microRNA information: hsa-miR-98-5p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-98-5p | miRbase |
| Accession: | MIMAT0000096 | miRbase |
| Precursor name: | hsa-mir-98 | miRbase |
| Precursor accession: | MI0000100 | miRbase |
| Symbol: | MIR98 | HGNC |
| RefSeq ID: | NR_029513 | GenBank |
| Sequence: | UGAGGUAGUAAGUUGUAUUGUU |
Reported expression in cancers: hsa-miR-98-5p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-98-5p | breast cancer | upregulation | "Over expression of miR 98 in FFPE tissues might se ......" | 24696733 | qPCR |
| hsa-miR-98-5p | colon cancer | deregulation | "The expression of 384 miRNAs was determined by usi ......" | 26556872 | Microarray |
| hsa-miR-98-5p | gastric cancer | deregulation | "The results from the miRNA microarray analysis wer ......" | 21475928 | qPCR; Microarray |
| hsa-miR-98-5p | liver cancer | downregulation | "The expression of serum microRNAs was measured usi ......" | 24697119 | Reverse transcription PCR; qPCR |
| hsa-miR-98-5p | liver cancer | downregulation | "MicroRNAs miRs are involved in the development and ......" | 27677076 | |
| hsa-miR-98-5p | prostate cancer | downregulation | "We investigated the expression profiles of 6 micro ......" | 20873592 | Microarray; in situ hybridization |
| hsa-miR-98-5p | thyroid cancer | deregulation | "Quantitative real-time PCR qRT-PCR was used to val ......" | 26380656 | qPCR |
Reported cancer pathway affected by hsa-miR-98-5p
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-98-5p | esophageal cancer | Apoptosis pathway | "Upregulation of microRNA 98 increases radiosensiti ......" | 27422937 | Flow cytometry; Transwell assay; Luciferase |
| hsa-miR-98-5p | liver cancer | Epithelial mesenchymal transition pathway | "MicroRNA 98 acts as a tumor suppressor in hepatoce ......" | 27677076 | |
| hsa-miR-98-5p | lung cancer | Apoptosis pathway | "MicroRNA 98 sensitizes cisplatin resistant human l ......" | 23700794 | |
| hsa-miR-98-5p | lung cancer | Apoptosis pathway | "MiR 98 inhibits cell proliferation and invasion of ......" | 26884926 |
Reported cancer prognosis affected by hsa-miR-98-5p
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-98-5p | breast cancer | poor survival | "To investigate the global expression profile of mi ......" | 18812439 | |
| hsa-miR-98-5p | esophageal cancer | staging; metastasis | "MicroRNA 98 and microRNA 214 post transcriptionall ......" | 22867052 | Western blot |
| hsa-miR-98-5p | esophageal cancer | drug resistance; metastasis | "Upregulation of microRNA 98 increases radiosensiti ......" | 27422937 | Flow cytometry; Transwell assay; Luciferase |
| hsa-miR-98-5p | liver cancer | progression; metastasis; poor survival; tumor size; malignant trasformation | "MicroRNA 98 acts as a tumor suppressor in hepatoce ......" | 27677076 | |
| hsa-miR-98-5p | melanoma | metastasis; staging; poor survival; cell migration; tumor size | "miR 98 suppresses melanoma metastasis through a ne ......" | 25277211 | |
| hsa-miR-98-5p | prostate cancer | drug resistance | "Identification of microRNA 98 as a therapeutic tar ......" | 23188821 |
Reported gene related to hsa-miR-98-5p
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-98-5p | esophageal cancer | EZH2 | "In Eca109 cells overexpression of miR-98 and miR-2 ......" | 22867052 |
| hsa-miR-98-5p | ovarian cancer | EZH2 | "EZH2 specific microRNA 98 inhibits human ovarian c ......" | 24771265 |
| hsa-miR-98-5p | head and neck cancer | HMGA2 | "Here we report that HMGA2 expression in head and n ......" | 17222355 |
| hsa-miR-98-5p | lung cancer | HMGA2 | "MicroRNA 98 sensitizes cisplatin resistant human l ......" | 23700794 |
| hsa-miR-98-5p | prostate cancer | CCNJ | "And CCNJ a protein controlling cell mitosis is dow ......" | 23188821 |
| hsa-miR-98-5p | esophageal cancer | CDC37 | "MicroRNA 98 and microRNA 214 post transcriptionall ......" | 22867052 |
| hsa-miR-98-5p | lung squamous cell cancer | FZD3 | "Our data analysis also showed that miR-98 and miR- ......" | 25521201 |
| hsa-miR-98-5p | melanoma | IL6 | "miR 98 suppresses melanoma metastasis through a ne ......" | 25277211 |
| hsa-miR-98-5p | lung squamous cell cancer | ITGB3 | "miR 98 targets ITGB3 to inhibit proliferation migr ......" | 26445551 |
| hsa-miR-98-5p | prostate cancer | LIN28A | "Mechanistic dissection revealed that 125-VD-induce ......" | 23188821 |
| hsa-miR-98-5p | esophageal cancer | MPP2 | "MicroRNA 98 and microRNA 214 post transcriptionall ......" | 22867052 |
| hsa-miR-98-5p | lung cancer | PAK1 | "MiR 98 inhibits cell proliferation and invasion of ......" | 26884926 |
| hsa-miR-98-5p | lung squamous cell cancer | RPS6KA3 | "Our data analysis also showed that miR-98 and miR- ......" | 25521201 |
| hsa-miR-98-5p | liver cancer | SALL4 | "MicroRNA 98 acts as a tumor suppressor in hepatoce ......" | 27677076 |
| hsa-miR-98-5p | prostate cancer | VDR | "Mechanistic dissection revealed that 125-VD-induce ......" | 23188821 |
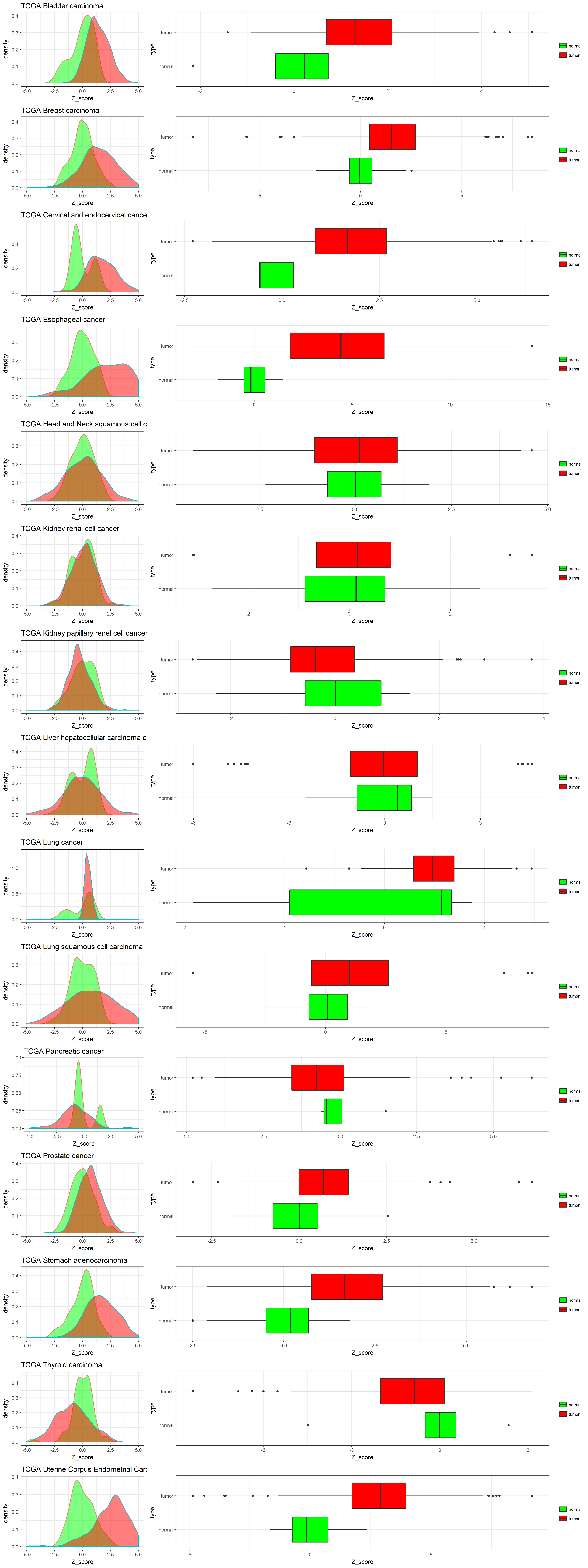
Expression profile in cancer corhorts:
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-98-5p | A2M | 9 cancers: BLCA; BRCA; CESC; COAD; LUAD; LUSC; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.549; TCGA BRCA -0.168; TCGA CESC -0.217; TCGA COAD -0.463; TCGA LUAD -0.234; TCGA LUSC -0.663; TCGA SARC -0.734; TCGA STAD -0.625; TCGA UCEC -0.328 |
hsa-miR-98-5p | AKAP2 | 9 cancers: BLCA; BRCA; COAD; ESCA; LUAD; LUSC; PRAD; SARC; UCEC | miRNAWalker2 validate | TCGA BLCA -0.925; TCGA BRCA -0.282; TCGA COAD -0.457; TCGA ESCA -0.425; TCGA LUAD -0.215; TCGA LUSC -0.785; TCGA PRAD -0.407; TCGA SARC -0.55; TCGA UCEC -0.697 |
hsa-miR-98-5p | ARSJ | 9 cancers: BLCA; BRCA; COAD; KIRP; LUSC; PAAD; PRAD; THCA; STAD | miRNAWalker2 validate | TCGA BLCA -1.073; TCGA BRCA -0.276; TCGA COAD -0.448; TCGA KIRP -0.485; TCGA LUSC -0.279; TCGA PAAD -0.35; TCGA PRAD -0.271; TCGA THCA -1.315; TCGA STAD -0.429 |
hsa-miR-98-5p | C3 | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; LUAD; LUSC; PAAD; THCA; STAD | miRNAWalker2 validate | TCGA BLCA -1.12; TCGA BRCA -0.265; TCGA CESC -0.524; TCGA COAD -0.819; TCGA ESCA -0.529; TCGA LUAD -0.289; TCGA LUSC -0.656; TCGA PAAD -0.839; TCGA THCA -1.684; TCGA STAD -0.404 |
hsa-miR-98-5p | CDC42BPA | 9 cancers: BLCA; BRCA; CESC; COAD; LIHC; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.39; TCGA BRCA -0.089; TCGA CESC -0.395; TCGA COAD -0.266; TCGA LIHC -0.107; TCGA PRAD -0.217; TCGA SARC -0.236; TCGA STAD -0.223; TCGA UCEC -0.331 |
hsa-miR-98-5p | CHST3 | 12 cancers: BLCA; COAD; ESCA; KIRC; KIRP; LGG; LUSC; PAAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.494; TCGA COAD -0.655; TCGA ESCA -0.365; TCGA KIRC -0.244; TCGA KIRP -0.273; TCGA LGG -0.203; TCGA LUSC -0.212; TCGA PAAD -0.468; TCGA SARC -0.154; TCGA THCA -0.152; TCGA STAD -0.586; TCGA UCEC -0.272 |
hsa-miR-98-5p | CYB5R3 | 10 cancers: BLCA; BRCA; ESCA; KIRP; LUAD; OV; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.182; TCGA BRCA -0.102; TCGA ESCA -0.163; TCGA KIRP -0.218; TCGA LUAD -0.164; TCGA OV -0.069; TCGA SARC -0.228; TCGA THCA -0.12; TCGA STAD -0.267; TCGA UCEC -0.145 |
hsa-miR-98-5p | DOCK5 | 9 cancers: BLCA; COAD; ESCA; LUAD; LUSC; PAAD; PRAD; SARC; UCEC | miRNAWalker2 validate | TCGA BLCA -0.23; TCGA COAD -0.288; TCGA ESCA -0.234; TCGA LUAD -0.204; TCGA LUSC -0.178; TCGA PAAD -1.05; TCGA PRAD -0.37; TCGA SARC -0.419; TCGA UCEC -0.206 |
hsa-miR-98-5p | ECM2 | 10 cancers: BLCA; BRCA; CESC; COAD; ESCA; LIHC; LUSC; PAAD; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.661; TCGA BRCA -0.651; TCGA CESC -0.255; TCGA COAD -0.434; TCGA ESCA -0.233; TCGA LIHC -0.183; TCGA LUSC -0.918; TCGA PAAD -0.383; TCGA STAD -0.541; TCGA UCEC -0.698 |
hsa-miR-98-5p | FAS | 10 cancers: BLCA; BRCA; COAD; HNSC; LIHC; LUSC; PAAD; SARC; THCA; UCEC | miRNAWalker2 validate | TCGA BLCA -0.771; TCGA BRCA -0.148; TCGA COAD -0.378; TCGA HNSC -0.14; TCGA LIHC -0.184; TCGA LUSC -0.523; TCGA PAAD -0.462; TCGA SARC -0.306; TCGA THCA -0.772; TCGA UCEC -0.516 |
hsa-miR-98-5p | FNDC3A | 9 cancers: BLCA; BRCA; CESC; ESCA; LGG; LUSC; OV; PRAD; UCEC | miRNAWalker2 validate; MirTarget | TCGA BLCA -0.105; TCGA BRCA -0.267; TCGA CESC -0.252; TCGA ESCA -0.16; TCGA LGG -0.173; TCGA LUSC -0.244; TCGA OV -0.099; TCGA PRAD -0.222; TCGA UCEC -0.38 |
hsa-miR-98-5p | FOXO1 | 13 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; OV; PRAD; SARC; UCEC | miRNAWalker2 validate | TCGA BLCA -0.253; TCGA BRCA -0.418; TCGA CESC -0.235; TCGA COAD -0.167; TCGA ESCA -0.277; TCGA HNSC -0.147; TCGA KIRC -0.179; TCGA LUAD -0.163; TCGA LUSC -0.283; TCGA OV -0.117; TCGA PRAD -0.234; TCGA SARC -0.184; TCGA UCEC -0.416 |
hsa-miR-98-5p | FRMD4B | 10 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRP; LUSC; PAAD; PRAD; STAD | miRNAWalker2 validate | TCGA BLCA -0.494; TCGA BRCA -0.31; TCGA COAD -0.218; TCGA ESCA -0.209; TCGA HNSC -0.156; TCGA KIRP -0.172; TCGA LUSC -0.233; TCGA PAAD -0.452; TCGA PRAD -0.167; TCGA STAD -0.268 |
hsa-miR-98-5p | HEG1 | 9 cancers: BLCA; BRCA; COAD; ESCA; LUAD; LUSC; PAAD; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.517; TCGA BRCA -0.308; TCGA COAD -0.539; TCGA ESCA -0.217; TCGA LUAD -0.232; TCGA LUSC -0.524; TCGA PAAD -0.291; TCGA STAD -0.386; TCGA UCEC -0.385 |
hsa-miR-98-5p | HSPB7 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; LUSC; OV; PAAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -1.373; TCGA BRCA -0.746; TCGA CESC -0.406; TCGA COAD -1.12; TCGA ESCA -0.835; TCGA KIRC -1.478; TCGA LUSC -0.756; TCGA OV -0.143; TCGA PAAD -0.7; TCGA SARC -1.573; TCGA STAD -2.316; TCGA UCEC -1.15 |
hsa-miR-98-5p | IL6R | 11 cancers: BLCA; BRCA; COAD; ESCA; LIHC; LUSC; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.469; TCGA BRCA -0.214; TCGA COAD -0.231; TCGA ESCA -0.258; TCGA LIHC -0.18; TCGA LUSC -0.213; TCGA PRAD -0.351; TCGA SARC -0.503; TCGA THCA -0.568; TCGA STAD -0.505; TCGA UCEC -0.352 |
hsa-miR-98-5p | IRS2 | 9 cancers: BLCA; BRCA; KIRC; LUSC; OV; PRAD; THCA; STAD; UCEC | miRNAWalker2 validate; MirTarget | TCGA BLCA -0.368; TCGA BRCA -0.41; TCGA KIRC -0.455; TCGA LUSC -0.393; TCGA OV -0.148; TCGA PRAD -0.197; TCGA THCA -0.315; TCGA STAD -0.442; TCGA UCEC -0.616 |
hsa-miR-98-5p | ITGA5 | 10 cancers: BLCA; COAD; ESCA; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.94; TCGA COAD -0.442; TCGA ESCA -0.331; TCGA LUAD -0.195; TCGA LUSC -0.401; TCGA PAAD -0.626; TCGA PRAD -0.303; TCGA SARC -0.352; TCGA STAD -0.394; TCGA UCEC -0.209 |
hsa-miR-98-5p | ITGB3 | 12 cancers: BLCA; BRCA; COAD; ESCA; KIRP; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate; MirTarget | TCGA BLCA -0.835; TCGA BRCA -0.411; TCGA COAD -0.511; TCGA ESCA -0.457; TCGA KIRP -0.393; TCGA LUSC -0.564; TCGA PAAD -0.462; TCGA PRAD -0.406; TCGA SARC -0.605; TCGA THCA -0.214; TCGA STAD -0.546; TCGA UCEC -0.643 |
hsa-miR-98-5p | KLF13 | 10 cancers: BLCA; BRCA; CESC; COAD; KIRC; KIRP; LUAD; LUSC; PAAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.558; TCGA BRCA -0.169; TCGA CESC -0.214; TCGA COAD -0.217; TCGA KIRC -0.194; TCGA KIRP -0.273; TCGA LUAD -0.218; TCGA LUSC -0.307; TCGA PAAD -0.203; TCGA UCEC -0.254 |
hsa-miR-98-5p | MITF | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; KIRP; LUSC; PAAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.862; TCGA BRCA -0.395; TCGA CESC -0.524; TCGA COAD -0.678; TCGA ESCA -0.481; TCGA KIRC -0.25; TCGA KIRP -0.468; TCGA LUSC -0.571; TCGA PAAD -0.944; TCGA SARC -0.688; TCGA STAD -0.883; TCGA UCEC -0.606 |
hsa-miR-98-5p | NAV2 | 10 cancers: BLCA; BRCA; COAD; KIRP; LGG; LUSC; PAAD; SARC; THCA; STAD | miRNAWalker2 validate | TCGA BLCA -0.261; TCGA BRCA -0.325; TCGA COAD -0.172; TCGA KIRP -0.221; TCGA LGG -0.179; TCGA LUSC -0.281; TCGA PAAD -0.3; TCGA SARC -0.847; TCGA THCA -0.159; TCGA STAD -0.328 |
hsa-miR-98-5p | NEK3 | 9 cancers: BLCA; CESC; HNSC; KIRP; LUAD; LUSC; OV; THCA; UCEC | miRNAWalker2 validate | TCGA BLCA -0.19; TCGA CESC -0.353; TCGA HNSC -0.181; TCGA KIRP -0.301; TCGA LUAD -0.122; TCGA LUSC -0.299; TCGA OV -0.153; TCGA THCA -0.096; TCGA UCEC -0.198 |
hsa-miR-98-5p | OSBPL10 | 10 cancers: BLCA; BRCA; CESC; HNSC; KIRP; LUAD; LUSC; PAAD; SARC; THCA | miRNAWalker2 validate | TCGA BLCA -0.445; TCGA BRCA -0.163; TCGA CESC -0.214; TCGA HNSC -0.14; TCGA KIRP -0.339; TCGA LUAD -0.094; TCGA LUSC -0.139; TCGA PAAD -0.239; TCGA SARC -0.504; TCGA THCA -0.441 |
hsa-miR-98-5p | OSMR | 9 cancers: BLCA; BRCA; COAD; ESCA; PAAD; PRAD; SARC; THCA; UCEC | miRNAWalker2 validate | TCGA BLCA -1.169; TCGA BRCA -0.26; TCGA COAD -0.652; TCGA ESCA -0.329; TCGA PAAD -0.755; TCGA PRAD -0.326; TCGA SARC -0.34; TCGA THCA -1.011; TCGA UCEC -0.259 |
hsa-miR-98-5p | PALM2-AKAP2 | 9 cancers: BLCA; BRCA; COAD; ESCA; LUAD; LUSC; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.615; TCGA BRCA -0.228; TCGA COAD -0.377; TCGA ESCA -0.255; TCGA LUAD -0.159; TCGA LUSC -0.642; TCGA SARC -0.278; TCGA STAD -0.507; TCGA UCEC -0.525 |
hsa-miR-98-5p | PPARGC1A | 10 cancers: BLCA; BRCA; KIRC; KIRP; LUSC; OV; PAAD; PRAD; SARC; STAD | miRNAWalker2 validate | TCGA BLCA -0.961; TCGA BRCA -0.429; TCGA KIRC -0.365; TCGA KIRP -0.509; TCGA LUSC -0.577; TCGA OV -0.535; TCGA PAAD -0.515; TCGA PRAD -0.54; TCGA SARC -1.435; TCGA STAD -0.513 |
hsa-miR-98-5p | RAB30 | 9 cancers: BLCA; BRCA; COAD; OV; PAAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.286; TCGA BRCA -0.33; TCGA COAD -0.211; TCGA OV -0.093; TCGA PAAD -0.267; TCGA PRAD -0.273; TCGA SARC -0.301; TCGA STAD -0.328; TCGA UCEC -0.144 |
hsa-miR-98-5p | RASAL2 | 9 cancers: BLCA; COAD; KIRP; LUAD; LUSC; PAAD; SARC; THCA; UCEC | miRNAWalker2 validate | TCGA BLCA -0.13; TCGA COAD -0.157; TCGA KIRP -0.116; TCGA LUAD -0.077; TCGA LUSC -0.228; TCGA PAAD -0.225; TCGA SARC -0.133; TCGA THCA -0.294; TCGA UCEC -0.244 |
hsa-miR-98-5p | RBPMS | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRP; LUSC; PAAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.155; TCGA BRCA -0.236; TCGA CESC -0.341; TCGA COAD -0.324; TCGA ESCA -0.317; TCGA KIRP -0.23; TCGA LUSC -0.23; TCGA PAAD -0.612; TCGA SARC -1.175; TCGA THCA -0.073; TCGA STAD -0.835; TCGA UCEC -0.363 |
hsa-miR-98-5p | ROR1 | 9 cancers: BLCA; BRCA; COAD; ESCA; LUSC; PRAD; SARC; THCA; STAD | miRNAWalker2 validate | TCGA BLCA -1.066; TCGA BRCA -0.297; TCGA COAD -1.008; TCGA ESCA -0.429; TCGA LUSC -0.884; TCGA PRAD -0.581; TCGA SARC -0.504; TCGA THCA -1.368; TCGA STAD -0.634 |
hsa-miR-98-5p | SGMS1 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; LGG; PRAD; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.103; TCGA BRCA -0.129; TCGA CESC -0.133; TCGA COAD -0.14; TCGA ESCA -0.15; TCGA KIRC -0.143; TCGA LGG -0.111; TCGA PRAD -0.183; TCGA THCA -0.415; TCGA STAD -0.202; TCGA UCEC -0.194 |
hsa-miR-98-5p | SNX24 | 9 cancers: BLCA; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; THCA; STAD | miRNAWalker2 validate | TCGA BLCA -0.175; TCGA COAD -0.36; TCGA ESCA -0.243; TCGA HNSC -0.215; TCGA KIRC -0.094; TCGA LIHC -0.109; TCGA LUAD -0.062; TCGA THCA -0.105; TCGA STAD -0.221 |
hsa-miR-98-5p | SPARCL1 | 9 cancers: BLCA; BRCA; COAD; ESCA; LUAD; OV; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.71; TCGA BRCA -0.626; TCGA COAD -0.665; TCGA ESCA -0.401; TCGA LUAD -0.158; TCGA OV -0.097; TCGA SARC -0.71; TCGA STAD -1.305; TCGA UCEC -0.861 |
hsa-miR-98-5p | ST5 | 15 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LGG; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.529; TCGA BRCA -0.145; TCGA CESC -0.33; TCGA COAD -0.432; TCGA ESCA -0.241; TCGA HNSC -0.136; TCGA KIRC -0.343; TCGA KIRP -0.143; TCGA LGG -0.066; TCGA LUSC -0.211; TCGA PAAD -0.283; TCGA PRAD -0.166; TCGA SARC -0.553; TCGA STAD -0.483; TCGA UCEC -0.476 |
hsa-miR-98-5p | TEAD1 | 12 cancers: BLCA; BRCA; COAD; ESCA; LGG; LUAD; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.276; TCGA BRCA -0.245; TCGA COAD -0.161; TCGA ESCA -0.143; TCGA LGG -0.232; TCGA LUAD -0.109; TCGA PAAD -0.163; TCGA PRAD -0.2; TCGA SARC -0.28; TCGA THCA -0.243; TCGA STAD -0.523; TCGA UCEC -0.218 |
hsa-miR-98-5p | TES | 10 cancers: BLCA; CESC; ESCA; KIRC; KIRP; PAAD; PRAD; SARC; THCA; STAD | miRNAWalker2 validate | TCGA BLCA -0.341; TCGA CESC -0.117; TCGA ESCA -0.159; TCGA KIRC -0.15; TCGA KIRP -0.523; TCGA PAAD -0.559; TCGA PRAD -0.266; TCGA SARC -0.752; TCGA THCA -0.154; TCGA STAD -0.39 |
hsa-miR-98-5p | TGFBR1 | 9 cancers: BLCA; BRCA; COAD; KIRC; LUSC; PAAD; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.402; TCGA BRCA -0.154; TCGA COAD -0.161; TCGA KIRC -0.208; TCGA LUSC -0.259; TCGA PAAD -0.186; TCGA THCA -0.571; TCGA STAD -0.27; TCGA UCEC -0.232 |
hsa-miR-98-5p | THBS1 | 9 cancers: BLCA; BRCA; COAD; ESCA; LUSC; PAAD; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.724; TCGA BRCA -0.196; TCGA COAD -0.471; TCGA ESCA -0.28; TCGA LUSC -0.611; TCGA PAAD -0.824; TCGA THCA -0.864; TCGA STAD -0.509; TCGA UCEC -0.708 |
hsa-miR-98-5p | TNFSF10 | 10 cancers: BLCA; BRCA; COAD; HNSC; KIRP; LIHC; LUSC; PAAD; PRAD; THCA | miRNAWalker2 validate | TCGA BLCA -0.62; TCGA BRCA -0.372; TCGA COAD -0.162; TCGA HNSC -0.372; TCGA KIRP -0.434; TCGA LIHC -0.19; TCGA LUSC -0.211; TCGA PAAD -0.588; TCGA PRAD -0.383; TCGA THCA -0.554 |
hsa-miR-98-5p | TNS1 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; LUAD; LUSC; PAAD; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.961; TCGA BRCA -0.555; TCGA CESC -0.294; TCGA COAD -0.79; TCGA ESCA -0.505; TCGA LUAD -0.18; TCGA LUSC -0.604; TCGA PAAD -0.459; TCGA PRAD -0.416; TCGA SARC -0.902; TCGA STAD -1.416; TCGA UCEC -0.571 |
hsa-miR-98-5p | ZFP36L1 | 9 cancers: BLCA; BRCA; CESC; COAD; ESCA; PAAD; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.357; TCGA BRCA -0.204; TCGA CESC -0.274; TCGA COAD -0.155; TCGA ESCA -0.24; TCGA PAAD -0.772; TCGA THCA -0.616; TCGA STAD -0.461; TCGA UCEC -0.182 |
hsa-miR-98-5p | MASP1 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRP; LGG; LUAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.309; TCGA BRCA -0.67; TCGA CESC -0.637; TCGA COAD -1.06; TCGA ESCA -0.61; TCGA KIRP -0.414; TCGA LGG -0.331; TCGA LUAD -0.282; TCGA SARC -1.657; TCGA STAD -1.188; TCGA UCEC -1.089 |
hsa-miR-98-5p | CPEB1 | 10 cancers: BLCA; BRCA; COAD; ESCA; LUAD; LUSC; OV; SARC; STAD; UCEC | MirTarget | TCGA BLCA -1.14; TCGA BRCA -0.527; TCGA COAD -1.01; TCGA ESCA -0.735; TCGA LUAD -0.303; TCGA LUSC -0.373; TCGA OV -0.239; TCGA SARC -0.723; TCGA STAD -1.653; TCGA UCEC -0.606 |
hsa-miR-98-5p | CGNL1 | 9 cancers: BLCA; BRCA; COAD; KIRC; LUSC; PAAD; PRAD; STAD; UCEC | MirTarget | TCGA BLCA -0.384; TCGA BRCA -0.473; TCGA COAD -0.323; TCGA KIRC -0.438; TCGA LUSC -0.642; TCGA PAAD -0.277; TCGA PRAD -0.471; TCGA STAD -0.839; TCGA UCEC -0.494 |
hsa-miR-98-5p | PITPNM3 | 10 cancers: BLCA; CESC; COAD; ESCA; KIRP; LUAD; PAAD; PRAD; SARC; STAD | MirTarget | TCGA BLCA -0.438; TCGA CESC -0.448; TCGA COAD -0.381; TCGA ESCA -0.313; TCGA KIRP -0.514; TCGA LUAD -0.161; TCGA PAAD -0.342; TCGA PRAD -0.411; TCGA SARC -0.757; TCGA STAD -1.14 |
hsa-miR-98-5p | ADAMTS15 | 9 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; LUAD; LUSC; STAD | MirTarget | TCGA BLCA -1.161; TCGA BRCA -1.065; TCGA CESC -0.478; TCGA COAD -1.005; TCGA ESCA -0.967; TCGA KIRC -0.372; TCGA LUAD -0.302; TCGA LUSC -0.934; TCGA STAD -0.698 |
hsa-miR-98-5p | SLC25A12 | 9 cancers: BLCA; BRCA; ESCA; KIRC; KIRP; LIHC; PRAD; SARC; STAD | MirTarget | TCGA BLCA -0.352; TCGA BRCA -0.178; TCGA ESCA -0.191; TCGA KIRC -0.186; TCGA KIRP -0.209; TCGA LIHC -0.245; TCGA PRAD -0.138; TCGA SARC -0.355; TCGA STAD -0.413 |
hsa-miR-98-5p | IGF1R | 10 cancers: BLCA; BRCA; COAD; ESCA; LGG; OV; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.31; TCGA BRCA -0.354; TCGA COAD -0.138; TCGA ESCA -0.44; TCGA LGG -0.151; TCGA OV -0.11; TCGA PRAD -0.528; TCGA SARC -0.467; TCGA STAD -0.554; TCGA UCEC -0.17 |
hsa-miR-98-5p | CD59 | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; KIRP; LUSC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.301; TCGA BRCA -0.098; TCGA COAD -0.137; TCGA ESCA -0.342; TCGA HNSC -0.149; TCGA KIRC -0.137; TCGA KIRP -0.177; TCGA LUSC -0.367; TCGA THCA -0.229; TCGA STAD -0.445; TCGA UCEC -0.187 |
hsa-miR-98-5p | PRRX1 | 10 cancers: BLCA; BRCA; COAD; KIRC; LGG; LUSC; PAAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -1.029; TCGA BRCA -0.199; TCGA COAD -0.559; TCGA KIRC -0.355; TCGA LGG -0.342; TCGA LUSC -0.6; TCGA PAAD -0.827; TCGA THCA -0.556; TCGA STAD -0.368; TCGA UCEC -0.906 |
hsa-miR-98-5p | PAPPA | 10 cancers: BLCA; BRCA; COAD; LGG; LUSC; OV; PAAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.581; TCGA BRCA -0.353; TCGA COAD -0.475; TCGA LGG -0.571; TCGA LUSC -0.754; TCGA OV -0.182; TCGA PAAD -0.641; TCGA THCA -1.011; TCGA STAD -0.415; TCGA UCEC -1.003 |
hsa-miR-98-5p | CRY2 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; LGG; LUAD; LUSC; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.237; TCGA BRCA -0.399; TCGA CESC -0.173; TCGA COAD -0.131; TCGA ESCA -0.309; TCGA LGG -0.155; TCGA LUAD -0.081; TCGA LUSC -0.162; TCGA PRAD -0.132; TCGA SARC -0.133; TCGA STAD -0.414; TCGA UCEC -0.227 |
hsa-miR-98-5p | HIF1AN | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; LUAD; PRAD; SARC; THCA; UCEC | MirTarget | TCGA BLCA -0.062; TCGA BRCA -0.138; TCGA CESC -0.074; TCGA COAD -0.074; TCGA ESCA -0.121; TCGA KIRC -0.088; TCGA LUAD -0.071; TCGA PRAD -0.14; TCGA SARC -0.071; TCGA THCA -0.07; TCGA UCEC -0.115 |
hsa-miR-98-5p | RBMS2 | 10 cancers: BLCA; BRCA; COAD; KIRP; LUAD; LUSC; PAAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.329; TCGA BRCA -0.198; TCGA COAD -0.168; TCGA KIRP -0.204; TCGA LUAD -0.15; TCGA LUSC -0.274; TCGA PAAD -0.413; TCGA THCA -0.072; TCGA STAD -0.325; TCGA UCEC -0.268 |
hsa-miR-98-5p | DCN | 10 cancers: BLCA; BRCA; COAD; ESCA; KIRC; LUSC; PAAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -1.025; TCGA BRCA -0.691; TCGA COAD -0.535; TCGA ESCA -0.304; TCGA KIRC -0.927; TCGA LUSC -0.601; TCGA PAAD -0.708; TCGA THCA -1.186; TCGA STAD -0.931; TCGA UCEC -0.857 |
hsa-miR-98-5p | CHRD | 9 cancers: BLCA; BRCA; COAD; ESCA; LIHC; LUSC; PAAD; STAD; UCEC | MirTarget | TCGA BLCA -0.669; TCGA BRCA -0.734; TCGA COAD -0.659; TCGA ESCA -0.34; TCGA LIHC -0.273; TCGA LUSC -0.584; TCGA PAAD -0.49; TCGA STAD -0.6; TCGA UCEC -0.55 |
hsa-miR-98-5p | TSPAN18 | 10 cancers: BLCA; BRCA; COAD; LUAD; LUSC; OV; PAAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.672; TCGA BRCA -0.147; TCGA COAD -0.428; TCGA LUAD -0.278; TCGA LUSC -0.281; TCGA OV -0.212; TCGA PAAD -0.414; TCGA SARC -0.279; TCGA STAD -0.462; TCGA UCEC -0.553 |
hsa-miR-98-5p | KLF9 | 12 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRP; LIHC; LUSC; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BLCA -0.776; TCGA BRCA -0.453; TCGA CESC -0.205; TCGA COAD -0.562; TCGA ESCA -0.459; TCGA KIRP -0.209; TCGA LIHC -0.265; TCGA LUSC -0.48; TCGA PRAD -0.188; TCGA SARC -0.221; TCGA STAD -0.96; TCGA UCEC -0.498 |
hsa-miR-98-5p | ATP8B4 | 9 cancers: BLCA; BRCA; CESC; COAD; LUSC; SARC; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.678; TCGA BRCA -0.203; TCGA CESC -0.199; TCGA COAD -0.237; TCGA LUSC -0.447; TCGA SARC -0.698; TCGA THCA -0.578; TCGA STAD -0.354; TCGA UCEC -0.471 |
hsa-miR-98-5p | BACH1 | 10 cancers: BLCA; BRCA; ESCA; LGG; LUSC; PAAD; PRAD; THCA; STAD; UCEC | MirTarget | TCGA BLCA -0.206; TCGA BRCA -0.117; TCGA ESCA -0.124; TCGA LGG -0.083; TCGA LUSC -0.149; TCGA PAAD -0.509; TCGA PRAD -0.124; TCGA THCA -0.354; TCGA STAD -0.116; TCGA UCEC -0.26 |
hsa-miR-98-5p | ADRB2 | 13 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LIHC; LUAD; LUSC; PRAD; SARC; THCA; STAD | MirTarget | TCGA BLCA -0.571; TCGA BRCA -0.718; TCGA COAD -0.503; TCGA ESCA -0.484; TCGA HNSC -0.425; TCGA KIRC -0.367; TCGA LIHC -0.414; TCGA LUAD -0.17; TCGA LUSC -0.692; TCGA PRAD -0.247; TCGA SARC -0.513; TCGA THCA -0.548; TCGA STAD -1.371 |
hsa-miR-98-5p | EEF2K | 9 cancers: BLCA; BRCA; CESC; LIHC; LUSC; PAAD; PRAD; SARC; THCA | MirTarget | TCGA BLCA -0.099; TCGA BRCA -0.23; TCGA CESC -0.131; TCGA LIHC -0.065; TCGA LUSC -0.123; TCGA PAAD -0.192; TCGA PRAD -0.211; TCGA SARC -0.109; TCGA THCA -0.104 |
hsa-miR-98-5p | N4BP1 | 11 cancers: BLCA; BRCA; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.126; TCGA BRCA -0.058; TCGA COAD -0.136; TCGA ESCA -0.23; TCGA HNSC -0.148; TCGA KIRC -0.07; TCGA LUAD -0.101; TCGA LUSC -0.213; TCGA PRAD -0.148; TCGA STAD -0.173; TCGA UCEC -0.194 |
hsa-miR-98-5p | KIAA0513 | 9 cancers: BLCA; COAD; ESCA; HNSC; KIRC; LUAD; LUSC; PRAD; STAD | mirMAP | TCGA BLCA -0.481; TCGA COAD -0.561; TCGA ESCA -0.546; TCGA HNSC -0.158; TCGA KIRC -0.193; TCGA LUAD -0.116; TCGA LUSC -0.219; TCGA PRAD -0.161; TCGA STAD -0.485 |
hsa-miR-98-5p | NTRK3 | 10 cancers: BLCA; BRCA; COAD; LGG; LUAD; LUSC; OV; PRAD; SARC; STAD | mirMAP | TCGA BLCA -0.755; TCGA BRCA -0.377; TCGA COAD -0.917; TCGA LGG -0.168; TCGA LUAD -0.248; TCGA LUSC -0.872; TCGA OV -0.428; TCGA PRAD -0.326; TCGA SARC -0.647; TCGA STAD -1.596 |
hsa-miR-98-5p | GPR107 | 11 cancers: BLCA; BRCA; CESC; COAD; ESCA; KIRC; KIRP; LUSC; PRAD; SARC; UCEC | mirMAP | TCGA BLCA -0.134; TCGA BRCA -0.113; TCGA CESC -0.126; TCGA COAD -0.13; TCGA ESCA -0.127; TCGA KIRC -0.104; TCGA KIRP -0.099; TCGA LUSC -0.105; TCGA PRAD -0.128; TCGA SARC -0.072; TCGA UCEC -0.136 |
hsa-miR-98-5p | KCNA6 | 9 cancers: BLCA; BRCA; COAD; ESCA; LUAD; LUSC; OV; STAD; UCEC | mirMAP | TCGA BLCA -0.702; TCGA BRCA -0.633; TCGA COAD -0.835; TCGA ESCA -0.495; TCGA LUAD -0.176; TCGA LUSC -0.354; TCGA OV -0.269; TCGA STAD -1.337; TCGA UCEC -0.32 |
hsa-miR-98-5p | ATXN1L | 10 cancers: BLCA; BRCA; COAD; ESCA; LUAD; LUSC; PAAD; PRAD; STAD; UCEC | mirMAP | TCGA BLCA -0.096; TCGA BRCA -0.106; TCGA COAD -0.163; TCGA ESCA -0.159; TCGA LUAD -0.059; TCGA LUSC -0.091; TCGA PAAD -0.154; TCGA PRAD -0.172; TCGA STAD -0.145; TCGA UCEC -0.227 |
hsa-miR-98-5p | CLINT1 | 9 cancers: BRCA; CESC; COAD; HNSC; KIRC; LGG; PAAD; PRAD; STAD | miRNAWalker2 validate | TCGA BRCA -0.06; TCGA CESC -0.266; TCGA COAD -0.113; TCGA HNSC -0.128; TCGA KIRC -0.169; TCGA LGG -0.06; TCGA PAAD -0.329; TCGA PRAD -0.11; TCGA STAD -0.13 |
hsa-miR-98-5p | MAST4 | 9 cancers: BRCA; COAD; ESCA; LUAD; LUSC; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BRCA -0.725; TCGA COAD -0.49; TCGA ESCA -0.477; TCGA LUAD -0.13; TCGA LUSC -0.185; TCGA PRAD -0.378; TCGA SARC -0.302; TCGA STAD -0.556; TCGA UCEC -0.195 |
hsa-miR-98-5p | MTUS1 | 10 cancers: BRCA; CESC; COAD; ESCA; KIRC; LIHC; PAAD; PRAD; THCA; UCEC | miRNAWalker2 validate | TCGA BRCA -0.424; TCGA CESC -0.251; TCGA COAD -0.222; TCGA ESCA -0.378; TCGA KIRC -0.232; TCGA LIHC -0.097; TCGA PAAD -0.3; TCGA PRAD -0.254; TCGA THCA -0.114; TCGA UCEC -0.186 |
hsa-miR-98-5p | SEMA3C | 11 cancers: BRCA; CESC; COAD; ESCA; KIRC; LUSC; PAAD; PRAD; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BRCA -0.649; TCGA CESC -0.26; TCGA COAD -0.236; TCGA ESCA -0.5; TCGA KIRC -0.751; TCGA LUSC -0.243; TCGA PAAD -1.184; TCGA PRAD -0.582; TCGA THCA -1.413; TCGA STAD -0.864; TCGA UCEC -0.424 |
hsa-miR-98-5p | VGLL3 | 11 cancers: BRCA; COAD; KIRC; LUAD; LUSC; PAAD; PRAD; SARC; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BRCA -0.422; TCGA COAD -0.708; TCGA KIRC -0.577; TCGA LUAD -0.231; TCGA LUSC -0.828; TCGA PAAD -0.886; TCGA PRAD -0.567; TCGA SARC -0.733; TCGA THCA -0.648; TCGA STAD -0.642; TCGA UCEC -0.906 |
hsa-miR-98-5p | ZFAND5 | 9 cancers: BRCA; ESCA; HNSC; LIHC; LUSC; PRAD; SARC; STAD; UCEC | miRNAWalker2 validate | TCGA BRCA -0.077; TCGA ESCA -0.178; TCGA HNSC -0.071; TCGA LIHC -0.122; TCGA LUSC -0.105; TCGA PRAD -0.105; TCGA SARC -0.089; TCGA STAD -0.261; TCGA UCEC -0.199 |
hsa-miR-98-5p | DZIP1L | 9 cancers: BRCA; COAD; KIRP; LGG; LUAD; LUSC; PAAD; SARC; STAD | MirTarget | TCGA BRCA -0.211; TCGA COAD -0.438; TCGA KIRP -0.269; TCGA LGG -0.204; TCGA LUAD -0.236; TCGA LUSC -0.388; TCGA PAAD -0.563; TCGA SARC -0.479; TCGA STAD -0.278 |
hsa-miR-98-5p | AMT | 11 cancers: BRCA; CESC; HNSC; KIRC; KIRP; LIHC; LUAD; PRAD; SARC; STAD; UCEC | MirTarget | TCGA BRCA -0.278; TCGA CESC -0.448; TCGA HNSC -0.421; TCGA KIRC -0.349; TCGA KIRP -0.543; TCGA LIHC -0.217; TCGA LUAD -0.156; TCGA PRAD -0.232; TCGA SARC -0.398; TCGA STAD -0.324; TCGA UCEC -0.328 |
hsa-miR-98-5p | SESTD1 | 9 cancers: BRCA; COAD; ESCA; KIRP; OV; PAAD; SARC; THCA; STAD | MirTarget | TCGA BRCA -0.103; TCGA COAD -0.171; TCGA ESCA -0.236; TCGA KIRP -0.359; TCGA OV -0.196; TCGA PAAD -0.213; TCGA SARC -0.233; TCGA THCA -0.445; TCGA STAD -0.243 |
hsa-miR-98-5p | GCNT4 | 9 cancers: BRCA; COAD; ESCA; HNSC; LUSC; SARC; THCA; STAD; UCEC | MirTarget | TCGA BRCA -0.609; TCGA COAD -0.522; TCGA ESCA -0.883; TCGA HNSC -0.27; TCGA LUSC -0.246; TCGA SARC -0.966; TCGA THCA -0.608; TCGA STAD -1.193; TCGA UCEC -0.365 |
hsa-miR-98-5p | FZD4 | 10 cancers: BRCA; COAD; ESCA; KIRP; LIHC; LUAD; LUSC; PRAD; STAD; UCEC | MirTarget | TCGA BRCA -0.528; TCGA COAD -0.369; TCGA ESCA -0.284; TCGA KIRP -0.205; TCGA LIHC -0.104; TCGA LUAD -0.192; TCGA LUSC -0.243; TCGA PRAD -0.175; TCGA STAD -0.396; TCGA UCEC -0.344 |
hsa-miR-98-5p | TRIOBP | 14 cancers: BRCA; CESC; COAD; ESCA; HNSC; KIRP; LGG; LUAD; PAAD; PRAD; SARC; THCA; STAD; UCEC | mirMAP | TCGA BRCA -0.134; TCGA CESC -0.185; TCGA COAD -0.158; TCGA ESCA -0.228; TCGA HNSC -0.132; TCGA KIRP -0.37; TCGA LGG -0.091; TCGA LUAD -0.168; TCGA PAAD -0.272; TCGA PRAD -0.077; TCGA SARC -0.161; TCGA THCA -0.175; TCGA STAD -0.198; TCGA UCEC -0.235 |
hsa-miR-98-5p | TRIM66 | 9 cancers: BRCA; CESC; HNSC; KIRC; KIRP; LIHC; LUAD; SARC; UCEC | mirMAP | TCGA BRCA -0.317; TCGA CESC -0.243; TCGA HNSC -0.25; TCGA KIRC -0.166; TCGA KIRP -0.369; TCGA LIHC -0.151; TCGA LUAD -0.196; TCGA SARC -0.333; TCGA UCEC -0.101 |
hsa-miR-98-5p | SREBF2 | 9 cancers: CESC; ESCA; KIRC; LGG; LUAD; PRAD; SARC; THCA; UCEC | MirTarget | TCGA CESC -0.126; TCGA ESCA -0.156; TCGA KIRC -0.311; TCGA LGG -0.076; TCGA LUAD -0.122; TCGA PRAD -0.34; TCGA SARC -0.261; TCGA THCA -0.163; TCGA UCEC -0.164 |
Enriched cancer pathways of putative targets