.063; TCGA LUAD -0.064; TCGA OV -0.052; TCGA PAAD -0.107; TCGA PRAD -0.148; TCGA SARC -0.138; TCGA UCEC -0.055 |
hsa-miR-218-5p | HAPLN1 | 12 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; THCA; STAD | MirTarget; miRNATAP | TCGA BRCA -0.915; TCGA CESC -0.261; TCGA COAD -0.301; TCGA ESCA -0.264; TCGA HNSC -0.236; TCGA KIRC -0.91; TCGA KIRP -0.258; TCGA LUAD -0.27; TCGA LUSC -0.355; TCGA PAAD -1.009; TCGA THCA -0.743; TCGA STAD -0.617 |
hsa-miR-218-5p | TTC39A | 10 cancers: BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; PRAD; THCA; STAD | mirMAP | TCGA BRCA -0.325; TCGA COAD -0.233; TCGA ESCA -0.501; TCGA HNSC -0.195; TCGA KIRC -0.274; TCGA KIRP -0.096; TCGA LUAD -0.127; TCGA PRAD -0.134; TCGA THCA -0.501; TCGA STAD -0.302 |
hsa-miR-218-5p | RFFL | 9 cancers: BRCA; COAD; ESCA; KIRP; LIHC; LUAD; PRAD; SARC; STAD | mirMAP | TCGA BRCA -0.057; TCGA COAD -0.099; TCGA ESCA -0.124; TCGA KIRP -0.062; TCGA LIHC -0.104; TCGA LUAD -0.117; TCGA PRAD -0.168; TCGA SARC -0.121; TCGA STAD -0.071 |
hsa-miR-218-5p | BCCIP | 10 cancers: BRCA; COAD; ESCA; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; STAD | mirMAP | TCGA BRCA -0.146; TCGA COAD -0.146; TCGA ESCA -0.128; TCGA KIRP -0.087; TCGA LGG -0.054; TCGA LIHC -0.085; TCGA LUAD -0.119; TCGA LUSC -0.152; TCGA PAAD -0.083; TCGA STAD -0.103 |
hsa-miR-218-5p | SLC7A11 | 9 cancers: BRCA; COAD; KIRP; LUAD; LUSC; PAAD; PRAD; THCA; STAD | mirMAP | TCGA BRCA -0.296; TCGA COAD -0.232; TCGA KIRP -0.332; TCGA LUAD -0.284; TCGA LUSC -0.476; TCGA PAAD -0.44; TCGA PRAD -0.369; TCGA THCA -0.176; TCGA STAD -0.554 |
hsa-miR-218-5p | OVOL1 | 10 cancers: BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; STAD | mirMAP | TCGA BRCA -0.392; TCGA CESC -0.492; TCGA COAD -0.116; TCGA ESCA -0.353; TCGA HNSC -0.393; TCGA LUAD -0.259; TCGA LUSC -0.697; TCGA PAAD -0.657; TCGA PRAD -0.275; TCGA STAD -0.489 |
hsa-miR-218-5p | BNIP3 | 9 cancers: BRCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PRAD; THCA | mirMAP | TCGA BRCA -0.221; TCGA HNSC -0.126; TCGA KIRC -0.256; TCGA KIRP -0.057; TCGA LIHC -0.214; TCGA LUAD -0.196; TCGA LUSC -0.201; TCGA PRAD -0.118; TCGA THCA -0.089 |
hsa-miR-218-5p | REPIN1 | 10 cancers: BRCA; COAD; ESCA; KIRC; LGG; LIHC; LUAD; PRAD; SARC; STAD | mirMAP | TCGA BRCA -0.163; TCGA COAD -0.077; TCGA ESCA -0.145; TCGA KIRC -0.054; TCGA LGG -0.11; TCGA LIHC -0.085; TCGA LUAD -0.118; TCGA PRAD -0.067; TCGA SARC -0.058; TCGA STAD -0.109 |
hsa-miR-218-5p | OXSR1 | 10 cancers: BRCA; CESC; COAD; ESCA; HNSC; PAAD; PRAD; SARC; THCA; STAD | miRNATAP | TCGA BRCA -0.068; TCGA CESC -0.091; TCGA COAD -0.105; TCGA ESCA -0.121; TCGA HNSC -0.145; TCGA PAAD -0.126; TCGA PRAD -0.068; TCGA SARC -0.072; TCGA THCA -0.053; TCGA STAD -0.162 |
hsa-miR-218-5p | PPP1CC | 11 cancers: BRCA; CESC; COAD; ESCA; KIRP; LGG; LIHC; LUAD; LUSC; PRAD; STAD | miRNATAP | TCGA BRCA -0.086; TCGA CESC -0.054; TCGA COAD -0.158; TCGA ESCA -0.068; TCGA KIRP -0.054; TCGA LGG -0.056; TCGA LIHC -0.062; TCGA LUAD -0.131; TCGA LUSC -0.114; TCGA PRAD -0.135; TCGA STAD -0.082 |
hsa-miR-218-5p | TCF20 | 9 cancers: BRCA; COAD; HNSC; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BRCA -0.068; TCGA COAD -0.1; TCGA HNSC -0.051; TCGA LUSC -0.156; TCGA PAAD -0.091; TCGA PRAD -0.112; TCGA SARC -0.053; TCGA STAD -0.147; TCGA UCEC -0.056 |
hsa-miR-218-5p | NUP50 | 9 cancers: BRCA; CESC; COAD; HNSC; LUAD; LUSC; PRAD; STAD; UCEC | miRNATAP | TCGA BRCA -0.068; TCGA CESC -0.091; TCGA COAD -0.129; TCGA HNSC -0.074; TCGA LUAD -0.055; TCGA LUSC -0.122; TCGA PRAD -0.099; TCGA STAD -0.209; TCGA UCEC -0.077 |
hsa-miR-218-5p | DBNDD1 | 9 cancers: BRCA; ESCA; KIRC; LUAD; LUSC; PAAD; SARC; THCA; STAD | miRNATAP | TCGA BRCA -0.442; TCGA ESCA -0.174; TCGA KIRC -0.112; TCGA LUAD -0.172; TCGA LUSC -0.138; TCGA PAAD -0.228; TCGA SARC -0.318; TCGA THCA -0.097; TCGA STAD -0.171 |
hsa-miR-218-5p | NUDT5 | 15 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; THCA; STAD | miRNATAP | TCGA BRCA -0.225; TCGA CESC -0.074; TCGA COAD -0.092; TCGA ESCA -0.191; TCGA HNSC -0.053; TCGA KIRC -0.087; TCGA KIRP -0.147; TCGA LGG -0.114; TCGA LIHC -0.144; TCGA LUAD -0.107; TCGA LUSC -0.187; TCGA PAAD -0.129; TCGA PRAD -0.069; TCGA THCA -0.139; TCGA STAD -0.163 |
hsa-miR-218-5p | TTYH3 | 9 cancers: BRCA; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; THCA; STAD | miRNATAP | TCGA BRCA -0.087; TCGA HNSC -0.108; TCGA KIRC -0.205; TCGA LUAD -0.078; TCGA LUSC -0.311; TCGA PAAD -0.17; TCGA PRAD -0.221; TCGA THCA -0.134; TCGA STAD -0.176 |
hsa-miR-218-5p | RPL37 | 9 cancers: CESC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; THCA | mirMAP | TCGA CESC -0.102; TCGA KIRC -0.108; TCGA KIRP -0.091; TCGA LGG -0.067; TCGA LIHC -0.128; TCGA LUAD -0.111; TCGA LUSC -0.189; TCGA PAAD -0.13; TCGA THCA -0.064 |
hsa-miR-218-5p | RPL12 | 9 cancers: COAD; KIRC; KIRP; LGG; LIHC; LUSC; PAAD; STAD; UCEC | miRNAWalker2 validate | TCGA COAD -0.134; TCGA KIRC -0.07; TCGA KIRP -0.077; TCGA LGG -0.103; TCGA LIHC -0.082; TCGA LUSC -0.108; TCGA PAAD -0.114; TCGA STAD -0.089; TCGA UCEC -0.076 |
hsa-miR-218-5p | TXNDC5 | 10 cancers: ESCA; KIRC; KIRP; LGG; LUAD; LUSC; OV; PRAD; THCA; UCEC | MirTarget | TCGA ESCA -0.097; TCGA KIRC -0.085; TCGA KIRP -0.054; TCGA LGG -0.052; TCGA LUAD -0.068; TCGA LUSC -0.099; TCGA OV -0.068; TCGA PRAD -0.136; TCGA THCA -0.105; TCGA UCEC -0.099 |
Enriched cancer pathways of putative targets