microRNA information: hsa-miR-376c-3p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-376c-3p | miRbase |
| Accession: | MIMAT0000720 | miRbase |
| Precursor name: | hsa-mir-376c | miRbase |
| Precursor accession: | MI0000776 | miRbase |
| Symbol: | MIR376C | HGNC |
| RefSeq ID: | NR_029861 | GenBank |
| Sequence: | AACAUAGAGGAAAUUCCACGU |
Reported expression in cancers: hsa-miR-376c-3p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-376c-3p | breast cancer | upregulation | "Selected candidates identified in the initial scre ......" | 22927033 | qPCR |
| hsa-miR-376c-3p | cervical and endocervical cancer | deregulation | "Quantitative RT-PCR was used to measure miR-376c e ......" | 27345009 | qPCR |
| hsa-miR-376c-3p | glioblastoma | deregulation | "Hypoxic signature of microRNAs in glioblastoma: in ......" | 25129238 | RNA-Seq |
| hsa-miR-376c-3p | lung squamous cell cancer | upregulation | "MicroRNA 376c suppresses non small cell lung cance ......" | 27049310 | qPCR |
| hsa-miR-376c-3p | melanoma | upregulation | "Silencing of a large microRNA cluster on human chr ......" | 22747855 | |
| hsa-miR-376c-3p | prostate cancer | downregulation | "Sustaining a role of miR-376c in the regulation of ......" | 26385605 |
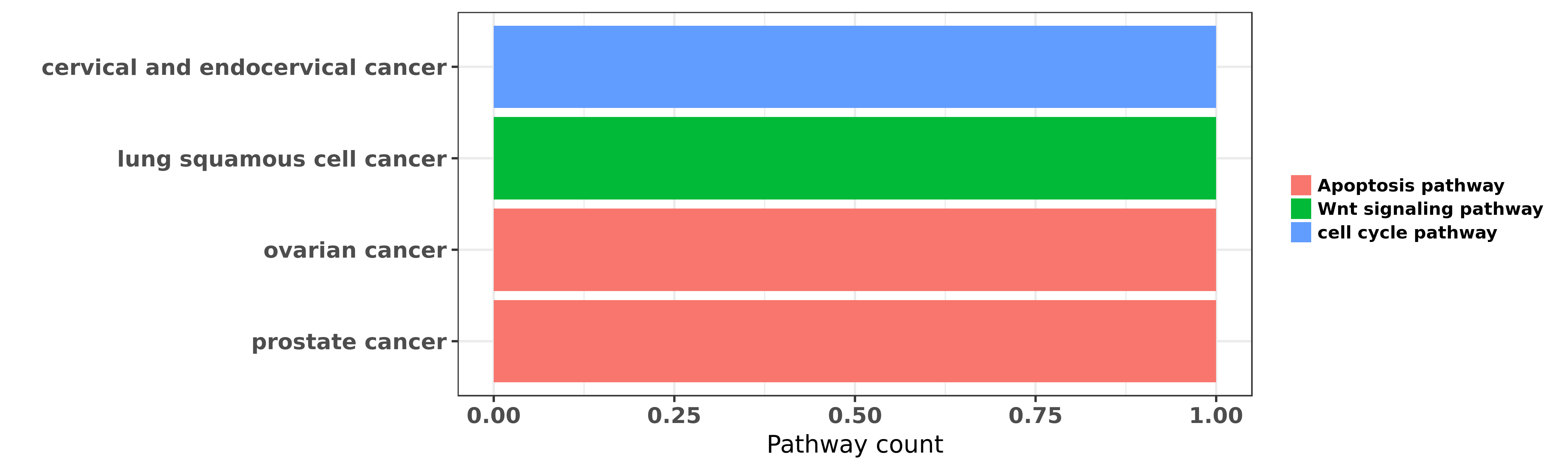
Reported cancer pathway affected by hsa-miR-376c-3p
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-376c-3p | cervical and endocervical cancer | cell cycle pathway | "miR 376c inhibits cervical cancer cell proliferati ......" | 27345009 | Luciferase |
| hsa-miR-376c-3p | lung squamous cell cancer | Wnt signaling pathway | "MicroRNA 376c suppresses non small cell lung cance ......" | 27049310 | Western blot; Luciferase |
| hsa-miR-376c-3p | ovarian cancer | Apoptosis pathway | "MicroRNA 376c enhances ovarian cancer cell surviva ......" | 21224400 | |
| hsa-miR-376c-3p | prostate cancer | Apoptosis pathway | "MicroRNAs miR 154 miR 299 5p miR 376a miR 376c miR ......" | 24166498 |
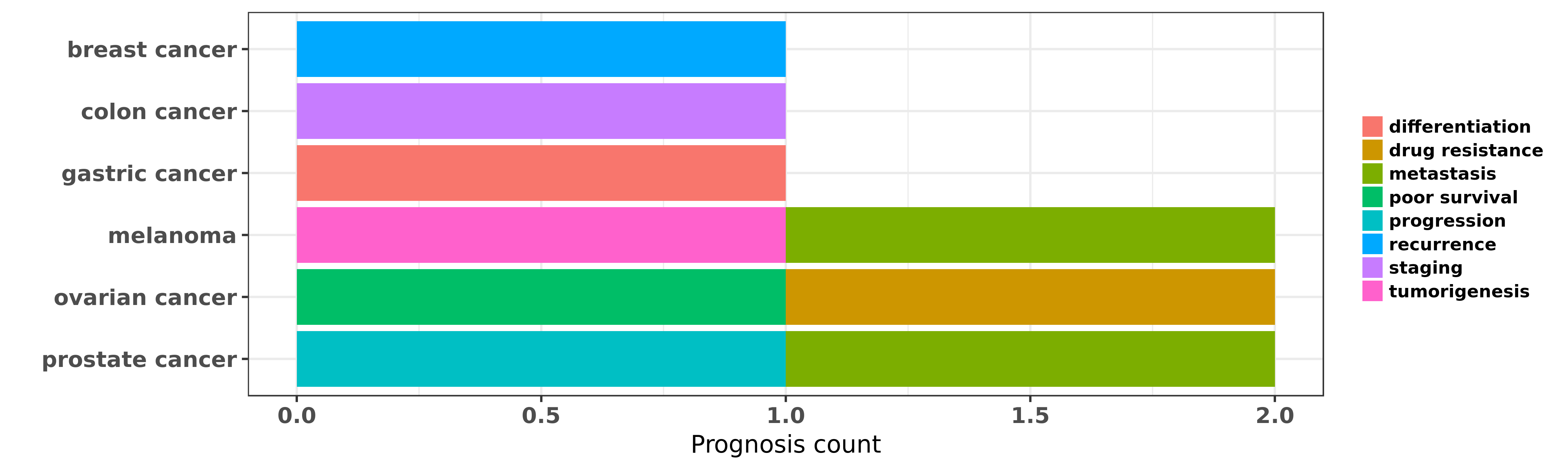
Reported cancer prognosis affected by hsa-miR-376c-3p
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-376c-3p | breast cancer | recurrence | "We identified seven miRNAs that were differentiall ......" | 27409424 | |
| hsa-miR-376c-3p | colon cancer | staging | "Large-scale microRNA expression profiling was perf ......" | 27485599 | |
| hsa-miR-376c-3p | gastric cancer | differentiation | "Differential miRNAs were identified in serum pools ......" | 22432036 | |
| hsa-miR-376c-3p | melanoma | metastasis; tumorigenesis | "Silencing of a large microRNA cluster on human chr ......" | 22747855 | Luciferase |
| hsa-miR-376c-3p | ovarian cancer | poor survival; drug resistance | "MicroRNA 376c enhances ovarian cancer cell surviva ......" | 21224400 | |
| hsa-miR-376c-3p | prostate cancer | metastasis; progression | "Reporter vector assays in HEK293 cells initially v ......" | 26385605 |
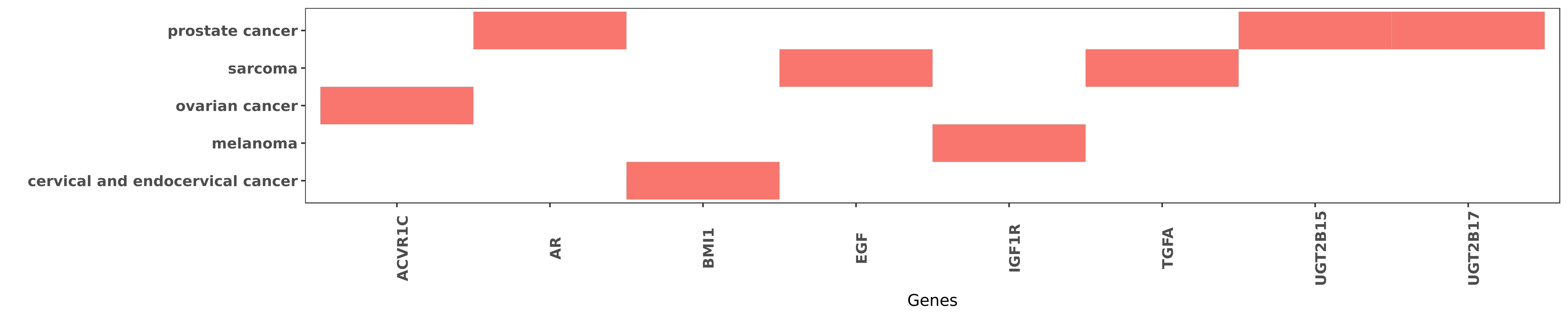
Reported gene related to hsa-miR-376c-3p
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-376c-3p | prostate cancer | UGT2B15 | "Ectopic miR-376c expression in prostate cancer cel ......" | 26385605 |
| hsa-miR-376c-3p | prostate cancer | UGT2B15 | "Regulation of Human UGT2B15 and UGT2B17 by miR 376 ......" | 26163549 |
| hsa-miR-376c-3p | prostate cancer | UGT2B17 | "Ectopic miR-376c expression in prostate cancer cel ......" | 26385605 |
| hsa-miR-376c-3p | prostate cancer | UGT2B17 | "Regulation of Human UGT2B15 and UGT2B17 by miR 376 ......" | 26163549 |
| hsa-miR-376c-3p | ovarian cancer | ACVR1C | "In this study we examined the regulation of ALK7 b ......" | 21224400 |
| hsa-miR-376c-3p | prostate cancer | AR | "Consistent with reduced androgen inactivation ecto ......" | 26385605 |
| hsa-miR-376c-3p | cervical and endocervical cancer | BMI1 | "miR 376c inhibits cervical cancer cell proliferati ......" | 27345009 |
| hsa-miR-376c-3p | sarcoma | EGF | "Overexpression of miR-376c suppressed TGFA express ......" | 23631646 |
| hsa-miR-376c-3p | melanoma | IGF1R | "Our results suggest that down-regulation of mir-37 ......" | 22747855 |
| hsa-miR-376c-3p | sarcoma | TGFA | "Overexpression of miR-376c suppressed TGFA express ......" | 23631646 |
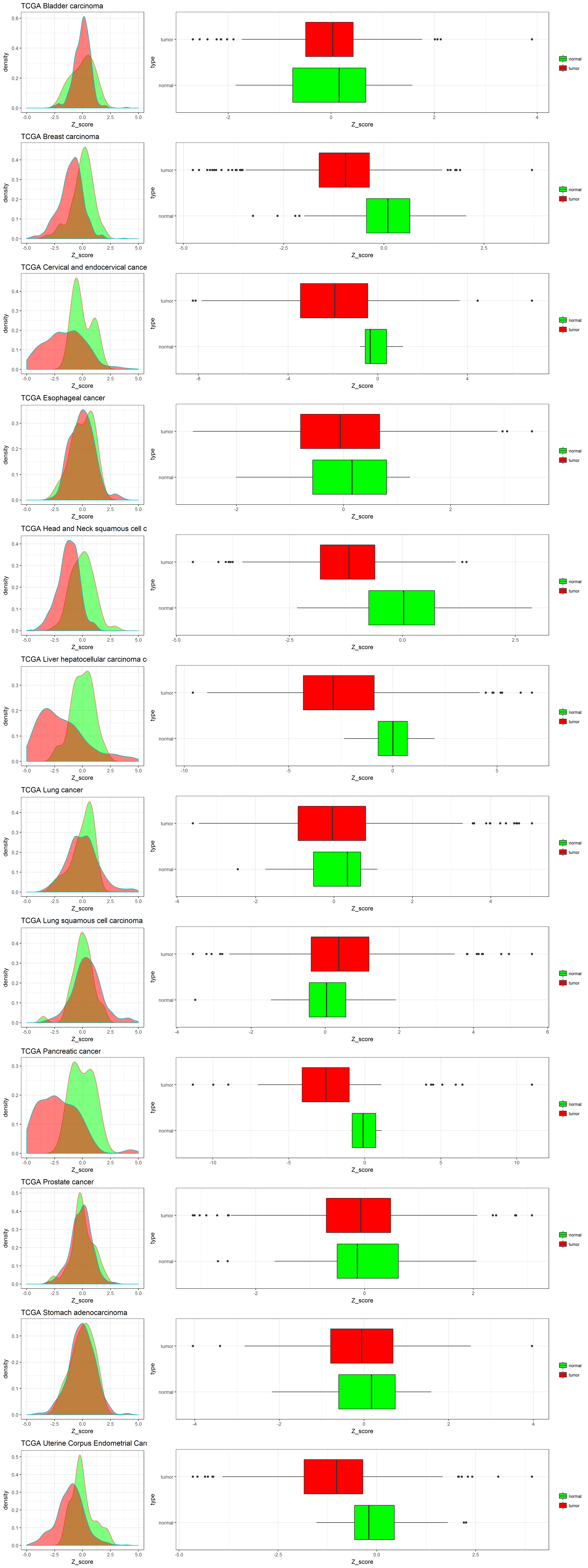
Expression profile in cancer corhorts:
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-376c-3p | ZFP62 | 9 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LGG; LUSC; PRAD; STAD | MirTarget | TCGA BLCA -0.116; TCGA BRCA -0.073; TCGA CESC -0.055; TCGA ESCA -0.091; TCGA HNSC -0.057; TCGA LGG -0.078; TCGA LUSC -0.056; TCGA PRAD -0.107; TCGA STAD -0.09 |
hsa-miR-376c-3p | FBXO28 | 9 cancers: BLCA; BRCA; COAD; ESCA; LGG; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.052; TCGA BRCA -0.084; TCGA COAD -0.064; TCGA ESCA -0.112; TCGA LGG -0.064; TCGA PAAD -0.122; TCGA PRAD -0.132; TCGA STAD -0.096; TCGA UCEC -0.083 |
hsa-miR-376c-3p | FAM199X | 9 cancers: BLCA; BRCA; COAD; ESCA; HNSC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.062; TCGA BRCA -0.102; TCGA COAD -0.091; TCGA ESCA -0.116; TCGA HNSC -0.094; TCGA PAAD -0.114; TCGA PRAD -0.226; TCGA STAD -0.121; TCGA UCEC -0.15 |
hsa-miR-376c-3p | MOSPD2 | 9 cancers: BLCA; BRCA; COAD; HNSC; LUSC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.081; TCGA BRCA -0.052; TCGA COAD -0.139; TCGA HNSC -0.058; TCGA LUSC -0.11; TCGA PAAD -0.114; TCGA PRAD -0.195; TCGA STAD -0.107; TCGA UCEC -0.081 |
hsa-miR-376c-3p | GABPB2 | 9 cancers: BLCA; BRCA; ESCA; LIHC; LUSC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.082; TCGA BRCA -0.099; TCGA ESCA -0.147; TCGA LIHC -0.052; TCGA LUSC -0.064; TCGA PAAD -0.117; TCGA PRAD -0.1; TCGA STAD -0.133; TCGA UCEC -0.076 |
hsa-miR-376c-3p | LCOR | 9 cancers: BLCA; BRCA; COAD; HNSC; LUAD; LUSC; PAAD; PRAD; UCEC | mirMAP | TCGA BLCA -0.117; TCGA BRCA -0.084; TCGA COAD -0.238; TCGA HNSC -0.111; TCGA LUAD -0.062; TCGA LUSC -0.101; TCGA PAAD -0.217; TCGA PRAD -0.264; TCGA UCEC -0.113 |
hsa-miR-376c-3p | CDK12 | 9 cancers: BRCA; COAD; ESCA; HNSC; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BRCA -0.085; TCGA COAD -0.086; TCGA ESCA -0.094; TCGA HNSC -0.085; TCGA LUSC -0.051; TCGA PAAD -0.063; TCGA PRAD -0.162; TCGA STAD -0.119; TCGA UCEC -0.069 |
Enriched cancer pathways of putative targets