microRNA information: hsa-miR-505-5p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-505-5p | miRbase |
| Accession: | MIMAT0004776 | miRbase |
| Precursor name: | hsa-mir-505 | miRbase |
| Precursor accession: | MI0003190 | miRbase |
| Symbol: | MIR505 | HGNC |
| RefSeq ID: | NR_030230 | GenBank |
| Sequence: | GGGAGCCAGGAAGUAUUGAUGU |
Reported expression in cancers: hsa-miR-505-5p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-505-5p | breast cancer | downregulation | "Here we performed an integrated genomic analysis c ......" | 22051041 | Microarray |
| hsa-miR-505-5p | endometrial cancer | downregulation | "MicroRNA 505 functions as a tumor suppressor in en ......" | 26832151 | Reverse transcription PCR |
Reported cancer pathway affected by hsa-miR-505-5p
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-505-5p | breast cancer | Apoptosis pathway | "Here we performed an integrated genomic analysis c ......" | 22051041 | |
| hsa-miR-505-5p | endometrial cancer | Apoptosis pathway | "MicroRNA 505 functions as a tumor suppressor in en ......" | 26832151 | Luciferase; Western blot |
Reported cancer prognosis affected by hsa-miR-505-5p
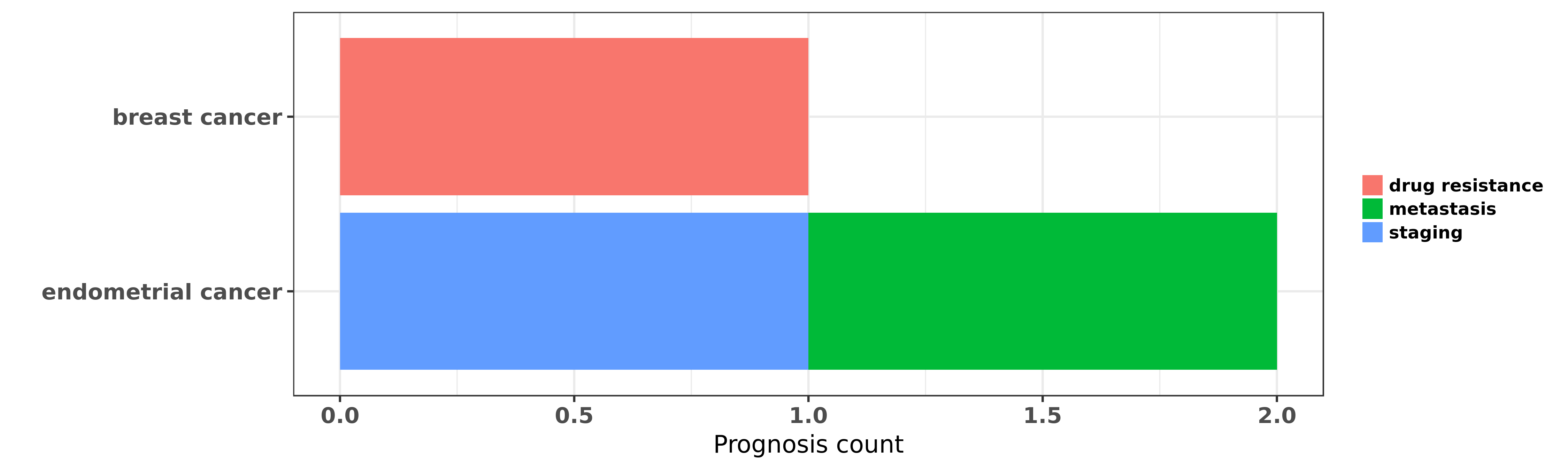
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-505-5p | breast cancer | drug resistance | "Here we performed an integrated genomic analysis c ......" | 22051041 | |
| hsa-miR-505-5p | endometrial cancer | staging; metastasis | "MicroRNA 505 functions as a tumor suppressor in en ......" | 26832151 | Luciferase; Western blot |
Reported gene related to hsa-miR-505-5p
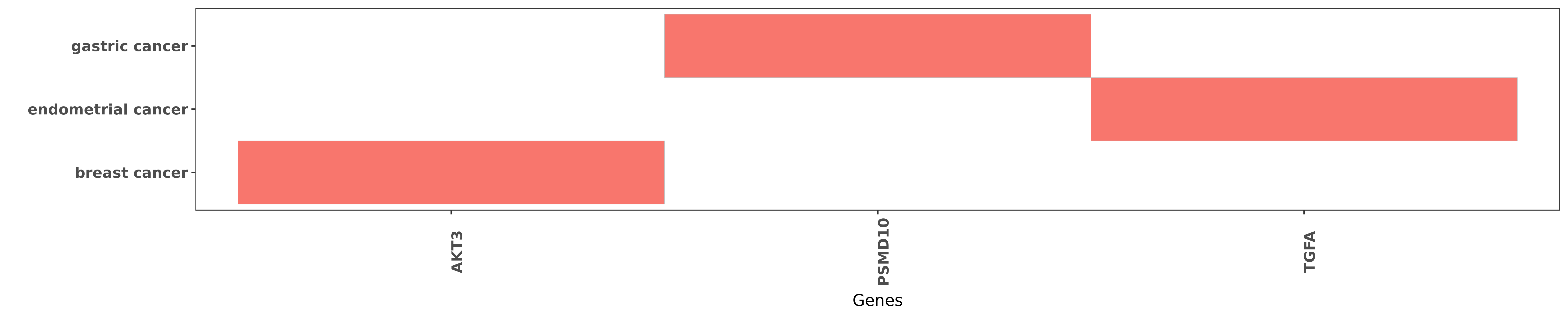
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-505-5p | breast cancer | AKT3 | "We also find that Akt3 correlate inversely with mi ......" | 22051041 |
| hsa-miR-505-5p | gastric cancer | PSMD10 | "Association between functional PSMD10 Rs111638916 ......" | 26394032 |
| hsa-miR-505-5p | endometrial cancer | TGFA | "A miR-505 binding site was identified in the 3' un ......" | 26832151 |
Expression profile in cancer corhorts:
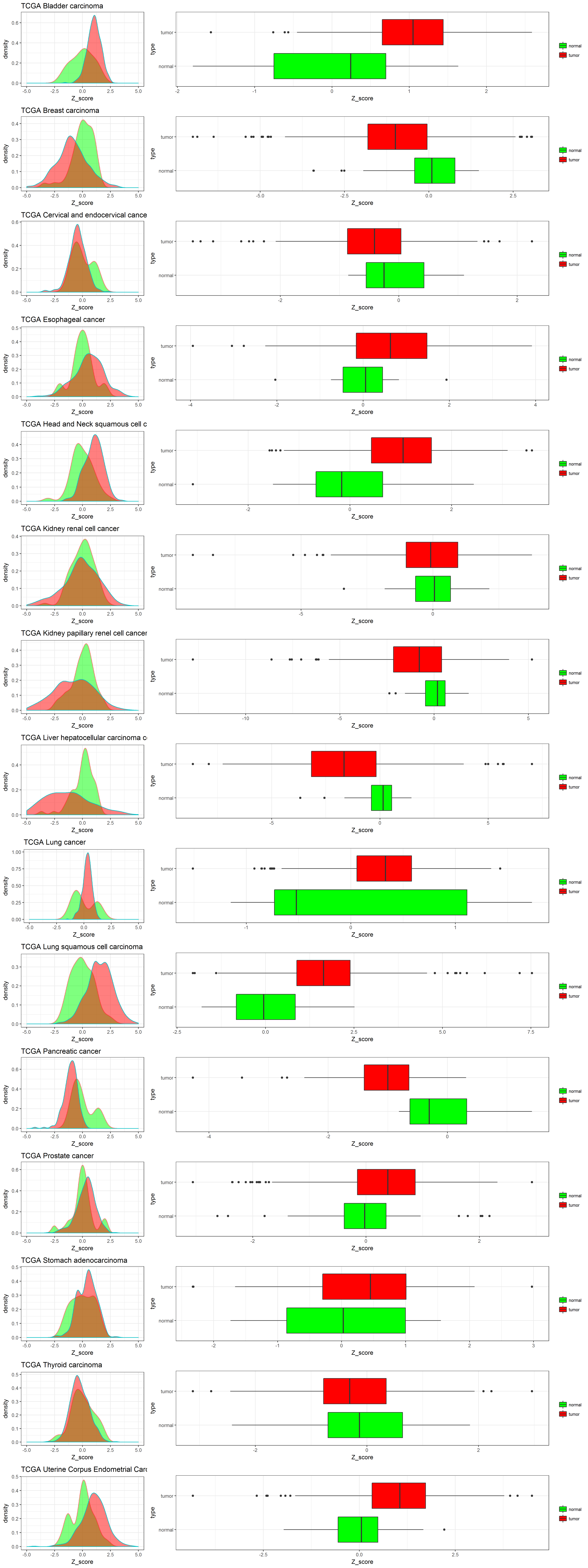
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-505-5p | ATP1B2 | 9 cancers: BLCA; COAD; ESCA; HNSC; LUAD; LUSC; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.243; TCGA COAD -0.317; TCGA ESCA -0.259; TCGA HNSC -0.149; TCGA LUAD -0.143; TCGA LUSC -0.248; TCGA PRAD -0.113; TCGA STAD -0.417; TCGA UCEC -0.173 |
hsa-miR-505-5p | HBP1 | 9 cancers: BLCA; CESC; COAD; HNSC; LUAD; LUSC; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.079; TCGA CESC -0.064; TCGA COAD -0.086; TCGA HNSC -0.145; TCGA LUAD -0.058; TCGA LUSC -0.056; TCGA SARC -0.064; TCGA STAD -0.064; TCGA UCEC -0.057 |
hsa-miR-505-5p | CTSO | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LGG; LUAD; LUSC; STAD; UCEC | MirTarget | TCGA BLCA -0.135; TCGA BRCA -0.089; TCGA COAD -0.184; TCGA ESCA -0.172; TCGA HNSC -0.112; TCGA LGG -0.105; TCGA LUAD -0.093; TCGA LUSC -0.286; TCGA STAD -0.146; TCGA UCEC -0.19 |
hsa-miR-505-5p | FMOD | 9 cancers: BLCA; BRCA; COAD; HNSC; LUAD; LUSC; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.461; TCGA BRCA -0.174; TCGA COAD -0.392; TCGA HNSC -0.114; TCGA LUAD -0.087; TCGA LUSC -0.352; TCGA PRAD -0.176; TCGA STAD -0.154; TCGA UCEC -0.294 |
hsa-miR-505-5p | STAM2 | 9 cancers: BLCA; BRCA; HNSC; KIRP; LGG; LUAD; LUSC; STAD; UCEC | MirTarget | TCGA BLCA -0.073; TCGA BRCA -0.053; TCGA HNSC -0.136; TCGA KIRP -0.059; TCGA LGG -0.057; TCGA LUAD -0.067; TCGA LUSC -0.097; TCGA STAD -0.07; TCGA UCEC -0.086 |
hsa-miR-505-5p | NAV1 | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; OV; STAD | MirTarget | TCGA BLCA -0.197; TCGA BRCA -0.07; TCGA CESC -0.186; TCGA COAD -0.26; TCGA ESCA -0.241; TCGA HNSC -0.128; TCGA LGG -0.131; TCGA LIHC -0.083; TCGA OV -0.148; TCGA STAD -0.086 |
hsa-miR-505-5p | ASH1L | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LUAD; PRAD; SARC; STAD | MirTarget | TCGA BLCA -0.161; TCGA CESC -0.141; TCGA COAD -0.073; TCGA ESCA -0.085; TCGA HNSC -0.211; TCGA KIRP -0.132; TCGA LUAD -0.092; TCGA PRAD -0.09; TCGA SARC -0.1; TCGA STAD -0.099 |
hsa-miR-505-5p | CRMP1 | 11 cancers: BLCA; BRCA; COAD; ESCA; KIRP; LGG; LIHC; OV; PAAD; PRAD; STAD | MirTarget | TCGA BLCA -0.186; TCGA BRCA -0.092; TCGA COAD -0.297; TCGA ESCA -0.311; TCGA KIRP -0.18; TCGA LGG -0.155; TCGA LIHC -0.306; TCGA OV -0.328; TCGA PAAD -0.528; TCGA PRAD -0.065; TCGA STAD -0.201 |
hsa-miR-505-5p | MAPT | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; OV; PAAD; SARC; STAD | MirTarget | TCGA BLCA -0.175; TCGA BRCA -0.751; TCGA CESC -0.33; TCGA COAD -0.392; TCGA ESCA -0.378; TCGA HNSC -0.339; TCGA LGG -0.166; TCGA LIHC -0.381; TCGA OV -0.233; TCGA PAAD -0.427; TCGA SARC -0.304; TCGA STAD -0.526 |
hsa-miR-505-5p | WDR31 | 9 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LUAD; PRAD; SARC; STAD | MirTarget | TCGA BLCA -0.199; TCGA BRCA -0.113; TCGA CESC -0.257; TCGA ESCA -0.216; TCGA HNSC -0.303; TCGA LUAD -0.074; TCGA PRAD -0.176; TCGA SARC -0.239; TCGA STAD -0.135 |
hsa-miR-505-5p | SEMA3G | 9 cancers: BLCA; BRCA; COAD; HNSC; LUAD; LUSC; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.236; TCGA BRCA -0.15; TCGA COAD -0.29; TCGA HNSC -0.164; TCGA LUAD -0.114; TCGA LUSC -0.336; TCGA SARC -0.175; TCGA STAD -0.133; TCGA UCEC -0.183 |
hsa-miR-505-5p | PJA2 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LUAD; LUSC; OV; PAAD; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.272; TCGA BRCA -0.074; TCGA CESC -0.089; TCGA COAD -0.139; TCGA ESCA -0.15; TCGA HNSC -0.214; TCGA LGG -0.086; TCGA LUAD -0.137; TCGA LUSC -0.154; TCGA OV -0.071; TCGA PAAD -0.135; TCGA PRAD -0.081; TCGA SARC -0.147; TCGA STAD -0.208; TCGA UCEC -0.189 |
hsa-miR-505-5p | MRVI1 | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LGG; LUAD; LUSC; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.603; TCGA BRCA -0.075; TCGA COAD -0.443; TCGA ESCA -0.464; TCGA HNSC -0.224; TCGA LGG -0.368; TCGA LUAD -0.101; TCGA LUSC -0.401; TCGA PRAD -0.139; TCGA SARC -0.756; TCGA STAD -0.479; TCGA UCEC -0.383 |
hsa-miR-505-5p | SOBP | 9 cancers: BLCA; COAD; ESCA; LGG; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.397; TCGA COAD -0.23; TCGA ESCA -0.215; TCGA LGG -0.116; TCGA LUSC -0.149; TCGA PAAD -0.192; TCGA PRAD -0.125; TCGA STAD -0.408; TCGA UCEC -0.167 |
hsa-miR-505-5p | DLG2 | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LGG; LUSC; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.376; TCGA BRCA -0.146; TCGA COAD -0.562; TCGA ESCA -0.467; TCGA HNSC -0.452; TCGA LGG -0.235; TCGA LUSC -0.2; TCGA SARC -0.428; TCGA STAD -0.57; TCGA UCEC -0.223 |
hsa-miR-505-5p | ETV1 | 12 cancers: BLCA; COAD; HNSC; KIRP; LGG; LUAD; LUSC; OV; PAAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.294; TCGA COAD -0.228; TCGA HNSC -0.212; TCGA KIRP -0.493; TCGA LGG -0.172; TCGA LUAD -0.126; TCGA LUSC -0.327; TCGA OV -0.194; TCGA PAAD -0.242; TCGA THCA -0.118; TCGA STAD -0.254; TCGA UCEC -0.338 |
hsa-miR-505-5p | ARHGEF12 | 11 cancers: BLCA; BRCA; CESC; HNSC; KIRP; LUAD; LUSC; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.119; TCGA BRCA -0.058; TCGA CESC -0.108; TCGA HNSC -0.213; TCGA KIRP -0.089; TCGA LUAD -0.14; TCGA LUSC -0.089; TCGA PRAD -0.051; TCGA SARC -0.156; TCGA STAD -0.111; TCGA UCEC -0.079 |
hsa-miR-505-5p | KATNAL1 | 10 cancers: BLCA; COAD; ESCA; HNSC; LUAD; LUSC; OV; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.399; TCGA COAD -0.2; TCGA ESCA -0.181; TCGA HNSC -0.074; TCGA LUAD -0.079; TCGA LUSC -0.16; TCGA OV -0.143; TCGA SARC -0.113; TCGA STAD -0.322; TCGA UCEC -0.182 |
hsa-miR-505-5p | TUB | 9 cancers: BLCA; COAD; ESCA; HNSC; LGG; OV; PAAD; SARC; STAD | MirTarget; miRNATAP | TCGA BLCA -0.344; TCGA COAD -0.305; TCGA ESCA -0.433; TCGA HNSC -0.174; TCGA LGG -0.283; TCGA OV -0.207; TCGA PAAD -0.309; TCGA SARC -0.258; TCGA STAD -0.371 |
hsa-miR-505-5p | SCN2B | 12 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LGG; LUSC; OV; PRAD; SARC; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.708; TCGA BRCA -0.274; TCGA COAD -0.449; TCGA ESCA -0.577; TCGA HNSC -0.311; TCGA LGG -0.35; TCGA LUSC -0.554; TCGA OV -0.375; TCGA PRAD -0.198; TCGA SARC -0.419; TCGA STAD -0.664; TCGA UCEC -0.311 |
hsa-miR-505-5p | NFIX | 9 cancers: BLCA; CESC; HNSC; LGG; LUAD; LUSC; OV; PRAD; STAD | mirMAP | TCGA BLCA -0.437; TCGA CESC -0.131; TCGA HNSC -0.216; TCGA LGG -0.058; TCGA LUAD -0.098; TCGA LUSC -0.186; TCGA OV -0.137; TCGA PRAD -0.066; TCGA STAD -0.094 |
hsa-miR-505-5p | SPTB | 9 cancers: BLCA; CESC; HNSC; LGG; LIHC; OV; PAAD; SARC; STAD | mirMAP | TCGA BLCA -0.12; TCGA CESC -0.145; TCGA HNSC -0.137; TCGA LGG -0.212; TCGA LIHC -0.102; TCGA OV -0.218; TCGA PAAD -0.289; TCGA SARC -0.479; TCGA STAD -0.106 |
hsa-miR-505-5p | ZBTB47 | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LGG; LUSC; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.178; TCGA BRCA -0.1; TCGA COAD -0.216; TCGA ESCA -0.185; TCGA HNSC -0.104; TCGA KIRC -0.207; TCGA LGG -0.116; TCGA LUSC -0.231; TCGA SARC -0.211; TCGA STAD -0.227; TCGA UCEC -0.134 |
hsa-miR-505-5p | KIAA0513 | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; LIHC; LUSC; PRAD; STAD | mirMAP | TCGA BLCA -0.348; TCGA CESC -0.152; TCGA COAD -0.176; TCGA ESCA -0.209; TCGA HNSC -0.151; TCGA LIHC -0.106; TCGA LUSC -0.305; TCGA PRAD -0.071; TCGA STAD -0.145 |
hsa-miR-505-5p | DAB2IP | 10 cancers: BLCA; HNSC; KIRC; KIRP; LGG; LIHC; LUSC; OV; PRAD; UCEC | mirMAP | TCGA BLCA -0.134; TCGA HNSC -0.061; TCGA KIRC -0.117; TCGA KIRP -0.086; TCGA LGG -0.05; TCGA LIHC -0.116; TCGA LUSC -0.176; TCGA OV -0.125; TCGA PRAD -0.09; TCGA UCEC -0.077 |
hsa-miR-505-5p | FTO | 10 cancers: BLCA; COAD; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; SARC; STAD | mirMAP | TCGA BLCA -0.133; TCGA COAD -0.059; TCGA ESCA -0.124; TCGA HNSC -0.118; TCGA LUAD -0.095; TCGA LUSC -0.084; TCGA PAAD -0.13; TCGA PRAD -0.058; TCGA SARC -0.092; TCGA STAD -0.165 |
hsa-miR-505-5p | NFIC | 10 cancers: BLCA; BRCA; CESC; COAD; HNSC; LUAD; LUSC; PRAD; SARC; STAD | mirMAP | TCGA BLCA -0.373; TCGA BRCA -0.108; TCGA CESC -0.145; TCGA COAD -0.202; TCGA HNSC -0.376; TCGA LUAD -0.061; TCGA LUSC -0.081; TCGA PRAD -0.12; TCGA SARC -0.242; TCGA STAD -0.146 |
hsa-miR-505-5p | PRKCA | 9 cancers: BLCA; HNSC; LGG; LIHC; LUSC; OV; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.171; TCGA HNSC -0.109; TCGA LGG -0.103; TCGA LIHC -0.072; TCGA LUSC -0.213; TCGA OV -0.157; TCGA PRAD -0.131; TCGA STAD -0.096; TCGA UCEC -0.277 |
hsa-miR-505-5p | SH3PXD2B | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; LIHC; LUSC; OV; PRAD; STAD | mirMAP | TCGA BLCA -0.197; TCGA CESC -0.187; TCGA COAD -0.115; TCGA ESCA -0.168; TCGA HNSC -0.152; TCGA LIHC -0.167; TCGA LUSC -0.122; TCGA OV -0.13; TCGA PRAD -0.129; TCGA STAD -0.153 |
hsa-miR-505-5p | TOM1L2 | 11 cancers: BLCA; BRCA; CESC; HNSC; LGG; LIHC; LUSC; PAAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.082; TCGA BRCA -0.175; TCGA CESC -0.232; TCGA HNSC -0.149; TCGA LGG -0.155; TCGA LIHC -0.072; TCGA LUSC -0.17; TCGA PAAD -0.136; TCGA SARC -0.279; TCGA STAD -0.126; TCGA UCEC -0.084 |
hsa-miR-505-5p | OLFML2A | 10 cancers: BLCA; BRCA; COAD; LIHC; LUAD; LUSC; OV; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.325; TCGA BRCA -0.101; TCGA COAD -0.189; TCGA LIHC -0.198; TCGA LUAD -0.086; TCGA LUSC -0.185; TCGA OV -0.301; TCGA PRAD -0.235; TCGA STAD -0.207; TCGA UCEC -0.301 |
hsa-miR-505-5p | CYS1 | 9 cancers: BLCA; BRCA; COAD; ESCA; HNSC; LUSC; OV; STAD; UCEC | mirMAP | TCGA BLCA -0.623; TCGA BRCA -0.193; TCGA COAD -0.407; TCGA ESCA -0.366; TCGA HNSC -0.205; TCGA LUSC -0.567; TCGA OV -0.277; TCGA STAD -0.426; TCGA UCEC -0.584 |
hsa-miR-505-5p | PCDHGC5 | 10 cancers: BLCA; CESC; HNSC; KIRP; LUSC; OV; PRAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.455; TCGA CESC -0.432; TCGA HNSC -0.423; TCGA KIRP -0.432; TCGA LUSC -0.292; TCGA OV -0.211; TCGA PRAD -0.19; TCGA SARC -0.623; TCGA STAD -0.183; TCGA UCEC -0.201 |
hsa-miR-505-5p | PCDHGA12 | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; PRAD; SARC | mirMAP | TCGA BLCA -0.576; TCGA CESC -0.323; TCGA COAD -0.441; TCGA ESCA -0.256; TCGA HNSC -0.444; TCGA LUAD -0.106; TCGA LUSC -0.516; TCGA PRAD -0.258; TCGA SARC -0.482 |
hsa-miR-505-5p | PCDHGB6 | 9 cancers: BLCA; COAD; ESCA; HNSC; LUSC; OV; PRAD; SARC; UCEC | mirMAP | TCGA BLCA -0.394; TCGA COAD -0.312; TCGA ESCA -0.274; TCGA HNSC -0.401; TCGA LUSC -0.44; TCGA OV -0.237; TCGA PRAD -0.275; TCGA SARC -0.275; TCGA UCEC -0.231 |
hsa-miR-505-5p | PCDHGA7 | 9 cancers: BLCA; BRCA; CESC; COAD; HNSC; LGG; LUSC; PAAD; PRAD | mirMAP | TCGA BLCA -0.241; TCGA BRCA -0.113; TCGA CESC -0.325; TCGA COAD -0.288; TCGA HNSC -0.376; TCGA LGG -0.194; TCGA LUSC -0.213; TCGA PAAD -0.351; TCGA PRAD -0.262 |
hsa-miR-505-5p | PCDHGA6 | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUSC; OV; PRAD; SARC | mirMAP | TCGA BLCA -0.407; TCGA CESC -0.33; TCGA COAD -0.351; TCGA ESCA -0.168; TCGA HNSC -0.379; TCGA LUSC -0.321; TCGA OV -0.206; TCGA PRAD -0.2; TCGA SARC -0.353 |
hsa-miR-505-5p | PCDHGA9 | 11 cancers: BLCA; CESC; COAD; ESCA; HNSC; LGG; LUAD; LUSC; OV; PRAD; SARC | mirMAP | TCGA BLCA -0.575; TCGA CESC -0.48; TCGA COAD -0.393; TCGA ESCA -0.213; TCGA HNSC -0.456; TCGA LGG -0.305; TCGA LUAD -0.165; TCGA LUSC -0.418; TCGA OV -0.279; TCGA PRAD -0.316; TCGA SARC -0.263 |
hsa-miR-505-5p | PLXNA4 | 11 cancers: BLCA; COAD; ESCA; HNSC; LGG; LUAD; LUSC; OV; PRAD; SARC; STAD | miRNATAP | TCGA BLCA -0.662; TCGA COAD -0.443; TCGA ESCA -0.41; TCGA HNSC -0.242; TCGA LGG -0.263; TCGA LUAD -0.12; TCGA LUSC -0.362; TCGA OV -0.356; TCGA PRAD -0.103; TCGA SARC -0.532; TCGA STAD -0.386 |
hsa-miR-505-5p | KLF7 | 9 cancers: BLCA; CESC; ESCA; HNSC; KIRP; LUSC; PRAD; SARC; STAD | miRNATAP | TCGA BLCA -0.303; TCGA CESC -0.281; TCGA ESCA -0.241; TCGA HNSC -0.129; TCGA KIRP -0.178; TCGA LUSC -0.213; TCGA PRAD -0.147; TCGA SARC -0.247; TCGA STAD -0.194 |
hsa-miR-505-5p | EHD2 | 10 cancers: BLCA; BRCA; COAD; ESCA; KIRP; LUAD; LUSC; PRAD; STAD; UCEC | miRNATAP | TCGA BLCA -0.362; TCGA BRCA -0.093; TCGA COAD -0.37; TCGA ESCA -0.25; TCGA KIRP -0.114; TCGA LUAD -0.109; TCGA LUSC -0.38; TCGA PRAD -0.124; TCGA STAD -0.216; TCGA UCEC -0.2 |
hsa-miR-505-5p | NFASC | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; LGG; LUSC; SARC; STAD | miRNATAP | TCGA BLCA -0.514; TCGA CESC -0.329; TCGA COAD -0.322; TCGA ESCA -0.505; TCGA HNSC -0.248; TCGA LGG -0.121; TCGA LUSC -0.297; TCGA SARC -0.877; TCGA STAD -0.497 |
hsa-miR-505-5p | UTRN | 10 cancers: BLCA; ESCA; HNSC; KIRP; LUAD; LUSC; PRAD; SARC; STAD; UCEC | miRNATAP | TCGA BLCA -0.29; TCGA ESCA -0.219; TCGA HNSC -0.27; TCGA KIRP -0.134; TCGA LUAD -0.101; TCGA LUSC -0.245; TCGA PRAD -0.111; TCGA SARC -0.119; TCGA STAD -0.186; TCGA UCEC -0.166 |
hsa-miR-505-5p | SLC22A15 | 9 cancers: BRCA; CESC; HNSC; KIRC; KIRP; LIHC; LUSC; PRAD; STAD | MirTarget | TCGA BRCA -0.143; TCGA CESC -0.259; TCGA HNSC -0.147; TCGA KIRC -0.396; TCGA KIRP -0.164; TCGA LIHC -0.237; TCGA LUSC -0.182; TCGA PRAD -0.094; TCGA STAD -0.187 |
hsa-miR-505-5p | SYNGR1 | 9 cancers: BRCA; COAD; HNSC; KIRC; LGG; LIHC; OV; PAAD; PRAD | mirMAP | TCGA BRCA -0.125; TCGA COAD -0.437; TCGA HNSC -0.234; TCGA KIRC -0.176; TCGA LGG -0.117; TCGA LIHC -0.449; TCGA OV -0.229; TCGA PAAD -0.193; TCGA PRAD -0.083 |
hsa-miR-505-5p | N4BP3 | 10 cancers: BRCA; CESC; COAD; ESCA; KIRC; KIRP; LIHC; LUSC; OV; STAD | mirMAP | TCGA BRCA -0.371; TCGA CESC -0.213; TCGA COAD -0.216; TCGA ESCA -0.165; TCGA KIRC -0.264; TCGA KIRP -0.161; TCGA LIHC -0.32; TCGA LUSC -0.143; TCGA OV -0.34; TCGA STAD -0.121 |
hsa-miR-505-5p | KLHL3 | 9 cancers: BRCA; HNSC; KIRC; KIRP; OV; PAAD; PRAD; SARC; STAD | mirMAP | TCGA BRCA -0.085; TCGA HNSC -0.127; TCGA KIRC -0.262; TCGA KIRP -0.23; TCGA OV -0.177; TCGA PAAD -0.29; TCGA PRAD -0.111; TCGA SARC -0.165; TCGA STAD -0.27 |
hsa-miR-505-5p | ARHGAP23 | 11 cancers: CESC; COAD; ESCA; HNSC; LGG; LUSC; OV; PRAD; SARC; STAD; UCEC | MirTarget | TCGA CESC -0.381; TCGA COAD -0.171; TCGA ESCA -0.199; TCGA HNSC -0.133; TCGA LGG -0.083; TCGA LUSC -0.157; TCGA OV -0.211; TCGA PRAD -0.242; TCGA SARC -0.128; TCGA STAD -0.175; TCGA UCEC -0.128 |
hsa-miR-505-5p | ATP9A | 9 cancers: CESC; HNSC; KIRP; LGG; LIHC; LUSC; PAAD; PRAD; SARC | mirMAP | TCGA CESC -0.128; TCGA HNSC -0.253; TCGA KIRP -0.21; TCGA LGG -0.178; TCGA LIHC -0.143; TCGA LUSC -0.108; TCGA PAAD -0.268; TCGA PRAD -0.066; TCGA SARC -0.324 |
Enriched cancer pathways of putative targets