microRNA information: hsa-miR-28-3p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-28-3p | miRbase |
| Accession: | MIMAT0004502 | miRbase |
| Precursor name: | hsa-mir-28 | miRbase |
| Precursor accession: | MI0000086 | miRbase |
| Symbol: | MIR28 | HGNC |
| RefSeq ID: | NR_029502 | GenBank |
| Sequence: | CACUAGAUUGUGAGCUCCUGGA |
Reported expression in cancers: hsa-miR-28-3p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-28-3p | bladder cancer | downregulation | "In the first - discovery - phase microarray cards ......" | 27468885 | Microarray; qPCR |
| hsa-miR-28-3p | colorectal cancer | downregulation | "We measured levels of miR-28-5p and miR-28-3p expr ......" | 22240480 | qPCR |
| hsa-miR-28-3p | esophageal cancer | deregulation | "The expression profiles of miRNAs in paired EC and ......" | 23761828 | Microarray; qPCR |
Reported cancer pathway affected by hsa-miR-28-3p
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|
Reported cancer prognosis affected by hsa-miR-28-3p

| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-28-3p | B cell lymphoma | malignant trasformation | "MicroRNA 28 controls cell proliferation and is dow ......" | 24843176 | |
| hsa-miR-28-3p | colorectal cancer | cell migration; metastasis | "Strand specific miR 28 5p and miR 28 3p have disti ......" | 22240480 | |
| hsa-miR-28-3p | kidney renal cell cancer | metastasis; malignant trasformation | "To obtain accurate results in miRNA expression cha ......" | 21741950 | |
| hsa-miR-28-3p | liver cancer | metastasis | "miR 28 5p IL 34 macrophage feedback loop modulates ......" | 26754294 |
Reported gene related to hsa-miR-28-3p

| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-28-3p | B cell lymphoma | BCL6 | "Expressions of bcl 6 lpp and miR 28 genes in diffu ......" | 19236753 |
| hsa-miR-28-3p | liver cancer | IL34 | "miR 28 5p IL 34 macrophage feedback loop modulates ......" | 26754294 |
| hsa-miR-28-3p | B cell lymphoma | LPP | "Expressions of bcl 6 lpp and miR 28 genes in diffu ......" | 19236753 |
| hsa-miR-28-3p | B cell lymphoma | MYC | "We identify the oncogene MYC as a negative regulat ......" | 24843176 |
| hsa-miR-28-3p | B cell lymphoma | NPR2 | "A comparative analysis of microRNAs expressed in n ......" | 24843176 |
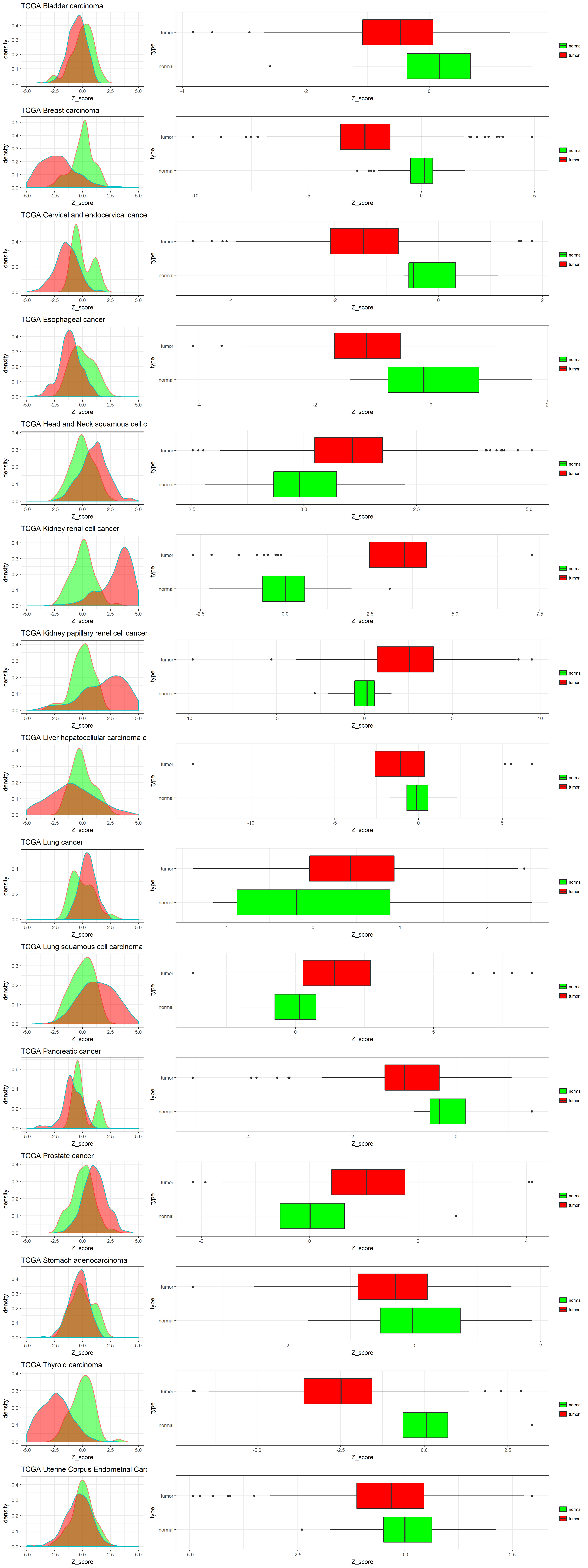
Expression profile in cancer corhorts:
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-28-3p | VPS53 | 9 cancers: BLCA; CESC; HNSC; KIRC; LGG; LIHC; PAAD; SARC; STAD | miRNAWalker2 validate | TCGA BLCA -0.113; TCGA CESC -0.156; TCGA HNSC -0.215; TCGA KIRC -0.265; TCGA LGG -0.128; TCGA LIHC -0.171; TCGA PAAD -0.231; TCGA SARC -0.22; TCGA STAD -0.131 |
hsa-miR-28-3p | INPP5E | 10 cancers: BLCA; BRCA; CESC; LGG; LIHC; OV; PAAD; PRAD; STAD; UCEC | PITA; miRNATAP | TCGA BLCA -0.095; TCGA BRCA -0.199; TCGA CESC -0.102; TCGA LGG -0.067; TCGA LIHC -0.205; TCGA OV -0.116; TCGA PAAD -0.163; TCGA PRAD -0.114; TCGA STAD -0.113; TCGA UCEC -0.118 |
hsa-miR-28-3p | RAP2C | 9 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRP; PRAD; SARC; STAD | PITA; miRNATAP | TCGA BLCA -0.159; TCGA BRCA -0.206; TCGA CESC -0.116; TCGA ESCA -0.116; TCGA HNSC -0.126; TCGA KIRP -0.082; TCGA PRAD -0.087; TCGA SARC -0.154; TCGA STAD -0.105 |
hsa-miR-28-3p | GATAD2B | 9 cancers: BLCA; KIRC; KIRP; LGG; LUAD; PAAD; PRAD; THCA; UCEC | mirMAP | TCGA BLCA -0.091; TCGA KIRC -0.129; TCGA KIRP -0.182; TCGA LGG -0.235; TCGA LUAD -0.132; TCGA PAAD -0.114; TCGA PRAD -0.166; TCGA THCA -0.114; TCGA UCEC -0.102 |
hsa-miR-28-3p | CXXC5 | 12 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LUSC; OV; PRAD; THCA; UCEC | miRNATAP | TCGA BLCA -0.271; TCGA BRCA -0.74; TCGA CESC -0.351; TCGA ESCA -0.29; TCGA HNSC -0.562; TCGA KIRC -0.244; TCGA KIRP -0.39; TCGA LUSC -0.75; TCGA OV -0.145; TCGA PRAD -0.12; TCGA THCA -0.465; TCGA UCEC -0.329 |
hsa-miR-28-3p | ADSS | 9 cancers: BLCA; BRCA; KIRC; KIRP; LUAD; PRAD; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.085; TCGA BRCA -0.075; TCGA KIRC -0.342; TCGA KIRP -0.232; TCGA LUAD -0.089; TCGA PRAD -0.156; TCGA THCA -0.31; TCGA STAD -0.161; TCGA UCEC -0.277 |
hsa-miR-28-3p | MAP9 | 9 cancers: BRCA; KIRC; KIRP; LUSC; OV; PAAD; PRAD; SARC; UCEC | MirTarget | TCGA BRCA -0.196; TCGA KIRC -0.613; TCGA KIRP -0.295; TCGA LUSC -0.332; TCGA OV -0.142; TCGA PAAD -0.43; TCGA PRAD -0.143; TCGA SARC -0.388; TCGA UCEC -0.289 |
hsa-miR-28-3p | LRP11 | 9 cancers: BRCA; ESCA; HNSC; KIRC; KIRP; LGG; PAAD; THCA; STAD | MirTarget | TCGA BRCA -0.1; TCGA ESCA -0.308; TCGA HNSC -0.257; TCGA KIRC -0.451; TCGA KIRP -0.279; TCGA LGG -0.152; TCGA PAAD -0.508; TCGA THCA -0.393; TCGA STAD -0.224 |
hsa-miR-28-3p | HMGCR | 9 cancers: BRCA; CESC; HNSC; KIRC; KIRP; LGG; PAAD; SARC; STAD | PITA | TCGA BRCA -0.092; TCGA CESC -0.124; TCGA HNSC -0.273; TCGA KIRC -0.554; TCGA KIRP -0.229; TCGA LGG -0.453; TCGA PAAD -0.153; TCGA SARC -0.147; TCGA STAD -0.274 |
hsa-miR-28-3p | N4BP3 | 11 cancers: BRCA; CESC; COAD; KIRC; KIRP; LGG; LIHC; LUSC; PAAD; SARC; THCA | mirMAP | TCGA BRCA -0.799; TCGA CESC -0.359; TCGA COAD -0.498; TCGA KIRC -0.606; TCGA KIRP -0.569; TCGA LGG -0.163; TCGA LIHC -0.216; TCGA LUSC -0.24; TCGA PAAD -0.148; TCGA SARC -0.275; TCGA THCA -0.389 |
hsa-miR-28-3p | PRRC1 | 9 cancers: BRCA; CESC; ESCA; HNSC; LGG; LUSC; PRAD; SARC; STAD | mirMAP | TCGA BRCA -0.092; TCGA CESC -0.162; TCGA ESCA -0.155; TCGA HNSC -0.152; TCGA LGG -0.068; TCGA LUSC -0.096; TCGA PRAD -0.117; TCGA SARC -0.101; TCGA STAD -0.112 |
hsa-miR-28-3p | LIAS | 9 cancers: BRCA; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; PAAD; SARC | miRNATAP | TCGA BRCA -0.081; TCGA ESCA -0.129; TCGA HNSC -0.108; TCGA KIRC -0.188; TCGA KIRP -0.119; TCGA LGG -0.099; TCGA LIHC -0.141; TCGA PAAD -0.257; TCGA SARC -0.08 |
hsa-miR-28-3p | AHCYL2 | 10 cancers: BRCA; HNSC; KIRC; KIRP; LUAD; LUSC; OV; PRAD; SARC; UCEC | miRNATAP | TCGA BRCA -0.177; TCGA HNSC -0.301; TCGA KIRC -0.502; TCGA KIRP -0.273; TCGA LUAD -0.242; TCGA LUSC -0.42; TCGA OV -0.145; TCGA PRAD -0.146; TCGA SARC -0.246; TCGA UCEC -0.189 |
hsa-miR-28-3p | SLC39A9 | 11 cancers: BRCA; HNSC; KIRC; KIRP; LGG; OV; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BRCA -0.195; TCGA HNSC -0.156; TCGA KIRC -0.385; TCGA KIRP -0.219; TCGA LGG -0.166; TCGA OV -0.136; TCGA PAAD -0.249; TCGA PRAD -0.152; TCGA SARC -0.151; TCGA STAD -0.172; TCGA UCEC -0.077 |
hsa-miR-28-3p | LMO7 | 9 cancers: CESC; ESCA; HNSC; KIRC; KIRP; LUSC; THCA; STAD; UCEC | PITA; miRNATAP | TCGA CESC -0.222; TCGA ESCA -0.345; TCGA HNSC -0.481; TCGA KIRC -0.939; TCGA KIRP -0.312; TCGA LUSC -0.365; TCGA THCA -0.235; TCGA STAD -0.349; TCGA UCEC -0.25 |
hsa-miR-28-3p | GRIN3A | 10 cancers: CESC; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; PRAD; SARC; STAD | mirMAP | TCGA CESC -0.463; TCGA ESCA -0.219; TCGA HNSC -0.196; TCGA LGG -1.007; TCGA LIHC -0.216; TCGA LUAD -0.162; TCGA LUSC -0.306; TCGA PRAD -0.844; TCGA SARC -0.313; TCGA STAD -0.207 |
hsa-miR-28-3p | FNDC3A | 9 cancers: HNSC; KIRC; LUAD; LUSC; OV; PAAD; SARC; STAD; UCEC | PITA; miRNATAP | TCGA HNSC -0.143; TCGA KIRC -0.188; TCGA LUAD -0.13; TCGA LUSC -0.272; TCGA OV -0.148; TCGA PAAD -0.394; TCGA SARC -0.252; TCGA STAD -0.107; TCGA UCEC -0.207 |
Enriched cancer pathways of putative targets