microRNA information: hsa-miR-374b-3p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-374b-3p | miRbase |
| Accession: | MIMAT0004956 | miRbase |
| Precursor name: | hsa-mir-374b | miRbase |
| Precursor accession: | MI0005566 | miRbase |
| Symbol: | MIR374B | HGNC |
| RefSeq ID: | NR_030620 | GenBank |
| Sequence: | CUUAGCAGGUUGUAUUAUCAUU |
Reported expression in cancers: hsa-miR-374b-3p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-374b-3p | lymphoma | downregulation | "The aim of the present study was to investigate th ......" | 26100275 | |
| hsa-miR-374b-3p | pancreatic cancer | downregulation | "Employing a hidden Markov model HMM algorithm we i ......" | 27229158 | |
| hsa-miR-374b-3p | prostate cancer | downregulation | "To further investigate the global association betw ......" | 24191917 | Microarray; qPCR |
Reported cancer pathway affected by hsa-miR-374b-3p

| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-374b-3p | lymphoma | Apoptosis pathway | "MicroRNA 374b Suppresses Proliferation and Promote ......" | 26100275 | Luciferase |
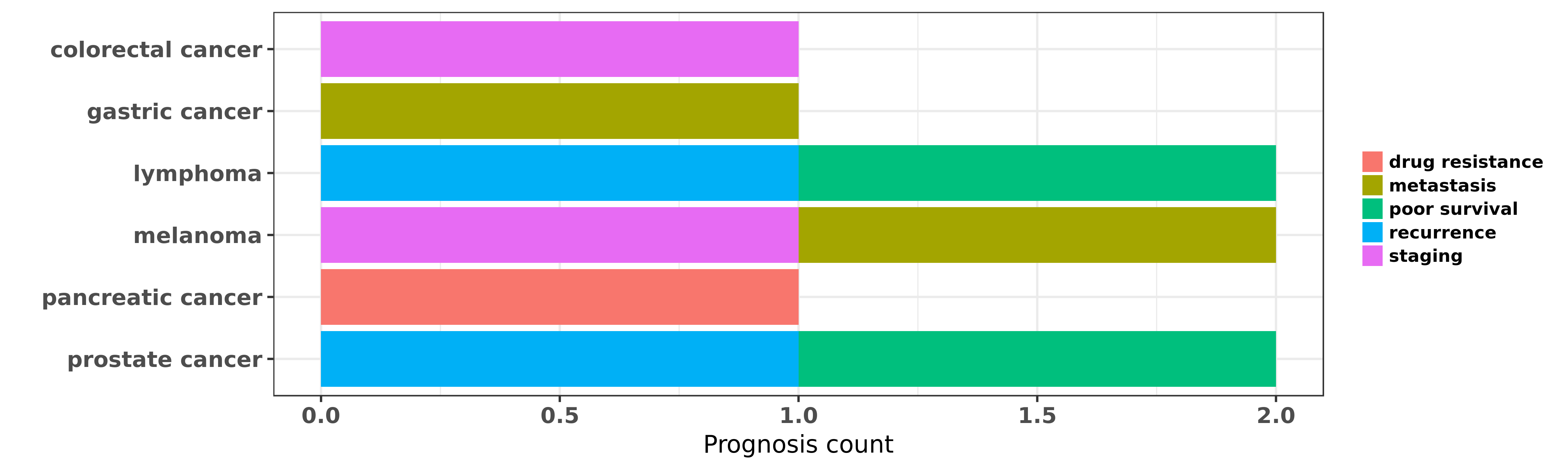
Reported cancer prognosis affected by hsa-miR-374b-3p
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-374b-3p | colorectal cancer | staging | "The potential value of miR 1 and miR 374b as bioma ......" | 26045793 | |
| hsa-miR-374b-3p | gastric cancer | metastasis | "MiR 374b 5p suppresses RECK expression and promote ......" | 25516656 | |
| hsa-miR-374b-3p | lymphoma | poor survival; recurrence | "MicroRNA 374b Suppresses Proliferation and Promote ......" | 26100275 | Luciferase |
| hsa-miR-374b-3p | melanoma | staging; metastasis | "Our analyses identified a 4-microRNA miR-150-5p mi ......" | 26089374 | |
| hsa-miR-374b-3p | pancreatic cancer | drug resistance | "Evidence for the role of microRNA 374b in acquired ......" | 27229158 | |
| hsa-miR-374b-3p | prostate cancer | poor survival; recurrence | "To further investigate the global association betw ......" | 24191917 |
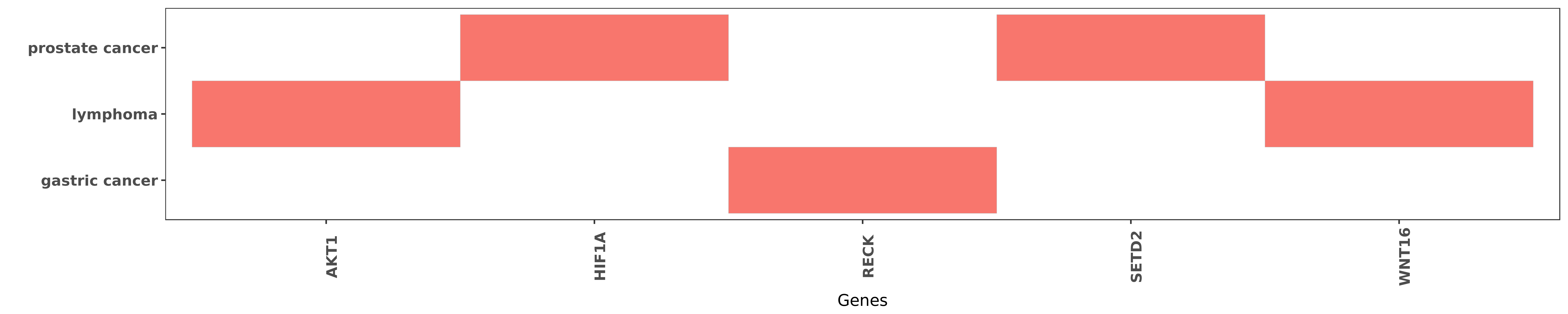
Reported gene related to hsa-miR-374b-3p
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-374b-3p | lymphoma | AKT1 | "MicroRNA 374b Suppresses Proliferation and Promote ......" | 26100275 |
| hsa-miR-374b-3p | prostate cancer | HIF1A | "Also overexpression of miR3195 or miR374b reduced ......" | 25553085 |
| hsa-miR-374b-3p | gastric cancer | RECK | "MiR 374b 5p suppresses RECK expression and promote ......" | 25516656 |
| hsa-miR-374b-3p | prostate cancer | SETD2 | "Also overexpression of miR3195 or miR374b reduced ......" | 25553085 |
| hsa-miR-374b-3p | lymphoma | WNT16 | "Real-time quantitative PCR and immunohistochemistr ......" | 26100275 |
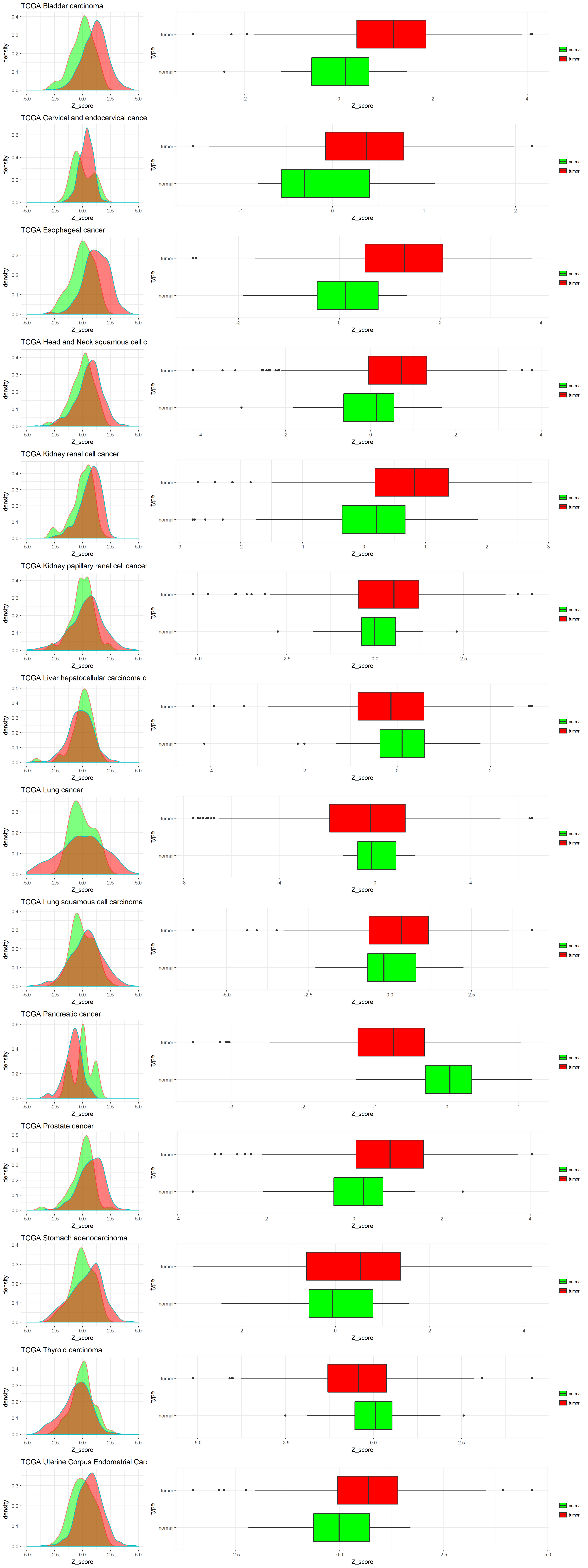
Expression profile in cancer corhorts:
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-374b-3p | NPTN | 9 cancers: BLCA; COAD; ESCA; KIRC; LUSC; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.162; TCGA COAD -0.076; TCGA ESCA -0.08; TCGA KIRC -0.098; TCGA LUSC -0.074; TCGA PRAD -0.114; TCGA SARC -0.24; TCGA STAD -0.056; TCGA UCEC -0.089 |
hsa-miR-374b-3p | PCMTD1 | 10 cancers: BLCA; CESC; ESCA; HNSC; KIRC; KIRP; LUSC; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.078; TCGA CESC -0.099; TCGA ESCA -0.119; TCGA HNSC -0.136; TCGA KIRC -0.114; TCGA KIRP -0.062; TCGA LUSC -0.1; TCGA SARC -0.24; TCGA STAD -0.23; TCGA UCEC -0.108 |
hsa-miR-374b-3p | FOXO1 | 10 cancers: BLCA; CESC; COAD; HNSC; KIRC; KIRP; LUSC; PRAD; SARC; UCEC | MirTarget | TCGA BLCA -0.219; TCGA CESC -0.109; TCGA COAD -0.252; TCGA HNSC -0.171; TCGA KIRC -0.151; TCGA KIRP -0.202; TCGA LUSC -0.183; TCGA PRAD -0.153; TCGA SARC -0.139; TCGA UCEC -0.186 |
hsa-miR-374b-3p | ARHGAP20 | 10 cancers: BLCA; COAD; ESCA; HNSC; LGG; LIHC; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.508; TCGA COAD -0.673; TCGA ESCA -0.439; TCGA HNSC -0.331; TCGA LGG -0.214; TCGA LIHC -0.234; TCGA PRAD -0.222; TCGA SARC -0.407; TCGA STAD -0.376; TCGA UCEC -0.311 |
hsa-miR-374b-3p | DCN | 9 cancers: BLCA; CESC; COAD; ESCA; KIRC; LIHC; LUSC; PAAD; UCEC | MirTarget | TCGA BLCA -0.6; TCGA CESC -0.284; TCGA COAD -0.612; TCGA ESCA -0.358; TCGA KIRC -0.633; TCGA LIHC -0.444; TCGA LUSC -0.352; TCGA PAAD -0.277; TCGA UCEC -0.389 |
hsa-miR-374b-3p | CCDC80 | 9 cancers: BLCA; COAD; ESCA; LIHC; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.477; TCGA COAD -0.792; TCGA ESCA -0.434; TCGA LIHC -0.361; TCGA LUSC -0.36; TCGA PAAD -0.333; TCGA PRAD -0.197; TCGA STAD -0.278; TCGA UCEC -0.194 |
hsa-miR-374b-3p | ASH1L | 9 cancers: BLCA; CESC; ESCA; HNSC; KIRC; KIRP; LUAD; SARC; STAD | MirTarget | TCGA BLCA -0.129; TCGA CESC -0.087; TCGA ESCA -0.097; TCGA HNSC -0.1; TCGA KIRC -0.118; TCGA KIRP -0.131; TCGA LUAD -0.113; TCGA SARC -0.189; TCGA STAD -0.116 |
hsa-miR-374b-3p | IL6ST | 9 cancers: BLCA; COAD; ESCA; HNSC; LUAD; LUSC; PRAD; SARC; STAD | MirTarget | TCGA BLCA -0.435; TCGA COAD -0.407; TCGA ESCA -0.302; TCGA HNSC -0.37; TCGA LUAD -0.195; TCGA LUSC -0.217; TCGA PRAD -0.336; TCGA SARC -0.262; TCGA STAD -0.229 |
hsa-miR-374b-3p | SLC24A3 | 12 cancers: BLCA; CESC; COAD; HNSC; KIRP; LGG; LIHC; LUSC; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.677; TCGA CESC -0.382; TCGA COAD -0.66; TCGA HNSC -0.377; TCGA KIRP -0.36; TCGA LGG -0.345; TCGA LIHC -0.352; TCGA LUSC -0.446; TCGA PRAD -0.173; TCGA SARC -1.142; TCGA STAD -0.199; TCGA UCEC -0.476 |
hsa-miR-374b-3p | MCC | 9 cancers: BLCA; COAD; HNSC; KIRC; LGG; LUSC; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.22; TCGA COAD -0.461; TCGA HNSC -0.136; TCGA KIRC -0.142; TCGA LGG -0.284; TCGA LUSC -0.251; TCGA PRAD -0.202; TCGA STAD -0.235; TCGA UCEC -0.178 |
hsa-miR-374b-3p | CAB39 | 9 cancers: BLCA; ESCA; HNSC; KIRC; LUSC; PAAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.094; TCGA ESCA -0.135; TCGA HNSC -0.137; TCGA KIRC -0.101; TCGA LUSC -0.104; TCGA PAAD -0.085; TCGA SARC -0.085; TCGA STAD -0.05; TCGA UCEC -0.093 |
hsa-miR-374b-3p | LCN2 | 9 cancers: BLCA; CESC; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; THCA | MirTarget | TCGA BLCA -0.483; TCGA CESC -0.475; TCGA HNSC -0.561; TCGA KIRC -0.639; TCGA KIRP -0.531; TCGA LUAD -0.296; TCGA LUSC -0.443; TCGA PAAD -0.916; TCGA THCA -0.703 |
hsa-miR-374b-3p | PGR | 9 cancers: BLCA; COAD; ESCA; KIRC; LUAD; LUSC; PRAD; SARC; STAD | mirMAP | TCGA BLCA -0.368; TCGA COAD -0.747; TCGA ESCA -0.402; TCGA KIRC -0.228; TCGA LUAD -0.209; TCGA LUSC -0.346; TCGA PRAD -0.238; TCGA SARC -0.93; TCGA STAD -0.52 |
hsa-miR-374b-3p | ENTPD1 | 10 cancers: BLCA; COAD; KIRP; LIHC; LUSC; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.156; TCGA COAD -0.24; TCGA KIRP -0.074; TCGA LIHC -0.129; TCGA LUSC -0.197; TCGA PRAD -0.068; TCGA SARC -0.281; TCGA THCA -0.201; TCGA STAD -0.128; TCGA UCEC -0.094 |
hsa-miR-374b-3p | GFRA1 | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.558; TCGA CESC -0.312; TCGA COAD -0.852; TCGA ESCA -0.501; TCGA HNSC -0.358; TCGA LUAD -0.365; TCGA LUSC -0.366; TCGA PRAD -0.194; TCGA STAD -0.537; TCGA UCEC -0.323 |
hsa-miR-374b-3p | JMY | 10 cancers: BLCA; CESC; COAD; HNSC; KIRC; KIRP; LUAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.102; TCGA CESC -0.127; TCGA COAD -0.141; TCGA HNSC -0.147; TCGA KIRC -0.202; TCGA KIRP -0.102; TCGA LUAD -0.077; TCGA SARC -0.206; TCGA STAD -0.21; TCGA UCEC -0.113 |
hsa-miR-374b-3p | SHROOM4 | 10 cancers: BLCA; CESC; HNSC; LIHC; LUAD; LUSC; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.184; TCGA CESC -0.215; TCGA HNSC -0.168; TCGA LIHC -0.179; TCGA LUAD -0.214; TCGA LUSC -0.203; TCGA PRAD -0.24; TCGA THCA -0.149; TCGA STAD -0.13; TCGA UCEC -0.135 |
hsa-miR-374b-3p | UBXN10 | 9 cancers: BLCA; CESC; ESCA; KIRP; LGG; LUAD; LUSC; PRAD; STAD | mirMAP | TCGA BLCA -0.262; TCGA CESC -0.444; TCGA ESCA -0.475; TCGA KIRP -0.315; TCGA LGG -0.458; TCGA LUAD -0.366; TCGA LUSC -0.31; TCGA PRAD -0.245; TCGA STAD -0.275 |
hsa-miR-374b-3p | ANTXR2 | 10 cancers: BLCA; COAD; ESCA; LIHC; LUAD; LUSC; PAAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.391; TCGA COAD -0.253; TCGA ESCA -0.297; TCGA LIHC -0.11; TCGA LUAD -0.127; TCGA LUSC -0.322; TCGA PAAD -0.201; TCGA SARC -0.145; TCGA STAD -0.286; TCGA UCEC -0.349 |
hsa-miR-374b-3p | TSHZ2 | 9 cancers: BLCA; CESC; COAD; HNSC; KIRC; KIRP; LUAD; PRAD; STAD | mirMAP | TCGA BLCA -0.289; TCGA CESC -0.332; TCGA COAD -0.763; TCGA HNSC -0.27; TCGA KIRC -0.197; TCGA KIRP -0.338; TCGA LUAD -0.218; TCGA PRAD -0.243; TCGA STAD -0.215 |
hsa-miR-374b-3p | SLIT3 | 9 cancers: BLCA; CESC; COAD; ESCA; LIHC; LUAD; LUSC; PRAD; SARC | mirMAP | TCGA BLCA -0.32; TCGA CESC -0.242; TCGA COAD -0.687; TCGA ESCA -0.298; TCGA LIHC -0.212; TCGA LUAD -0.216; TCGA LUSC -0.443; TCGA PRAD -0.247; TCGA SARC -0.245 |
hsa-miR-374b-3p | PARVA | 11 cancers: BLCA; COAD; ESCA; LGG; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.206; TCGA COAD -0.115; TCGA ESCA -0.161; TCGA LGG -0.169; TCGA LUAD -0.099; TCGA LUSC -0.103; TCGA PAAD -0.131; TCGA PRAD -0.131; TCGA SARC -0.162; TCGA STAD -0.225; TCGA UCEC -0.077 |
hsa-miR-374b-3p | PLXNA4 | 10 cancers: BLCA; CESC; COAD; KIRC; LGG; LIHC; LUSC; PRAD; SARC; STAD | mirMAP | TCGA BLCA -0.414; TCGA CESC -0.277; TCGA COAD -0.833; TCGA KIRC -0.457; TCGA LGG -0.323; TCGA LIHC -0.382; TCGA LUSC -0.349; TCGA PRAD -0.293; TCGA SARC -0.715; TCGA STAD -0.245 |
hsa-miR-374b-3p | NIPAL3 | 9 cancers: CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; STAD | MirTarget | TCGA CESC -0.084; TCGA COAD -0.091; TCGA ESCA -0.131; TCGA HNSC -0.077; TCGA KIRC -0.156; TCGA KIRP -0.267; TCGA LIHC -0.076; TCGA LUAD -0.116; TCGA STAD -0.097 |
hsa-miR-374b-3p | QSOX1 | 12 cancers: CESC; COAD; ESCA; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; THCA | mirMAP | TCGA CESC -0.163; TCGA COAD -0.232; TCGA ESCA -0.331; TCGA HNSC -0.127; TCGA KIRP -0.142; TCGA LGG -0.085; TCGA LIHC -0.233; TCGA LUAD -0.134; TCGA LUSC -0.124; TCGA PAAD -0.189; TCGA PRAD -0.173; TCGA THCA -0.143 |
hsa-miR-374b-3p | TP53INP1 | 9 cancers: CESC; COAD; ESCA; HNSC; LIHC; LUSC; SARC; STAD; UCEC | mirMAP | TCGA CESC -0.127; TCGA COAD -0.231; TCGA ESCA -0.206; TCGA HNSC -0.161; TCGA LIHC -0.131; TCGA LUSC -0.124; TCGA SARC -0.178; TCGA STAD -0.181; TCGA UCEC -0.124 |
hsa-miR-374b-3p | TMEM30A | 9 cancers: COAD; ESCA; HNSC; KIRC; KIRP; LUAD; SARC; STAD; UCEC | MirTarget | TCGA COAD -0.094; TCGA ESCA -0.088; TCGA HNSC -0.115; TCGA KIRC -0.061; TCGA KIRP -0.067; TCGA LUAD -0.074; TCGA SARC -0.12; TCGA STAD -0.111; TCGA UCEC -0.083 |
Enriched cancer pathways of putative targets