microRNA information: hsa-miR-425-3p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-425-3p | miRbase |
| Accession: | MIMAT0001343 | miRbase |
| Precursor name: | hsa-mir-425 | miRbase |
| Precursor accession: | MI0001448 | miRbase |
| Symbol: | MIR425 | HGNC |
| RefSeq ID: | NR_029948 | GenBank |
| Sequence: | AUCGGGAAUGUCGUGUCCGCCC |
Reported expression in cancers: hsa-miR-425-3p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-425-3p | breast cancer | deregulation | "Finally eight upregulated miRNAs including hsa-miR ......" | 27446395 | |
| hsa-miR-425-3p | gastric cancer | upregulation | "And the miRecords algorithm and luciferase reporte ......" | 25154996 |
Reported cancer pathway affected by hsa-miR-425-3p

| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-425-3p | gastric cancer | Apoptosis pathway | "NF kappaB dependent microRNA 425 upregulation prom ......" | 24571667 | |
| hsa-miR-425-3p | gastric cancer | cell cycle pathway | "In this study the expression of miR-191 and miR-42 ......" | 24603541 | |
| hsa-miR-425-3p | gastric cancer | Apoptosis pathway | "MiR 425 up regulation induced by interleukin 1β p ......" | 25154996 | Luciferase; Cell proliferation assay; Cell Proliferation Assay; Flow cytometry |
| hsa-miR-425-3p | glioblastoma | cell cycle pathway | "An unbiased functional microRNA screen identified ......" | 27399532 | |
| hsa-miR-425-3p | melanoma | Apoptosis pathway | "miR 425 inhibits melanoma metastasis through repre ......" | 26463631 | Luciferase |
Reported cancer prognosis affected by hsa-miR-425-3p

| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-425-3p | breast cancer | metastasis; tumorigenesis | "MiR-191/425 locus characterization revealed that t ......" | 23505378 | |
| hsa-miR-425-3p | breast cancer | staging | "Global miRNA analysis was performed on serum from ......" | 24694649 | |
| hsa-miR-425-3p | colorectal cancer | drug resistance | "MicroRNA 425 5p regulates chemoresistance in color ......" | 26647742 | |
| hsa-miR-425-3p | esophageal cancer | tumorigenesis; metastasis | "Enhanced Expression of miR 425 Promotes Esophageal ......" | 26674378 | Colony formation |
| hsa-miR-425-3p | gastric cancer | poor survival | "NF kappaB dependent microRNA 425 upregulation prom ......" | 24571667 | |
| hsa-miR-425-3p | gastric cancer | progression; cell migration; tumorigenesis | "In this study the expression of miR-191 and miR-42 ......" | 24603541 | |
| hsa-miR-425-3p | gastric cancer | metastasis | "microRNA 425 5p is upregulated in human gastric ca ......" | 26136868 | |
| hsa-miR-425-3p | glioblastoma | differentiation | "We examined the microRNA profiles of Glioblastoma ......" | 18765229 | |
| hsa-miR-425-3p | glioblastoma | drug resistance | "An unbiased functional microRNA screen identified ......" | 27399532 | |
| hsa-miR-425-3p | kidney renal cell cancer | poor survival; recurrence | "In the survival analyses the expression levels of ......" | 25981392 | |
| hsa-miR-425-3p | liver cancer | drug resistance; motility; staging | "MicroRNA 425 3p predicts response to sorafenib the ......" | 25040368 | |
| hsa-miR-425-3p | lung cancer | staging | "Based on the results from the screening and valida ......" | 27036025 | |
| hsa-miR-425-3p | lung squamous cell cancer | progression; staging | "With the use of next-generation deep sequencing te ......" | 25813410 | |
| hsa-miR-425-3p | melanoma | staging; poor survival; recurrence | "We recently reported our array-based discovery of ......" | 25155861 | |
| hsa-miR-425-3p | melanoma | metastasis; progression | "miR 425 inhibits melanoma metastasis through repre ......" | 26463631 | Luciferase |
Reported gene related to hsa-miR-425-3p
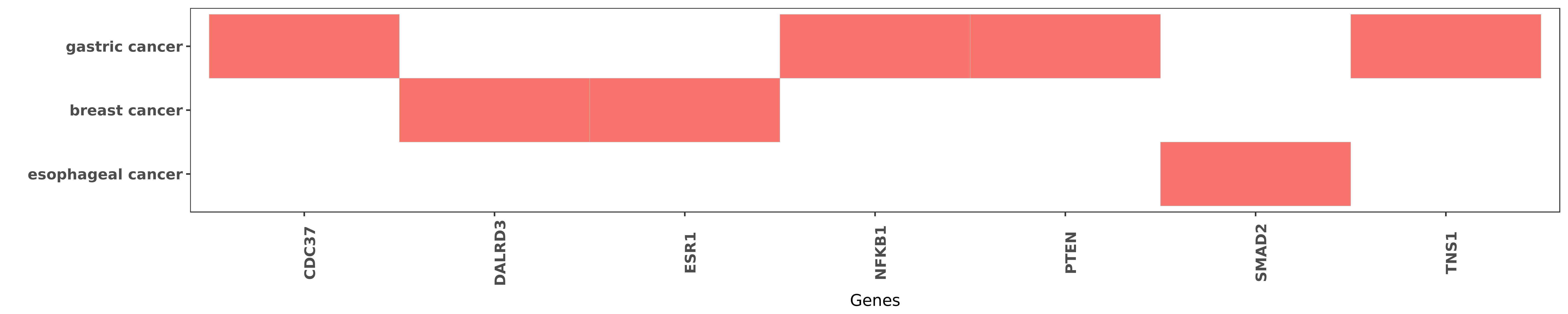
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-425-3p | gastric cancer | NFKB1 | "An increase in miR-425 depended upon IL-1β-induce ......" | 25154996 |
| hsa-miR-425-3p | gastric cancer | NFKB1 | "An increase in miR-425 depends on IL-1β-induced N ......" | 24571667 |
| hsa-miR-425-3p | gastric cancer | CDC37 | "Here we show that IL-1β induces the upregulation ......" | 24571667 |
| hsa-miR-425-3p | breast cancer | DALRD3 | "MiR-191/425 locus characterization revealed that t ......" | 23505378 |
| hsa-miR-425-3p | breast cancer | ESR1 | "MiR-191/425 locus characterization revealed that t ......" | 23505378 |
| hsa-miR-425-3p | gastric cancer | PTEN | "An increase in miR-425 depended upon IL-1β-induce ......" | 25154996 |
| hsa-miR-425-3p | esophageal cancer | SMAD2 | "Enhanced Expression of miR 425 Promotes Esophageal ......" | 26674378 |
| hsa-miR-425-3p | gastric cancer | TNS1 | "Here we show that IL-1β induces the upregulation ......" | 24571667 |
Expression profile in cancer corhorts:

Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-425-3p | MECP2 | 10 cancers: BLCA; BRCA; KIRC; KIRP; LIHC; OV; SARC; THCA; STAD; UCEC | miRNAWalker2 validate; miRNATAP | TCGA BLCA -0.073; TCGA BRCA -0.107; TCGA KIRC -0.057; TCGA KIRP -0.109; TCGA LIHC -0.062; TCGA OV -0.124; TCGA SARC -0.131; TCGA THCA -0.062; TCGA STAD -0.145; TCGA UCEC -0.146 |
hsa-miR-425-3p | DUSP1 | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; STAD; UCEC | MirTarget | TCGA BLCA -0.656; TCGA BRCA -0.399; TCGA COAD -0.341; TCGA ESCA -0.405; TCGA HNSC -0.192; TCGA KIRC -0.334; TCGA KIRP -0.196; TCGA LIHC -0.327; TCGA LUAD -0.342; TCGA LUSC -0.194; TCGA STAD -0.378; TCGA UCEC -0.511 |
hsa-miR-425-3p | FIBIN | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; OV; PAAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.873; TCGA BRCA -0.285; TCGA CESC -0.72; TCGA COAD -0.872; TCGA ESCA -0.701; TCGA HNSC -0.217; TCGA LUAD -0.36; TCGA LUSC -0.466; TCGA OV -0.359; TCGA PAAD -0.557; TCGA SARC -0.3; TCGA STAD -0.451; TCGA UCEC -0.5 |
hsa-miR-425-3p | PER2 | 9 cancers: BLCA; BRCA; ESCA; KIRC; LIHC; OV; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.27; TCGA BRCA -0.238; TCGA ESCA -0.212; TCGA KIRC -0.322; TCGA LIHC -0.097; TCGA OV -0.175; TCGA SARC -0.186; TCGA STAD -0.124; TCGA UCEC -0.186 |
hsa-miR-425-3p | CRISPLD2 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LUAD; LUSC; OV; PAAD; STAD; UCEC | MirTarget | TCGA BLCA -0.615; TCGA BRCA -0.226; TCGA CESC -0.342; TCGA COAD -0.602; TCGA ESCA -0.416; TCGA HNSC -0.265; TCGA LGG -0.158; TCGA LUAD -0.21; TCGA LUSC -0.359; TCGA OV -0.414; TCGA PAAD -0.387; TCGA STAD -0.414; TCGA UCEC -0.448 |
hsa-miR-425-3p | NFIX | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; KIRP; LGG; LIHC; LUAD; PAAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.475; TCGA BRCA -0.234; TCGA CESC -0.272; TCGA COAD -0.122; TCGA ESCA -0.189; TCGA KIRC -0.119; TCGA KIRP -0.217; TCGA LGG -0.093; TCGA LIHC -0.365; TCGA LUAD -0.432; TCGA PAAD -0.28; TCGA SARC -0.187; TCGA THCA -0.142; TCGA STAD -0.331; TCGA UCEC -0.295 |
hsa-miR-425-3p | MARVELD1 | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; OV; STAD; UCEC | mirMAP | TCGA BLCA -0.431; TCGA BRCA -0.124; TCGA COAD -0.218; TCGA ESCA -0.357; TCGA HNSC -0.117; TCGA KIRP -0.254; TCGA LUAD -0.175; TCGA LUSC -0.131; TCGA OV -0.193; TCGA STAD -0.27; TCGA UCEC -0.259 |
hsa-miR-425-3p | KCNS3 | 10 cancers: CESC; COAD; ESCA; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA CESC -0.356; TCGA COAD -0.223; TCGA ESCA -0.619; TCGA LUAD -0.234; TCGA LUSC -0.162; TCGA PAAD -0.422; TCGA PRAD -0.117; TCGA SARC -0.31; TCGA STAD -0.338; TCGA UCEC -0.15 |
Enriched cancer pathways of putative targets