microRNA information: hsa-miR-424-5p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-424-5p | miRbase |
| Accession: | MIMAT0001341 | miRbase |
| Precursor name: | hsa-mir-424 | miRbase |
| Precursor accession: | MI0001446 | miRbase |
| Symbol: | MIR424 | HGNC |
| RefSeq ID: | NR_029946 | GenBank |
| Sequence: | CAGCAGCAAUUCAUGUUUUGAA |
Reported expression in cancers: hsa-miR-424-5p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-424-5p | B cell lymphoma | deregulation | "Here we report a comprehensive miRNA-profiling stu ......" | 21062812 | |
| hsa-miR-424-5p | cervical and endocervical cancer | downregulation | "We previously identified a different characteristi ......" | 22469983 | qPCR |
| hsa-miR-424-5p | colorectal cancer | downregulation | "The in vivo significance of expression of miR-16/1 ......" | 21390519 | qPCR |
| hsa-miR-424-5p | colorectal cancer | upregulation | "Similarly to miR-195 the members of the same miR f ......" | 22710713 | |
| hsa-miR-424-5p | gastric cancer | upregulation | "Recently miR-424-5p was reported to play important ......" | 27655675 | |
| hsa-miR-424-5p | liver cancer | deregulation | "In this study we demonstrated the down-regulation ......" | 24675898 | qPCR |
Reported cancer pathway affected by hsa-miR-424-5p

| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-424-5p | cervical and endocervical cancer | Apoptosis pathway | "Suppressed miR 424 expression via upregulation of ......" | 22469983 | Luciferase; RNAi |
| hsa-miR-424-5p | endometrial cancer | PI3K/Akt signaling pathway | "MicroRNA 424 suppresses estradiol induced cell pro ......" | 26638889 | Luciferase |
| hsa-miR-424-5p | gastric cancer | cell cycle pathway | "miR 424 5p promotes proliferation of gastric cance ......" | 27655675 | Western blot; Luciferase |
| hsa-miR-424-5p | pancreatic cancer | Apoptosis pathway | "MicroRNA 424 5p suppresses the expression of SOCS6 ......" | 23653113 | Luciferase; Flow cytometry |
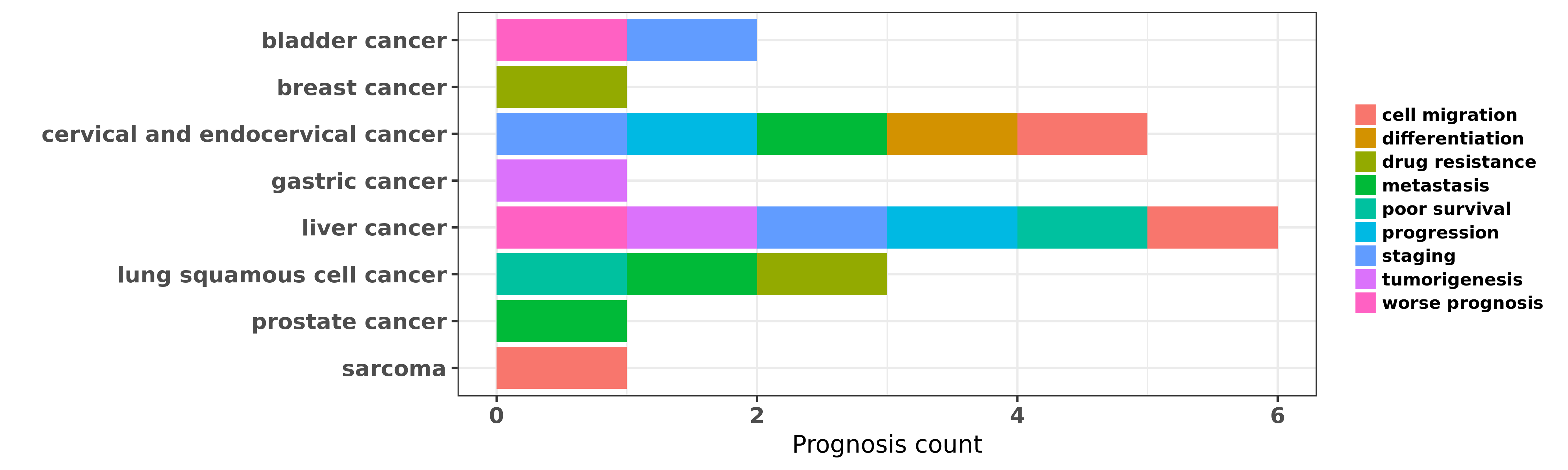
Reported cancer prognosis affected by hsa-miR-424-5p
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-424-5p | bladder cancer | staging; worse prognosis | "Our results revealed that the miR-424 level is sig ......" | 26090723 | |
| hsa-miR-424-5p | breast cancer | drug resistance | "To gain further insight into the molecular mechani ......" | 25633049 | |
| hsa-miR-424-5p | cervical and endocervical cancer | progression; staging; metastasis; differentiation; cell migration | "Suppressed miR 424 expression via upregulation of ......" | 22469983 | Luciferase; RNAi |
| hsa-miR-424-5p | gastric cancer | tumorigenesis | "miR 424 5p promotes proliferation of gastric cance ......" | 27655675 | Western blot; Luciferase |
| hsa-miR-424-5p | liver cancer | cell migration; tumorigenesis | "MicroRNA 424 is down regulated in hepatocellular c ......" | 24675898 | |
| hsa-miR-424-5p | liver cancer | progression; worse prognosis; poor survival | "MicroRNA 424 inhibits Akt3/E2F3 axis and tumor gro ......" | 26315541 | |
| hsa-miR-424-5p | liver cancer | worse prognosis; progression; poor survival; staging | "Decreased expression of serum miR 424 correlates w ......" | 26823812 | |
| hsa-miR-424-5p | lung squamous cell cancer | metastasis | "The miRs were quantified by microarray hybridizati ......" | 22295063 | |
| hsa-miR-424-5p | lung squamous cell cancer | poor survival | "Using a linear combination of the miR CT values wi ......" | 24007627 | |
| hsa-miR-424-5p | lung squamous cell cancer | drug resistance | "Loss of MiR 424 3p not miR 424 5p confers chemores ......" | 27500472 | Transwell assay |
| hsa-miR-424-5p | prostate cancer | metastasis | "Regulation of epithelial plasticity by miR 424 and ......" | 24193225 | |
| hsa-miR-424-5p | sarcoma | cell migration | "Tumor suppressive microRNA 424 inhibits osteosarco ......" | 23599729 | Luciferase; Western blot |
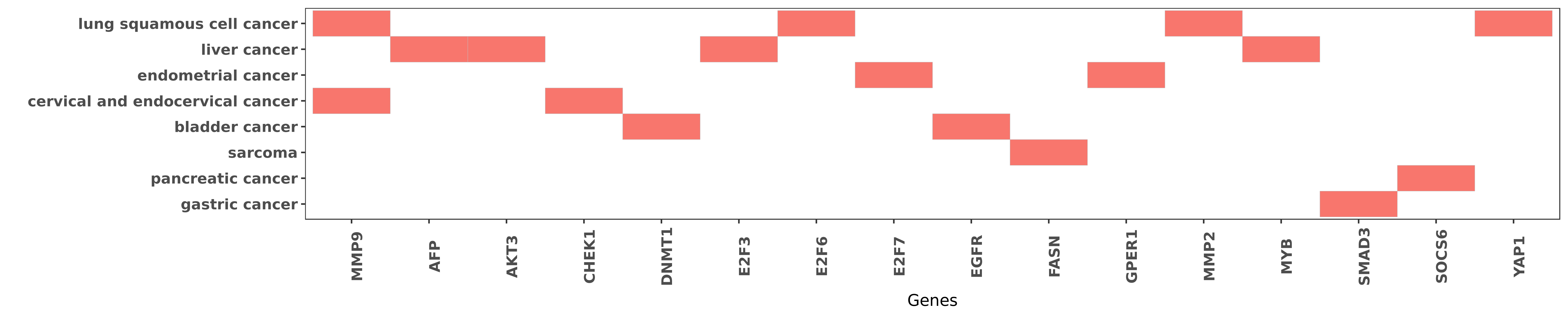
Reported gene related to hsa-miR-424-5p
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-424-5p | cervical and endocervical cancer | MMP9 | "Furthermore RNAi-mediated knockdown of Chk1 decrea ......" | 22469983 |
| hsa-miR-424-5p | lung squamous cell cancer | MMP9 | "After transfection of miR-424 the proliferation an ......" | 27666545 |
| hsa-miR-424-5p | liver cancer | AFP | "Reduced expression of serum miR-424 was associated ......" | 26823812 |
| hsa-miR-424-5p | liver cancer | AKT3 | "Silencing Akt3 and E2F3 by siRNA pheno-copied the ......" | 26315541 |
| hsa-miR-424-5p | cervical and endocervical cancer | CHEK1 | "Suppressed miR 424 expression via upregulation of ......" | 22469983 |
| hsa-miR-424-5p | bladder cancer | DNMT1 | "Our results revealed that the miR-424 level is sig ......" | 26090723 |
| hsa-miR-424-5p | liver cancer | E2F3 | "Silencing Akt3 and E2F3 by siRNA pheno-copied the ......" | 26315541 |
| hsa-miR-424-5p | lung squamous cell cancer | E2F6 | "The 3'UTR of E2F6 was cloned into luciferase repor ......" | 27666545 |
| hsa-miR-424-5p | endometrial cancer | E2F7 | "MicroRNA 424 may function as a tumor suppressor in ......" | 25708247 |
| hsa-miR-424-5p | bladder cancer | EGFR | "Furthermore the EGFR pathway plays a role in the t ......" | 26090723 |
| hsa-miR-424-5p | sarcoma | FASN | "Tumor suppressive microRNA 424 inhibits osteosarco ......" | 23599729 |
| hsa-miR-424-5p | endometrial cancer | GPER1 | "MicroRNA 424 suppresses estradiol induced cell pro ......" | 26638889 |
| hsa-miR-424-5p | lung squamous cell cancer | MMP2 | "After transfection of miR-424 the proliferation an ......" | 27666545 |
| hsa-miR-424-5p | liver cancer | MYB | "MicroRNA 424 is down regulated in hepatocellular c ......" | 24675898 |
| hsa-miR-424-5p | gastric cancer | SMAD3 | "miR 424 5p promotes proliferation of gastric cance ......" | 27655675 |
| hsa-miR-424-5p | pancreatic cancer | SOCS6 | "MicroRNA 424 5p suppresses the expression of SOCS6 ......" | 23653113 |
| hsa-miR-424-5p | lung squamous cell cancer | YAP1 | "Additionally it is miR-424-3p but not miR-424-5p t ......" | 27500472 |
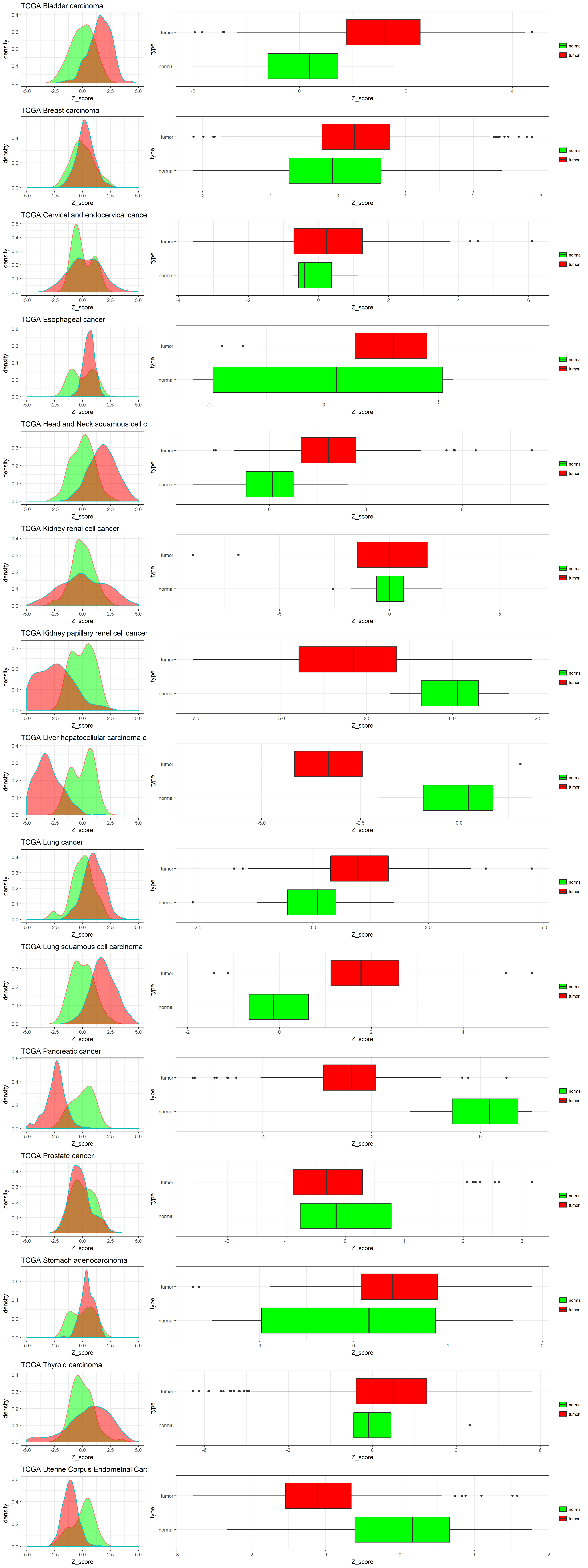
Expression profile in cancer corhorts:
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-424-5p | ITPR1 | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUAD; LUSC; PRAD; SARC; STAD | miRNAWalker2 validate; MirTarget; miRNATAP | TCGA BLCA -0.227; TCGA BRCA -0.085; TCGA COAD -0.31; TCGA ESCA -0.574; TCGA HNSC -0.219; TCGA LUAD -0.155; TCGA LUSC -0.38; TCGA PRAD -0.142; TCGA SARC -0.557; TCGA STAD -0.288 |
hsa-miR-424-5p | PIAS1 | 9 cancers: BLCA; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; SARC; STAD | miRNAWalker2 validate | TCGA BLCA -0.056; TCGA COAD -0.051; TCGA ESCA -0.085; TCGA HNSC -0.07; TCGA LIHC -0.053; TCGA LUAD -0.074; TCGA LUSC -0.081; TCGA SARC -0.086; TCGA STAD -0.088 |
hsa-miR-424-5p | MASP1 | 9 cancers: BLCA; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; SARC; STAD | MirTarget | TCGA BLCA -0.347; TCGA COAD -0.766; TCGA ESCA -0.472; TCGA HNSC -0.264; TCGA KIRP -0.21; TCGA LUAD -0.394; TCGA LUSC -0.413; TCGA SARC -0.73; TCGA STAD -0.379 |
hsa-miR-424-5p | AFF4 | 9 cancers: BLCA; COAD; ESCA; HNSC; LUAD; LUSC; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.084; TCGA COAD -0.089; TCGA ESCA -0.186; TCGA HNSC -0.074; TCGA LUAD -0.105; TCGA LUSC -0.071; TCGA SARC -0.096; TCGA STAD -0.17; TCGA UCEC -0.057 |
hsa-miR-424-5p | DCAF8 | 11 cancers: BLCA; BRCA; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; PAAD; SARC; STAD | MirTarget | TCGA BLCA -0.089; TCGA BRCA -0.062; TCGA ESCA -0.084; TCGA HNSC -0.057; TCGA LGG -0.082; TCGA LIHC -0.087; TCGA LUAD -0.063; TCGA LUSC -0.102; TCGA PAAD -0.112; TCGA SARC -0.117; TCGA STAD -0.093 |
hsa-miR-424-5p | ADCY5 | 9 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUAD; LUSC; SARC; STAD | MirTarget; miRNATAP | TCGA BLCA -0.843; TCGA BRCA -0.181; TCGA COAD -0.54; TCGA ESCA -1.128; TCGA HNSC -0.58; TCGA LUAD -0.226; TCGA LUSC -0.232; TCGA SARC -0.929; TCGA STAD -0.69 |
hsa-miR-424-5p | REEP1 | 11 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; SARC; STAD | MirTarget | TCGA BLCA -0.471; TCGA BRCA -0.204; TCGA CESC -0.194; TCGA ESCA -0.498; TCGA HNSC -0.188; TCGA LUAD -0.237; TCGA LUSC -0.448; TCGA PAAD -0.317; TCGA PRAD -0.109; TCGA SARC -0.546; TCGA STAD -0.617 |
hsa-miR-424-5p | THRB | 10 cancers: BLCA; COAD; ESCA; HNSC; LGG; LUAD; LUSC; SARC; THCA; STAD | MirTarget | TCGA BLCA -0.294; TCGA COAD -0.407; TCGA ESCA -0.479; TCGA HNSC -0.224; TCGA LGG -0.159; TCGA LUAD -0.242; TCGA LUSC -0.311; TCGA SARC -0.208; TCGA THCA -0.072; TCGA STAD -0.324 |
hsa-miR-424-5p | NBR1 | 9 cancers: BLCA; ESCA; HNSC; LGG; LUAD; LUSC; PAAD; SARC; STAD | MirTarget | TCGA BLCA -0.106; TCGA ESCA -0.113; TCGA HNSC -0.083; TCGA LGG -0.052; TCGA LUAD -0.067; TCGA LUSC -0.069; TCGA PAAD -0.1; TCGA SARC -0.052; TCGA STAD -0.084 |
hsa-miR-424-5p | SLC25A12 | 9 cancers: BLCA; ESCA; HNSC; KIRC; KIRP; LGG; PAAD; SARC; STAD | MirTarget | TCGA BLCA -0.185; TCGA ESCA -0.154; TCGA HNSC -0.1; TCGA KIRC -0.055; TCGA KIRP -0.057; TCGA LGG -0.11; TCGA PAAD -0.117; TCGA SARC -0.209; TCGA STAD -0.284 |
hsa-miR-424-5p | THSD4 | 9 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUAD; PRAD; SARC; STAD | MirTarget | TCGA BLCA -0.158; TCGA BRCA -0.179; TCGA COAD -0.132; TCGA ESCA -0.341; TCGA HNSC -0.119; TCGA LUAD -0.171; TCGA PRAD -0.242; TCGA SARC -0.363; TCGA STAD -0.2 |
hsa-miR-424-5p | PDK4 | 10 cancers: BLCA; COAD; ESCA; HNSC; LGG; LUAD; LUSC; PAAD; SARC; STAD | MirTarget; miRNATAP | TCGA BLCA -0.454; TCGA COAD -0.266; TCGA ESCA -0.917; TCGA HNSC -0.214; TCGA LGG -0.155; TCGA LUAD -0.359; TCGA LUSC -0.924; TCGA PAAD -0.315; TCGA SARC -0.286; TCGA STAD -0.657 |
hsa-miR-424-5p | TSPYL2 | 10 cancers: BLCA; BRCA; ESCA; HNSC; LGG; LUAD; LUSC; PAAD; SARC; STAD | MirTarget; miRNATAP | TCGA BLCA -0.213; TCGA BRCA -0.066; TCGA ESCA -0.185; TCGA HNSC -0.068; TCGA LGG -0.289; TCGA LUAD -0.071; TCGA LUSC -0.099; TCGA PAAD -0.159; TCGA SARC -0.226; TCGA STAD -0.313 |
hsa-miR-424-5p | SYNJ1 | 9 cancers: BLCA; COAD; ESCA; HNSC; LGG; LUAD; LUSC; SARC; STAD | MirTarget; miRNATAP | TCGA BLCA -0.059; TCGA COAD -0.101; TCGA ESCA -0.137; TCGA HNSC -0.059; TCGA LGG -0.107; TCGA LUAD -0.079; TCGA LUSC -0.155; TCGA SARC -0.074; TCGA STAD -0.111 |
hsa-miR-424-5p | AXIN2 | 11 cancers: BLCA; ESCA; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; SARC | MirTarget; miRNATAP | TCGA BLCA -0.264; TCGA ESCA -0.315; TCGA HNSC -0.104; TCGA KIRP -0.202; TCGA LGG -0.147; TCGA LIHC -0.192; TCGA LUAD -0.198; TCGA LUSC -0.256; TCGA PAAD -0.217; TCGA PRAD -0.084; TCGA SARC -0.124 |
hsa-miR-424-5p | ARHGAP5 | 10 cancers: BLCA; COAD; ESCA; HNSC; LGG; LUAD; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.073; TCGA COAD -0.08; TCGA ESCA -0.16; TCGA HNSC -0.069; TCGA LGG -0.082; TCGA LUAD -0.055; TCGA PRAD -0.052; TCGA SARC -0.079; TCGA STAD -0.176; TCGA UCEC -0.089 |
hsa-miR-424-5p | ZNF662 | 9 cancers: BLCA; HNSC; KIRC; KIRP; LUAD; LUSC; PRAD; SARC; STAD | MirTarget | TCGA BLCA -0.432; TCGA HNSC -0.264; TCGA KIRC -0.158; TCGA KIRP -0.289; TCGA LUAD -0.231; TCGA LUSC -0.193; TCGA PRAD -0.12; TCGA SARC -0.17; TCGA STAD -0.459 |
hsa-miR-424-5p | MLLT6 | 12 cancers: BLCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PRAD; SARC | MirTarget | TCGA BLCA -0.15; TCGA CESC -0.068; TCGA ESCA -0.202; TCGA HNSC -0.139; TCGA KIRC -0.113; TCGA KIRP -0.078; TCGA LGG -0.071; TCGA LIHC -0.148; TCGA LUAD -0.112; TCGA LUSC -0.133; TCGA PRAD -0.07; TCGA SARC -0.153 |
hsa-miR-424-5p | BTAF1 | 10 cancers: BLCA; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; SARC; STAD | MirTarget | TCGA BLCA -0.088; TCGA ESCA -0.106; TCGA HNSC -0.058; TCGA KIRC -0.074; TCGA KIRP -0.107; TCGA LGG -0.086; TCGA LUAD -0.094; TCGA LUSC -0.063; TCGA SARC -0.06; TCGA STAD -0.14 |
hsa-miR-424-5p | TNFAIP8L3 | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LUSC; SARC; THCA; STAD | MirTarget | TCGA BLCA -0.198; TCGA BRCA -0.072; TCGA COAD -0.184; TCGA ESCA -0.451; TCGA HNSC -0.119; TCGA KIRC -0.195; TCGA LUSC -0.351; TCGA SARC -0.283; TCGA THCA -0.164; TCGA STAD -0.403 |
hsa-miR-424-5p | ASH1L | 11 cancers: BLCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; SARC; STAD | MirTarget; miRNATAP | TCGA BLCA -0.144; TCGA CESC -0.087; TCGA COAD -0.075; TCGA ESCA -0.189; TCGA HNSC -0.169; TCGA LGG -0.058; TCGA LIHC -0.083; TCGA LUAD -0.126; TCGA LUSC -0.079; TCGA SARC -0.138; TCGA STAD -0.166 |
hsa-miR-424-5p | RABGAP1 | 10 cancers: BLCA; COAD; ESCA; HNSC; LGG; LUAD; LUSC; PAAD; SARC; STAD | MirTarget | TCGA BLCA -0.116; TCGA COAD -0.054; TCGA ESCA -0.181; TCGA HNSC -0.077; TCGA LGG -0.054; TCGA LUAD -0.081; TCGA LUSC -0.059; TCGA PAAD -0.14; TCGA SARC -0.222; TCGA STAD -0.247 |
hsa-miR-424-5p | RPRD2 | 9 cancers: BLCA; CESC; ESCA; HNSC; LIHC; LUAD; PAAD; SARC; STAD | MirTarget | TCGA BLCA -0.1; TCGA CESC -0.083; TCGA ESCA -0.061; TCGA HNSC -0.135; TCGA LIHC -0.091; TCGA LUAD -0.088; TCGA PAAD -0.138; TCGA SARC -0.118; TCGA STAD -0.113 |
hsa-miR-424-5p | LSM11 | 9 cancers: BLCA; CESC; ESCA; KIRC; KIRP; LGG; LIHC; SARC; STAD | MirTarget | TCGA BLCA -0.191; TCGA CESC -0.087; TCGA ESCA -0.134; TCGA KIRC -0.055; TCGA KIRP -0.083; TCGA LGG -0.083; TCGA LIHC -0.183; TCGA SARC -0.241; TCGA STAD -0.172 |
hsa-miR-424-5p | SPRYD3 | 9 cancers: BLCA; BRCA; ESCA; KIRC; KIRP; LGG; LIHC; LUAD; PAAD | MirTarget; miRNATAP | TCGA BLCA -0.096; TCGA BRCA -0.132; TCGA ESCA -0.095; TCGA KIRC -0.056; TCGA KIRP -0.052; TCGA LGG -0.124; TCGA LIHC -0.079; TCGA LUAD -0.089; TCGA PAAD -0.124 |
hsa-miR-424-5p | CCDC6 | 10 cancers: BLCA; CESC; ESCA; HNSC; LGG; PRAD; SARC; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.063; TCGA CESC -0.059; TCGA ESCA -0.148; TCGA HNSC -0.123; TCGA LGG -0.074; TCGA PRAD -0.145; TCGA SARC -0.183; TCGA THCA -0.077; TCGA STAD -0.155; TCGA UCEC -0.137 |
hsa-miR-424-5p | CASZ1 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD | MirTarget | TCGA BLCA -0.259; TCGA BRCA -0.09; TCGA CESC -0.176; TCGA COAD -0.072; TCGA ESCA -0.277; TCGA HNSC -0.192; TCGA LUAD -0.184; TCGA LUSC -0.239; TCGA PAAD -0.227; TCGA PRAD -0.093; TCGA SARC -0.406; TCGA THCA -0.088; TCGA STAD -0.175 |
hsa-miR-424-5p | MYRIP | 10 cancers: BLCA; BRCA; ESCA; HNSC; LGG; LUAD; LUSC; PRAD; SARC; STAD | MirTarget | TCGA BLCA -0.629; TCGA BRCA -0.196; TCGA ESCA -0.677; TCGA HNSC -0.603; TCGA LGG -0.132; TCGA LUAD -0.356; TCGA LUSC -0.771; TCGA PRAD -0.114; TCGA SARC -0.466; TCGA STAD -0.398 |
hsa-miR-424-5p | RELN | 11 cancers: BLCA; COAD; ESCA; HNSC; KIRP; LGG; LUAD; LUSC; SARC; THCA; STAD | MirTarget; miRNATAP | TCGA BLCA -0.419; TCGA COAD -0.548; TCGA ESCA -0.628; TCGA HNSC -0.18; TCGA KIRP -0.607; TCGA LGG -0.379; TCGA LUAD -0.17; TCGA LUSC -0.644; TCGA SARC -0.395; TCGA THCA -0.281; TCGA STAD -0.466 |
hsa-miR-424-5p | PLXNA2 | 9 cancers: BLCA; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; SARC; STAD | MirTarget; miRNATAP | TCGA BLCA -0.15; TCGA ESCA -0.251; TCGA HNSC -0.101; TCGA LIHC -0.145; TCGA LUAD -0.191; TCGA LUSC -0.233; TCGA PAAD -0.271; TCGA SARC -0.091; TCGA STAD -0.075 |
hsa-miR-424-5p | BCL2L2 | 9 cancers: BLCA; BRCA; COAD; ESCA; KIRC; LGG; LUAD; PAAD; SARC | MirTarget; miRNATAP | TCGA BLCA -0.132; TCGA BRCA -0.079; TCGA COAD -0.103; TCGA ESCA -0.123; TCGA KIRC -0.098; TCGA LGG -0.115; TCGA LUAD -0.137; TCGA PAAD -0.161; TCGA SARC -0.174 |
hsa-miR-424-5p | PRKAB2 | 9 cancers: BLCA; COAD; KIRC; LGG; LIHC; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.078; TCGA COAD -0.089; TCGA KIRC -0.054; TCGA LGG -0.06; TCGA LIHC -0.197; TCGA SARC -0.175; TCGA THCA -0.067; TCGA STAD -0.276; TCGA UCEC -0.095 |
hsa-miR-424-5p | ARHGEF12 | 10 cancers: BLCA; COAD; ESCA; HNSC; LGG; LUAD; LUSC; PRAD; SARC; STAD | MirTarget | TCGA BLCA -0.091; TCGA COAD -0.076; TCGA ESCA -0.099; TCGA HNSC -0.099; TCGA LGG -0.066; TCGA LUAD -0.125; TCGA LUSC -0.085; TCGA PRAD -0.052; TCGA SARC -0.09; TCGA STAD -0.144 |
hsa-miR-424-5p | ZNF654 | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; LGG; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.083; TCGA CESC -0.057; TCGA COAD -0.069; TCGA ESCA -0.144; TCGA HNSC -0.116; TCGA LGG -0.066; TCGA SARC -0.105; TCGA STAD -0.144; TCGA UCEC -0.082 |
hsa-miR-424-5p | ATP8A1 | 10 cancers: BLCA; ESCA; HNSC; KIRP; LGG; LUAD; LUSC; PRAD; SARC; STAD | MirTarget | TCGA BLCA -0.352; TCGA ESCA -0.42; TCGA HNSC -0.264; TCGA KIRP -0.144; TCGA LGG -0.218; TCGA LUAD -0.269; TCGA LUSC -0.626; TCGA PRAD -0.076; TCGA SARC -0.176; TCGA STAD -0.124 |
hsa-miR-424-5p | SH3GL2 | 9 cancers: BLCA; ESCA; LGG; LUAD; LUSC; PRAD; SARC; THCA; STAD | MirTarget; miRNATAP | TCGA BLCA -0.329; TCGA ESCA -0.649; TCGA LGG -0.502; TCGA LUAD -0.363; TCGA LUSC -0.758; TCGA PRAD -0.392; TCGA SARC -0.598; TCGA THCA -0.301; TCGA STAD -0.386 |
hsa-miR-424-5p | RCOR3 | 9 cancers: BLCA; BRCA; ESCA; HNSC; LGG; LIHC; PAAD; SARC; STAD | MirTarget; miRNATAP | TCGA BLCA -0.117; TCGA BRCA -0.062; TCGA ESCA -0.071; TCGA HNSC -0.076; TCGA LGG -0.072; TCGA LIHC -0.158; TCGA PAAD -0.115; TCGA SARC -0.163; TCGA STAD -0.185 |
hsa-miR-424-5p | FGF2 | 9 cancers: BLCA; COAD; ESCA; HNSC; KIRP; LGG; LUAD; LUSC; STAD | MirTarget; miRNATAP | TCGA BLCA -0.199; TCGA COAD -0.317; TCGA ESCA -0.473; TCGA HNSC -0.246; TCGA KIRP -0.099; TCGA LGG -0.138; TCGA LUAD -0.12; TCGA LUSC -0.256; TCGA STAD -0.421 |
hsa-miR-424-5p | MTHFR | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PRAD | MirTarget | TCGA BLCA -0.099; TCGA CESC -0.11; TCGA COAD -0.088; TCGA ESCA -0.205; TCGA HNSC -0.12; TCGA LIHC -0.077; TCGA LUAD -0.119; TCGA LUSC -0.172; TCGA PRAD -0.068 |
hsa-miR-424-5p | KIAA2022 | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LGG; LUAD; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.595; TCGA BRCA -0.143; TCGA COAD -0.55; TCGA ESCA -0.796; TCGA HNSC -0.69; TCGA LGG -0.219; TCGA LUAD -0.293; TCGA PRAD -0.091; TCGA SARC -0.589; TCGA STAD -0.756; TCGA UCEC -0.142 |
hsa-miR-424-5p | ANK2 | 9 cancers: BLCA; COAD; ESCA; HNSC; LGG; LUAD; LUSC; THCA; STAD | MirTarget; miRNATAP | TCGA BLCA -0.288; TCGA COAD -0.457; TCGA ESCA -0.834; TCGA HNSC -0.368; TCGA LGG -0.124; TCGA LUAD -0.141; TCGA LUSC -0.496; TCGA THCA -0.134; TCGA STAD -0.616 |
hsa-miR-424-5p | COL4A3BP | 11 cancers: BLCA; CESC; COAD; ESCA; HNSC; LGG; LUAD; LUSC; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.106; TCGA CESC -0.075; TCGA COAD -0.067; TCGA ESCA -0.146; TCGA HNSC -0.105; TCGA LGG -0.067; TCGA LUAD -0.13; TCGA LUSC -0.186; TCGA SARC -0.073; TCGA STAD -0.142; TCGA UCEC -0.068 |
hsa-miR-424-5p | NISCH | 12 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LGG; LIHC; LUAD; LUSC; SARC; STAD | MirTarget | TCGA BLCA -0.123; TCGA BRCA -0.113; TCGA CESC -0.067; TCGA ESCA -0.185; TCGA HNSC -0.071; TCGA KIRC -0.13; TCGA LGG -0.073; TCGA LIHC -0.143; TCGA LUAD -0.117; TCGA LUSC -0.095; TCGA SARC -0.064; TCGA STAD -0.121 |
hsa-miR-424-5p | CBFA2T3 | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LGG; LUAD; LUSC; PRAD; THCA; STAD | MirTarget; miRNATAP | TCGA BLCA -0.26; TCGA BRCA -0.125; TCGA COAD -0.262; TCGA ESCA -0.224; TCGA HNSC -0.215; TCGA KIRC -0.104; TCGA LGG -0.199; TCGA LUAD -0.192; TCGA LUSC -0.255; TCGA PRAD -0.113; TCGA THCA -0.08; TCGA STAD -0.174 |
hsa-miR-424-5p | EZH1 | 14 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; SARC; STAD | MirTarget; miRNATAP | TCGA BLCA -0.184; TCGA BRCA -0.118; TCGA CESC -0.064; TCGA ESCA -0.317; TCGA HNSC -0.114; TCGA KIRC -0.107; TCGA KIRP -0.06; TCGA LGG -0.121; TCGA LIHC -0.155; TCGA LUAD -0.143; TCGA LUSC -0.179; TCGA PAAD -0.086; TCGA SARC -0.213; TCGA STAD -0.267 |
hsa-miR-424-5p | SARM1 | 11 cancers: BLCA; CESC; ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; PRAD; SARC; STAD | mirMAP | TCGA BLCA -0.233; TCGA CESC -0.124; TCGA ESCA -0.202; TCGA HNSC -0.242; TCGA KIRP -0.073; TCGA LUAD -0.151; TCGA LUSC -0.139; TCGA PAAD -0.179; TCGA PRAD -0.067; TCGA SARC -0.195; TCGA STAD -0.085 |
hsa-miR-424-5p | NFIX | 10 cancers: BLCA; BRCA; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; SARC; STAD | mirMAP | TCGA BLCA -0.134; TCGA BRCA -0.111; TCGA ESCA -0.263; TCGA HNSC -0.264; TCGA LGG -0.11; TCGA LIHC -0.08; TCGA LUAD -0.221; TCGA LUSC -0.294; TCGA SARC -0.088; TCGA STAD -0.174 |
hsa-miR-424-5p | IGSF9B | 10 cancers: BLCA; BRCA; COAD; ESCA; KIRC; LUAD; PAAD; PRAD; SARC; STAD | mirMAP | TCGA BLCA -0.643; TCGA BRCA -0.235; TCGA COAD -0.29; TCGA ESCA -0.909; TCGA KIRC -0.258; TCGA LUAD -0.191; TCGA PAAD -0.313; TCGA PRAD -0.227; TCGA SARC -0.815; TCGA STAD -0.438 |
hsa-miR-424-5p | GNAO1 | 9 cancers: BLCA; COAD; ESCA; LGG; LUAD; PAAD; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.46; TCGA COAD -0.456; TCGA ESCA -0.467; TCGA LGG -0.128; TCGA LUAD -0.164; TCGA PAAD -0.301; TCGA SARC -0.595; TCGA THCA -0.104; TCGA STAD -0.61 |
hsa-miR-424-5p | ABLIM1 | 10 cancers: BLCA; CESC; ESCA; HNSC; LGG; LUAD; LUSC; PAAD; THCA; STAD | mirMAP | TCGA BLCA -0.185; TCGA CESC -0.148; TCGA ESCA -0.314; TCGA HNSC -0.286; TCGA LGG -0.181; TCGA LUAD -0.238; TCGA LUSC -0.214; TCGA PAAD -0.148; TCGA THCA -0.055; TCGA STAD -0.081 |
hsa-miR-424-5p | C1orf21 | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; SARC; STAD | mirMAP; miRNATAP | TCGA BLCA -0.237; TCGA BRCA -0.156; TCGA COAD -0.117; TCGA ESCA -0.23; TCGA HNSC -0.074; TCGA LUAD -0.125; TCGA LUSC -0.195; TCGA PAAD -0.177; TCGA SARC -0.08; TCGA STAD -0.29 |
hsa-miR-424-5p | TMEM108 | 9 cancers: BLCA; COAD; ESCA; HNSC; LGG; LUAD; LUSC; SARC; STAD | mirMAP | TCGA BLCA -0.584; TCGA COAD -0.206; TCGA ESCA -0.325; TCGA HNSC -0.194; TCGA LGG -0.078; TCGA LUAD -0.234; TCGA LUSC -0.38; TCGA SARC -0.38; TCGA STAD -0.276 |
hsa-miR-424-5p | PTCHD1 | 10 cancers: BLCA; COAD; ESCA; HNSC; LUAD; LUSC; PRAD; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.516; TCGA COAD -0.625; TCGA ESCA -0.55; TCGA HNSC -0.312; TCGA LUAD -0.246; TCGA LUSC -0.636; TCGA PRAD -0.301; TCGA SARC -0.889; TCGA THCA -0.173; TCGA STAD -1.073 |
hsa-miR-424-5p | AR | 9 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUAD; LUSC; SARC; STAD | mirMAP; miRNATAP | TCGA BLCA -0.655; TCGA BRCA -0.137; TCGA COAD -0.387; TCGA ESCA -0.654; TCGA HNSC -0.494; TCGA LUAD -0.28; TCGA LUSC -0.581; TCGA SARC -0.51; TCGA STAD -0.52 |
hsa-miR-424-5p | KCNK3 | 9 cancers: BLCA; COAD; ESCA; LGG; LUAD; LUSC; PRAD; SARC; STAD | mirMAP | TCGA BLCA -0.386; TCGA COAD -0.5; TCGA ESCA -0.759; TCGA LGG -0.345; TCGA LUAD -0.309; TCGA LUSC -0.721; TCGA PRAD -0.169; TCGA SARC -0.728; TCGA STAD -0.651 |
hsa-miR-424-5p | ARNT2 | 10 cancers: BLCA; BRCA; COAD; HNSC; LGG; LUAD; LUSC; PRAD; SARC; STAD | mirMAP | TCGA BLCA -0.205; TCGA BRCA -0.097; TCGA COAD -0.318; TCGA HNSC -0.331; TCGA LGG -0.072; TCGA LUAD -0.273; TCGA LUSC -0.213; TCGA PRAD -0.172; TCGA SARC -0.332; TCGA STAD -0.206 |
hsa-miR-424-5p | TOM1L2 | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; PAAD; SARC; STAD | mirMAP | TCGA BLCA -0.122; TCGA BRCA -0.105; TCGA COAD -0.082; TCGA ESCA -0.235; TCGA HNSC -0.127; TCGA LGG -0.12; TCGA LIHC -0.117; TCGA LUAD -0.146; TCGA LUSC -0.165; TCGA PAAD -0.173; TCGA SARC -0.167; TCGA STAD -0.182 |
hsa-miR-424-5p | MDM4 | 10 cancers: BLCA; CESC; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; SARC; STAD | mirMAP | TCGA BLCA -0.109; TCGA CESC -0.079; TCGA ESCA -0.097; TCGA HNSC -0.078; TCGA KIRC -0.123; TCGA KIRP -0.114; TCGA LIHC -0.146; TCGA LUAD -0.074; TCGA SARC -0.109; TCGA STAD -0.205 |
hsa-miR-424-5p | CACNB4 | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; PRAD; SARC; STAD | miRNATAP | TCGA BLCA -0.436; TCGA CESC -0.213; TCGA COAD -0.18; TCGA ESCA -0.316; TCGA HNSC -0.619; TCGA LGG -0.12; TCGA LIHC -0.407; TCGA LUAD -0.257; TCGA LUSC -0.413; TCGA PRAD -0.232; TCGA SARC -0.157; TCGA STAD -0.333 |
hsa-miR-424-5p | RUNX1T1 | 9 cancers: BLCA; COAD; ESCA; HNSC; LGG; LUAD; LUSC; SARC; STAD | miRNATAP | TCGA BLCA -0.241; TCGA COAD -0.374; TCGA ESCA -0.456; TCGA HNSC -0.169; TCGA LGG -0.176; TCGA LUAD -0.208; TCGA LUSC -0.498; TCGA SARC -0.123; TCGA STAD -0.422 |
hsa-miR-424-5p | SCN4B | 9 cancers: BLCA; BRCA; COAD; ESCA; LIHC; LUAD; LUSC; SARC; STAD | miRNATAP | TCGA BLCA -0.248; TCGA BRCA -0.092; TCGA COAD -0.349; TCGA ESCA -0.275; TCGA LIHC -0.157; TCGA LUAD -0.376; TCGA LUSC -0.362; TCGA SARC -0.216; TCGA STAD -0.316 |
hsa-miR-424-5p | SON | 9 cancers: BLCA; COAD; ESCA; HNSC; LUAD; LUSC; PRAD; SARC; STAD | miRNATAP | TCGA BLCA -0.084; TCGA COAD -0.073; TCGA ESCA -0.109; TCGA HNSC -0.09; TCGA LUAD -0.091; TCGA LUSC -0.1; TCGA PRAD -0.072; TCGA SARC -0.057; TCGA STAD -0.117 |
hsa-miR-424-5p | CNKSR2 | 9 cancers: BLCA; ESCA; HNSC; LGG; LUAD; LUSC; SARC; THCA; STAD | miRNATAP | TCGA BLCA -0.317; TCGA ESCA -0.591; TCGA HNSC -0.388; TCGA LGG -0.209; TCGA LUAD -0.199; TCGA LUSC -0.447; TCGA SARC -0.501; TCGA THCA -0.137; TCGA STAD -0.809 |
hsa-miR-424-5p | USP19 | 9 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LIHC; LUAD; PAAD; SARC | miRNATAP | TCGA BLCA -0.104; TCGA BRCA -0.082; TCGA CESC -0.065; TCGA ESCA -0.105; TCGA HNSC -0.09; TCGA LIHC -0.07; TCGA LUAD -0.054; TCGA PAAD -0.072; TCGA SARC -0.09 |
hsa-miR-424-5p | NLGN1 | 10 cancers: BLCA; COAD; ESCA; HNSC; KIRP; LGG; PAAD; SARC; THCA; STAD | miRNATAP | TCGA BLCA -0.436; TCGA COAD -0.524; TCGA ESCA -0.476; TCGA HNSC -0.547; TCGA KIRP -0.149; TCGA LGG -0.067; TCGA PAAD -0.335; TCGA SARC -0.353; TCGA THCA -0.237; TCGA STAD -0.69 |
hsa-miR-424-5p | CEP350 | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; LIHC; LUSC; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.052; TCGA CESC -0.051; TCGA COAD -0.054; TCGA ESCA -0.094; TCGA HNSC -0.109; TCGA LIHC -0.103; TCGA LUSC -0.051; TCGA SARC -0.06; TCGA STAD -0.083; TCGA UCEC -0.072 |
hsa-miR-424-5p | ZER1 | 10 cancers: BLCA; BRCA; ESCA; HNSC; LGG; LUAD; LUSC; PAAD; SARC; STAD | miRNATAP | TCGA BLCA -0.107; TCGA BRCA -0.069; TCGA ESCA -0.169; TCGA HNSC -0.095; TCGA LGG -0.07; TCGA LUAD -0.063; TCGA LUSC -0.073; TCGA PAAD -0.181; TCGA SARC -0.101; TCGA STAD -0.11 |
hsa-miR-424-5p | FGF18 | 9 cancers: BLCA; BRCA; ESCA; HNSC; LUAD; LUSC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.262; TCGA BRCA -0.118; TCGA ESCA -0.447; TCGA HNSC -0.411; TCGA LUAD -0.209; TCGA LUSC -0.591; TCGA THCA -0.147; TCGA STAD -0.165; TCGA UCEC -0.304 |
hsa-miR-424-5p | FGF7 | 10 cancers: BLCA; COAD; ESCA; HNSC; LGG; LUAD; LUSC; SARC; THCA; STAD | miRNATAP | TCGA BLCA -0.233; TCGA COAD -0.323; TCGA ESCA -0.685; TCGA HNSC -0.321; TCGA LGG -0.09; TCGA LUAD -0.106; TCGA LUSC -0.454; TCGA SARC -0.258; TCGA THCA -0.109; TCGA STAD -0.387 |
hsa-miR-424-5p | SYDE2 | 10 cancers: BLCA; COAD; ESCA; HNSC; LGG; LUAD; LUSC; PAAD; SARC; STAD | miRNATAP | TCGA BLCA -0.253; TCGA COAD -0.126; TCGA ESCA -0.369; TCGA HNSC -0.331; TCGA LGG -0.14; TCGA LUAD -0.09; TCGA LUSC -0.229; TCGA PAAD -0.286; TCGA SARC -0.195; TCGA STAD -0.45 |
hsa-miR-424-5p | ZC3H12B | 9 cancers: BLCA; ESCA; HNSC; LGG; LUAD; LUSC; PAAD; SARC; STAD | miRNATAP | TCGA BLCA -0.32; TCGA ESCA -0.36; TCGA HNSC -0.102; TCGA LGG -0.165; TCGA LUAD -0.151; TCGA LUSC -0.315; TCGA PAAD -0.177; TCGA SARC -0.204; TCGA STAD -0.314 |
hsa-miR-424-5p | MAN2A2 | 11 cancers: BRCA; CESC; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD | MirTarget | TCGA BRCA -0.068; TCGA CESC -0.081; TCGA ESCA -0.136; TCGA HNSC -0.165; TCGA LIHC -0.076; TCGA LUAD -0.113; TCGA LUSC -0.083; TCGA PAAD -0.099; TCGA PRAD -0.056; TCGA SARC -0.086; TCGA STAD -0.062 |
hsa-miR-424-5p | LRIG1 | 9 cancers: BRCA; ESCA; HNSC; LGG; LUAD; LUSC; PRAD; SARC; STAD | MirTarget | TCGA BRCA -0.126; TCGA ESCA -0.266; TCGA HNSC -0.16; TCGA LGG -0.228; TCGA LUAD -0.188; TCGA LUSC -0.095; TCGA PRAD -0.128; TCGA SARC -0.251; TCGA STAD -0.218 |
hsa-miR-424-5p | INPP5J | 9 cancers: BRCA; CESC; HNSC; KIRC; LGG; LUAD; PAAD; PRAD; SARC | MirTarget; miRNATAP | TCGA BRCA -0.241; TCGA CESC -0.17; TCGA HNSC -0.308; TCGA KIRC -0.265; TCGA LGG -0.192; TCGA LUAD -0.134; TCGA PAAD -0.284; TCGA PRAD -0.097; TCGA SARC -0.484 |
hsa-miR-424-5p | ANKRD29 | 10 cancers: BRCA; ESCA; KIRP; LGG; LIHC; LUAD; LUSC; PRAD; SARC; STAD | MirTarget | TCGA BRCA -0.104; TCGA ESCA -0.261; TCGA KIRP -0.085; TCGA LGG -0.179; TCGA LIHC -0.275; TCGA LUAD -0.342; TCGA LUSC -0.39; TCGA PRAD -0.159; TCGA SARC -0.327; TCGA STAD -0.32 |
hsa-miR-424-5p | NEK8 | 9 cancers: BRCA; KIRC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; SARC | mirMAP | TCGA BRCA -0.052; TCGA KIRC -0.079; TCGA KIRP -0.153; TCGA LIHC -0.108; TCGA LUAD -0.063; TCGA LUSC -0.095; TCGA PAAD -0.15; TCGA PRAD -0.054; TCGA SARC -0.116 |
hsa-miR-424-5p | MTMR3 | 9 cancers: CESC; COAD; ESCA; HNSC; KIRC; LGG; LUAD; PRAD; STAD | MirTarget | TCGA CESC -0.076; TCGA COAD -0.062; TCGA ESCA -0.122; TCGA HNSC -0.11; TCGA KIRC -0.051; TCGA LGG -0.05; TCGA LUAD -0.094; TCGA PRAD -0.077; TCGA STAD -0.09 |
hsa-miR-424-5p | ZDHHC21 | 9 cancers: CESC; HNSC; LGG; LUAD; LUSC; PRAD; SARC; STAD; UCEC | MirTarget | TCGA CESC -0.11; TCGA HNSC -0.391; TCGA LGG -0.083; TCGA LUAD -0.068; TCGA LUSC -0.284; TCGA PRAD -0.092; TCGA SARC -0.123; TCGA STAD -0.164; TCGA UCEC -0.075 |
hsa-miR-424-5p | NOTCH2 | 9 cancers: CESC; COAD; ESCA; HNSC; KIRP; LUSC; PRAD; THCA; STAD | MirTarget | TCGA CESC -0.088; TCGA COAD -0.157; TCGA ESCA -0.218; TCGA HNSC -0.134; TCGA KIRP -0.089; TCGA LUSC -0.163; TCGA PRAD -0.091; TCGA THCA -0.054; TCGA STAD -0.127 |
hsa-miR-424-5p | TMEM161B | 9 cancers: CESC; HNSC; KIRC; LGG; LUAD; LUSC; PRAD; SARC; UCEC | MirTarget | TCGA CESC -0.074; TCGA HNSC -0.077; TCGA KIRC -0.065; TCGA LGG -0.101; TCGA LUAD -0.055; TCGA LUSC -0.085; TCGA PRAD -0.059; TCGA SARC -0.071; TCGA UCEC -0.073 |
hsa-miR-424-5p | UNC119B | 9 cancers: CESC; COAD; HNSC; LIHC; LUAD; PAAD; PRAD; SARC; STAD | MirTarget | TCGA CESC -0.13; TCGA COAD -0.151; TCGA HNSC -0.118; TCGA LIHC -0.25; TCGA LUAD -0.148; TCGA PAAD -0.142; TCGA PRAD -0.078; TCGA SARC -0.053; TCGA STAD -0.06 |
hsa-miR-424-5p | RASGEF1B | 9 cancers: CESC; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; PRAD; THCA | MirTarget | TCGA CESC -0.171; TCGA COAD -0.175; TCGA ESCA -0.141; TCGA HNSC -0.171; TCGA KIRP -0.076; TCGA LUAD -0.113; TCGA LUSC -0.378; TCGA PRAD -0.104; TCGA THCA -0.121 |
hsa-miR-424-5p | CAMSAP1 | 9 cancers: CESC; COAD; ESCA; HNSC; KIRP; LUAD; PAAD; SARC; STAD | MirTarget; miRNATAP | TCGA CESC -0.085; TCGA COAD -0.057; TCGA ESCA -0.103; TCGA HNSC -0.15; TCGA KIRP -0.113; TCGA LUAD -0.053; TCGA PAAD -0.087; TCGA SARC -0.08; TCGA STAD -0.103 |
hsa-miR-424-5p | PDPK1 | 9 cancers: CESC; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; PRAD; SARC | mirMAP | TCGA CESC -0.107; TCGA ESCA -0.125; TCGA HNSC -0.081; TCGA LGG -0.064; TCGA LIHC -0.106; TCGA LUAD -0.114; TCGA LUSC -0.111; TCGA PRAD -0.07; TCGA SARC -0.082 |
hsa-miR-424-5p | RSPO3 | 9 cancers: COAD; ESCA; HNSC; LGG; LUSC; PRAD; SARC; THCA; STAD | MirTarget | TCGA COAD -0.385; TCGA ESCA -0.843; TCGA HNSC -0.397; TCGA LGG -0.36; TCGA LUSC -0.395; TCGA PRAD -0.356; TCGA SARC -0.39; TCGA THCA -0.338; TCGA STAD -0.38 |
Enriched cancer pathways of putative targets