microRNA information: hsa-miR-452-3p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-452-3p | miRbase |
| Accession: | MIMAT0001636 | miRbase |
| Precursor name: | hsa-mir-452 | miRbase |
| Precursor accession: | MI0001733 | miRbase |
| Symbol: | MIR452 | HGNC |
| RefSeq ID: | NR_029973 | GenBank |
| Sequence: | CUCAUCUGCAAAGAAGUAAGUG |
Reported expression in cancers: hsa-miR-452-3p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-452-3p | colorectal cancer | downregulation | "Decreased miR 452 expression in human colorectal c ......" | 27323070 | qPCR |
| hsa-miR-452-3p | esophageal cancer | deregulation | "The expression profiles of miRNAs in paired EC and ......" | 23761828 | Microarray; qPCR |
| hsa-miR-452-3p | kidney renal cell cancer | deregulation | "This study aims to profile dysregulated microRNA m ......" | 25938468 | Microarray |
| hsa-miR-452-3p | liver cancer | upregulation | "We examined the transcript expression of miR-224 a ......" | 22459148 | |
| hsa-miR-452-3p | prostate cancer | downregulation | "Multiple tumor-suppressive miRNAs were downregulat ......" | 22719071 | |
| hsa-miR-452-3p | prostate cancer | downregulation | "MicroRNA-224 miR-224 and microRNA-452 miR-452 are ......" | 27070713 |
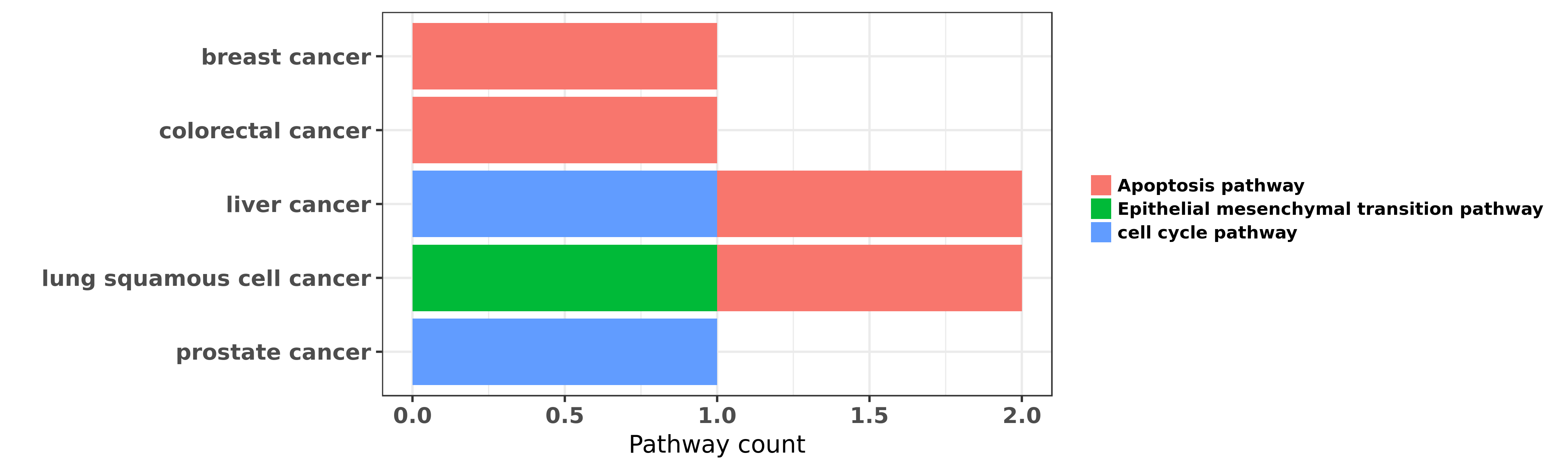
Reported cancer pathway affected by hsa-miR-452-3p
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-452-3p | breast cancer | Apoptosis pathway | "In the present study microRNA-452 miR-452 was foun ......" | 25040964 | Western blot |
| hsa-miR-452-3p | colorectal cancer | Apoptosis pathway | "Decreased miR 452 expression in human colorectal c ......" | 27323070 | |
| hsa-miR-452-3p | liver cancer | cell cycle pathway; Apoptosis pathway | "MicroRNA 452 promotes tumorigenesis in hepatocellu ......" | 24381057 | |
| hsa-miR-452-3p | lung squamous cell cancer | Apoptosis pathway; Epithelial mesenchymal transition pathway | "Expression of microRNA 452 via adenoviral vector i ......" | 26718215 | |
| hsa-miR-452-3p | prostate cancer | cell cycle pathway | "GABRE∼miR-452∼miR-224 promoter methylation was ......" | 24737792 |
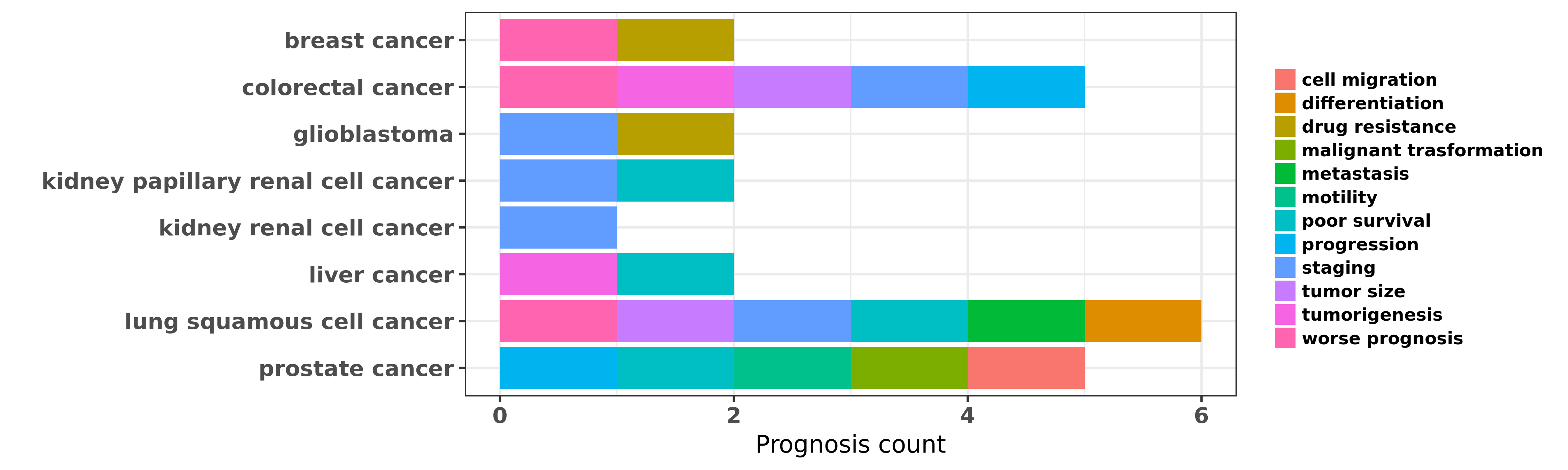
Reported cancer prognosis affected by hsa-miR-452-3p
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-452-3p | breast cancer | drug resistance | "MicroRNA 452 contributes to the docetaxel resistan ......" | 24648265 | Flow cytometry; MTT assay; Western blot |
| hsa-miR-452-3p | breast cancer | drug resistance | "In the present study microRNA-452 miR-452 was foun ......" | 25040964 | Western blot |
| hsa-miR-452-3p | breast cancer | drug resistance | "MTT-cytotoxic miRNA microarray Real-time quantitat ......" | 25562151 | Western blot; Luciferase |
| hsa-miR-452-3p | breast cancer | worse prognosis; drug resistance | "The aim of this study was to investigate the role ......" | 27746365 | |
| hsa-miR-452-3p | colorectal cancer | progression; tumorigenesis; staging; tumor size; worse prognosis | "Decreased miR 452 expression in human colorectal c ......" | 27323070 | |
| hsa-miR-452-3p | glioblastoma | drug resistance; staging | "The SOX2 response program in glioblastoma multifor ......" | 21211035 | Colony formation |
| hsa-miR-452-3p | kidney papillary renal cell cancer | staging; poor survival | "In the training stage the expression levels of 12 ......" | 25906110 | |
| hsa-miR-452-3p | kidney renal cell cancer | staging | "This study aims to profile dysregulated microRNA m ......" | 25938468 | |
| hsa-miR-452-3p | liver cancer | tumorigenesis | "MicroRNA 452 promotes tumorigenesis in hepatocellu ......" | 24381057 | |
| hsa-miR-452-3p | liver cancer | tumorigenesis; poor survival | "MicroRNA 452 promotes stem like cells of hepatocel ......" | 27058905 | |
| hsa-miR-452-3p | lung squamous cell cancer | metastasis; staging | "Up Regulation of MiR 452 Inhibits Metastasis of No ......" | 26316085 | Transwell assay; Western blot; Luciferase |
| hsa-miR-452-3p | lung squamous cell cancer | metastasis; poor survival | "Expression of microRNA 452 via adenoviral vector i ......" | 26718215 | |
| hsa-miR-452-3p | lung squamous cell cancer | worse prognosis; poor survival; metastasis; differentiation; tumor size | "Down regulation of miR 452 is associated with poor ......" | 27162664 | |
| hsa-miR-452-3p | prostate cancer | malignant trasformation; motility | "GABRE∼miR-452∼miR-224 promoter methylation was ......" | 24737792 | |
| hsa-miR-452-3p | prostate cancer | cell migration; poor survival; progression | "Regulation of E3 ubiquitin ligase 1 WWP1 by microR ......" | 27070713 | Luciferase |
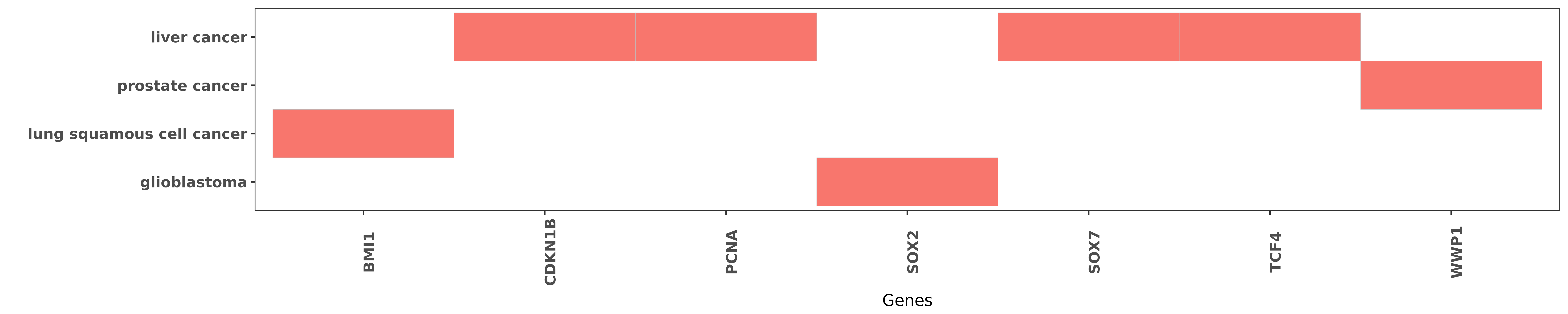
Reported gene related to hsa-miR-452-3p
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-452-3p | lung squamous cell cancer | BMI1 | "Up Regulation of MiR 452 Inhibits Metastasis of No ......" | 26316085 |
| hsa-miR-452-3p | liver cancer | CDKN1B | "Silencing CDKN1B by small interfering RNA resemble ......" | 24381057 |
| hsa-miR-452-3p | liver cancer | PCNA | "MicroRNA 452 promotes tumorigenesis in hepatocellu ......" | 24381057 |
| hsa-miR-452-3p | glioblastoma | SOX2 | "Using next generation sequencing we identified 105 ......" | 21211035 |
| hsa-miR-452-3p | liver cancer | SOX7 | "MicroRNA 452 promotes stem like cells of hepatocel ......" | 27058905 |
| hsa-miR-452-3p | liver cancer | TCF4 | "Further Sox7 was identified as the direct target o ......" | 27058905 |
| hsa-miR-452-3p | prostate cancer | WWP1 | "Regulation of E3 ubiquitin ligase 1 WWP1 by microR ......" | 27070713 |
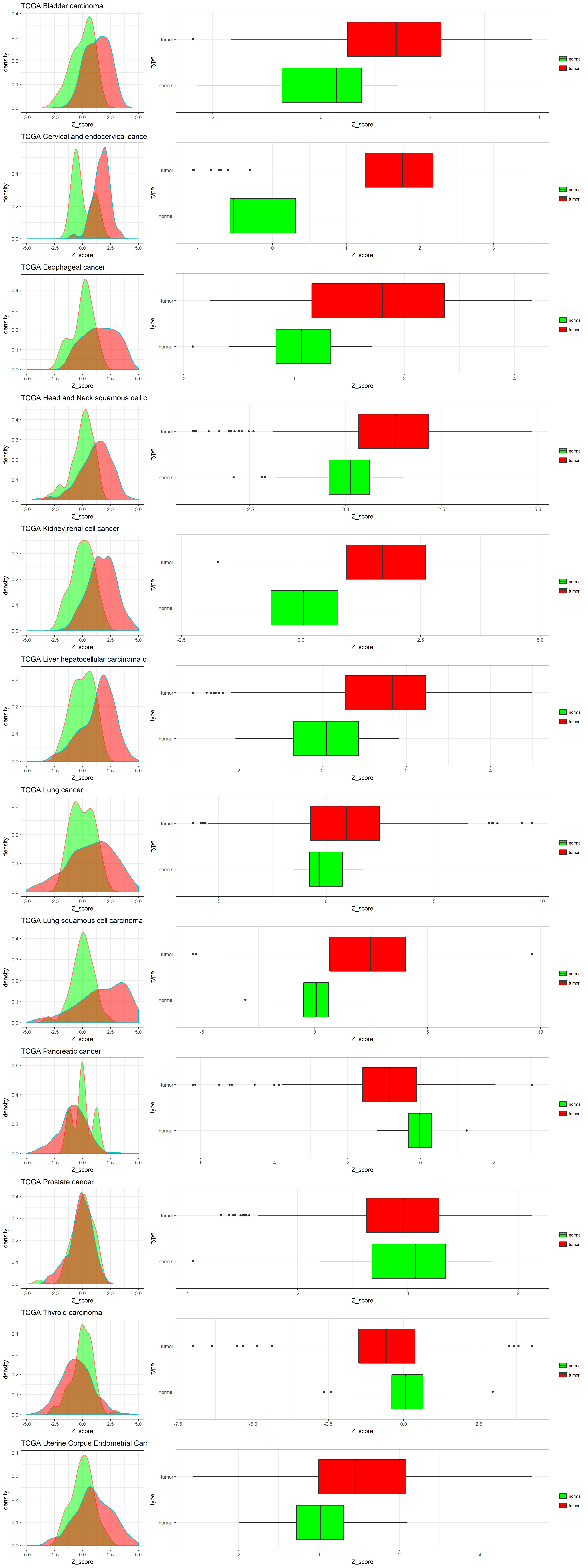
Expression profile in cancer corhorts:
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-452-3p | PRKD1 | 10 cancers: BLCA; CESC; COAD; HNSC; KIRC; LUAD; LUSC; PAAD; PRAD; SARC | MirTarget | TCGA BLCA -0.223; TCGA CESC -0.203; TCGA COAD -0.513; TCGA HNSC -0.365; TCGA KIRC -0.219; TCGA LUAD -0.085; TCGA LUSC -0.215; TCGA PAAD -0.353; TCGA PRAD -0.091; TCGA SARC -0.131 |
hsa-miR-452-3p | SMAD9 | 11 cancers: BLCA; CESC; COAD; ESCA; HNSC; LIHC; LUAD; LUSC; PAAD; THCA; UCEC | MirTarget | TCGA BLCA -0.226; TCGA CESC -0.269; TCGA COAD -0.28; TCGA ESCA -0.194; TCGA HNSC -0.305; TCGA LIHC -0.175; TCGA LUAD -0.212; TCGA LUSC -0.176; TCGA PAAD -0.352; TCGA THCA -0.176; TCGA UCEC -0.161 |
hsa-miR-452-3p | RNF125 | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; LIHC; LUSC; THCA; UCEC | MirTarget | TCGA BLCA -0.093; TCGA CESC -0.166; TCGA COAD -0.242; TCGA ESCA -0.378; TCGA HNSC -0.271; TCGA LIHC -0.165; TCGA LUSC -0.209; TCGA THCA -0.122; TCGA UCEC -0.122 |
hsa-miR-452-3p | ANK2 | 11 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; PAAD; SARC | MirTarget | TCGA BLCA -0.281; TCGA CESC -0.214; TCGA COAD -0.608; TCGA ESCA -0.314; TCGA HNSC -0.507; TCGA KIRC -0.335; TCGA LIHC -0.109; TCGA LUAD -0.119; TCGA LUSC -0.414; TCGA PAAD -0.619; TCGA SARC -0.138 |
hsa-miR-452-3p | TRAK2 | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LUSC; PRAD; SARC | MirTarget | TCGA BLCA -0.06; TCGA CESC -0.122; TCGA COAD -0.071; TCGA ESCA -0.131; TCGA HNSC -0.161; TCGA KIRC -0.11; TCGA LUSC -0.122; TCGA PRAD -0.07; TCGA SARC -0.06 |
hsa-miR-452-3p | ATP2A3 | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; SARC; THCA | MirTarget | TCGA BLCA -0.201; TCGA CESC -0.172; TCGA COAD -0.163; TCGA ESCA -0.542; TCGA HNSC -0.337; TCGA LUAD -0.2; TCGA LUSC -0.277; TCGA PAAD -0.191; TCGA SARC -0.112; TCGA THCA -0.199 |
hsa-miR-452-3p | KLF9 | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; LUSC; PAAD | MirTarget | TCGA BLCA -0.14; TCGA CESC -0.23; TCGA COAD -0.336; TCGA ESCA -0.27; TCGA HNSC -0.221; TCGA KIRC -0.077; TCGA LIHC -0.163; TCGA LUSC -0.255; TCGA PAAD -0.184 |
hsa-miR-452-3p | PPP1R12B | 10 cancers: BLCA; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; SARC; UCEC | mirMAP | TCGA BLCA -0.218; TCGA COAD -0.385; TCGA ESCA -0.276; TCGA HNSC -0.218; TCGA KIRC -0.093; TCGA LUAD -0.069; TCGA LUSC -0.199; TCGA PAAD -0.18; TCGA SARC -0.708; TCGA UCEC -0.177 |
hsa-miR-452-3p | SYT11 | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; SARC | mirMAP | TCGA BLCA -0.216; TCGA CESC -0.177; TCGA COAD -0.4; TCGA ESCA -0.176; TCGA HNSC -0.254; TCGA LUAD -0.058; TCGA LUSC -0.298; TCGA PAAD -0.332; TCGA SARC -0.157 |
hsa-miR-452-3p | ALPK3 | 9 cancers: BLCA; CESC; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; SARC | mirMAP | TCGA BLCA -0.154; TCGA CESC -0.248; TCGA ESCA -0.256; TCGA HNSC -0.252; TCGA KIRC -0.21; TCGA LUAD -0.162; TCGA LUSC -0.299; TCGA PAAD -0.393; TCGA SARC -0.35 |
hsa-miR-452-3p | PRDM16 | 9 cancers: BLCA; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; SARC; UCEC | mirMAP | TCGA BLCA -0.325; TCGA COAD -0.243; TCGA ESCA -0.605; TCGA HNSC -0.132; TCGA KIRC -0.796; TCGA LUAD -0.188; TCGA LUSC -0.179; TCGA SARC -0.346; TCGA UCEC -0.187 |
hsa-miR-452-3p | AR | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; LIHC; LUSC; PRAD; SARC; UCEC | mirMAP | TCGA BLCA -0.204; TCGA CESC -0.354; TCGA COAD -0.543; TCGA ESCA -0.149; TCGA HNSC -0.43; TCGA LIHC -0.214; TCGA LUSC -0.28; TCGA PRAD -0.199; TCGA SARC -0.479; TCGA UCEC -0.219 |
hsa-miR-452-3p | PARM1 | 11 cancers: BLCA; CESC; ESCA; HNSC; KIRC; LUAD; LUSC; PAAD; SARC; THCA; UCEC | mirMAP | TCGA BLCA -0.151; TCGA CESC -0.148; TCGA ESCA -0.326; TCGA HNSC -0.388; TCGA KIRC -0.206; TCGA LUAD -0.178; TCGA LUSC -0.258; TCGA PAAD -0.188; TCGA SARC -0.368; TCGA THCA -0.153; TCGA UCEC -0.112 |
hsa-miR-452-3p | LONRF2 | 11 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; PAAD; PRAD; SARC; UCEC | mirMAP | TCGA BLCA -0.467; TCGA CESC -0.395; TCGA COAD -0.887; TCGA ESCA -0.31; TCGA HNSC -0.581; TCGA KIRC -0.537; TCGA LIHC -0.177; TCGA PAAD -0.535; TCGA PRAD -0.104; TCGA SARC -0.457; TCGA UCEC -0.126 |
hsa-miR-452-3p | FAM46C | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; PAAD; SARC; UCEC | mirMAP | TCGA BLCA -0.142; TCGA CESC -0.267; TCGA COAD -0.194; TCGA ESCA -0.424; TCGA HNSC -0.385; TCGA KIRC -0.269; TCGA LIHC -0.116; TCGA LUAD -0.161; TCGA LUSC -0.388; TCGA PAAD -0.247; TCGA SARC -0.317; TCGA UCEC -0.194 |
hsa-miR-452-3p | LDOC1L | 9 cancers: BLCA; CESC; COAD; HNSC; KIRC; LIHC; LUAD; PAAD; SARC | mirMAP | TCGA BLCA -0.07; TCGA CESC -0.082; TCGA COAD -0.092; TCGA HNSC -0.082; TCGA KIRC -0.103; TCGA LIHC -0.094; TCGA LUAD -0.064; TCGA PAAD -0.168; TCGA SARC -0.067 |
hsa-miR-452-3p | RGAG4 | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; PAAD | mirMAP | TCGA BLCA -0.244; TCGA CESC -0.148; TCGA COAD -0.347; TCGA ESCA -0.12; TCGA HNSC -0.375; TCGA KIRC -0.236; TCGA LIHC -0.291; TCGA LUAD -0.109; TCGA LUSC -0.138; TCGA PAAD -0.409 |
hsa-miR-452-3p | APPL1 | 9 cancers: CESC; ESCA; HNSC; KIRC; LUAD; PAAD; PRAD; THCA; UCEC | MirTarget | TCGA CESC -0.065; TCGA ESCA -0.092; TCGA HNSC -0.07; TCGA KIRC -0.108; TCGA LUAD -0.061; TCGA PAAD -0.078; TCGA PRAD -0.052; TCGA THCA -0.058; TCGA UCEC -0.076 |
hsa-miR-452-3p | POLK | 11 cancers: CESC; COAD; ESCA; HNSC; LIHC; LUSC; PAAD; PRAD; SARC; THCA; UCEC | MirTarget | TCGA CESC -0.065; TCGA COAD -0.056; TCGA ESCA -0.055; TCGA HNSC -0.087; TCGA LIHC -0.052; TCGA LUSC -0.077; TCGA PAAD -0.082; TCGA PRAD -0.098; TCGA SARC -0.088; TCGA THCA -0.065; TCGA UCEC -0.07 |
hsa-miR-452-3p | LPIN2 | 10 cancers: CESC; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; PRAD; THCA | MirTarget | TCGA CESC -0.18; TCGA COAD -0.066; TCGA ESCA -0.42; TCGA HNSC -0.143; TCGA KIRC -0.181; TCGA LIHC -0.113; TCGA LUAD -0.087; TCGA LUSC -0.211; TCGA PRAD -0.076; TCGA THCA -0.057 |
hsa-miR-452-3p | ARL15 | 10 cancers: CESC; COAD; ESCA; HNSC; LIHC; LUAD; PAAD; PRAD; THCA; UCEC | MirTarget | TCGA CESC -0.09; TCGA COAD -0.091; TCGA ESCA -0.061; TCGA HNSC -0.08; TCGA LIHC -0.07; TCGA LUAD -0.054; TCGA PAAD -0.236; TCGA PRAD -0.11; TCGA THCA -0.082; TCGA UCEC -0.098 |
Enriched cancer pathways of putative targets