microRNA information: hsa-miR-99b-3p
| Section | ID | link |
|---|---|---|
| Mature name: | hsa-miR-99b-3p | miRbase |
| Accession: | MIMAT0004678 | miRbase |
| Precursor name: | hsa-mir-99b | miRbase |
| Precursor accession: | MI0000746 | miRbase |
| Symbol: | MIR99B | HGNC |
| RefSeq ID: | NR_029843 | GenBank |
| Sequence: | CAAGCUCGUGUCUGUGGGUCCG |
Reported expression in cancers: hsa-miR-99b-3p
| miRNA | cancer | regulation | reporting | PUBMED | method |
|---|---|---|---|---|---|
| hsa-miR-99b-3p | esophageal cancer | deregulation | "The expression profiles of miRNAs in paired EC and ......" | 23761828 | Microarray; qPCR |
| hsa-miR-99b-3p | lung squamous cell cancer | downregulation | "microRNA 99b acts as a tumor suppressor in non sma ......" | 22969861 | Microarray |
| hsa-miR-99b-3p | prostate cancer | downregulation | "PCR array was performed in FFPE PCa tissues 5 Cauc ......" | 24167554 | qPCR |
| hsa-miR-99b-3p | sarcoma | upregulation | "There were 21 significantly up-regulated miRNAs in ......" | 21213367 | |
| hsa-miR-99b-3p | thyroid cancer | upregulation | "The aim of this study was to assess miRNA expressi ......" | 23427895 | Microarray |
Reported cancer pathway affected by hsa-miR-99b-3p
| miRNA | cancer | pathway | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-99b-3p | head and neck cancer | mTOR signaling pathway | "Selected microRNAs including members of miR-99 fam ......" | 22425712 | |
| hsa-miR-99b-3p | melanoma | Epithelial mesenchymal transition pathway | "An exploratory miRNA analysis of 666 miRs by low d ......" | 20302635 |
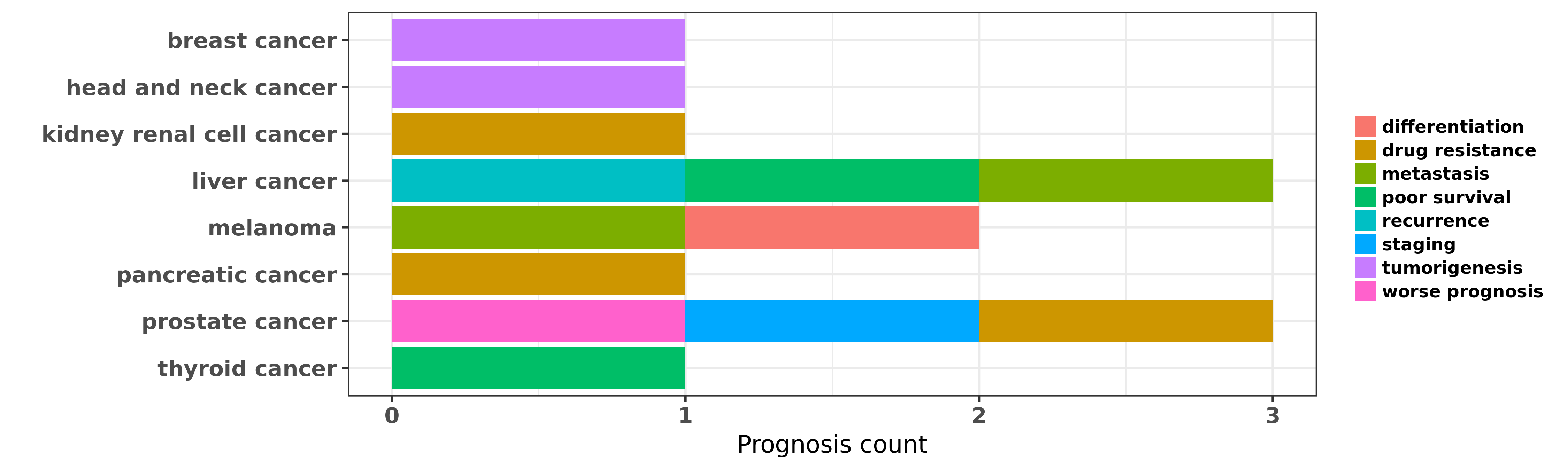
Reported cancer prognosis affected by hsa-miR-99b-3p
| miRNA | cancer | prognosis | reporting | PUBMED | functional study |
|---|---|---|---|---|---|
| hsa-miR-99b-3p | breast cancer | tumorigenesis | "Microarray-based techniques are being useful to ob ......" | 22167321 | |
| hsa-miR-99b-3p | head and neck cancer | tumorigenesis | "Selected microRNAs including members of miR-99 fam ......" | 22425712 | |
| hsa-miR-99b-3p | kidney renal cell cancer | drug resistance | "MiR 99b 5p expression and response to tyrosine kin ......" | 27738339 | |
| hsa-miR-99b-3p | liver cancer | metastasis; poor survival; recurrence | "miR 99b promotes metastasis of hepatocellular carc ......" | 26134929 | Luciferase |
| hsa-miR-99b-3p | melanoma | metastasis; differentiation | "An exploratory miRNA analysis of 666 miRs by low d ......" | 20302635 | |
| hsa-miR-99b-3p | pancreatic cancer | drug resistance | "miR 99b targeted mTOR induction contributes to irr ......" | 23886294 | Western blot |
| hsa-miR-99b-3p | prostate cancer | staging; worse prognosis | "miR 99 family of MicroRNAs suppresses the expressi ......" | 21212412 | |
| hsa-miR-99b-3p | prostate cancer | drug resistance | "The miR-99 family has been shown to play an import ......" | 27340920 | |
| hsa-miR-99b-3p | thyroid cancer | poor survival | "The aim of this study was to assess miRNA expressi ......" | 23427895 |
Reported gene related to hsa-miR-99b-3p
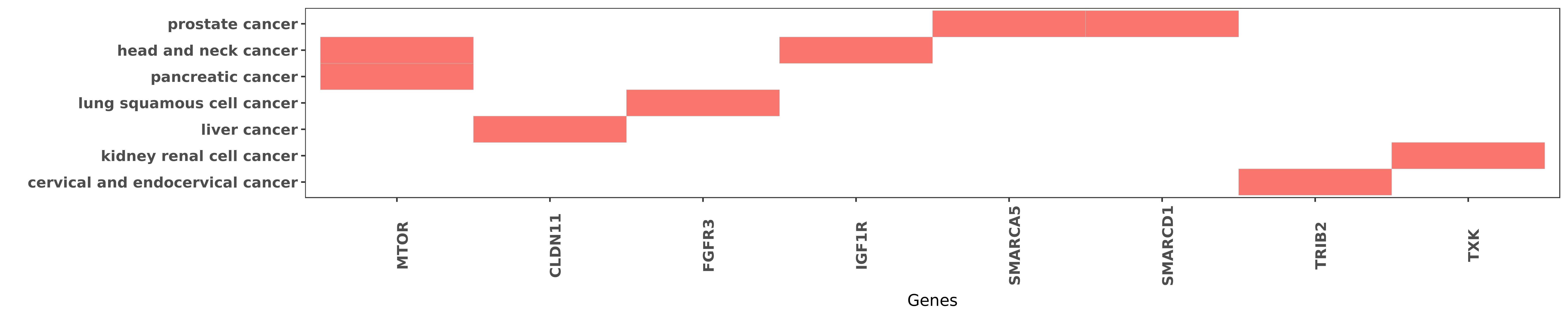
| miRNA | cancer | gene | reporting | PUBMED |
|---|---|---|---|---|
| hsa-miR-99b-3p | head and neck cancer | MTOR | "Furthermore ectopic transfection of miR-99 family ......" | 22425712 |
| hsa-miR-99b-3p | pancreatic cancer | MTOR | "miR 99b targeted mTOR induction contributes to irr ......" | 23886294 |
| hsa-miR-99b-3p | prostate cancer | SMARCA5 | "The miR-99 family has been shown to play an import ......" | 27340920 |
| hsa-miR-99b-3p | prostate cancer | SMARCA5 | "We determined that PSA is posttranscriptionally re ......" | 21212412 |
| hsa-miR-99b-3p | liver cancer | CLDN11 | "Furthermore using a dual‑luciferase assay we dem ......" | 26134929 |
| hsa-miR-99b-3p | lung squamous cell cancer | FGFR3 | "miR-99b was downregulated and FGFR3 was upregulate ......" | 22969861 |
| hsa-miR-99b-3p | head and neck cancer | IGF1R | "Furthermore ectopic transfection of miR-99 family ......" | 22425712 |
| hsa-miR-99b-3p | prostate cancer | SMARCD1 | "The miR-99 family has been shown to play an import ......" | 27340920 |
| hsa-miR-99b-3p | cervical and endocervical cancer | TRIB2 | "miR 99 inhibits cervical carcinoma cell proliferat ......" | 24137458 |
| hsa-miR-99b-3p | kidney renal cell cancer | TXK | "MiR 99b 5p expression and response to tyrosine kin ......" | 27738339 |
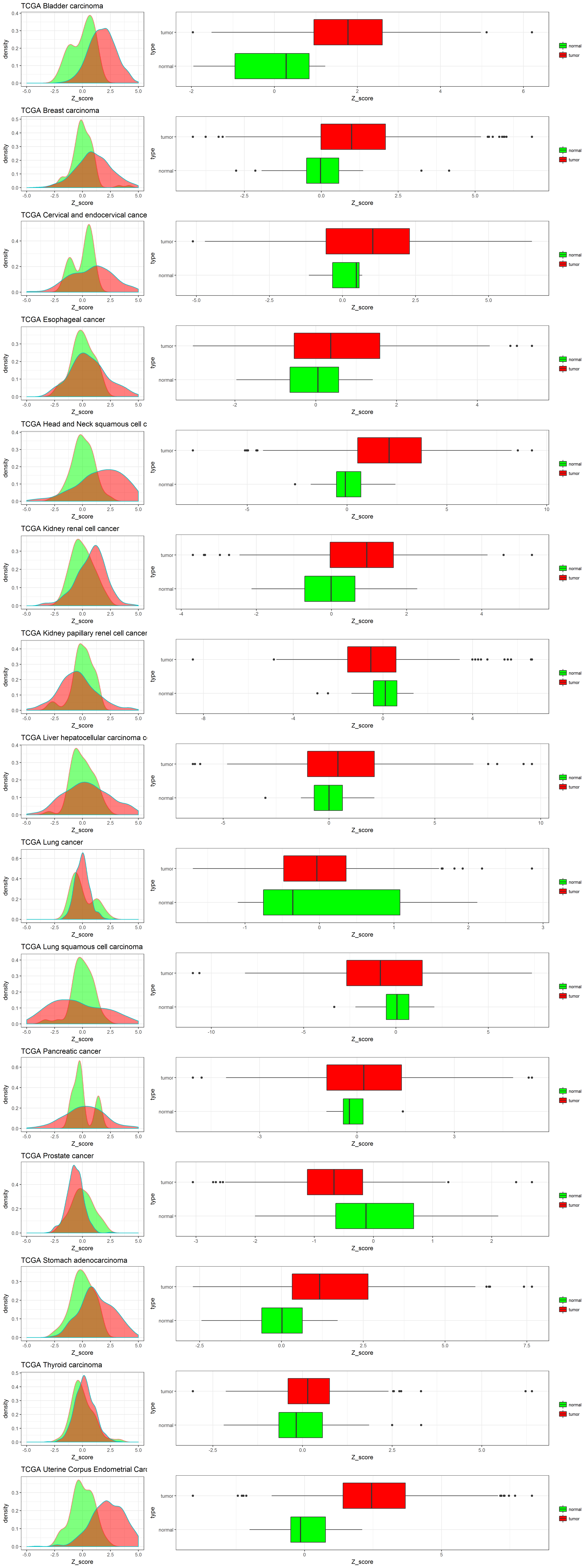
Expression profile in cancer corhorts:
Putative target regulations
| miRNA | Gene | Cancer | Interaction | Correlation beta |
|---|---|---|---|---|
hsa-miR-99b-3p | EEF2 | 16 cancers: BLCA; BRCA; CESC; COAD; ESCA; HNSC; LGG; LIHC; LUAD; LUSC; OV; PAAD; PRAD; THCA; STAD; UCEC | miRNAWalker2 validate | TCGA BLCA -0.154; TCGA BRCA -0.145; TCGA CESC -0.127; TCGA COAD -0.096; TCGA ESCA -0.125; TCGA HNSC -0.097; TCGA LGG -0.238; TCGA LIHC -0.1; TCGA LUAD -0.121; TCGA LUSC -0.132; TCGA OV -0.209; TCGA PAAD -0.15; TCGA PRAD -0.165; TCGA THCA -0.16; TCGA STAD -0.227; TCGA UCEC -0.196 |
hsa-miR-99b-3p | RPL27A | 12 cancers: BLCA; BRCA; HNSC; KIRP; LGG; LUAD; LUSC; OV; PAAD; PRAD; THCA; STAD | miRNAWalker2 validate | TCGA BLCA -0.07; TCGA BRCA -0.054; TCGA HNSC -0.192; TCGA KIRP -0.134; TCGA LGG -0.213; TCGA LUAD -0.078; TCGA LUSC -0.126; TCGA OV -0.165; TCGA PAAD -0.343; TCGA PRAD -0.329; TCGA THCA -0.084; TCGA STAD -0.149 |
hsa-miR-99b-3p | SYK | 11 cancers: BLCA; CESC; COAD; ESCA; HNSC; KIRC; KIRP; LUAD; LUSC; PAAD; THCA | miRNAWalker2 validate | TCGA BLCA -0.227; TCGA CESC -0.304; TCGA COAD -0.12; TCGA ESCA -0.304; TCGA HNSC -0.27; TCGA KIRC -0.244; TCGA KIRP -0.223; TCGA LUAD -0.106; TCGA LUSC -0.166; TCGA PAAD -0.395; TCGA THCA -0.097 |
hsa-miR-99b-3p | GIMAP1 | 9 cancers: BLCA; BRCA; CESC; HNSC; KIRP; LUAD; SARC; THCA; UCEC | MirTarget | TCGA BLCA -0.382; TCGA BRCA -0.366; TCGA CESC -0.244; TCGA HNSC -0.18; TCGA KIRP -0.207; TCGA LUAD -0.174; TCGA SARC -0.239; TCGA THCA -0.129; TCGA UCEC -0.333 |
hsa-miR-99b-3p | CBX7 | 14 cancers: BLCA; BRCA; CESC; ESCA; HNSC; KIRC; KIRP; LIHC; LUAD; OV; SARC; THCA; STAD; UCEC | MirTarget; miRNATAP | TCGA BLCA -0.328; TCGA BRCA -0.455; TCGA CESC -0.292; TCGA ESCA -0.187; TCGA HNSC -0.263; TCGA KIRC -0.185; TCGA KIRP -0.185; TCGA LIHC -0.201; TCGA LUAD -0.168; TCGA OV -0.3; TCGA SARC -0.716; TCGA THCA -0.085; TCGA STAD -0.36; TCGA UCEC -0.427 |
hsa-miR-99b-3p | STX1B | 9 cancers: BLCA; BRCA; KIRP; LGG; LIHC; LUAD; SARC; THCA; UCEC | mirMAP | TCGA BLCA -0.34; TCGA BRCA -0.345; TCGA KIRP -0.63; TCGA LGG -0.145; TCGA LIHC -0.343; TCGA LUAD -0.336; TCGA SARC -0.24; TCGA THCA -0.312; TCGA UCEC -0.139 |
hsa-miR-99b-3p | AGFG2 | 11 cancers: BRCA; HNSC; KIRC; KIRP; LIHC; OV; PAAD; PRAD; SARC; THCA; UCEC | MirTarget | TCGA BRCA -0.19; TCGA HNSC -0.258; TCGA KIRC -0.184; TCGA KIRP -0.202; TCGA LIHC -0.227; TCGA OV -0.259; TCGA PAAD -0.152; TCGA PRAD -0.119; TCGA SARC -0.095; TCGA THCA -0.07; TCGA UCEC -0.142 |
hsa-miR-99b-3p | TAZ | 11 cancers: BRCA; ESCA; HNSC; KIRC; KIRP; LGG; LUSC; OV; PAAD; PRAD; SARC | mirMAP | TCGA BRCA -0.068; TCGA ESCA -0.089; TCGA HNSC -0.137; TCGA KIRC -0.233; TCGA KIRP -0.262; TCGA LGG -0.137; TCGA LUSC -0.168; TCGA OV -0.101; TCGA PAAD -0.196; TCGA PRAD -0.133; TCGA SARC -0.105 |
hsa-miR-99b-3p | DDA1 | 10 cancers: CESC; ESCA; KIRC; KIRP; LUAD; LUSC; PAAD; SARC; THCA; STAD | mirMAP | TCGA CESC -0.059; TCGA ESCA -0.093; TCGA KIRC -0.109; TCGA KIRP -0.093; TCGA LUAD -0.057; TCGA LUSC -0.142; TCGA PAAD -0.143; TCGA SARC -0.119; TCGA THCA -0.084; TCGA STAD -0.091 |
hsa-miR-99b-3p | USMG5 | 9 cancers: HNSC; KIRC; KIRP; LGG; LIHC; LUSC; PAAD; PRAD; STAD | miRNAWalker2 validate | TCGA HNSC -0.181; TCGA KIRC -0.205; TCGA KIRP -0.217; TCGA LGG -0.059; TCGA LIHC -0.15; TCGA LUSC -0.117; TCGA PAAD -0.189; TCGA PRAD -0.303; TCGA STAD -0.123 |
Enriched cancer pathways of putative targets