microRNA information: hsa-miR-450b-5p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-450b-5p | miRbase |
| Accession: | MIMAT0004909 | miRbase |
| Precursor name: | hsa-mir-450b | miRbase |
| Precursor accession: | MI0005531 | miRbase |
| Symbol: | MIR450B | HGNC |
| RefSeq ID: | NR_030587 | GenBank |
| Sequence: | UUUUGCAAUAUGUUCCUGAAUA |
Reported expression in cancers: hsa-miR-450b-5p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-450b-5p | colorectal cancer | downregulation | "To investigate the role of miR-450b-5p a newly ide ......" | 26845645 | |
| hsa-miR-450b-5p | colorectal cancer | upregulation | "Analyses of published microarray datasets revealed ......" | 27494869 | Microarray |
Reported cancer pathway affected by hsa-miR-450b-5p
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-450b-5p | colorectal cancer | Apoptosis pathway | "miR 450b 5p induced by oncogenic KRAS is required ......" | 27494869 |
Reported cancer prognosis affected by hsa-miR-450b-5p
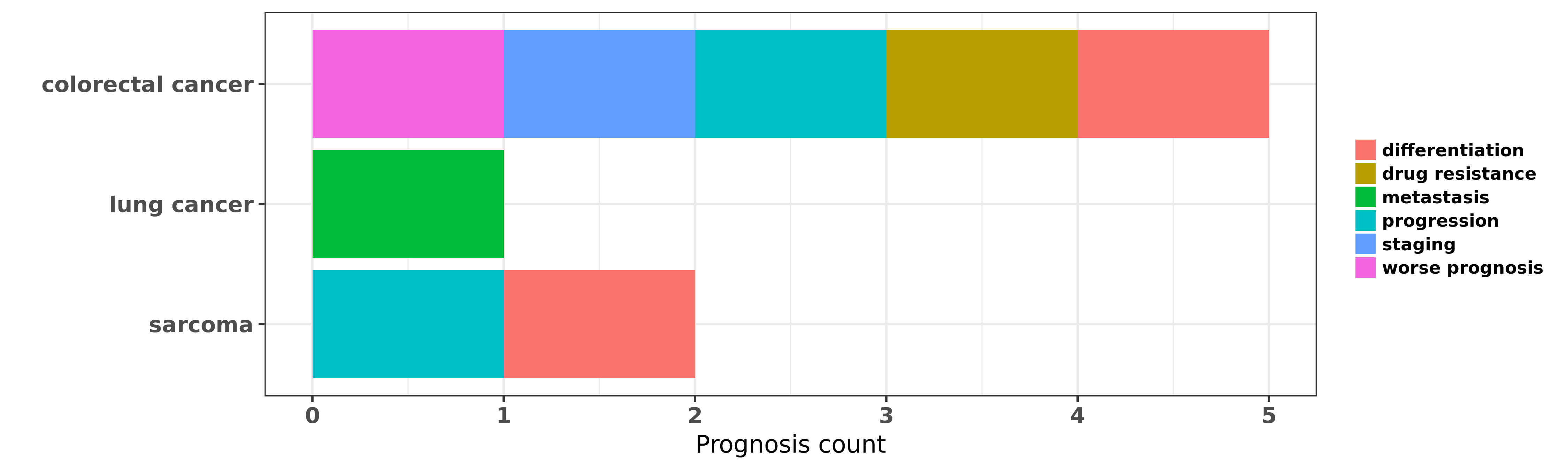
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-450b-5p | colorectal cancer | drug resistance | "miR 450b 5p Suppresses Stemness and the Developmen ......" | 26845645 | |
| hsa-miR-450b-5p | colorectal cancer | progression; staging; worse prognosis; differentiation | "miR 450b 5p induced by oncogenic KRAS is required ......" | 27494869 | |
| hsa-miR-450b-5p | lung cancer | metastasis | "By developing an integrated bioinformatics analysi ......" | 26075299 | |
| hsa-miR-450b-5p | sarcoma | differentiation | "TGF β1 suppression of microRNA 450b 5p expression ......" | 23665678 | Western blot |
| hsa-miR-450b-5p | sarcoma | progression; differentiation | "Therefore in this review we focus on the regulatio ......" | 24831881 |
Reported gene related to hsa-miR-450b-5p
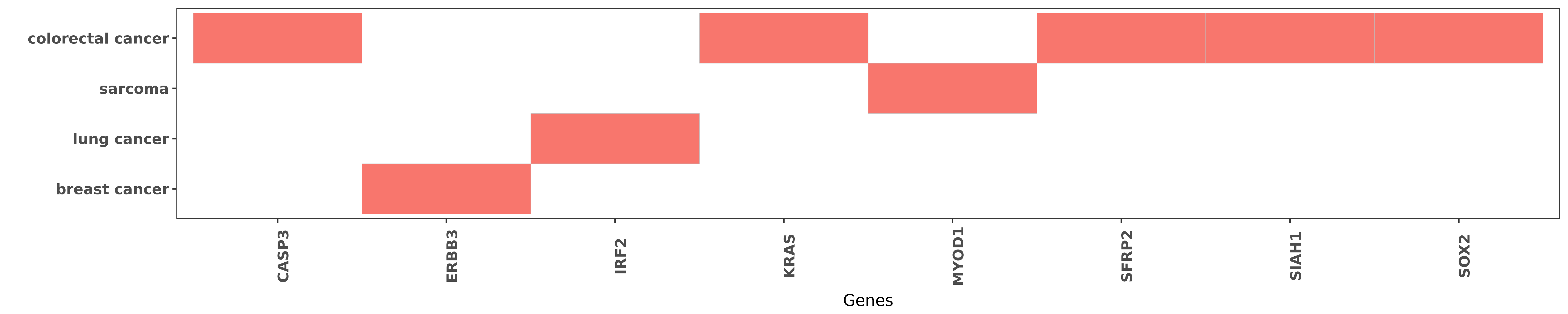
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-450b-5p | colorectal cancer | CASP3 | "Importantly overexpression of miR-450b-5p in 5-FU- ......" | 26845645 |
| hsa-miR-450b-5p | breast cancer | ERBB3 | "Targeting HER3 with miR 450b 3p suppresses breast ......" | 25046105 |
| hsa-miR-450b-5p | lung cancer | IRF2 | "miR-450 targets IRF2 and thus supresses lung cance ......" | 27246609 |
| hsa-miR-450b-5p | colorectal cancer | KRAS | "miR 450b 5p induced by oncogenic KRAS is required ......" | 27494869 |
| hsa-miR-450b-5p | sarcoma | MYOD1 | "Both in cultured cells and tumor implants miR-450b ......" | 23665678 |
| hsa-miR-450b-5p | colorectal cancer | SFRP2 | "Furthermore we demonstrated that miR-450b-5p direc ......" | 27494869 |
| hsa-miR-450b-5p | colorectal cancer | SIAH1 | "Furthermore we demonstrated that miR-450b-5p direc ......" | 27494869 |
| hsa-miR-450b-5p | colorectal cancer | SOX2 | "miR 450b 5p Suppresses Stemness and the Developmen ......" | 26845645 |
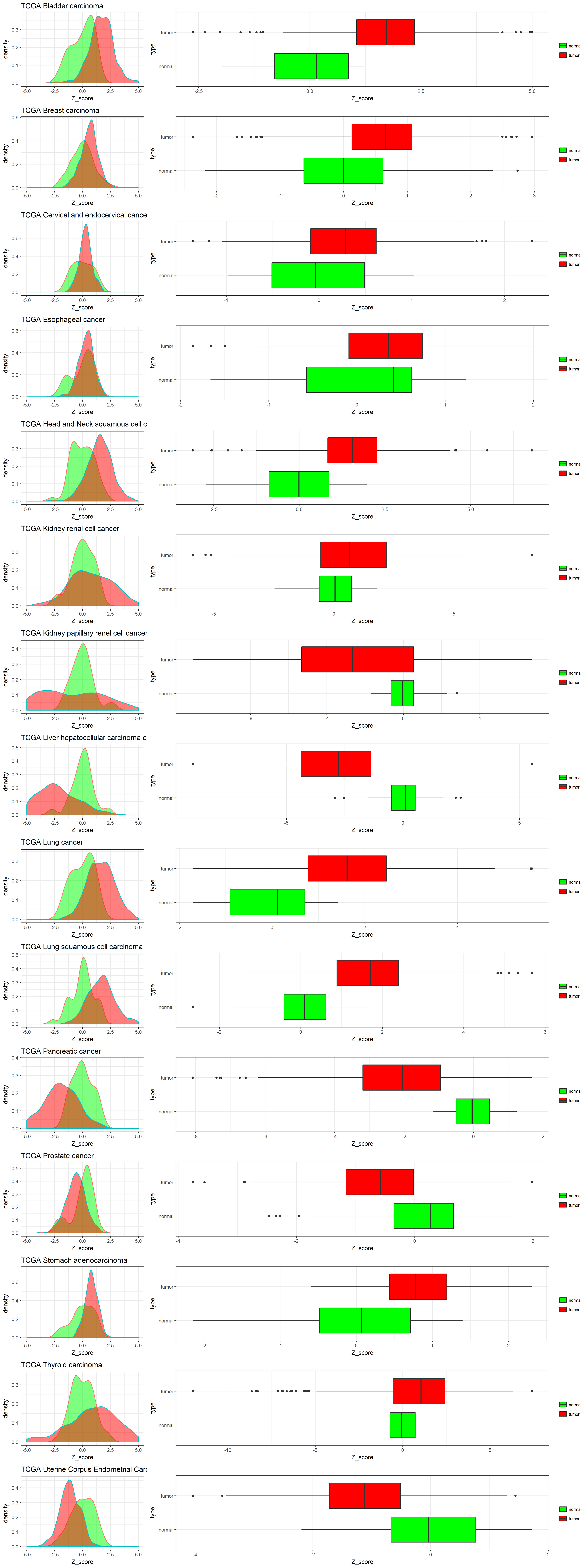
Expression profile in cancer corhorts:
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-450b-5p | RIC3 | 9 cancers: BLCA; COAD; HNSC; KIRC; KIRP; LGG; LUAD; SARC; STAD | MirTarget; mirMAP | TCGA BLCA -0.594; TCGA COAD -0.575; TCGA HNSC -0.335; TCGA KIRC -0.522; TCGA KIRP -0.139; TCGA LGG -0.098; TCGA LUAD -0.224; TCGA SARC -0.624; TCGA STAD -0.689 |
hsa-miR-450b-5p | ADH1B | 9 cancers: BLCA; COAD; ESCA; HNSC; KIRC; LGG; LUAD; LUSC; STAD | MirTarget | TCGA BLCA -0.545; TCGA COAD -1.004; TCGA ESCA -1.076; TCGA HNSC -1.082; TCGA KIRC -0.415; TCGA LGG -0.511; TCGA LUAD -0.613; TCGA LUSC -1.096; TCGA STAD -0.654 |
hsa-miR-450b-5p | IRS2 | 9 cancers: BLCA; BRCA; KIRC; LUAD; LUSC; PAAD; SARC; THCA; STAD | MirTarget; PITA; mirMAP; miRNATAP | TCGA BLCA -0.204; TCGA BRCA -0.216; TCGA KIRC -0.155; TCGA LUAD -0.381; TCGA LUSC -0.318; TCGA PAAD -0.153; TCGA SARC -0.124; TCGA THCA -0.066; TCGA STAD -0.175 |
hsa-miR-450b-5p | HSPB8 | 9 cancers: BLCA; COAD; HNSC; LGG; LUAD; LUSC; SARC; THCA; STAD | MirTarget; miRNATAP | TCGA BLCA -0.279; TCGA COAD -0.486; TCGA HNSC -0.316; TCGA LGG -0.389; TCGA LUAD -0.101; TCGA LUSC -0.492; TCGA SARC -0.453; TCGA THCA -0.196; TCGA STAD -0.879 |
hsa-miR-450b-5p | MLLT6 | 9 cancers: BLCA; ESCA; HNSC; KIRC; KIRP; LGG; LIHC; PRAD; SARC | MirTarget; PITA; miRNATAP | TCGA BLCA -0.062; TCGA ESCA -0.111; TCGA HNSC -0.15; TCGA KIRC -0.098; TCGA KIRP -0.072; TCGA LGG -0.052; TCGA LIHC -0.125; TCGA PRAD -0.072; TCGA SARC -0.139 |
hsa-miR-450b-5p | LNPEP | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUAD; SARC; THCA; STAD; UCEC | MirTarget; PITA; miRNATAP | TCGA BLCA -0.162; TCGA BRCA -0.106; TCGA COAD -0.171; TCGA ESCA -0.114; TCGA HNSC -0.157; TCGA LUAD -0.148; TCGA SARC -0.061; TCGA THCA -0.086; TCGA STAD -0.156; TCGA UCEC -0.094 |
hsa-miR-450b-5p | FAM189A2 | 9 cancers: BLCA; COAD; ESCA; HNSC; KIRC; LGG; LUSC; SARC; STAD | MirTarget; miRNATAP | TCGA BLCA -0.238; TCGA COAD -0.522; TCGA ESCA -0.372; TCGA HNSC -0.577; TCGA KIRC -0.307; TCGA LGG -0.292; TCGA LUSC -0.365; TCGA SARC -0.315; TCGA STAD -0.581 |
hsa-miR-450b-5p | ADRB2 | 11 cancers: BLCA; CESC; COAD; KIRP; LGG; LUAD; LUSC; PRAD; SARC; THCA; STAD | MirTarget | TCGA BLCA -0.333; TCGA CESC -0.302; TCGA COAD -0.459; TCGA KIRP -0.17; TCGA LGG -0.148; TCGA LUAD -0.153; TCGA LUSC -0.527; TCGA PRAD -0.114; TCGA SARC -0.219; TCGA THCA -0.192; TCGA STAD -0.462 |
hsa-miR-450b-5p | MYRIP | 10 cancers: BLCA; BRCA; HNSC; LGG; LUAD; LUSC; PAAD; PRAD; SARC; STAD | MirTarget; PITA | TCGA BLCA -0.26; TCGA BRCA -0.191; TCGA HNSC -0.545; TCGA LGG -0.21; TCGA LUAD -0.362; TCGA LUSC -0.567; TCGA PAAD -0.337; TCGA PRAD -0.094; TCGA SARC -0.426; TCGA STAD -0.505 |
hsa-miR-450b-5p | GCOM1 | 10 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LUAD; LUSC; SARC; THCA; STAD | MirTarget | TCGA BLCA -0.252; TCGA BRCA -0.219; TCGA ESCA -0.435; TCGA HNSC -0.464; TCGA KIRC -0.38; TCGA LUAD -0.333; TCGA LUSC -0.603; TCGA SARC -0.134; TCGA THCA -0.259; TCGA STAD -0.201 |
hsa-miR-450b-5p | PBX1 | 9 cancers: BLCA; COAD; HNSC; KIRC; LUAD; LUSC; PAAD; SARC; STAD | MirTarget | TCGA BLCA -0.134; TCGA COAD -0.224; TCGA HNSC -0.327; TCGA KIRC -0.159; TCGA LUAD -0.142; TCGA LUSC -0.148; TCGA PAAD -0.13; TCGA SARC -0.316; TCGA STAD -0.326 |
hsa-miR-450b-5p | SLU7 | 10 cancers: BLCA; BRCA; CESC; HNSC; LGG; LIHC; LUAD; LUSC; PAAD; STAD | MirTarget | TCGA BLCA -0.057; TCGA BRCA -0.065; TCGA CESC -0.08; TCGA HNSC -0.073; TCGA LGG -0.062; TCGA LIHC -0.15; TCGA LUAD -0.084; TCGA LUSC -0.072; TCGA PAAD -0.116; TCGA STAD -0.075 |
hsa-miR-450b-5p | ANO5 | 9 cancers: BLCA; COAD; HNSC; KIRC; LUAD; LUSC; PAAD; SARC; STAD | MirTarget | TCGA BLCA -0.557; TCGA COAD -0.798; TCGA HNSC -0.247; TCGA KIRC -0.387; TCGA LUAD -0.264; TCGA LUSC -0.47; TCGA PAAD -0.366; TCGA SARC -0.532; TCGA STAD -0.638 |
hsa-miR-450b-5p | DIXDC1 | 9 cancers: BLCA; BRCA; COAD; ESCA; LGG; LUAD; LUSC; SARC; STAD | MirTarget; PITA; miRNATAP | TCGA BLCA -0.272; TCGA BRCA -0.098; TCGA COAD -0.289; TCGA ESCA -0.168; TCGA LGG -0.088; TCGA LUAD -0.123; TCGA LUSC -0.284; TCGA SARC -0.306; TCGA STAD -0.521 |
hsa-miR-450b-5p | ADAMTS8 | 9 cancers: BLCA; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; SARC; STAD | MirTarget; PITA; miRNATAP | TCGA BLCA -0.487; TCGA COAD -0.506; TCGA ESCA -0.592; TCGA HNSC -0.317; TCGA KIRC -0.256; TCGA LUAD -0.384; TCGA LUSC -0.86; TCGA SARC -0.589; TCGA STAD -0.599 |
hsa-miR-450b-5p | CCNDBP1 | 9 cancers: BLCA; BRCA; HNSC; KIRC; LGG; LUAD; LUSC; SARC; STAD | MirTarget | TCGA BLCA -0.126; TCGA BRCA -0.129; TCGA HNSC -0.068; TCGA KIRC -0.072; TCGA LGG -0.075; TCGA LUAD -0.141; TCGA LUSC -0.162; TCGA SARC -0.09; TCGA STAD -0.12 |
hsa-miR-450b-5p | CREBL2 | 9 cancers: BLCA; BRCA; COAD; HNSC; LGG; LUAD; LUSC; PAAD; STAD | MirTarget; PITA; miRNATAP | TCGA BLCA -0.115; TCGA BRCA -0.119; TCGA COAD -0.101; TCGA HNSC -0.064; TCGA LGG -0.1; TCGA LUAD -0.169; TCGA LUSC -0.164; TCGA PAAD -0.16; TCGA STAD -0.126 |
hsa-miR-450b-5p | HIPK3 | 9 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUAD; LUSC; SARC; STAD | MirTarget; mirMAP | TCGA BLCA -0.189; TCGA BRCA -0.121; TCGA COAD -0.165; TCGA ESCA -0.119; TCGA HNSC -0.222; TCGA LUAD -0.168; TCGA LUSC -0.111; TCGA SARC -0.144; TCGA STAD -0.221 |
hsa-miR-450b-5p | PURA | 9 cancers: BLCA; BRCA; COAD; HNSC; KIRC; LUAD; LUSC; PAAD; STAD | MirTarget; PITA; miRNATAP | TCGA BLCA -0.083; TCGA BRCA -0.053; TCGA COAD -0.071; TCGA HNSC -0.073; TCGA KIRC -0.055; TCGA LUAD -0.104; TCGA LUSC -0.064; TCGA PAAD -0.111; TCGA STAD -0.119 |
hsa-miR-450b-5p | NR3C2 | 9 cancers: BLCA; HNSC; KIRC; LGG; LUAD; LUSC; PAAD; SARC; STAD | PITA; mirMAP | TCGA BLCA -0.418; TCGA HNSC -0.576; TCGA KIRC -0.276; TCGA LGG -0.258; TCGA LUAD -0.192; TCGA LUSC -0.263; TCGA PAAD -0.221; TCGA SARC -0.172; TCGA STAD -0.319 |
hsa-miR-450b-5p | KCNMA1 | 9 cancers: BLCA; COAD; ESCA; LGG; LUAD; PAAD; SARC; THCA; STAD | PITA; mirMAP; miRNATAP | TCGA BLCA -0.309; TCGA COAD -0.817; TCGA ESCA -0.569; TCGA LGG -0.109; TCGA LUAD -0.144; TCGA PAAD -0.27; TCGA SARC -0.491; TCGA THCA -0.17; TCGA STAD -0.9 |
hsa-miR-450b-5p | PRKCA | 9 cancers: BLCA; COAD; ESCA; KIRP; LGG; LIHC; LUSC; THCA; STAD | PITA; mirMAP; miRNATAP | TCGA BLCA -0.165; TCGA COAD -0.165; TCGA ESCA -0.173; TCGA KIRP -0.07; TCGA LGG -0.063; TCGA LIHC -0.225; TCGA LUSC -0.139; TCGA THCA -0.14; TCGA STAD -0.125 |
hsa-miR-450b-5p | CALM1 | 9 cancers: BLCA; BRCA; COAD; LGG; LUAD; LUSC; PAAD; SARC; STAD | PITA; miRNATAP | TCGA BLCA -0.075; TCGA BRCA -0.057; TCGA COAD -0.054; TCGA LGG -0.112; TCGA LUAD -0.052; TCGA LUSC -0.092; TCGA PAAD -0.141; TCGA SARC -0.081; TCGA STAD -0.202 |
hsa-miR-450b-5p | TLE4 | 9 cancers: BLCA; COAD; HNSC; KIRC; KIRP; LUAD; SARC; THCA; STAD | PITA | TCGA BLCA -0.214; TCGA COAD -0.223; TCGA HNSC -0.096; TCGA KIRC -0.131; TCGA KIRP -0.088; TCGA LUAD -0.107; TCGA SARC -0.093; TCGA THCA -0.085; TCGA STAD -0.236 |
hsa-miR-450b-5p | SLMAP | 9 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LUAD; SARC; STAD | PITA; mirMAP; miRNATAP | TCGA BLCA -0.215; TCGA BRCA -0.084; TCGA COAD -0.128; TCGA ESCA -0.208; TCGA HNSC -0.093; TCGA KIRC -0.125; TCGA LUAD -0.072; TCGA SARC -0.429; TCGA STAD -0.433 |
hsa-miR-450b-5p | OGN | 9 cancers: BLCA; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; THCA; STAD | PITA | TCGA BLCA -0.363; TCGA COAD -0.634; TCGA ESCA -0.61; TCGA HNSC -0.264; TCGA KIRC -0.334; TCGA LUAD -0.214; TCGA LUSC -0.825; TCGA THCA -0.301; TCGA STAD -0.666 |
hsa-miR-450b-5p | CPEB3 | 12 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LGG; LUAD; LUSC; PAAD; SARC; STAD | PITA; mirMAP; miRNATAP | TCGA BLCA -0.2; TCGA BRCA -0.096; TCGA CESC -0.126; TCGA ESCA -0.195; TCGA HNSC -0.403; TCGA KIRC -0.129; TCGA LGG -0.226; TCGA LUAD -0.118; TCGA LUSC -0.186; TCGA PAAD -0.176; TCGA SARC -0.067; TCGA STAD -0.176 |
hsa-miR-450b-5p | FOXP1 | 11 cancers: BLCA; ESCA; KIRC; KIRP; LGG; LIHC; LUAD; LUSC; PRAD; SARC; STAD | PITA; mirMAP | TCGA BLCA -0.07; TCGA ESCA -0.158; TCGA KIRC -0.104; TCGA KIRP -0.08; TCGA LGG -0.131; TCGA LIHC -0.104; TCGA LUAD -0.166; TCGA LUSC -0.252; TCGA PRAD -0.082; TCGA SARC -0.104; TCGA STAD -0.155 |
hsa-miR-450b-5p | RERG | 9 cancers: BLCA; BRCA; COAD; HNSC; LGG; LUAD; LUSC; SARC; STAD | PITA | TCGA BLCA -0.174; TCGA BRCA -0.195; TCGA COAD -0.268; TCGA HNSC -0.172; TCGA LGG -0.268; TCGA LUAD -0.134; TCGA LUSC -0.222; TCGA SARC -0.381; TCGA STAD -0.545 |
hsa-miR-450b-5p | PRKAR2B | 9 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LUSC; SARC; THCA; STAD | mirMAP; miRNATAP | TCGA BLCA -0.137; TCGA BRCA -0.157; TCGA ESCA -0.254; TCGA HNSC -0.22; TCGA KIRC -0.191; TCGA LUSC -0.206; TCGA SARC -0.119; TCGA THCA -0.16; TCGA STAD -0.389 |
hsa-miR-450b-5p | AKAP11 | 10 cancers: BLCA; ESCA; HNSC; KIRC; LGG; LUAD; LUSC; PAAD; SARC; STAD | mirMAP | TCGA BLCA -0.1; TCGA ESCA -0.114; TCGA HNSC -0.06; TCGA KIRC -0.08; TCGA LGG -0.086; TCGA LUAD -0.143; TCGA LUSC -0.141; TCGA PAAD -0.115; TCGA SARC -0.068; TCGA STAD -0.09 |
hsa-miR-450b-5p | ABCC9 | 9 cancers: BLCA; BRCA; COAD; ESCA; LGG; LUAD; LUSC; SARC; STAD | mirMAP | TCGA BLCA -0.219; TCGA BRCA -0.097; TCGA COAD -0.337; TCGA ESCA -0.35; TCGA LGG -0.165; TCGA LUAD -0.21; TCGA LUSC -0.38; TCGA SARC -0.304; TCGA STAD -0.458 |
hsa-miR-450b-5p | CYBRD1 | 10 cancers: BLCA; BRCA; COAD; HNSC; KIRC; LGG; LUAD; LUSC; PRAD; STAD | mirMAP | TCGA BLCA -0.167; TCGA BRCA -0.199; TCGA COAD -0.241; TCGA HNSC -0.151; TCGA KIRC -0.133; TCGA LGG -0.129; TCGA LUAD -0.194; TCGA LUSC -0.325; TCGA PRAD -0.069; TCGA STAD -0.264 |
hsa-miR-450b-5p | FBXW11 | 9 cancers: BLCA; BRCA; COAD; HNSC; LGG; LUAD; LUSC; PAAD; STAD | mirMAP | TCGA BLCA -0.084; TCGA BRCA -0.055; TCGA COAD -0.063; TCGA HNSC -0.122; TCGA LGG -0.052; TCGA LUAD -0.057; TCGA LUSC -0.087; TCGA PAAD -0.144; TCGA STAD -0.079 |
hsa-miR-450b-5p | WDR7 | 10 cancers: BLCA; BRCA; COAD; HNSC; LGG; LUAD; LUSC; PAAD; SARC; STAD | mirMAP | TCGA BLCA -0.119; TCGA BRCA -0.057; TCGA COAD -0.183; TCGA HNSC -0.154; TCGA LGG -0.06; TCGA LUAD -0.143; TCGA LUSC -0.107; TCGA PAAD -0.178; TCGA SARC -0.072; TCGA STAD -0.101 |
hsa-miR-450b-5p | FBXO9 | 9 cancers: BLCA; BRCA; HNSC; KIRC; LGG; LIHC; PAAD; SARC; STAD | mirMAP | TCGA BLCA -0.054; TCGA BRCA -0.062; TCGA HNSC -0.065; TCGA KIRC -0.076; TCGA LGG -0.054; TCGA LIHC -0.088; TCGA PAAD -0.199; TCGA SARC -0.065; TCGA STAD -0.095 |
hsa-miR-450b-5p | COL4A3BP | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LGG; LUAD; LUSC; PAAD; SARC; STAD | mirMAP | TCGA BLCA -0.097; TCGA BRCA -0.108; TCGA COAD -0.06; TCGA ESCA -0.152; TCGA HNSC -0.172; TCGA LGG -0.051; TCGA LUAD -0.114; TCGA LUSC -0.152; TCGA PAAD -0.114; TCGA SARC -0.061; TCGA STAD -0.131 |
hsa-miR-450b-5p | PDE4D | 9 cancers: BLCA; COAD; HNSC; KIRC; LUAD; LUSC; PAAD; SARC; STAD | mirMAP | TCGA BLCA -0.254; TCGA COAD -0.195; TCGA HNSC -0.176; TCGA KIRC -0.116; TCGA LUAD -0.322; TCGA LUSC -0.227; TCGA PAAD -0.216; TCGA SARC -0.314; TCGA STAD -0.248 |
hsa-miR-450b-5p | PDE1A | 9 cancers: BLCA; ESCA; HNSC; KIRC; LGG; LUAD; LUSC; SARC; STAD | mirMAP | TCGA BLCA -0.236; TCGA ESCA -0.248; TCGA HNSC -0.141; TCGA KIRC -0.244; TCGA LGG -0.154; TCGA LUAD -0.128; TCGA LUSC -0.278; TCGA SARC -0.177; TCGA STAD -0.271 |
hsa-miR-450b-5p | ESYT2 | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LUSC; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.12; TCGA BRCA -0.066; TCGA COAD -0.069; TCGA ESCA -0.233; TCGA HNSC -0.077; TCGA KIRP -0.094; TCGA LUSC -0.069; TCGA SARC -0.154; TCGA THCA -0.063; TCGA STAD -0.106 |
hsa-miR-450b-5p | LDB3 | 9 cancers: BLCA; BRCA; COAD; ESCA; LGG; LUAD; LUSC; SARC; STAD | mirMAP | TCGA BLCA -0.327; TCGA BRCA -0.166; TCGA COAD -0.634; TCGA ESCA -0.569; TCGA LGG -0.226; TCGA LUAD -0.1; TCGA LUSC -0.443; TCGA SARC -0.593; TCGA STAD -1.11 |
hsa-miR-450b-5p | ATP8A1 | 9 cancers: BLCA; HNSC; KIRP; LGG; LUAD; LUSC; PAAD; PRAD; SARC | mirMAP | TCGA BLCA -0.316; TCGA HNSC -0.256; TCGA KIRP -0.111; TCGA LGG -0.158; TCGA LUAD -0.171; TCGA LUSC -0.416; TCGA PAAD -0.302; TCGA PRAD -0.108; TCGA SARC -0.136 |
hsa-miR-450b-5p | KIF1C | 10 cancers: BLCA; BRCA; COAD; HNSC; KIRC; KIRP; LGG; LUAD; LUSC; STAD | mirMAP | TCGA BLCA -0.084; TCGA BRCA -0.076; TCGA COAD -0.116; TCGA HNSC -0.117; TCGA KIRC -0.088; TCGA KIRP -0.061; TCGA LGG -0.08; TCGA LUAD -0.136; TCGA LUSC -0.186; TCGA STAD -0.095 |
hsa-miR-450b-5p | CASZ1 | 13 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PRAD; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.116; TCGA CESC -0.146; TCGA COAD -0.123; TCGA ESCA -0.16; TCGA HNSC -0.196; TCGA KIRC -0.153; TCGA KIRP -0.075; TCGA LUAD -0.114; TCGA LUSC -0.17; TCGA PRAD -0.063; TCGA SARC -0.369; TCGA THCA -0.062; TCGA STAD -0.217 |
hsa-miR-450b-5p | PCBD2 | 9 cancers: BLCA; BRCA; CESC; HNSC; LIHC; LUAD; PAAD; SARC; STAD | mirMAP | TCGA BLCA -0.065; TCGA BRCA -0.058; TCGA CESC -0.08; TCGA HNSC -0.189; TCGA LIHC -0.063; TCGA LUAD -0.073; TCGA PAAD -0.101; TCGA SARC -0.117; TCGA STAD -0.112 |
hsa-miR-450b-5p | FGF2 | 9 cancers: BLCA; BRCA; COAD; HNSC; LGG; LUAD; LUSC; THCA; STAD | mirMAP | TCGA BLCA -0.301; TCGA BRCA -0.146; TCGA COAD -0.376; TCGA HNSC -0.227; TCGA LGG -0.118; TCGA LUAD -0.166; TCGA LUSC -0.146; TCGA THCA -0.09; TCGA STAD -0.448 |
hsa-miR-450b-5p | GNAL | 9 cancers: BLCA; COAD; HNSC; KIRC; LIHC; LUAD; LUSC; SARC; STAD | mirMAP | TCGA BLCA -0.465; TCGA COAD -0.358; TCGA HNSC -0.167; TCGA KIRC -0.119; TCGA LIHC -0.184; TCGA LUAD -0.173; TCGA LUSC -0.173; TCGA SARC -0.309; TCGA STAD -0.501 |
hsa-miR-450b-5p | RPRD1A | 9 cancers: BLCA; BRCA; HNSC; KIRC; LIHC; LUAD; PAAD; SARC; STAD | mirMAP; miRNATAP | TCGA BLCA -0.084; TCGA BRCA -0.075; TCGA HNSC -0.084; TCGA KIRC -0.06; TCGA LIHC -0.05; TCGA LUAD -0.077; TCGA PAAD -0.241; TCGA SARC -0.159; TCGA STAD -0.104 |
hsa-miR-450b-5p | SMAD4 | 10 cancers: BLCA; BRCA; COAD; HNSC; KIRC; LUAD; LUSC; PAAD; SARC; STAD | mirMAP | TCGA BLCA -0.086; TCGA BRCA -0.088; TCGA COAD -0.196; TCGA HNSC -0.113; TCGA KIRC -0.085; TCGA LUAD -0.125; TCGA LUSC -0.06; TCGA PAAD -0.165; TCGA SARC -0.115; TCGA STAD -0.116 |
hsa-miR-450b-5p | RABL3 | 9 cancers: BLCA; BRCA; HNSC; KIRC; LIHC; LUAD; LUSC; PAAD; STAD | mirMAP; miRNATAP | TCGA BLCA -0.061; TCGA BRCA -0.066; TCGA HNSC -0.062; TCGA KIRC -0.072; TCGA LIHC -0.059; TCGA LUAD -0.061; TCGA LUSC -0.062; TCGA PAAD -0.093; TCGA STAD -0.067 |
hsa-miR-450b-5p | SETD7 | 9 cancers: BLCA; BRCA; ESCA; HNSC; LGG; LUAD; LUSC; THCA; STAD | mirMAP | TCGA BLCA -0.084; TCGA BRCA -0.109; TCGA ESCA -0.09; TCGA HNSC -0.113; TCGA LGG -0.083; TCGA LUAD -0.057; TCGA LUSC -0.153; TCGA THCA -0.102; TCGA STAD -0.196 |
hsa-miR-450b-5p | FBXL17 | 10 cancers: BLCA; BRCA; COAD; HNSC; KIRC; LIHC; LUAD; LUSC; PAAD; STAD | mirMAP | TCGA BLCA -0.134; TCGA BRCA -0.088; TCGA COAD -0.11; TCGA HNSC -0.21; TCGA KIRC -0.089; TCGA LIHC -0.056; TCGA LUAD -0.129; TCGA LUSC -0.163; TCGA PAAD -0.165; TCGA STAD -0.115 |
hsa-miR-450b-5p | FBXO25 | 10 cancers: BLCA; BRCA; CESC; HNSC; LGG; LUAD; LUSC; PAAD; SARC; UCEC | mirMAP | TCGA BLCA -0.076; TCGA BRCA -0.08; TCGA CESC -0.098; TCGA HNSC -0.106; TCGA LGG -0.057; TCGA LUAD -0.126; TCGA LUSC -0.122; TCGA PAAD -0.147; TCGA SARC -0.081; TCGA UCEC -0.087 |
hsa-miR-450b-5p | FBXO32 | 9 cancers: BLCA; CESC; COAD; ESCA; LGG; LIHC; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.174; TCGA CESC -0.15; TCGA COAD -0.257; TCGA ESCA -0.271; TCGA LGG -0.098; TCGA LIHC -0.269; TCGA SARC -0.478; TCGA THCA -0.146; TCGA STAD -0.582 |
hsa-miR-450b-5p | KCNB1 | 9 cancers: BLCA; COAD; ESCA; KIRP; LGG; LUAD; PAAD; SARC; STAD | mirMAP | TCGA BLCA -0.332; TCGA COAD -0.569; TCGA ESCA -0.782; TCGA KIRP -0.171; TCGA LGG -0.16; TCGA LUAD -0.25; TCGA PAAD -0.603; TCGA SARC -0.295; TCGA STAD -1.004 |
hsa-miR-450b-5p | CDC42SE2 | 9 cancers: BLCA; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; PRAD | mirMAP | TCGA BLCA -0.083; TCGA COAD -0.09; TCGA ESCA -0.093; TCGA HNSC -0.088; TCGA LGG -0.075; TCGA LIHC -0.064; TCGA LUAD -0.087; TCGA LUSC -0.087; TCGA PRAD -0.082 |
hsa-miR-450b-5p | EIF4E3 | 10 cancers: BLCA; BRCA; COAD; HNSC; LGG; LUAD; LUSC; PAAD; SARC; STAD | mirMAP | TCGA BLCA -0.13; TCGA BRCA -0.156; TCGA COAD -0.194; TCGA HNSC -0.205; TCGA LGG -0.104; TCGA LUAD -0.171; TCGA LUSC -0.279; TCGA PAAD -0.122; TCGA SARC -0.087; TCGA STAD -0.332 |
hsa-miR-450b-5p | ATXN7 | 9 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; LUAD; LUSC; PRAD; SARC | mirMAP; miRNATAP | TCGA BLCA -0.066; TCGA BRCA -0.06; TCGA ESCA -0.138; TCGA HNSC -0.119; TCGA KIRC -0.075; TCGA LUAD -0.06; TCGA LUSC -0.106; TCGA PRAD -0.062; TCGA SARC -0.057 |
hsa-miR-450b-5p | FYCO1 | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; SARC; STAD | mirMAP | TCGA BLCA -0.188; TCGA BRCA -0.115; TCGA COAD -0.174; TCGA ESCA -0.239; TCGA HNSC -0.256; TCGA KIRC -0.091; TCGA LUAD -0.104; TCGA LUSC -0.218; TCGA SARC -0.282; TCGA STAD -0.333 |
hsa-miR-450b-5p | LONRF2 | 9 cancers: BLCA; COAD; HNSC; KIRC; LGG; LUAD; PAAD; SARC; STAD | mirMAP | TCGA BLCA -0.265; TCGA COAD -0.859; TCGA HNSC -0.421; TCGA KIRC -0.431; TCGA LGG -0.076; TCGA LUAD -0.161; TCGA PAAD -0.308; TCGA SARC -0.76; TCGA STAD -0.893 |
hsa-miR-450b-5p | KCND3 | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; SARC; STAD | mirMAP | TCGA BLCA -0.595; TCGA BRCA -0.182; TCGA CESC -0.295; TCGA COAD -0.707; TCGA ESCA -0.369; TCGA HNSC -0.212; TCGA LUAD -0.473; TCGA LUSC -0.494; TCGA SARC -0.792; TCGA STAD -0.386 |
hsa-miR-450b-5p | BCL2 | 9 cancers: BLCA; BRCA; COAD; HNSC; KIRP; LGG; LUAD; SARC; STAD | mirMAP | TCGA BLCA -0.195; TCGA BRCA -0.23; TCGA COAD -0.241; TCGA HNSC -0.303; TCGA KIRP -0.096; TCGA LGG -0.075; TCGA LUAD -0.197; TCGA SARC -0.131; TCGA STAD -0.233 |
hsa-miR-450b-5p | NRXN1 | 10 cancers: BLCA; COAD; ESCA; HNSC; KIRC; LUAD; PAAD; PRAD; SARC; STAD | mirMAP; miRNATAP | TCGA BLCA -0.655; TCGA COAD -0.993; TCGA ESCA -0.467; TCGA HNSC -0.337; TCGA KIRC -0.515; TCGA LUAD -0.288; TCGA PAAD -0.396; TCGA PRAD -0.123; TCGA SARC -0.397; TCGA STAD -0.984 |
hsa-miR-450b-5p | ZADH2 | 9 cancers: BLCA; BRCA; COAD; HNSC; KIRC; LUAD; LUSC; PAAD; STAD | mirMAP | TCGA BLCA -0.087; TCGA BRCA -0.083; TCGA COAD -0.075; TCGA HNSC -0.17; TCGA KIRC -0.113; TCGA LUAD -0.135; TCGA LUSC -0.053; TCGA PAAD -0.148; TCGA STAD -0.077 |
hsa-miR-450b-5p | LYNX1 | 10 cancers: BLCA; COAD; HNSC; KIRC; LGG; LIHC; LUSC; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.347; TCGA COAD -0.225; TCGA HNSC -0.305; TCGA KIRC -0.168; TCGA LGG -0.305; TCGA LIHC -0.149; TCGA LUSC -0.339; TCGA SARC -0.528; TCGA THCA -0.089; TCGA STAD -0.632 |
hsa-miR-450b-5p | PCGF5 | 11 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; LGG; LUAD; LUSC; PAAD; STAD | mirMAP | TCGA BLCA -0.149; TCGA BRCA -0.09; TCGA CESC -0.086; TCGA ESCA -0.137; TCGA HNSC -0.206; TCGA KIRC -0.072; TCGA LGG -0.083; TCGA LUAD -0.14; TCGA LUSC -0.164; TCGA PAAD -0.153; TCGA STAD -0.052 |
hsa-miR-450b-5p | GREM2 | 11 cancers: BLCA; COAD; ESCA; HNSC; KIRC; LGG; LUSC; PRAD; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.248; TCGA COAD -0.597; TCGA ESCA -0.66; TCGA HNSC -0.393; TCGA KIRC -0.351; TCGA LGG -0.261; TCGA LUSC -0.347; TCGA PRAD -0.144; TCGA SARC -0.346; TCGA THCA -0.142; TCGA STAD -0.691 |
hsa-miR-450b-5p | PRELP | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LGG; LUAD; LUSC; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.491; TCGA BRCA -0.185; TCGA COAD -0.55; TCGA ESCA -0.578; TCGA HNSC -0.471; TCGA LGG -0.186; TCGA LUAD -0.241; TCGA LUSC -0.586; TCGA SARC -0.441; TCGA THCA -0.182; TCGA STAD -0.65 |
hsa-miR-450b-5p | PEG3 | 10 cancers: BLCA; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; SARC; STAD | mirMAP | TCGA BLCA -0.329; TCGA COAD -0.349; TCGA ESCA -0.247; TCGA HNSC -0.29; TCGA KIRC -0.21; TCGA LUAD -0.161; TCGA LUSC -0.484; TCGA PAAD -0.312; TCGA SARC -0.239; TCGA STAD -0.612 |
hsa-miR-450b-5p | SKP1 | 11 cancers: BLCA; BRCA; COAD; HNSC; KIRC; LGG; LIHC; LUSC; PAAD; SARC; STAD | miRNATAP | TCGA BLCA -0.093; TCGA BRCA -0.061; TCGA COAD -0.069; TCGA HNSC -0.064; TCGA KIRC -0.057; TCGA LGG -0.098; TCGA LIHC -0.105; TCGA LUSC -0.051; TCGA PAAD -0.114; TCGA SARC -0.053; TCGA STAD -0.123 |
hsa-miR-450b-5p | CLEC3B | 9 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LGG; LUAD; LUSC; STAD | miRNATAP | TCGA BLCA -0.329; TCGA BRCA -0.217; TCGA COAD -0.22; TCGA ESCA -0.25; TCGA HNSC -0.346; TCGA LGG -0.351; TCGA LUAD -0.316; TCGA LUSC -0.706; TCGA STAD -0.445 |
hsa-miR-450b-5p | FBXL5 | 9 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUSC; PAAD; SARC; STAD | miRNATAP | TCGA BLCA -0.08; TCGA BRCA -0.103; TCGA COAD -0.096; TCGA ESCA -0.104; TCGA HNSC -0.127; TCGA LUSC -0.11; TCGA PAAD -0.097; TCGA SARC -0.061; TCGA STAD -0.192 |
hsa-miR-450b-5p | PRKCB | 9 cancers: BLCA; COAD; ESCA; HNSC; LGG; LUAD; LUSC; THCA; STAD | miRNATAP | TCGA BLCA -0.3; TCGA COAD -0.473; TCGA ESCA -0.291; TCGA HNSC -0.164; TCGA LGG -0.162; TCGA LUAD -0.183; TCGA LUSC -0.447; TCGA THCA -0.152; TCGA STAD -0.38 |
hsa-miR-450b-5p | TCEAL1 | 9 cancers: BLCA; BRCA; ESCA; LGG; LIHC; LUSC; PAAD; SARC; STAD | miRNATAP | TCGA BLCA -0.117; TCGA BRCA -0.086; TCGA ESCA -0.142; TCGA LGG -0.084; TCGA LIHC -0.106; TCGA LUSC -0.076; TCGA PAAD -0.126; TCGA SARC -0.348; TCGA STAD -0.288 |
hsa-miR-450b-5p | BCL6 | 9 cancers: BLCA; BRCA; COAD; HNSC; KIRP; LUAD; SARC; THCA; STAD | miRNATAP | TCGA BLCA -0.09; TCGA BRCA -0.109; TCGA COAD -0.153; TCGA HNSC -0.226; TCGA KIRP -0.141; TCGA LUAD -0.054; TCGA SARC -0.079; TCGA THCA -0.117; TCGA STAD -0.259 |
hsa-miR-450b-5p | SAP30L | 10 cancers: BLCA; BRCA; CESC; COAD; HNSC; LIHC; LUAD; LUSC; PAAD; STAD | miRNATAP | TCGA BLCA -0.082; TCGA BRCA -0.081; TCGA CESC -0.073; TCGA COAD -0.076; TCGA HNSC -0.094; TCGA LIHC -0.144; TCGA LUAD -0.084; TCGA LUSC -0.158; TCGA PAAD -0.07; TCGA STAD -0.082 |
hsa-miR-450b-5p | STAT5B | 9 cancers: BLCA; BRCA; ESCA; LIHC; LUAD; LUSC; PAAD; SARC; STAD | miRNATAP | TCGA BLCA -0.095; TCGA BRCA -0.095; TCGA ESCA -0.118; TCGA LIHC -0.058; TCGA LUAD -0.092; TCGA LUSC -0.131; TCGA PAAD -0.089; TCGA SARC -0.16; TCGA STAD -0.156 |
hsa-miR-450b-5p | ARHGEF3 | 11 cancers: BLCA; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; THCA; STAD | miRNATAP | TCGA BLCA -0.112; TCGA COAD -0.131; TCGA ESCA -0.128; TCGA HNSC -0.154; TCGA KIRC -0.099; TCGA LUAD -0.094; TCGA LUSC -0.234; TCGA PAAD -0.126; TCGA PRAD -0.067; TCGA THCA -0.066; TCGA STAD -0.093 |
hsa-miR-450b-5p | ABLIM1 | 10 cancers: BLCA; CESC; ESCA; HNSC; LGG; LUAD; LUSC; PAAD; THCA; STAD | miRNATAP | TCGA BLCA -0.141; TCGA CESC -0.159; TCGA ESCA -0.234; TCGA HNSC -0.325; TCGA LGG -0.182; TCGA LUAD -0.254; TCGA LUSC -0.223; TCGA PAAD -0.129; TCGA THCA -0.076; TCGA STAD -0.147 |
hsa-miR-450b-5p | ARL6IP5 | 9 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LGG; LUSC; STAD | miRNATAP | TCGA BLCA -0.132; TCGA BRCA -0.089; TCGA COAD -0.094; TCGA ESCA -0.183; TCGA HNSC -0.081; TCGA KIRP -0.084; TCGA LGG -0.079; TCGA LUSC -0.213; TCGA STAD -0.121 |
hsa-miR-450b-5p | THRB | 10 cancers: BRCA; COAD; HNSC; KIRC; LGG; LUAD; LUSC; SARC; THCA; STAD | MirTarget; PITA; miRNATAP | TCGA BRCA -0.1; TCGA COAD -0.326; TCGA HNSC -0.136; TCGA KIRC -0.225; TCGA LGG -0.15; TCGA LUAD -0.201; TCGA LUSC -0.277; TCGA SARC -0.184; TCGA THCA -0.078; TCGA STAD -0.305 |
hsa-miR-450b-5p | AFF1 | 9 cancers: BRCA; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; SARC; STAD | MirTarget; mirMAP; miRNATAP | TCGA BRCA -0.053; TCGA COAD -0.078; TCGA ESCA -0.137; TCGA HNSC -0.156; TCGA KIRC -0.08; TCGA LUAD -0.094; TCGA LUSC -0.138; TCGA SARC -0.098; TCGA STAD -0.116 |
hsa-miR-450b-5p | CDHR3 | 9 cancers: BRCA; HNSC; LGG; LIHC; LUAD; LUSC; PAAD; PRAD; STAD | MirTarget; miRNATAP | TCGA BRCA -0.193; TCGA HNSC -0.179; TCGA LGG -0.148; TCGA LIHC -0.318; TCGA LUAD -0.46; TCGA LUSC -0.459; TCGA PAAD -0.484; TCGA PRAD -0.139; TCGA STAD -0.264 |
hsa-miR-450b-5p | TMEM56 | 9 cancers: BRCA; HNSC; KIRC; LGG; LUSC; PAAD; PRAD; SARC; STAD | MirTarget; mirMAP; miRNATAP | TCGA BRCA -0.199; TCGA HNSC -0.248; TCGA KIRC -0.137; TCGA LGG -0.277; TCGA LUSC -0.149; TCGA PAAD -0.418; TCGA PRAD -0.067; TCGA SARC -0.364; TCGA STAD -0.218 |
hsa-miR-450b-5p | HIGD1A | 10 cancers: BRCA; ESCA; HNSC; KIRC; LGG; LUAD; LUSC; SARC; STAD; UCEC | PITA | TCGA BRCA -0.121; TCGA ESCA -0.101; TCGA HNSC -0.089; TCGA KIRC -0.174; TCGA LGG -0.111; TCGA LUAD -0.071; TCGA LUSC -0.069; TCGA SARC -0.116; TCGA STAD -0.091; TCGA UCEC -0.066 |
hsa-miR-450b-5p | GKAP1 | 9 cancers: BRCA; HNSC; KIRC; LGG; LIHC; LUAD; PAAD; SARC; STAD | PITA | TCGA BRCA -0.158; TCGA HNSC -0.291; TCGA KIRC -0.11; TCGA LGG -0.081; TCGA LIHC -0.138; TCGA LUAD -0.072; TCGA PAAD -0.434; TCGA SARC -0.343; TCGA STAD -0.233 |
hsa-miR-450b-5p | NEDD4L | 10 cancers: BRCA; COAD; HNSC; KIRC; LIHC; LUAD; LUSC; PAAD; PRAD; UCEC | mirMAP; miRNATAP | TCGA BRCA -0.103; TCGA COAD -0.113; TCGA HNSC -0.111; TCGA KIRC -0.145; TCGA LIHC -0.231; TCGA LUAD -0.159; TCGA LUSC -0.124; TCGA PAAD -0.228; TCGA PRAD -0.078; TCGA UCEC -0.089 |
hsa-miR-450b-5p | SORT1 | 10 cancers: BRCA; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; SARC; STAD; UCEC | mirMAP | TCGA BRCA -0.109; TCGA ESCA -0.176; TCGA HNSC -0.29; TCGA LIHC -0.178; TCGA LUAD -0.055; TCGA LUSC -0.182; TCGA PAAD -0.112; TCGA SARC -0.31; TCGA STAD -0.085; TCGA UCEC -0.121 |
hsa-miR-450b-5p | CDKN2B | 11 cancers: BRCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; SARC; THCA | mirMAP | TCGA BRCA -0.155; TCGA CESC -0.27; TCGA COAD -0.289; TCGA ESCA -0.326; TCGA HNSC -0.269; TCGA LGG -0.174; TCGA LIHC -0.178; TCGA LUAD -0.104; TCGA LUSC -0.203; TCGA SARC -0.141; TCGA THCA -0.154 |
hsa-miR-450b-5p | HNMT | 9 cancers: BRCA; ESCA; HNSC; LGG; LUAD; LUSC; SARC; THCA; STAD | miRNATAP | TCGA BRCA -0.073; TCGA ESCA -0.21; TCGA HNSC -0.195; TCGA LGG -0.069; TCGA LUAD -0.099; TCGA LUSC -0.298; TCGA SARC -0.126; TCGA THCA -0.1; TCGA STAD -0.1 |
hsa-miR-450b-5p | MAPT | 10 cancers: COAD; ESCA; HNSC; LGG; LIHC; LUAD; PAAD; PRAD; SARC; STAD | mirMAP | TCGA COAD -0.59; TCGA ESCA -0.292; TCGA HNSC -0.309; TCGA LGG -0.158; TCGA LIHC -0.407; TCGA LUAD -0.202; TCGA PAAD -0.582; TCGA PRAD -0.067; TCGA SARC -0.491; TCGA STAD -0.68 |
hsa-miR-450b-5p | TC2N | 9 cancers: ESCA; HNSC; KIRC; KIRP; LUAD; PRAD; SARC; THCA; UCEC | MirTarget; mirMAP | TCGA ESCA -0.208; TCGA HNSC -0.298; TCGA KIRC -0.18; TCGA KIRP -0.073; TCGA LUAD -0.139; TCGA PRAD -0.08; TCGA SARC -0.243; TCGA THCA -0.119; TCGA UCEC -0.104 |
hsa-miR-450b-5p | NEBL | 9 cancers: ESCA; HNSC; KIRP; LGG; LUAD; LUSC; PAAD; PRAD; STAD | MirTarget; miRNATAP | TCGA ESCA -0.275; TCGA HNSC -0.487; TCGA KIRP -0.149; TCGA LGG -0.317; TCGA LUAD -0.119; TCGA LUSC -0.287; TCGA PAAD -0.262; TCGA PRAD -0.082; TCGA STAD -0.247 |
Enriched cancer pathways of putative targets