microRNA information: hsa-miR-425-5p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-425-5p | miRbase |
| Accession: | MIMAT0003393 | miRbase |
| Precursor name: | hsa-mir-425 | miRbase |
| Precursor accession: | MI0001448 | miRbase |
| Symbol: | MIR425 | HGNC |
| RefSeq ID: | NR_029948 | GenBank |
| Sequence: | AAUGACACGAUCACUCCCGUUGA |
Reported expression in cancers: hsa-miR-425-5p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-425-5p | breast cancer | deregulation | "Finally eight upregulated miRNAs including hsa-miR ......" | 27446395 | |
| hsa-miR-425-5p | gastric cancer | upregulation | "And the miRecords algorithm and luciferase reporte ......" | 25154996 | |
| hsa-miR-425-5p | gastric cancer | downregulation | "The aim of the current study was to explore the po ......" | 26136868 |
Reported cancer pathway affected by hsa-miR-425-5p

| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-425-5p | gastric cancer | Apoptosis pathway | "NF kappaB dependent microRNA 425 upregulation prom ......" | 24571667 | |
| hsa-miR-425-5p | gastric cancer | cell cycle pathway | "In this study the expression of miR-191 and miR-42 ......" | 24603541 | |
| hsa-miR-425-5p | gastric cancer | Apoptosis pathway | "MiR 425 up regulation induced by interleukin 1β p ......" | 25154996 | Luciferase; Cell proliferation assay; Cell Proliferation Assay; Flow cytometry |
| hsa-miR-425-5p | gastric cancer | Apoptosis pathway | "microRNA 425 5p is upregulated in human gastric ca ......" | 26136868 | Flow cytometry; Colony formation |
| hsa-miR-425-5p | glioblastoma | cell cycle pathway | "An unbiased functional microRNA screen identified ......" | 27399532 | |
| hsa-miR-425-5p | melanoma | Apoptosis pathway | "miR 425 inhibits melanoma metastasis through repre ......" | 26463631 | Luciferase |
Reported cancer prognosis affected by hsa-miR-425-5p
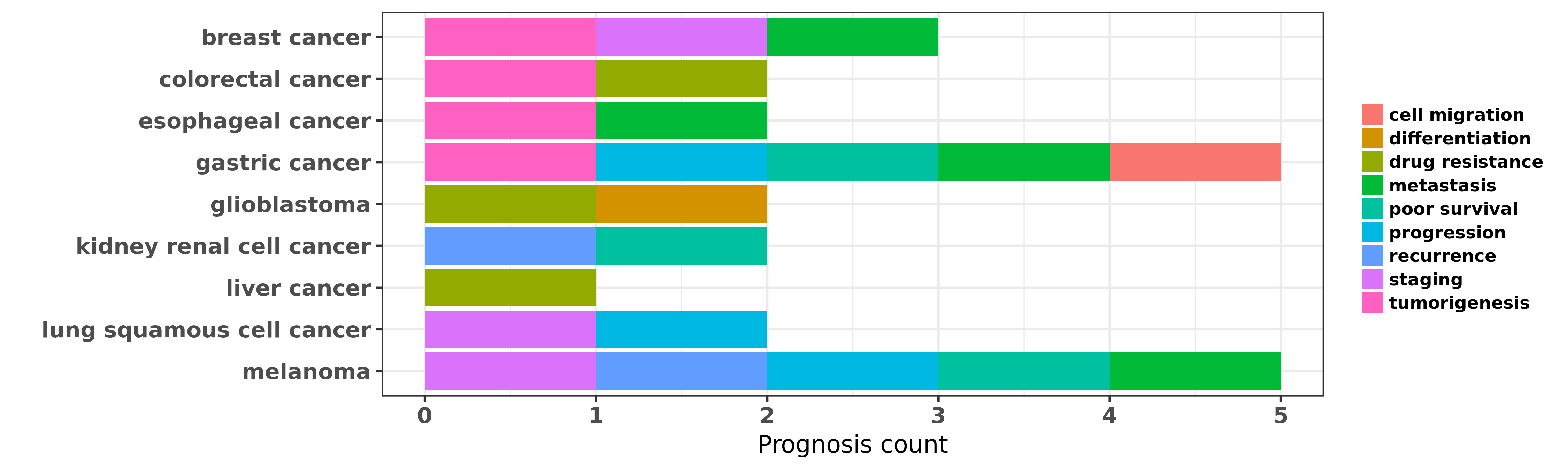
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-425-5p | breast cancer | metastasis; tumorigenesis | "MiR-191/425 locus characterization revealed that t ......" | 23505378 | |
| hsa-miR-425-5p | breast cancer | staging | "Global miRNA analysis was performed on serum from ......" | 24694649 | |
| hsa-miR-425-5p | colorectal cancer | drug resistance; tumorigenesis | "MicroRNA 425 5p regulates chemoresistance in color ......" | 26647742 | Western blot; Luciferase |
| hsa-miR-425-5p | esophageal cancer | tumorigenesis; metastasis | "Enhanced Expression of miR 425 Promotes Esophageal ......" | 26674378 | Colony formation |
| hsa-miR-425-5p | gastric cancer | poor survival | "NF kappaB dependent microRNA 425 upregulation prom ......" | 24571667 | |
| hsa-miR-425-5p | gastric cancer | progression; cell migration; tumorigenesis | "In this study the expression of miR-191 and miR-42 ......" | 24603541 | |
| hsa-miR-425-5p | gastric cancer | metastasis | "microRNA 425 5p is upregulated in human gastric ca ......" | 26136868 | Flow cytometry; Colony formation |
| hsa-miR-425-5p | glioblastoma | differentiation | "We examined the microRNA profiles of Glioblastoma ......" | 18765229 | |
| hsa-miR-425-5p | glioblastoma | drug resistance | "An unbiased functional microRNA screen identified ......" | 27399532 | |
| hsa-miR-425-5p | kidney renal cell cancer | poor survival; recurrence | "In the survival analyses the expression levels of ......" | 25981392 | |
| hsa-miR-425-5p | liver cancer | drug resistance | "MicroRNA 425 3p predicts response to sorafenib the ......" | 25040368 | |
| hsa-miR-425-5p | lung squamous cell cancer | progression; staging | "With the use of next-generation deep sequencing te ......" | 25813410 | |
| hsa-miR-425-5p | melanoma | staging; poor survival; recurrence | "We recently reported our array-based discovery of ......" | 25155861 | |
| hsa-miR-425-5p | melanoma | metastasis; progression | "miR 425 inhibits melanoma metastasis through repre ......" | 26463631 | Luciferase |
Reported gene related to hsa-miR-425-5p

| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-425-5p | gastric cancer | NFKB1 | "An increase in miR-425 depended upon IL-1β-induce ......" | 25154996 |
| hsa-miR-425-5p | gastric cancer | NFKB1 | "An increase in miR-425 depends on IL-1β-induced N ......" | 24571667 |
| hsa-miR-425-5p | gastric cancer | CDC37 | "Here we show that IL-1β induces the upregulation ......" | 24571667 |
| hsa-miR-425-5p | breast cancer | DALRD3 | "MiR-191/425 locus characterization revealed that t ......" | 23505378 |
| hsa-miR-425-5p | breast cancer | ESR1 | "MiR-191/425 locus characterization revealed that t ......" | 23505378 |
| hsa-miR-425-5p | colorectal cancer | PDCD10 | "Programmed cell death 10 PDCD10 is the direct targ ......" | 26647742 |
| hsa-miR-425-5p | gastric cancer | PTEN | "An increase in miR-425 depended upon IL-1β-induce ......" | 25154996 |
| hsa-miR-425-5p | esophageal cancer | SMAD2 | "Enhanced Expression of miR 425 Promotes Esophageal ......" | 26674378 |
| hsa-miR-425-5p | gastric cancer | TNS1 | "Here we show that IL-1β induces the upregulation ......" | 24571667 |
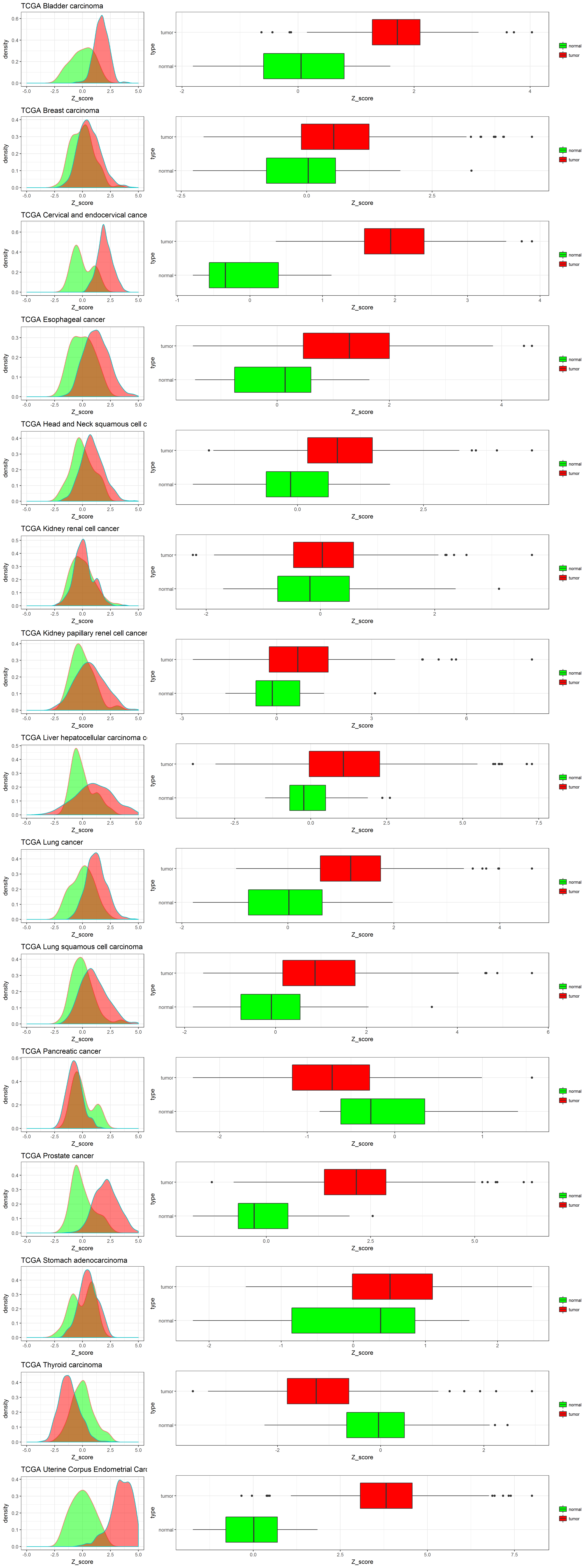
Expression profile in cancer corhorts:
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-425-5p | CELF2 | 13 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LUAD; LUSC; OV; PAAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate; miRNATAP | TCGA BLCA -0.655; TCGA BRCA -0.14; TCGA CESC -0.162; TCGA ESCA -0.537; TCGA HNSC -0.218; TCGA LUAD -0.457; TCGA LUSC -0.401; TCGA OV -0.162; TCGA PAAD -0.384; TCGA PRAD -0.445; TCGA SARC -0.21; TCGA STAD -0.482; TCGA UCEC -0.372 |
hsa-miR-425-5p | CLU | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.683; TCGA BRCA -0.193; TCGA CESC -0.308; TCGA COAD -0.639; TCGA ESCA -0.355; TCGA HNSC -0.404; TCGA KIRC -0.176; TCGA LIHC -0.313; TCGA LUAD -0.302; TCGA LUSC -0.333; TCGA PAAD -0.496; TCGA PRAD -0.453; TCGA SARC -0.761; TCGA STAD -0.417; TCGA UCEC -0.266 |
hsa-miR-425-5p | DPYSL2 | 11 cancers: BLCA; BRCA; COAD; LIHC; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.447; TCGA BRCA -0.168; TCGA COAD -0.238; TCGA LIHC -0.108; TCGA LUAD -0.415; TCGA LUSC -0.382; TCGA OV -0.272; TCGA PAAD -0.154; TCGA PRAD -0.158; TCGA STAD -0.212; TCGA UCEC -0.277 |
hsa-miR-425-5p | FAM122A | 15 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.119; TCGA BRCA -0.084; TCGA CESC -0.063; TCGA ESCA -0.139; TCGA HNSC -0.055; TCGA KIRC -0.074; TCGA KIRP -0.176; TCGA LIHC -0.209; TCGA LUAD -0.118; TCGA LUSC -0.1; TCGA OV -0.116; TCGA PAAD -0.1; TCGA PRAD -0.126; TCGA STAD -0.093; TCGA UCEC -0.17 |
hsa-miR-425-5p | GNB5 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; KIRP; LUAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.191; TCGA BRCA -0.128; TCGA CESC -0.095; TCGA COAD -0.287; TCGA ESCA -0.176; TCGA KIRC -0.086; TCGA KIRP -0.2; TCGA LUAD -0.053; TCGA PRAD -0.194; TCGA SARC -0.183; TCGA STAD -0.197; TCGA UCEC -0.093 |
hsa-miR-425-5p | PPP2CB | 14 cancers: BLCA; BRCA; CESC; COAD; KIRC; KIRP; LIHC; LUAD; LUSC; OV; PAAD; PRAD; SARC; UCEC | miRNAWalker2 validate | TCGA BLCA -0.185; TCGA BRCA -0.143; TCGA CESC -0.111; TCGA COAD -0.081; TCGA KIRC -0.066; TCGA KIRP -0.132; TCGA LIHC -0.21; TCGA LUAD -0.23; TCGA LUSC -0.118; TCGA OV -0.231; TCGA PAAD -0.111; TCGA PRAD -0.117; TCGA SARC -0.115; TCGA UCEC -0.175 |
hsa-miR-425-5p | QKI | 16 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.132; TCGA BRCA -0.092; TCGA CESC -0.107; TCGA COAD -0.453; TCGA ESCA -0.27; TCGA HNSC -0.072; TCGA KIRP -0.169; TCGA LGG -0.12; TCGA LIHC -0.086; TCGA LUAD -0.17; TCGA LUSC -0.177; TCGA OV -0.106; TCGA PAAD -0.193; TCGA PRAD -0.072; TCGA STAD -0.294; TCGA UCEC -0.159 |
hsa-miR-425-5p | TAOK1 | 9 cancers: BLCA; COAD; HNSC; KIRP; LGG; LUAD; PRAD; SARC; STAD | miRNAWalker2 validate | TCGA BLCA -0.106; TCGA COAD -0.23; TCGA HNSC -0.208; TCGA KIRP -0.503; TCGA LGG -0.186; TCGA LUAD -0.103; TCGA PRAD -0.358; TCGA SARC -0.254; TCGA STAD -0.086 |
hsa-miR-425-5p | WASF2 | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; OV; PRAD; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.19; TCGA BRCA -0.089; TCGA COAD -0.097; TCGA ESCA -0.165; TCGA HNSC -0.051; TCGA KIRC -0.053; TCGA KIRP -0.105; TCGA LUAD -0.063; TCGA OV -0.091; TCGA PRAD -0.084; TCGA STAD -0.12; TCGA UCEC -0.162 |
hsa-miR-425-5p | AFF4 | 13 cancers: BLCA; COAD; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; OV; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.1; TCGA COAD -0.071; TCGA HNSC -0.093; TCGA KIRP -0.201; TCGA LGG -0.113; TCGA LIHC -0.184; TCGA LUAD -0.104; TCGA LUSC -0.098; TCGA OV -0.081; TCGA PRAD -0.13; TCGA SARC -0.171; TCGA STAD -0.144; TCGA UCEC -0.104 |
hsa-miR-425-5p | CBL | 10 cancers: BLCA; COAD; KIRP; LGG; LUAD; OV; PRAD; THCA; STAD; UCEC | MirTarget; mirMAP | TCGA BLCA -0.079; TCGA COAD -0.081; TCGA KIRP -0.135; TCGA LGG -0.096; TCGA LUAD -0.086; TCGA OV -0.078; TCGA PRAD -0.085; TCGA THCA -0.061; TCGA STAD -0.091; TCGA UCEC -0.101 |
hsa-miR-425-5p | RNF11 | 11 cancers: BLCA; COAD; ESCA; KIRP; LIHC; LUAD; LUSC; PRAD; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.082; TCGA COAD -0.128; TCGA ESCA -0.14; TCGA KIRP -0.129; TCGA LIHC -0.125; TCGA LUAD -0.14; TCGA LUSC -0.051; TCGA PRAD -0.078; TCGA THCA -0.066; TCGA STAD -0.164; TCGA UCEC -0.148 |
hsa-miR-425-5p | HERC2 | 11 cancers: BLCA; BRCA; HNSC; KIRC; KIRP; LIHC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.08; TCGA BRCA -0.066; TCGA HNSC -0.097; TCGA KIRC -0.085; TCGA KIRP -0.14; TCGA LIHC -0.09; TCGA PAAD -0.127; TCGA PRAD -0.095; TCGA SARC -0.13; TCGA STAD -0.052; TCGA UCEC -0.058 |
hsa-miR-425-5p | KIAA2026 | 14 cancers: BLCA; BRCA; ESCA; KIRC; KIRP; LGG; LIHC; LUAD; OV; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.068; TCGA BRCA -0.054; TCGA ESCA -0.117; TCGA KIRC -0.053; TCGA KIRP -0.19; TCGA LGG -0.127; TCGA LIHC -0.128; TCGA LUAD -0.111; TCGA OV -0.132; TCGA PAAD -0.132; TCGA PRAD -0.15; TCGA SARC -0.119; TCGA STAD -0.094; TCGA UCEC -0.122 |
hsa-miR-425-5p | EDIL3 | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.457; TCGA BRCA -0.197; TCGA CESC -0.328; TCGA COAD -0.685; TCGA ESCA -0.292; TCGA HNSC -0.528; TCGA KIRP -0.453; TCGA LUAD -0.45; TCGA LUSC -0.652; TCGA PAAD -0.342; TCGA PRAD -0.55; TCGA THCA -0.21; TCGA STAD -0.268; TCGA UCEC -0.517 |
hsa-miR-425-5p | TBCEL | 13 cancers: BLCA; COAD; ESCA; HNSC; KIRP; LGG; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.083; TCGA COAD -0.2; TCGA ESCA -0.227; TCGA HNSC -0.076; TCGA KIRP -0.196; TCGA LGG -0.128; TCGA LUAD -0.179; TCGA LUSC -0.125; TCGA PAAD -0.336; TCGA PRAD -0.099; TCGA SARC -0.16; TCGA STAD -0.167; TCGA UCEC -0.067 |
hsa-miR-425-5p | ANKRD40 | 11 cancers: BLCA; ESCA; HNSC; KIRP; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.069; TCGA ESCA -0.077; TCGA HNSC -0.086; TCGA KIRP -0.091; TCGA LUAD -0.111; TCGA LUSC -0.062; TCGA PAAD -0.103; TCGA PRAD -0.079; TCGA SARC -0.138; TCGA STAD -0.172; TCGA UCEC -0.118 |
hsa-miR-425-5p | CYGB | 11 cancers: BLCA; BRCA; COAD; LIHC; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.222; TCGA BRCA -0.195; TCGA COAD -0.261; TCGA LIHC -0.304; TCGA LUAD -0.125; TCGA LUSC -0.256; TCGA OV -0.172; TCGA PAAD -0.206; TCGA PRAD -0.073; TCGA STAD -0.109; TCGA UCEC -0.146 |
hsa-miR-425-5p | ITGB3 | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.675; TCGA BRCA -0.162; TCGA COAD -0.614; TCGA ESCA -0.221; TCGA HNSC -0.335; TCGA KIRP -0.458; TCGA LUAD -0.336; TCGA LUSC -0.482; TCGA PRAD -0.6; TCGA SARC -0.287; TCGA STAD -0.324; TCGA UCEC -0.329 |
hsa-miR-425-5p | RYR2 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.7; TCGA BRCA -0.155; TCGA CESC -0.226; TCGA COAD -0.621; TCGA ESCA -0.844; TCGA HNSC -0.209; TCGA LUAD -0.456; TCGA LUSC -0.198; TCGA PAAD -0.642; TCGA PRAD -0.303; TCGA SARC -0.704; TCGA STAD -0.744; TCGA UCEC -0.266 |
hsa-miR-425-5p | KIAA0430 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; OV; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.115; TCGA BRCA -0.063; TCGA CESC -0.087; TCGA COAD -0.1; TCGA ESCA -0.124; TCGA HNSC -0.103; TCGA KIRP -0.112; TCGA LIHC -0.192; TCGA LUAD -0.155; TCGA LUSC -0.068; TCGA OV -0.083; TCGA PRAD -0.153; TCGA SARC -0.209; TCGA STAD -0.173; TCGA UCEC -0.147 |
hsa-miR-425-5p | ZNF423 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.152; TCGA BRCA -0.233; TCGA CESC -0.196; TCGA COAD -0.296; TCGA ESCA -0.433; TCGA HNSC -0.446; TCGA LGG -0.178; TCGA LIHC -0.329; TCGA LUAD -0.341; TCGA LUSC -0.505; TCGA OV -0.329; TCGA PAAD -0.477; TCGA PRAD -0.375; TCGA STAD -0.487; TCGA UCEC -0.181 |
hsa-miR-425-5p | PAFAH1B1 | 14 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.137; TCGA BRCA -0.06; TCGA COAD -0.136; TCGA ESCA -0.168; TCGA HNSC -0.051; TCGA KIRP -0.144; TCGA LIHC -0.197; TCGA LUAD -0.14; TCGA LUSC -0.121; TCGA PAAD -0.154; TCGA PRAD -0.13; TCGA SARC -0.073; TCGA STAD -0.098; TCGA UCEC -0.095 |
hsa-miR-425-5p | RGS13 | 9 cancers: BLCA; BRCA; COAD; ESCA; LUAD; LUSC; PRAD; SARC; STAD | MirTarget | TCGA BLCA -0.592; TCGA BRCA -0.194; TCGA COAD -0.369; TCGA ESCA -0.29; TCGA LUAD -0.502; TCGA LUSC -0.458; TCGA PRAD -0.41; TCGA SARC -0.354; TCGA STAD -0.3 |
hsa-miR-425-5p | AMPH | 11 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; PAAD; PRAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.452; TCGA CESC -0.498; TCGA COAD -0.665; TCGA ESCA -0.416; TCGA HNSC -0.257; TCGA KIRC -0.24; TCGA KIRP -0.523; TCGA PAAD -0.607; TCGA PRAD -0.3; TCGA STAD -0.416; TCGA UCEC -0.457 |
hsa-miR-425-5p | DIP2C | 16 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUSC; OV; PAAD; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.173; TCGA BRCA -0.078; TCGA CESC -0.112; TCGA COAD -0.379; TCGA ESCA -0.254; TCGA HNSC -0.182; TCGA KIRC -0.187; TCGA KIRP -0.163; TCGA LIHC -0.453; TCGA LUSC -0.147; TCGA OV -0.223; TCGA PAAD -0.314; TCGA PRAD -0.168; TCGA SARC -0.27; TCGA STAD -0.32; TCGA UCEC -0.227 |
hsa-miR-425-5p | PCDHB5 | 9 cancers: BLCA; BRCA; CESC; COAD; HNSC; LUSC; PAAD; STAD; UCEC | MirTarget | TCGA BLCA -0.211; TCGA BRCA -0.21; TCGA CESC -0.207; TCGA COAD -0.647; TCGA HNSC -0.416; TCGA LUSC -0.216; TCGA PAAD -0.408; TCGA STAD -0.542; TCGA UCEC -0.423 |
hsa-miR-425-5p | HSPB8 | 13 cancers: BLCA; CESC; COAD; ESCA; HNSC; LGG; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.757; TCGA CESC -0.224; TCGA COAD -0.94; TCGA ESCA -0.789; TCGA HNSC -0.275; TCGA LGG -0.142; TCGA LUAD -0.316; TCGA LUSC -0.362; TCGA PAAD -0.33; TCGA PRAD -0.742; TCGA SARC -0.92; TCGA STAD -0.882; TCGA UCEC -0.448 |
hsa-miR-425-5p | DNAJC27 | 10 cancers: BLCA; BRCA; KIRC; KIRP; LIHC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.073; TCGA BRCA -0.085; TCGA KIRC -0.131; TCGA KIRP -0.269; TCGA LIHC -0.207; TCGA PAAD -0.196; TCGA PRAD -0.119; TCGA SARC -0.173; TCGA STAD -0.156; TCGA UCEC -0.152 |
hsa-miR-425-5p | GRIA3 | 11 cancers: BLCA; BRCA; CESC; ESCA; LGG; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.509; TCGA BRCA -0.211; TCGA CESC -0.23; TCGA ESCA -0.38; TCGA LGG -0.176; TCGA LUSC -0.35; TCGA PAAD -0.657; TCGA PRAD -0.621; TCGA SARC -0.649; TCGA STAD -0.298; TCGA UCEC -0.583 |
hsa-miR-425-5p | KLHL13 | 11 cancers: BLCA; BRCA; COAD; ESCA; KIRP; LGG; PAAD; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.623; TCGA BRCA -0.259; TCGA COAD -0.324; TCGA ESCA -0.449; TCGA KIRP -0.689; TCGA LGG -0.208; TCGA PAAD -0.289; TCGA PRAD -0.383; TCGA SARC -0.618; TCGA STAD -0.46; TCGA UCEC -0.269 |
hsa-miR-425-5p | MEF2C | 14 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.414; TCGA BRCA -0.154; TCGA CESC -0.327; TCGA COAD -0.436; TCGA ESCA -0.217; TCGA HNSC -0.378; TCGA LIHC -0.178; TCGA LUAD -0.414; TCGA LUSC -0.398; TCGA PAAD -0.337; TCGA PRAD -0.217; TCGA SARC -0.229; TCGA STAD -0.318; TCGA UCEC -0.455 |
hsa-miR-425-5p | PNMAL1 | 13 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.554; TCGA BRCA -0.199; TCGA COAD -0.816; TCGA ESCA -0.439; TCGA HNSC -0.462; TCGA KIRP -0.51; TCGA LUAD -0.235; TCGA LUSC -0.374; TCGA OV -0.18; TCGA PAAD -0.616; TCGA PRAD -0.462; TCGA STAD -0.626; TCGA UCEC -0.149 |
hsa-miR-425-5p | CYYR1 | 14 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; LIHC; LUAD; LUSC; PAAD; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.21; TCGA BRCA -0.217; TCGA CESC -0.233; TCGA COAD -0.42; TCGA HNSC -0.154; TCGA KIRC -0.14; TCGA LIHC -0.326; TCGA LUAD -0.322; TCGA LUSC -0.307; TCGA PAAD -0.38; TCGA PRAD -0.163; TCGA THCA -0.145; TCGA STAD -0.24; TCGA UCEC -0.379 |
hsa-miR-425-5p | KCNT2 | 13 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LGG; LUSC; OV; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.394; TCGA BRCA -0.229; TCGA CESC -0.224; TCGA ESCA -0.828; TCGA HNSC -0.335; TCGA LGG -0.295; TCGA LUSC -0.41; TCGA OV -0.354; TCGA PAAD -0.542; TCGA PRAD -0.404; TCGA SARC -0.293; TCGA STAD -0.601; TCGA UCEC -0.42 |
hsa-miR-425-5p | PPM1L | 9 cancers: BLCA; COAD; ESCA; HNSC; KIRP; LGG; PRAD; SARC; STAD | MirTarget | TCGA BLCA -0.159; TCGA COAD -0.154; TCGA ESCA -0.263; TCGA HNSC -0.32; TCGA KIRP -0.342; TCGA LGG -0.158; TCGA PRAD -0.404; TCGA SARC -0.36; TCGA STAD -0.27 |
hsa-miR-425-5p | IFFO2 | 10 cancers: BLCA; BRCA; ESCA; LUAD; LUSC; OV; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.189; TCGA BRCA -0.198; TCGA ESCA -0.313; TCGA LUAD -0.157; TCGA LUSC -0.255; TCGA OV -0.205; TCGA PRAD -0.132; TCGA THCA -0.134; TCGA STAD -0.115; TCGA UCEC -0.13 |
hsa-miR-425-5p | PDS5B | 10 cancers: BLCA; HNSC; KIRP; LIHC; LUAD; LUSC; OV; PAAD; PRAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.124; TCGA HNSC -0.073; TCGA KIRP -0.236; TCGA LIHC -0.07; TCGA LUAD -0.088; TCGA LUSC -0.127; TCGA OV -0.16; TCGA PAAD -0.108; TCGA PRAD -0.165; TCGA UCEC -0.184 |
hsa-miR-425-5p | WWC3 | 10 cancers: BLCA; COAD; ESCA; HNSC; LUAD; LUSC; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.233; TCGA COAD -0.084; TCGA ESCA -0.321; TCGA HNSC -0.078; TCGA LUAD -0.16; TCGA LUSC -0.175; TCGA PRAD -0.177; TCGA THCA -0.053; TCGA STAD -0.296; TCGA UCEC -0.177 |
hsa-miR-425-5p | MAN1A1 | 10 cancers: BLCA; BRCA; HNSC; KIRP; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.137; TCGA BRCA -0.08; TCGA HNSC -0.281; TCGA KIRP -0.282; TCGA LUAD -0.181; TCGA LUSC -0.453; TCGA PAAD -0.342; TCGA PRAD -0.255; TCGA STAD -0.126; TCGA UCEC -0.306 |
hsa-miR-425-5p | NECAB1 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRP; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.477; TCGA BRCA -0.19; TCGA CESC -0.436; TCGA COAD -0.722; TCGA ESCA -0.702; TCGA KIRP -0.478; TCGA LUAD -0.417; TCGA LUSC -0.309; TCGA PAAD -0.368; TCGA PRAD -0.601; TCGA SARC -0.315; TCGA STAD -0.821; TCGA UCEC -0.647 |
hsa-miR-425-5p | TNC | 11 cancers: BLCA; BRCA; COAD; ESCA; KIRP; LUAD; LUSC; OV; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.773; TCGA BRCA -0.118; TCGA COAD -0.851; TCGA ESCA -0.773; TCGA KIRP -0.511; TCGA LUAD -0.18; TCGA LUSC -0.332; TCGA OV -0.221; TCGA PRAD -0.325; TCGA STAD -0.497; TCGA UCEC -0.126 |
hsa-miR-425-5p | ZMAT3 | 13 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.225; TCGA BRCA -0.118; TCGA COAD -0.271; TCGA ESCA -0.202; TCGA HNSC -0.249; TCGA LGG -0.176; TCGA LIHC -0.339; TCGA LUAD -0.24; TCGA LUSC -0.22; TCGA PAAD -0.333; TCGA PRAD -0.311; TCGA STAD -0.159; TCGA UCEC -0.081 |
hsa-miR-425-5p | ITPR1 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.351; TCGA BRCA -0.152; TCGA CESC -0.2; TCGA COAD -0.458; TCGA ESCA -0.473; TCGA HNSC -0.202; TCGA KIRP -0.323; TCGA LIHC -0.163; TCGA LUAD -0.195; TCGA LUSC -0.338; TCGA PAAD -0.253; TCGA PRAD -0.58; TCGA SARC -0.511; TCGA STAD -0.378; TCGA UCEC -0.428 |
hsa-miR-425-5p | AJAP1 | 9 cancers: BLCA; BRCA; KIRC; LUAD; LUSC; OV; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.193; TCGA BRCA -0.155; TCGA KIRC -0.202; TCGA LUAD -0.289; TCGA LUSC -0.568; TCGA OV -0.513; TCGA PRAD -0.452; TCGA STAD -0.222; TCGA UCEC -0.452 |
hsa-miR-425-5p | ANTXR1 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.406; TCGA BRCA -0.068; TCGA CESC -0.245; TCGA COAD -0.814; TCGA ESCA -0.354; TCGA HNSC -0.221; TCGA LGG -0.212; TCGA LIHC -0.217; TCGA LUAD -0.265; TCGA LUSC -0.457; TCGA OV -0.305; TCGA PAAD -0.288; TCGA PRAD -0.289; TCGA STAD -0.347; TCGA UCEC -0.333 |
hsa-miR-425-5p | LDOC1L | 16 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; OV; PAAD; PRAD; SARC; STAD; UCEC | MirTarget; mirMAP | TCGA BLCA -0.194; TCGA BRCA -0.076; TCGA COAD -0.12; TCGA ESCA -0.133; TCGA HNSC -0.143; TCGA KIRC -0.085; TCGA KIRP -0.184; TCGA LGG -0.136; TCGA LIHC -0.143; TCGA LUAD -0.115; TCGA OV -0.121; TCGA PAAD -0.274; TCGA PRAD -0.106; TCGA SARC -0.161; TCGA STAD -0.053; TCGA UCEC -0.147 |
hsa-miR-425-5p | ATG2B | 10 cancers: BLCA; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.074; TCGA COAD -0.076; TCGA ESCA -0.106; TCGA HNSC -0.099; TCGA KIRC -0.085; TCGA KIRP -0.166; TCGA LIHC -0.117; TCGA SARC -0.081; TCGA STAD -0.054; TCGA UCEC -0.105 |
hsa-miR-425-5p | TNS1 | 16 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; OV; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.887; TCGA BRCA -0.24; TCGA CESC -0.254; TCGA COAD -0.88; TCGA ESCA -0.818; TCGA HNSC -0.461; TCGA KIRC -0.13; TCGA KIRP -0.257; TCGA LUAD -0.449; TCGA LUSC -0.524; TCGA OV -0.231; TCGA PAAD -0.473; TCGA PRAD -0.597; TCGA SARC -0.731; TCGA STAD -0.76; TCGA UCEC -0.498 |
hsa-miR-425-5p | ERC1 | 12 cancers: BLCA; BRCA; COAD; ESCA; KIRP; LGG; LIHC; LUAD; PAAD; PRAD; SARC; STAD | mirMAP | TCGA BLCA -0.291; TCGA BRCA -0.074; TCGA COAD -0.118; TCGA ESCA -0.225; TCGA KIRP -0.129; TCGA LGG -0.107; TCGA LIHC -0.086; TCGA LUAD -0.103; TCGA PAAD -0.159; TCGA PRAD -0.243; TCGA SARC -0.155; TCGA STAD -0.161 |
hsa-miR-425-5p | WDR7 | 14 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.124; TCGA CESC -0.097; TCGA COAD -0.179; TCGA ESCA -0.158; TCGA HNSC -0.118; TCGA KIRP -0.071; TCGA LIHC -0.186; TCGA LUAD -0.099; TCGA LUSC -0.087; TCGA PAAD -0.233; TCGA PRAD -0.172; TCGA SARC -0.118; TCGA STAD -0.106; TCGA UCEC -0.084 |
hsa-miR-425-5p | XYLT1 | 12 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LGG; LIHC; LUSC; OV; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.272; TCGA BRCA -0.102; TCGA CESC -0.263; TCGA ESCA -0.174; TCGA HNSC -0.357; TCGA LGG -0.108; TCGA LIHC -0.141; TCGA LUSC -0.416; TCGA OV -0.25; TCGA PRAD -0.18; TCGA STAD -0.155; TCGA UCEC -0.468 |
hsa-miR-425-5p | KCTD20 | 10 cancers: BLCA; COAD; HNSC; KIRP; LGG; LUAD; LUSC; PAAD; STAD; UCEC | mirMAP | TCGA BLCA -0.177; TCGA COAD -0.098; TCGA HNSC -0.147; TCGA KIRP -0.193; TCGA LGG -0.127; TCGA LUAD -0.094; TCGA LUSC -0.105; TCGA PAAD -0.186; TCGA STAD -0.128; TCGA UCEC -0.122 |
hsa-miR-425-5p | VPS13B | 10 cancers: BLCA; COAD; ESCA; HNSC; KIRC; KIRP; LGG; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.064; TCGA COAD -0.08; TCGA ESCA -0.083; TCGA HNSC -0.087; TCGA KIRC -0.079; TCGA KIRP -0.251; TCGA LGG -0.148; TCGA SARC -0.194; TCGA STAD -0.071; TCGA UCEC -0.144 |
hsa-miR-425-5p | CHL1 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.483; TCGA BRCA -0.414; TCGA CESC -0.379; TCGA COAD -0.833; TCGA ESCA -0.726; TCGA HNSC -0.376; TCGA KIRP -0.415; TCGA PAAD -0.645; TCGA PRAD -0.271; TCGA STAD -0.494; TCGA UCEC -0.241 |
hsa-miR-425-5p | DTNA | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.825; TCGA CESC -0.204; TCGA COAD -0.714; TCGA ESCA -0.691; TCGA HNSC -0.324; TCGA KIRP -0.24; TCGA LUSC -0.212; TCGA PAAD -0.567; TCGA PRAD -0.347; TCGA SARC -0.504; TCGA STAD -0.711; TCGA UCEC -0.391 |
hsa-miR-425-5p | KIAA0513 | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; OV; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.404; TCGA CESC -0.122; TCGA COAD -0.155; TCGA ESCA -0.396; TCGA HNSC -0.133; TCGA LIHC -0.104; TCGA LUAD -0.175; TCGA LUSC -0.335; TCGA OV -0.12; TCGA PRAD -0.265; TCGA STAD -0.24; TCGA UCEC -0.127 |
hsa-miR-425-5p | KIAA1614 | 14 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LUSC; OV; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.487; TCGA BRCA -0.296; TCGA COAD -0.384; TCGA ESCA -0.517; TCGA HNSC -0.202; TCGA KIRC -0.22; TCGA KIRP -0.422; TCGA LUSC -0.206; TCGA OV -0.127; TCGA PAAD -0.385; TCGA PRAD -0.502; TCGA SARC -0.313; TCGA STAD -0.375; TCGA UCEC -0.255 |
hsa-miR-425-5p | SMAD4 | 16 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.082; TCGA BRCA -0.074; TCGA CESC -0.096; TCGA COAD -0.131; TCGA ESCA -0.105; TCGA HNSC -0.066; TCGA KIRC -0.057; TCGA KIRP -0.136; TCGA LGG -0.11; TCGA LIHC -0.055; TCGA LUAD -0.093; TCGA PAAD -0.267; TCGA PRAD -0.142; TCGA SARC -0.162; TCGA STAD -0.084; TCGA UCEC -0.149 |
hsa-miR-425-5p | WTIP | 11 cancers: BLCA; BRCA; COAD; ESCA; KIRP; LUAD; LUSC; OV; PAAD; STAD; UCEC | mirMAP | TCGA BLCA -0.114; TCGA BRCA -0.199; TCGA COAD -0.428; TCGA ESCA -0.229; TCGA KIRP -0.164; TCGA LUAD -0.193; TCGA LUSC -0.156; TCGA OV -0.176; TCGA PAAD -0.219; TCGA STAD -0.268; TCGA UCEC -0.139 |
hsa-miR-425-5p | SSBP2 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.309; TCGA BRCA -0.053; TCGA CESC -0.214; TCGA COAD -0.475; TCGA ESCA -0.461; TCGA HNSC -0.099; TCGA KIRP -0.309; TCGA LUAD -0.219; TCGA PRAD -0.17; TCGA SARC -0.288; TCGA THCA -0.202; TCGA STAD -0.428; TCGA UCEC -0.23 |
hsa-miR-425-5p | GPR107 | 9 cancers: BLCA; COAD; ESCA; HNSC; KIRC; KIRP; LUSC; SARC; UCEC | mirMAP | TCGA BLCA -0.052; TCGA COAD -0.064; TCGA ESCA -0.079; TCGA HNSC -0.086; TCGA KIRC -0.057; TCGA KIRP -0.134; TCGA LUSC -0.119; TCGA SARC -0.071; TCGA UCEC -0.056 |
hsa-miR-425-5p | EIF4EBP2 | 11 cancers: BLCA; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; OV; PAAD; PRAD; STAD | mirMAP | TCGA BLCA -0.076; TCGA KIRC -0.088; TCGA KIRP -0.104; TCGA LGG -0.094; TCGA LIHC -0.081; TCGA LUAD -0.162; TCGA LUSC -0.127; TCGA OV -0.078; TCGA PAAD -0.078; TCGA PRAD -0.098; TCGA STAD -0.068 |
hsa-miR-425-5p | KCTD15 | 12 cancers: BLCA; CESC; COAD; ESCA; KIRC; LGG; LIHC; OV; PAAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.402; TCGA CESC -0.186; TCGA COAD -0.388; TCGA ESCA -0.402; TCGA KIRC -0.12; TCGA LGG -0.05; TCGA LIHC -0.231; TCGA OV -0.151; TCGA PAAD -0.131; TCGA SARC -0.308; TCGA STAD -0.291; TCGA UCEC -0.33 |
hsa-miR-425-5p | HHIP | 9 cancers: BLCA; COAD; HNSC; KIRP; LIHC; LUAD; LUSC; OV; PRAD | mirMAP | TCGA BLCA -0.34; TCGA COAD -0.484; TCGA HNSC -0.322; TCGA KIRP -0.42; TCGA LIHC -0.79; TCGA LUAD -0.869; TCGA LUSC -0.568; TCGA OV -0.302; TCGA PRAD -0.562 |
hsa-miR-425-5p | MECP2 | 15 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; OV; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.104; TCGA BRCA -0.093; TCGA COAD -0.055; TCGA ESCA -0.124; TCGA HNSC -0.068; TCGA KIRC -0.086; TCGA KIRP -0.184; TCGA LIHC -0.052; TCGA LUAD -0.077; TCGA OV -0.147; TCGA PAAD -0.086; TCGA PRAD -0.128; TCGA SARC -0.16; TCGA STAD -0.163; TCGA UCEC -0.162 |
hsa-miR-425-5p | LONRF2 | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LIHC; PAAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BLCA -0.329; TCGA CESC -0.607; TCGA COAD -1.407; TCGA ESCA -0.683; TCGA HNSC -0.366; TCGA KIRP -0.781; TCGA LIHC -0.42; TCGA PAAD -0.582; TCGA SARC -1.253; TCGA THCA -0.4; TCGA STAD -0.775; TCGA UCEC -0.494 |
hsa-miR-425-5p | LRRC15 | 9 cancers: BLCA; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; OV; UCEC | mirMAP | TCGA BLCA -0.596; TCGA COAD -0.649; TCGA ESCA -0.511; TCGA HNSC -0.362; TCGA LIHC -0.288; TCGA LUAD -0.257; TCGA LUSC -0.671; TCGA OV -0.409; TCGA UCEC -0.333 |
hsa-miR-425-5p | SCN4B | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.426; TCGA BRCA -0.375; TCGA CESC -0.301; TCGA COAD -0.544; TCGA ESCA -0.749; TCGA HNSC -0.154; TCGA KIRC -0.214; TCGA LIHC -0.186; TCGA LUAD -0.544; TCGA LUSC -0.33; TCGA PAAD -0.562; TCGA PRAD -0.305; TCGA SARC -0.352; TCGA STAD -0.507; TCGA UCEC -0.531 |
hsa-miR-425-5p | WSCD1 | 12 cancers: BLCA; BRCA; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.307; TCGA BRCA -0.224; TCGA HNSC -0.3; TCGA KIRC -0.296; TCGA KIRP -0.259; TCGA LGG -0.139; TCGA LUAD -0.141; TCGA LUSC -0.224; TCGA PAAD -0.216; TCGA PRAD -0.095; TCGA STAD -0.223; TCGA UCEC -0.331 |
hsa-miR-425-5p | TEAD1 | 15 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.303; TCGA BRCA -0.105; TCGA COAD -0.116; TCGA ESCA -0.141; TCGA HNSC -0.142; TCGA KIRP -0.192; TCGA LGG -0.222; TCGA LIHC -0.167; TCGA LUAD -0.131; TCGA LUSC -0.102; TCGA PAAD -0.199; TCGA PRAD -0.32; TCGA SARC -0.184; TCGA STAD -0.368; TCGA UCEC -0.17 |
hsa-miR-425-5p | ADARB1 | 12 cancers: BLCA; BRCA; COAD; ESCA; KIRP; LIHC; LUAD; LUSC; OV; PAAD; STAD; UCEC | mirMAP | TCGA BLCA -0.44; TCGA BRCA -0.075; TCGA COAD -0.283; TCGA ESCA -0.212; TCGA KIRP -0.231; TCGA LIHC -0.184; TCGA LUAD -0.273; TCGA LUSC -0.352; TCGA OV -0.109; TCGA PAAD -0.169; TCGA STAD -0.342; TCGA UCEC -0.229 |
hsa-miR-425-5p | GIGYF2 | 12 cancers: BLCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; OV; PAAD; PRAD; SARC; UCEC | mirMAP | TCGA BLCA -0.119; TCGA HNSC -0.1; TCGA KIRC -0.078; TCGA KIRP -0.154; TCGA LGG -0.081; TCGA LIHC -0.091; TCGA LUAD -0.107; TCGA OV -0.074; TCGA PAAD -0.129; TCGA PRAD -0.079; TCGA SARC -0.135; TCGA UCEC -0.069 |
hsa-miR-425-5p | PTEN | 11 cancers: BLCA; ESCA; KIRP; LIHC; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.139; TCGA ESCA -0.137; TCGA KIRP -0.081; TCGA LIHC -0.144; TCGA LUAD -0.169; TCGA LUSC -0.24; TCGA OV -0.24; TCGA PAAD -0.108; TCGA PRAD -0.193; TCGA STAD -0.107; TCGA UCEC -0.167 |
hsa-miR-425-5p | CPEB2 | 11 cancers: BLCA; BRCA; ESCA; KIRP; LIHC; LUAD; LUSC; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.301; TCGA BRCA -0.07; TCGA ESCA -0.247; TCGA KIRP -0.186; TCGA LIHC -0.185; TCGA LUAD -0.167; TCGA LUSC -0.17; TCGA PRAD -0.104; TCGA SARC -0.17; TCGA STAD -0.165; TCGA UCEC -0.207 |
hsa-miR-425-5p | FOXN3 | 17 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; OV; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.255; TCGA BRCA -0.107; TCGA CESC -0.166; TCGA COAD -0.169; TCGA ESCA -0.349; TCGA HNSC -0.167; TCGA KIRP -0.115; TCGA LGG -0.076; TCGA LIHC -0.124; TCGA LUAD -0.204; TCGA LUSC -0.135; TCGA OV -0.169; TCGA PAAD -0.206; TCGA PRAD -0.162; TCGA SARC -0.287; TCGA STAD -0.288; TCGA UCEC -0.238 |
hsa-miR-425-5p | CFL2 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.552; TCGA BRCA -0.103; TCGA CESC -0.096; TCGA COAD -0.481; TCGA ESCA -0.382; TCGA HNSC -0.191; TCGA KIRP -0.111; TCGA LUSC -0.232; TCGA PAAD -0.23; TCGA PRAD -0.365; TCGA SARC -0.326; TCGA STAD -0.63; TCGA UCEC -0.287 |
hsa-miR-425-5p | CHD9 | 17 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.177; TCGA BRCA -0.104; TCGA CESC -0.176; TCGA COAD -0.07; TCGA ESCA -0.217; TCGA HNSC -0.099; TCGA KIRC -0.094; TCGA KIRP -0.236; TCGA LGG -0.202; TCGA LIHC -0.305; TCGA LUAD -0.146; TCGA LUSC -0.138; TCGA OV -0.116; TCGA PAAD -0.143; TCGA PRAD -0.249; TCGA STAD -0.152; TCGA UCEC -0.223 |
hsa-miR-425-5p | SAMD4A | 13 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PRAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.55; TCGA BRCA -0.137; TCGA COAD -0.391; TCGA ESCA -0.204; TCGA HNSC -0.156; TCGA LIHC -0.123; TCGA LUAD -0.189; TCGA LUSC -0.332; TCGA PRAD -0.231; TCGA SARC -0.202; TCGA THCA -0.151; TCGA STAD -0.417; TCGA UCEC -0.37 |
hsa-miR-425-5p | DGKG | 10 cancers: BLCA; CESC; ESCA; HNSC; LGG; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.505; TCGA CESC -0.232; TCGA ESCA -0.785; TCGA HNSC -0.278; TCGA LGG -0.207; TCGA PAAD -0.28; TCGA PRAD -0.56; TCGA SARC -0.606; TCGA STAD -0.508; TCGA UCEC -0.159 |
hsa-miR-425-5p | CBX6 | 14 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LUSC; OV; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.255; TCGA BRCA -0.243; TCGA COAD -0.483; TCGA ESCA -0.271; TCGA HNSC -0.284; TCGA KIRC -0.119; TCGA KIRP -0.28; TCGA LUSC -0.222; TCGA OV -0.272; TCGA PAAD -0.349; TCGA PRAD -0.332; TCGA SARC -0.303; TCGA STAD -0.403; TCGA UCEC -0.308 |
hsa-miR-425-5p | ATRX | 12 cancers: BLCA; BRCA; HNSC; KIRP; LIHC; LUAD; OV; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.071; TCGA BRCA -0.05; TCGA HNSC -0.085; TCGA KIRP -0.23; TCGA LIHC -0.073; TCGA LUAD -0.125; TCGA OV -0.064; TCGA PAAD -0.147; TCGA PRAD -0.145; TCGA SARC -0.232; TCGA STAD -0.117; TCGA UCEC -0.096 |
hsa-miR-425-5p | PDE4A | 11 cancers: BLCA; BRCA; COAD; HNSC; KIRP; LUAD; LUSC; OV; PAAD; PRAD; UCEC | miRNATAP | TCGA BLCA -0.183; TCGA BRCA -0.108; TCGA COAD -0.128; TCGA HNSC -0.137; TCGA KIRP -0.193; TCGA LUAD -0.245; TCGA LUSC -0.311; TCGA OV -0.16; TCGA PAAD -0.122; TCGA PRAD -0.38; TCGA UCEC -0.123 |
hsa-miR-425-5p | NCAM1 | 16 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRP; LGG; LIHC; LUAD; OV; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -1.143; TCGA BRCA -0.263; TCGA CESC -0.491; TCGA COAD -0.529; TCGA ESCA -0.923; TCGA HNSC -0.399; TCGA KIRP -0.259; TCGA LGG -0.17; TCGA LIHC -0.373; TCGA LUAD -0.209; TCGA OV -0.305; TCGA PAAD -0.577; TCGA PRAD -0.655; TCGA SARC -0.564; TCGA STAD -0.701; TCGA UCEC -0.714 |
hsa-miR-425-5p | C3orf70 | 13 cancers: BLCA; CESC; COAD; HNSC; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNATAP | TCGA BLCA -0.347; TCGA CESC -0.249; TCGA COAD -0.232; TCGA HNSC -0.205; TCGA LIHC -0.122; TCGA LUAD -0.175; TCGA LUSC -0.225; TCGA PAAD -0.301; TCGA PRAD -0.557; TCGA SARC -0.652; TCGA THCA -0.157; TCGA STAD -0.518; TCGA UCEC -0.705 |
hsa-miR-425-5p | OSBPL8 | 12 cancers: BLCA; COAD; HNSC; KIRP; LGG; LIHC; LUAD; LUSC; OV; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.112; TCGA COAD -0.112; TCGA HNSC -0.061; TCGA KIRP -0.2; TCGA LGG -0.162; TCGA LIHC -0.075; TCGA LUAD -0.141; TCGA LUSC -0.064; TCGA OV -0.08; TCGA PRAD -0.1; TCGA STAD -0.065; TCGA UCEC -0.166 |
hsa-miR-425-5p | RABGAP1L | 11 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; LUSC; PRAD; SARC; UCEC | miRNATAP | TCGA BLCA -0.107; TCGA CESC -0.114; TCGA COAD -0.162; TCGA ESCA -0.08; TCGA HNSC -0.068; TCGA KIRP -0.15; TCGA LUAD -0.15; TCGA LUSC -0.102; TCGA PRAD -0.196; TCGA SARC -0.202; TCGA UCEC -0.145 |
hsa-miR-425-5p | KIAA2022 | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.566; TCGA CESC -0.555; TCGA COAD -1.003; TCGA ESCA -1.1; TCGA HNSC -0.274; TCGA KIRP -0.769; TCGA LUAD -0.628; TCGA PAAD -0.875; TCGA PRAD -0.237; TCGA SARC -0.821; TCGA STAD -0.95; TCGA UCEC -0.204 |
hsa-miR-425-5p | RAB8B | 9 cancers: BLCA; CESC; COAD; LUAD; LUSC; OV; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.053; TCGA CESC -0.091; TCGA COAD -0.247; TCGA LUAD -0.193; TCGA LUSC -0.222; TCGA OV -0.078; TCGA PRAD -0.165; TCGA STAD -0.123; TCGA UCEC -0.262 |
hsa-miR-425-5p | BEX4 | 13 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; PAAD; PRAD; THCA; STAD; UCEC | miRNAWalker2 validate; MirTarget | TCGA BRCA -0.09; TCGA CESC -0.306; TCGA COAD -0.401; TCGA ESCA -0.363; TCGA HNSC -0.244; TCGA KIRC -0.126; TCGA KIRP -0.122; TCGA LUAD -0.117; TCGA PAAD -0.36; TCGA PRAD -0.061; TCGA THCA -0.063; TCGA STAD -0.536; TCGA UCEC -0.185 |
hsa-miR-425-5p | BHLHB9 | 11 cancers: BRCA; CESC; HNSC; KIRC; KIRP; LIHC; PAAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate; MirTarget | TCGA BRCA -0.117; TCGA CESC -0.091; TCGA HNSC -0.091; TCGA KIRC -0.122; TCGA KIRP -0.207; TCGA LIHC -0.16; TCGA PAAD -0.247; TCGA PRAD -0.152; TCGA SARC -0.169; TCGA STAD -0.147; TCGA UCEC -0.119 |
hsa-miR-425-5p | IGSF1 | 9 cancers: BRCA; COAD; HNSC; KIRP; PAAD; PRAD; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BRCA -0.277; TCGA COAD -0.315; TCGA HNSC -0.224; TCGA KIRP -0.516; TCGA PAAD -0.635; TCGA PRAD -1.104; TCGA THCA -0.358; TCGA STAD -0.426; TCGA UCEC -0.253 |
hsa-miR-425-5p | ZNF417 | 10 cancers: BRCA; HNSC; KIRC; KIRP; LUAD; LUSC; OV; PRAD; SARC; STAD | miRNAWalker2 validate | TCGA BRCA -0.075; TCGA HNSC -0.128; TCGA KIRC -0.096; TCGA KIRP -0.214; TCGA LUAD -0.099; TCGA LUSC -0.066; TCGA OV -0.084; TCGA PRAD -0.07; TCGA SARC -0.116; TCGA STAD -0.081 |
hsa-miR-425-5p | FARP1 | 10 cancers: BRCA; HNSC; KIRC; KIRP; LIHC; LUAD; LUSC; OV; PAAD; STAD | MirTarget | TCGA BRCA -0.081; TCGA HNSC -0.148; TCGA KIRC -0.12; TCGA KIRP -0.092; TCGA LIHC -0.09; TCGA LUAD -0.12; TCGA LUSC -0.209; TCGA OV -0.145; TCGA PAAD -0.162; TCGA STAD -0.083 |
hsa-miR-425-5p | ANK3 | 10 cancers: BRCA; CESC; ESCA; HNSC; KIRC; LUAD; LUSC; SARC; STAD; UCEC | MirTarget | TCGA BRCA -0.085; TCGA CESC -0.228; TCGA ESCA -0.159; TCGA HNSC -0.119; TCGA KIRC -0.264; TCGA LUAD -0.336; TCGA LUSC -0.192; TCGA SARC -0.311; TCGA STAD -0.097; TCGA UCEC -0.074 |
hsa-miR-425-5p | THRB | 10 cancers: BRCA; COAD; HNSC; KIRC; KIRP; LUAD; LUSC; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BRCA -0.133; TCGA COAD -0.35; TCGA HNSC -0.229; TCGA KIRC -0.247; TCGA KIRP -0.227; TCGA LUAD -0.304; TCGA LUSC -0.181; TCGA SARC -0.45; TCGA STAD -0.414; TCGA UCEC -0.317 |
hsa-miR-425-5p | MAN1C1 | 13 cancers: BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; OV; PAAD; SARC; STAD; UCEC | MirTarget | TCGA BRCA -0.082; TCGA CESC -0.316; TCGA COAD -0.484; TCGA ESCA -0.437; TCGA HNSC -0.21; TCGA LIHC -0.367; TCGA LUAD -0.325; TCGA LUSC -0.275; TCGA OV -0.133; TCGA PAAD -0.453; TCGA SARC -0.256; TCGA STAD -0.354; TCGA UCEC -0.335 |
hsa-miR-425-5p | NOVA2 | 11 cancers: BRCA; COAD; HNSC; LIHC; LUAD; LUSC; OV; PAAD; THCA; STAD; UCEC | mirMAP | TCGA BRCA -0.084; TCGA COAD -0.31; TCGA HNSC -0.076; TCGA LIHC -0.139; TCGA LUAD -0.114; TCGA LUSC -0.299; TCGA OV -0.211; TCGA PAAD -0.24; TCGA THCA -0.262; TCGA STAD -0.111; TCGA UCEC -0.236 |
hsa-miR-425-5p | PSD3 | 11 cancers: BRCA; CESC; COAD; HNSC; LIHC; LUAD; LUSC; OV; THCA; STAD; UCEC | mirMAP | TCGA BRCA -0.198; TCGA CESC -0.225; TCGA COAD -0.183; TCGA HNSC -0.251; TCGA LIHC -0.48; TCGA LUAD -0.121; TCGA LUSC -0.141; TCGA OV -0.196; TCGA THCA -0.216; TCGA STAD -0.106; TCGA UCEC -0.275 |
hsa-miR-425-5p | PTCHD1 | 12 cancers: BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; OV; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BRCA -0.357; TCGA CESC -0.366; TCGA COAD -0.889; TCGA ESCA -0.658; TCGA HNSC -0.356; TCGA LUAD -0.239; TCGA LUSC -0.405; TCGA OV -0.28; TCGA PRAD -0.775; TCGA SARC -1.102; TCGA STAD -1.173; TCGA UCEC -0.616 |
hsa-miR-425-5p | ZNF561 | 11 cancers: BRCA; ESCA; KIRP; LGG; LIHC; LUAD; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BRCA -0.065; TCGA ESCA -0.129; TCGA KIRP -0.058; TCGA LGG -0.104; TCGA LIHC -0.126; TCGA LUAD -0.09; TCGA PAAD -0.074; TCGA PRAD -0.131; TCGA SARC -0.27; TCGA STAD -0.054; TCGA UCEC -0.077 |
hsa-miR-425-5p | PTPRT | 12 cancers: BRCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; OV; PAAD; SARC; STAD; UCEC | mirMAP | TCGA BRCA -0.403; TCGA CESC -0.459; TCGA COAD -0.535; TCGA ESCA -0.494; TCGA HNSC -0.326; TCGA LIHC -0.315; TCGA LUAD -0.672; TCGA OV -0.748; TCGA PAAD -0.776; TCGA SARC -0.6; TCGA STAD -0.269; TCGA UCEC -0.267 |
hsa-miR-425-5p | ZNF677 | 13 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LUSC; OV; PAAD; STAD; UCEC | mirMAP | TCGA BRCA -0.167; TCGA CESC -0.27; TCGA COAD -0.553; TCGA ESCA -0.505; TCGA HNSC -0.313; TCGA KIRC -0.142; TCGA KIRP -0.423; TCGA LGG -0.118; TCGA LUSC -0.442; TCGA OV -0.173; TCGA PAAD -0.331; TCGA STAD -0.497; TCGA UCEC -0.279 |
hsa-miR-425-5p | SYNJ1 | 12 cancers: CESC; COAD; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA CESC -0.071; TCGA COAD -0.102; TCGA HNSC -0.069; TCGA KIRP -0.262; TCGA LIHC -0.094; TCGA LUAD -0.183; TCGA LUSC -0.104; TCGA PAAD -0.168; TCGA PRAD -0.083; TCGA SARC -0.149; TCGA STAD -0.07; TCGA UCEC -0.135 |
hsa-miR-425-5p | NOS1 | 11 cancers: COAD; ESCA; HNSC; KIRP; LUAD; LUSC; OV; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA COAD -1.012; TCGA ESCA -1.502; TCGA HNSC -0.568; TCGA KIRP -0.543; TCGA LUAD -0.485; TCGA LUSC -0.591; TCGA OV -0.36; TCGA PAAD -0.667; TCGA PRAD -1.117; TCGA STAD -1.113; TCGA UCEC -0.23 |
hsa-miR-425-5p | ZBTB10 | 9 cancers: HNSC; KIRC; KIRP; LGG; LIHC; LUSC; PAAD; SARC; STAD | miRNAWalker2 validate | TCGA HNSC -0.283; TCGA KIRC -0.137; TCGA KIRP -0.372; TCGA LGG -0.189; TCGA LIHC -0.109; TCGA LUSC -0.134; TCGA PAAD -0.176; TCGA SARC -0.183; TCGA STAD -0.172 |
hsa-miR-425-5p | ZNF148 | 10 cancers: HNSC; KIRP; LGG; LIHC; LUAD; OV; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate; miRNATAP | TCGA HNSC -0.085; TCGA KIRP -0.191; TCGA LGG -0.085; TCGA LIHC -0.065; TCGA LUAD -0.076; TCGA OV -0.085; TCGA PRAD -0.093; TCGA SARC -0.1; TCGA STAD -0.067; TCGA UCEC -0.081 |
hsa-miR-425-5p | WDR35 | 10 cancers: HNSC; KIRC; KIRP; LGG; OV; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA HNSC -0.083; TCGA KIRC -0.081; TCGA KIRP -0.216; TCGA LGG -0.12; TCGA OV -0.11; TCGA PAAD -0.199; TCGA PRAD -0.068; TCGA SARC -0.109; TCGA STAD -0.202; TCGA UCEC -0.126 |
Enriched cancer pathways of putative targets