microRNA information: hsa-miR-338-5p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-338-5p | miRbase |
| Accession: | MIMAT0004701 | miRbase |
| Precursor name: | hsa-mir-338 | miRbase |
| Precursor accession: | MI0000814 | miRbase |
| Symbol: | MIR338 | HGNC |
| RefSeq ID: | NR_029897 | GenBank |
| Sequence: | AACAAUAUCCUGGUGCUGAGUG |
Reported expression in cancers: hsa-miR-338-5p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-338-5p | gastric cancer | upregulation | "MicroRNA microarray was applied to assess the miRN ......" | 24054006 | Microarray |
| hsa-miR-338-5p | gastric cancer | upregulation | "Anticancer bioactive peptide 3 inhibits human gast ......" | 27688872 | Microarray |
| hsa-miR-338-5p | liver cancer | downregulation | "Bead based microarray analysis of microRNA express ......" | 19473441 | Microarray; Reverse transcription PCR; qPCR |
| hsa-miR-338-5p | lung cancer | downregulation | "In the present study we found that miR-338 was str ......" | 27431198 |
Reported cancer pathway affected by hsa-miR-338-5p
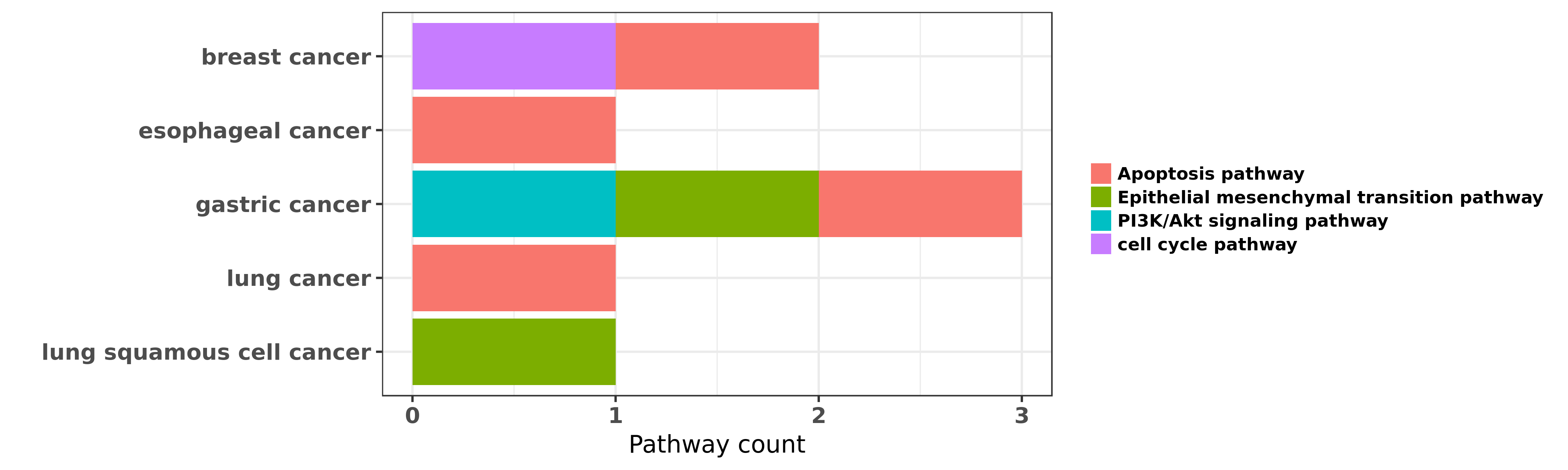
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-338-5p | breast cancer | cell cycle pathway; Apoptosis pathway | "MicroRNA 338 3p functions as tumor suppressor in b ......" | 26252944 | Western blot; Luciferase; Colony formation |
| hsa-miR-338-5p | esophageal cancer | Apoptosis pathway | "MicroRNA 338 3p suppresses tumor growth of esophag ......" | 26004521 | Colony formation |
| hsa-miR-338-5p | gastric cancer | Apoptosis pathway | "MicroRNA 338 inhibits growth invasion and metastas ......" | 24736504 | |
| hsa-miR-338-5p | gastric cancer | PI3K/Akt signaling pathway; Epithelial mesenchymal transition pathway | "MiR 338 3p inhibits epithelial mesenchymal transit ......" | 25945841 | |
| hsa-miR-338-5p | gastric cancer | Apoptosis pathway | "Anticancer bioactive peptide 3 inhibits human gast ......" | 27688872 | |
| hsa-miR-338-5p | lung cancer | Apoptosis pathway | "miR 338 inhibits the metastasis of lung cancer by ......" | 27431198 | |
| hsa-miR-338-5p | lung squamous cell cancer | Epithelial mesenchymal transition pathway | "miR 338 3p suppresses epithelial mesenchymal trans ......" | 27453416 | Transwell assay |
Reported cancer prognosis affected by hsa-miR-338-5p
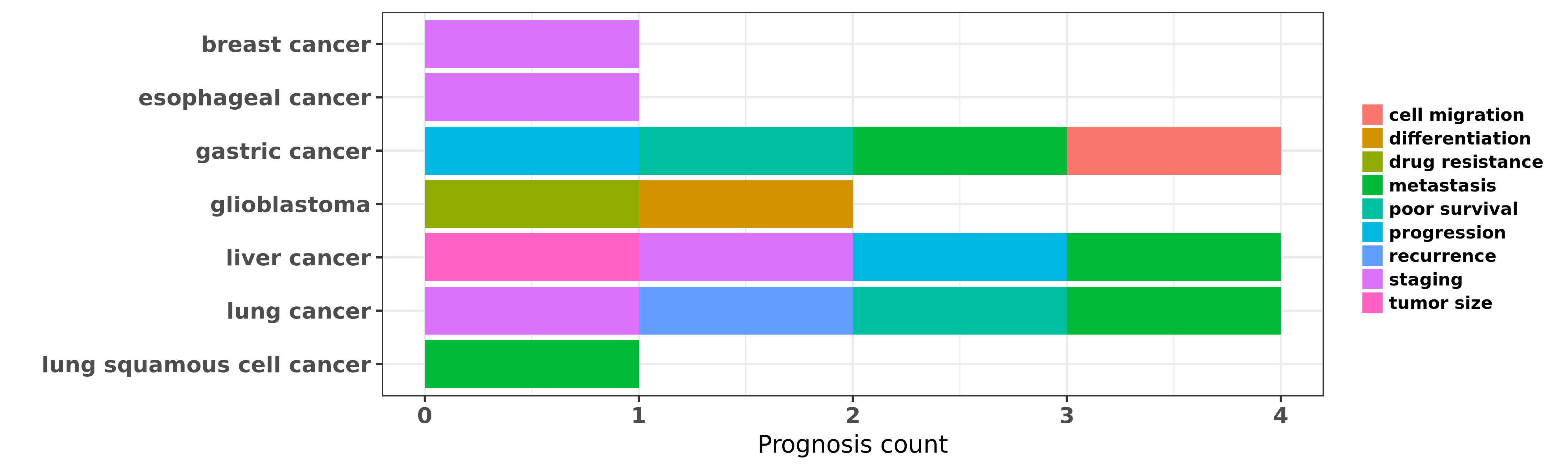
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-338-5p | breast cancer | staging | "MicroRNA 338 3p functions as tumor suppressor in b ......" | 26252944 | Western blot; Luciferase; Colony formation |
| hsa-miR-338-5p | esophageal cancer | staging | "MicroRNA 338 3p suppresses tumor growth of esophag ......" | 26004521 | Colony formation |
| hsa-miR-338-5p | gastric cancer | poor survival | "Several microarray studies have reported microRNA ......" | 19951901 | |
| hsa-miR-338-5p | gastric cancer | progression | "miR 338 3p suppresses gastric cancer progression t ......" | 24375644 | |
| hsa-miR-338-5p | gastric cancer | metastasis; cell migration | "MicroRNA 338 inhibits growth invasion and metastas ......" | 24736504 | |
| hsa-miR-338-5p | glioblastoma | drug resistance; differentiation | "MiR 338 5p sensitizes glioblastoma cells to radiat ......" | 26692101 | |
| hsa-miR-338-5p | liver cancer | staging; metastasis; tumor size | "Bead based microarray analysis of microRNA express ......" | 19473441 | |
| hsa-miR-338-5p | liver cancer | metastasis | "Identification of metastasis related microRNAs of ......" | 19912688 | |
| hsa-miR-338-5p | liver cancer | progression | "Mineralocorticoid receptor suppresses cancer progr ......" | 26082033 | |
| hsa-miR-338-5p | liver cancer | tumor size | "Plasma miR 15b 5p miR 338 5p and miR 764 as Biomar ......" | 26119771 | |
| hsa-miR-338-5p | lung cancer | metastasis | "MicroRNA 338 3p suppresses metastasis of lung canc ......" | 27186391 | |
| hsa-miR-338-5p | lung cancer | metastasis; staging; poor survival; recurrence | "miR 338 inhibits the metastasis of lung cancer by ......" | 27431198 | |
| hsa-miR-338-5p | lung cancer | metastasis | "Erratum: MicroRNA 338 3p suppresses metastasis of ......" | 27508100 | |
| hsa-miR-338-5p | lung squamous cell cancer | metastasis | "miR 338 3p suppresses epithelial mesenchymal trans ......" | 27453416 | Transwell assay |
Reported gene related to hsa-miR-338-5p
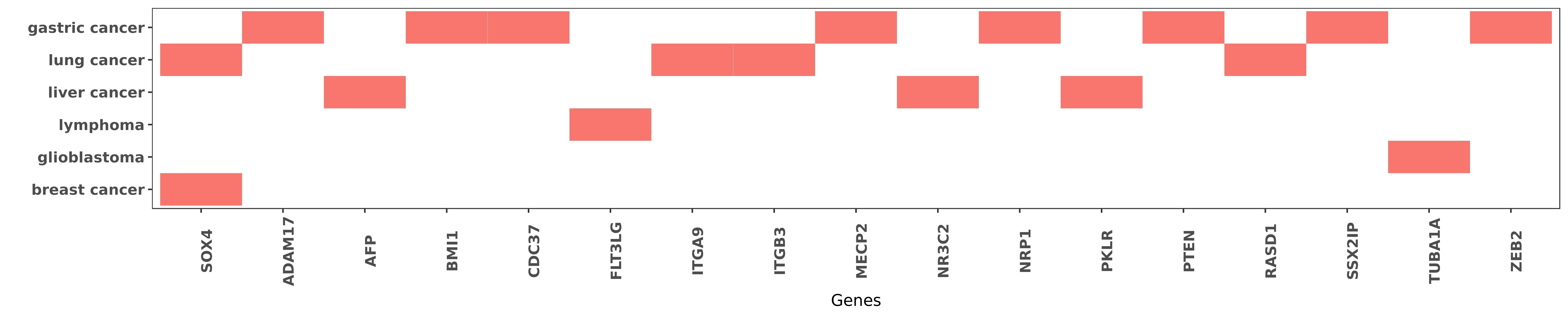
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-338-5p | breast cancer | SOX4 | "MicroRNA 338 3p functions as tumor suppressor in b ......" | 26252944 |
| hsa-miR-338-5p | lung cancer | SOX4 | "Erratum: MicroRNA 338 3p suppresses metastasis of ......" | 27508100 |
| hsa-miR-338-5p | lung cancer | SOX4 | "MicroRNA 338 3p suppresses metastasis of lung canc ......" | 27186391 |
| hsa-miR-338-5p | gastric cancer | ADAM17 | "MiR 338 3p inhibits the proliferation and migratio ......" | 26617808 |
| hsa-miR-338-5p | liver cancer | AFP | "The expression level of miR-338-5p was negatively ......" | 26119771 |
| hsa-miR-338-5p | gastric cancer | BMI1 | "Furthermore miR-338-5p can suppress GC cell growth ......" | 27166996 |
| hsa-miR-338-5p | gastric cancer | CDC37 | "Furthermore miR-338-5p can suppress GC cell growth ......" | 27166996 |
| hsa-miR-338-5p | lymphoma | FLT3LG | "The array indicated that miR-451 showed the greate ......" | 25019040 |
| hsa-miR-338-5p | lung cancer | ITGA9 | "miR 338 inhibits the metastasis of lung cancer by ......" | 27431198 |
| hsa-miR-338-5p | lung cancer | ITGB3 | "Furthermore we also identified a metastasis relate ......" | 27431198 |
| hsa-miR-338-5p | gastric cancer | MECP2 | "MECP2 promotes the growth of gastric cancer cells ......" | 27166996 |
| hsa-miR-338-5p | liver cancer | NR3C2 | "Mineralocorticoid receptor suppresses cancer progr ......" | 26082033 |
| hsa-miR-338-5p | gastric cancer | NRP1 | "MicroRNA 338 inhibits growth invasion and metastas ......" | 24736504 |
| hsa-miR-338-5p | liver cancer | PKLR | "Mineralocorticoid receptor suppresses cancer progr ......" | 26082033 |
| hsa-miR-338-5p | gastric cancer | PTEN | "miR 338 3p suppresses gastric cancer progression t ......" | 24375644 |
| hsa-miR-338-5p | lung cancer | RASD1 | "microRNA 338 3p functions as a tumor suppressor in ......" | 25374067 |
| hsa-miR-338-5p | gastric cancer | SSX2IP | "Epigenetic silencing of miR 338 3p contributes to ......" | 23826132 |
| hsa-miR-338-5p | glioblastoma | TUBA1A | "MiR-338 is a brain-specific miRNA which has been d ......" | 26692101 |
| hsa-miR-338-5p | gastric cancer | ZEB2 | "MiR 338 3p inhibits epithelial mesenchymal transit ......" | 25945841 |
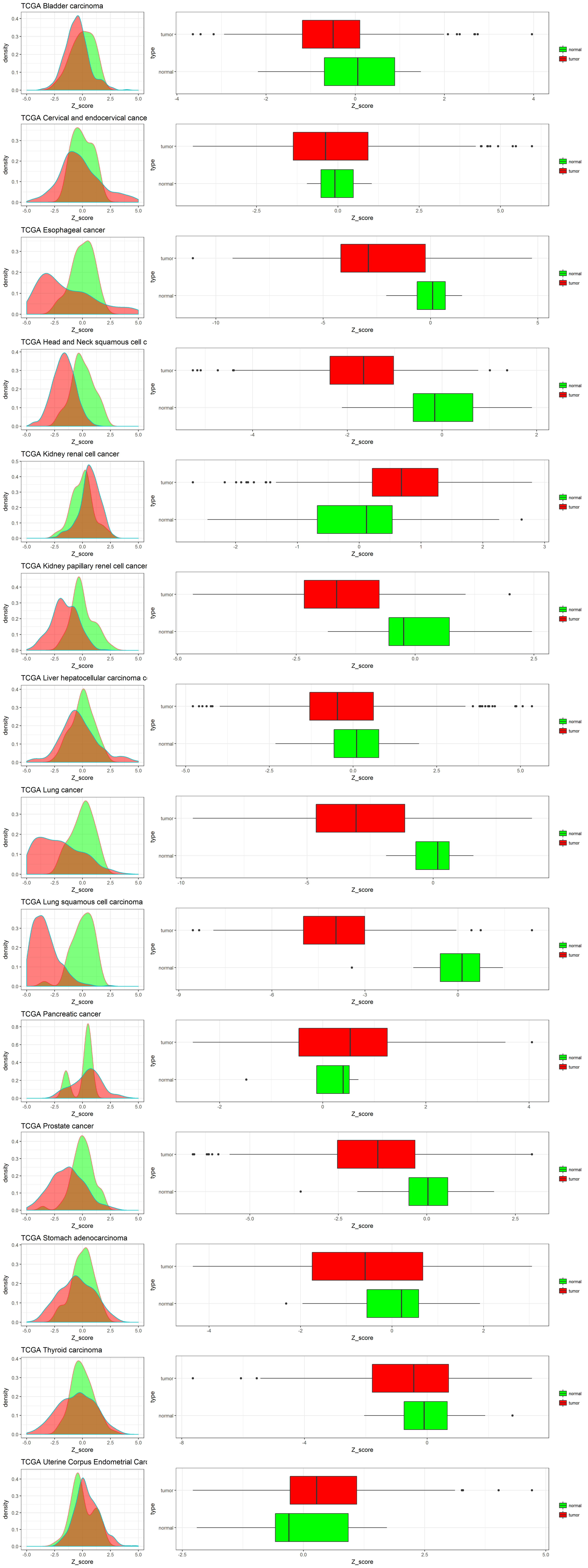
Expression profile in cancer corhorts:
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-338-5p | PFDN4 | 11 cancers: BLCA; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; THCA; STAD | MirTarget; PITA; miRNATAP | TCGA BLCA -0.075; TCGA COAD -0.095; TCGA ESCA -0.083; TCGA HNSC -0.089; TCGA KIRC -0.103; TCGA LUAD -0.102; TCGA LUSC -0.152; TCGA PAAD -0.105; TCGA PRAD -0.082; TCGA THCA -0.057; TCGA STAD -0.111 |
hsa-miR-338-5p | GRHL2 | 9 cancers: BLCA; CESC; COAD; ESCA; KIRC; LUAD; LUSC; PAAD; PRAD | MirTarget; PITA | TCGA BLCA -0.31; TCGA CESC -0.083; TCGA COAD -0.098; TCGA ESCA -0.228; TCGA KIRC -0.994; TCGA LUAD -0.073; TCGA LUSC -0.263; TCGA PAAD -0.689; TCGA PRAD -0.059 |
hsa-miR-338-5p | CXCL14 | 9 cancers: BLCA; CESC; COAD; ESCA; LUSC; PAAD; PRAD; THCA; UCEC | MirTarget; PITA; miRNATAP | TCGA BLCA -0.344; TCGA CESC -0.445; TCGA COAD -0.288; TCGA ESCA -0.406; TCGA LUSC -0.589; TCGA PAAD -0.417; TCGA PRAD -0.219; TCGA THCA -0.454; TCGA UCEC -0.465 |
hsa-miR-338-5p | COL4A5 | 10 cancers: BLCA; COAD; ESCA; HNSC; KIRC; LUSC; PAAD; SARC; STAD; UCEC | PITA; miRNATAP | TCGA BLCA -0.346; TCGA COAD -0.299; TCGA ESCA -0.439; TCGA HNSC -0.172; TCGA KIRC -0.362; TCGA LUSC -0.312; TCGA PAAD -0.283; TCGA SARC -0.444; TCGA STAD -0.271; TCGA UCEC -0.166 |
hsa-miR-338-5p | LYPLA1 | 9 cancers: BLCA; COAD; KIRC; LGG; LUAD; LUSC; PAAD; PRAD; THCA | PITA; mirMAP | TCGA BLCA -0.076; TCGA COAD -0.075; TCGA KIRC -0.191; TCGA LGG -0.079; TCGA LUAD -0.073; TCGA LUSC -0.11; TCGA PAAD -0.133; TCGA PRAD -0.15; TCGA THCA -0.064 |
hsa-miR-338-5p | STAU2 | 9 cancers: BLCA; COAD; ESCA; KIRC; LUAD; LUSC; PRAD; SARC; STAD | PITA | TCGA BLCA -0.055; TCGA COAD -0.105; TCGA ESCA -0.067; TCGA KIRC -0.101; TCGA LUAD -0.088; TCGA LUSC -0.064; TCGA PRAD -0.069; TCGA SARC -0.061; TCGA STAD -0.052 |
hsa-miR-338-5p | MARK1 | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; STAD; UCEC | PITA; mirMAP; miRNATAP | TCGA BLCA -0.183; TCGA CESC -0.353; TCGA COAD -0.332; TCGA ESCA -0.577; TCGA HNSC -0.114; TCGA LUAD -0.216; TCGA LUSC -0.483; TCGA STAD -0.261; TCGA UCEC -0.176 |
hsa-miR-338-5p | SLC38A2 | 11 cancers: BLCA; CESC; COAD; ESCA; KIRP; LIHC; LUSC; PAAD; SARC; THCA; STAD | PITA; mirMAP | TCGA BLCA -0.129; TCGA CESC -0.129; TCGA COAD -0.057; TCGA ESCA -0.258; TCGA KIRP -0.068; TCGA LIHC -0.139; TCGA LUSC -0.073; TCGA PAAD -0.129; TCGA SARC -0.076; TCGA THCA -0.069; TCGA STAD -0.113 |
hsa-miR-338-5p | OXR1 | 9 cancers: BLCA; CESC; COAD; KIRC; KIRP; PAAD; SARC; STAD; UCEC | PITA; mirMAP; miRNATAP | TCGA BLCA -0.099; TCGA CESC -0.062; TCGA COAD -0.078; TCGA KIRC -0.107; TCGA KIRP -0.238; TCGA PAAD -0.13; TCGA SARC -0.1; TCGA STAD -0.066; TCGA UCEC -0.061 |
hsa-miR-338-5p | TIA1 | 9 cancers: BLCA; COAD; ESCA; HNSC; KIRP; LGG; LUSC; SARC; STAD | mirMAP | TCGA BLCA -0.173; TCGA COAD -0.074; TCGA ESCA -0.07; TCGA HNSC -0.094; TCGA KIRP -0.221; TCGA LGG -0.071; TCGA LUSC -0.125; TCGA SARC -0.058; TCGA STAD -0.083 |
hsa-miR-338-5p | SIX1 | 10 cancers: BLCA; COAD; ESCA; LGG; LUAD; LUSC; PAAD; PRAD; THCA; STAD | mirMAP | TCGA BLCA -0.119; TCGA COAD -0.222; TCGA ESCA -0.576; TCGA LGG -0.186; TCGA LUAD -0.372; TCGA LUSC -0.363; TCGA PAAD -0.424; TCGA PRAD -0.184; TCGA THCA -0.138; TCGA STAD -0.254 |
hsa-miR-338-5p | MNX1 | 9 cancers: BLCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; PRAD; UCEC | miRNATAP | TCGA BLCA -0.26; TCGA HNSC -0.223; TCGA KIRC -0.494; TCGA KIRP -0.493; TCGA LGG -0.25; TCGA LUAD -0.155; TCGA LUSC -0.315; TCGA PRAD -0.287; TCGA UCEC -0.314 |
hsa-miR-338-5p | TMEM159 | 9 cancers: BLCA; CESC; ESCA; KIRP; LUSC; PAAD; PRAD; SARC; UCEC | miRNATAP | TCGA BLCA -0.146; TCGA CESC -0.154; TCGA ESCA -0.148; TCGA KIRP -0.09; TCGA LUSC -0.116; TCGA PAAD -0.257; TCGA PRAD -0.079; TCGA SARC -0.203; TCGA UCEC -0.081 |
hsa-miR-338-5p | MARCKS | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LGG; LUAD; LUSC; STAD | miRNATAP | TCGA BLCA -0.12; TCGA CESC -0.144; TCGA COAD -0.107; TCGA ESCA -0.165; TCGA HNSC -0.08; TCGA KIRP -0.156; TCGA LGG -0.086; TCGA LUAD -0.083; TCGA LUSC -0.16; TCGA STAD -0.134 |
hsa-miR-338-5p | ADM | 9 cancers: CESC; ESCA; HNSC; LGG; LUAD; LUSC; PAAD; PRAD; UCEC | MirTarget; PITA; miRNATAP | TCGA CESC -0.162; TCGA ESCA -0.276; TCGA HNSC -0.169; TCGA LGG -0.142; TCGA LUAD -0.213; TCGA LUSC -0.291; TCGA PAAD -0.364; TCGA PRAD -0.101; TCGA UCEC -0.128 |
hsa-miR-338-5p | PSD3 | 9 cancers: CESC; KIRC; LIHC; LUAD; LUSC; PAAD; THCA; STAD; UCEC | MirTarget; PITA; miRNATAP | TCGA CESC -0.186; TCGA KIRC -0.281; TCGA LIHC -0.155; TCGA LUAD -0.053; TCGA LUSC -0.18; TCGA PAAD -0.141; TCGA THCA -0.085; TCGA STAD -0.122; TCGA UCEC -0.085 |
hsa-miR-338-5p | CAV2 | 9 cancers: CESC; COAD; ESCA; KIRP; PAAD; SARC; THCA; STAD; UCEC | MirTarget; PITA | TCGA CESC -0.206; TCGA COAD -0.12; TCGA ESCA -0.377; TCGA KIRP -0.153; TCGA PAAD -0.225; TCGA SARC -0.13; TCGA THCA -0.224; TCGA STAD -0.167; TCGA UCEC -0.084 |
hsa-miR-338-5p | PALLD | 10 cancers: CESC; ESCA; LGG; LUAD; LUSC; PAAD; SARC; THCA; STAD; UCEC | MirTarget; PITA | TCGA CESC -0.107; TCGA ESCA -0.249; TCGA LGG -0.075; TCGA LUAD -0.08; TCGA LUSC -0.088; TCGA PAAD -0.125; TCGA SARC -0.422; TCGA THCA -0.123; TCGA STAD -0.187; TCGA UCEC -0.234 |
hsa-miR-338-5p | ENAH | 11 cancers: CESC; COAD; ESCA; KIRC; LGG; LUAD; LUSC; PAAD; SARC; THCA; STAD | PITA; mirMAP; miRNATAP | TCGA CESC -0.087; TCGA COAD -0.185; TCGA ESCA -0.237; TCGA KIRC -0.124; TCGA LGG -0.058; TCGA LUAD -0.111; TCGA LUSC -0.148; TCGA PAAD -0.148; TCGA SARC -0.254; TCGA THCA -0.178; TCGA STAD -0.139 |
hsa-miR-338-5p | AMMECR1 | 10 cancers: CESC; ESCA; KIRC; KIRP; LGG; LUAD; LUSC; PAAD; THCA; STAD | PITA; mirMAP; miRNATAP | TCGA CESC -0.092; TCGA ESCA -0.127; TCGA KIRC -0.122; TCGA KIRP -0.09; TCGA LGG -0.082; TCGA LUAD -0.079; TCGA LUSC -0.16; TCGA PAAD -0.151; TCGA THCA -0.057; TCGA STAD -0.096 |
hsa-miR-338-5p | DPYSL3 | 10 cancers: CESC; COAD; ESCA; LGG; LUAD; PAAD; SARC; THCA; STAD; UCEC | PITA | TCGA CESC -0.174; TCGA COAD -0.224; TCGA ESCA -0.275; TCGA LGG -0.1; TCGA LUAD -0.189; TCGA PAAD -0.201; TCGA SARC -0.289; TCGA THCA -0.123; TCGA STAD -0.336; TCGA UCEC -0.298 |
hsa-miR-338-5p | PCNA | 11 cancers: CESC; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; PAAD; PRAD; THCA | PITA | TCGA CESC -0.081; TCGA ESCA -0.114; TCGA HNSC -0.127; TCGA KIRC -0.114; TCGA KIRP -0.05; TCGA LGG -0.105; TCGA LUAD -0.112; TCGA LUSC -0.304; TCGA PAAD -0.089; TCGA PRAD -0.067; TCGA THCA -0.122 |
hsa-miR-338-5p | CDK5R1 | 9 cancers: CESC; COAD; ESCA; KIRP; LUAD; LUSC; PRAD; THCA; STAD | PITA; miRNATAP | TCGA CESC -0.24; TCGA COAD -0.072; TCGA ESCA -0.44; TCGA KIRP -0.123; TCGA LUAD -0.19; TCGA LUSC -0.429; TCGA PRAD -0.084; TCGA THCA -0.14; TCGA STAD -0.283 |
hsa-miR-338-5p | HOXA9 | 9 cancers: CESC; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; PRAD | PITA; miRNATAP | TCGA CESC -0.227; TCGA ESCA -0.218; TCGA HNSC -0.24; TCGA KIRC -0.188; TCGA KIRP -0.226; TCGA LGG -0.316; TCGA LUAD -0.172; TCGA LUSC -0.269; TCGA PRAD -0.102 |
hsa-miR-338-5p | C18orf54 | 9 cancers: CESC; ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; SARC; STAD | mirMAP | TCGA CESC -0.186; TCGA ESCA -0.254; TCGA HNSC -0.136; TCGA KIRP -0.134; TCGA LUAD -0.152; TCGA LUSC -0.17; TCGA PAAD -0.107; TCGA SARC -0.06; TCGA STAD -0.115 |
hsa-miR-338-5p | WDHD1 | 9 cancers: CESC; ESCA; HNSC; KIRP; LGG; LUAD; LUSC; PAAD; THCA | mirMAP | TCGA CESC -0.089; TCGA ESCA -0.264; TCGA HNSC -0.136; TCGA KIRP -0.1; TCGA LGG -0.087; TCGA LUAD -0.239; TCGA LUSC -0.39; TCGA PAAD -0.127; TCGA THCA -0.196 |
hsa-miR-338-5p | ZNF286B | 10 cancers: CESC; COAD; ESCA; HNSC; KIRP; LGG; LUSC; SARC; STAD; UCEC | mirMAP | TCGA CESC -0.094; TCGA COAD -0.088; TCGA ESCA -0.086; TCGA HNSC -0.129; TCGA KIRP -0.087; TCGA LGG -0.074; TCGA LUSC -0.144; TCGA SARC -0.118; TCGA STAD -0.155; TCGA UCEC -0.102 |
hsa-miR-338-5p | NSMCE2 | 9 cancers: CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; STAD | miRNATAP | TCGA CESC -0.083; TCGA COAD -0.108; TCGA ESCA -0.077; TCGA HNSC -0.093; TCGA LUAD -0.091; TCGA LUSC -0.062; TCGA PAAD -0.077; TCGA PRAD -0.128; TCGA STAD -0.09 |
hsa-miR-338-5p | WDYHV1 | 10 cancers: CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PRAD; THCA; STAD | miRNATAP | TCGA CESC -0.058; TCGA COAD -0.062; TCGA ESCA -0.091; TCGA HNSC -0.134; TCGA KIRC -0.082; TCGA LUAD -0.088; TCGA LUSC -0.186; TCGA PRAD -0.091; TCGA THCA -0.106; TCGA STAD -0.057 |
hsa-miR-338-5p | CLIC4 | 10 cancers: CESC; COAD; ESCA; HNSC; KIRP; LIHC; PAAD; SARC; THCA; STAD | miRNATAP | TCGA CESC -0.068; TCGA COAD -0.128; TCGA ESCA -0.206; TCGA HNSC -0.061; TCGA KIRP -0.11; TCGA LIHC -0.065; TCGA PAAD -0.136; TCGA SARC -0.167; TCGA THCA -0.056; TCGA STAD -0.242 |
hsa-miR-338-5p | CBX3 | 9 cancers: COAD; HNSC; KIRP; LGG; LUAD; LUSC; PAAD; PRAD; THCA | MirTarget; PITA; miRNATAP | TCGA COAD -0.052; TCGA HNSC -0.121; TCGA KIRP -0.098; TCGA LGG -0.075; TCGA LUAD -0.093; TCGA LUSC -0.176; TCGA PAAD -0.102; TCGA PRAD -0.106; TCGA THCA -0.087 |
hsa-miR-338-5p | KDELC2 | 9 cancers: COAD; ESCA; LGG; LUAD; LUSC; PAAD; SARC; STAD; UCEC | MirTarget; mirMAP | TCGA COAD -0.105; TCGA ESCA -0.082; TCGA LGG -0.061; TCGA LUAD -0.098; TCGA LUSC -0.088; TCGA PAAD -0.179; TCGA SARC -0.149; TCGA STAD -0.204; TCGA UCEC -0.082 |
hsa-miR-338-5p | SLC4A7 | 9 cancers: COAD; ESCA; KIRC; LGG; LUAD; LUSC; PAAD; SARC; THCA | MirTarget; PITA; miRNATAP | TCGA COAD -0.102; TCGA ESCA -0.102; TCGA KIRC -0.132; TCGA LGG -0.098; TCGA LUAD -0.087; TCGA LUSC -0.066; TCGA PAAD -0.132; TCGA SARC -0.173; TCGA THCA -0.164 |
hsa-miR-338-5p | STK3 | 9 cancers: COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; SARC; STAD | PITA | TCGA COAD -0.067; TCGA ESCA -0.15; TCGA HNSC -0.082; TCGA KIRC -0.099; TCGA LUAD -0.064; TCGA LUSC -0.051; TCGA PAAD -0.13; TCGA SARC -0.13; TCGA STAD -0.09 |
hsa-miR-338-5p | MCM10 | 9 cancers: ESCA; HNSC; KIRP; LGG; LUAD; LUSC; PAAD; PRAD; THCA | MirTarget | TCGA ESCA -0.191; TCGA HNSC -0.14; TCGA KIRP -0.186; TCGA LGG -0.232; TCGA LUAD -0.365; TCGA LUSC -0.677; TCGA PAAD -0.283; TCGA PRAD -0.231; TCGA THCA -0.226 |
hsa-miR-338-5p | HMGB3 | 10 cancers: ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; PRAD; THCA; STAD | MirTarget; PITA; miRNATAP | TCGA ESCA -0.179; TCGA HNSC -0.23; TCGA KIRC -0.247; TCGA KIRP -0.261; TCGA LGG -0.054; TCGA LUAD -0.205; TCGA LUSC -0.306; TCGA PRAD -0.108; TCGA THCA -0.127; TCGA STAD -0.119 |
hsa-miR-338-5p | FKBP14 | 9 cancers: ESCA; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; THCA; STAD | MirTarget | TCGA ESCA -0.107; TCGA KIRP -0.075; TCGA LGG -0.093; TCGA LIHC -0.078; TCGA LUAD -0.09; TCGA LUSC -0.115; TCGA PAAD -0.141; TCGA THCA -0.052; TCGA STAD -0.124 |
hsa-miR-338-5p | FAP | 9 cancers: ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; THCA; STAD; UCEC | PITA; miRNATAP | TCGA ESCA -0.477; TCGA HNSC -0.258; TCGA LUAD -0.228; TCGA LUSC -0.196; TCGA PAAD -0.435; TCGA PRAD -0.29; TCGA THCA -0.625; TCGA STAD -0.33; TCGA UCEC -0.232 |
hsa-miR-338-5p | LAMC2 | 9 cancers: ESCA; HNSC; KIRC; KIRP; LUSC; PAAD; SARC; THCA; UCEC | mirMAP | TCGA ESCA -0.281; TCGA HNSC -0.167; TCGA KIRC -0.463; TCGA KIRP -0.337; TCGA LUSC -0.261; TCGA PAAD -0.474; TCGA SARC -0.155; TCGA THCA -0.325; TCGA UCEC -0.154 |
hsa-miR-338-5p | SULF1 | 9 cancers: ESCA; HNSC; LUAD; LUSC; PAAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA ESCA -0.368; TCGA HNSC -0.14; TCGA LUAD -0.317; TCGA LUSC -0.303; TCGA PAAD -0.377; TCGA SARC -0.204; TCGA THCA -0.111; TCGA STAD -0.268; TCGA UCEC -0.159 |
hsa-miR-338-5p | TPBG | 10 cancers: ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; THCA; STAD; UCEC | mirMAP | TCGA ESCA -0.243; TCGA HNSC -0.112; TCGA KIRC -0.141; TCGA KIRP -0.204; TCGA LUAD -0.171; TCGA LUSC -0.318; TCGA PAAD -0.348; TCGA THCA -0.18; TCGA STAD -0.176; TCGA UCEC -0.109 |
hsa-miR-338-5p | COL5A2 | 9 cancers: ESCA; HNSC; LGG; LUAD; LUSC; PAAD; PRAD; THCA; UCEC | mirMAP | TCGA ESCA -0.252; TCGA HNSC -0.267; TCGA LGG -0.145; TCGA LUAD -0.221; TCGA LUSC -0.297; TCGA PAAD -0.356; TCGA PRAD -0.103; TCGA THCA -0.282; TCGA UCEC -0.148 |
hsa-miR-338-5p | COL5A1 | 9 cancers: ESCA; HNSC; LGG; LUAD; LUSC; PAAD; THCA; STAD; UCEC | miRNATAP | TCGA ESCA -0.281; TCGA HNSC -0.244; TCGA LGG -0.143; TCGA LUAD -0.188; TCGA LUSC -0.238; TCGA PAAD -0.419; TCGA THCA -0.431; TCGA STAD -0.137; TCGA UCEC -0.17 |
hsa-miR-338-5p | ROR2 | 9 cancers: ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; SARC; STAD; UCEC | miRNATAP | TCGA ESCA -0.353; TCGA HNSC -0.144; TCGA KIRC -0.5; TCGA LUAD -0.103; TCGA LUSC -0.13; TCGA PAAD -0.335; TCGA SARC -0.229; TCGA STAD -0.343; TCGA UCEC -0.147 |
hsa-miR-338-5p | DIAPH3 | 9 cancers: HNSC; KIRC; LGG; LUAD; LUSC; PAAD; PRAD; SARC; THCA | MirTarget | TCGA HNSC -0.085; TCGA KIRC -0.265; TCGA LGG -0.109; TCGA LUAD -0.155; TCGA LUSC -0.167; TCGA PAAD -0.311; TCGA PRAD -0.313; TCGA SARC -0.101; TCGA THCA -0.491 |
Enriched cancer pathways of putative targets