microRNA information: hsa-miR-664a-5p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-664a-5p | miRbase |
| Accession: | MIMAT0005948 | miRbase |
| Precursor name: | hsa-mir-664a | miRbase |
| Precursor accession: | MI0006442 | miRbase |
| Symbol: | NA | HGNC |
| RefSeq ID: | NA | GenBank |
| Sequence: | ACUGGCUAGGGAAAAUGAUUGGAU |
Reported expression in cancers: hsa-miR-664a-5p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-664a-5p | cervical and endocervical cancer | downregulation | "Down regulation of miR 664 in cervical cancer is a ......" | 27212165 | qPCR |
| hsa-miR-664a-5p | lymphoma | deregulation | "We found miR-34a miR-146a and miR-193b to be up-re ......" | 25862860 |
Reported cancer pathway affected by hsa-miR-664a-5p
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-664a-5p | melanoma | cell cycle pathway | "Loss of MiR 664 Expression Enhances Cutaneous Mali ......" | 26287415 |
Reported cancer prognosis affected by hsa-miR-664a-5p
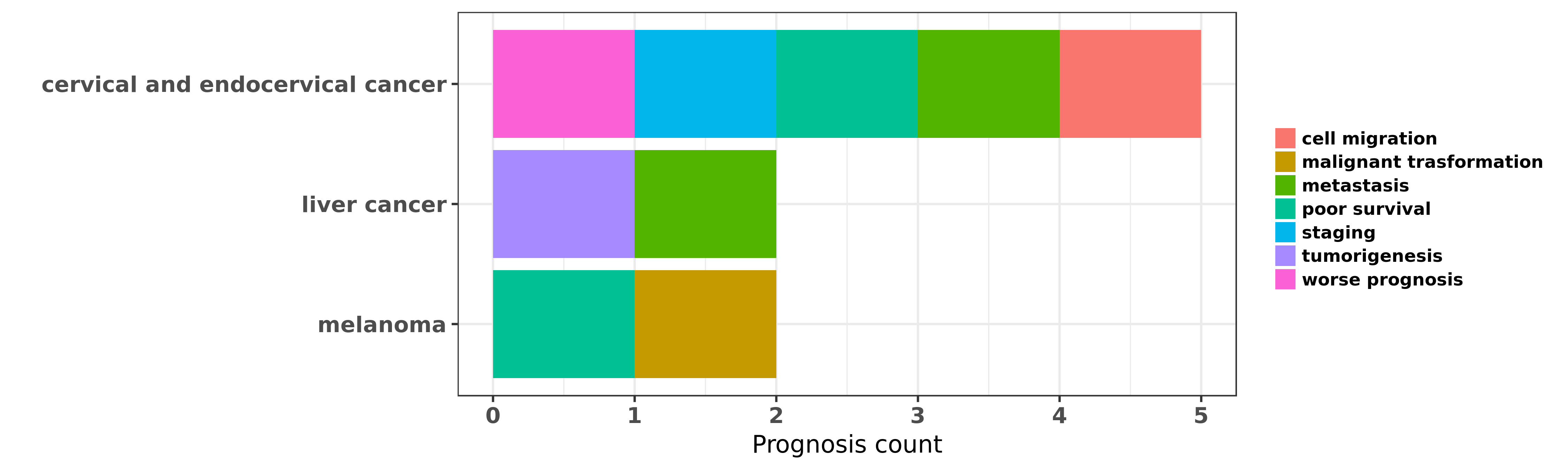
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-664a-5p | cervical and endocervical cancer | cell migration | "In the present study we investigated the role of m ......" | 26770409 | Western blot |
| hsa-miR-664a-5p | cervical and endocervical cancer | poor survival; staging; metastasis; worse prognosis | "Down regulation of miR 664 in cervical cancer is a ......" | 27212165 | |
| hsa-miR-664a-5p | liver cancer | metastasis; tumorigenesis | "Expression of miR-664 miR-485-3p and miR-495 poten ......" | 23241961 | |
| hsa-miR-664a-5p | melanoma | malignant trasformation; poor survival | "Loss of MiR 664 Expression Enhances Cutaneous Mali ......" | 26287415 |
Reported gene related to hsa-miR-664a-5p
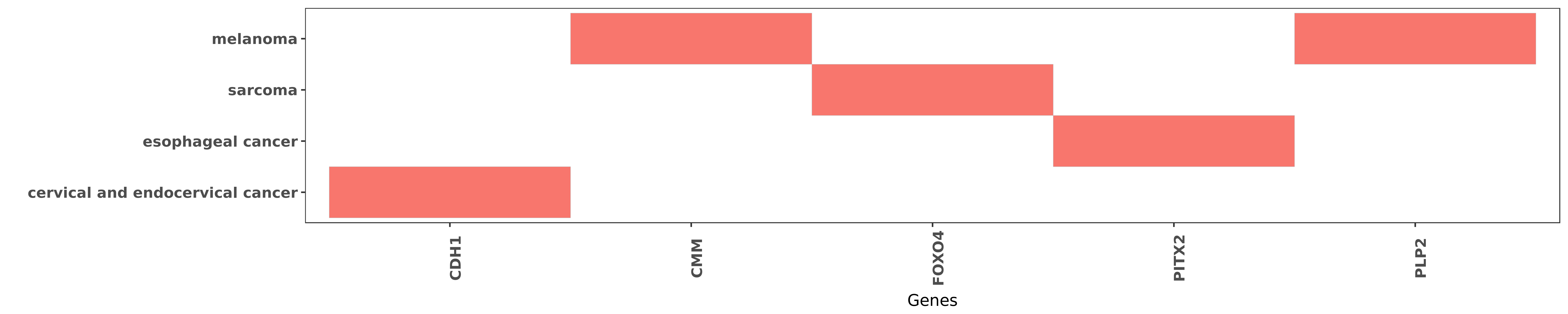
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-664a-5p | cervical and endocervical cancer | CDH1 | "Furthermore the effect of miR-664 up-regulation on ......" | 26770409 |
| hsa-miR-664a-5p | melanoma | CMM | "However the regulatory mechanisms of PLP2 in cance ......" | 26287415 |
| hsa-miR-664a-5p | sarcoma | FOXO4 | "Mir 664 promotes osteosarcoma cells proliferation ......" | 26463624 |
| hsa-miR-664a-5p | esophageal cancer | PITX2 | "Mechanistic studies revealed that miR-664a attenua ......" | 27407092 |
| hsa-miR-664a-5p | melanoma | PLP2 | "Loss of MiR 664 Expression Enhances Cutaneous Mali ......" | 26287415 |
Expression profile in cancer corhorts:
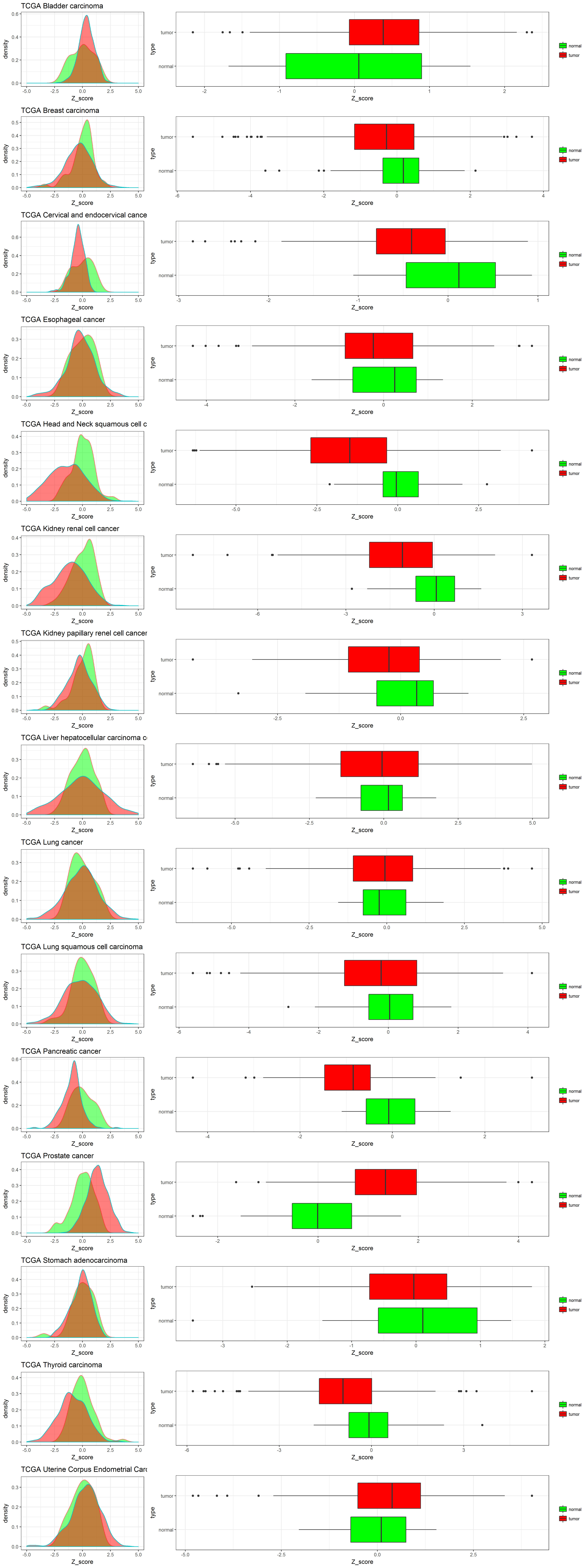
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-664a-5p | KPNA1 | 10 cancers: BLCA; CESC; ESCA; HNSC; LUAD; OV; PAAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.054; TCGA CESC -0.062; TCGA ESCA -0.091; TCGA HNSC -0.063; TCGA LUAD -0.131; TCGA OV -0.076; TCGA PAAD -0.073; TCGA SARC -0.06; TCGA STAD -0.062; TCGA UCEC -0.075 |
hsa-miR-664a-5p | ATP1B3 | 11 cancers: BLCA; BRCA; ESCA; HNSC; LIHC; LUAD; PAAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.15; TCGA BRCA -0.167; TCGA ESCA -0.329; TCGA HNSC -0.175; TCGA LIHC -0.174; TCGA LUAD -0.172; TCGA PAAD -0.191; TCGA SARC -0.128; TCGA THCA -0.186; TCGA STAD -0.104; TCGA UCEC -0.105 |
hsa-miR-664a-5p | RNGTT | 11 cancers: BLCA; CESC; COAD; ESCA; LIHC; LUAD; LUSC; OV; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.106; TCGA CESC -0.075; TCGA COAD -0.06; TCGA ESCA -0.094; TCGA LIHC -0.066; TCGA LUAD -0.067; TCGA LUSC -0.064; TCGA OV -0.082; TCGA SARC -0.075; TCGA STAD -0.085; TCGA UCEC -0.127 |
hsa-miR-664a-5p | BCL9L | 11 cancers: BLCA; CESC; ESCA; HNSC; KIRC; KIRP; LGG; LUAD; PAAD; PRAD; THCA | MirTarget | TCGA BLCA -0.125; TCGA CESC -0.152; TCGA ESCA -0.101; TCGA HNSC -0.083; TCGA KIRC -0.241; TCGA KIRP -0.139; TCGA LGG -0.125; TCGA LUAD -0.129; TCGA PAAD -0.2; TCGA PRAD -0.084; TCGA THCA -0.206 |
hsa-miR-664a-5p | SEC23A | 13 cancers: BLCA; BRCA; CESC; ESCA; HNSC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.265; TCGA BRCA -0.056; TCGA CESC -0.09; TCGA ESCA -0.185; TCGA HNSC -0.105; TCGA LUAD -0.179; TCGA LUSC -0.128; TCGA PAAD -0.26; TCGA PRAD -0.337; TCGA SARC -0.164; TCGA THCA -0.216; TCGA STAD -0.12; TCGA UCEC -0.142 |
hsa-miR-664a-5p | RALB | 10 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUAD; PAAD; PRAD; THCA; UCEC | MirTarget | TCGA BLCA -0.163; TCGA CESC -0.085; TCGA COAD -0.06; TCGA ESCA -0.264; TCGA HNSC -0.151; TCGA LUAD -0.103; TCGA PAAD -0.216; TCGA PRAD -0.063; TCGA THCA -0.112; TCGA UCEC -0.109 |
hsa-miR-664a-5p | PANK3 | 11 cancers: BLCA; CESC; COAD; HNSC; LUAD; LUSC; OV; PAAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.166; TCGA CESC -0.142; TCGA COAD -0.137; TCGA HNSC -0.146; TCGA LUAD -0.116; TCGA LUSC -0.079; TCGA OV -0.137; TCGA PAAD -0.118; TCGA SARC -0.106; TCGA STAD -0.092; TCGA UCEC -0.163 |
hsa-miR-664a-5p | POLR3G | 9 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LUAD; STAD; UCEC | MirTarget | TCGA BLCA -0.403; TCGA BRCA -0.078; TCGA CESC -0.206; TCGA COAD -0.216; TCGA ESCA -0.246; TCGA HNSC -0.217; TCGA LUAD -0.174; TCGA STAD -0.233; TCGA UCEC -0.159 |
hsa-miR-664a-5p | NUDCD1 | 10 cancers: BLCA; CESC; ESCA; HNSC; KIRP; LGG; LUAD; LUSC; PAAD; UCEC | MirTarget | TCGA BLCA -0.089; TCGA CESC -0.069; TCGA ESCA -0.146; TCGA HNSC -0.07; TCGA KIRP -0.118; TCGA LGG -0.057; TCGA LUAD -0.07; TCGA LUSC -0.114; TCGA PAAD -0.106; TCGA UCEC -0.217 |
hsa-miR-664a-5p | HMGA2 | 11 cancers: BLCA; BRCA; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; PAAD; THCA; UCEC | MirTarget | TCGA BLCA -1.306; TCGA BRCA -0.367; TCGA ESCA -0.547; TCGA HNSC -0.557; TCGA KIRP -0.868; TCGA LIHC -0.355; TCGA LUAD -0.727; TCGA LUSC -0.397; TCGA PAAD -0.917; TCGA THCA -1.205; TCGA UCEC -0.256 |
hsa-miR-664a-5p | PHF19 | 11 cancers: BLCA; BRCA; COAD; KIRC; KIRP; LIHC; LUAD; PRAD; SARC; THCA; UCEC | mirMAP | TCGA BLCA -0.483; TCGA BRCA -0.206; TCGA COAD -0.077; TCGA KIRC -0.135; TCGA KIRP -0.11; TCGA LIHC -0.363; TCGA LUAD -0.233; TCGA PRAD -0.222; TCGA SARC -0.222; TCGA THCA -0.164; TCGA UCEC -0.191 |
hsa-miR-664a-5p | TRAF3 | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; PRAD; UCEC | mirMAP | TCGA BLCA -0.159; TCGA BRCA -0.071; TCGA COAD -0.137; TCGA ESCA -0.197; TCGA HNSC -0.056; TCGA KIRC -0.135; TCGA LIHC -0.078; TCGA LUAD -0.077; TCGA PRAD -0.101; TCGA UCEC -0.056 |
hsa-miR-664a-5p | DST | 9 cancers: BLCA; ESCA; LGG; LUAD; PAAD; PRAD; SARC; THCA; STAD | mirMAP | TCGA BLCA -0.128; TCGA ESCA -0.243; TCGA LGG -0.164; TCGA LUAD -0.15; TCGA PAAD -0.138; TCGA PRAD -0.334; TCGA SARC -0.233; TCGA THCA -0.417; TCGA STAD -0.159 |
hsa-miR-664a-5p | AMOTL1 | 11 cancers: BLCA; BRCA; ESCA; HNSC; KIRC; KIRP; LGG; PAAD; SARC; STAD; UCEC | mirMAP | TCGA BLCA -0.545; TCGA BRCA -0.132; TCGA ESCA -0.229; TCGA HNSC -0.078; TCGA KIRC -0.135; TCGA KIRP -0.158; TCGA LGG -0.201; TCGA PAAD -0.173; TCGA SARC -0.123; TCGA STAD -0.158; TCGA UCEC -0.105 |
hsa-miR-664a-5p | SPSB1 | 9 cancers: BLCA; BRCA; COAD; ESCA; KIRC; LGG; PAAD; PRAD; THCA | mirMAP | TCGA BLCA -0.138; TCGA BRCA -0.172; TCGA COAD -0.095; TCGA ESCA -0.169; TCGA KIRC -0.238; TCGA LGG -0.213; TCGA PAAD -0.268; TCGA PRAD -0.229; TCGA THCA -0.276 |
hsa-miR-664a-5p | SH3PXD2B | 11 cancers: BLCA; BRCA; CESC; ESCA; KIRC; LUAD; LUSC; PAAD; PRAD; THCA; STAD | mirMAP | TCGA BLCA -0.307; TCGA BRCA -0.135; TCGA CESC -0.102; TCGA ESCA -0.16; TCGA KIRC -0.193; TCGA LUAD -0.198; TCGA LUSC -0.125; TCGA PAAD -0.363; TCGA PRAD -0.355; TCGA THCA -0.118; TCGA STAD -0.115 |
hsa-miR-664a-5p | LMNB2 | 13 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; OV; PAAD; THCA; UCEC | mirMAP | TCGA BLCA -0.202; TCGA BRCA -0.328; TCGA COAD -0.136; TCGA ESCA -0.303; TCGA HNSC -0.096; TCGA KIRC -0.095; TCGA LIHC -0.206; TCGA LUAD -0.377; TCGA LUSC -0.143; TCGA OV -0.079; TCGA PAAD -0.106; TCGA THCA -0.097; TCGA UCEC -0.226 |
hsa-miR-664a-5p | SOX11 | 12 cancers: BLCA; BRCA; CESC; HNSC; KIRC; KIRP; LGG; LIHC; OV; PAAD; THCA; STAD | mirMAP | TCGA BLCA -0.524; TCGA BRCA -0.563; TCGA CESC -0.411; TCGA HNSC -0.275; TCGA KIRC -1.083; TCGA KIRP -0.329; TCGA LGG -0.515; TCGA LIHC -0.334; TCGA OV -0.688; TCGA PAAD -0.648; TCGA THCA -0.765; TCGA STAD -0.463 |
hsa-miR-664a-5p | PTRF | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; PAAD; PRAD; THCA; UCEC | mirMAP | TCGA BLCA -0.645; TCGA BRCA -0.16; TCGA COAD -0.157; TCGA ESCA -0.358; TCGA HNSC -0.203; TCGA KIRC -0.188; TCGA KIRP -0.243; TCGA PAAD -0.252; TCGA PRAD -0.406; TCGA THCA -0.184; TCGA UCEC -0.127 |
hsa-miR-664a-5p | CCDC85C | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LUAD; OV; UCEC | mirMAP | TCGA BLCA -0.24; TCGA BRCA -0.051; TCGA CESC -0.141; TCGA COAD -0.133; TCGA ESCA -0.228; TCGA HNSC -0.163; TCGA LGG -0.173; TCGA LUAD -0.112; TCGA OV -0.099; TCGA UCEC -0.11 |
hsa-miR-664a-5p | FAT1 | 10 cancers: BRCA; CESC; COAD; ESCA; KIRP; LGG; LUAD; LUSC; PAAD; STAD | MirTarget | TCGA BRCA -0.178; TCGA CESC -0.16; TCGA COAD -0.144; TCGA ESCA -0.229; TCGA KIRP -0.274; TCGA LGG -0.146; TCGA LUAD -0.14; TCGA LUSC -0.191; TCGA PAAD -0.407; TCGA STAD -0.174 |
hsa-miR-664a-5p | SH3PXD2A | 10 cancers: BRCA; ESCA; HNSC; KIRC; KIRP; LIHC; PAAD; PRAD; THCA; UCEC | mirMAP | TCGA BRCA -0.095; TCGA ESCA -0.175; TCGA HNSC -0.098; TCGA KIRC -0.271; TCGA KIRP -0.14; TCGA LIHC -0.15; TCGA PAAD -0.157; TCGA PRAD -0.335; TCGA THCA -0.23; TCGA UCEC -0.084 |
hsa-miR-664a-5p | FRMD8 | 9 cancers: BRCA; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; PAAD; THCA | mirMAP | TCGA BRCA -0.103; TCGA COAD -0.09; TCGA ESCA -0.164; TCGA HNSC -0.122; TCGA KIRC -0.28; TCGA LIHC -0.051; TCGA LUAD -0.104; TCGA PAAD -0.111; TCGA THCA -0.06 |
Enriched cancer pathways of putative targets