microRNA information: hsa-miR-769-3p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-769-3p | miRbase |
| Accession: | MIMAT0003887 | miRbase |
| Precursor name: | hsa-mir-769 | miRbase |
| Precursor accession: | MI0003834 | miRbase |
| Symbol: | MIR769 | HGNC |
| RefSeq ID: | NR_030412 | GenBank |
| Sequence: | CUGGGAUCUCCGGGGUCUUGGUU |
Reported expression in cancers: hsa-miR-769-3p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|
Reported cancer pathway affected by hsa-miR-769-3p
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|
Reported cancer prognosis affected by hsa-miR-769-3p
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|
Reported gene related to hsa-miR-769-3p

| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-769-3p | melanoma | GSK3B | "MiR 769 promoted cell proliferation in human melan ......" | 27470346 |
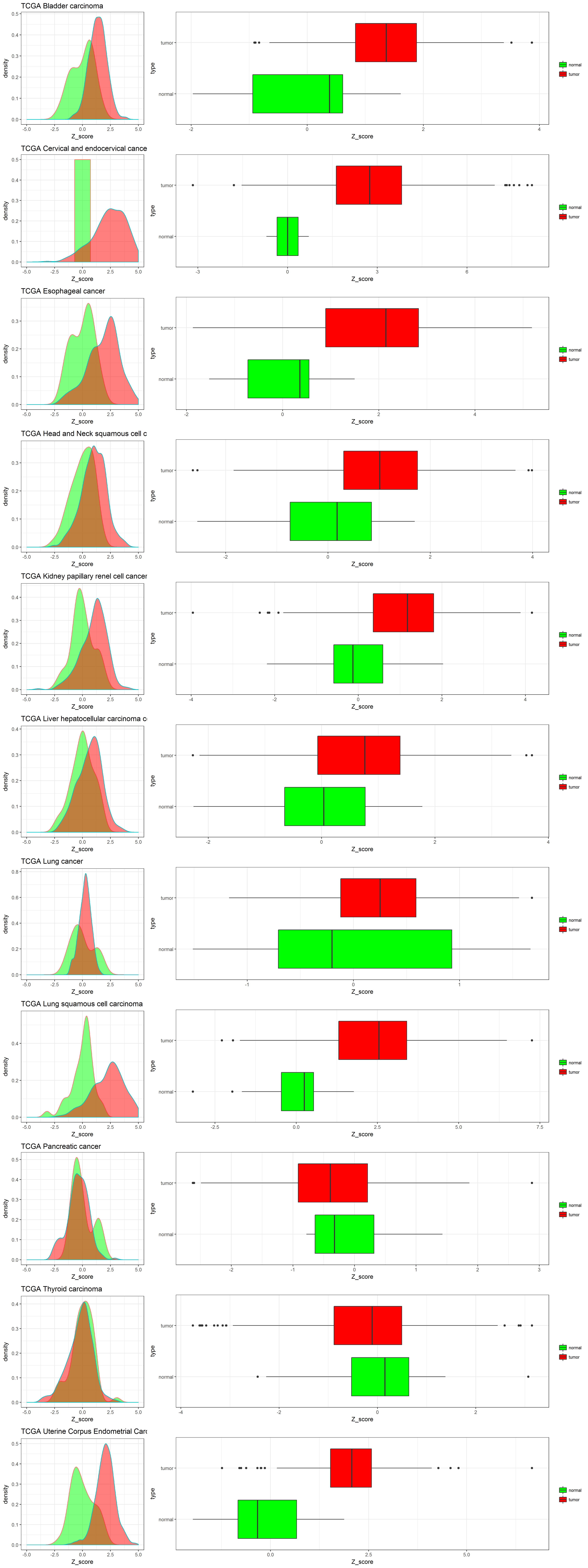
Expression profile in cancer corhorts:
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-769-3p | EEF2 | 9 cancers: BLCA; ESCA; HNSC; KIRP; LGG; LIHC; OV; THCA; UCEC | miRNAWalker2 validate | TCGA BLCA -0.101; TCGA ESCA -0.096; TCGA HNSC -0.113; TCGA KIRP -0.067; TCGA LGG -0.159; TCGA LIHC -0.063; TCGA OV -0.079; TCGA THCA -0.095; TCGA UCEC -0.072 |
hsa-miR-769-3p | HMCN1 | 10 cancers: BLCA; COAD; ESCA; HNSC; KIRP; LGG; LUAD; LUSC; PAAD; THCA | MirTarget; PITA; miRNATAP | TCGA BLCA -0.321; TCGA COAD -0.476; TCGA ESCA -0.43; TCGA HNSC -0.332; TCGA KIRP -0.512; TCGA LGG -0.115; TCGA LUAD -0.201; TCGA LUSC -0.408; TCGA PAAD -0.269; TCGA THCA -0.23 |
hsa-miR-769-3p | IKZF1 | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; THCA; UCEC | MirTarget | TCGA BLCA -0.353; TCGA CESC -0.356; TCGA COAD -0.178; TCGA ESCA -0.225; TCGA HNSC -0.21; TCGA LUAD -0.212; TCGA LUSC -0.404; TCGA THCA -0.376; TCGA UCEC -0.124 |
hsa-miR-769-3p | GPRASP1 | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LIHC; LUSC; UCEC | MirTarget | TCGA BLCA -0.308; TCGA CESC -0.211; TCGA COAD -0.268; TCGA ESCA -0.346; TCGA HNSC -0.204; TCGA KIRP -0.167; TCGA LIHC -0.14; TCGA LUSC -0.245; TCGA UCEC -0.581 |
hsa-miR-769-3p | TSHZ3 | 9 cancers: BLCA; COAD; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; UCEC | MirTarget; PITA; miRNATAP | TCGA BLCA -0.516; TCGA COAD -0.355; TCGA ESCA -0.188; TCGA HNSC -0.191; TCGA KIRP -0.185; TCGA LIHC -0.104; TCGA LUAD -0.093; TCGA LUSC -0.18; TCGA UCEC -0.511 |
hsa-miR-769-3p | MEIS2 | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LGG; PAAD; UCEC | PITA | TCGA BLCA -0.265; TCGA CESC -0.213; TCGA COAD -0.312; TCGA ESCA -0.167; TCGA HNSC -0.093; TCGA KIRP -0.191; TCGA LGG -0.051; TCGA PAAD -0.169; TCGA UCEC -0.273 |
hsa-miR-769-3p | ZBTB4 | 10 cancers: BLCA; COAD; ESCA; HNSC; KIRP; LIHC; LUAD; LUSC; OV; UCEC | PITA; miRNATAP | TCGA BLCA -0.145; TCGA COAD -0.117; TCGA ESCA -0.171; TCGA HNSC -0.107; TCGA KIRP -0.059; TCGA LIHC -0.073; TCGA LUAD -0.08; TCGA LUSC -0.144; TCGA OV -0.086; TCGA UCEC -0.249 |
hsa-miR-769-3p | ENTPD1 | 12 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LGG; LUAD; LUSC; OV; PAAD; UCEC | mirMAP | TCGA BLCA -0.263; TCGA CESC -0.089; TCGA COAD -0.176; TCGA ESCA -0.135; TCGA HNSC -0.063; TCGA KIRP -0.097; TCGA LGG -0.051; TCGA LUAD -0.079; TCGA LUSC -0.221; TCGA OV -0.062; TCGA PAAD -0.122; TCGA UCEC -0.24 |
hsa-miR-769-3p | NFIC | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRP; LIHC; THCA; UCEC | mirMAP | TCGA BLCA -0.239; TCGA CESC -0.182; TCGA COAD -0.113; TCGA ESCA -0.229; TCGA HNSC -0.334; TCGA KIRP -0.316; TCGA LIHC -0.29; TCGA THCA -0.143; TCGA UCEC -0.218 |
hsa-miR-769-3p | DOCK2 | 9 cancers: BLCA; CESC; COAD; ESCA; HNSC; LUAD; LUSC; THCA; UCEC | miRNATAP | TCGA BLCA -0.5; TCGA CESC -0.342; TCGA COAD -0.224; TCGA ESCA -0.356; TCGA HNSC -0.236; TCGA LUAD -0.227; TCGA LUSC -0.533; TCGA THCA -0.396; TCGA UCEC -0.118 |
hsa-miR-769-3p | FOXP1 | 9 cancers: BLCA; COAD; ESCA; HNSC; LUAD; LUSC; OV; PAAD; UCEC | miRNATAP | TCGA BLCA -0.127; TCGA COAD -0.079; TCGA ESCA -0.178; TCGA HNSC -0.194; TCGA LUAD -0.079; TCGA LUSC -0.249; TCGA OV -0.088; TCGA PAAD -0.148; TCGA UCEC -0.227 |
Enriched cancer pathways of putative targets