microRNA information: hsa-miR-616-5p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-616-5p | miRbase |
| Accession: | MIMAT0003284 | miRbase |
| Precursor name: | hsa-mir-616 | miRbase |
| Precursor accession: | MI0003629 | miRbase |
| Symbol: | MIR616 | HGNC |
| RefSeq ID: | NR_030346 | GenBank |
| Sequence: | ACUCAAAACCCUUCAGUGACUU |
Reported expression in cancers: hsa-miR-616-5p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|
Reported cancer pathway affected by hsa-miR-616-5p
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|
Reported cancer prognosis affected by hsa-miR-616-5p
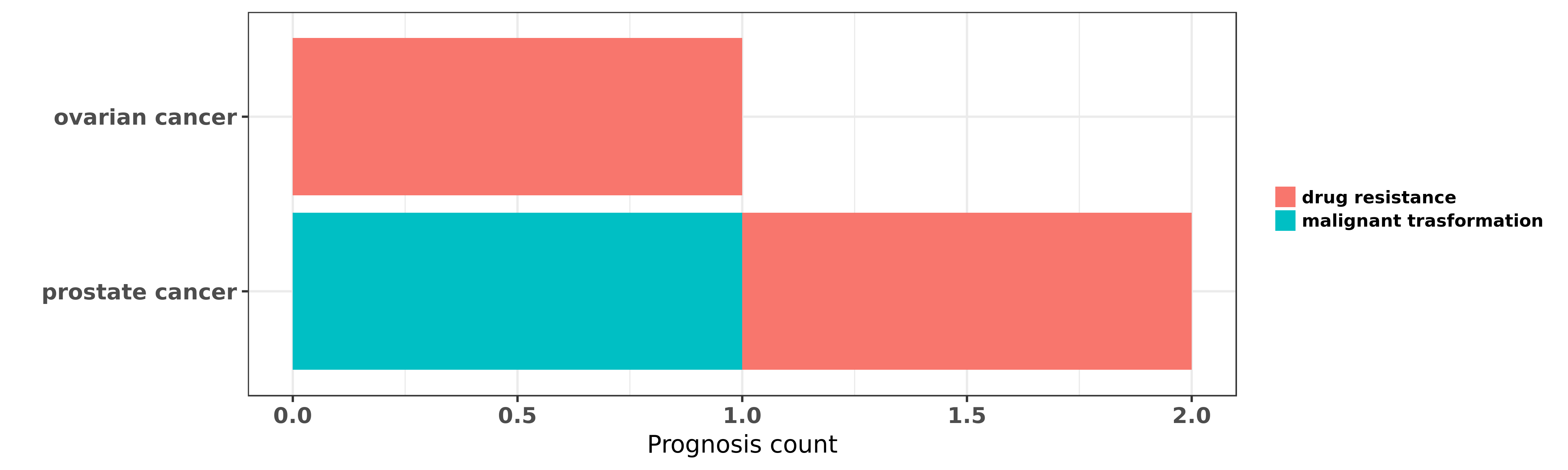
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-616-5p | ovarian cancer | drug resistance | "Of these 10 miRNAs miR-193a-5p miR-375 miR-339-3p ......" | 26485143 | |
| hsa-miR-616-5p | prostate cancer | malignant trasformation; drug resistance | "MicroRNA 616 induces androgen independent growth o ......" | 21224345 |
Reported gene related to hsa-miR-616-5p
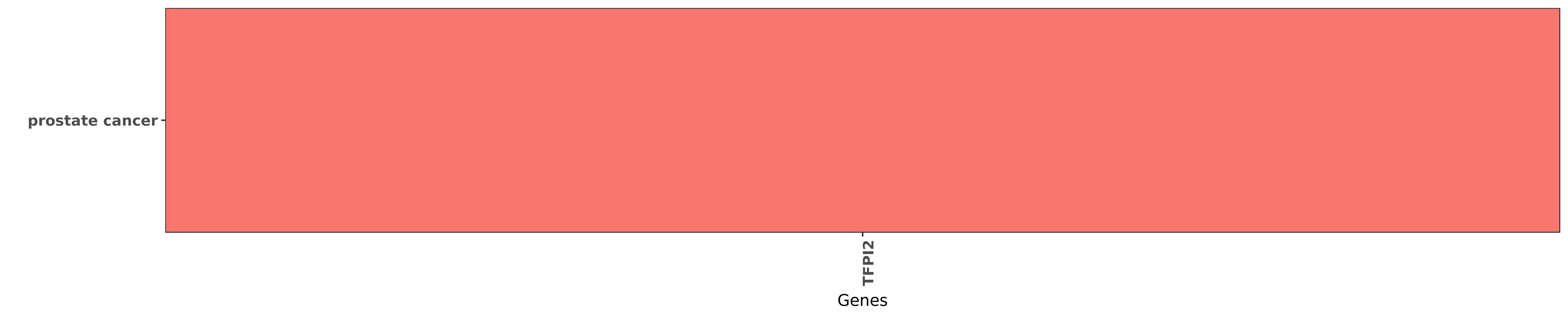
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-616-5p | prostate cancer | TFPI2 | "Microarray profiling and bioinformatics analysis i ......" | 21224345 |
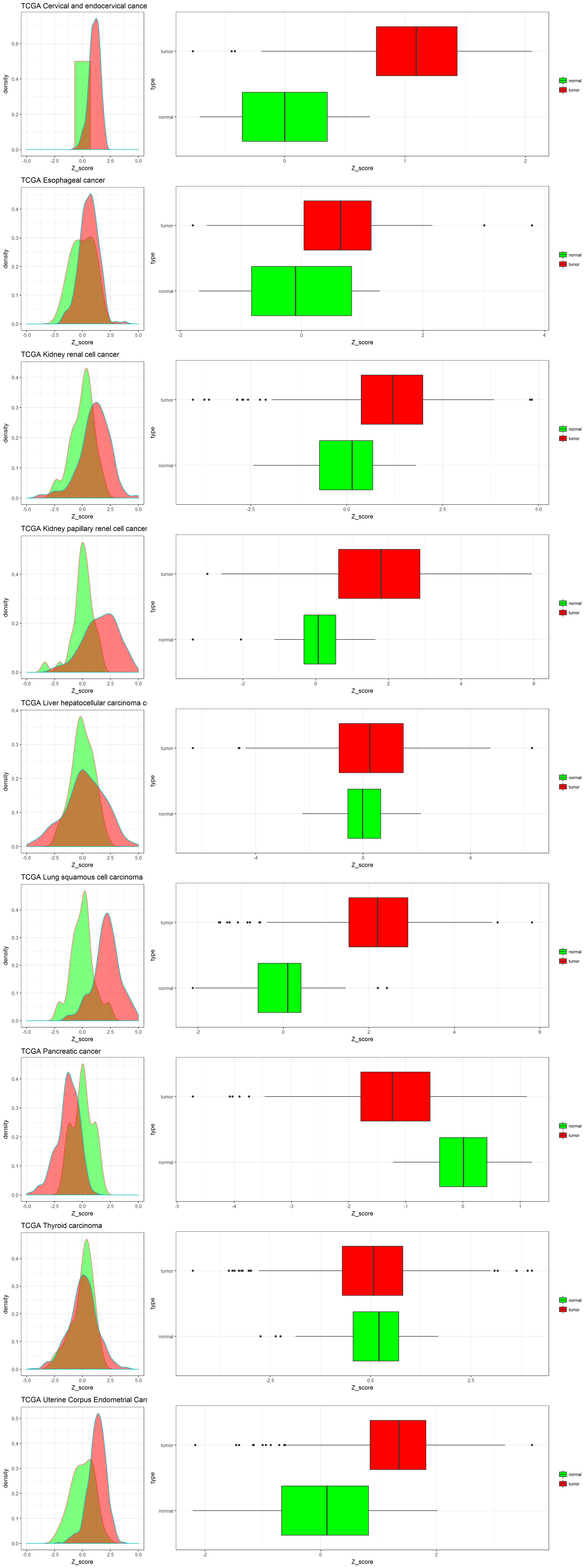
Expression profile in cancer corhorts:
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-616-5p | LRCH2 | 9 cancers: CESC; ESCA; KIRC; KIRP; LIHC; LUSC; SARC; THCA; UCEC | MirTarget; mirMAP | TCGA CESC -0.282; TCGA ESCA -0.258; TCGA KIRC -0.133; TCGA KIRP -0.249; TCGA LIHC -0.119; TCGA LUSC -0.335; TCGA SARC -0.157; TCGA THCA -0.327; TCGA UCEC -0.461 |
hsa-miR-616-5p | ARID5B | 10 cancers: CESC; ESCA; KIRC; KIRP; LIHC; LUSC; PAAD; SARC; THCA; UCEC | MirTarget; miRNATAP | TCGA CESC -0.073; TCGA ESCA -0.133; TCGA KIRC -0.122; TCGA KIRP -0.091; TCGA LIHC -0.086; TCGA LUSC -0.06; TCGA PAAD -0.127; TCGA SARC -0.105; TCGA THCA -0.227; TCGA UCEC -0.173 |
hsa-miR-616-5p | KCTD12 | 9 cancers: CESC; ESCA; KIRC; KIRP; LIHC; LUSC; OV; THCA; UCEC | MirTarget; miRNATAP | TCGA CESC -0.15; TCGA ESCA -0.175; TCGA KIRC -0.084; TCGA KIRP -0.186; TCGA LIHC -0.271; TCGA LUSC -0.354; TCGA OV -0.103; TCGA THCA -0.214; TCGA UCEC -0.388 |
hsa-miR-616-5p | FAM180A | 9 cancers: CESC; ESCA; KIRC; KIRP; LIHC; LUSC; PAAD; THCA; UCEC | MirTarget | TCGA CESC -0.319; TCGA ESCA -0.362; TCGA KIRC -0.509; TCGA KIRP -0.235; TCGA LIHC -0.453; TCGA LUSC -0.469; TCGA PAAD -0.214; TCGA THCA -0.35; TCGA UCEC -0.365 |
hsa-miR-616-5p | SLC8A1 | 9 cancers: CESC; ESCA; KIRC; KIRP; LIHC; LUSC; SARC; THCA; UCEC | MirTarget | TCGA CESC -0.179; TCGA ESCA -0.152; TCGA KIRC -0.127; TCGA KIRP -0.273; TCGA LIHC -0.225; TCGA LUSC -0.269; TCGA SARC -0.323; TCGA THCA -0.136; TCGA UCEC -0.333 |
hsa-miR-616-5p | MBNL2 | 9 cancers: CESC; ESCA; KIRC; KIRP; LUSC; OV; PAAD; SARC; UCEC | MirTarget; mirMAP; miRNATAP | TCGA CESC -0.127; TCGA ESCA -0.125; TCGA KIRC -0.075; TCGA KIRP -0.069; TCGA LUSC -0.219; TCGA OV -0.159; TCGA PAAD -0.09; TCGA SARC -0.117; TCGA UCEC -0.216 |
hsa-miR-616-5p | CHRDL1 | 9 cancers: CESC; ESCA; KIRC; KIRP; LIHC; LUSC; SARC; THCA; UCEC | MirTarget; mirMAP | TCGA CESC -0.424; TCGA ESCA -1.085; TCGA KIRC -0.419; TCGA KIRP -0.44; TCGA LIHC -0.248; TCGA LUSC -0.493; TCGA SARC -0.402; TCGA THCA -0.581; TCGA UCEC -0.794 |
hsa-miR-616-5p | STON1 | 9 cancers: CESC; ESCA; KIRC; KIRP; LIHC; LUSC; SARC; THCA; UCEC | MirTarget | TCGA CESC -0.344; TCGA ESCA -0.408; TCGA KIRC -0.265; TCGA KIRP -0.156; TCGA LIHC -0.261; TCGA LUSC -0.141; TCGA SARC -0.312; TCGA THCA -0.199; TCGA UCEC -0.516 |
hsa-miR-616-5p | PPP1R12B | 9 cancers: CESC; ESCA; KIRC; KIRP; LIHC; LUSC; OV; SARC; UCEC | mirMAP | TCGA CESC -0.176; TCGA ESCA -0.401; TCGA KIRC -0.243; TCGA KIRP -0.117; TCGA LIHC -0.144; TCGA LUSC -0.203; TCGA OV -0.141; TCGA SARC -0.742; TCGA UCEC -0.446 |
hsa-miR-616-5p | SMOC2 | 10 cancers: CESC; ESCA; KIRC; LIHC; LUSC; OV; PAAD; SARC; THCA; UCEC | mirMAP | TCGA CESC -0.198; TCGA ESCA -0.379; TCGA KIRC -0.212; TCGA LIHC -0.3; TCGA LUSC -0.151; TCGA OV -0.217; TCGA PAAD -0.357; TCGA SARC -0.463; TCGA THCA -0.151; TCGA UCEC -0.47 |
hsa-miR-616-5p | DPYSL3 | 9 cancers: CESC; ESCA; KIRP; LIHC; LUSC; PAAD; SARC; THCA; UCEC | mirMAP | TCGA CESC -0.284; TCGA ESCA -0.418; TCGA KIRP -0.145; TCGA LIHC -0.385; TCGA LUSC -0.177; TCGA PAAD -0.189; TCGA SARC -0.165; TCGA THCA -0.227; TCGA UCEC -0.308 |
hsa-miR-616-5p | IGFBP5 | 9 cancers: CESC; ESCA; KIRC; KIRP; LIHC; OV; SARC; THCA; UCEC | mirMAP | TCGA CESC -0.235; TCGA ESCA -0.375; TCGA KIRC -0.354; TCGA KIRP -0.257; TCGA LIHC -0.383; TCGA OV -0.207; TCGA SARC -0.259; TCGA THCA -0.479; TCGA UCEC -0.347 |
hsa-miR-616-5p | EMP1 | 9 cancers: CESC; KIRC; KIRP; LIHC; LUSC; OV; PAAD; THCA; UCEC | mirMAP | TCGA CESC -0.298; TCGA KIRC -0.268; TCGA KIRP -0.336; TCGA LIHC -0.328; TCGA LUSC -0.164; TCGA OV -0.14; TCGA PAAD -0.234; TCGA THCA -0.148; TCGA UCEC -0.24 |
hsa-miR-616-5p | YAP1 | 9 cancers: CESC; ESCA; KIRC; KIRP; LIHC; LUSC; OV; SARC; UCEC | mirMAP | TCGA CESC -0.137; TCGA ESCA -0.1; TCGA KIRC -0.107; TCGA KIRP -0.143; TCGA LIHC -0.101; TCGA LUSC -0.077; TCGA OV -0.099; TCGA SARC -0.258; TCGA UCEC -0.069 |
hsa-miR-616-5p | IGF1R | 9 cancers: CESC; KIRC; KIRP; LIHC; OV; PAAD; SARC; THCA; UCEC | mirMAP | TCGA CESC -0.123; TCGA KIRC -0.092; TCGA KIRP -0.066; TCGA LIHC -0.342; TCGA OV -0.13; TCGA PAAD -0.106; TCGA SARC -0.131; TCGA THCA -0.124; TCGA UCEC -0.102 |
hsa-miR-616-5p | ADCYAP1 | 9 cancers: CESC; ESCA; KIRC; KIRP; LIHC; LUSC; SARC; THCA; UCEC | mirMAP | TCGA CESC -0.351; TCGA ESCA -0.605; TCGA KIRC -0.208; TCGA KIRP -0.364; TCGA LIHC -0.407; TCGA LUSC -0.258; TCGA SARC -0.417; TCGA THCA -0.281; TCGA UCEC -0.513 |
hsa-miR-616-5p | GFRA1 | 9 cancers: CESC; ESCA; KIRC; KIRP; LUSC; OV; PAAD; THCA; UCEC | mirMAP | TCGA CESC -0.322; TCGA ESCA -0.506; TCGA KIRC -0.231; TCGA KIRP -0.347; TCGA LUSC -0.453; TCGA OV -0.291; TCGA PAAD -0.288; TCGA THCA -0.298; TCGA UCEC -0.645 |
hsa-miR-616-5p | RBMS1 | 9 cancers: CESC; ESCA; KIRC; KIRP; LIHC; LUSC; OV; THCA; UCEC | mirMAP | TCGA CESC -0.175; TCGA ESCA -0.201; TCGA KIRC -0.067; TCGA KIRP -0.055; TCGA LIHC -0.107; TCGA LUSC -0.093; TCGA OV -0.053; TCGA THCA -0.054; TCGA UCEC -0.185 |
hsa-miR-616-5p | JAM2 | 9 cancers: CESC; ESCA; KIRC; KIRP; LIHC; LUSC; OV; THCA; UCEC | mirMAP | TCGA CESC -0.261; TCGA ESCA -0.419; TCGA KIRC -0.287; TCGA KIRP -0.124; TCGA LIHC -0.364; TCGA LUSC -0.452; TCGA OV -0.173; TCGA THCA -0.092; TCGA UCEC -0.416 |
hsa-miR-616-5p | ERG | 9 cancers: CESC; ESCA; KIRC; KIRP; LIHC; LUSC; OV; THCA; UCEC | mirMAP | TCGA CESC -0.218; TCGA ESCA -0.135; TCGA KIRC -0.187; TCGA KIRP -0.177; TCGA LIHC -0.216; TCGA LUSC -0.372; TCGA OV -0.116; TCGA THCA -0.068; TCGA UCEC -0.275 |
hsa-miR-616-5p | BTG2 | 9 cancers: CESC; ESCA; KIRC; KIRP; LIHC; LUSC; SARC; THCA; UCEC | mirMAP | TCGA CESC -0.134; TCGA ESCA -0.244; TCGA KIRC -0.258; TCGA KIRP -0.097; TCGA LIHC -0.18; TCGA LUSC -0.207; TCGA SARC -0.229; TCGA THCA -0.122; TCGA UCEC -0.192 |
hsa-miR-616-5p | C1QTNF7 | 10 cancers: CESC; ESCA; KIRC; KIRP; LIHC; LUSC; OV; SARC; THCA; UCEC | mirMAP | TCGA CESC -0.43; TCGA ESCA -0.506; TCGA KIRC -0.599; TCGA KIRP -0.298; TCGA LIHC -0.455; TCGA LUSC -0.647; TCGA OV -0.296; TCGA SARC -0.241; TCGA THCA -0.276; TCGA UCEC -0.518 |
hsa-miR-616-5p | NFASC | 9 cancers: CESC; ESCA; KIRC; KIRP; LIHC; LUSC; SARC; THCA; UCEC | mirMAP | TCGA CESC -0.325; TCGA ESCA -0.388; TCGA KIRC -0.496; TCGA KIRP -0.308; TCGA LIHC -0.263; TCGA LUSC -0.191; TCGA SARC -0.707; TCGA THCA -0.082; TCGA UCEC -0.391 |
hsa-miR-616-5p | PTRF | 9 cancers: CESC; ESCA; KIRP; LIHC; LUSC; PAAD; SARC; THCA; UCEC | mirMAP | TCGA CESC -0.269; TCGA ESCA -0.249; TCGA KIRP -0.283; TCGA LIHC -0.245; TCGA LUSC -0.279; TCGA PAAD -0.126; TCGA SARC -0.093; TCGA THCA -0.184; TCGA UCEC -0.374 |
hsa-miR-616-5p | TCF4 | 9 cancers: CESC; ESCA; KIRC; KIRP; LIHC; LUSC; OV; THCA; UCEC | mirMAP | TCGA CESC -0.264; TCGA ESCA -0.134; TCGA KIRC -0.105; TCGA KIRP -0.128; TCGA LIHC -0.218; TCGA LUSC -0.168; TCGA OV -0.118; TCGA THCA -0.092; TCGA UCEC -0.247 |
hsa-miR-616-5p | DACT3 | 9 cancers: CESC; ESCA; KIRC; KIRP; LIHC; LUSC; SARC; THCA; UCEC | mirMAP | TCGA CESC -0.303; TCGA ESCA -0.497; TCGA KIRC -0.228; TCGA KIRP -0.126; TCGA LIHC -0.414; TCGA LUSC -0.365; TCGA SARC -0.516; TCGA THCA -0.13; TCGA UCEC -0.571 |
hsa-miR-616-5p | DIO3 | 9 cancers: CESC; ESCA; KIRC; LIHC; LUSC; OV; PAAD; THCA; UCEC | mirMAP | TCGA CESC -0.434; TCGA ESCA -0.286; TCGA KIRC -0.259; TCGA LIHC -0.521; TCGA LUSC -0.423; TCGA OV -0.205; TCGA PAAD -0.399; TCGA THCA -0.375; TCGA UCEC -0.464 |
Enriched cancer pathways of putative targets