microRNA information: hsa-miR-495-3p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-495-3p | miRbase |
| Accession: | MIMAT0002817 | miRbase |
| Precursor name: | hsa-mir-495 | miRbase |
| Precursor accession: | MI0003135 | miRbase |
| Symbol: | MIR495 | HGNC |
| RefSeq ID: | NR_030175 | GenBank |
| Sequence: | AAACAAACAUGGUGCACUUCUU |
Reported expression in cancers: hsa-miR-495-3p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-495-3p | breast cancer | upregulation | "Coincidently high upregulation of miR-495 was also ......" | 21258409 | |
| hsa-miR-495-3p | breast cancer | upregulation | "In the present study we found that miR-495 was mar ......" | 25070379 | qPCR |
| hsa-miR-495-3p | glioblastoma | downregulation | "This study revealed miR-495 is down-regulated in g ......" | 23594394 | |
| hsa-miR-495-3p | lung cancer | upregulation | "The expression of 319 miRNAs was evaluated by Exiq ......" | 20818338 | Microarray |
| hsa-miR-495-3p | lung cancer | downregulation | "MicroRNA 495 mimics delivery inhibits lung tumor p ......" | 25286762 |
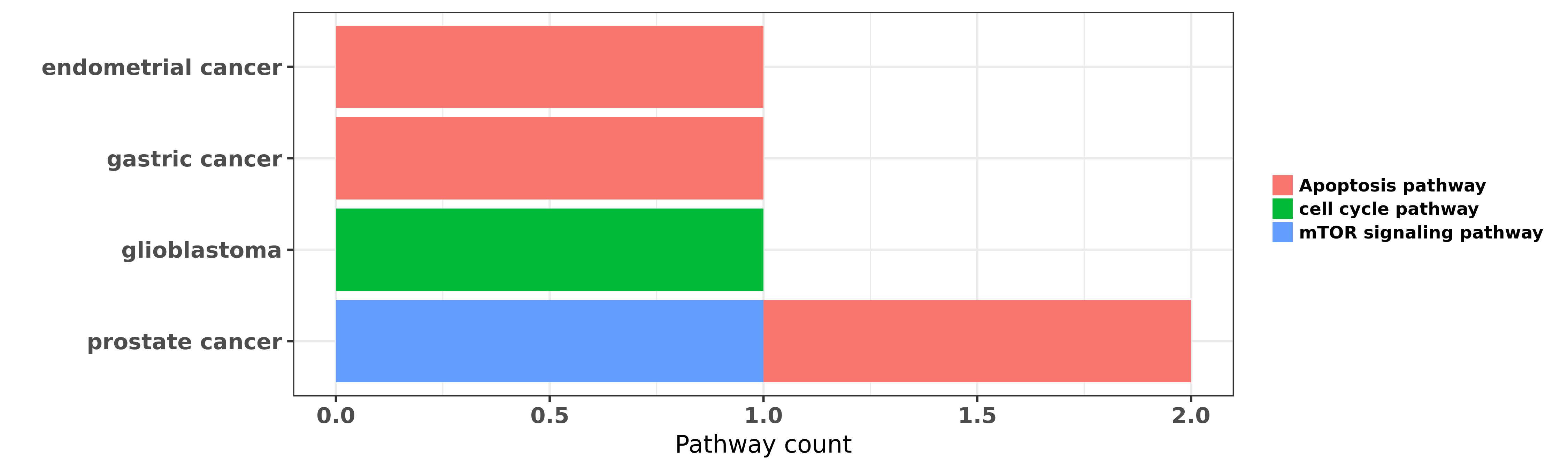
Reported cancer pathway affected by hsa-miR-495-3p
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-495-3p | endometrial cancer | Apoptosis pathway | "MicroRNA 495 downregulates FOXC1 expression to sup ......" | 26198045 | |
| hsa-miR-495-3p | gastric cancer | Apoptosis pathway | "Targeting of RUNX3 by miR 130a and miR 495 coopera ......" | 26375442 | Luciferase |
| hsa-miR-495-3p | glioblastoma | cell cycle pathway | "MicroRNA 495 inhibits proliferation of glioblastom ......" | 23594394 | Western blot |
| hsa-miR-495-3p | prostate cancer | Apoptosis pathway | "MicroRNAs miR 154 miR 299 5p miR 376a miR 376c miR ......" | 24166498 | |
| hsa-miR-495-3p | prostate cancer | mTOR signaling pathway | "MicroRNA 495 Regulates Migration and Invasion in P ......" | 27031291 |
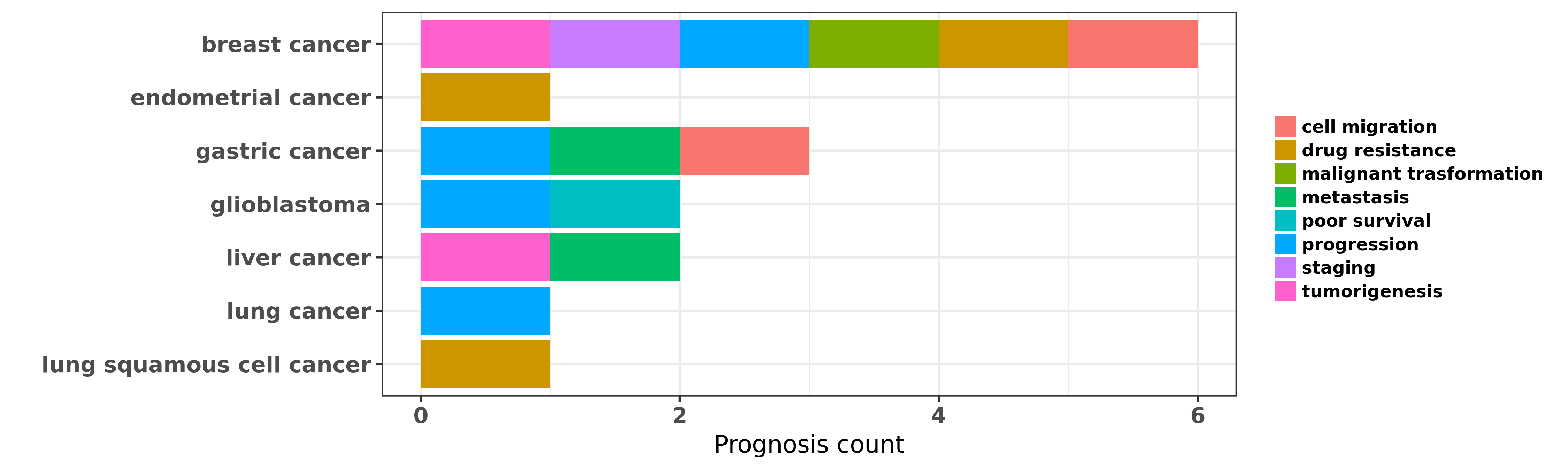
Reported cancer prognosis affected by hsa-miR-495-3p
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-495-3p | breast cancer | drug resistance; tumorigenesis | "miR 495 is upregulated by E12/E47 in breast cancer ......" | 21258409 | Colony formation |
| hsa-miR-495-3p | breast cancer | cell migration; progression | "MicroRNA 495 induces breast cancer cell migration ......" | 25070379 | Western blot; Luciferase |
| hsa-miR-495-3p | breast cancer | progression; malignant trasformation | "Downregulated miR 495 Corrected Inhibits the G1 S ......" | 26020378 | Flow cytometry; Luciferase; Colony formation |
| hsa-miR-495-3p | breast cancer | staging | "Results showed differential pattern of expressions ......" | 26276721 | |
| hsa-miR-495-3p | endometrial cancer | drug resistance | "MicroRNA 495 downregulates FOXC1 expression to sup ......" | 26198045 | |
| hsa-miR-495-3p | gastric cancer | cell migration | "miR 495 and miR 551a inhibit the migration and inv ......" | 22469786 | Western blot |
| hsa-miR-495-3p | gastric cancer | metastasis | "Methylation associated silencing of miR 495 inhibi ......" | 25475733 | |
| hsa-miR-495-3p | gastric cancer | progression | "Targeting of RUNX3 by miR 130a and miR 495 coopera ......" | 26375442 | Luciferase |
| hsa-miR-495-3p | glioblastoma | poor survival; progression | "MicroRNA 495 inhibits proliferation of glioblastom ......" | 23594394 | Western blot |
| hsa-miR-495-3p | liver cancer | metastasis; tumorigenesis | "Expression of miR-664 miR-485-3p and miR-495 poten ......" | 23241961 | |
| hsa-miR-495-3p | lung cancer | progression | "MicroRNA 495 mimics delivery inhibits lung tumor p ......" | 25286762 | |
| hsa-miR-495-3p | lung squamous cell cancer | drug resistance | "To better understand the molecular mechanisms of m ......" | 20371173 | |
| hsa-miR-495-3p | lung squamous cell cancer | drug resistance | "miR 495 enhances the sensitivity of non small cell ......" | 24038379 | Western blot |
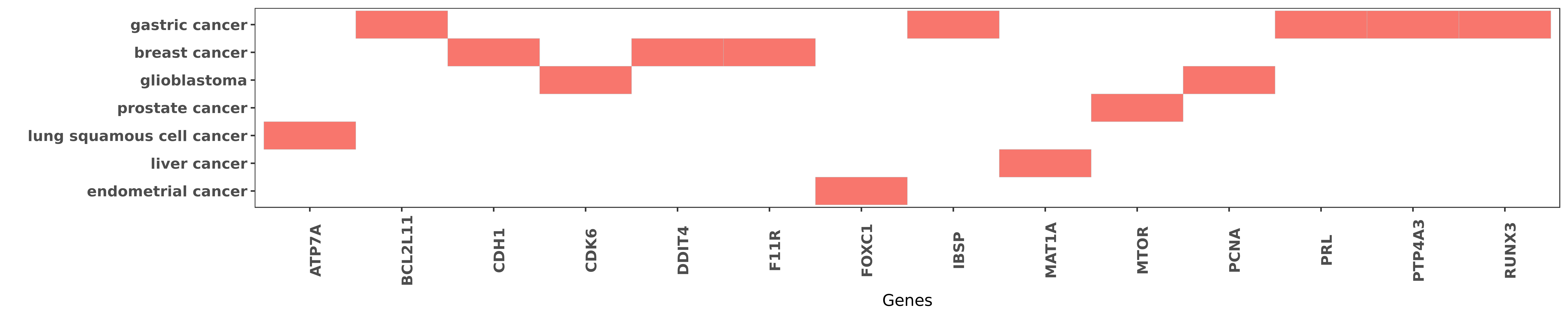
Reported gene related to hsa-miR-495-3p
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-495-3p | gastric cancer | PRL | "Methylation associated silencing of miR 495 inhibi ......" | 25475733 |
| hsa-miR-495-3p | gastric cancer | PRL | "miR 495 and miR 551a inhibit the migration and inv ......" | 22469786 |
| hsa-miR-495-3p | gastric cancer | PTP4A3 | "Methylation associated silencing of miR 495 inhibi ......" | 25475733 |
| hsa-miR-495-3p | gastric cancer | PTP4A3 | "miR 495 and miR 551a inhibit the migration and inv ......" | 22469786 |
| hsa-miR-495-3p | lung squamous cell cancer | ATP7A | "miR 495 enhances the sensitivity of non small cell ......" | 24038379 |
| hsa-miR-495-3p | gastric cancer | BCL2L11 | "Combination of miR-130a and miR-495 significantly ......" | 26375442 |
| hsa-miR-495-3p | breast cancer | CDH1 | "miR 495 is upregulated by E12/E47 in breast cancer ......" | 21258409 |
| hsa-miR-495-3p | glioblastoma | CDK6 | "Expression of CDK6 was also analyzed after over-ex ......" | 23594394 |
| hsa-miR-495-3p | breast cancer | DDIT4 | "miR 495 is upregulated by E12/E47 in breast cancer ......" | 21258409 |
| hsa-miR-495-3p | breast cancer | F11R | "MicroRNA 495 induces breast cancer cell migration ......" | 25070379 |
| hsa-miR-495-3p | endometrial cancer | FOXC1 | "MicroRNA 495 downregulates FOXC1 expression to sup ......" | 26198045 |
| hsa-miR-495-3p | gastric cancer | IBSP | "Methylation specific PCR MSP and sodium bisulfite ......" | 25475733 |
| hsa-miR-495-3p | liver cancer | MAT1A | "Blocking MAT1A induction significantly reduced the ......" | 23241961 |
| hsa-miR-495-3p | prostate cancer | MTOR | "MicroRNA 495 Regulates Migration and Invasion in P ......" | 27031291 |
| hsa-miR-495-3p | glioblastoma | PCNA | "MicroRNA 495 inhibits proliferation of glioblastom ......" | 23594394 |
| hsa-miR-495-3p | gastric cancer | RUNX3 | "Targeting of RUNX3 by miR 130a and miR 495 coopera ......" | 26375442 |
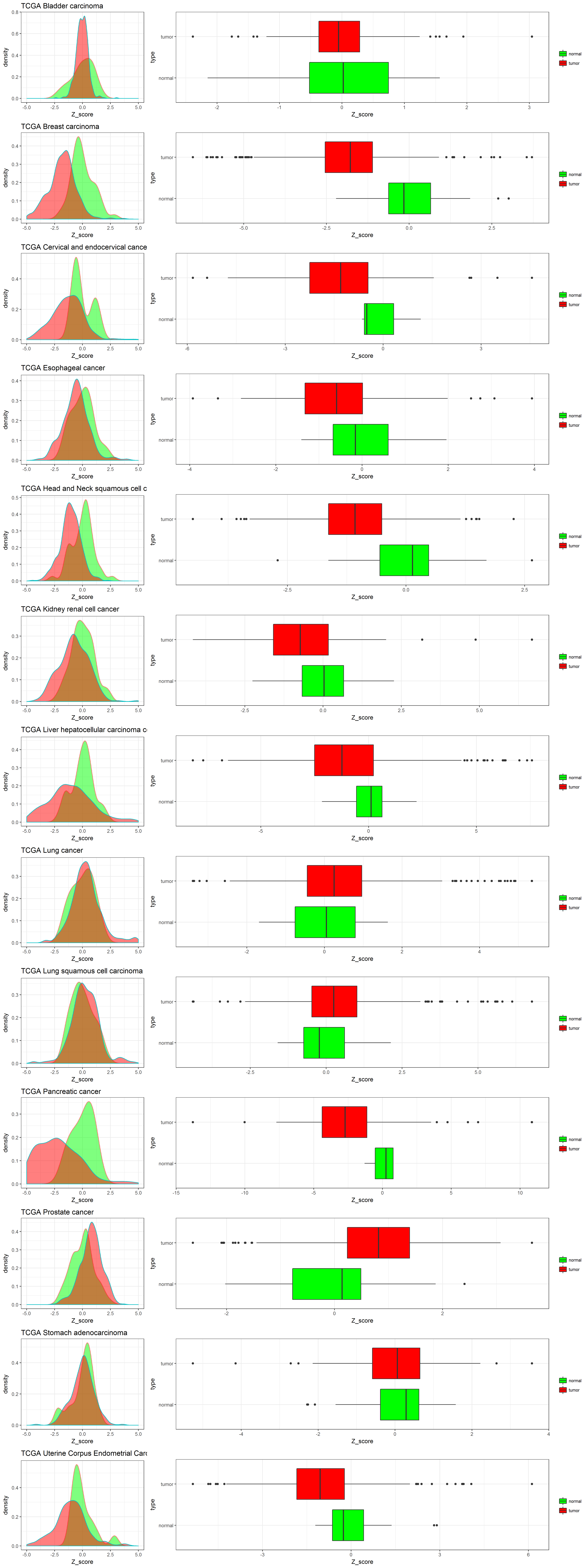
Expression profile in cancer corhorts:
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-495-3p | RPRD1A | 9 cancers: BLCA; BRCA; COAD; HNSC; LUAD; LUSC; PRAD; SARC; STAD | MirTarget; mirMAP | TCGA BLCA -0.127; TCGA BRCA -0.11; TCGA COAD -0.117; TCGA HNSC -0.079; TCGA LUAD -0.052; TCGA LUSC -0.082; TCGA PRAD -0.148; TCGA SARC -0.077; TCGA STAD -0.137 |
hsa-miR-495-3p | PHC3 | 9 cancers: BLCA; COAD; ESCA; HNSC; KIRC; LGG; PRAD; SARC; STAD | MirTarget | TCGA BLCA -0.103; TCGA COAD -0.075; TCGA ESCA -0.093; TCGA HNSC -0.08; TCGA KIRC -0.054; TCGA LGG -0.061; TCGA PRAD -0.083; TCGA SARC -0.086; TCGA STAD -0.138 |
hsa-miR-495-3p | ATL2 | 10 cancers: BLCA; COAD; HNSC; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.07; TCGA COAD -0.123; TCGA HNSC -0.078; TCGA LUAD -0.083; TCGA LUSC -0.067; TCGA PAAD -0.08; TCGA PRAD -0.137; TCGA SARC -0.092; TCGA STAD -0.143; TCGA UCEC -0.057 |
hsa-miR-495-3p | MYNN | 9 cancers: BLCA; COAD; ESCA; HNSC; LGG; LUAD; PAAD; PRAD; STAD | MirTarget | TCGA BLCA -0.073; TCGA COAD -0.121; TCGA ESCA -0.105; TCGA HNSC -0.083; TCGA LGG -0.07; TCGA LUAD -0.055; TCGA PAAD -0.054; TCGA PRAD -0.071; TCGA STAD -0.073 |
hsa-miR-495-3p | RBM41 | 10 cancers: BLCA; COAD; HNSC; KIRC; LIHC; PAAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.137; TCGA COAD -0.139; TCGA HNSC -0.078; TCGA KIRC -0.079; TCGA LIHC -0.075; TCGA PAAD -0.143; TCGA PRAD -0.093; TCGA SARC -0.053; TCGA STAD -0.159; TCGA UCEC -0.062 |
hsa-miR-495-3p | ZBTB41 | 9 cancers: BLCA; BRCA; COAD; ESCA; LIHC; LUSC; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.066; TCGA BRCA -0.116; TCGA COAD -0.105; TCGA ESCA -0.105; TCGA LIHC -0.08; TCGA LUSC -0.064; TCGA PRAD -0.125; TCGA STAD -0.161; TCGA UCEC -0.069 |
hsa-miR-495-3p | POLD3 | 10 cancers: BLCA; BRCA; CESC; COAD; HNSC; LGG; PAAD; PRAD; SARC; STAD | mirMAP | TCGA BLCA -0.069; TCGA BRCA -0.066; TCGA CESC -0.067; TCGA COAD -0.096; TCGA HNSC -0.064; TCGA LGG -0.064; TCGA PAAD -0.069; TCGA PRAD -0.055; TCGA SARC -0.1; TCGA STAD -0.097 |
hsa-miR-495-3p | WDR76 | 10 cancers: BLCA; BRCA; CESC; HNSC; LGG; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.098; TCGA BRCA -0.193; TCGA CESC -0.114; TCGA HNSC -0.21; TCGA LGG -0.132; TCGA PAAD -0.14; TCGA PRAD -0.082; TCGA SARC -0.144; TCGA STAD -0.165; TCGA UCEC -0.071 |
hsa-miR-495-3p | FMR1 | 9 cancers: BLCA; BRCA; COAD; HNSC; LIHC; LUSC; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.111; TCGA BRCA -0.058; TCGA COAD -0.105; TCGA HNSC -0.098; TCGA LIHC -0.091; TCGA LUSC -0.066; TCGA SARC -0.055; TCGA STAD -0.167; TCGA UCEC -0.053 |
hsa-miR-495-3p | MDM1 | 9 cancers: BLCA; BRCA; CESC; COAD; HNSC; KIRC; LGG; PRAD; STAD | mirMAP | TCGA BLCA -0.108; TCGA BRCA -0.062; TCGA CESC -0.069; TCGA COAD -0.114; TCGA HNSC -0.081; TCGA KIRC -0.072; TCGA LGG -0.086; TCGA PRAD -0.058; TCGA STAD -0.172 |
hsa-miR-495-3p | KPNA1 | 9 cancers: BLCA; BRCA; COAD; ESCA; HNSC; PAAD; PRAD; SARC; STAD | mirMAP | TCGA BLCA -0.062; TCGA BRCA -0.054; TCGA COAD -0.066; TCGA ESCA -0.09; TCGA HNSC -0.067; TCGA PAAD -0.051; TCGA PRAD -0.058; TCGA SARC -0.064; TCGA STAD -0.084 |
hsa-miR-495-3p | FAM199X | 9 cancers: BLCA; BRCA; COAD; HNSC; LIHC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.09; TCGA BRCA -0.074; TCGA COAD -0.148; TCGA HNSC -0.115; TCGA LIHC -0.052; TCGA PAAD -0.09; TCGA PRAD -0.143; TCGA STAD -0.165; TCGA UCEC -0.105 |
hsa-miR-495-3p | INTS7 | 9 cancers: BLCA; BRCA; COAD; HNSC; LGG; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.057; TCGA BRCA -0.187; TCGA COAD -0.117; TCGA HNSC -0.102; TCGA LGG -0.074; TCGA PRAD -0.151; TCGA SARC -0.075; TCGA STAD -0.075; TCGA UCEC -0.099 |
hsa-miR-495-3p | FBXO28 | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; PAAD; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.057; TCGA BRCA -0.066; TCGA COAD -0.078; TCGA ESCA -0.118; TCGA HNSC -0.06; TCGA PAAD -0.117; TCGA PRAD -0.081; TCGA SARC -0.051; TCGA STAD -0.1; TCGA UCEC -0.055 |
hsa-miR-495-3p | GTF2E1 | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LIHC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.052; TCGA BRCA -0.064; TCGA COAD -0.078; TCGA ESCA -0.094; TCGA HNSC -0.082; TCGA LIHC -0.053; TCGA PAAD -0.077; TCGA PRAD -0.083; TCGA STAD -0.1; TCGA UCEC -0.052 |
hsa-miR-495-3p | DTX3L | 10 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LGG; PAAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.073; TCGA BRCA -0.105; TCGA CESC -0.126; TCGA ESCA -0.128; TCGA HNSC -0.149; TCGA LGG -0.123; TCGA PAAD -0.172; TCGA SARC -0.101; TCGA STAD -0.117; TCGA UCEC -0.101 |
hsa-miR-495-3p | PRRG4 | 10 cancers: BRCA; CESC; COAD; HNSC; KIRC; LUSC; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BRCA -0.151; TCGA CESC -0.146; TCGA COAD -0.249; TCGA HNSC -0.25; TCGA KIRC -0.102; TCGA LUSC -0.106; TCGA PRAD -0.204; TCGA SARC -0.252; TCGA STAD -0.155; TCGA UCEC -0.142 |
hsa-miR-495-3p | TUFT1 | 10 cancers: BRCA; CESC; HNSC; KIRC; LUAD; LUSC; PAAD; SARC; STAD; UCEC | MirTarget | TCGA BRCA -0.192; TCGA CESC -0.12; TCGA HNSC -0.131; TCGA KIRC -0.088; TCGA LUAD -0.085; TCGA LUSC -0.112; TCGA PAAD -0.147; TCGA SARC -0.065; TCGA STAD -0.108; TCGA UCEC -0.108 |
hsa-miR-495-3p | MOCOS | 10 cancers: BRCA; CESC; ESCA; HNSC; LIHC; LUSC; PAAD; PRAD; SARC; UCEC | mirMAP | TCGA BRCA -0.114; TCGA CESC -0.128; TCGA ESCA -0.203; TCGA HNSC -0.142; TCGA LIHC -0.095; TCGA LUSC -0.115; TCGA PAAD -0.494; TCGA PRAD -0.134; TCGA SARC -0.071; TCGA UCEC -0.123 |
hsa-miR-495-3p | TAF5L | 9 cancers: BRCA; COAD; ESCA; HNSC; LGG; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BRCA -0.086; TCGA COAD -0.062; TCGA ESCA -0.105; TCGA HNSC -0.06; TCGA LGG -0.081; TCGA PRAD -0.065; TCGA SARC -0.078; TCGA STAD -0.056; TCGA UCEC -0.068 |
hsa-miR-495-3p | UBXN2B | 9 cancers: BRCA; COAD; HNSC; KIRC; LIHC; PAAD; PRAD; SARC; STAD | mirMAP | TCGA BRCA -0.09; TCGA COAD -0.15; TCGA HNSC -0.054; TCGA KIRC -0.054; TCGA LIHC -0.056; TCGA PAAD -0.067; TCGA PRAD -0.08; TCGA SARC -0.062; TCGA STAD -0.131 |
Enriched cancer pathways of putative targets