microRNA information: hsa-miR-378a-5p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-378a-5p | miRbase |
| Accession: | MIMAT0000731 | miRbase |
| Precursor name: | hsa-mir-378a | miRbase |
| Precursor accession: | MI0000786 | miRbase |
| Symbol: | NA | HGNC |
| RefSeq ID: | NA | GenBank |
| Sequence: | CUCCUGACUCCAGGUCCUGUGU |
Reported expression in cancers: hsa-miR-378a-5p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-378a-5p | acute myeloid leukemia | upregulation | "Overexpression of miR 378 is frequent and may affe ......" | 23582927 | qPCR |
| hsa-miR-378a-5p | bladder cancer | deregulation | "In other recruited 17 patients with BUC who were d ......" | 22886973 | Reverse transcription PCR; Microarray |
| hsa-miR-378a-5p | breast cancer | upregulation | "Association between mir 24 and mir 378 in formalin ......" | 25120807 | qPCR |
| hsa-miR-378a-5p | colorectal cancer | downregulation | "MiR 378 is an independent prognostic factor and in ......" | 24555885 | |
| hsa-miR-378a-5p | gastric cancer | upregulation | "The most highly expressed miRNAs in non-tumorous t ......" | 19175831 | |
| hsa-miR-378a-5p | gastric cancer | deregulation | "The results from the miRNA microarray analysis wer ......" | 21475928 | qPCR; Microarray |
| hsa-miR-378a-5p | gastric cancer | downregulation | "The low expression of miR-195 and miR-378 was clos ......" | 23333942 | |
| hsa-miR-378a-5p | gastric cancer | upregulation | "We identified five miRNAs that were most consisten ......" | 24040025 | qPCR |
| hsa-miR-378a-5p | gastric cancer | downregulation | "Currently a large number of miRNAs have been repor ......" | 24139413 | |
| hsa-miR-378a-5p | liver cancer | upregulation | "miR-378 regulates osteoblast differentiation and p ......" | 25562172 | qPCR |
| hsa-miR-378a-5p | lung cancer | upregulation | "The effect of miR-378 upregulation on tumor growth ......" | 26781643 | |
| hsa-miR-378a-5p | lung squamous cell cancer | deregulation | "The most potently down-regulated was miR-378. Over ......" | 23617628 | |
| hsa-miR-378a-5p | prostate cancer | downregulation | "Thereafter RNA was polyadenylated and reverse tran ......" | 25153390 | qPCR |
| hsa-miR-378a-5p | prostate cancer | downregulation | "Here we evaluated the anti-proliferative role of m ......" | 26346167 |
Reported cancer pathway affected by hsa-miR-378a-5p
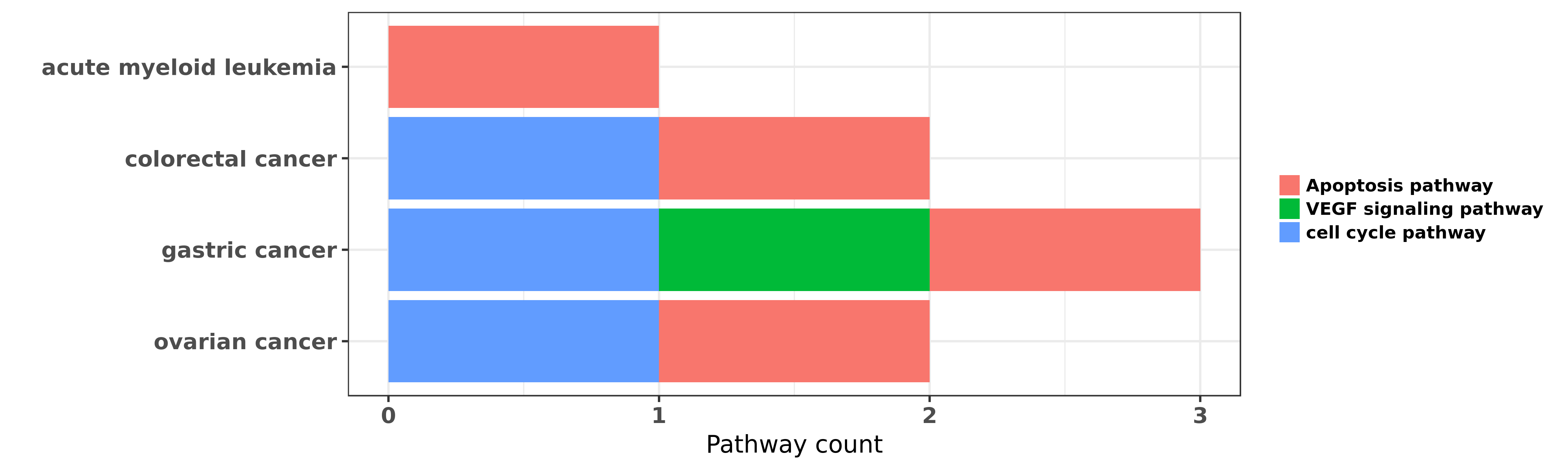
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-378a-5p | acute myeloid leukemia | Apoptosis pathway | "Overexpression of miR 378 is frequent and may affe ......" | 23582927 | |
| hsa-miR-378a-5p | colorectal cancer | cell cycle pathway; Apoptosis pathway | "Clinical and biological significance of miR 378a 3 ......" | 24412052 | Colony formation |
| hsa-miR-378a-5p | colorectal cancer | Apoptosis pathway | "MiR-378 is frequently downregulated in colorectal ......" | 25328987 | Luciferase |
| hsa-miR-378a-5p | gastric cancer | VEGF signaling pathway | "We found that microRNA-195 miR-195 and microRNA-37 ......" | 23333942 | |
| hsa-miR-378a-5p | gastric cancer | cell cycle pathway; Apoptosis pathway | "MiR 378 inhibits progression of human gastric canc ......" | 24139413 | |
| hsa-miR-378a-5p | ovarian cancer | cell cycle pathway; Apoptosis pathway | "MiR 378 as a biomarker for response to anti angiog ......" | 24680769 |
Reported cancer prognosis affected by hsa-miR-378a-5p
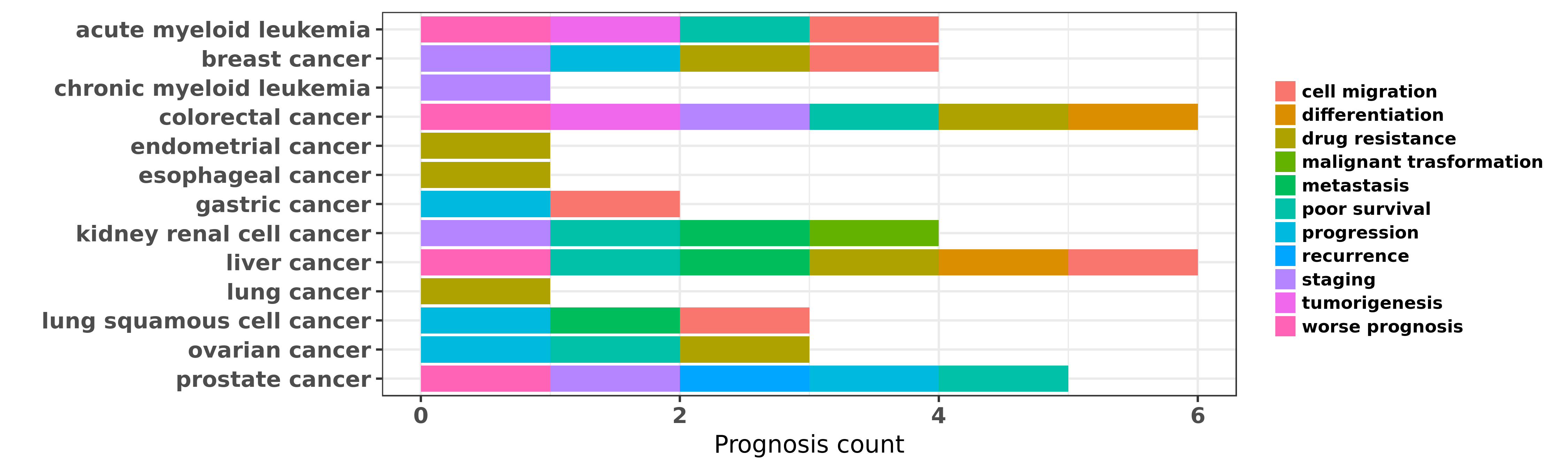
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-378a-5p | acute myeloid leukemia | tumorigenesis; poor survival; cell migration; worse prognosis | "Overexpression of miR 378 is frequent and may affe ......" | 23582927 | |
| hsa-miR-378a-5p | acute myeloid leukemia | poor survival | "The 5' flanking region of miR 378 is hypomethylate ......" | 26191124 | |
| hsa-miR-378a-5p | breast cancer | drug resistance | "Cell-based high-throughput screen identified miR-3 ......" | 25268374 | Western blot |
| hsa-miR-378a-5p | breast cancer | staging; progression; cell migration | "Microarray profiling of the MMTV-PyMT model reveal ......" | 26749280 | Luciferase |
| hsa-miR-378a-5p | chronic myeloid leukemia | staging | "Methylation of miR 378 in Chronic Myeloid Leukemia ......" | 26913395 | |
| hsa-miR-378a-5p | colorectal cancer | staging; differentiation; poor survival; tumorigenesis | "Clinical and biological significance of miR 378a 3 ......" | 24412052 | Colony formation |
| hsa-miR-378a-5p | colorectal cancer | poor survival; worse prognosis | "MiR 378 is an independent prognostic factor and in ......" | 24555885 | Luciferase |
| hsa-miR-378a-5p | colorectal cancer | worse prognosis; drug resistance | "MiR-378 is frequently downregulated in colorectal ......" | 25328987 | Luciferase |
| hsa-miR-378a-5p | colorectal cancer | drug resistance | "Previous studies have connected higher expression ......" | 26496897 | |
| hsa-miR-378a-5p | endometrial cancer | drug resistance | "Expression levels of ERα and PR and their respons ......" | 21472251 | |
| hsa-miR-378a-5p | esophageal cancer | drug resistance | "An in-vitro model of acquired chemotherapy resista ......" | 25356050 | |
| hsa-miR-378a-5p | gastric cancer | progression; cell migration | "MiR 378 inhibits progression of human gastric canc ......" | 24139413 | |
| hsa-miR-378a-5p | kidney renal cell cancer | malignant trasformation; staging; metastasis | "Analysis of serum microRNAs miR 26a 2* miR 191 miR ......" | 22542158 | |
| hsa-miR-378a-5p | kidney renal cell cancer | staging | "Markedly dysregulated miRNAs in RCC cases were sub ......" | 25556603 | |
| hsa-miR-378a-5p | kidney renal cell cancer | staging; poor survival | "Combination of MiR 378 and MiR 210 Serum Levels En ......" | 26426010 | |
| hsa-miR-378a-5p | liver cancer | worse prognosis; poor survival | "A genetic variant in primary miR 378 is associated ......" | 24751683 | |
| hsa-miR-378a-5p | liver cancer | differentiation; drug resistance; cell migration; metastasis | "MiR 378 promotes the migration of liver cancer cel ......" | 25562172 | |
| hsa-miR-378a-5p | lung cancer | drug resistance | "The objectives of the study were to determine the ......" | 26781643 | Luciferase |
| hsa-miR-378a-5p | lung squamous cell cancer | metastasis; cell migration | "In this study using RT-PCR and further northern bl ......" | 22052152 | |
| hsa-miR-378a-5p | lung squamous cell cancer | metastasis; progression | "Interplay between heme oxygenase 1 and miR 378 aff ......" | 23617628 | |
| hsa-miR-378a-5p | ovarian cancer | drug resistance; progression; poor survival | "MiR 378 as a biomarker for response to anti angiog ......" | 24680769 | |
| hsa-miR-378a-5p | prostate cancer | staging; worse prognosis; poor survival; recurrence; progression | "Loss of miR 378 in prostate cancer a common regula ......" | 25153390 |
Reported gene related to hsa-miR-378a-5p
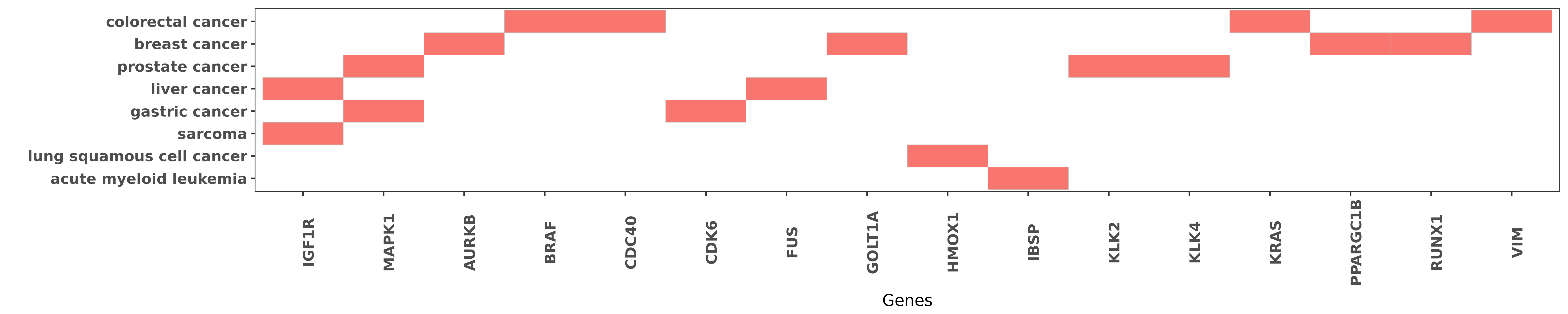
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-378a-5p | liver cancer | IGF1R | "The insulin-like growth factor 1 receptor IGF1R wa ......" | 24119742 |
| hsa-miR-378a-5p | sarcoma | IGF1R | "Interestingly members of the miR-378 family presen ......" | 25427715 |
| hsa-miR-378a-5p | gastric cancer | MAPK1 | "MiR 378 inhibits progression of human gastric canc ......" | 24139413 |
| hsa-miR-378a-5p | prostate cancer | MAPK1 | "MiR 378 suppresses prostate cancer cell growth thr ......" | 26346167 |
| hsa-miR-378a-5p | breast cancer | AURKB | "Moreover excess miR-378a-5p triggered receptor tyr ......" | 25268374 |
| hsa-miR-378a-5p | colorectal cancer | BRAF | "Previous studies have connected higher expression ......" | 26496897 |
| hsa-miR-378a-5p | colorectal cancer | CDC40 | "Bioinformatics analysis further deduced that CDC40 ......" | 25328987 |
| hsa-miR-378a-5p | gastric cancer | CDK6 | "Expression of cyclin-dependent kinase 6 and vascul ......" | 23333942 |
| hsa-miR-378a-5p | liver cancer | FUS | "MiR 378 promotes the migration of liver cancer cel ......" | 25562172 |
| hsa-miR-378a-5p | breast cancer | GOLT1A | "miR 378a 3p modulates tamoxifen sensitivity in bre ......" | 26255816 |
| hsa-miR-378a-5p | lung squamous cell cancer | HMOX1 | "Interplay between heme oxygenase 1 and miR 378 aff ......" | 23617628 |
| hsa-miR-378a-5p | acute myeloid leukemia | IBSP | "Methylation status of miR-378 5'-flanking region w ......" | 26191124 |
| hsa-miR-378a-5p | prostate cancer | KLK2 | "Loss of miR 378 in prostate cancer a common regula ......" | 25153390 |
| hsa-miR-378a-5p | prostate cancer | KLK4 | "Loss of miR 378 in prostate cancer a common regula ......" | 25153390 |
| hsa-miR-378a-5p | colorectal cancer | KRAS | "Previous studies have connected higher expression ......" | 26496897 |
| hsa-miR-378a-5p | breast cancer | PPARGC1B | "Depletion of Runx1 in late-stage breast cancer cel ......" | 26749280 |
| hsa-miR-378a-5p | breast cancer | RUNX1 | "One of these miR-378 was inversely correlated with ......" | 26749280 |
| hsa-miR-378a-5p | colorectal cancer | VIM | "Luciferase assay was performed to assess miR-378 b ......" | 24555885 |
Expression profile in cancer corhorts:
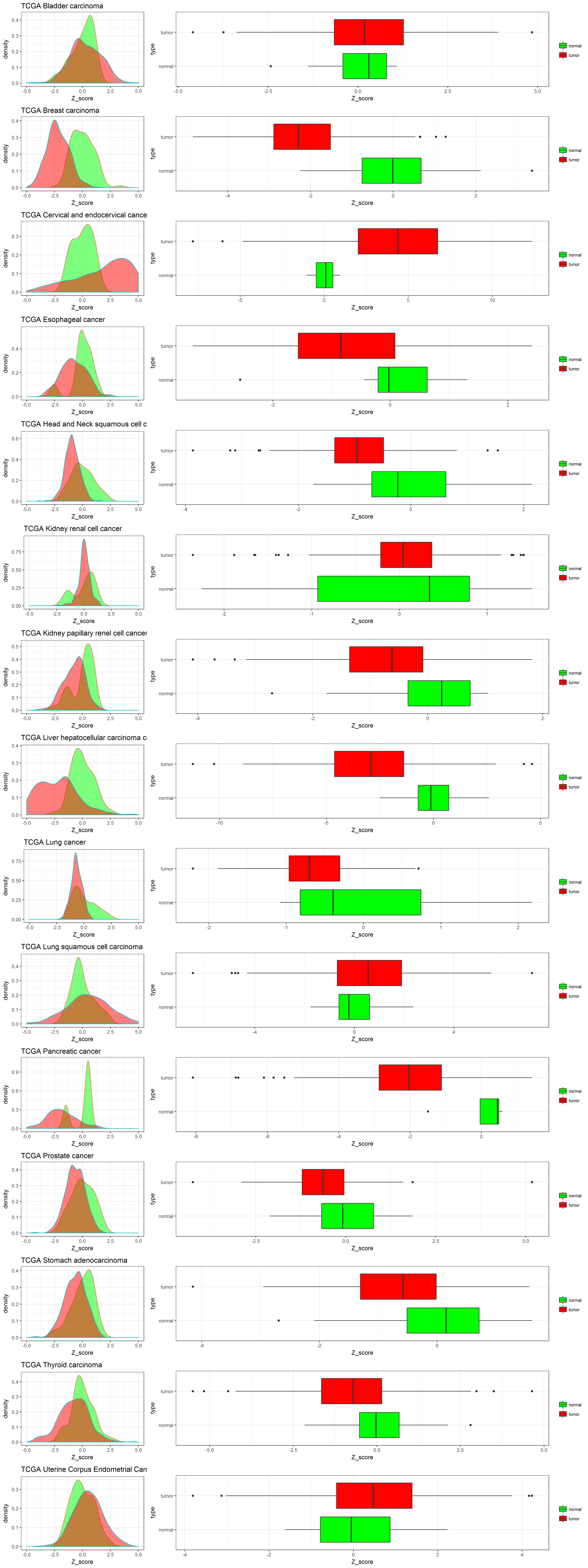
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-378a-5p | IL13RA1 | 11 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRP; LUSC; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.163; TCGA BRCA -0.068; TCGA CESC -0.118; TCGA ESCA -0.118; TCGA HNSC -0.082; TCGA KIRP -0.086; TCGA LUSC -0.134; TCGA PRAD -0.074; TCGA THCA -0.094; TCGA STAD -0.112; TCGA UCEC -0.065 |
hsa-miR-378a-5p | ARHGAP29 | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUSC; PRAD; STAD | MirTarget | TCGA BLCA -0.231; TCGA CESC -0.563; TCGA COAD -0.285; TCGA ESCA -0.266; TCGA HNSC -0.198; TCGA KIRC -0.227; TCGA KIRP -0.355; TCGA LUSC -0.381; TCGA PRAD -0.117; TCGA STAD -0.266 |
hsa-miR-378a-5p | ANKH | 9 cancers: BLCA; CESC; ESCA; KIRC; LUSC; PAAD; PRAD; THCA; STAD | MirTarget | TCGA BLCA -0.443; TCGA CESC -0.19; TCGA ESCA -0.228; TCGA KIRC -0.057; TCGA LUSC -0.274; TCGA PAAD -0.225; TCGA PRAD -0.094; TCGA THCA -0.112; TCGA STAD -0.301 |
hsa-miR-378a-5p | CSRP1 | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LUSC; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.334; TCGA CESC -0.232; TCGA COAD -0.273; TCGA ESCA -0.249; TCGA HNSC -0.066; TCGA KIRC -0.097; TCGA LUSC -0.215; TCGA SARC -0.475; TCGA STAD -0.324; TCGA UCEC -0.151 |
hsa-miR-378a-5p | NUAK1 | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; LUSC; STAD; UCEC | MirTarget | TCGA BLCA -0.435; TCGA CESC -0.264; TCGA COAD -0.392; TCGA ESCA -0.258; TCGA HNSC -0.224; TCGA KIRC -0.182; TCGA LIHC -0.165; TCGA LUSC -0.359; TCGA STAD -0.301; TCGA UCEC -0.197 |
hsa-miR-378a-5p | ATP2B4 | 9 cancers: BLCA; COAD; ESCA; KIRP; LIHC; LUSC; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.297; TCGA COAD -0.387; TCGA ESCA -0.248; TCGA KIRP -0.145; TCGA LIHC -0.141; TCGA LUSC -0.094; TCGA SARC -0.173; TCGA STAD -0.313; TCGA UCEC -0.081 |
hsa-miR-378a-5p | LMO3 | 9 cancers: BLCA; COAD; ESCA; KIRC; LUAD; LUSC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.326; TCGA COAD -1.383; TCGA ESCA -0.851; TCGA KIRC -0.281; TCGA LUAD -0.202; TCGA LUSC -0.367; TCGA THCA -0.24; TCGA STAD -0.71; TCGA UCEC -0.258 |
hsa-miR-378a-5p | CACNA1C | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; LUSC; OV; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.438; TCGA CESC -0.363; TCGA COAD -0.399; TCGA ESCA -0.527; TCGA HNSC -0.309; TCGA KIRC -0.121; TCGA LIHC -0.186; TCGA LUSC -0.513; TCGA OV -0.246; TCGA SARC -0.337; TCGA STAD -0.569; TCGA UCEC -0.438 |
hsa-miR-378a-5p | VAMP4 | 14 cancers: BLCA; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; OV; PAAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.069; TCGA COAD -0.147; TCGA ESCA -0.203; TCGA HNSC -0.062; TCGA KIRC -0.055; TCGA KIRP -0.114; TCGA LIHC -0.063; TCGA LUAD -0.061; TCGA LUSC -0.097; TCGA OV -0.126; TCGA PAAD -0.135; TCGA THCA -0.063; TCGA STAD -0.131; TCGA UCEC -0.163 |
hsa-miR-378a-5p | BMPR2 | 9 cancers: BLCA; CESC; COAD; ESCA; KIRP; LUSC; OV; STAD; UCEC | MirTarget | TCGA BLCA -0.106; TCGA CESC -0.089; TCGA COAD -0.25; TCGA ESCA -0.157; TCGA KIRP -0.068; TCGA LUSC -0.134; TCGA OV -0.084; TCGA STAD -0.212; TCGA UCEC -0.152 |
hsa-miR-378a-5p | PTGFR | 9 cancers: BLCA; CESC; COAD; ESCA; KIRC; KIRP; LUSC; STAD; UCEC | MirTarget | TCGA BLCA -0.74; TCGA CESC -0.407; TCGA COAD -0.606; TCGA ESCA -0.544; TCGA KIRC -0.267; TCGA KIRP -0.294; TCGA LUSC -0.246; TCGA STAD -0.493; TCGA UCEC -0.55 |
hsa-miR-378a-5p | ITGB5 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LGG; LIHC; LUSC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.34; TCGA BRCA -0.137; TCGA CESC -0.118; TCGA COAD -0.168; TCGA ESCA -0.246; TCGA HNSC -0.223; TCGA KIRC -0.07; TCGA LGG -0.091; TCGA LIHC -0.122; TCGA LUSC -0.13; TCGA THCA -0.098; TCGA STAD -0.257; TCGA UCEC -0.101 |
hsa-miR-378a-5p | STXBP1 | 9 cancers: BLCA; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; STAD; UCEC | MirTarget | TCGA BLCA -0.403; TCGA COAD -0.275; TCGA ESCA -0.349; TCGA HNSC -0.229; TCGA LUAD -0.153; TCGA LUSC -0.14; TCGA PAAD -0.306; TCGA STAD -0.441; TCGA UCEC -0.107 |
hsa-miR-378a-5p | ZBTB47 | 11 cancers: BLCA; CESC; COAD; ESCA; KIRC; LGG; LUSC; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.139; TCGA CESC -0.195; TCGA COAD -0.278; TCGA ESCA -0.233; TCGA KIRC -0.129; TCGA LGG -0.101; TCGA LUSC -0.211; TCGA SARC -0.088; TCGA THCA -0.052; TCGA STAD -0.31; TCGA UCEC -0.085 |
hsa-miR-378a-5p | IGF1R | 14 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRP; LIHC; LUAD; OV; PAAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.128; TCGA BRCA -0.366; TCGA CESC -0.174; TCGA COAD -0.175; TCGA HNSC -0.093; TCGA KIRP -0.089; TCGA LIHC -0.265; TCGA LUAD -0.111; TCGA OV -0.187; TCGA PAAD -0.263; TCGA SARC -0.168; TCGA THCA -0.198; TCGA STAD -0.26; TCGA UCEC -0.093 |
hsa-miR-378a-5p | DLG2 | 9 cancers: BLCA; CESC; COAD; ESCA; LUSC; OV; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.217; TCGA CESC -0.289; TCGA COAD -0.759; TCGA ESCA -0.648; TCGA LUSC -0.496; TCGA OV -0.242; TCGA SARC -0.285; TCGA STAD -0.638; TCGA UCEC -0.462 |
hsa-miR-378a-5p | NMNAT2 | 9 cancers: BLCA; CESC; COAD; HNSC; LIHC; LUSC; PAAD; STAD; UCEC | mirMAP | TCGA BLCA -0.612; TCGA CESC -0.325; TCGA COAD -0.453; TCGA HNSC -0.163; TCGA LIHC -0.14; TCGA LUSC -0.603; TCGA PAAD -0.377; TCGA STAD -0.533; TCGA UCEC -0.284 |
hsa-miR-378a-5p | ELFN2 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LIHC; LUSC; PAAD; PRAD; THCA; STAD | mirMAP | TCGA BLCA -0.307; TCGA BRCA -0.282; TCGA CESC -0.289; TCGA COAD -0.54; TCGA ESCA -0.579; TCGA HNSC -0.549; TCGA KIRP -0.263; TCGA LIHC -0.673; TCGA LUSC -0.3; TCGA PAAD -0.547; TCGA PRAD -0.347; TCGA THCA -0.484; TCGA STAD -0.383 |
hsa-miR-378a-5p | LRP10 | 10 cancers: BLCA; BRCA; CESC; KIRC; LGG; LIHC; LUSC; SARC; THCA; UCEC | mirMAP | TCGA BLCA -0.103; TCGA BRCA -0.11; TCGA CESC -0.067; TCGA KIRC -0.063; TCGA LGG -0.091; TCGA LIHC -0.073; TCGA LUSC -0.087; TCGA SARC -0.123; TCGA THCA -0.073; TCGA UCEC -0.074 |
hsa-miR-378a-5p | ADARB1 | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUSC; STAD | mirMAP | TCGA BLCA -0.318; TCGA CESC -0.182; TCGA COAD -0.153; TCGA ESCA -0.258; TCGA HNSC -0.208; TCGA KIRC -0.081; TCGA KIRP -0.17; TCGA LUSC -0.389; TCGA STAD -0.282 |
hsa-miR-378a-5p | IPO9 | 11 cancers: BLCA; BRCA; KIRC; LIHC; LUAD; OV; PAAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.088; TCGA BRCA -0.066; TCGA KIRC -0.055; TCGA LIHC -0.183; TCGA LUAD -0.113; TCGA OV -0.053; TCGA PAAD -0.07; TCGA SARC -0.056; TCGA THCA -0.07; TCGA STAD -0.074; TCGA UCEC -0.068 |
hsa-miR-378a-5p | PLXNA4 | 11 cancers: BLCA; CESC; COAD; ESCA; KIRC; LUSC; OV; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.789; TCGA CESC -0.424; TCGA COAD -0.831; TCGA ESCA -0.999; TCGA KIRC -0.199; TCGA LUSC -0.491; TCGA OV -0.375; TCGA SARC -0.493; TCGA THCA -0.23; TCGA STAD -0.741; TCGA UCEC -0.25 |
hsa-miR-378a-5p | DTNA | 10 cancers: BLCA; COAD; ESCA; KIRP; LGG; LUSC; OV; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.513; TCGA COAD -0.976; TCGA ESCA -0.406; TCGA KIRP -0.129; TCGA LGG -0.114; TCGA LUSC -0.269; TCGA OV -0.287; TCGA SARC -0.29; TCGA STAD -0.678; TCGA UCEC -0.155 |
hsa-miR-378a-5p | FANCF | 9 cancers: BRCA; HNSC; KIRP; LIHC; LUAD; OV; PAAD; PRAD; THCA | MirTarget | TCGA BRCA -0.236; TCGA HNSC -0.096; TCGA KIRP -0.068; TCGA LIHC -0.134; TCGA LUAD -0.126; TCGA OV -0.081; TCGA PAAD -0.09; TCGA PRAD -0.142; TCGA THCA -0.062 |
hsa-miR-378a-5p | ZNF441 | 10 cancers: BRCA; COAD; ESCA; KIRC; KIRP; LUSC; PAAD; SARC; THCA; STAD | MirTarget | TCGA BRCA -0.07; TCGA COAD -0.085; TCGA ESCA -0.174; TCGA KIRC -0.141; TCGA KIRP -0.211; TCGA LUSC -0.165; TCGA PAAD -0.152; TCGA SARC -0.154; TCGA THCA -0.13; TCGA STAD -0.153 |
hsa-miR-378a-5p | PBX1 | 10 cancers: BRCA; COAD; ESCA; LGG; LUSC; OV; SARC; THCA; STAD; UCEC | MirTarget | TCGA BRCA -0.311; TCGA COAD -0.37; TCGA ESCA -0.251; TCGA LGG -0.081; TCGA LUSC -0.143; TCGA OV -0.092; TCGA SARC -0.221; TCGA THCA -0.106; TCGA STAD -0.383; TCGA UCEC -0.15 |
hsa-miR-378a-5p | SRCIN1 | 11 cancers: BRCA; KIRC; KIRP; LIHC; LUAD; LUSC; OV; PAAD; PRAD; THCA; STAD | mirMAP | TCGA BRCA -0.489; TCGA KIRC -0.33; TCGA KIRP -0.451; TCGA LIHC -0.167; TCGA LUAD -0.311; TCGA LUSC -0.257; TCGA OV -0.271; TCGA PAAD -0.605; TCGA PRAD -0.278; TCGA THCA -0.421; TCGA STAD -0.256 |
hsa-miR-378a-5p | RAB11FIP3 | 9 cancers: BRCA; ESCA; OV; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BRCA -0.312; TCGA ESCA -0.14; TCGA OV -0.069; TCGA PAAD -0.113; TCGA PRAD -0.07; TCGA SARC -0.074; TCGA THCA -0.119; TCGA STAD -0.117; TCGA UCEC -0.062 |
hsa-miR-378a-5p | BMF | 14 cancers: BRCA; COAD; HNSC; KIRC; LGG; LIHC; LUAD; LUSC; OV; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BRCA -0.115; TCGA COAD -0.177; TCGA HNSC -0.135; TCGA KIRC -0.131; TCGA LGG -0.166; TCGA LIHC -0.395; TCGA LUAD -0.132; TCGA LUSC -0.308; TCGA OV -0.177; TCGA PRAD -0.173; TCGA SARC -0.151; TCGA THCA -0.206; TCGA STAD -0.189; TCGA UCEC -0.162 |
hsa-miR-378a-5p | HDAC11 | 9 cancers: BRCA; KIRC; KIRP; LIHC; LUSC; PAAD; SARC; THCA; STAD | mirMAP | TCGA BRCA -0.363; TCGA KIRC -0.119; TCGA KIRP -0.086; TCGA LIHC -0.258; TCGA LUSC -0.108; TCGA PAAD -0.29; TCGA SARC -0.164; TCGA THCA -0.124; TCGA STAD -0.113 |
hsa-miR-378a-5p | KSR2 | 9 cancers: BRCA; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; THCA | mirMAP | TCGA BRCA -0.385; TCGA KIRC -0.258; TCGA KIRP -0.279; TCGA LIHC -0.169; TCGA LUAD -0.253; TCGA LUSC -0.331; TCGA PAAD -0.521; TCGA PRAD -0.218; TCGA THCA -0.106 |
hsa-miR-378a-5p | ZNF449 | 9 cancers: CESC; COAD; KIRC; KIRP; LIHC; LUAD; LUSC; STAD; UCEC | MirTarget | TCGA CESC -0.094; TCGA COAD -0.118; TCGA KIRC -0.077; TCGA KIRP -0.134; TCGA LIHC -0.125; TCGA LUAD -0.054; TCGA LUSC -0.068; TCGA STAD -0.222; TCGA UCEC -0.051 |
hsa-miR-378a-5p | VGLL3 | 9 cancers: CESC; COAD; ESCA; HNSC; KIRC; LUSC; SARC; STAD; UCEC | MirTarget | TCGA CESC -0.63; TCGA COAD -0.714; TCGA ESCA -0.692; TCGA HNSC -0.296; TCGA KIRC -0.221; TCGA LUSC -0.593; TCGA SARC -0.422; TCGA STAD -0.464; TCGA UCEC -0.353 |
hsa-miR-378a-5p | FOXP4 | 9 cancers: CESC; KIRP; LIHC; LUAD; LUSC; OV; PAAD; THCA; UCEC | mirMAP | TCGA CESC -0.077; TCGA KIRP -0.074; TCGA LIHC -0.116; TCGA LUAD -0.146; TCGA LUSC -0.087; TCGA OV -0.222; TCGA PAAD -0.285; TCGA THCA -0.208; TCGA UCEC -0.058 |
hsa-miR-378a-5p | ZC3H6 | 10 cancers: COAD; ESCA; KIRC; KIRP; LUAD; LUSC; SARC; THCA; STAD; UCEC | MirTarget | TCGA COAD -0.134; TCGA ESCA -0.206; TCGA KIRC -0.098; TCGA KIRP -0.187; TCGA LUAD -0.091; TCGA LUSC -0.086; TCGA SARC -0.073; TCGA THCA -0.084; TCGA STAD -0.182; TCGA UCEC -0.12 |
hsa-miR-378a-5p | PHF2 | 10 cancers: COAD; KIRC; KIRP; LGG; OV; PAAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA COAD -0.073; TCGA KIRC -0.088; TCGA KIRP -0.081; TCGA LGG -0.075; TCGA OV -0.062; TCGA PAAD -0.106; TCGA SARC -0.075; TCGA THCA -0.112; TCGA STAD -0.145; TCGA UCEC -0.126 |
Enriched cancer pathways of putative targets