microRNA information: hsa-miR-28-5p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-28-5p | miRbase |
| Accession: | MIMAT0000085 | miRbase |
| Precursor name: | hsa-mir-28 | miRbase |
| Precursor accession: | MI0000086 | miRbase |
| Symbol: | MIR28 | HGNC |
| RefSeq ID: | NR_029502 | GenBank |
| Sequence: | AAGGAGCUCACAGUCUAUUGAG |
Reported expression in cancers: hsa-miR-28-5p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-28-5p | colorectal cancer | downregulation | "We measured levels of miR-28-5p and miR-28-3p expr ......" | 22240480 | qPCR |
| hsa-miR-28-5p | colorectal cancer | downregulation | "Secondly validation of the results was carried out ......" | 27365381 | qPCR |
| hsa-miR-28-5p | liver cancer | downregulation | "In this study we employed miRNA-sequencing and ide ......" | 26754294 | RNA-Seq |
Reported cancer pathway affected by hsa-miR-28-5p
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|
Reported cancer prognosis affected by hsa-miR-28-5p
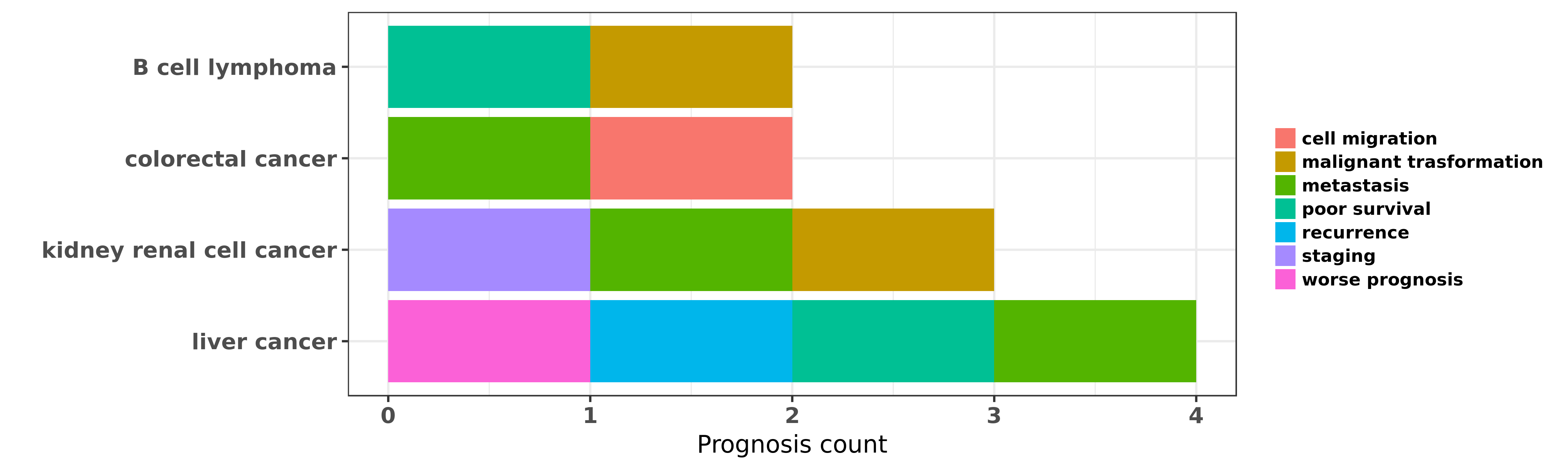
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-28-5p | B cell lymphoma | malignant trasformation | "MicroRNA 28 controls cell proliferation and is dow ......" | 24843176 | |
| hsa-miR-28-5p | B cell lymphoma | poor survival | "Comprehensive miRNA sequence analysis reveals surv ......" | 25723320 | |
| hsa-miR-28-5p | colorectal cancer | cell migration; metastasis | "Strand specific miR 28 5p and miR 28 3p have disti ......" | 22240480 | |
| hsa-miR-28-5p | colorectal cancer | metastasis | "Secondly validation of the results was carried out ......" | 27365381 | |
| hsa-miR-28-5p | kidney renal cell cancer | metastasis; malignant trasformation | "To obtain accurate results in miRNA expression cha ......" | 21741950 | |
| hsa-miR-28-5p | kidney renal cell cancer | staging | "Markedly dysregulated miRNAs in RCC cases were sub ......" | 25556603 | |
| hsa-miR-28-5p | liver cancer | metastasis; worse prognosis; poor survival; recurrence | "miR 28 5p IL 34 macrophage feedback loop modulates ......" | 26754294 |
Reported gene related to hsa-miR-28-5p
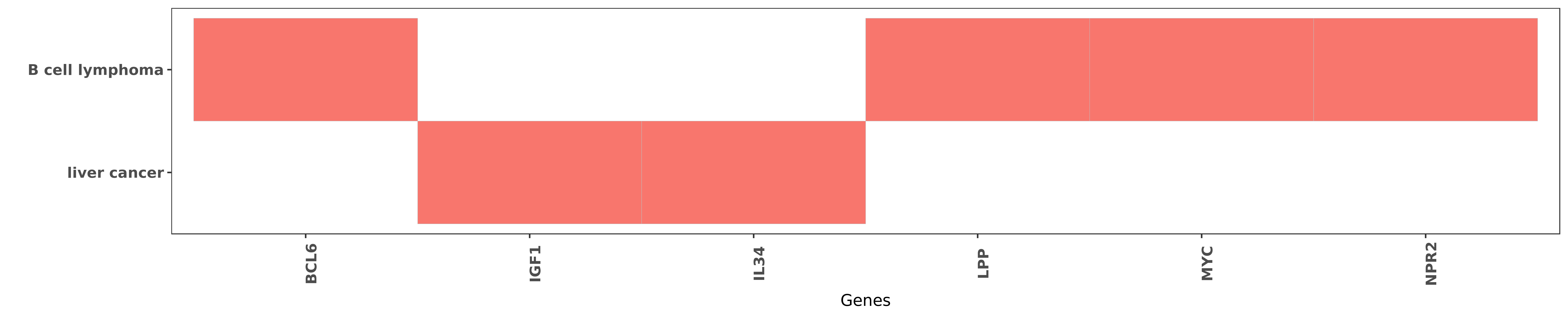
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-28-5p | B cell lymphoma | BCL6 | "Expressions of bcl 6 lpp and miR 28 genes in diffu ......" | 19236753 |
| hsa-miR-28-5p | liver cancer | IGF1 | "Luciferase report in HCC cells expressing miR-28-5 ......" | 26160280 |
| hsa-miR-28-5p | liver cancer | IL34 | "miR 28 5p IL 34 macrophage feedback loop modulates ......" | 26754294 |
| hsa-miR-28-5p | B cell lymphoma | LPP | "Expressions of bcl 6 lpp and miR 28 genes in diffu ......" | 19236753 |
| hsa-miR-28-5p | B cell lymphoma | MYC | "We identify the oncogene MYC as a negative regulat ......" | 24843176 |
| hsa-miR-28-5p | B cell lymphoma | NPR2 | "A comparative analysis of microRNAs expressed in n ......" | 24843176 |
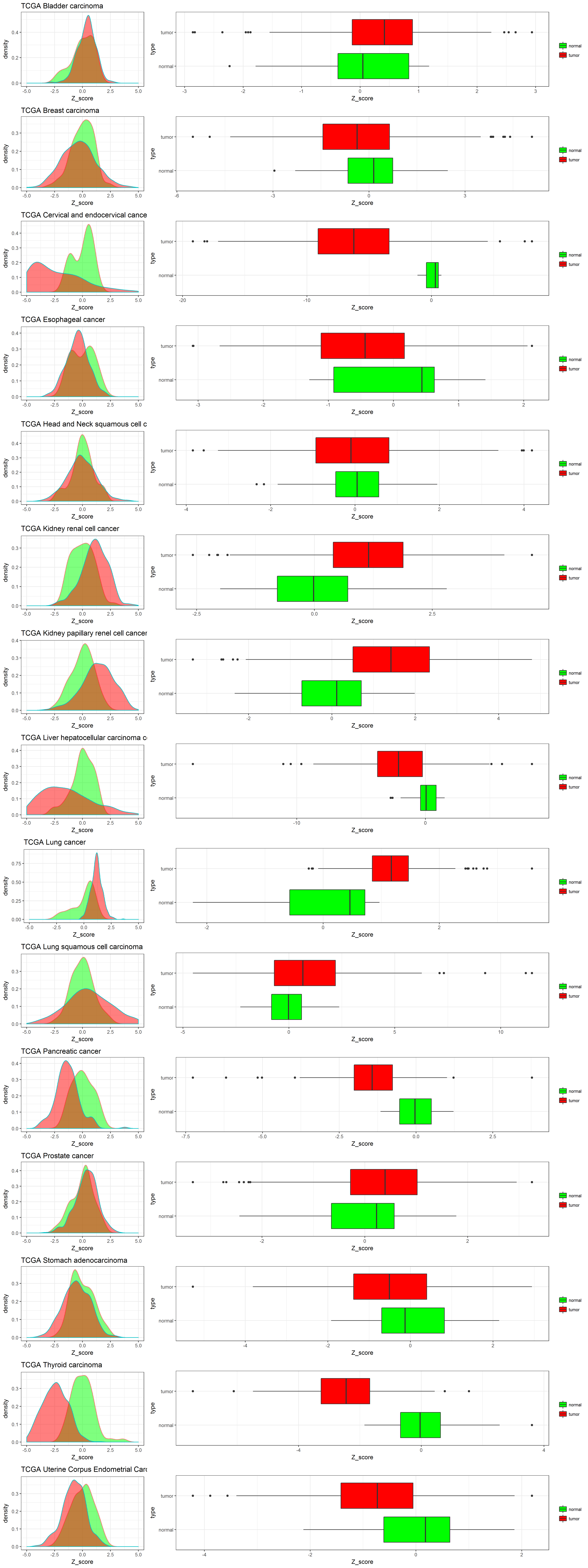
Expression profile in cancer corhorts:
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-28-5p | CDKN1A | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LUAD; LUSC; PRAD; THCA; UCEC | miRNAWalker2 validate; miRTarBase; MirTarget; miRanda; miRNATAP | TCGA BLCA -0.557; TCGA BRCA -0.294; TCGA CESC -0.546; TCGA ESCA -0.269; TCGA HNSC -0.739; TCGA LUAD -0.202; TCGA LUSC -0.488; TCGA PRAD -0.34; TCGA THCA -0.394; TCGA UCEC -0.256 |
hsa-miR-28-5p | ZBTB43 | 10 cancers: BLCA; CESC; ESCA; HNSC; KIRC; LGG; LUAD; SARC; THCA; STAD | MirTarget; miRanda | TCGA BLCA -0.154; TCGA CESC -0.275; TCGA ESCA -0.245; TCGA HNSC -0.131; TCGA KIRC -0.256; TCGA LGG -0.083; TCGA LUAD -0.124; TCGA SARC -0.14; TCGA THCA -0.109; TCGA STAD -0.299 |
hsa-miR-28-5p | TRIM23 | 9 cancers: BLCA; BRCA; CESC; COAD; LGG; LUSC; OV; PAAD; THCA | MirTarget; miRanda | TCGA BLCA -0.215; TCGA BRCA -0.241; TCGA CESC -0.1; TCGA COAD -0.19; TCGA LGG -0.196; TCGA LUSC -0.125; TCGA OV -0.134; TCGA PAAD -0.226; TCGA THCA -0.096 |
hsa-miR-28-5p | SLC35E1 | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRP; LIHC; SARC; THCA; STAD | MirTarget | TCGA BLCA -0.098; TCGA BRCA -0.119; TCGA CESC -0.19; TCGA COAD -0.124; TCGA ESCA -0.114; TCGA KIRP -0.258; TCGA LIHC -0.122; TCGA SARC -0.102; TCGA THCA -0.143; TCGA STAD -0.099 |
hsa-miR-28-5p | ETF1 | 11 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LIHC; LUAD; PRAD; STAD; UCEC | MirTarget; PITA; miRanda; miRNATAP | TCGA BLCA -0.31; TCGA BRCA -0.057; TCGA CESC -0.227; TCGA HNSC -0.231; TCGA KIRC -0.146; TCGA KIRP -0.158; TCGA LIHC -0.12; TCGA LUAD -0.126; TCGA PRAD -0.13; TCGA STAD -0.132; TCGA UCEC -0.074 |
hsa-miR-28-5p | TMEM167A | 13 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUSC; PAAD; SARC; STAD; UCEC | MirTarget; miRanda | TCGA BLCA -0.132; TCGA BRCA -0.112; TCGA ESCA -0.281; TCGA HNSC -0.089; TCGA KIRC -0.176; TCGA KIRP -0.09; TCGA LGG -0.06; TCGA LIHC -0.075; TCGA LUSC -0.104; TCGA PAAD -0.225; TCGA SARC -0.27; TCGA STAD -0.229; TCGA UCEC -0.121 |
hsa-miR-28-5p | LUZP1 | 10 cancers: BLCA; CESC; ESCA; HNSC; KIRC; LIHC; LUSC; PRAD; SARC; THCA | PITA; mirMAP; miRNATAP | TCGA BLCA -0.189; TCGA CESC -0.252; TCGA ESCA -0.202; TCGA HNSC -0.236; TCGA KIRC -0.151; TCGA LIHC -0.141; TCGA LUSC -0.126; TCGA PRAD -0.152; TCGA SARC -0.224; TCGA THCA -0.097 |
hsa-miR-28-5p | EIF4A1 | 9 cancers: BLCA; CESC; COAD; HNSC; LIHC; PRAD; SARC; THCA; STAD | miRanda | TCGA BLCA -0.245; TCGA CESC -0.173; TCGA COAD -0.22; TCGA HNSC -0.357; TCGA LIHC -0.333; TCGA PRAD -0.123; TCGA SARC -0.401; TCGA THCA -0.087; TCGA STAD -0.145 |
hsa-miR-28-5p | JKAMP | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LUSC; OV; PAAD; SARC; STAD | miRanda | TCGA BLCA -0.203; TCGA BRCA -0.25; TCGA COAD -0.299; TCGA ESCA -0.289; TCGA HNSC -0.243; TCGA KIRC -0.134; TCGA LUSC -0.196; TCGA OV -0.093; TCGA PAAD -0.299; TCGA SARC -0.088; TCGA STAD -0.161 |
hsa-miR-28-5p | DLST | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LGG; LUSC; SARC; THCA | miRanda | TCGA BLCA -0.263; TCGA BRCA -0.087; TCGA CESC -0.102; TCGA ESCA -0.194; TCGA HNSC -0.247; TCGA KIRC -0.479; TCGA LGG -0.052; TCGA LUSC -0.162; TCGA SARC -0.081; TCGA THCA -0.114 |
hsa-miR-28-5p | MED6 | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; LGG; OV; SARC; STAD | miRanda | TCGA BLCA -0.211; TCGA CESC -0.175; TCGA COAD -0.187; TCGA ESCA -0.169; TCGA HNSC -0.332; TCGA LGG -0.068; TCGA OV -0.089; TCGA SARC -0.127; TCGA STAD -0.237 |
hsa-miR-28-5p | PPP6C | 9 cancers: BLCA; BRCA; CESC; KIRC; KIRP; LUAD; PAAD; PRAD; STAD | miRanda | TCGA BLCA -0.151; TCGA BRCA -0.099; TCGA CESC -0.109; TCGA KIRC -0.166; TCGA KIRP -0.122; TCGA LUAD -0.056; TCGA PAAD -0.164; TCGA PRAD -0.066; TCGA STAD -0.131 |
hsa-miR-28-5p | PFKFB2 | 10 cancers: BLCA; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; PRAD; STAD; UCEC | miRanda | TCGA BLCA -0.204; TCGA ESCA -0.627; TCGA HNSC -0.417; TCGA KIRC -0.874; TCGA LIHC -0.214; TCGA LUAD -0.127; TCGA LUSC -0.196; TCGA PRAD -0.157; TCGA STAD -0.526; TCGA UCEC -0.267 |
hsa-miR-28-5p | SLCO4A1 | 11 cancers: BLCA; CESC; ESCA; HNSC; KIRP; LIHC; LUSC; SARC; THCA; STAD; UCEC | miRanda | TCGA BLCA -0.634; TCGA CESC -0.398; TCGA ESCA -0.639; TCGA HNSC -0.554; TCGA KIRP -0.947; TCGA LIHC -0.751; TCGA LUSC -0.698; TCGA SARC -0.466; TCGA THCA -0.795; TCGA STAD -0.458; TCGA UCEC -0.415 |
hsa-miR-28-5p | DDX31 | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; PAAD; SARC; STAD | miRanda | TCGA BLCA -0.176; TCGA CESC -0.176; TCGA COAD -0.143; TCGA ESCA -0.198; TCGA HNSC -0.088; TCGA LGG -0.064; TCGA LIHC -0.123; TCGA PAAD -0.208; TCGA SARC -0.189; TCGA STAD -0.409 |
hsa-miR-28-5p | PTP4A1 | 10 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; KIRP; LGG; LUSC; PRAD; STAD | miRanda | TCGA BLCA -0.424; TCGA BRCA -0.077; TCGA ESCA -0.363; TCGA HNSC -0.228; TCGA KIRC -0.234; TCGA KIRP -0.234; TCGA LGG -0.107; TCGA LUSC -0.164; TCGA PRAD -0.23; TCGA STAD -0.202 |
hsa-miR-28-5p | GRPEL2 | 9 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRP; LGG; LIHC; STAD | miRanda | TCGA BLCA -0.136; TCGA BRCA -0.06; TCGA CESC -0.261; TCGA ESCA -0.274; TCGA HNSC -0.548; TCGA KIRP -0.128; TCGA LGG -0.097; TCGA LIHC -0.221; TCGA STAD -0.425 |
hsa-miR-28-5p | RAB14 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; PRAD; SARC; THCA; STAD | miRanda; miRNATAP | TCGA BLCA -0.14; TCGA BRCA -0.104; TCGA CESC -0.13; TCGA COAD -0.155; TCGA ESCA -0.237; TCGA HNSC -0.09; TCGA KIRC -0.218; TCGA KIRP -0.179; TCGA LGG -0.106; TCGA LUAD -0.116; TCGA PRAD -0.087; TCGA SARC -0.176; TCGA THCA -0.17; TCGA STAD -0.279 |
hsa-miR-28-5p | GNAI1 | 10 cancers: BLCA; CESC; COAD; HNSC; KIRC; KIRP; LGG; OV; PAAD; PRAD | miRanda | TCGA BLCA -0.371; TCGA CESC -0.363; TCGA COAD -0.438; TCGA HNSC -0.525; TCGA KIRC -0.594; TCGA KIRP -0.262; TCGA LGG -0.128; TCGA OV -0.153; TCGA PAAD -0.429; TCGA PRAD -0.204 |
hsa-miR-28-5p | METAP2 | 10 cancers: BLCA; COAD; HNSC; KIRC; KIRP; LGG; OV; PAAD; PRAD; THCA | miRanda | TCGA BLCA -0.208; TCGA COAD -0.219; TCGA HNSC -0.208; TCGA KIRC -0.227; TCGA KIRP -0.274; TCGA LGG -0.064; TCGA OV -0.103; TCGA PAAD -0.121; TCGA PRAD -0.164; TCGA THCA -0.087 |
hsa-miR-28-5p | FCHO2 | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LUAD; LUSC; THCA; STAD | miRanda | TCGA BLCA -0.158; TCGA BRCA -0.141; TCGA CESC -0.216; TCGA ESCA -0.41; TCGA HNSC -0.153; TCGA KIRC -0.145; TCGA LUAD -0.114; TCGA LUSC -0.345; TCGA THCA -0.208; TCGA STAD -0.26 |
hsa-miR-28-5p | PAQR5 | 11 cancers: BLCA; CESC; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; OV; SARC | miRanda | TCGA BLCA -0.462; TCGA CESC -0.679; TCGA ESCA -0.637; TCGA HNSC -1.451; TCGA KIRC -0.653; TCGA KIRP -0.791; TCGA LIHC -0.617; TCGA LUAD -0.316; TCGA LUSC -0.346; TCGA OV -0.208; TCGA SARC -0.29 |
hsa-miR-28-5p | TSNAX | 9 cancers: BLCA; BRCA; COAD; ESCA; KIRC; KIRP; OV; PRAD; STAD | miRanda | TCGA BLCA -0.147; TCGA BRCA -0.125; TCGA COAD -0.186; TCGA ESCA -0.118; TCGA KIRC -0.256; TCGA KIRP -0.165; TCGA OV -0.13; TCGA PRAD -0.164; TCGA STAD -0.162 |
hsa-miR-28-5p | ZNF410 | 11 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; LGG; OV; PRAD; THCA | miRanda | TCGA BLCA -0.206; TCGA BRCA -0.087; TCGA CESC -0.094; TCGA COAD -0.331; TCGA HNSC -0.214; TCGA KIRC -0.152; TCGA KIRP -0.075; TCGA LGG -0.074; TCGA OV -0.085; TCGA PRAD -0.052; TCGA THCA -0.118 |
hsa-miR-28-5p | RAP1GDS1 | 11 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; KIRP; LIHC; PAAD; PRAD; THCA | miRanda | TCGA BLCA -0.194; TCGA BRCA -0.18; TCGA CESC -0.218; TCGA COAD -0.196; TCGA HNSC -0.312; TCGA KIRC -0.198; TCGA KIRP -0.217; TCGA LIHC -0.203; TCGA PAAD -0.1; TCGA PRAD -0.108; TCGA THCA -0.156 |
hsa-miR-28-5p | ZFYVE26 | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LGG; LUAD; LUSC; SARC | miRanda | TCGA BLCA -0.309; TCGA BRCA -0.124; TCGA CESC -0.264; TCGA ESCA -0.151; TCGA HNSC -0.28; TCGA KIRC -0.121; TCGA LGG -0.067; TCGA LUAD -0.126; TCGA LUSC -0.208; TCGA SARC -0.141 |
hsa-miR-28-5p | CANT1 | 11 cancers: BLCA; BRCA; ESCA; HNSC; KIRP; LIHC; OV; SARC; THCA; STAD; UCEC | miRanda; miRNATAP | TCGA BLCA -0.159; TCGA BRCA -0.314; TCGA ESCA -0.266; TCGA HNSC -0.109; TCGA KIRP -0.155; TCGA LIHC -0.12; TCGA OV -0.081; TCGA SARC -0.189; TCGA THCA -0.33; TCGA STAD -0.255; TCGA UCEC -0.121 |
hsa-miR-28-5p | ITGA3 | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LIHC; LUSC; PRAD; THCA; UCEC | miRanda | TCGA BLCA -0.299; TCGA BRCA -0.361; TCGA CESC -0.398; TCGA ESCA -0.456; TCGA HNSC -0.786; TCGA LIHC -0.655; TCGA LUSC -0.758; TCGA PRAD -0.39; TCGA THCA -1.008; TCGA UCEC -0.277 |
hsa-miR-28-5p | OTUD4 | 9 cancers: BLCA; CESC; COAD; ESCA; KIRC; KIRP; LUAD; LUSC; STAD | miRanda | TCGA BLCA -0.154; TCGA CESC -0.238; TCGA COAD -0.154; TCGA ESCA -0.311; TCGA KIRC -0.241; TCGA KIRP -0.132; TCGA LUAD -0.112; TCGA LUSC -0.209; TCGA STAD -0.17 |
hsa-miR-28-5p | BHLHE40 | 9 cancers: BLCA; BRCA; CESC; KIRC; KIRP; LUSC; PRAD; SARC; THCA | miRanda | TCGA BLCA -0.402; TCGA BRCA -0.47; TCGA CESC -0.431; TCGA KIRC -0.188; TCGA KIRP -0.311; TCGA LUSC -0.551; TCGA PRAD -0.346; TCGA SARC -0.331; TCGA THCA -0.828 |
hsa-miR-28-5p | STC1 | 9 cancers: BLCA; BRCA; COAD; ESCA; KIRC; KIRP; LIHC; LUSC; SARC | miRanda | TCGA BLCA -0.441; TCGA BRCA -0.42; TCGA COAD -0.629; TCGA ESCA -0.429; TCGA KIRC -0.263; TCGA KIRP -1.255; TCGA LIHC -0.278; TCGA LUSC -0.389; TCGA SARC -0.329 |
hsa-miR-28-5p | GALNT6 | 9 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LIHC; PRAD; SARC; UCEC | miRanda | TCGA BLCA -0.637; TCGA BRCA -0.691; TCGA ESCA -0.519; TCGA HNSC -1.036; TCGA KIRC -0.439; TCGA LIHC -0.321; TCGA PRAD -0.227; TCGA SARC -0.372; TCGA UCEC -0.237 |
hsa-miR-28-5p | SHB | 9 cancers: BLCA; HNSC; KIRP; LGG; LUSC; PRAD; SARC; THCA; UCEC | miRanda | TCGA BLCA -0.259; TCGA HNSC -0.476; TCGA KIRP -0.138; TCGA LGG -0.097; TCGA LUSC -0.376; TCGA PRAD -0.367; TCGA SARC -0.165; TCGA THCA -0.287; TCGA UCEC -0.193 |
hsa-miR-28-5p | HIF1A | 9 cancers: BLCA; CESC; COAD; KIRC; LUSC; PRAD; THCA; STAD; UCEC | miRanda | TCGA BLCA -0.411; TCGA CESC -0.253; TCGA COAD -0.421; TCGA KIRC -0.418; TCGA LUSC -0.256; TCGA PRAD -0.155; TCGA THCA -0.245; TCGA STAD -0.292; TCGA UCEC -0.132 |
hsa-miR-28-5p | FHOD1 | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LIHC; LUAD; LUSC; SARC; THCA | miRanda | TCGA BLCA -0.144; TCGA BRCA -0.096; TCGA CESC -0.253; TCGA ESCA -0.21; TCGA HNSC -0.544; TCGA LIHC -0.328; TCGA LUAD -0.154; TCGA LUSC -0.558; TCGA SARC -0.403; TCGA THCA -0.662 |
hsa-miR-28-5p | PMPCA | 9 cancers: BLCA; BRCA; ESCA; HNSC; PAAD; SARC; THCA; STAD; UCEC | miRanda | TCGA BLCA -0.166; TCGA BRCA -0.119; TCGA ESCA -0.154; TCGA HNSC -0.347; TCGA PAAD -0.205; TCGA SARC -0.151; TCGA THCA -0.156; TCGA STAD -0.226; TCGA UCEC -0.089 |
hsa-miR-28-5p | NUAK2 | 10 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LUSC; PAAD; STAD; UCEC | miRanda | TCGA BLCA -0.846; TCGA BRCA -0.376; TCGA CESC -0.413; TCGA HNSC -0.461; TCGA KIRC -0.648; TCGA KIRP -0.674; TCGA LUSC -0.315; TCGA PAAD -0.341; TCGA STAD -0.661; TCGA UCEC -0.244 |
hsa-miR-28-5p | CPSF2 | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; LGG; OV; PRAD; STAD | miRanda | TCGA BLCA -0.246; TCGA BRCA -0.09; TCGA CESC -0.187; TCGA COAD -0.219; TCGA ESCA -0.219; TCGA KIRC -0.203; TCGA LGG -0.078; TCGA OV -0.086; TCGA PRAD -0.108; TCGA STAD -0.251 |
hsa-miR-28-5p | PNP | 13 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUSC; PRAD; SARC; THCA; STAD; UCEC | miRanda | TCGA BLCA -0.467; TCGA CESC -0.346; TCGA COAD -0.246; TCGA ESCA -0.384; TCGA HNSC -0.726; TCGA KIRC -0.293; TCGA KIRP -0.2; TCGA LUSC -0.302; TCGA PRAD -0.196; TCGA SARC -0.254; TCGA THCA -0.791; TCGA STAD -0.227; TCGA UCEC -0.29 |
hsa-miR-28-5p | IL13RA1 | 10 cancers: BLCA; BRCA; ESCA; HNSC; KIRP; LUSC; PRAD; SARC; THCA; STAD | miRanda | TCGA BLCA -0.263; TCGA BRCA -0.147; TCGA ESCA -0.207; TCGA HNSC -0.319; TCGA KIRP -0.108; TCGA LUSC -0.263; TCGA PRAD -0.088; TCGA SARC -0.171; TCGA THCA -0.399; TCGA STAD -0.209 |
hsa-miR-28-5p | SPTLC1 | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; PRAD; SARC; THCA; STAD | miRanda | TCGA BLCA -0.169; TCGA CESC -0.231; TCGA COAD -0.163; TCGA ESCA -0.236; TCGA HNSC -0.125; TCGA KIRC -0.197; TCGA KIRP -0.093; TCGA LGG -0.055; TCGA PRAD -0.089; TCGA SARC -0.223; TCGA THCA -0.156; TCGA STAD -0.335 |
hsa-miR-28-5p | IPO5 | 9 cancers: BLCA; CESC; KIRC; KIRP; LGG; OV; PRAD; THCA; STAD | miRanda | TCGA BLCA -0.21; TCGA CESC -0.123; TCGA KIRC -0.195; TCGA KIRP -0.18; TCGA LGG -0.054; TCGA OV -0.184; TCGA PRAD -0.119; TCGA THCA -0.124; TCGA STAD -0.341 |
hsa-miR-28-5p | LRP10 | 9 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LIHC; LUSC; THCA | miRanda | TCGA BLCA -0.238; TCGA BRCA -0.232; TCGA CESC -0.241; TCGA HNSC -0.197; TCGA KIRC -0.174; TCGA KIRP -0.18; TCGA LIHC -0.2; TCGA LUSC -0.296; TCGA THCA -0.225 |
hsa-miR-28-5p | PELO | 9 cancers: BLCA; BRCA; COAD; HNSC; KIRP; LIHC; LUAD; LUSC; PRAD | miRanda | TCGA BLCA -0.244; TCGA BRCA -0.09; TCGA COAD -0.176; TCGA HNSC -0.098; TCGA KIRP -0.138; TCGA LIHC -0.162; TCGA LUAD -0.087; TCGA LUSC -0.122; TCGA PRAD -0.076 |
hsa-miR-28-5p | ANGEL1 | 10 cancers: BLCA; BRCA; ESCA; KIRC; KIRP; LGG; OV; PAAD; SARC; THCA | miRanda | TCGA BLCA -0.205; TCGA BRCA -0.141; TCGA ESCA -0.199; TCGA KIRC -0.331; TCGA KIRP -0.159; TCGA LGG -0.107; TCGA OV -0.125; TCGA PAAD -0.657; TCGA SARC -0.115; TCGA THCA -0.062 |
hsa-miR-28-5p | HES2 | 9 cancers: BLCA; CESC; HNSC; LIHC; LUAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -1.401; TCGA CESC -1.501; TCGA HNSC -1.238; TCGA LIHC -0.637; TCGA LUAD -0.283; TCGA SARC -0.71; TCGA THCA -1.383; TCGA STAD -1.274; TCGA UCEC -0.567 |
hsa-miR-28-5p | SLC7A1 | 9 cancers: BLCA; CESC; HNSC; KIRC; KIRP; LGG; LIHC; PAAD; STAD | mirMAP | TCGA BLCA -0.65; TCGA CESC -0.187; TCGA HNSC -0.362; TCGA KIRC -0.598; TCGA KIRP -0.783; TCGA LGG -0.197; TCGA LIHC -0.935; TCGA PAAD -0.305; TCGA STAD -0.449 |
hsa-miR-28-5p | VPS53 | 12 cancers: BLCA; CESC; HNSC; KIRC; LGG; LIHC; LUAD; PAAD; PRAD; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.128; TCGA CESC -0.168; TCGA HNSC -0.17; TCGA KIRC -0.216; TCGA LGG -0.062; TCGA LIHC -0.199; TCGA LUAD -0.077; TCGA PAAD -0.276; TCGA PRAD -0.097; TCGA SARC -0.277; TCGA THCA -0.103; TCGA STAD -0.183 |
hsa-miR-28-5p | BDKRB2 | 10 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LIHC; LUSC; PRAD; SARC | mirMAP | TCGA BLCA -0.477; TCGA BRCA -0.174; TCGA CESC -0.435; TCGA HNSC -0.748; TCGA KIRC -1.543; TCGA KIRP -1.022; TCGA LIHC -0.529; TCGA LUSC -0.642; TCGA PRAD -0.308; TCGA SARC -0.411 |
hsa-miR-28-5p | SACM1L | 9 cancers: BRCA; ESCA; KIRC; KIRP; LUAD; LUSC; OV; PAAD; STAD | MirTarget; PITA; miRanda | TCGA BRCA -0.096; TCGA ESCA -0.286; TCGA KIRC -0.543; TCGA KIRP -0.117; TCGA LUAD -0.11; TCGA LUSC -0.104; TCGA OV -0.097; TCGA PAAD -0.143; TCGA STAD -0.162 |
hsa-miR-28-5p | EFR3B | 10 cancers: BRCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; OV; PAAD; THCA | MirTarget; miRNATAP | TCGA BRCA -0.183; TCGA HNSC -0.449; TCGA KIRC -0.444; TCGA KIRP -0.705; TCGA LGG -0.157; TCGA LIHC -0.247; TCGA LUAD -0.465; TCGA OV -0.228; TCGA PAAD -1.108; TCGA THCA -0.276 |
hsa-miR-28-5p | CYTH2 | 9 cancers: BRCA; ESCA; HNSC; LIHC; LUSC; OV; SARC; THCA; UCEC | MirTarget; miRanda; miRNATAP | TCGA BRCA -0.196; TCGA ESCA -0.14; TCGA HNSC -0.226; TCGA LIHC -0.198; TCGA LUSC -0.199; TCGA OV -0.094; TCGA SARC -0.104; TCGA THCA -0.089; TCGA UCEC -0.171 |
hsa-miR-28-5p | CPD | 9 cancers: BRCA; KIRC; KIRP; LIHC; PRAD; SARC; THCA; STAD; UCEC | PITA; miRanda | TCGA BRCA -0.26; TCGA KIRC -0.282; TCGA KIRP -0.229; TCGA LIHC -0.159; TCGA PRAD -0.27; TCGA SARC -0.242; TCGA THCA -0.319; TCGA STAD -0.354; TCGA UCEC -0.26 |
hsa-miR-28-5p | ARHGAP32 | 9 cancers: BRCA; CESC; ESCA; HNSC; KIRC; LGG; SARC; THCA; STAD | miRanda | TCGA BRCA -0.358; TCGA CESC -0.471; TCGA ESCA -0.291; TCGA HNSC -0.376; TCGA KIRC -0.126; TCGA LGG -0.096; TCGA SARC -0.308; TCGA THCA -0.091; TCGA STAD -0.396 |
hsa-miR-28-5p | DHRS2 | 9 cancers: BRCA; ESCA; HNSC; KIRC; LGG; OV; PAAD; SARC; THCA | miRanda | TCGA BRCA -1.422; TCGA ESCA -0.833; TCGA HNSC -0.547; TCGA KIRC -1.005; TCGA LGG -0.602; TCGA OV -0.42; TCGA PAAD -1.767; TCGA SARC -0.601; TCGA THCA -0.727 |
hsa-miR-28-5p | GIPR | 12 cancers: BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; THCA; STAD | miRanda | TCGA BRCA -0.685; TCGA CESC -0.433; TCGA ESCA -0.93; TCGA HNSC -0.413; TCGA KIRC -0.684; TCGA KIRP -0.461; TCGA LIHC -0.439; TCGA LUAD -0.261; TCGA LUSC -0.658; TCGA PAAD -1.367; TCGA THCA -0.684; TCGA STAD -0.62 |
hsa-miR-28-5p | PUS7L | 10 cancers: BRCA; CESC; COAD; ESCA; KIRC; KIRP; LGG; OV; SARC; STAD | miRanda | TCGA BRCA -0.115; TCGA CESC -0.156; TCGA COAD -0.381; TCGA ESCA -0.181; TCGA KIRC -0.318; TCGA KIRP -0.216; TCGA LGG -0.115; TCGA OV -0.139; TCGA SARC -0.263; TCGA STAD -0.533 |
hsa-miR-28-5p | AAGAB | 10 cancers: BRCA; CESC; ESCA; HNSC; KIRC; LIHC; PRAD; SARC; STAD; UCEC | miRanda | TCGA BRCA -0.261; TCGA CESC -0.117; TCGA ESCA -0.178; TCGA HNSC -0.211; TCGA KIRC -0.281; TCGA LIHC -0.093; TCGA PRAD -0.072; TCGA SARC -0.138; TCGA STAD -0.309; TCGA UCEC -0.143 |
hsa-miR-28-5p | SEC16A | 10 cancers: BRCA; CESC; ESCA; KIRP; LGG; PAAD; SARC; THCA; STAD; UCEC | miRanda | TCGA BRCA -0.339; TCGA CESC -0.095; TCGA ESCA -0.168; TCGA KIRP -0.125; TCGA LGG -0.071; TCGA PAAD -0.142; TCGA SARC -0.079; TCGA THCA -0.131; TCGA STAD -0.286; TCGA UCEC -0.104 |
hsa-miR-28-5p | BAG1 | 9 cancers: BRCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; PRAD; THCA | miRanda | TCGA BRCA -0.308; TCGA HNSC -0.397; TCGA KIRC -0.544; TCGA KIRP -0.331; TCGA LGG -0.092; TCGA LIHC -0.11; TCGA LUAD -0.145; TCGA PRAD -0.133; TCGA THCA -0.212 |
hsa-miR-28-5p | PCSK6 | 9 cancers: BRCA; CESC; HNSC; KIRP; OV; PRAD; SARC; STAD; UCEC | miRanda | TCGA BRCA -0.325; TCGA CESC -0.407; TCGA HNSC -0.363; TCGA KIRP -0.952; TCGA OV -0.307; TCGA PRAD -0.255; TCGA SARC -0.798; TCGA STAD -0.302; TCGA UCEC -0.262 |
hsa-miR-28-5p | QSOX1 | 9 cancers: BRCA; HNSC; KIRP; LIHC; LUSC; PRAD; SARC; THCA; UCEC | miRanda | TCGA BRCA -0.153; TCGA HNSC -0.317; TCGA KIRP -0.247; TCGA LIHC -0.589; TCGA LUSC -0.273; TCGA PRAD -0.264; TCGA SARC -0.294; TCGA THCA -0.835; TCGA UCEC -0.188 |
hsa-miR-28-5p | FAM151B | 10 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; SARC; UCEC | miRanda | TCGA BRCA -0.356; TCGA CESC -0.313; TCGA COAD -0.337; TCGA ESCA -0.379; TCGA HNSC -0.284; TCGA KIRC -0.274; TCGA LUAD -0.138; TCGA LUSC -0.45; TCGA SARC -0.236; TCGA UCEC -0.235 |
hsa-miR-28-5p | CNKSR1 | 9 cancers: BRCA; CESC; ESCA; HNSC; KIRC; KIRP; THCA; STAD; UCEC | miRanda | TCGA BRCA -0.137; TCGA CESC -0.161; TCGA ESCA -0.34; TCGA HNSC -0.476; TCGA KIRC -1.885; TCGA KIRP -1.229; TCGA THCA -0.293; TCGA STAD -0.357; TCGA UCEC -0.313 |
hsa-miR-28-5p | GPBP1 | 9 cancers: BRCA; ESCA; KIRC; KIRP; LGG; LIHC; OV; PAAD; STAD | miRanda | TCGA BRCA -0.127; TCGA ESCA -0.152; TCGA KIRC -0.128; TCGA KIRP -0.154; TCGA LGG -0.066; TCGA LIHC -0.091; TCGA OV -0.119; TCGA PAAD -0.162; TCGA STAD -0.135 |
hsa-miR-28-5p | DHX29 | 10 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; STAD | miRanda | TCGA BRCA -0.227; TCGA CESC -0.113; TCGA COAD -0.163; TCGA ESCA -0.158; TCGA HNSC -0.199; TCGA KIRC -0.258; TCGA KIRP -0.138; TCGA LUAD -0.095; TCGA LUSC -0.129; TCGA STAD -0.182 |
hsa-miR-28-5p | DALRD3 | 11 cancers: BRCA; HNSC; KIRC; LIHC; LUSC; OV; PAAD; PRAD; SARC; THCA; UCEC | miRanda | TCGA BRCA -0.448; TCGA HNSC -0.193; TCGA KIRC -0.191; TCGA LIHC -0.124; TCGA LUSC -0.117; TCGA OV -0.156; TCGA PAAD -0.169; TCGA PRAD -0.109; TCGA SARC -0.153; TCGA THCA -0.21; TCGA UCEC -0.102 |
hsa-miR-28-5p | MARVELD2 | 9 cancers: BRCA; ESCA; KIRC; KIRP; OV; PAAD; PRAD; STAD; UCEC | miRanda | TCGA BRCA -0.411; TCGA ESCA -0.416; TCGA KIRC -1.113; TCGA KIRP -0.332; TCGA OV -0.114; TCGA PAAD -0.433; TCGA PRAD -0.203; TCGA STAD -0.597; TCGA UCEC -0.41 |
hsa-miR-28-5p | TTC8 | 9 cancers: BRCA; COAD; ESCA; KIRC; LUSC; OV; PAAD; SARC; THCA | miRanda | TCGA BRCA -0.381; TCGA COAD -0.216; TCGA ESCA -0.196; TCGA KIRC -0.192; TCGA LUSC -0.142; TCGA OV -0.169; TCGA PAAD -0.628; TCGA SARC -0.129; TCGA THCA -0.117 |
hsa-miR-28-5p | TRNT1 | 9 cancers: BRCA; ESCA; KIRC; LGG; LIHC; LUSC; PRAD; STAD; UCEC | miRanda | TCGA BRCA -0.057; TCGA ESCA -0.282; TCGA KIRC -0.405; TCGA LGG -0.109; TCGA LIHC -0.12; TCGA LUSC -0.105; TCGA PRAD -0.179; TCGA STAD -0.159; TCGA UCEC -0.131 |
hsa-miR-28-5p | RRM2B | 10 cancers: BRCA; CESC; ESCA; KIRC; KIRP; LIHC; LUSC; PAAD; THCA; STAD | miRanda | TCGA BRCA -0.182; TCGA CESC -0.126; TCGA ESCA -0.347; TCGA KIRC -0.306; TCGA KIRP -0.267; TCGA LIHC -0.141; TCGA LUSC -0.171; TCGA PAAD -0.159; TCGA THCA -0.078; TCGA STAD -0.323 |
hsa-miR-28-5p | TMEM229B | 9 cancers: BRCA; ESCA; HNSC; LGG; LIHC; LUSC; PAAD; PRAD; UCEC | miRanda | TCGA BRCA -0.43; TCGA ESCA -0.427; TCGA HNSC -0.553; TCGA LGG -0.111; TCGA LIHC -0.443; TCGA LUSC -0.651; TCGA PAAD -0.262; TCGA PRAD -0.219; TCGA UCEC -0.207 |
hsa-miR-28-5p | COG3 | 11 cancers: BRCA; CESC; ESCA; HNSC; KIRC; LGG; LUSC; OV; PAAD; SARC; STAD | miRanda | TCGA BRCA -0.075; TCGA CESC -0.158; TCGA ESCA -0.295; TCGA HNSC -0.097; TCGA KIRC -0.119; TCGA LGG -0.053; TCGA LUSC -0.096; TCGA OV -0.081; TCGA PAAD -0.231; TCGA SARC -0.085; TCGA STAD -0.406 |
hsa-miR-28-5p | SCAMP4 | 10 cancers: BRCA; HNSC; KIRC; KIRP; LIHC; LUAD; PAAD; SARC; THCA; UCEC | miRanda | TCGA BRCA -0.228; TCGA HNSC -0.147; TCGA KIRC -0.231; TCGA KIRP -0.243; TCGA LIHC -0.12; TCGA LUAD -0.088; TCGA PAAD -0.404; TCGA SARC -0.18; TCGA THCA -0.273; TCGA UCEC -0.196 |
hsa-miR-28-5p | TANC2 | 10 cancers: BRCA; COAD; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; SARC; THCA | miRanda | TCGA BRCA -0.242; TCGA COAD -0.639; TCGA KIRC -0.435; TCGA KIRP -0.368; TCGA LGG -0.091; TCGA LIHC -0.596; TCGA LUAD -0.185; TCGA LUSC -0.226; TCGA SARC -0.615; TCGA THCA -0.419 |
hsa-miR-28-5p | TOM1L2 | 9 cancers: BRCA; CESC; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; PAAD | mirMAP | TCGA BRCA -0.303; TCGA CESC -0.349; TCGA HNSC -0.524; TCGA KIRC -0.354; TCGA KIRP -0.212; TCGA LGG -0.14; TCGA LIHC -0.164; TCGA LUAD -0.311; TCGA PAAD -0.484 |
hsa-miR-28-5p | AMMECR1 | 9 cancers: CESC; ESCA; HNSC; KIRC; PRAD; SARC; THCA; STAD; UCEC | PITA; miRanda | TCGA CESC -0.329; TCGA ESCA -0.282; TCGA HNSC -0.362; TCGA KIRC -0.137; TCGA PRAD -0.237; TCGA SARC -0.369; TCGA THCA -0.179; TCGA STAD -0.494; TCGA UCEC -0.176 |
hsa-miR-28-5p | OSBPL3 | 10 cancers: CESC; ESCA; HNSC; KIRC; LIHC; LUSC; SARC; THCA; STAD; UCEC | miRanda | TCGA CESC -0.51; TCGA ESCA -0.477; TCGA HNSC -0.329; TCGA KIRC -0.526; TCGA LIHC -0.454; TCGA LUSC -0.424; TCGA SARC -0.487; TCGA THCA -0.142; TCGA STAD -0.506; TCGA UCEC -0.213 |
hsa-miR-28-5p | KLHL18 | 9 cancers: CESC; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; THCA; STAD | miRanda | TCGA CESC -0.249; TCGA ESCA -0.221; TCGA HNSC -0.384; TCGA KIRC -0.248; TCGA LIHC -0.091; TCGA LUAD -0.092; TCGA LUSC -0.112; TCGA THCA -0.07; TCGA STAD -0.136 |
hsa-miR-28-5p | TDG | 10 cancers: CESC; COAD; ESCA; HNSC; LGG; OV; PRAD; SARC; STAD; UCEC | miRanda | TCGA CESC -0.172; TCGA COAD -0.235; TCGA ESCA -0.202; TCGA HNSC -0.189; TCGA LGG -0.06; TCGA OV -0.101; TCGA PRAD -0.125; TCGA SARC -0.102; TCGA STAD -0.466; TCGA UCEC -0.136 |
hsa-miR-28-5p | TGFA | 10 cancers: CESC; ESCA; HNSC; KIRP; LIHC; PRAD; SARC; THCA; STAD; UCEC | miRanda | TCGA CESC -0.796; TCGA ESCA -0.379; TCGA HNSC -0.717; TCGA KIRP -0.458; TCGA LIHC -0.351; TCGA PRAD -0.342; TCGA SARC -0.35; TCGA THCA -1.199; TCGA STAD -0.508; TCGA UCEC -0.56 |
hsa-miR-28-5p | MDFI | 10 cancers: CESC; COAD; HNSC; KIRP; LGG; LIHC; LUSC; PRAD; SARC; THCA | mirMAP | TCGA CESC -0.582; TCGA COAD -0.407; TCGA HNSC -0.984; TCGA KIRP -0.945; TCGA LGG -0.187; TCGA LIHC -0.754; TCGA LUSC -0.336; TCGA PRAD -0.983; TCGA SARC -1.017; TCGA THCA -0.692 |
hsa-miR-28-5p | PDXK | 12 cancers: CESC; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PRAD; SARC; THCA; STAD | mirMAP | TCGA CESC -0.175; TCGA HNSC -0.321; TCGA KIRC -0.463; TCGA KIRP -0.298; TCGA LGG -0.06; TCGA LIHC -0.251; TCGA LUAD -0.173; TCGA LUSC -0.37; TCGA PRAD -0.161; TCGA SARC -0.215; TCGA THCA -0.218; TCGA STAD -0.176 |
hsa-miR-28-5p | DMKN | 10 cancers: CESC; COAD; HNSC; KIRC; KIRP; LIHC; OV; PAAD; PRAD; SARC | miRNATAP | TCGA CESC -0.725; TCGA COAD -0.87; TCGA HNSC -2.068; TCGA KIRC -1.509; TCGA KIRP -0.774; TCGA LIHC -0.999; TCGA OV -0.23; TCGA PAAD -0.504; TCGA PRAD -0.454; TCGA SARC -0.981 |
hsa-miR-28-5p | SLC4A11 | 9 cancers: COAD; KIRC; KIRP; LIHC; PRAD; SARC; THCA; STAD; UCEC | PITA; miRanda; miRNATAP | TCGA COAD -0.791; TCGA KIRC -1.847; TCGA KIRP -0.873; TCGA LIHC -1.061; TCGA PRAD -0.541; TCGA SARC -0.53; TCGA THCA -0.282; TCGA STAD -0.969; TCGA UCEC -0.3 |
hsa-miR-28-5p | FAM60A | 9 cancers: COAD; KIRC; KIRP; LGG; LIHC; OV; PRAD; THCA; STAD | miRanda | TCGA COAD -0.222; TCGA KIRC -0.531; TCGA KIRP -0.152; TCGA LGG -0.164; TCGA LIHC -0.305; TCGA OV -0.169; TCGA PRAD -0.123; TCGA THCA -0.308; TCGA STAD -0.672 |
Enriched cancer pathways of putative targets