microRNA information: hsa-miR-378c
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-378c | miRbase |
| Accession: | MIMAT0016847 | miRbase |
| Precursor name: | hsa-mir-378c | miRbase |
| Precursor accession: | MI0015825 | miRbase |
| Symbol: | MIR378C | HGNC |
| RefSeq ID: | NR_036180 | GenBank |
| Sequence: | ACUGGACUUGGAGUCAGAAGAGUGG |
Reported expression in cancers: hsa-miR-378c
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|
Reported cancer pathway affected by hsa-miR-378c
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|
Reported cancer prognosis affected by hsa-miR-378c
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|
Reported gene related to hsa-miR-378c
| miRNA | cancer | gene | reporting | PUBMED |
|---|
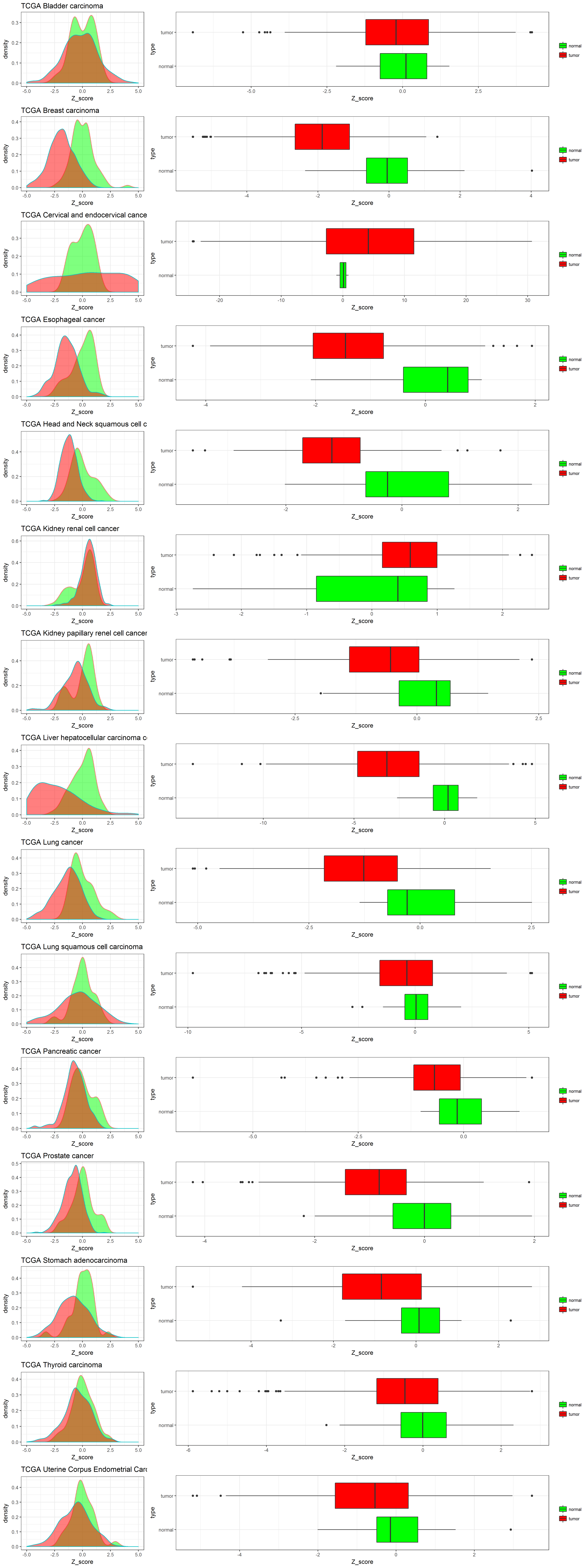
Expression profile in cancer corhorts:
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-378c | SULF1 | 13 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.759; TCGA BRCA -0.295; TCGA CESC -0.387; TCGA ESCA -0.756; TCGA HNSC -0.147; TCGA KIRP -0.421; TCGA LGG -0.28; TCGA LIHC -0.233; TCGA LUAD -0.183; TCGA LUSC -0.345; TCGA SARC -0.222; TCGA STAD -0.559; TCGA UCEC -0.275 |
hsa-miR-378c | UHRF1 | 11 cancers: BLCA; BRCA; ESCA; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; PRAD; UCEC | MirTarget | TCGA BLCA -0.148; TCGA BRCA -0.393; TCGA ESCA -0.378; TCGA HNSC -0.081; TCGA KIRP -0.108; TCGA LGG -0.16; TCGA LIHC -0.491; TCGA LUAD -0.178; TCGA LUSC -0.182; TCGA PRAD -0.417; TCGA UCEC -0.15 |
hsa-miR-378c | CNTD2 | 9 cancers: BLCA; BRCA; HNSC; KIRC; KIRP; LUAD; LUSC; PRAD; THCA | MirTarget | TCGA BLCA -0.189; TCGA BRCA -0.553; TCGA HNSC -0.485; TCGA KIRC -0.739; TCGA KIRP -0.395; TCGA LUAD -0.743; TCGA LUSC -0.343; TCGA PRAD -0.21; TCGA THCA -0.169 |
hsa-miR-378c | SLC2A1 | 9 cancers: BLCA; BRCA; ESCA; HNSC; LIHC; LUSC; SARC; THCA; STAD | MirTarget; miRNATAP | TCGA BLCA -0.149; TCGA BRCA -0.171; TCGA ESCA -0.794; TCGA HNSC -0.358; TCGA LIHC -0.226; TCGA LUSC -0.266; TCGA SARC -0.187; TCGA THCA -0.068; TCGA STAD -0.226 |
hsa-miR-378c | TMEM130 | 9 cancers: BLCA; BRCA; ESCA; KIRC; KIRP; LIHC; PAAD; PRAD; STAD | MirTarget | TCGA BLCA -0.361; TCGA BRCA -0.129; TCGA ESCA -0.443; TCGA KIRC -0.498; TCGA KIRP -0.788; TCGA LIHC -0.395; TCGA PAAD -0.45; TCGA PRAD -0.427; TCGA STAD -0.366 |
hsa-miR-378c | EFR3B | 9 cancers: BLCA; BRCA; CESC; KIRC; LIHC; LUAD; PAAD; STAD; UCEC | mirMAP | TCGA BLCA -0.176; TCGA BRCA -0.271; TCGA CESC -0.3; TCGA KIRC -0.265; TCGA LIHC -0.151; TCGA LUAD -0.187; TCGA PAAD -0.456; TCGA STAD -0.241; TCGA UCEC -0.212 |
hsa-miR-378c | ABL2 | 9 cancers: BLCA; CESC; ESCA; HNSC; KIRP; LIHC; LUSC; STAD; UCEC | mirMAP | TCGA BLCA -0.123; TCGA CESC -0.19; TCGA ESCA -0.248; TCGA HNSC -0.132; TCGA KIRP -0.068; TCGA LIHC -0.212; TCGA LUSC -0.084; TCGA STAD -0.187; TCGA UCEC -0.107 |
hsa-miR-378c | CBX2 | 11 cancers: BLCA; ESCA; HNSC; KIRC; LGG; LIHC; LUAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.273; TCGA ESCA -0.522; TCGA HNSC -0.144; TCGA KIRC -0.231; TCGA LGG -0.19; TCGA LIHC -0.311; TCGA LUAD -0.353; TCGA PRAD -0.428; TCGA SARC -0.14; TCGA STAD -0.389; TCGA UCEC -0.13 |
hsa-miR-378c | PLEKHG2 | 10 cancers: BLCA; CESC; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; STAD; UCEC | miRNATAP | TCGA BLCA -0.19; TCGA CESC -0.166; TCGA HNSC -0.093; TCGA KIRP -0.13; TCGA LGG -0.066; TCGA LIHC -0.318; TCGA LUAD -0.299; TCGA LUSC -0.133; TCGA STAD -0.272; TCGA UCEC -0.116 |
hsa-miR-378c | SPEG | 11 cancers: BLCA; CESC; ESCA; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; SARC; STAD | miRNATAP | TCGA BLCA -0.425; TCGA CESC -0.418; TCGA ESCA -0.791; TCGA KIRC -0.196; TCGA KIRP -0.342; TCGA LIHC -0.263; TCGA LUAD -0.39; TCGA LUSC -0.329; TCGA PAAD -0.28; TCGA SARC -0.453; TCGA STAD -0.716 |
hsa-miR-378c | IPO9 | 9 cancers: BLCA; BRCA; ESCA; LIHC; LUAD; LUSC; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.104; TCGA BRCA -0.086; TCGA ESCA -0.135; TCGA LIHC -0.181; TCGA LUAD -0.064; TCGA LUSC -0.065; TCGA SARC -0.077; TCGA STAD -0.107; TCGA UCEC -0.117 |
hsa-miR-378c | ELFN2 | 9 cancers: BLCA; BRCA; CESC; KIRP; LIHC; PAAD; PRAD; THCA; STAD | miRNATAP | TCGA BLCA -0.234; TCGA BRCA -0.234; TCGA CESC -0.309; TCGA KIRP -0.193; TCGA LIHC -0.577; TCGA PAAD -0.395; TCGA PRAD -0.493; TCGA THCA -0.378; TCGA STAD -0.381 |
hsa-miR-378c | CELSR3 | 9 cancers: BRCA; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; UCEC | MirTarget | TCGA BRCA -0.293; TCGA HNSC -0.125; TCGA KIRP -0.211; TCGA LIHC -0.162; TCGA LUAD -0.406; TCGA LUSC -0.211; TCGA PAAD -0.37; TCGA PRAD -0.575; TCGA UCEC -0.108 |
hsa-miR-378c | ZNF260 | 10 cancers: BRCA; CESC; ESCA; KIRC; LGG; LIHC; LUAD; PRAD; STAD; UCEC | MirTarget | TCGA BRCA -0.068; TCGA CESC -0.072; TCGA ESCA -0.174; TCGA KIRC -0.059; TCGA LGG -0.097; TCGA LIHC -0.125; TCGA LUAD -0.09; TCGA PRAD -0.115; TCGA STAD -0.137; TCGA UCEC -0.067 |
hsa-miR-378c | QSOX1 | 9 cancers: BRCA; CESC; HNSC; KIRC; KIRP; LIHC; SARC; THCA; UCEC | mirMAP | TCGA BRCA -0.215; TCGA CESC -0.102; TCGA HNSC -0.11; TCGA KIRC -0.137; TCGA KIRP -0.088; TCGA LIHC -0.437; TCGA SARC -0.109; TCGA THCA -0.128; TCGA UCEC -0.13 |
hsa-miR-378c | BRD3 | 9 cancers: BRCA; ESCA; KIRC; KIRP; LGG; LIHC; SARC; STAD; UCEC | miRNATAP | TCGA BRCA -0.096; TCGA ESCA -0.105; TCGA KIRC -0.079; TCGA KIRP -0.117; TCGA LGG -0.107; TCGA LIHC -0.124; TCGA SARC -0.078; TCGA STAD -0.147; TCGA UCEC -0.079 |
hsa-miR-378c | RIT1 | 9 cancers: BRCA; CESC; ESCA; HNSC; LGG; LIHC; LUAD; SARC; UCEC | miRNATAP | TCGA BRCA -0.06; TCGA CESC -0.089; TCGA ESCA -0.18; TCGA HNSC -0.072; TCGA LGG -0.072; TCGA LIHC -0.195; TCGA LUAD -0.109; TCGA SARC -0.078; TCGA UCEC -0.1 |
hsa-miR-378c | GRIK2 | 11 cancers: BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; STAD; UCEC | miRNATAP | TCGA BRCA -0.18; TCGA CESC -0.323; TCGA ESCA -0.6; TCGA HNSC -0.213; TCGA KIRC -0.343; TCGA KIRP -0.353; TCGA LUAD -0.25; TCGA LUSC -0.802; TCGA PAAD -0.648; TCGA STAD -0.418; TCGA UCEC -0.208 |
hsa-miR-378c | KCND1 | 9 cancers: ESCA; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; SARC; STAD | miRNATAP | TCGA ESCA -0.195; TCGA KIRC -0.28; TCGA KIRP -0.113; TCGA LIHC -0.099; TCGA LUAD -0.13; TCGA LUSC -0.106; TCGA PAAD -0.17; TCGA SARC -0.149; TCGA STAD -0.293 |
Enriched cancer pathways of putative targets