microRNA information: hsa-miR-3913-5p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-3913-5p | miRbase |
| Accession: | MIMAT0018187 | miRbase |
| Precursor name: | hsa-mir-3913-1 | miRbase |
| Precursor accession: | MI0016417 | miRbase |
| Symbol: | MIR3913-1 | HGNC |
| RefSeq ID: | NR_037475 | GenBank |
| Sequence: | UUUGGGACUGAUCUUGAUGUCU |
Reported expression in cancers: hsa-miR-3913-5p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|
Reported cancer pathway affected by hsa-miR-3913-5p
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|
Reported cancer prognosis affected by hsa-miR-3913-5p
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|
Reported gene related to hsa-miR-3913-5p
| miRNA | cancer | gene | reporting | PUBMED |
|---|
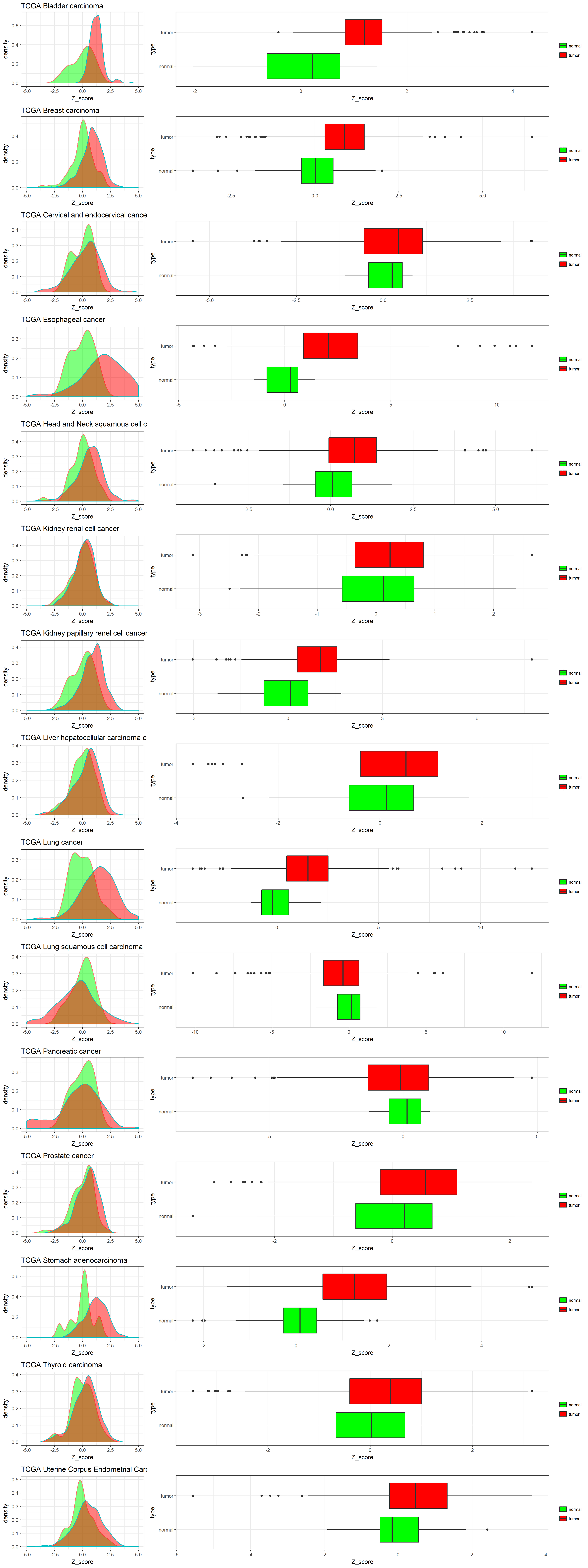
Expression profile in cancer corhorts:
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-3913-5p | PJA2 | 9 cancers: BLCA; BRCA; ESCA; KIRC; KIRP; LIHC; PRAD; SARC; STAD | MirTarget; miRNATAP | TCGA BLCA -0.25; TCGA BRCA -0.175; TCGA ESCA -0.109; TCGA KIRC -0.099; TCGA KIRP -0.177; TCGA LIHC -0.08; TCGA PRAD -0.143; TCGA SARC -0.057; TCGA STAD -0.196 |
hsa-miR-3913-5p | DUSP3 | 13 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LIHC; LUAD; LUSC; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.236; TCGA BRCA -0.124; TCGA CESC -0.111; TCGA ESCA -0.155; TCGA HNSC -0.136; TCGA LIHC -0.143; TCGA LUAD -0.113; TCGA LUSC -0.118; TCGA PRAD -0.079; TCGA SARC -0.187; TCGA THCA -0.06; TCGA STAD -0.234; TCGA UCEC -0.119 |
hsa-miR-3913-5p | GBP1 | 9 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LIHC; SARC; THCA | MirTarget | TCGA BLCA -0.404; TCGA BRCA -0.168; TCGA CESC -0.368; TCGA ESCA -0.215; TCGA HNSC -0.243; TCGA KIRC -0.176; TCGA LIHC -0.192; TCGA SARC -0.255; TCGA THCA -0.26 |
hsa-miR-3913-5p | RABGAP1L | 9 cancers: BLCA; BRCA; ESCA; KIRP; LUSC; PRAD; SARC; THCA; UCEC | MirTarget | TCGA BLCA -0.127; TCGA BRCA -0.14; TCGA ESCA -0.116; TCGA KIRP -0.201; TCGA LUSC -0.079; TCGA PRAD -0.111; TCGA SARC -0.089; TCGA THCA -0.309; TCGA UCEC -0.15 |
hsa-miR-3913-5p | ARRDC4 | 12 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PRAD; THCA; STAD | MirTarget | TCGA BLCA -0.105; TCGA BRCA -0.186; TCGA CESC -0.228; TCGA ESCA -0.223; TCGA HNSC -0.205; TCGA KIRC -0.153; TCGA KIRP -0.174; TCGA LUAD -0.113; TCGA LUSC -0.129; TCGA PRAD -0.064; TCGA THCA -0.148; TCGA STAD -0.259 |
hsa-miR-3913-5p | NCOA4 | 12 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LGG; LIHC; LUSC; PAAD; PRAD; STAD | MirTarget | TCGA BLCA -0.081; TCGA BRCA -0.143; TCGA CESC -0.127; TCGA ESCA -0.097; TCGA HNSC -0.111; TCGA KIRC -0.107; TCGA LGG -0.077; TCGA LIHC -0.151; TCGA LUSC -0.081; TCGA PAAD -0.098; TCGA PRAD -0.106; TCGA STAD -0.149 |
hsa-miR-3913-5p | WWTR1 | 10 cancers: BLCA; BRCA; HNSC; KIRC; KIRP; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.411; TCGA BRCA -0.204; TCGA HNSC -0.097; TCGA KIRC -0.142; TCGA KIRP -0.188; TCGA PRAD -0.143; TCGA SARC -0.077; TCGA THCA -0.253; TCGA STAD -0.159; TCGA UCEC -0.173 |
hsa-miR-3913-5p | KIF1B | 10 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LIHC; PAAD; PRAD; SARC | mirMAP | TCGA BLCA -0.118; TCGA BRCA -0.207; TCGA CESC -0.18; TCGA HNSC -0.15; TCGA KIRC -0.102; TCGA KIRP -0.122; TCGA LIHC -0.191; TCGA PAAD -0.124; TCGA PRAD -0.064; TCGA SARC -0.051 |
hsa-miR-3913-5p | TMCC3 | 9 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LUSC; STAD | mirMAP | TCGA BLCA -0.431; TCGA BRCA -0.31; TCGA CESC -0.487; TCGA ESCA -0.422; TCGA HNSC -0.27; TCGA KIRC -0.144; TCGA KIRP -0.184; TCGA LUSC -0.204; TCGA STAD -0.244 |
hsa-miR-3913-5p | GNA11 | 9 cancers: BLCA; BRCA; CESC; ESCA; KIRC; KIRP; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.142; TCGA BRCA -0.085; TCGA CESC -0.194; TCGA ESCA -0.108; TCGA KIRC -0.118; TCGA KIRP -0.309; TCGA SARC -0.153; TCGA STAD -0.17; TCGA UCEC -0.157 |
hsa-miR-3913-5p | MAPK1 | 11 cancers: BLCA; BRCA; CESC; ESCA; KIRC; KIRP; LGG; LUAD; LUSC; PAAD; PRAD | mirMAP | TCGA BLCA -0.086; TCGA BRCA -0.146; TCGA CESC -0.108; TCGA ESCA -0.077; TCGA KIRC -0.09; TCGA KIRP -0.113; TCGA LGG -0.062; TCGA LUAD -0.093; TCGA LUSC -0.083; TCGA PAAD -0.086; TCGA PRAD -0.072 |
hsa-miR-3913-5p | CBX7 | 12 cancers: BLCA; BRCA; ESCA; KIRC; KIRP; LGG; LIHC; LUAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.173; TCGA BRCA -0.13; TCGA ESCA -0.182; TCGA KIRC -0.102; TCGA KIRP -0.426; TCGA LGG -0.164; TCGA LIHC -0.244; TCGA LUAD -0.113; TCGA PRAD -0.103; TCGA SARC -0.309; TCGA STAD -0.452; TCGA UCEC -0.18 |
hsa-miR-3913-5p | TRIP11 | 9 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LUSC; PRAD; STAD | mirMAP | TCGA BLCA -0.089; TCGA BRCA -0.17; TCGA CESC -0.14; TCGA ESCA -0.118; TCGA HNSC -0.108; TCGA KIRC -0.117; TCGA LUSC -0.087; TCGA PRAD -0.113; TCGA STAD -0.061 |
hsa-miR-3913-5p | HLF | 9 cancers: BLCA; BRCA; KIRC; LGG; LIHC; LUAD; PRAD; THCA; STAD | mirMAP | TCGA BLCA -0.577; TCGA BRCA -0.228; TCGA KIRC -0.265; TCGA LGG -0.257; TCGA LIHC -0.453; TCGA LUAD -0.268; TCGA PRAD -0.17; TCGA THCA -0.987; TCGA STAD -0.651 |
hsa-miR-3913-5p | KCTD10 | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LUAD; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.13; TCGA BRCA -0.098; TCGA CESC -0.128; TCGA ESCA -0.133; TCGA HNSC -0.083; TCGA LUAD -0.068; TCGA PRAD -0.08; TCGA THCA -0.125; TCGA STAD -0.077; TCGA UCEC -0.056 |
hsa-miR-3913-5p | PRKAR2A | 12 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRP; LUSC; PAAD; PRAD; SARC; THCA; UCEC | mirMAP | TCGA BLCA -0.202; TCGA BRCA -0.287; TCGA CESC -0.246; TCGA ESCA -0.158; TCGA HNSC -0.136; TCGA KIRP -0.306; TCGA LUSC -0.095; TCGA PAAD -0.282; TCGA PRAD -0.167; TCGA SARC -0.14; TCGA THCA -0.266; TCGA UCEC -0.197 |
hsa-miR-3913-5p | RAP1A | 11 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LUAD; PRAD; SARC; STAD | mirMAP | TCGA BLCA -0.191; TCGA BRCA -0.122; TCGA CESC -0.111; TCGA ESCA -0.191; TCGA HNSC -0.051; TCGA KIRC -0.054; TCGA KIRP -0.121; TCGA LUAD -0.062; TCGA PRAD -0.072; TCGA SARC -0.165; TCGA STAD -0.271 |
hsa-miR-3913-5p | KIAA0513 | 11 cancers: BLCA; BRCA; ESCA; HNSC; KIRP; LGG; PAAD; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.243; TCGA BRCA -0.095; TCGA ESCA -0.202; TCGA HNSC -0.135; TCGA KIRP -0.136; TCGA LGG -0.244; TCGA PAAD -0.201; TCGA PRAD -0.156; TCGA THCA -0.221; TCGA STAD -0.352; TCGA UCEC -0.344 |
hsa-miR-3913-5p | NFIC | 10 cancers: BLCA; BRCA; ESCA; KIRC; KIRP; LIHC; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.347; TCGA BRCA -0.135; TCGA ESCA -0.167; TCGA KIRC -0.131; TCGA KIRP -0.335; TCGA LIHC -0.214; TCGA SARC -0.158; TCGA THCA -0.209; TCGA STAD -0.246; TCGA UCEC -0.192 |
hsa-miR-3913-5p | LPP | 9 cancers: BLCA; BRCA; KIRC; LUSC; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.283; TCGA BRCA -0.27; TCGA KIRC -0.144; TCGA LUSC -0.15; TCGA PRAD -0.252; TCGA SARC -0.391; TCGA THCA -0.375; TCGA STAD -0.305; TCGA UCEC -0.263 |
hsa-miR-3913-5p | STOM | 11 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LIHC; LUSC; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.469; TCGA BRCA -0.173; TCGA CESC -0.231; TCGA ESCA -0.276; TCGA HNSC -0.087; TCGA KIRC -0.108; TCGA LIHC -0.168; TCGA LUSC -0.116; TCGA PRAD -0.12; TCGA STAD -0.215; TCGA UCEC -0.135 |
hsa-miR-3913-5p | KLHL21 | 10 cancers: BLCA; BRCA; CESC; HNSC; KIRP; LIHC; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.215; TCGA BRCA -0.111; TCGA CESC -0.239; TCGA HNSC -0.079; TCGA KIRP -0.268; TCGA LIHC -0.254; TCGA PRAD -0.095; TCGA SARC -0.241; TCGA STAD -0.114; TCGA UCEC -0.115 |
hsa-miR-3913-5p | NFASC | 10 cancers: BLCA; ESCA; HNSC; KIRC; KIRP; PAAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.346; TCGA ESCA -0.372; TCGA HNSC -0.296; TCGA KIRC -0.386; TCGA KIRP -0.753; TCGA PAAD -0.335; TCGA SARC -0.497; TCGA THCA -0.469; TCGA STAD -0.467; TCGA UCEC -0.199 |
hsa-miR-3913-5p | GAPVD1 | 9 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRP; LUSC; PAAD; UCEC | mirMAP | TCGA BLCA -0.084; TCGA BRCA -0.058; TCGA CESC -0.135; TCGA ESCA -0.103; TCGA HNSC -0.098; TCGA KIRP -0.071; TCGA LUSC -0.127; TCGA PAAD -0.091; TCGA UCEC -0.053 |
hsa-miR-3913-5p | MARCH8 | 10 cancers: BLCA; BRCA; KIRC; KIRP; LIHC; PAAD; PRAD; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.162; TCGA BRCA -0.226; TCGA KIRC -0.174; TCGA KIRP -0.314; TCGA LIHC -0.182; TCGA PAAD -0.202; TCGA PRAD -0.167; TCGA SARC -0.072; TCGA THCA -0.158; TCGA STAD -0.115 |
hsa-miR-3913-5p | FAM102A | 9 cancers: BLCA; CESC; ESCA; KIRP; LGG; LUSC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.115; TCGA CESC -0.136; TCGA ESCA -0.163; TCGA KIRP -0.317; TCGA LGG -0.051; TCGA LUSC -0.309; TCGA THCA -0.105; TCGA STAD -0.13; TCGA UCEC -0.187 |
hsa-miR-3913-5p | LRRC28 | 11 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; PAAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.059; TCGA BRCA -0.086; TCGA CESC -0.082; TCGA ESCA -0.114; TCGA HNSC -0.07; TCGA KIRC -0.081; TCGA KIRP -0.236; TCGA PAAD -0.086; TCGA THCA -0.062; TCGA STAD -0.092; TCGA UCEC -0.09 |
hsa-miR-3913-5p | KCNK3 | 10 cancers: BLCA; BRCA; KIRP; LGG; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.443; TCGA BRCA -0.185; TCGA KIRP -0.487; TCGA LGG -0.741; TCGA PAAD -0.512; TCGA PRAD -0.339; TCGA SARC -0.463; TCGA THCA -0.364; TCGA STAD -0.668; TCGA UCEC -0.251 |
hsa-miR-3913-5p | SYNPO | 11 cancers: BLCA; BRCA; CESC; ESCA; KIRC; KIRP; LUAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.351; TCGA BRCA -0.197; TCGA CESC -0.188; TCGA ESCA -0.22; TCGA KIRC -0.151; TCGA KIRP -0.575; TCGA LUAD -0.15; TCGA PRAD -0.107; TCGA SARC -0.128; TCGA STAD -0.209; TCGA UCEC -0.126 |
hsa-miR-3913-5p | ZHX3 | 10 cancers: BLCA; BRCA; ESCA; HNSC; KIRP; LIHC; PAAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.202; TCGA BRCA -0.107; TCGA ESCA -0.145; TCGA HNSC -0.081; TCGA KIRP -0.156; TCGA LIHC -0.132; TCGA PAAD -0.168; TCGA THCA -0.181; TCGA STAD -0.191; TCGA UCEC -0.163 |
hsa-miR-3913-5p | DENND2C | 11 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LUSC; PRAD; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.225; TCGA BRCA -0.312; TCGA CESC -0.631; TCGA HNSC -0.317; TCGA KIRC -0.141; TCGA KIRP -0.254; TCGA LUSC -0.324; TCGA PRAD -0.167; TCGA THCA -0.16; TCGA STAD -0.117; TCGA UCEC -0.192 |
hsa-miR-3913-5p | LYNX1 | 9 cancers: BLCA; BRCA; HNSC; KIRP; LGG; LIHC; PRAD; SARC; STAD | mirMAP | TCGA BLCA -0.38; TCGA BRCA -0.2; TCGA HNSC -0.691; TCGA KIRP -0.294; TCGA LGG -0.201; TCGA LIHC -0.278; TCGA PRAD -0.122; TCGA SARC -0.299; TCGA STAD -0.348 |
hsa-miR-3913-5p | SLC8A1 | 10 cancers: BLCA; BRCA; ESCA; KIRC; KIRP; PAAD; PRAD; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.362; TCGA BRCA -0.265; TCGA ESCA -0.213; TCGA KIRC -0.384; TCGA KIRP -0.493; TCGA PAAD -0.327; TCGA PRAD -0.251; TCGA SARC -0.245; TCGA THCA -0.211; TCGA STAD -0.305 |
hsa-miR-3913-5p | CBX6 | 9 cancers: BLCA; BRCA; KIRP; LGG; LUSC; PAAD; PRAD; SARC; UCEC | mirMAP | TCGA BLCA -0.128; TCGA BRCA -0.104; TCGA KIRP -0.2; TCGA LGG -0.121; TCGA LUSC -0.168; TCGA PAAD -0.239; TCGA PRAD -0.175; TCGA SARC -0.122; TCGA UCEC -0.168 |
hsa-miR-3913-5p | C3orf70 | 9 cancers: BLCA; KIRC; KIRP; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.121; TCGA KIRC -0.272; TCGA KIRP -0.548; TCGA PAAD -0.277; TCGA PRAD -0.226; TCGA SARC -0.258; TCGA THCA -0.324; TCGA STAD -0.269; TCGA UCEC -0.29 |
hsa-miR-3913-5p | FITM2 | 11 cancers: BLCA; BRCA; HNSC; LGG; LIHC; LUSC; PAAD; PRAD; SARC; THCA; UCEC | mirMAP | TCGA BLCA -0.13; TCGA BRCA -0.145; TCGA HNSC -0.125; TCGA LGG -0.086; TCGA LIHC -0.236; TCGA LUSC -0.11; TCGA PAAD -0.243; TCGA PRAD -0.125; TCGA SARC -0.117; TCGA THCA -0.241; TCGA UCEC -0.175 |
hsa-miR-3913-5p | LRP10 | 11 cancers: BLCA; CESC; ESCA; HNSC; KIRP; LUAD; LUSC; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.201; TCGA CESC -0.199; TCGA ESCA -0.256; TCGA HNSC -0.192; TCGA KIRP -0.135; TCGA LUAD -0.08; TCGA LUSC -0.133; TCGA PRAD -0.079; TCGA SARC -0.118; TCGA STAD -0.294; TCGA UCEC -0.146 |
hsa-miR-3913-5p | MIER1 | 9 cancers: BLCA; BRCA; ESCA; KIRC; LIHC; LUAD; PRAD; SARC; STAD | mirMAP | TCGA BLCA -0.059; TCGA BRCA -0.138; TCGA ESCA -0.084; TCGA KIRC -0.085; TCGA LIHC -0.075; TCGA LUAD -0.053; TCGA PRAD -0.072; TCGA SARC -0.057; TCGA STAD -0.076 |
hsa-miR-3913-5p | SORBS1 | 10 cancers: BLCA; BRCA; ESCA; KIRC; LIHC; LUAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.585; TCGA BRCA -0.408; TCGA ESCA -0.348; TCGA KIRC -0.155; TCGA LIHC -0.17; TCGA LUAD -0.211; TCGA PRAD -0.318; TCGA SARC -0.599; TCGA STAD -0.577; TCGA UCEC -0.219 |
hsa-miR-3913-5p | RAB14 | 9 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRP; LGG; LIHC; LUSC | miRNATAP | TCGA BLCA -0.067; TCGA BRCA -0.082; TCGA CESC -0.129; TCGA ESCA -0.109; TCGA HNSC -0.078; TCGA KIRP -0.091; TCGA LGG -0.066; TCGA LIHC -0.061; TCGA LUSC -0.058 |
Enriched cancer pathways of putative targets