microRNA information: hsa-miR-27b-3p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-27b-3p | miRbase |
| Accession: | MIMAT0000419 | miRbase |
| Precursor name: | hsa-mir-27b | miRbase |
| Precursor accession: | MI0000440 | miRbase |
| Symbol: | MIR27B | HGNC |
| RefSeq ID: | NR_029665 | GenBank |
| Sequence: | UUCACAGUGGCUAAGUUCUGC |
Reported expression in cancers: hsa-miR-27b-3p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-27b-3p | breast cancer | downregulation | "These miRNAs include miR-21 and miR-27 that were f ......" | 20952761 | |
| hsa-miR-27b-3p | breast cancer | upregulation | "We identified miR-23b and miR-27b as miRNAs that a ......" | 23338610 | |
| hsa-miR-27b-3p | cervical and endocervical cancer | upregulation | "In the present study we report that HPV16 E7 upreg ......" | 26397063 | |
| hsa-miR-27b-3p | cervical and endocervical cancer | upregulation | "MicroRNA 27b up regulated by human papillomavirus ......" | 26910911 | |
| hsa-miR-27b-3p | colorectal cancer | upregulation | "To address this issue we profiled 742 microRNAs in ......" | 23506979 | |
| hsa-miR-27b-3p | colorectal cancer | upregulation | "High expression of miR-27b miR-148a and miR-326 we ......" | 24119443 | |
| hsa-miR-27b-3p | gastric cancer | upregulation | "In this study we found that ectopic miR-27b is suf ......" | 26623719 | |
| hsa-miR-27b-3p | kidney renal cell cancer | downregulation | "Conversely decreased expression of miR-106b miR-99 ......" | 26416448 | |
| hsa-miR-27b-3p | liver cancer | upregulation | "Up-regulation of miR-15a miR-16 miR-27b miR-30b mi ......" | 24314246 | qPCR |
| hsa-miR-27b-3p | lymphoma | upregulation | "The up-regulation of miR-155 miR-27b miR-30c and m ......" | 22776000 | Microarray |
| hsa-miR-27b-3p | prostate cancer | downregulation | "The expression of miR-23b and miR-27b which are en ......" | 23300597 | |
| hsa-miR-27b-3p | prostate cancer | downregulation | "Kaplan-Meier survival curves showed that low expre ......" | 25115396 |
Reported cancer pathway affected by hsa-miR-27b-3p
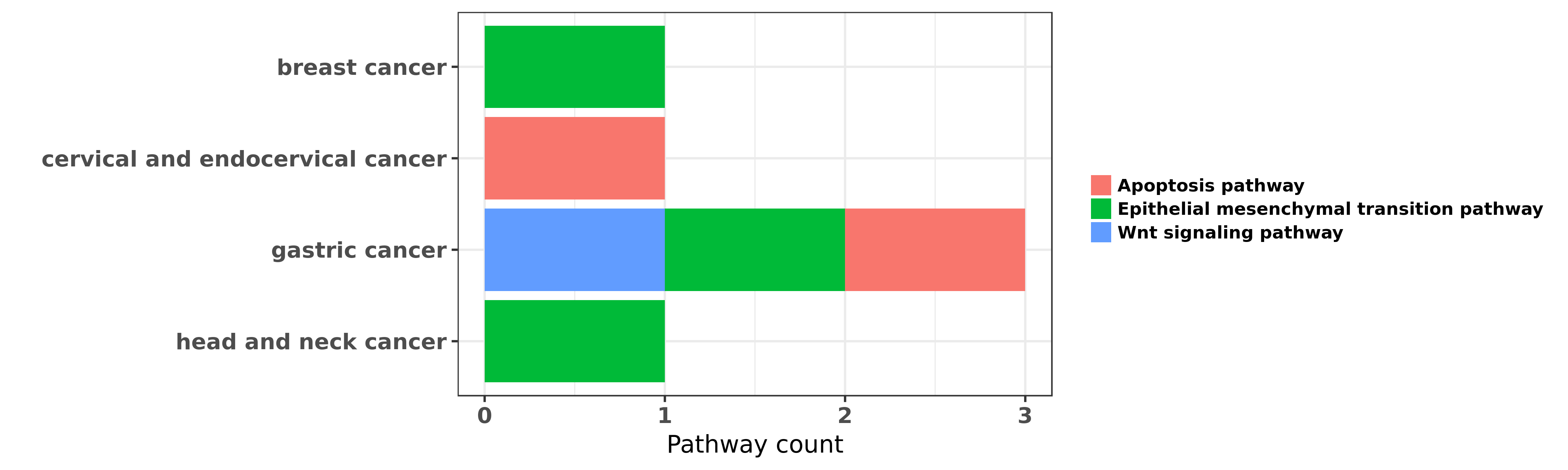
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-27b-3p | breast cancer | Epithelial mesenchymal transition pathway | "MiR 27b is epigenetically downregulated in tamoxif ......" | 27363334 | Western blot; Luciferase |
| hsa-miR-27b-3p | cervical and endocervical cancer | Apoptosis pathway | "MicroRNA 27b up regulated by human papillomavirus ......" | 26910911 | Luciferase |
| hsa-miR-27b-3p | gastric cancer | Epithelial mesenchymal transition pathway | "miR 27 promotes human gastric cancer cell metastas ......" | 22018270 | Western blot; Luciferase |
| hsa-miR-27b-3p | gastric cancer | Wnt signaling pathway | "MicroRNA 27b suppresses Helicobacter pylori induce ......" | 26780940 | |
| hsa-miR-27b-3p | gastric cancer | Apoptosis pathway | "Long Non Coding RNA LncRNA Urothelial Carcinoma As ......" | 27694794 | Western blot |
| hsa-miR-27b-3p | head and neck cancer | Epithelial mesenchymal transition pathway | "Among several miRNAs in which the expression was a ......" | 21899661 |
Reported cancer prognosis affected by hsa-miR-27b-3p
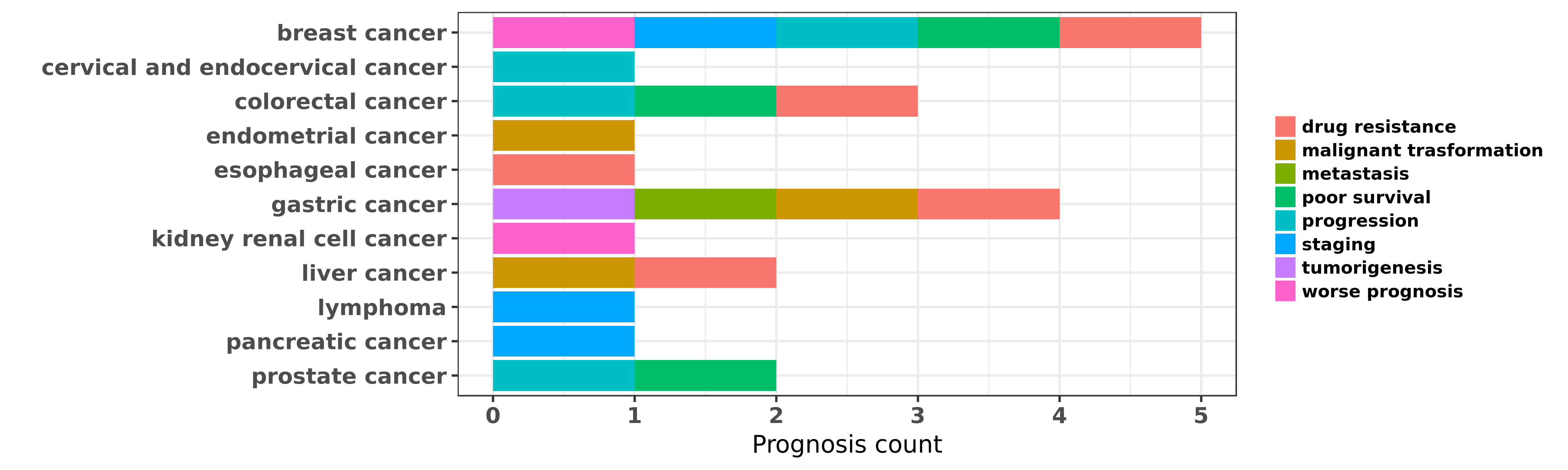
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-27b-3p | breast cancer | progression; poor survival | "MiR 27 as a prognostic marker for breast cancer pr ......" | 23240057 | |
| hsa-miR-27b-3p | breast cancer | worse prognosis | "A prognostic model of triple negative breast cance ......" | 24945253 | |
| hsa-miR-27b-3p | breast cancer | drug resistance | "MiR 27b is epigenetically downregulated in tamoxif ......" | 27363334 | Western blot; Luciferase |
| hsa-miR-27b-3p | breast cancer | staging | "Our results showed significant increase of miR-17 ......" | 27596294 | |
| hsa-miR-27b-3p | cervical and endocervical cancer | progression | "Elevation of miR 27b by HPV16 E7 inhibits PPARγ e ......" | 26397063 | |
| hsa-miR-27b-3p | colorectal cancer | drug resistance | "To address this issue we profiled 742 microRNAs in ......" | 23506979 | |
| hsa-miR-27b-3p | colorectal cancer | progression | "A tumor suppressor role for miR-27b has recently b ......" | 23593282 | Colony formation |
| hsa-miR-27b-3p | colorectal cancer | progression; poor survival | "High expression of miR-27b miR-148a and miR-326 we ......" | 24119443 | |
| hsa-miR-27b-3p | endometrial cancer | malignant trasformation | "Expression profiling of normal and malignant endom ......" | 20028871 | |
| hsa-miR-27b-3p | esophageal cancer | drug resistance | "An in-vitro model of acquired chemotherapy resista ......" | 25356050 | |
| hsa-miR-27b-3p | esophageal cancer | drug resistance | "miR 27 is associated with chemoresistance in esoph ......" | 26026166 | |
| hsa-miR-27b-3p | gastric cancer | metastasis | "miR 27 promotes human gastric cancer cell metastas ......" | 22018270 | Western blot; Luciferase |
| hsa-miR-27b-3p | gastric cancer | malignant trasformation | "miR 27b 3p suppresses cell proliferation through t ......" | 26576539 | Western blot; Luciferase; Colony formation |
| hsa-miR-27b-3p | gastric cancer | drug resistance | "The miR27b CCNG1 P53 miR 508 5p axis regulates mul ......" | 26623719 | |
| hsa-miR-27b-3p | gastric cancer | tumorigenesis | "MicroRNA 27b suppresses Helicobacter pylori induce ......" | 26780940 | |
| hsa-miR-27b-3p | gastric cancer | drug resistance | "Long Non Coding RNA LncRNA Urothelial Carcinoma As ......" | 27694794 | Western blot |
| hsa-miR-27b-3p | kidney renal cell cancer | worse prognosis | "Conversely decreased expression of miR-106b miR-99 ......" | 26416448 | |
| hsa-miR-27b-3p | liver cancer | drug resistance | "The cell viability MTT assay was used to detect dr ......" | 23229111 | MTT assay |
| hsa-miR-27b-3p | liver cancer | malignant trasformation | "Using gene expression microarrays and high-through ......" | 26646011 | |
| hsa-miR-27b-3p | lymphoma | staging | "The miRNA expression profiles of skin biopsies fro ......" | 22776000 | |
| hsa-miR-27b-3p | pancreatic cancer | staging | "Based on relevance to cancer a seven-miRNA signatu ......" | 25184537 | |
| hsa-miR-27b-3p | prostate cancer | progression; poor survival | "The expression of miR-23b and miR-27b which are en ......" | 23300597 | Colony formation |
| hsa-miR-27b-3p | prostate cancer | progression; poor survival | "Kaplan-Meier survival curves showed that low expre ......" | 25115396 |
Reported gene related to hsa-miR-27b-3p
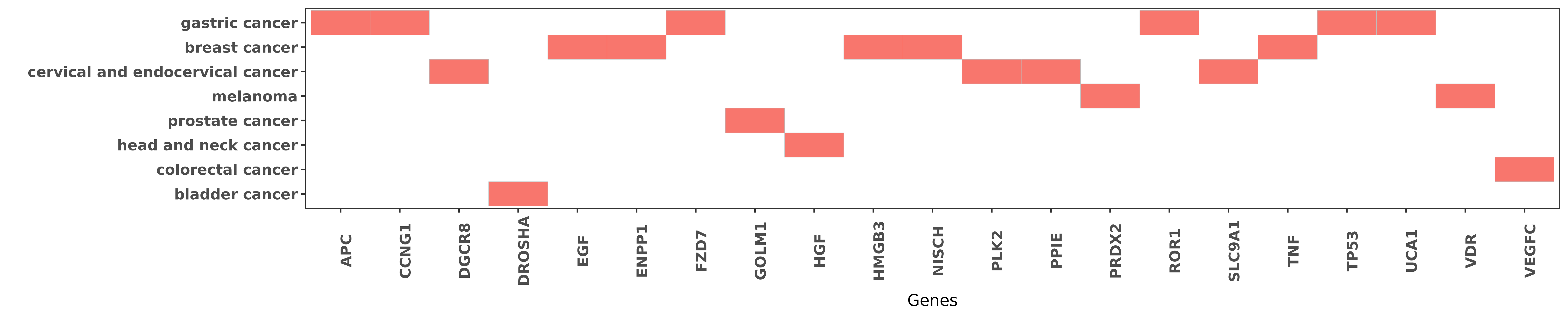
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-27b-3p | gastric cancer | APC | "Finally the APC gene was identified as the direct ......" | 22018270 |
| hsa-miR-27b-3p | gastric cancer | CCNG1 | "Moreover miR-27b directly targets the 3' untransla ......" | 26623719 |
| hsa-miR-27b-3p | cervical and endocervical cancer | DGCR8 | "Furthermore DGCR8 was found to mediate the up-regu ......" | 26910911 |
| hsa-miR-27b-3p | bladder cancer | DROSHA | "Genetic variation in DROSHA 3'UTR regulated by hsa ......" | 24312312 |
| hsa-miR-27b-3p | breast cancer | EGF | "Prooncogenic factors miR 23b and miR 27b are regul ......" | 23338610 |
| hsa-miR-27b-3p | breast cancer | ENPP1 | "Loss of microRNA 27b contributes to breast cancer ......" | 26065921 |
| hsa-miR-27b-3p | gastric cancer | FZD7 | "Restoration of FZD7 expression significantly atten ......" | 26780940 |
| hsa-miR-27b-3p | prostate cancer | GOLM1 | "GOLM1 was directly regulated by miR-27b in PCa cel ......" | 25115396 |
| hsa-miR-27b-3p | head and neck cancer | HGF | "Among several miRNAs in which the expression was a ......" | 21899661 |
| hsa-miR-27b-3p | breast cancer | HMGB3 | "MiR 27b is epigenetically downregulated in tamoxif ......" | 27363334 |
| hsa-miR-27b-3p | breast cancer | NISCH | "Nischarin NISCH expression was augmented by knockd ......" | 23338610 |
| hsa-miR-27b-3p | cervical and endocervical cancer | PLK2 | "Dual-luciferase experiment confirmed miR-27b down- ......" | 26910911 |
| hsa-miR-27b-3p | cervical and endocervical cancer | PPIE | "The results showed that PPARγ as a target of miR- ......" | 26397063 |
| hsa-miR-27b-3p | melanoma | PRDX2 | "Treatment with 125OH2D3 and/or epigenetic drugs 5- ......" | 22213330 |
| hsa-miR-27b-3p | gastric cancer | ROR1 | "Bioinformatic prediction luciferase reporter assay ......" | 26576539 |
| hsa-miR-27b-3p | cervical and endocervical cancer | SLC9A1 | "The results showed that PPARγ as a target of miR- ......" | 26397063 |
| hsa-miR-27b-3p | breast cancer | TNF | "Prooncogenic factors miR 23b and miR 27b are regul ......" | 23338610 |
| hsa-miR-27b-3p | gastric cancer | TP53 | "Moreover miR-27b directly targets the 3' untransla ......" | 26623719 |
| hsa-miR-27b-3p | gastric cancer | UCA1 | "Long Non Coding RNA LncRNA Urothelial Carcinoma As ......" | 27694794 |
| hsa-miR-27b-3p | melanoma | VDR | "The expression of VDR mRNA protein and two candida ......" | 22213330 |
| hsa-miR-27b-3p | colorectal cancer | VEGFC | "We identified vascular endothelial growth factor C ......" | 23593282 |
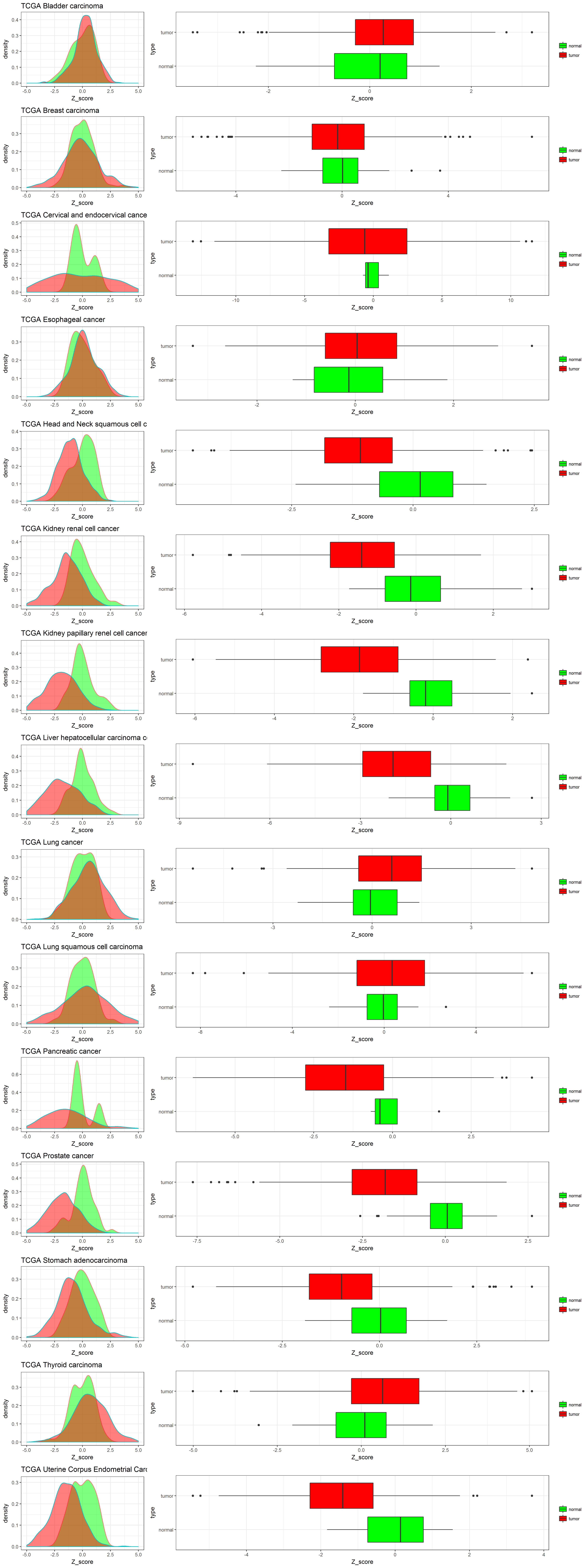
Expression profile in cancer corhorts:
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-27b-3p | CALM3 | 9 cancers: BLCA; CESC; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC | miRNAWalker2 validate; miRNATAP | TCGA BLCA -0.103; TCGA CESC -0.135; TCGA ESCA -0.231; TCGA HNSC -0.194; TCGA KIRC -0.094; TCGA KIRP -0.094; TCGA LIHC -0.107; TCGA LUAD -0.173; TCGA LUSC -0.222 |
hsa-miR-27b-3p | CCND3 | 9 cancers: BLCA; CESC; ESCA; HNSC; KIRP; LGG; LIHC; LUSC; SARC | miRNAWalker2 validate | TCGA BLCA -0.134; TCGA CESC -0.142; TCGA ESCA -0.177; TCGA HNSC -0.132; TCGA KIRP -0.154; TCGA LGG -0.053; TCGA LIHC -0.237; TCGA LUSC -0.263; TCGA SARC -0.231 |
hsa-miR-27b-3p | CUL4B | 11 cancers: BLCA; BRCA; COAD; KIRC; KIRP; LGG; LIHC; LUAD; PAAD; PRAD; STAD | miRNAWalker2 validate | TCGA BLCA -0.079; TCGA BRCA -0.062; TCGA COAD -0.137; TCGA KIRC -0.156; TCGA KIRP -0.075; TCGA LGG -0.068; TCGA LIHC -0.106; TCGA LUAD -0.132; TCGA PAAD -0.124; TCGA PRAD -0.147; TCGA STAD -0.113 |
hsa-miR-27b-3p | UTP14A | 10 cancers: BLCA; COAD; HNSC; KIRC; KIRP; LGG; LIHC; PAAD; PRAD; STAD | miRNAWalker2 validate | TCGA BLCA -0.06; TCGA COAD -0.238; TCGA HNSC -0.087; TCGA KIRC -0.139; TCGA KIRP -0.108; TCGA LGG -0.078; TCGA LIHC -0.165; TCGA PAAD -0.105; TCGA PRAD -0.209; TCGA STAD -0.127 |
hsa-miR-27b-3p | ABL2 | 10 cancers: BLCA; ESCA; HNSC; KIRC; KIRP; LGG; PAAD; PRAD; THCA; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.12; TCGA ESCA -0.132; TCGA HNSC -0.314; TCGA KIRC -0.351; TCGA KIRP -0.086; TCGA LGG -0.119; TCGA PAAD -0.248; TCGA PRAD -0.118; TCGA THCA -0.117; TCGA UCEC -0.09 |
hsa-miR-27b-3p | RSAD2 | 10 cancers: BLCA; COAD; HNSC; KIRC; LUAD; LUSC; PAAD; SARC; THCA; STAD | MirTarget | TCGA BLCA -0.366; TCGA COAD -0.666; TCGA HNSC -0.693; TCGA KIRC -0.251; TCGA LUAD -0.175; TCGA LUSC -0.411; TCGA PAAD -0.486; TCGA SARC -0.23; TCGA THCA -0.254; TCGA STAD -0.252 |
hsa-miR-27b-3p | GOLM1 | 10 cancers: BLCA; CESC; ESCA; HNSC; LGG; LIHC; LUSC; PRAD; SARC; THCA | MirTarget | TCGA BLCA -0.148; TCGA CESC -0.58; TCGA ESCA -1.022; TCGA HNSC -0.288; TCGA LGG -0.051; TCGA LIHC -0.594; TCGA LUSC -0.194; TCGA PRAD -0.748; TCGA SARC -0.24; TCGA THCA -0.202 |
hsa-miR-27b-3p | GRB2 | 11 cancers: BLCA; BRCA; KIRC; KIRP; LIHC; LUAD; PAAD; PRAD; SARC; THCA; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.052; TCGA BRCA -0.059; TCGA KIRC -0.085; TCGA KIRP -0.105; TCGA LIHC -0.088; TCGA LUAD -0.113; TCGA PAAD -0.202; TCGA PRAD -0.057; TCGA SARC -0.091; TCGA THCA -0.103; TCGA UCEC -0.06 |
hsa-miR-27b-3p | TNPO1 | 10 cancers: BLCA; BRCA; COAD; ESCA; KIRP; LUAD; PRAD; SARC; THCA; STAD | MirTarget | TCGA BLCA -0.076; TCGA BRCA -0.084; TCGA COAD -0.087; TCGA ESCA -0.096; TCGA KIRP -0.089; TCGA LUAD -0.149; TCGA PRAD -0.139; TCGA SARC -0.069; TCGA THCA -0.084; TCGA STAD -0.179 |
hsa-miR-27b-3p | BRPF3 | 9 cancers: BLCA; BRCA; CESC; ESCA; LGG; LUAD; LUSC; PRAD; STAD | MirTarget; miRNATAP | TCGA BLCA -0.128; TCGA BRCA -0.145; TCGA CESC -0.173; TCGA ESCA -0.296; TCGA LGG -0.136; TCGA LUAD -0.094; TCGA LUSC -0.157; TCGA PRAD -0.234; TCGA STAD -0.121 |
hsa-miR-27b-3p | PLXND1 | 9 cancers: BLCA; CESC; ESCA; HNSC; KIRC; LIHC; LUSC; PAAD; SARC | MirTarget; miRNATAP | TCGA BLCA -0.209; TCGA CESC -0.168; TCGA ESCA -0.326; TCGA HNSC -0.169; TCGA KIRC -0.286; TCGA LIHC -0.189; TCGA LUSC -0.419; TCGA PAAD -0.241; TCGA SARC -0.142 |
hsa-miR-27b-3p | PDHX | 9 cancers: BLCA; BRCA; ESCA; KIRP; LGG; LUAD; PRAD; THCA; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.11; TCGA BRCA -0.081; TCGA ESCA -0.115; TCGA KIRP -0.105; TCGA LGG -0.063; TCGA LUAD -0.126; TCGA PRAD -0.226; TCGA THCA -0.064; TCGA UCEC -0.138 |
hsa-miR-27b-3p | SLFN11 | 11 cancers: BLCA; CESC; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; THCA; STAD | MirTarget | TCGA BLCA -0.285; TCGA CESC -0.52; TCGA ESCA -0.538; TCGA HNSC -0.219; TCGA KIRC -0.65; TCGA KIRP -0.168; TCGA LUAD -0.256; TCGA LUSC -0.511; TCGA PAAD -0.348; TCGA THCA -0.21; TCGA STAD -0.34 |
hsa-miR-27b-3p | ARHGEF6 | 10 cancers: BLCA; CESC; ESCA; KIRC; KIRP; LUSC; PAAD; SARC; THCA; STAD | MirTarget | TCGA BLCA -0.246; TCGA CESC -0.18; TCGA ESCA -0.459; TCGA KIRC -0.529; TCGA KIRP -0.142; TCGA LUSC -0.623; TCGA PAAD -0.609; TCGA SARC -0.091; TCGA THCA -0.429; TCGA STAD -0.226 |
hsa-miR-27b-3p | IQGAP2 | 12 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LUAD; LUSC; OV; PRAD; SARC; THCA; STAD | MirTarget | TCGA BLCA -0.302; TCGA BRCA -0.219; TCGA CESC -0.417; TCGA ESCA -1.271; TCGA HNSC -0.241; TCGA LUAD -0.189; TCGA LUSC -0.672; TCGA OV -0.161; TCGA PRAD -0.333; TCGA SARC -0.369; TCGA THCA -0.484; TCGA STAD -0.303 |
hsa-miR-27b-3p | PSMA1 | 10 cancers: BLCA; COAD; HNSC; KIRC; KIRP; LIHC; LUAD; OV; PRAD; UCEC | MirTarget | TCGA BLCA -0.118; TCGA COAD -0.136; TCGA HNSC -0.137; TCGA KIRC -0.237; TCGA KIRP -0.176; TCGA LIHC -0.119; TCGA LUAD -0.106; TCGA OV -0.074; TCGA PRAD -0.211; TCGA UCEC -0.081 |
hsa-miR-27b-3p | AQP11 | 9 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LGG; LUAD; OV; PRAD | MirTarget; miRNATAP | TCGA BLCA -0.18; TCGA BRCA -0.289; TCGA CESC -0.337; TCGA ESCA -0.598; TCGA HNSC -0.154; TCGA LGG -0.106; TCGA LUAD -0.182; TCGA OV -0.233; TCGA PRAD -0.502 |
hsa-miR-27b-3p | LBR | 11 cancers: BLCA; CESC; COAD; ESCA; KIRC; LGG; LUAD; OV; PAAD; THCA; STAD | MirTarget | TCGA BLCA -0.211; TCGA CESC -0.165; TCGA COAD -0.189; TCGA ESCA -0.234; TCGA KIRC -0.093; TCGA LGG -0.125; TCGA LUAD -0.1; TCGA OV -0.285; TCGA PAAD -0.226; TCGA THCA -0.266; TCGA STAD -0.247 |
hsa-miR-27b-3p | MAD2L1BP | 12 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LGG; LIHC; LUAD; OV; PRAD; SARC; UCEC | MirTarget | TCGA BLCA -0.08; TCGA BRCA -0.06; TCGA CESC -0.11; TCGA ESCA -0.199; TCGA HNSC -0.12; TCGA LGG -0.093; TCGA LIHC -0.07; TCGA LUAD -0.088; TCGA OV -0.098; TCGA PRAD -0.214; TCGA SARC -0.064; TCGA UCEC -0.099 |
hsa-miR-27b-3p | SGTB | 11 cancers: BLCA; COAD; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; THCA; STAD | MirTarget | TCGA BLCA -0.164; TCGA COAD -0.176; TCGA HNSC -0.196; TCGA KIRC -0.163; TCGA KIRP -0.192; TCGA LIHC -0.161; TCGA LUAD -0.125; TCGA LUSC -0.134; TCGA PAAD -0.194; TCGA THCA -0.215; TCGA STAD -0.203 |
hsa-miR-27b-3p | PHLPP2 | 13 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; SARC; THCA; STAD | MirTarget; miRNATAP | TCGA BLCA -0.119; TCGA BRCA -0.175; TCGA CESC -0.266; TCGA ESCA -0.362; TCGA HNSC -0.166; TCGA KIRP -0.268; TCGA LGG -0.083; TCGA LIHC -0.141; TCGA LUAD -0.142; TCGA LUSC -0.144; TCGA SARC -0.126; TCGA THCA -0.103; TCGA STAD -0.159 |
hsa-miR-27b-3p | ATP11C | 9 cancers: BLCA; COAD; ESCA; KIRC; LGG; LUSC; PAAD; THCA; STAD | MirTarget; miRNATAP | TCGA BLCA -0.145; TCGA COAD -0.33; TCGA ESCA -0.232; TCGA KIRC -0.109; TCGA LGG -0.056; TCGA LUSC -0.211; TCGA PAAD -0.447; TCGA THCA -0.191; TCGA STAD -0.186 |
hsa-miR-27b-3p | FAM78A | 12 cancers: BLCA; CESC; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; PAAD; SARC; THCA; STAD | MirTarget; miRNATAP | TCGA BLCA -0.228; TCGA CESC -0.193; TCGA ESCA -0.524; TCGA HNSC -0.176; TCGA KIRC -0.609; TCGA LIHC -0.226; TCGA LUAD -0.183; TCGA LUSC -0.489; TCGA PAAD -0.652; TCGA SARC -0.375; TCGA THCA -0.477; TCGA STAD -0.301 |
hsa-miR-27b-3p | TNFRSF1B | 9 cancers: BLCA; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; SARC; STAD | mirMAP | TCGA BLCA -0.229; TCGA ESCA -0.714; TCGA HNSC -0.218; TCGA KIRC -0.366; TCGA LUAD -0.127; TCGA LUSC -0.621; TCGA PAAD -0.449; TCGA SARC -0.497; TCGA STAD -0.182 |
hsa-miR-27b-3p | EIF2AK2 | 12 cancers: BLCA; CESC; COAD; HNSC; KIRC; KIRP; LIHC; LUAD; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.165; TCGA CESC -0.102; TCGA COAD -0.276; TCGA HNSC -0.3; TCGA KIRC -0.192; TCGA KIRP -0.175; TCGA LIHC -0.152; TCGA LUAD -0.203; TCGA PRAD -0.261; TCGA THCA -0.226; TCGA STAD -0.242; TCGA UCEC -0.21 |
hsa-miR-27b-3p | CD84 | 12 cancers: BLCA; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.323; TCGA COAD -0.726; TCGA ESCA -0.594; TCGA HNSC -0.244; TCGA KIRC -1.073; TCGA KIRP -0.527; TCGA LUAD -0.316; TCGA LUSC -0.806; TCGA PAAD -0.983; TCGA SARC -0.279; TCGA THCA -0.696; TCGA STAD -0.581 |
hsa-miR-27b-3p | IQCE | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LIHC; LUSC; OV; PRAD; SARC | mirMAP | TCGA BLCA -0.116; TCGA BRCA -0.079; TCGA CESC -0.292; TCGA ESCA -0.352; TCGA HNSC -0.146; TCGA LIHC -0.325; TCGA LUSC -0.13; TCGA OV -0.193; TCGA PRAD -0.176; TCGA SARC -0.117 |
hsa-miR-27b-3p | GPR180 | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; OV; PRAD; THCA; UCEC | mirMAP | TCGA BLCA -0.118; TCGA CESC -0.115; TCGA COAD -0.156; TCGA ESCA -0.203; TCGA HNSC -0.174; TCGA KIRC -0.233; TCGA KIRP -0.2; TCGA LUAD -0.113; TCGA OV -0.151; TCGA PRAD -0.227; TCGA THCA -0.089; TCGA UCEC -0.108 |
hsa-miR-27b-3p | IKZF1 | 9 cancers: BLCA; ESCA; KIRC; KIRP; LUSC; PAAD; SARC; THCA; STAD | miRNATAP | TCGA BLCA -0.306; TCGA ESCA -0.39; TCGA KIRC -0.811; TCGA KIRP -0.227; TCGA LUSC -0.68; TCGA PAAD -0.872; TCGA SARC -0.338; TCGA THCA -0.789; TCGA STAD -0.307 |
hsa-miR-27b-3p | GNS | 15 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.114; TCGA BRCA -0.1; TCGA CESC -0.239; TCGA ESCA -0.263; TCGA HNSC -0.142; TCGA KIRP -0.186; TCGA LIHC -0.216; TCGA LUAD -0.181; TCGA LUSC -0.184; TCGA PAAD -0.152; TCGA PRAD -0.063; TCGA SARC -0.133; TCGA THCA -0.193; TCGA STAD -0.156; TCGA UCEC -0.072 |
hsa-miR-27b-3p | MMD | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; SARC; STAD | miRNATAP | TCGA BLCA -0.267; TCGA CESC -0.133; TCGA COAD -0.212; TCGA ESCA -0.232; TCGA HNSC -0.194; TCGA KIRC -0.275; TCGA KIRP -0.358; TCGA LUAD -0.195; TCGA LUSC -0.156; TCGA PAAD -0.349; TCGA SARC -0.208; TCGA STAD -0.193 |
hsa-miR-27b-3p | TMSB10 | 10 cancers: BLCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; OV; SARC; UCEC | miRNATAP | TCGA BLCA -0.19; TCGA HNSC -0.461; TCGA KIRC -0.645; TCGA KIRP -0.217; TCGA LIHC -0.316; TCGA LUAD -0.109; TCGA LUSC -0.241; TCGA OV -0.159; TCGA SARC -0.196; TCGA UCEC -0.125 |
hsa-miR-27b-3p | ZEB2 | 9 cancers: BLCA; COAD; ESCA; HNSC; KIRC; LUSC; PAAD; SARC; THCA | miRNATAP | TCGA BLCA -0.26; TCGA COAD -0.431; TCGA ESCA -0.336; TCGA HNSC -0.305; TCGA KIRC -0.324; TCGA LUSC -0.482; TCGA PAAD -0.617; TCGA SARC -0.137; TCGA THCA -0.281 |
hsa-miR-27b-3p | CD28 | 11 cancers: BLCA; ESCA; HNSC; KIRC; KIRP; LIHC; LUSC; PAAD; SARC; THCA; STAD | miRNATAP | TCGA BLCA -0.225; TCGA ESCA -0.583; TCGA HNSC -0.204; TCGA KIRC -0.762; TCGA KIRP -0.32; TCGA LIHC -0.207; TCGA LUSC -0.624; TCGA PAAD -0.825; TCGA SARC -0.42; TCGA THCA -0.848; TCGA STAD -0.308 |
hsa-miR-27b-3p | DLL4 | 9 cancers: BLCA; CESC; ESCA; HNSC; KIRC; LIHC; LUSC; SARC; STAD | miRNATAP | TCGA BLCA -0.124; TCGA CESC -0.147; TCGA ESCA -0.569; TCGA HNSC -0.215; TCGA KIRC -0.285; TCGA LIHC -0.153; TCGA LUSC -0.356; TCGA SARC -0.139; TCGA STAD -0.206 |
hsa-miR-27b-3p | SNX10 | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PRAD; THCA; STAD | miRNATAP | TCGA BLCA -0.476; TCGA BRCA -0.099; TCGA COAD -0.336; TCGA ESCA -0.253; TCGA HNSC -0.48; TCGA KIRC -0.499; TCGA KIRP -0.291; TCGA LUAD -0.302; TCGA LUSC -0.308; TCGA PRAD -0.281; TCGA THCA -0.389; TCGA STAD -0.513 |
hsa-miR-27b-3p | MARCH1 | 13 cancers: BLCA; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; PAAD; SARC; THCA; STAD | miRNATAP | TCGA BLCA -0.221; TCGA COAD -0.346; TCGA ESCA -0.248; TCGA HNSC -0.139; TCGA KIRC -0.727; TCGA KIRP -0.208; TCGA LGG -0.098; TCGA LUAD -0.252; TCGA LUSC -0.454; TCGA PAAD -0.755; TCGA SARC -0.23; TCGA THCA -0.38; TCGA STAD -0.288 |
hsa-miR-27b-3p | H2AFZ | 10 cancers: BLCA; KIRC; KIRP; LGG; LIHC; LUAD; OV; PRAD; THCA; UCEC | miRNATAP | TCGA BLCA -0.106; TCGA KIRC -0.2; TCGA KIRP -0.074; TCGA LGG -0.057; TCGA LIHC -0.287; TCGA LUAD -0.216; TCGA OV -0.131; TCGA PRAD -0.092; TCGA THCA -0.143; TCGA UCEC -0.143 |
hsa-miR-27b-3p | MAP3K12 | 10 cancers: BLCA; BRCA; CESC; COAD; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD | miRNATAP | TCGA BLCA -0.169; TCGA BRCA -0.142; TCGA CESC -0.221; TCGA COAD -0.321; TCGA KIRC -0.428; TCGA KIRP -0.162; TCGA LIHC -0.211; TCGA LUAD -0.111; TCGA LUSC -0.3; TCGA PAAD -0.255 |
hsa-miR-27b-3p | RALA | 9 cancers: BLCA; KIRC; LGG; LIHC; LUAD; OV; PRAD; SARC; THCA | miRNATAP | TCGA BLCA -0.09; TCGA KIRC -0.152; TCGA LGG -0.063; TCGA LIHC -0.128; TCGA LUAD -0.089; TCGA OV -0.11; TCGA PRAD -0.154; TCGA SARC -0.116; TCGA THCA -0.17 |
hsa-miR-27b-3p | E2F6 | 10 cancers: BLCA; COAD; KIRC; KIRP; LGG; LIHC; LUAD; OV; PRAD; STAD | miRNATAP | TCGA BLCA -0.06; TCGA COAD -0.168; TCGA KIRC -0.088; TCGA KIRP -0.121; TCGA LGG -0.062; TCGA LIHC -0.109; TCGA LUAD -0.108; TCGA OV -0.197; TCGA PRAD -0.051; TCGA STAD -0.152 |
hsa-miR-27b-3p | KIF21B | 11 cancers: BLCA; CESC; ESCA; HNSC; KIRC; LGG; LIHC; LUSC; PAAD; THCA; STAD | miRNATAP | TCGA BLCA -0.162; TCGA CESC -0.309; TCGA ESCA -0.659; TCGA HNSC -0.301; TCGA KIRC -0.712; TCGA LGG -0.245; TCGA LIHC -0.675; TCGA LUSC -0.705; TCGA PAAD -0.3; TCGA THCA -0.633; TCGA STAD -0.239 |
hsa-miR-27b-3p | RAP2C | 12 cancers: BLCA; BRCA; COAD; HNSC; KIRC; LUAD; LUSC; OV; PAAD; SARC; THCA; STAD | miRNATAP | TCGA BLCA -0.091; TCGA BRCA -0.132; TCGA COAD -0.288; TCGA HNSC -0.079; TCGA KIRC -0.215; TCGA LUAD -0.156; TCGA LUSC -0.09; TCGA OV -0.089; TCGA PAAD -0.132; TCGA SARC -0.096; TCGA THCA -0.099; TCGA STAD -0.184 |
hsa-miR-27b-3p | KLHDC3 | 9 cancers: BLCA; CESC; HNSC; LGG; LUAD; LUSC; OV; PRAD; UCEC | miRNATAP | TCGA BLCA -0.104; TCGA CESC -0.081; TCGA HNSC -0.068; TCGA LGG -0.083; TCGA LUAD -0.084; TCGA LUSC -0.097; TCGA OV -0.203; TCGA PRAD -0.144; TCGA UCEC -0.07 |
hsa-miR-27b-3p | TSR1 | 10 cancers: BRCA; KIRC; KIRP; LGG; LIHC; LUAD; PAAD; PRAD; THCA; UCEC | miRNAWalker2 validate | TCGA BRCA -0.056; TCGA KIRC -0.169; TCGA KIRP -0.131; TCGA LGG -0.051; TCGA LIHC -0.072; TCGA LUAD -0.122; TCGA PAAD -0.166; TCGA PRAD -0.14; TCGA THCA -0.099; TCGA UCEC -0.065 |
hsa-miR-27b-3p | B4GALT3 | 10 cancers: BRCA; ESCA; HNSC; KIRP; LGG; LIHC; OV; PRAD; SARC; UCEC | MirTarget; miRNATAP | TCGA BRCA -0.091; TCGA ESCA -0.151; TCGA HNSC -0.128; TCGA KIRP -0.073; TCGA LGG -0.13; TCGA LIHC -0.203; TCGA OV -0.174; TCGA PRAD -0.364; TCGA SARC -0.096; TCGA UCEC -0.215 |
hsa-miR-27b-3p | CCNG1 | 10 cancers: BRCA; CESC; ESCA; KIRP; LUAD; LUSC; PRAD; SARC; THCA; STAD | MirTarget; miRNATAP | TCGA BRCA -0.12; TCGA CESC -0.108; TCGA ESCA -0.285; TCGA KIRP -0.275; TCGA LUAD -0.172; TCGA LUSC -0.262; TCGA PRAD -0.302; TCGA SARC -0.132; TCGA THCA -0.094; TCGA STAD -0.177 |
hsa-miR-27b-3p | LPCAT1 | 12 cancers: BRCA; CESC; HNSC; KIRC; KIRP; LGG; LIHC; LUSC; OV; PRAD; THCA; UCEC | MirTarget | TCGA BRCA -0.069; TCGA CESC -0.291; TCGA HNSC -0.417; TCGA KIRC -0.79; TCGA KIRP -0.141; TCGA LGG -0.169; TCGA LIHC -0.601; TCGA LUSC -0.341; TCGA OV -0.134; TCGA PRAD -0.239; TCGA THCA -0.255; TCGA UCEC -0.108 |
hsa-miR-27b-3p | MOCS3 | 9 cancers: BRCA; KIRC; KIRP; LGG; LUAD; OV; PRAD; SARC; UCEC | MirTarget | TCGA BRCA -0.058; TCGA KIRC -0.161; TCGA KIRP -0.137; TCGA LGG -0.077; TCGA LUAD -0.078; TCGA OV -0.116; TCGA PRAD -0.182; TCGA SARC -0.077; TCGA UCEC -0.072 |
hsa-miR-27b-3p | ZNF100 | 11 cancers: BRCA; CESC; COAD; ESCA; KIRC; LGG; LUSC; OV; PRAD; SARC; STAD | MirTarget | TCGA BRCA -0.184; TCGA CESC -0.287; TCGA COAD -0.189; TCGA ESCA -0.448; TCGA KIRC -0.088; TCGA LGG -0.206; TCGA LUSC -0.194; TCGA OV -0.18; TCGA PRAD -0.18; TCGA SARC -0.127; TCGA STAD -0.248 |
hsa-miR-27b-3p | RPS6KB1 | 9 cancers: BRCA; COAD; KIRC; KIRP; LGG; LUAD; PAAD; PRAD; STAD | MirTarget; miRNATAP | TCGA BRCA -0.097; TCGA COAD -0.106; TCGA KIRC -0.173; TCGA KIRP -0.066; TCGA LGG -0.09; TCGA LUAD -0.084; TCGA PAAD -0.182; TCGA PRAD -0.251; TCGA STAD -0.134 |
hsa-miR-27b-3p | EPB41 | 10 cancers: BRCA; ESCA; KIRC; KIRP; LIHC; LUSC; OV; PRAD; THCA; STAD | MirTarget; miRNATAP | TCGA BRCA -0.129; TCGA ESCA -0.271; TCGA KIRC -0.306; TCGA KIRP -0.097; TCGA LIHC -0.092; TCGA LUSC -0.11; TCGA OV -0.099; TCGA PRAD -0.208; TCGA THCA -0.344; TCGA STAD -0.112 |
hsa-miR-27b-3p | RPGRIP1L | 10 cancers: BRCA; CESC; HNSC; KIRC; KIRP; LIHC; LUSC; PRAD; THCA; STAD | MirTarget; miRNATAP | TCGA BRCA -0.107; TCGA CESC -0.224; TCGA HNSC -0.078; TCGA KIRC -0.335; TCGA KIRP -0.238; TCGA LIHC -0.182; TCGA LUSC -0.142; TCGA PRAD -0.231; TCGA THCA -0.078; TCGA STAD -0.165 |
hsa-miR-27b-3p | SMCR8 | 9 cancers: BRCA; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; THCA; STAD | MirTarget | TCGA BRCA -0.16; TCGA KIRP -0.104; TCGA LIHC -0.113; TCGA LUAD -0.144; TCGA LUSC -0.105; TCGA PAAD -0.22; TCGA PRAD -0.182; TCGA THCA -0.123; TCGA STAD -0.113 |
hsa-miR-27b-3p | TMED5 | 13 cancers: BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; OV; PRAD; SARC; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BRCA -0.138; TCGA CESC -0.127; TCGA COAD -0.149; TCGA ESCA -0.305; TCGA HNSC -0.125; TCGA LUAD -0.193; TCGA LUSC -0.259; TCGA OV -0.098; TCGA PRAD -0.22; TCGA SARC -0.128; TCGA THCA -0.097; TCGA STAD -0.207; TCGA UCEC -0.127 |
hsa-miR-27b-3p | SEC24A | 13 cancers: BRCA; CESC; ESCA; KIRC; KIRP; LGG; LUAD; LUSC; PRAD; SARC; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BRCA -0.077; TCGA CESC -0.141; TCGA ESCA -0.317; TCGA KIRC -0.348; TCGA KIRP -0.138; TCGA LGG -0.109; TCGA LUAD -0.107; TCGA LUSC -0.291; TCGA PRAD -0.23; TCGA SARC -0.072; TCGA THCA -0.199; TCGA STAD -0.144; TCGA UCEC -0.145 |
hsa-miR-27b-3p | TMEM167A | 11 cancers: BRCA; ESCA; KIRC; KIRP; LGG; LIHC; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BRCA -0.055; TCGA ESCA -0.113; TCGA KIRC -0.141; TCGA KIRP -0.132; TCGA LGG -0.06; TCGA LIHC -0.147; TCGA PRAD -0.225; TCGA SARC -0.142; TCGA THCA -0.12; TCGA STAD -0.109; TCGA UCEC -0.066 |
hsa-miR-27b-3p | HYOU1 | 11 cancers: BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LIHC; OV; PRAD; THCA; UCEC | MirTarget | TCGA BRCA -0.062; TCGA CESC -0.187; TCGA ESCA -0.185; TCGA HNSC -0.083; TCGA KIRC -0.147; TCGA KIRP -0.177; TCGA LIHC -0.106; TCGA OV -0.093; TCGA PRAD -0.34; TCGA THCA -0.263; TCGA UCEC -0.183 |
hsa-miR-27b-3p | AAGAB | 9 cancers: BRCA; HNSC; LIHC; LUAD; OV; PRAD; SARC; THCA; UCEC | MirTarget | TCGA BRCA -0.157; TCGA HNSC -0.068; TCGA LIHC -0.126; TCGA LUAD -0.161; TCGA OV -0.133; TCGA PRAD -0.195; TCGA SARC -0.064; TCGA THCA -0.102; TCGA UCEC -0.13 |
hsa-miR-27b-3p | SSR1 | 11 cancers: BRCA; CESC; KIRP; LGG; LIHC; LUAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BRCA -0.061; TCGA CESC -0.144; TCGA KIRP -0.092; TCGA LGG -0.055; TCGA LIHC -0.08; TCGA LUAD -0.082; TCGA PRAD -0.257; TCGA SARC -0.09; TCGA THCA -0.118; TCGA STAD -0.139; TCGA UCEC -0.1 |
hsa-miR-27b-3p | DCP2 | 12 cancers: BRCA; CESC; ESCA; KIRC; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD | miRNATAP | TCGA BRCA -0.07; TCGA CESC -0.087; TCGA ESCA -0.173; TCGA KIRC -0.189; TCGA LIHC -0.08; TCGA LUAD -0.077; TCGA LUSC -0.19; TCGA PAAD -0.174; TCGA PRAD -0.093; TCGA SARC -0.081; TCGA THCA -0.093; TCGA STAD -0.265 |
hsa-miR-27b-3p | GXYLT1 | 12 cancers: BRCA; CESC; COAD; ESCA; KIRC; KIRP; LUAD; LUSC; OV; PRAD; SARC; STAD | miRNATAP | TCGA BRCA -0.141; TCGA CESC -0.159; TCGA COAD -0.125; TCGA ESCA -0.382; TCGA KIRC -0.222; TCGA KIRP -0.176; TCGA LUAD -0.219; TCGA LUSC -0.125; TCGA OV -0.117; TCGA PRAD -0.214; TCGA SARC -0.314; TCGA STAD -0.24 |
hsa-miR-27b-3p | UHRF1BP1 | 12 cancers: BRCA; CESC; COAD; KIRP; LGG; LIHC; LUAD; PAAD; PRAD; THCA; STAD; UCEC | miRNATAP | TCGA BRCA -0.119; TCGA CESC -0.082; TCGA COAD -0.127; TCGA KIRP -0.186; TCGA LGG -0.102; TCGA LIHC -0.108; TCGA LUAD -0.13; TCGA PAAD -0.148; TCGA PRAD -0.352; TCGA THCA -0.174; TCGA STAD -0.159; TCGA UCEC -0.152 |
hsa-miR-27b-3p | ARHGAP12 | 9 cancers: BRCA; CESC; ESCA; LGG; LIHC; LUSC; PRAD; SARC; UCEC | miRNATAP | TCGA BRCA -0.069; TCGA CESC -0.23; TCGA ESCA -0.548; TCGA LGG -0.137; TCGA LIHC -0.108; TCGA LUSC -0.159; TCGA PRAD -0.162; TCGA SARC -0.143; TCGA UCEC -0.071 |
hsa-miR-27b-3p | NLK | 9 cancers: BRCA; CESC; ESCA; KIRP; LUAD; OV; PRAD; STAD; UCEC | miRNATAP | TCGA BRCA -0.116; TCGA CESC -0.106; TCGA ESCA -0.358; TCGA KIRP -0.097; TCGA LUAD -0.118; TCGA OV -0.089; TCGA PRAD -0.188; TCGA STAD -0.097; TCGA UCEC -0.086 |
hsa-miR-27b-3p | GFPT1 | 10 cancers: BRCA; CESC; ESCA; KIRP; LGG; PRAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BRCA -0.151; TCGA CESC -0.356; TCGA ESCA -0.579; TCGA KIRP -0.25; TCGA LGG -0.095; TCGA PRAD -0.39; TCGA SARC -0.145; TCGA THCA -0.137; TCGA STAD -0.204; TCGA UCEC -0.133 |
hsa-miR-27b-3p | CPD | 12 cancers: BRCA; CESC; ESCA; KIRC; KIRP; LIHC; LUAD; OV; PRAD; SARC; THCA; UCEC | miRNATAP | TCGA BRCA -0.236; TCGA CESC -0.424; TCGA ESCA -0.269; TCGA KIRC -0.223; TCGA KIRP -0.337; TCGA LIHC -0.245; TCGA LUAD -0.175; TCGA OV -0.108; TCGA PRAD -0.302; TCGA SARC -0.132; TCGA THCA -0.149; TCGA UCEC -0.207 |
hsa-miR-27b-3p | FNDC3A | 9 cancers: BRCA; CESC; ESCA; HNSC; LGG; LUAD; LUSC; OV; SARC | miRNATAP | TCGA BRCA -0.142; TCGA CESC -0.273; TCGA ESCA -0.534; TCGA HNSC -0.14; TCGA LGG -0.078; TCGA LUAD -0.126; TCGA LUSC -0.341; TCGA OV -0.167; TCGA SARC -0.247 |
hsa-miR-27b-3p | SPATA2 | 9 cancers: BRCA; ESCA; KIRP; OV; PRAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BRCA -0.051; TCGA ESCA -0.219; TCGA KIRP -0.086; TCGA OV -0.073; TCGA PRAD -0.232; TCGA SARC -0.121; TCGA THCA -0.086; TCGA STAD -0.124; TCGA UCEC -0.059 |
hsa-miR-27b-3p | APAF1 | 10 cancers: BRCA; CESC; ESCA; KIRC; LUAD; LUSC; OV; SARC; THCA; STAD | miRNATAP | TCGA BRCA -0.091; TCGA CESC -0.158; TCGA ESCA -0.478; TCGA KIRC -0.319; TCGA LUAD -0.166; TCGA LUSC -0.262; TCGA OV -0.09; TCGA SARC -0.149; TCGA THCA -0.076; TCGA STAD -0.265 |
hsa-miR-27b-3p | SUZ12 | 9 cancers: BRCA; COAD; ESCA; LGG; LUAD; PRAD; THCA; STAD; UCEC | miRNATAP | TCGA BRCA -0.087; TCGA COAD -0.158; TCGA ESCA -0.186; TCGA LGG -0.079; TCGA LUAD -0.151; TCGA PRAD -0.178; TCGA THCA -0.056; TCGA STAD -0.145; TCGA UCEC -0.091 |
hsa-miR-27b-3p | DNAJB9 | 9 cancers: BRCA; COAD; ESCA; KIRC; LUSC; OV; PRAD; SARC; THCA | miRNATAP | TCGA BRCA -0.143; TCGA COAD -0.148; TCGA ESCA -0.213; TCGA KIRC -0.191; TCGA LUSC -0.163; TCGA OV -0.104; TCGA PRAD -0.151; TCGA SARC -0.125; TCGA THCA -0.15 |
hsa-miR-27b-3p | DMXL2 | 13 cancers: BRCA; CESC; ESCA; HNSC; KIRC; LGG; LIHC; LUAD; LUSC; OV; PRAD; SARC; STAD | miRNATAP | TCGA BRCA -0.151; TCGA CESC -0.159; TCGA ESCA -0.6; TCGA HNSC -0.228; TCGA KIRC -0.154; TCGA LGG -0.058; TCGA LIHC -0.123; TCGA LUAD -0.333; TCGA LUSC -0.338; TCGA OV -0.096; TCGA PRAD -0.194; TCGA SARC -0.069; TCGA STAD -0.301 |
hsa-miR-27b-3p | MANEAL | 10 cancers: BRCA; CESC; ESCA; KIRC; KIRP; LGG; LUSC; OV; PRAD; STAD | miRNATAP | TCGA BRCA -0.229; TCGA CESC -0.27; TCGA ESCA -0.409; TCGA KIRC -0.213; TCGA KIRP -0.501; TCGA LGG -0.107; TCGA LUSC -0.469; TCGA OV -0.174; TCGA PRAD -0.483; TCGA STAD -0.431 |
hsa-miR-27b-3p | RPN2 | 12 cancers: BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; OV; PRAD; SARC; UCEC | miRNATAP | TCGA BRCA -0.072; TCGA CESC -0.146; TCGA ESCA -0.215; TCGA HNSC -0.112; TCGA KIRC -0.212; TCGA KIRP -0.175; TCGA LIHC -0.155; TCGA LUAD -0.084; TCGA OV -0.132; TCGA PRAD -0.254; TCGA SARC -0.112; TCGA UCEC -0.21 |
hsa-miR-27b-3p | HSPD1 | 11 cancers: CESC; COAD; ESCA; KIRP; LGG; LIHC; LUAD; PRAD; THCA; STAD; UCEC | MirTarget | TCGA CESC -0.101; TCGA COAD -0.193; TCGA ESCA -0.235; TCGA KIRP -0.234; TCGA LGG -0.086; TCGA LIHC -0.084; TCGA LUAD -0.121; TCGA PRAD -0.501; TCGA THCA -0.127; TCGA STAD -0.234; TCGA UCEC -0.105 |
hsa-miR-27b-3p | BEST1 | 10 cancers: CESC; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; PAAD; SARC; THCA | MirTarget | TCGA CESC -0.257; TCGA ESCA -0.352; TCGA HNSC -0.155; TCGA KIRC -0.785; TCGA LIHC -0.248; TCGA LUAD -0.223; TCGA LUSC -0.252; TCGA PAAD -0.513; TCGA SARC -0.268; TCGA THCA -0.122 |
hsa-miR-27b-3p | NUS1 | 11 cancers: CESC; ESCA; HNSC; LGG; LUAD; OV; PAAD; PRAD; THCA; STAD; UCEC | MirTarget | TCGA CESC -0.081; TCGA ESCA -0.206; TCGA HNSC -0.088; TCGA LGG -0.065; TCGA LUAD -0.147; TCGA OV -0.082; TCGA PAAD -0.123; TCGA PRAD -0.209; TCGA THCA -0.06; TCGA STAD -0.162; TCGA UCEC -0.071 |
hsa-miR-27b-3p | NEK6 | 10 cancers: CESC; ESCA; HNSC; KIRC; LUSC; PAAD; SARC; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA CESC -0.365; TCGA ESCA -0.466; TCGA HNSC -0.334; TCGA KIRC -0.578; TCGA LUSC -0.35; TCGA PAAD -0.186; TCGA SARC -0.228; TCGA THCA -0.638; TCGA STAD -0.203; TCGA UCEC -0.167 |
hsa-miR-27b-3p | CCNC | 9 cancers: CESC; ESCA; LGG; LUAD; OV; PRAD; SARC; THCA; UCEC | MirTarget | TCGA CESC -0.113; TCGA ESCA -0.12; TCGA LGG -0.065; TCGA LUAD -0.113; TCGA OV -0.129; TCGA PRAD -0.243; TCGA SARC -0.092; TCGA THCA -0.097; TCGA UCEC -0.16 |
hsa-miR-27b-3p | YWHAB | 11 cancers: CESC; HNSC; KIRP; LIHC; LUAD; PAAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA CESC -0.064; TCGA HNSC -0.078; TCGA KIRP -0.096; TCGA LIHC -0.084; TCGA LUAD -0.063; TCGA PAAD -0.084; TCGA PRAD -0.055; TCGA SARC -0.064; TCGA THCA -0.062; TCGA STAD -0.096; TCGA UCEC -0.118 |
hsa-miR-27b-3p | ONECUT2 | 10 cancers: CESC; ESCA; KIRC; KIRP; LGG; OV; PRAD; THCA; STAD; UCEC | miRNATAP | TCGA CESC -0.43; TCGA ESCA -1.613; TCGA KIRC -0.484; TCGA KIRP -0.52; TCGA LGG -0.375; TCGA OV -0.443; TCGA PRAD -0.991; TCGA THCA -0.231; TCGA STAD -0.476; TCGA UCEC -0.271 |
hsa-miR-27b-3p | UBASH3B | 11 cancers: CESC; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; SARC; THCA; STAD | miRNATAP | TCGA CESC -0.329; TCGA HNSC -0.291; TCGA KIRC -0.353; TCGA KIRP -0.348; TCGA LIHC -0.253; TCGA LUAD -0.225; TCGA LUSC -0.488; TCGA PAAD -0.554; TCGA SARC -0.19; TCGA THCA -0.682; TCGA STAD -0.2 |
hsa-miR-27b-3p | TTYH3 | 9 cancers: CESC; HNSC; KIRC; LIHC; OV; PRAD; SARC; THCA; STAD | miRNATAP | TCGA CESC -0.259; TCGA HNSC -0.451; TCGA KIRC -0.57; TCGA LIHC -0.303; TCGA OV -0.117; TCGA PRAD -0.289; TCGA SARC -0.319; TCGA THCA -0.267; TCGA STAD -0.22 |
hsa-miR-27b-3p | FOXN2 | 9 cancers: COAD; ESCA; KIRC; LGG; LUAD; PAAD; PRAD; SARC; STAD | MirTarget | TCGA COAD -0.276; TCGA ESCA -0.163; TCGA KIRC -0.216; TCGA LGG -0.073; TCGA LUAD -0.144; TCGA PAAD -0.175; TCGA PRAD -0.162; TCGA SARC -0.124; TCGA STAD -0.234 |
hsa-miR-27b-3p | PRR16 | 9 cancers: COAD; ESCA; HNSC; KIRC; LIHC; LUSC; PAAD; PRAD; STAD | MirTarget | TCGA COAD -0.537; TCGA ESCA -0.355; TCGA HNSC -0.383; TCGA KIRC -0.648; TCGA LIHC -0.298; TCGA LUSC -0.299; TCGA PAAD -0.352; TCGA PRAD -0.864; TCGA STAD -0.185 |
hsa-miR-27b-3p | UBE2D1 | 10 cancers: COAD; ESCA; HNSC; KIRC; KIRP; LUSC; OV; PRAD; SARC; STAD | MirTarget | TCGA COAD -0.118; TCGA ESCA -0.106; TCGA HNSC -0.096; TCGA KIRC -0.195; TCGA KIRP -0.124; TCGA LUSC -0.191; TCGA OV -0.08; TCGA PRAD -0.193; TCGA SARC -0.092; TCGA STAD -0.169 |
hsa-miR-27b-3p | SKA3 | 11 cancers: COAD; KIRC; KIRP; LGG; LIHC; LUAD; OV; PRAD; THCA; STAD; UCEC | MirTarget | TCGA COAD -0.266; TCGA KIRC -0.956; TCGA KIRP -0.281; TCGA LGG -0.185; TCGA LIHC -0.618; TCGA LUAD -0.381; TCGA OV -0.214; TCGA PRAD -0.935; TCGA THCA -0.258; TCGA STAD -0.297; TCGA UCEC -0.367 |
hsa-miR-27b-3p | KBTBD8 | 10 cancers: COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; SARC; THCA; STAD | MirTarget | TCGA COAD -0.336; TCGA ESCA -0.465; TCGA HNSC -0.174; TCGA KIRC -0.367; TCGA LUAD -0.147; TCGA LUSC -0.442; TCGA PAAD -0.546; TCGA SARC -0.321; TCGA THCA -0.449; TCGA STAD -0.215 |
hsa-miR-27b-3p | RGS1 | 9 cancers: COAD; ESCA; HNSC; KIRC; KIRP; LUSC; PAAD; THCA; STAD | MirTarget; miRNATAP | TCGA COAD -0.381; TCGA ESCA -0.292; TCGA HNSC -0.222; TCGA KIRC -1.008; TCGA KIRP -0.454; TCGA LUSC -0.584; TCGA PAAD -0.549; TCGA THCA -0.464; TCGA STAD -0.395 |
hsa-miR-27b-3p | PCYT1A | 9 cancers: COAD; HNSC; KIRC; KIRP; LIHC; LUAD; PAAD; THCA; UCEC | mirMAP | TCGA COAD -0.224; TCGA HNSC -0.08; TCGA KIRC -0.081; TCGA KIRP -0.158; TCGA LIHC -0.07; TCGA LUAD -0.196; TCGA PAAD -0.267; TCGA THCA -0.218; TCGA UCEC -0.065 |
hsa-miR-27b-3p | DNAJC5B | 12 cancers: COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; SARC; THCA; STAD | miRNATAP | TCGA COAD -0.705; TCGA ESCA -0.853; TCGA HNSC -0.271; TCGA KIRC -1.544; TCGA KIRP -0.787; TCGA LIHC -0.22; TCGA LUAD -0.354; TCGA LUSC -0.795; TCGA PAAD -0.985; TCGA SARC -0.22; TCGA THCA -0.6; TCGA STAD -0.765 |
hsa-miR-27b-3p | CCNA2 | 9 cancers: COAD; KIRC; KIRP; LGG; LIHC; LUAD; PRAD; THCA; UCEC | miRNATAP | TCGA COAD -0.204; TCGA KIRC -0.993; TCGA KIRP -0.337; TCGA LGG -0.116; TCGA LIHC -0.541; TCGA LUAD -0.355; TCGA PRAD -0.661; TCGA THCA -0.224; TCGA UCEC -0.276 |
hsa-miR-27b-3p | SLC5A6 | 10 cancers: ESCA; KIRC; KIRP; LIHC; LUAD; OV; PRAD; SARC; THCA; UCEC | miRNAWalker2 validate | TCGA ESCA -0.167; TCGA KIRC -0.149; TCGA KIRP -0.135; TCGA LIHC -0.142; TCGA LUAD -0.149; TCGA OV -0.186; TCGA PRAD -0.324; TCGA SARC -0.152; TCGA THCA -0.174; TCGA UCEC -0.085 |
hsa-miR-27b-3p | CCNJ | 9 cancers: ESCA; KIRC; KIRP; LGG; LUSC; OV; PRAD; SARC; STAD | MirTarget; miRNATAP | TCGA ESCA -0.247; TCGA KIRC -0.174; TCGA KIRP -0.09; TCGA LGG -0.161; TCGA LUSC -0.18; TCGA OV -0.195; TCGA PRAD -0.266; TCGA SARC -0.14; TCGA STAD -0.145 |
hsa-miR-27b-3p | ICOS | 10 cancers: ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; PAAD; SARC; THCA; STAD | miRNATAP | TCGA ESCA -0.37; TCGA HNSC -0.399; TCGA KIRC -1.188; TCGA LIHC -0.333; TCGA LUAD -0.246; TCGA LUSC -0.537; TCGA PAAD -0.906; TCGA SARC -0.348; TCGA THCA -1.028; TCGA STAD -0.507 |
hsa-miR-27b-3p | CBFB | 9 cancers: HNSC; KIRC; KIRP; LGG; LIHC; PAAD; PRAD; SARC; STAD | MirTarget; miRNATAP | TCGA HNSC -0.054; TCGA KIRC -0.348; TCGA KIRP -0.189; TCGA LGG -0.091; TCGA LIHC -0.19; TCGA PAAD -0.117; TCGA PRAD -0.156; TCGA SARC -0.089; TCGA STAD -0.2 |
hsa-miR-27b-3p | CKAP4 | 11 cancers: HNSC; KIRC; KIRP; LGG; LIHC; LUAD; OV; PRAD; SARC; THCA; UCEC | miRNATAP | TCGA HNSC -0.231; TCGA KIRC -0.351; TCGA KIRP -0.192; TCGA LGG -0.064; TCGA LIHC -0.206; TCGA LUAD -0.133; TCGA OV -0.13; TCGA PRAD -0.185; TCGA SARC -0.171; TCGA THCA -0.155; TCGA UCEC -0.086 |
hsa-miR-27b-3p | NRAS | 10 cancers: HNSC; KIRC; LUAD; LUSC; OV; PRAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA HNSC -0.096; TCGA KIRC -0.134; TCGA LUAD -0.155; TCGA LUSC -0.11; TCGA OV -0.147; TCGA PRAD -0.166; TCGA SARC -0.085; TCGA THCA -0.174; TCGA STAD -0.123; TCGA UCEC -0.123 |
hsa-miR-27b-3p | CAPZA1 | 11 cancers: HNSC; KIRC; KIRP; LIHC; LUAD; OV; PAAD; PRAD; SARC; THCA; UCEC | miRNATAP | TCGA HNSC -0.077; TCGA KIRC -0.366; TCGA KIRP -0.111; TCGA LIHC -0.109; TCGA LUAD -0.128; TCGA OV -0.065; TCGA PAAD -0.211; TCGA PRAD -0.213; TCGA SARC -0.081; TCGA THCA -0.093; TCGA UCEC -0.101 |
hsa-miR-27b-3p | SLC36A1 | 9 cancers: KIRC; KIRP; LIHC; LUAD; PAAD; PRAD; SARC; THCA; UCEC | mirMAP | TCGA KIRC -0.372; TCGA KIRP -0.19; TCGA LIHC -0.327; TCGA LUAD -0.103; TCGA PAAD -0.344; TCGA PRAD -0.366; TCGA SARC -0.131; TCGA THCA -0.202; TCGA UCEC -0.085 |
hsa-miR-27b-3p | PPP1CC | 9 cancers: KIRC; KIRP; LGG; LIHC; LUAD; OV; PRAD; SARC; THCA | miRNATAP | TCGA KIRC -0.265; TCGA KIRP -0.123; TCGA LGG -0.096; TCGA LIHC -0.137; TCGA LUAD -0.148; TCGA OV -0.1; TCGA PRAD -0.225; TCGA SARC -0.085; TCGA THCA -0.07 |
hsa-miR-27b-3p | NEK2 | 9 cancers: KIRC; KIRP; LIHC; LUAD; OV; PRAD; THCA; STAD; UCEC | miRNATAP | TCGA KIRC -1.14; TCGA KIRP -0.566; TCGA LIHC -0.776; TCGA LUAD -0.309; TCGA OV -0.237; TCGA PRAD -1.064; TCGA THCA -0.217; TCGA STAD -0.296; TCGA UCEC -0.461 |
Enriched cancer pathways of putative targets