This regulatory network was inferred from the input dataset. The miRNAs and mRNAs are
presented as round and rectangle nodes respectively. The numerical value popped up upon mouse over the gene node is the log2 transformed fold-change of the gene expression between the two groups. All of the nodes are clickable, and the detailed information of the miRNAs/mRNAs and related cancer pathway will be displayed in another window. The edges between nodes are supported by both interactions (predicted or experimentally verified) and correlations learnt from cancer dataset. The numerical value popped up upon mouse over the edge is the correlation beat value (effect size) between the two nodes. The experimental evidences of the edges reported in previous cancer studies are highlighted by red/orange color. All of these information can be accessed by the "mouse-over" action. This network shows a full map of the miRNA-mRNA regulation of the input gene list(s), and the hub miRNAs (with the high network degree/betweenness centrality) would be the potential cancer drivers or tumor suppressors. The full result table can be accessed in the "Regulations" tab.
"miRNACancerMAP" is also a network visualization tool for users to draw their regulatory network by personal customization. Users can set the complexity of the network by limiting the number of nodes or edges. And the color of the nodes can be defined by different categories of the mRNAs and miRNAs, such as Gene-Ontology, pathway, and expression status. Users can also select to use network degree or network betweenness centrality to define the node size. And edges can be black or colored by the correlation. Purple edge means negative correlation (mostly found between miRNA and mRNA), and blue edge means positive correlation (found in PPI or miRNA-miRNA sponge effect). We can also add the protein-protein interactions (PPI) into the network. This result will show the cluster of genes regulated by some specific miRNAs. Additionally, miRNA-miRNA edges can be added by the "miRNA sponge" button, presenting some clusters of miRNAs that have the interactions via sponge effect.
miRNA-gene regulations
| Num | microRNA | Gene | miRNA log2FC | miRNA pvalue | Gene log2FC | Gene pvalue | Interaction | Correlation beta | Correlation P-value | PMID | Reported in cancer studies |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | hsa-miR-200c-3p | ABL2 | 3.5 | 0 | -0.32 | 0.04554 | MirTarget | -0.15 | 0 | NA | |
| 2 | hsa-miR-200c-3p | ACTC1 | 3.5 | 0 | -7.19 | 0 | MirTarget | -1.34 | 0 | NA | |
| 3 | hsa-miR-200c-3p | ADAMTS3 | 3.5 | 0 | -1.18 | 0.00448 | miRNATAP | -0.41 | 0 | NA | |
| 4 | hsa-miR-200c-3p | ADCY2 | 3.5 | 0 | -3.87 | 0 | MirTarget; miRNATAP | -0.44 | 0 | NA | |
| 5 | hsa-miR-200c-3p | AFF3 | 3.5 | 0 | -5 | 0 | MirTarget; miRNATAP | -0.59 | 0 | NA | |
| 6 | hsa-miR-200c-3p | AMOTL2 | 3.5 | 0 | -1.01 | 0.00015 | MirTarget; miRNATAP | -0.22 | 0 | NA | |
| 7 | hsa-miR-200c-3p | ANKH | 3.5 | 0 | -0.38 | 0.21654 | miRNATAP | -0.29 | 0 | NA | |
| 8 | hsa-miR-200c-3p | AP1S2 | 3.5 | 0 | -1.31 | 0 | MirTarget; miRNATAP | -0.38 | 0 | NA | |
| 9 | hsa-miR-200c-3p | ARHGAP20 | 3.5 | 0 | -3.52 | 0 | MirTarget; miRNATAP | -0.51 | 0 | NA | |
| 10 | hsa-miR-200c-3p | ARHGAP28 | 3.5 | 0 | -1.54 | 0.00053 | MirTarget | -0.2 | 1.0E-5 | NA | |
| 11 | hsa-miR-200c-3p | ARHGAP6 | 3.5 | 0 | -3.25 | 0 | MirTarget | -0.12 | 0.0192 | NA | |
| 12 | hsa-miR-200c-3p | ARHGEF17 | 3.5 | 0 | -1.19 | 0 | miRNATAP | -0.24 | 0 | NA | |
| 13 | hsa-miR-200c-3p | ARL10 | 3.5 | 0 | -1.12 | 0.00198 | mirMAP | -0.29 | 0 | NA | |
| 14 | hsa-miR-200c-3p | ASAP1 | 3.5 | 0 | 0.06 | 0.77944 | MirTarget; miRNATAP | -0.16 | 0 | NA | |
| 15 | hsa-miR-200c-3p | ATXN1 | 3.5 | 0 | -1.51 | 0 | MirTarget; miRNATAP | -0.24 | 0 | NA | |
| 16 | hsa-miR-200c-3p | BACH2 | 3.5 | 0 | -1.52 | 0.00134 | mirMAP | -0.54 | 0 | NA | |
| 17 | hsa-miR-200c-3p | BASP1 | 3.5 | 0 | -0.12 | 0.79725 | miRNATAP | -0.19 | 6.0E-5 | NA | |
| 18 | hsa-miR-200c-3p | BCL2 | 3.5 | 0 | -2.02 | 0 | miRNAWalker2 validate; miRTarBase; mirMAP | -0.19 | 0 | NA | |
| 19 | hsa-miR-200c-3p | BEND4 | 3.5 | 0 | -1.97 | 7.0E-5 | mirMAP | -0.32 | 0 | NA | |
| 20 | hsa-miR-200c-3p | BNC2 | 3.5 | 0 | -2.95 | 0 | MirTarget | -0.7 | 0 | NA | |
| 21 | hsa-miR-200c-3p | C11orf87 | 3.5 | 0 | -1.55 | 0.01186 | miRNATAP | -0.34 | 0 | NA | |
| 22 | hsa-miR-200c-3p | C17orf51 | 3.5 | 0 | -0.68 | 0.02254 | mirMAP | -0.28 | 0 | NA | |
| 23 | hsa-miR-200c-3p | CACNA1C | 3.5 | 0 | -2.53 | 0 | MirTarget | -0.44 | 0 | NA | |
| 24 | hsa-miR-200c-3p | CALU | 3.5 | 0 | -0.15 | 0.46318 | MirTarget | -0.23 | 0 | NA | |
| 25 | hsa-miR-200c-3p | CBL | 3.5 | 0 | -0.31 | 0.06484 | MirTarget; mirMAP | -0.11 | 0 | NA | |
| 26 | hsa-miR-200c-3p | CDK6 | 3.5 | 0 | -0.77 | 0.06479 | mirMAP | -0.32 | 0 | NA | |
| 27 | hsa-miR-200c-3p | CELF2 | 3.5 | 0 | -3.05 | 0 | mirMAP; miRNATAP | -0.51 | 0 | NA | |
| 28 | hsa-miR-200c-3p | CFL2 | 3.5 | 0 | -2.62 | 0 | MirTarget; miRNATAP | -0.48 | 0 | 23497265 | We characterized one of the target genes of miR-200c CFL2 and demonstrated that CFL2 is overexpressed in aggressive breast cancer cell lines and can be significantly down-regulated by exogenous miR-200c |
| 29 | hsa-miR-200c-3p | CHRDL1 | 3.5 | 0 | -6.15 | 0 | MirTarget | -0.99 | 0 | NA | |
| 30 | hsa-miR-200c-3p | CHST11 | 3.5 | 0 | 0.54 | 0.2311 | mirMAP | -0.4 | 0 | NA | |
| 31 | hsa-miR-200c-3p | CIITA | 3.5 | 0 | -0.02 | 0.95968 | mirMAP | -0.2 | 2.0E-5 | NA | |
| 32 | hsa-miR-200c-3p | CLIC4 | 3.5 | 0 | -1.72 | 0 | MirTarget; miRNATAP | -0.46 | 0 | NA | |
| 33 | hsa-miR-200c-3p | CLIP2 | 3.5 | 0 | -0.8 | 0.00158 | MirTarget; miRNATAP | -0.18 | 0 | NA | |
| 34 | hsa-miR-200c-3p | CNN3 | 3.5 | 0 | -0.66 | 0.00535 | MirTarget; miRNATAP | -0.2 | 0 | NA | |
| 35 | hsa-miR-200c-3p | CNTFR | 3.5 | 0 | -3.86 | 0 | miRNATAP | -0.44 | 0 | NA | |
| 36 | hsa-miR-200c-3p | CNTN1 | 3.5 | 0 | -4.98 | 0 | MirTarget; miRNATAP | -0.74 | 0 | NA | |
| 37 | hsa-miR-200c-3p | CNTN4 | 3.5 | 0 | -3.06 | 0 | miRNATAP | -0.19 | 5.0E-5 | NA | |
| 38 | hsa-miR-200c-3p | COL4A3 | 3.5 | 0 | -3.14 | 0 | miRNATAP | -0.32 | 0 | NA | |
| 39 | hsa-miR-200c-3p | COPS8 | 3.5 | 0 | -0.27 | 0.01391 | MirTarget | -0.11 | 0 | NA | |
| 40 | hsa-miR-200c-3p | CORO1C | 3.5 | 0 | -1.12 | 0 | MirTarget; miRNATAP | -0.29 | 0 | NA | |
| 41 | hsa-miR-200c-3p | CREB5 | 3.5 | 0 | -2.28 | 0 | miRNATAP | -0.47 | 0 | NA | |
| 42 | hsa-miR-200c-3p | CYP1B1 | 3.5 | 0 | -2.82 | 0 | miRNATAP | -0.55 | 0 | 25860934 | Loss of miR 200c up regulates CYP1B1 and confers docetaxel resistance in renal cell carcinoma; Additionally miR-200c which is significantly down-regulated in RCC regulates CYP1B1 expression and activity; An inverse association was also observed between the expression levels of miR-200c and CYP1B1 protein in RCC tissues; Restoration of docetaxel resistance by exogenous expression of CYP1B1 in miR-200c-over-expressing cells indicates that CYP1B1 is a functional target of miR-200c; These results suggest that CYP1B1 up-regulation mediated by low miR-200c is one of the mechanisms underlying resistance of RCC cells to docetaxel; Therefore expression of CYP1B1 and miR-200c in RCC may be useful as a prediction for docetaxel response |
| 43 | hsa-miR-200c-3p | CYTH3 | 3.5 | 0 | -0.27 | 0.19343 | MirTarget | -0.17 | 0 | NA | |
| 44 | hsa-miR-200c-3p | DACT1 | 3.5 | 0 | -1.78 | 2.0E-5 | miRNATAP | -0.47 | 0 | NA | |
| 45 | hsa-miR-200c-3p | DCBLD2 | 3.5 | 0 | -0.13 | 0.73597 | MirTarget | -0.31 | 0 | NA | |
| 46 | hsa-miR-200c-3p | DDIT4L | 3.5 | 0 | -1.9 | 0.00047 | MirTarget | -0.47 | 0 | NA | |
| 47 | hsa-miR-200c-3p | DENND5A | 3.5 | 0 | -1.19 | 0 | MirTarget | -0.25 | 0 | NA | |
| 48 | hsa-miR-200c-3p | DENND5B | 3.5 | 0 | -1.35 | 0.0001 | MirTarget; miRNATAP | -0.39 | 0 | NA | |
| 49 | hsa-miR-200c-3p | DIAPH2 | 3.5 | 0 | -0.89 | 0.00037 | mirMAP | -0.11 | 4.0E-5 | NA | |
| 50 | hsa-miR-200c-3p | DIXDC1 | 3.5 | 0 | -3.01 | 0 | MirTarget | -0.52 | 0 | NA | |
| 51 | hsa-miR-200c-3p | DLC1 | 3.5 | 0 | -1.8 | 0 | miRNATAP | -0.2 | 0 | NA | |
| 52 | hsa-miR-200c-3p | DMD | 3.5 | 0 | -3.69 | 0 | miRNATAP | -0.53 | 0 | NA | |
| 53 | hsa-miR-200c-3p | DNAJB5 | 3.5 | 0 | -2.65 | 0 | miRNATAP | -0.52 | 0 | NA | |
| 54 | hsa-miR-200c-3p | DOCK4 | 3.5 | 0 | -0.48 | 0.07912 | MirTarget | -0.22 | 0 | NA | |
| 55 | hsa-miR-200c-3p | DRP2 | 3.5 | 0 | -1.35 | 0.00023 | mirMAP | -0.31 | 0 | NA | |
| 56 | hsa-miR-200c-3p | DST | 3.5 | 0 | -1.12 | 9.0E-5 | mirMAP | -0.1 | 0.00054 | NA | |
| 57 | hsa-miR-200c-3p | DTNA | 3.5 | 0 | -4.06 | 0 | MirTarget | -0.65 | 0 | NA | |
| 58 | hsa-miR-200c-3p | DUSP1 | 3.5 | 0 | -3.47 | 0 | MirTarget; miRNATAP | -0.32 | 0 | NA | |
| 59 | hsa-miR-200c-3p | DYNC1I1 | 3.5 | 0 | -2.05 | 0.0001 | MirTarget | -0.2 | 0.00038 | NA | |
| 60 | hsa-miR-200c-3p | DZIP1 | 3.5 | 0 | -1.57 | 2.0E-5 | MirTarget | -0.34 | 0 | NA | |
| 61 | hsa-miR-200c-3p | EDNRA | 3.5 | 0 | -2.19 | 0 | miRNAWalker2 validate; miRNATAP | -0.43 | 0 | NA | |
| 62 | hsa-miR-200c-3p | ELL2 | 3.5 | 0 | -1.08 | 1.0E-5 | MirTarget | -0.13 | 0 | NA | |
| 63 | hsa-miR-200c-3p | ENTPD1 | 3.5 | 0 | -1.12 | 0 | MirTarget; mirMAP | -0.24 | 0 | NA | |
| 64 | hsa-miR-200c-3p | ERC1 | 3.5 | 0 | -1.1 | 0 | mirMAP | -0.17 | 0 | NA | |
| 65 | hsa-miR-200c-3p | ETS1 | 3.5 | 0 | -0.7 | 0.00228 | MirTarget; miRNATAP | -0.13 | 0 | NA | |
| 66 | hsa-miR-200c-3p | ETV5 | 3.5 | 0 | 0.24 | 0.42305 | MirTarget; miRNATAP | -0.2 | 0 | 27276064 | Another study suggested that ERM expression was regulated directly by miR-200c and had a critical role in miR-200c suppressing cell migration |
| 67 | hsa-miR-200c-3p | EXT1 | 3.5 | 0 | 0.13 | 0.57371 | mirMAP | -0.12 | 0 | NA | |
| 68 | hsa-miR-200c-3p | F2RL2 | 3.5 | 0 | -0.07 | 0.88286 | MirTarget | -0.4 | 0 | NA | |
| 69 | hsa-miR-200c-3p | FAT3 | 3.5 | 0 | -2.79 | 0 | MirTarget; miRNATAP | -0.23 | 0.00025 | NA | |
| 70 | hsa-miR-200c-3p | FBLN5 | 3.5 | 0 | -2.76 | 0 | miRNAWalker2 validate; miRTarBase | -0.4 | 0 | NA | |
| 71 | hsa-miR-200c-3p | FGD1 | 3.5 | 0 | -0.05 | 0.85964 | miRNATAP | -0.18 | 0 | NA | |
| 72 | hsa-miR-200c-3p | FGF18 | 3.5 | 0 | -1.42 | 0.00104 | miRNATAP | -0.26 | 0 | NA | |
| 73 | hsa-miR-200c-3p | FGF2 | 3.5 | 0 | -3.46 | 0 | mirMAP | -0.58 | 0 | NA | |
| 74 | hsa-miR-200c-3p | FHL1 | 3.5 | 0 | -4.79 | 0 | MirTarget | -0.78 | 0 | NA | |
| 75 | hsa-miR-200c-3p | FHOD1 | 3.5 | 0 | 0.61 | 0.00071 | miRNAWalker2 validate | -0.11 | 0 | 22144583 | MicroRNA 200c represses migration and invasion of breast cancer cells by targeting actin regulatory proteins FHOD1 and PPM1F; We identified FHOD1 and PPM1F direct regulators of the actin cytoskeleton as novel targets of miR-200c; Remarkably expression levels of FHOD1 and PPM1F were inversely correlated with the level of miR-200c in breast cancer cell lines breast cancer patient samples and 58 cancer cell lines of various origins |
| 76 | hsa-miR-200c-3p | FKBP14 | 3.5 | 0 | 0.03 | 0.86788 | mirMAP | -0.12 | 0 | NA | |
| 77 | hsa-miR-200c-3p | FLI1 | 3.5 | 0 | -1.11 | 7.0E-5 | MirTarget; miRNATAP | -0.32 | 0 | NA | |
| 78 | hsa-miR-200c-3p | FLNA | 3.5 | 0 | -2.63 | 0 | miRNAWalker2 validate | -0.57 | 0 | NA | |
| 79 | hsa-miR-200c-3p | FN1 | 3.5 | 0 | -0.41 | 0.45793 | miRNAWalker2 validate; miRTarBase; MirTarget; miRNATAP | -0.66 | 0 | NA | |
| 80 | hsa-miR-200c-3p | FOXF1 | 3.5 | 0 | -3.6 | 0 | MirTarget | -0.33 | 0 | NA | |
| 81 | hsa-miR-200c-3p | FSTL1 | 3.5 | 0 | -1.2 | 3.0E-5 | MirTarget | -0.36 | 0 | NA | |
| 82 | hsa-miR-200c-3p | FUNDC2 | 3.5 | 0 | -0.54 | 1.0E-5 | mirMAP | -0.12 | 0 | NA | |
| 83 | hsa-miR-200c-3p | FYN | 3.5 | 0 | -1.47 | 0 | miRNATAP | -0.35 | 0 | NA | |
| 84 | hsa-miR-200c-3p | FZD4 | 3.5 | 0 | -0.83 | 0.00068 | mirMAP | -0.11 | 1.0E-5 | NA | |
| 85 | hsa-miR-200c-3p | GATA4 | 3.5 | 0 | 0.21 | 0.73605 | MirTarget; miRNATAP | -0.25 | 0.00011 | NA | |
| 86 | hsa-miR-200c-3p | GEM | 3.5 | 0 | -3.69 | 0 | MirTarget; miRNATAP | -0.6 | 0 | NA | |
| 87 | hsa-miR-200c-3p | GFI1 | 3.5 | 0 | 0.35 | 0.37482 | miRNATAP | -0.14 | 0.00083 | NA | |
| 88 | hsa-miR-200c-3p | GFRA1 | 3.5 | 0 | -5 | 0 | mirMAP | -0.33 | 0 | NA | |
| 89 | hsa-miR-200c-3p | GJA3 | 3.5 | 0 | 0.72 | 0.20501 | mirMAP | -0.16 | 0.00815 | NA | |
| 90 | hsa-miR-200c-3p | GJC1 | 3.5 | 0 | -0.97 | 0.00222 | MirTarget; miRNATAP | -0.31 | 0 | NA | |
| 91 | hsa-miR-200c-3p | GLI3 | 3.5 | 0 | -0.85 | 0.02024 | miRNATAP | -0.22 | 0 | NA | |
| 92 | hsa-miR-200c-3p | GLIS2 | 3.5 | 0 | -0.61 | 0.05279 | miRNATAP | -0.31 | 0 | NA | |
| 93 | hsa-miR-200c-3p | GLRX | 3.5 | 0 | -0.28 | 0.31677 | MirTarget | -0.17 | 0 | NA | |
| 94 | hsa-miR-200c-3p | GNG4 | 3.5 | 0 | 0.22 | 0.74468 | mirMAP | -0.42 | 0 | NA | |
| 95 | hsa-miR-200c-3p | GPM6A | 3.5 | 0 | -4.43 | 0 | miRNATAP | -0.61 | 0 | NA | |
| 96 | hsa-miR-200c-3p | GPR146 | 3.5 | 0 | -1.49 | 0 | miRNATAP | -0.18 | 0 | NA | |
| 97 | hsa-miR-200c-3p | GPRASP2 | 3.5 | 0 | -1.46 | 0 | MirTarget | -0.18 | 0 | NA | |
| 98 | hsa-miR-200c-3p | GREM1 | 3.5 | 0 | 0.09 | 0.91453 | MirTarget | -0.65 | 0 | NA | |
| 99 | hsa-miR-200c-3p | HCFC2 | 3.5 | 0 | -0.81 | 0 | miRNATAP | -0.12 | 0 | NA | |
| 100 | hsa-miR-200c-3p | HDAC4 | 3.5 | 0 | -1.8 | 0 | miRNATAP | -0.26 | 0 | NA | |
| 101 | hsa-miR-200c-3p | HIPK3 | 3.5 | 0 | -1.75 | 0 | MirTarget | -0.18 | 0 | NA | |
| 102 | hsa-miR-200c-3p | HIVEP3 | 3.5 | 0 | 0.03 | 0.92033 | miRNATAP | -0.21 | 0 | NA | |
| 103 | hsa-miR-200c-3p | HLF | 3.5 | 0 | -5.48 | 0 | MirTarget; miRNATAP | -0.6 | 0 | NA | |
| 104 | hsa-miR-200c-3p | HPS5 | 3.5 | 0 | -0.39 | 0.01461 | MirTarget | -0.12 | 0 | NA | |
| 105 | hsa-miR-200c-3p | HS3ST3A1 | 3.5 | 0 | -0.13 | 0.83157 | MirTarget | -0.63 | 0 | NA | |
| 106 | hsa-miR-200c-3p | HUNK | 3.5 | 0 | -2.27 | 1.0E-5 | mirMAP | -0.23 | 2.0E-5 | NA | |
| 107 | hsa-miR-200c-3p | IFIT5 | 3.5 | 0 | -0.14 | 0.54951 | miRNATAP | -0.13 | 0 | NA | |
| 108 | hsa-miR-200c-3p | IGSF10 | 3.5 | 0 | -5.3 | 0 | MirTarget | -0.54 | 0 | NA | |
| 109 | hsa-miR-200c-3p | IL6ST | 3.5 | 0 | -2.1 | 2.0E-5 | mirMAP | -0.35 | 0 | NA | |
| 110 | hsa-miR-200c-3p | ITGA1 | 3.5 | 0 | -1.43 | 0 | MirTarget | -0.34 | 0 | NA | |
| 111 | hsa-miR-200c-3p | ITPR1 | 3.5 | 0 | -2.58 | 0 | miRNATAP | -0.31 | 0 | NA | |
| 112 | hsa-miR-200c-3p | ITPR2 | 3.5 | 0 | -0.18 | 0.43611 | MirTarget | -0.15 | 0 | NA | |
| 113 | hsa-miR-200c-3p | JAZF1 | 3.5 | 0 | -1.92 | 0 | MirTarget; miRNATAP | -0.42 | 0 | NA | |
| 114 | hsa-miR-200c-3p | JUN | 3.5 | 0 | -2.05 | 0 | MirTarget | -0.2 | 0 | NA | |
| 115 | hsa-miR-200c-3p | KCNJ2 | 3.5 | 0 | -0.28 | 0.36165 | miRNATAP | -0.12 | 0.00012 | NA | |
| 116 | hsa-miR-200c-3p | KCNK2 | 3.5 | 0 | -3.96 | 0 | miRNATAP | -0.5 | 0 | NA | |
| 117 | hsa-miR-200c-3p | KCNQ4 | 3.5 | 0 | -2.64 | 0 | miRNATAP | -0.36 | 0 | NA | |
| 118 | hsa-miR-200c-3p | KCTD15 | 3.5 | 0 | -1.07 | 0.00367 | MirTarget; miRNATAP | -0.22 | 0 | NA | |
| 119 | hsa-miR-200c-3p | KDR | 3.5 | 0 | -0.81 | 0.0014 | miRNATAP | -0.1 | 0.00017 | 24205206 | MiR 200c increases the radiosensitivity of non small cell lung cancer cell line A549 by targeting VEGF VEGFR2 pathway; MiR-200c at the nexus of epithelial-mesenchymal transition EMT is predicted to target VEGFR2; The purpose of this study is to test the hypothesis that regulation of VEGFR2 pathway by miR-200c could modulate the radiosensitivity of cancer cells; Bioinformatic analysis luciferase reporter assays and biochemical assays were carried out to validate VEGFR2 as a direct target of miR-200c; We identified VEGFR2 as a novel target of miR-200c |
| 120 | hsa-miR-200c-3p | KIAA1462 | 3.5 | 0 | -1.72 | 0 | MirTarget | -0.39 | 0 | NA | |
| 121 | hsa-miR-200c-3p | KIF13A | 3.5 | 0 | -1.22 | 0 | miRNATAP | -0.12 | 0 | NA | |
| 122 | hsa-miR-200c-3p | KLF10 | 3.5 | 0 | -1.13 | 0 | miRNATAP | -0.18 | 0 | NA | |
| 123 | hsa-miR-200c-3p | KLF11 | 3.5 | 0 | -0.89 | 1.0E-5 | miRNAWalker2 validate | -0.11 | 0 | NA | |
| 124 | hsa-miR-200c-3p | KLF13 | 3.5 | 0 | -1.16 | 1.0E-5 | miRNATAP | -0.22 | 0 | NA | |
| 125 | hsa-miR-200c-3p | KLF4 | 3.5 | 0 | -2.67 | 0 | MirTarget; miRNATAP | -0.11 | 0.00217 | NA | |
| 126 | hsa-miR-200c-3p | KLF9 | 3.5 | 0 | -2.79 | 0 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.4 | 0 | NA | |
| 127 | hsa-miR-200c-3p | KLHDC1 | 3.5 | 0 | -1.74 | 0 | MirTarget | -0.12 | 4.0E-5 | NA | |
| 128 | hsa-miR-200c-3p | LAMC1 | 3.5 | 0 | -0.82 | 0.0001 | MirTarget; miRNATAP | -0.19 | 0 | NA | |
| 129 | hsa-miR-200c-3p | LATS2 | 3.5 | 0 | -1.5 | 0 | miRNATAP | -0.26 | 0 | NA | |
| 130 | hsa-miR-200c-3p | LHFP | 3.5 | 0 | -1.83 | 0 | MirTarget; miRNATAP | -0.36 | 0 | NA | |
| 131 | hsa-miR-200c-3p | LIX1L | 3.5 | 0 | -1.29 | 0 | MirTarget | -0.34 | 0 | NA | |
| 132 | hsa-miR-200c-3p | LPAR1 | 3.5 | 0 | -1.92 | 0 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.18 | 0 | NA | |
| 133 | hsa-miR-200c-3p | LPIN1 | 3.5 | 0 | -0.9 | 0.00013 | MirTarget | -0.17 | 0 | NA | |
| 134 | hsa-miR-200c-3p | MAF | 3.5 | 0 | -1.28 | 0.00016 | miRNATAP | -0.36 | 0 | NA | |
| 135 | hsa-miR-200c-3p | MAP2 | 3.5 | 0 | -0.75 | 0.09708 | MirTarget; miRNATAP | -0.26 | 0 | NA | |
| 136 | hsa-miR-200c-3p | MARCH1 | 3.5 | 0 | 0.25 | 0.34759 | MirTarget | -0.19 | 0 | NA | |
| 137 | hsa-miR-200c-3p | MARCH8 | 3.5 | 0 | -0.88 | 0.00115 | MirTarget | -0.12 | 2.0E-5 | NA | |
| 138 | hsa-miR-200c-3p | MBNL1 | 3.5 | 0 | -1.04 | 0 | miRNATAP | -0.15 | 0 | NA | |
| 139 | hsa-miR-200c-3p | MCFD2 | 3.5 | 0 | -0.27 | 0.08021 | MirTarget | -0.13 | 0 | NA | |
| 140 | hsa-miR-200c-3p | MEF2D | 3.5 | 0 | -1.25 | 0 | mirMAP | -0.14 | 0 | NA | |
| 141 | hsa-miR-200c-3p | MEGF10 | 3.5 | 0 | -0.57 | 0.2307 | MirTarget | -0.15 | 0.00326 | NA | |
| 142 | hsa-miR-200c-3p | MEX3B | 3.5 | 0 | 0.13 | 0.71267 | miRNATAP | -0.21 | 0 | NA | |
| 143 | hsa-miR-200c-3p | MFAP5 | 3.5 | 0 | -4.35 | 0 | miRNATAP | -0.9 | 0 | NA | |
| 144 | hsa-miR-200c-3p | MITF | 3.5 | 0 | -2.11 | 0 | miRNATAP | -0.37 | 0 | NA | |
| 145 | hsa-miR-200c-3p | MMD | 3.5 | 0 | -0.05 | 0.87055 | miRNATAP | -0.31 | 0 | NA | |
| 146 | hsa-miR-200c-3p | MMGT1 | 3.5 | 0 | -0.69 | 1.0E-5 | MirTarget | -0.14 | 0 | NA | |
| 147 | hsa-miR-200c-3p | MPDZ | 3.5 | 0 | -1.47 | 0 | MirTarget | -0.21 | 0 | NA | |
| 148 | hsa-miR-200c-3p | MRVI1 | 3.5 | 0 | -3.2 | 0 | MirTarget | -0.43 | 0 | NA | |
| 149 | hsa-miR-200c-3p | MSN | 3.5 | 0 | -0.86 | 0.01366 | miRNAWalker2 validate; miRTarBase; MirTarget | -0.39 | 0 | NA | |
| 150 | hsa-miR-200c-3p | MSRB3 | 3.5 | 0 | -3.32 | 0 | mirMAP | -0.64 | 0 | NA |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | REGULATION OF MULTICELLULAR ORGANISMAL DEVELOPMENT | 78 | 1672 | 1.392e-20 | 6.479e-17 |
| 2 | CELL DEVELOPMENT | 66 | 1426 | 3.927e-17 | 9.135e-14 |
| 3 | REGULATION OF CELL DIFFERENTIATION | 67 | 1492 | 9.884e-17 | 1.533e-13 |
| 4 | REGULATION OF NEURON DIFFERENTIATION | 36 | 554 | 8.468e-14 | 9.85e-11 |
| 5 | REGULATION OF CELL DEVELOPMENT | 44 | 836 | 2.017e-13 | 1.877e-10 |
| 6 | REGULATION OF ANATOMICAL STRUCTURE MORPHOGENESIS | 48 | 1021 | 8.993e-13 | 6.387e-10 |
| 7 | POSITIVE REGULATION OF DEVELOPMENTAL PROCESS | 51 | 1142 | 1.098e-12 | 6.387e-10 |
| 8 | REGULATION OF CELLULAR COMPONENT MOVEMENT | 41 | 771 | 1.055e-12 | 6.387e-10 |
| 9 | TISSUE DEVELOPMENT | 60 | 1518 | 1.475e-12 | 7.628e-10 |
| 10 | REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 40 | 750 | 1.874e-12 | 8.197e-10 |
| 11 | NEUROGENESIS | 57 | 1402 | 1.938e-12 | 8.197e-10 |
| 12 | POSITIVE REGULATION OF CELL DIFFERENTIATION | 41 | 823 | 8.405e-12 | 3.259e-09 |
| 13 | RESPONSE TO WOUNDING | 33 | 563 | 1.615e-11 | 5.781e-09 |
| 14 | MOVEMENT OF CELL OR SUBCELLULAR COMPONENT | 51 | 1275 | 6.298e-11 | 2.093e-08 |
| 15 | NEGATIVE REGULATION OF CELL PROLIFERATION | 34 | 643 | 1.251e-10 | 3.881e-08 |
| 16 | CENTRAL NERVOUS SYSTEM DEVELOPMENT | 40 | 872 | 1.867e-10 | 5.43e-08 |
| 17 | REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 61 | 1784 | 3.683e-10 | 1.008e-07 |
| 18 | REGULATION OF NEURON PROJECTION DEVELOPMENT | 26 | 408 | 4.404e-10 | 1.124e-07 |
| 19 | CELLULAR COMPONENT MORPHOGENESIS | 40 | 900 | 4.732e-10 | 1.124e-07 |
| 20 | CARDIOVASCULAR SYSTEM DEVELOPMENT | 37 | 788 | 5.075e-10 | 1.124e-07 |
| 21 | CIRCULATORY SYSTEM DEVELOPMENT | 37 | 788 | 5.075e-10 | 1.124e-07 |
| 22 | CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 29 | 513 | 6.878e-10 | 1.455e-07 |
| 23 | POSITIVE REGULATION OF GENE EXPRESSION | 59 | 1733 | 9.057e-10 | 1.832e-07 |
| 24 | POSITIVE REGULATION OF NEURON DIFFERENTIATION | 22 | 306 | 1.101e-09 | 2.134e-07 |
| 25 | REGULATION OF CELL PROLIFERATION | 53 | 1496 | 1.975e-09 | 3.675e-07 |
| 26 | POSITIVE REGULATION OF CELL DEVELOPMENT | 27 | 472 | 2.134e-09 | 3.818e-07 |
| 27 | LOCOMOTION | 44 | 1114 | 2.314e-09 | 3.987e-07 |
| 28 | TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 34 | 724 | 2.711e-09 | 4.505e-07 |
| 29 | REGULATION OF CELL PROJECTION ORGANIZATION | 29 | 558 | 4.657e-09 | 7.471e-07 |
| 30 | BIOLOGICAL ADHESION | 41 | 1032 | 7.395e-09 | 1.147e-06 |
| 31 | POSITIVE REGULATION OF MOLECULAR FUNCTION | 58 | 1791 | 8.192e-09 | 1.23e-06 |
| 32 | POSITIVE REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 25 | 437 | 8.634e-09 | 1.255e-06 |
| 33 | CELLULAR RESPONSE TO ORGANIC SUBSTANCE | 59 | 1848 | 9.945e-09 | 1.402e-06 |
| 34 | POSITIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 49 | 1395 | 1.18e-08 | 1.615e-06 |
| 35 | NEGATIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 33 | 740 | 1.661e-08 | 2.209e-06 |
| 36 | FOREBRAIN DEVELOPMENT | 22 | 357 | 1.864e-08 | 2.344e-06 |
| 37 | NEGATIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 39 | 983 | 1.861e-08 | 2.344e-06 |
| 38 | HEAD DEVELOPMENT | 32 | 709 | 2.103e-08 | 2.575e-06 |
| 39 | HEART DEVELOPMENT | 25 | 466 | 3.067e-08 | 3.659e-06 |
| 40 | NEGATIVE REGULATION OF DEVELOPMENTAL PROCESS | 34 | 801 | 3.315e-08 | 3.857e-06 |
| 41 | RESPONSE TO ENDOGENOUS STIMULUS | 49 | 1450 | 3.992e-08 | 4.53e-06 |
| 42 | NEURON PROJECTION DEVELOPMENT | 27 | 545 | 4.385e-08 | 4.858e-06 |
| 43 | CELL JUNCTION ASSEMBLY | 13 | 129 | 6.138e-08 | 6.632e-06 |
| 44 | REGULATION OF CELL DEATH | 49 | 1472 | 6.363e-08 | 6.632e-06 |
| 45 | REGULATION OF CELL ADHESION | 29 | 629 | 6.414e-08 | 6.632e-06 |
| 46 | INTRACELLULAR SIGNAL TRANSDUCTION | 51 | 1572 | 7.392e-08 | 7.477e-06 |
| 47 | POSITIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 38 | 1004 | 9.795e-08 | 9.698e-06 |
| 48 | CELLULAR RESPONSE TO ENDOGENOUS STIMULUS | 38 | 1008 | 1.084e-07 | 1.051e-05 |
| 49 | POSITIVE REGULATION OF CELL PROJECTION ORGANIZATION | 19 | 303 | 1.362e-07 | 1.293e-05 |
| 50 | WOUND HEALING | 24 | 470 | 1.463e-07 | 1.347e-05 |
| 51 | EMBRYO DEVELOPMENT | 35 | 894 | 1.477e-07 | 1.347e-05 |
| 52 | NEURON PROJECTION MORPHOGENESIS | 22 | 402 | 1.509e-07 | 1.351e-05 |
| 53 | MESENCHYME DEVELOPMENT | 15 | 190 | 1.554e-07 | 1.364e-05 |
| 54 | NEGATIVE REGULATION OF NITROGEN COMPOUND METABOLIC PROCESS | 49 | 1517 | 1.593e-07 | 1.373e-05 |
| 55 | EPITHELIUM DEVELOPMENT | 36 | 945 | 1.881e-07 | 1.591e-05 |
| 56 | REGULATION OF CELLULAR RESPONSE TO GROWTH FACTOR STIMULUS | 16 | 229 | 3.194e-07 | 2.654e-05 |
| 57 | EPITHELIAL CELL DIFFERENTIATION | 24 | 495 | 3.76e-07 | 3.054e-05 |
| 58 | POSITIVE REGULATION OF NEURON PROJECTION DEVELOPMENT | 16 | 232 | 3.807e-07 | 3.054e-05 |
| 59 | NEURON DEVELOPMENT | 29 | 687 | 4.074e-07 | 3.107e-05 |
| 60 | POSITIVE REGULATION OF BIOSYNTHETIC PROCESS | 54 | 1805 | 3.951e-07 | 3.107e-05 |
| 61 | POSITIVE REGULATION OF CATALYTIC ACTIVITY | 48 | 1518 | 4.064e-07 | 3.107e-05 |
| 62 | EMBRYONIC MORPHOGENESIS | 25 | 539 | 4.86e-07 | 3.647e-05 |
| 63 | BLOOD VESSEL MORPHOGENESIS | 20 | 364 | 5.289e-07 | 3.898e-05 |
| 64 | VASCULATURE DEVELOPMENT | 23 | 469 | 5.445e-07 | 3.898e-05 |
| 65 | CELL PROJECTION ORGANIZATION | 34 | 902 | 5.403e-07 | 3.898e-05 |
| 66 | CELL MORPHOGENESIS INVOLVED IN NEURON DIFFERENTIATION | 20 | 368 | 6.278e-07 | 4.426e-05 |
| 67 | CELL JUNCTION ORGANIZATION | 14 | 185 | 6.769e-07 | 4.701e-05 |
| 68 | REGULATION OF CELL MORPHOGENESIS | 25 | 552 | 7.525e-07 | 5.149e-05 |
| 69 | REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 50 | 1656 | 8.842e-07 | 5.962e-05 |
| 70 | CELLULAR RESPONSE TO OXYGEN CONTAINING COMPOUND | 31 | 799 | 9.713e-07 | 6.405e-05 |
| 71 | ORGAN MORPHOGENESIS | 32 | 841 | 9.773e-07 | 6.405e-05 |
| 72 | ACTIN FILAMENT BASED PROCESS | 22 | 450 | 1.019e-06 | 6.583e-05 |
| 73 | REGULATION OF PHOSPHORUS METABOLIC PROCESS | 49 | 1618 | 1.064e-06 | 6.781e-05 |
| 74 | CONNECTIVE TISSUE DEVELOPMENT | 14 | 194 | 1.198e-06 | 7.531e-05 |
| 75 | SKELETAL SYSTEM DEVELOPMENT | 22 | 455 | 1.223e-06 | 7.587e-05 |
| 76 | REGULATION OF MUSCLE SYSTEM PROCESS | 14 | 195 | 1.273e-06 | 7.797e-05 |
| 77 | TELENCEPHALON DEVELOPMENT | 15 | 228 | 1.598e-06 | 9.656e-05 |
| 78 | REGULATION OF CELL SUBSTRATE ADHESION | 13 | 173 | 1.84e-06 | 0.0001098 |
| 79 | CARTILAGE DEVELOPMENT | 12 | 147 | 1.934e-06 | 0.0001139 |
| 80 | NEGATIVE REGULATION OF LOCOMOTION | 16 | 263 | 2.005e-06 | 0.0001166 |
| 81 | CELL SUBSTRATE JUNCTION ASSEMBLY | 7 | 41 | 2.07e-06 | 0.0001189 |
| 82 | NEURON DIFFERENTIATION | 32 | 874 | 2.217e-06 | 0.0001253 |
| 83 | REGULATION OF PROTEIN MODIFICATION PROCESS | 50 | 1710 | 2.235e-06 | 0.0001253 |
| 84 | CELL MOTILITY | 31 | 835 | 2.429e-06 | 0.000133 |
| 85 | LOCALIZATION OF CELL | 31 | 835 | 2.429e-06 | 0.000133 |
| 86 | NEGATIVE REGULATION OF CELL COMMUNICATION | 39 | 1192 | 2.546e-06 | 0.0001377 |
| 87 | CELL PART MORPHOGENESIS | 26 | 633 | 2.75e-06 | 0.0001462 |
| 88 | BEHAVIOR | 23 | 516 | 2.765e-06 | 0.0001462 |
| 89 | REGULATION OF CARTILAGE DEVELOPMENT | 8 | 63 | 3.905e-06 | 0.0002042 |
| 90 | HEMOSTASIS | 17 | 311 | 4.095e-06 | 0.0002117 |
| 91 | REGULATION OF CHONDROCYTE DIFFERENTIATION | 7 | 46 | 4.626e-06 | 0.0002366 |
| 92 | MUSCLE SYSTEM PROCESS | 16 | 282 | 4.917e-06 | 0.0002487 |
| 93 | ANATOMICAL STRUCTURE FORMATION INVOLVED IN MORPHOGENESIS | 33 | 957 | 5.463e-06 | 0.0002733 |
| 94 | RESPONSE TO OXYGEN CONTAINING COMPOUND | 42 | 1381 | 6.304e-06 | 0.000312 |
| 95 | TUBE MORPHOGENESIS | 17 | 323 | 6.764e-06 | 0.0003313 |
| 96 | NEGATIVE REGULATION OF RESPONSE TO STIMULUS | 41 | 1360 | 1.015e-05 | 0.0004921 |
| 97 | POSITIVE REGULATION OF PHOSPHATE METABOLIC PROCESS | 34 | 1036 | 1.113e-05 | 0.0005286 |
| 98 | POSITIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS | 34 | 1036 | 1.113e-05 | 0.0005286 |
| 99 | DIVALENT INORGANIC CATION TRANSPORT | 15 | 268 | 1.145e-05 | 0.0005382 |
| 100 | GLIOGENESIS | 12 | 175 | 1.172e-05 | 0.0005391 |
| 101 | REGULATION OF ORGAN GROWTH | 8 | 73 | 1.193e-05 | 0.0005391 |
| 102 | NEURON PROJECTION GUIDANCE | 13 | 205 | 1.182e-05 | 0.0005391 |
| 103 | REGULATION OF DENDRITE DEVELOPMENT | 10 | 120 | 1.164e-05 | 0.0005391 |
| 104 | EXTRACELLULAR STRUCTURE ORGANIZATION | 16 | 304 | 1.263e-05 | 0.0005653 |
| 105 | NEGATIVE REGULATION OF CELL DEATH | 30 | 872 | 1.566e-05 | 0.000694 |
| 106 | NEGATIVE REGULATION OF GENE EXPRESSION | 43 | 1493 | 1.837e-05 | 0.0008065 |
| 107 | TAXIS | 20 | 464 | 2.029e-05 | 0.0008823 |
| 108 | REGULATION OF MUSCLE TISSUE DEVELOPMENT | 9 | 103 | 2.184e-05 | 0.0009325 |
| 109 | RESPONSE TO ABIOTIC STIMULUS | 33 | 1024 | 2.182e-05 | 0.0009325 |
| 110 | REGULATION OF BODY FLUID LEVELS | 21 | 506 | 2.207e-05 | 0.0009334 |
| 111 | REGULATION OF SYSTEM PROCESS | 21 | 507 | 2.272e-05 | 0.0009523 |
| 112 | REGULATION OF EPITHELIAL CELL PROLIFERATION | 15 | 285 | 2.36e-05 | 0.000979 |
| 113 | REGULATION OF GTPASE ACTIVITY | 25 | 673 | 2.378e-05 | 0.000979 |
| 114 | RESPONSE TO HORMONE | 30 | 893 | 2.451e-05 | 0.001 |
| 115 | MUSCLE STRUCTURE DEVELOPMENT | 19 | 432 | 2.486e-05 | 0.001004 |
| 116 | VASCULOGENESIS | 7 | 59 | 2.504e-05 | 0.001004 |
| 117 | CELLULAR RESPONSE TO HORMONE STIMULUS | 22 | 552 | 2.605e-05 | 0.001036 |
| 118 | SECOND MESSENGER MEDIATED SIGNALING | 11 | 160 | 2.646e-05 | 0.001043 |
| 119 | STEM CELL DIFFERENTIATION | 12 | 190 | 2.667e-05 | 0.001043 |
| 120 | REGULATION OF DEVELOPMENTAL GROWTH | 15 | 289 | 2.774e-05 | 0.001076 |
| 121 | OLIGODENDROCYTE DIFFERENTIATION | 7 | 60 | 2.799e-05 | 0.001076 |
| 122 | REGULATION OF HYDROLASE ACTIVITY | 39 | 1327 | 3.019e-05 | 0.001151 |
| 123 | POSITIVE REGULATION OF HYDROLASE ACTIVITY | 30 | 905 | 3.142e-05 | 0.001188 |
| 124 | TISSUE MIGRATION | 8 | 84 | 3.365e-05 | 0.001263 |
| 125 | GLIAL CELL DIFFERENTIATION | 10 | 136 | 3.466e-05 | 0.00129 |
| 126 | HEART PROCESS | 8 | 85 | 3.667e-05 | 0.001354 |
| 127 | REGULATION OF HOMEOSTATIC PROCESS | 19 | 447 | 3.953e-05 | 0.001426 |
| 128 | MORPHOGENESIS OF A BRANCHING STRUCTURE | 11 | 167 | 3.929e-05 | 0.001426 |
| 129 | NEGATIVE REGULATION OF CELL DIFFERENTIATION | 23 | 609 | 3.938e-05 | 0.001426 |
| 130 | PIGMENTATION | 8 | 86 | 3.992e-05 | 0.001429 |
| 131 | POSITIVE REGULATION OF CELLULAR COMPONENT ORGANIZATION | 35 | 1152 | 4.107e-05 | 0.001459 |
| 132 | POSITIVE REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 29 | 876 | 4.386e-05 | 0.001546 |
| 133 | REGULATION OF CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 16 | 337 | 4.427e-05 | 0.001549 |
| 134 | REGULATION OF VASCULATURE DEVELOPMENT | 13 | 233 | 4.54e-05 | 0.001577 |
| 135 | REGULATION OF ION HOMEOSTASIS | 12 | 201 | 4.63e-05 | 0.001596 |
| 136 | TISSUE MORPHOGENESIS | 21 | 533 | 4.699e-05 | 0.001608 |
| 137 | SENSORY ORGAN DEVELOPMENT | 20 | 493 | 4.764e-05 | 0.001618 |
| 138 | POST EMBRYONIC DEVELOPMENT | 8 | 89 | 5.115e-05 | 0.001725 |
| 139 | REGULATION OF CARDIAC MUSCLE CONTRACTION | 7 | 66 | 5.234e-05 | 0.001752 |
| 140 | CELL MIGRATION INVOLVED IN SPROUTING ANGIOGENESIS | 4 | 15 | 5.569e-05 | 0.001838 |
| 141 | REGULATION OF CELL MATRIX ADHESION | 8 | 90 | 5.543e-05 | 0.001838 |
| 142 | SINGLE ORGANISM CELL ADHESION | 19 | 459 | 5.638e-05 | 0.001847 |
| 143 | POSITIVE REGULATION OF LOCOMOTION | 18 | 420 | 5.68e-05 | 0.001848 |
| 144 | POSITIVE REGULATION OF RESPONSE TO STIMULUS | 50 | 1929 | 5.771e-05 | 0.001865 |
| 145 | SYNAPSE ORGANIZATION | 10 | 145 | 5.984e-05 | 0.00192 |
| 146 | REGULATION OF CELL JUNCTION ASSEMBLY | 7 | 68 | 6.354e-05 | 0.002025 |
| 147 | REGULATION OF MUSCLE CONTRACTION | 10 | 147 | 6.717e-05 | 0.002126 |
| 148 | PROTEIN PHOSPHORYLATION | 30 | 944 | 6.782e-05 | 0.002132 |
| 149 | SYSTEM PROCESS | 47 | 1785 | 6.838e-05 | 0.002135 |
| 150 | POSITIVE REGULATION OF SMOOTH MUSCLE CELL MIGRATION | 5 | 30 | 7.107e-05 | 0.002176 |
| 151 | POSITIVE REGULATION OF PROTEIN MODIFICATION PROCESS | 34 | 1135 | 7.03e-05 | 0.002176 |
| 152 | RESPONSE TO MECHANICAL STIMULUS | 12 | 210 | 7.068e-05 | 0.002176 |
| 153 | TUBE DEVELOPMENT | 21 | 552 | 7.731e-05 | 0.002345 |
| 154 | REGULATION OF CHEMOTAXIS | 11 | 180 | 7.761e-05 | 0.002345 |
| 155 | REGULATION OF SMOOTH MUSCLE CELL MIGRATION | 6 | 49 | 8.031e-05 | 0.002411 |
| 156 | POSITIVE REGULATION OF PROTEIN METABOLIC PROCESS | 41 | 1492 | 8.417e-05 | 0.002511 |
| 157 | RESPONSE TO GROWTH FACTOR | 19 | 475 | 8.864e-05 | 0.002627 |
| 158 | REGULATION OF ADHERENS JUNCTION ORGANIZATION | 6 | 50 | 9.015e-05 | 0.002655 |
| 159 | ENZYME LINKED RECEPTOR PROTEIN SIGNALING PATHWAY | 24 | 689 | 9.463e-05 | 0.002759 |
| 160 | REGULATION OF PRI MIRNA TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 4 | 17 | 9.486e-05 | 0.002759 |
| 161 | OSSIFICATION | 13 | 251 | 9.678e-05 | 0.002797 |
| 162 | MORPHOGENESIS OF AN EPITHELIUM | 17 | 400 | 0.0001009 | 0.002898 |
| 163 | RESPONSE TO EXTERNAL STIMULUS | 47 | 1821 | 0.0001103 | 0.003147 |
| 164 | REGULATION OF HEART CONTRACTION | 12 | 221 | 0.0001149 | 0.003259 |
| 165 | RESPONSE TO LIPID | 28 | 888 | 0.0001363 | 0.003844 |
| 166 | REGULATION OF MUSCLE ORGAN DEVELOPMENT | 8 | 103 | 0.0001442 | 0.004017 |
| 167 | BRANCHING MORPHOGENESIS OF AN EPITHELIAL TUBE | 9 | 131 | 0.0001437 | 0.004017 |
| 168 | POSITIVE REGULATION OF CELL COMMUNICATION | 41 | 1532 | 0.0001491 | 0.004129 |
| 169 | CELLULAR RESPONSE TO LIPID | 18 | 457 | 0.0001642 | 0.004521 |
| 170 | REGULATION OF STRIATED MUSCLE CONTRACTION | 7 | 79 | 0.0001655 | 0.00453 |
| 171 | NEGATIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS | 20 | 541 | 0.0001677 | 0.004536 |
| 172 | NEGATIVE REGULATION OF PHOSPHATE METABOLIC PROCESS | 20 | 541 | 0.0001677 | 0.004536 |
| 173 | REGULATION OF EPITHELIAL CELL MIGRATION | 10 | 166 | 0.0001839 | 0.004946 |
| 174 | MELANOCYTE DIFFERENTIATION | 4 | 20 | 0.0001865 | 0.004987 |
| 175 | REGULATION OF SEQUESTERING OF CALCIUM ION | 8 | 107 | 0.0001879 | 0.004995 |
| 176 | REGULATION OF MEMBRANE POTENTIAL | 15 | 343 | 0.000189 | 0.004997 |
| 177 | REGULATION OF GROWTH | 22 | 633 | 0.0001914 | 0.005004 |
| 178 | NEGATIVE REGULATION OF PHOSPHORYLATION | 17 | 422 | 0.000191 | 0.005004 |
| 179 | REGULATION OF PHOSPHATIDYLINOSITOL 3 KINASE SIGNALING | 9 | 138 | 0.000213 | 0.005538 |
| 180 | MUSCLE CELL DIFFERENTIATION | 12 | 237 | 0.0002204 | 0.005696 |
| 181 | REGULATION OF CELL SHAPE | 9 | 139 | 0.0002249 | 0.005781 |
| 182 | REGULATION OF KINASE ACTIVITY | 25 | 776 | 0.0002269 | 0.0058 |
| 183 | RESPONSE TO ORGANIC CYCLIC COMPOUND | 28 | 917 | 0.0002314 | 0.005883 |
| 184 | REGULATION OF ACTIN FILAMENT BASED PROCESS | 14 | 312 | 0.000237 | 0.005994 |
| 185 | RESPONSE TO DRUG | 17 | 431 | 0.0002446 | 0.006152 |
| 186 | MUSCLE ORGAN DEVELOPMENT | 13 | 277 | 0.0002558 | 0.006377 |
| 187 | REGULATION OF PHOSPHOLIPASE C ACTIVITY | 5 | 39 | 0.0002576 | 0.006377 |
| 188 | MEMBRANE DEPOLARIZATION DURING ACTION POTENTIAL | 5 | 39 | 0.0002576 | 0.006377 |
| 189 | PLATELET ACTIVATION | 9 | 142 | 0.0002638 | 0.006462 |
| 190 | REGULATION OF SMALL GTPASE MEDIATED SIGNAL TRANSDUCTION | 13 | 278 | 0.0002649 | 0.006462 |
| 191 | REGULATION OF TRANSMEMBRANE RECEPTOR PROTEIN SERINE THREONINE KINASE SIGNALING PATHWAY | 11 | 207 | 0.0002653 | 0.006462 |
| 192 | REGULATION OF ORGAN MORPHOGENESIS | 12 | 242 | 0.0002669 | 0.006467 |
| 193 | MEMBRANE DEPOLARIZATION | 6 | 61 | 0.0002751 | 0.006631 |
| 194 | DEVELOPMENTAL PIGMENTATION | 5 | 40 | 0.0002909 | 0.006978 |
| 195 | POSITIVE REGULATION OF PHOSPHATIDYLINOSITOL 3 KINASE SIGNALING | 6 | 62 | 0.0003008 | 0.007178 |
| 196 | NEPHRON DEVELOPMENT | 8 | 115 | 0.0003081 | 0.007314 |
| 197 | POSITIVE REGULATION OF KINASE ACTIVITY | 18 | 482 | 0.0003135 | 0.007367 |
| 198 | REGULATION OF NUCLEOTIDE METABOLIC PROCESS | 11 | 211 | 0.0003125 | 0.007367 |
| 199 | REGULATION OF OSSIFICATION | 10 | 178 | 0.0003228 | 0.007547 |
| 200 | REGULATION OF MUSCLE ADAPTATION | 6 | 63 | 0.0003284 | 0.00764 |
| 201 | REGULATION OF MAPK CASCADE | 22 | 660 | 0.0003408 | 0.007889 |
| 202 | POSITIVE REGULATION OF DENDRITE DEVELOPMENT | 6 | 64 | 0.0003579 | 0.008245 |
| 203 | REGULATION OF SEQUENCE SPECIFIC DNA BINDING TRANSCRIPTION FACTOR ACTIVITY | 15 | 365 | 0.0003667 | 0.008375 |
| 204 | REGULATION OF HEART GROWTH | 5 | 42 | 0.0003672 | 0.008375 |
| 205 | EYE DEVELOPMENT | 14 | 326 | 0.0003696 | 0.008388 |
| 206 | COGNITION | 12 | 251 | 0.0003717 | 0.008396 |
| 207 | CIRCULATORY SYSTEM PROCESS | 15 | 366 | 0.0003774 | 0.008484 |
| 208 | BLOOD VESSEL ENDOTHELIAL CELL MIGRATION | 4 | 24 | 0.0003904 | 0.008692 |
| 209 | NEGATIVE REGULATION OF PEPTIDYL SERINE PHOSPHORYLATION | 4 | 24 | 0.0003904 | 0.008692 |
| 210 | SOMATIC STEM CELL POPULATION MAINTENANCE | 6 | 66 | 0.0004232 | 0.009378 |
| 211 | REGULATION OF MUSCLE CELL DIFFERENTIATION | 9 | 152 | 0.0004359 | 0.009608 |
| 212 | ANGIOGENESIS | 13 | 293 | 0.0004378 | 0.009608 |
| 213 | ACTIN FILAMENT BASED MOVEMENT | 7 | 93 | 0.0004541 | 0.009919 |
| 214 | PALLIUM DEVELOPMENT | 9 | 153 | 0.0004572 | 0.009942 |
| 215 | POSITIVE REGULATION OF CELL PROLIFERATION | 25 | 814 | 0.0004615 | 0.009948 |
| 216 | TRANSMEMBRANE RECEPTOR PROTEIN TYROSINE KINASE SIGNALING PATHWAY | 18 | 498 | 0.0004618 | 0.009948 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | REGULATORY REGION NUCLEIC ACID BINDING | 37 | 818 | 1.412e-09 | 1.312e-06 |
| 2 | RNA POLYMERASE II TRANSCRIPTION FACTOR ACTIVITY SEQUENCE SPECIFIC DNA BINDING | 31 | 629 | 4.661e-09 | 2.165e-06 |
| 3 | NUCLEIC ACID BINDING TRANSCRIPTION FACTOR ACTIVITY | 45 | 1199 | 7.129e-09 | 2.208e-06 |
| 4 | TRANSCRIPTIONAL ACTIVATOR ACTIVITY RNA POLYMERASE II TRANSCRIPTION REGULATORY REGION SEQUENCE SPECIFIC BINDING | 20 | 315 | 5.159e-08 | 1.198e-05 |
| 5 | TRANSCRIPTION FACTOR ACTIVITY RNA POLYMERASE II CORE PROMOTER PROXIMAL REGION SEQUENCE SPECIFIC BINDING | 20 | 328 | 9.998e-08 | 1.858e-05 |
| 6 | SEQUENCE SPECIFIC DNA BINDING | 38 | 1037 | 2.224e-07 | 3.443e-05 |
| 7 | TRANSCRIPTION FACTOR ACTIVITY RNA POLYMERASE II TRANSCRIPTION FACTOR BINDING | 12 | 133 | 6.647e-07 | 8.822e-05 |
| 8 | TRANSCRIPTIONAL ACTIVATOR ACTIVITY RNA POLYMERASE II CORE PROMOTER PROXIMAL REGION SEQUENCE SPECIFIC BINDING | 15 | 226 | 1.431e-06 | 0.0001583 |
| 9 | MACROMOLECULAR COMPLEX BINDING | 44 | 1399 | 1.534e-06 | 0.0001583 |
| 10 | ACTIVATING TRANSCRIPTION FACTOR BINDING | 8 | 57 | 1.799e-06 | 0.0001671 |
| 11 | TRANSCRIPTION FACTOR ACTIVITY PROTEIN BINDING | 24 | 588 | 7.488e-06 | 0.0006324 |
| 12 | MOLECULAR FUNCTION REGULATOR | 40 | 1353 | 2.065e-05 | 0.001599 |
| 13 | CYTOSKELETAL PROTEIN BINDING | 28 | 819 | 3.405e-05 | 0.002433 |
| 14 | CORE PROMOTER PROXIMAL REGION DNA BINDING | 17 | 371 | 4.003e-05 | 0.002656 |
| 15 | METAL ION TRANSMEMBRANE TRANSPORTER ACTIVITY | 18 | 417 | 5.18e-05 | 0.003208 |
| 16 | TRANSCRIPTION FACTOR ACTIVITY RNA POLYMERASE II DISTAL ENHANCER SEQUENCE SPECIFIC BINDING | 8 | 90 | 5.543e-05 | 0.003219 |
| 17 | DOUBLE STRANDED DNA BINDING | 26 | 764 | 7.061e-05 | 0.003859 |
| 18 | ACTIN BINDING | 17 | 393 | 8.145e-05 | 0.004204 |
| 19 | TRANSCRIPTION FACTOR BINDING | 20 | 524 | 0.0001096 | 0.005358 |
| 20 | TRANSCRIPTION COACTIVATOR ACTIVITY | 14 | 296 | 0.0001376 | 0.006391 |
| 21 | RAS GUANYL NUCLEOTIDE EXCHANGE FACTOR ACTIVITY | 12 | 228 | 0.0001539 | 0.006809 |
| 22 | GUANYL NUCLEOTIDE EXCHANGE FACTOR ACTIVITY | 14 | 303 | 0.0001754 | 0.007408 |
| 23 | DIVALENT INORGANIC CATION TRANSMEMBRANE TRANSPORTER ACTIVITY | 10 | 167 | 0.0001931 | 0.0078 |
| 24 | PROTEIN KINASE ACTIVITY | 22 | 640 | 0.0002231 | 0.008637 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | CELL PROJECTION | 60 | 1786 | 1.053e-09 | 6.151e-07 |
| 2 | NEURON PART | 47 | 1265 | 4.311e-09 | 1.135e-06 |
| 3 | NEURON PROJECTION | 39 | 942 | 5.829e-09 | 1.135e-06 |
| 4 | CELL JUNCTION | 42 | 1151 | 5.329e-08 | 7.78e-06 |
| 5 | MEMBRANE REGION | 41 | 1134 | 1.005e-07 | 1.174e-05 |
| 6 | SOMATODENDRITIC COMPARTMENT | 29 | 650 | 1.288e-07 | 1.253e-05 |
| 7 | CELL BODY | 23 | 494 | 1.328e-06 | 0.0001108 |
| 8 | DENDRITE | 21 | 451 | 3.872e-06 | 0.0002826 |
| 9 | MEMBRANE MICRODOMAIN | 16 | 288 | 6.421e-06 | 0.0004167 |
| 10 | SYNAPSE | 28 | 754 | 7.631e-06 | 0.0004457 |
| 11 | ACTIN CYTOSKELETON | 19 | 444 | 3.61e-05 | 0.001916 |
| 12 | CELL LEADING EDGE | 16 | 350 | 6.93e-05 | 0.003373 |
| 13 | CELL SUBSTRATE JUNCTION | 17 | 398 | 9.496e-05 | 0.004266 |
| 14 | RUFFLE | 10 | 156 | 0.0001104 | 0.004604 |
| 15 | PLASMA MEMBRANE PROTEIN COMPLEX | 19 | 510 | 0.000221 | 0.008606 |
Over-represented Pathway
| Num | Pathway | Pathview | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|---|
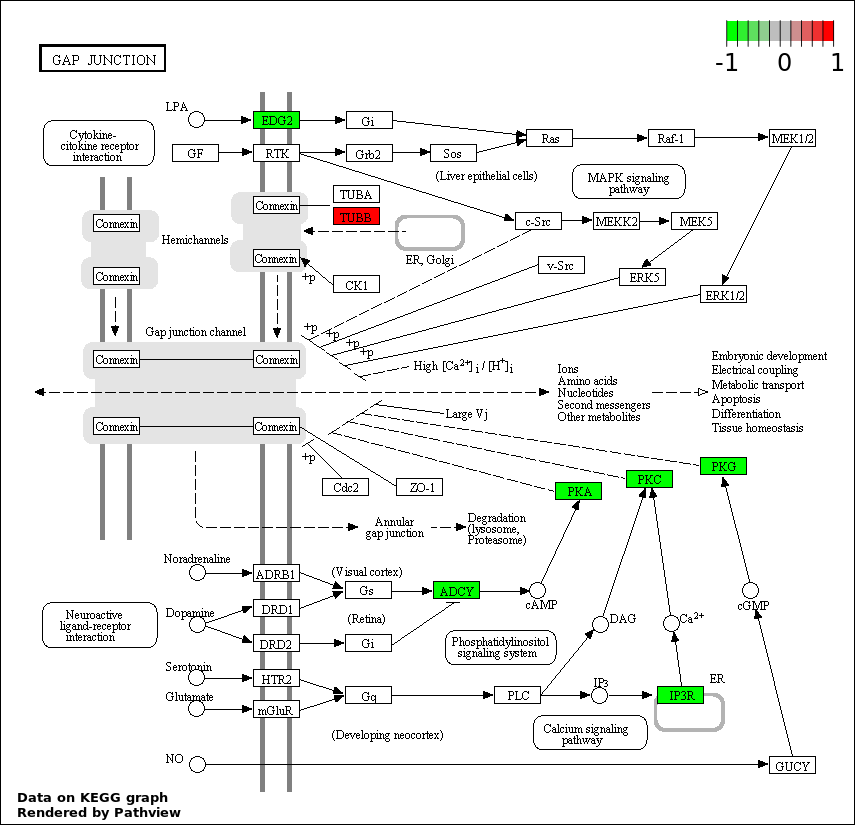
| 1 | hsa04540_Gap_junction | 10 | 90 | 8.46e-07 | 0.0001523 | |
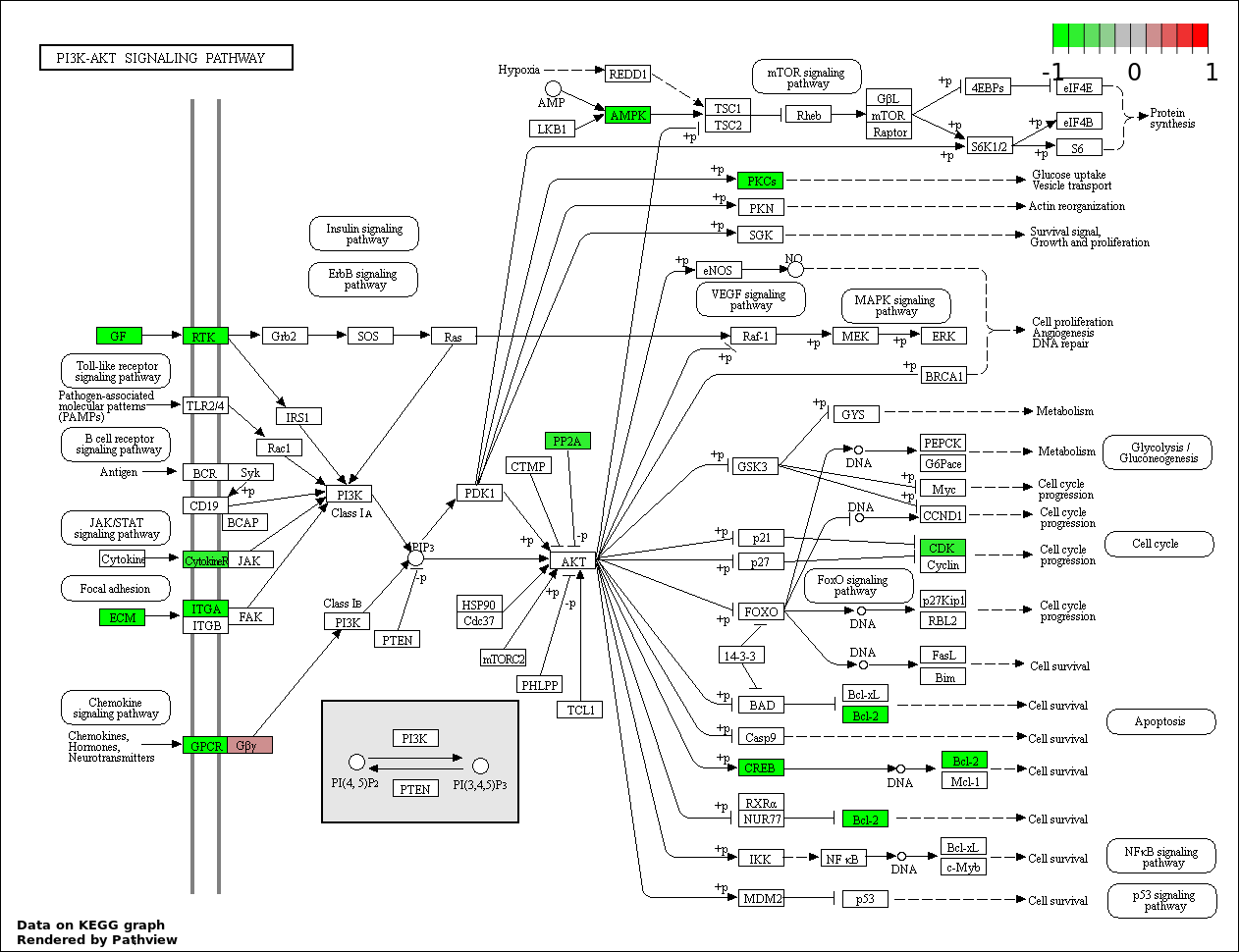
| 2 | hsa04151_PI3K_AKT_signaling_pathway | 18 | 351 | 5.191e-06 | 0.0004081 | |
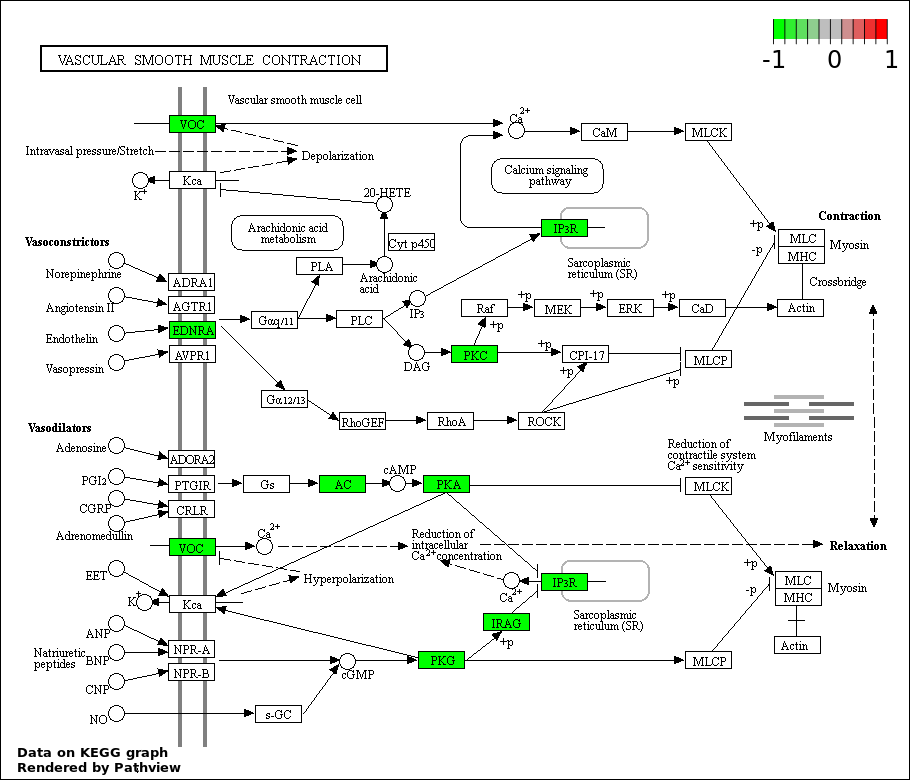
| 3 | hsa04270_Vascular_smooth_muscle_contraction | 10 | 116 | 8.614e-06 | 0.0004081 | |
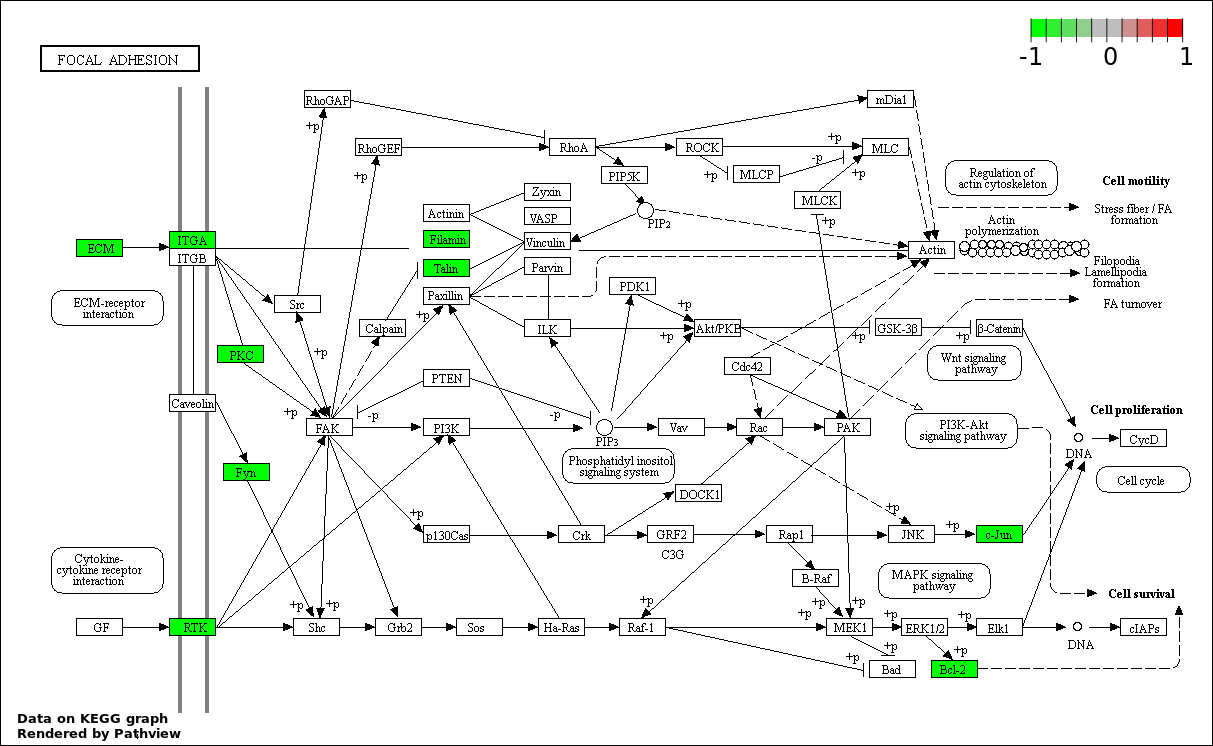
| 4 | hsa04510_Focal_adhesion | 13 | 200 | 9.068e-06 | 0.0004081 | |
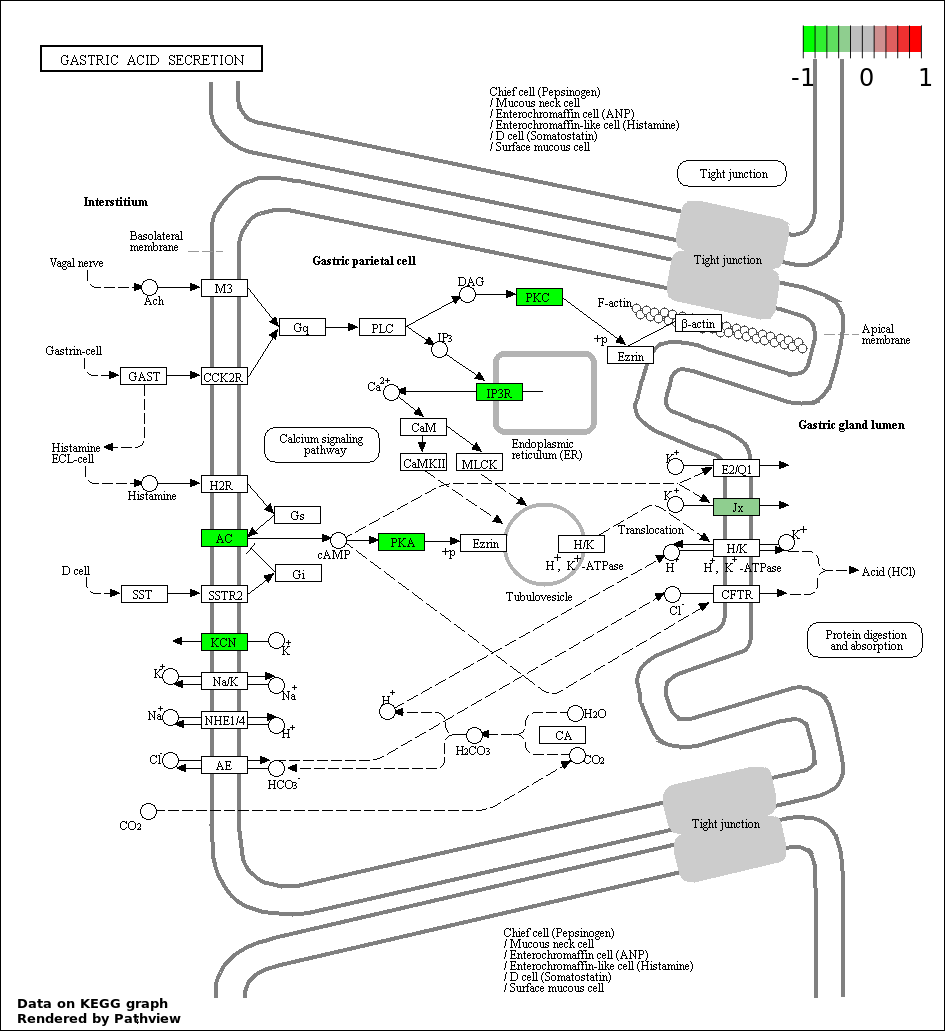
| 5 | hsa04971_Gastric_acid_secretion | 8 | 74 | 1.321e-05 | 0.0004756 | |
| 6 | hsa04720_Long.term_potentiation | 7 | 70 | 7.662e-05 | 0.002299 | |
| 7 | hsa04912_GnRH_signaling_pathway | 8 | 101 | 0.0001257 | 0.003232 | |
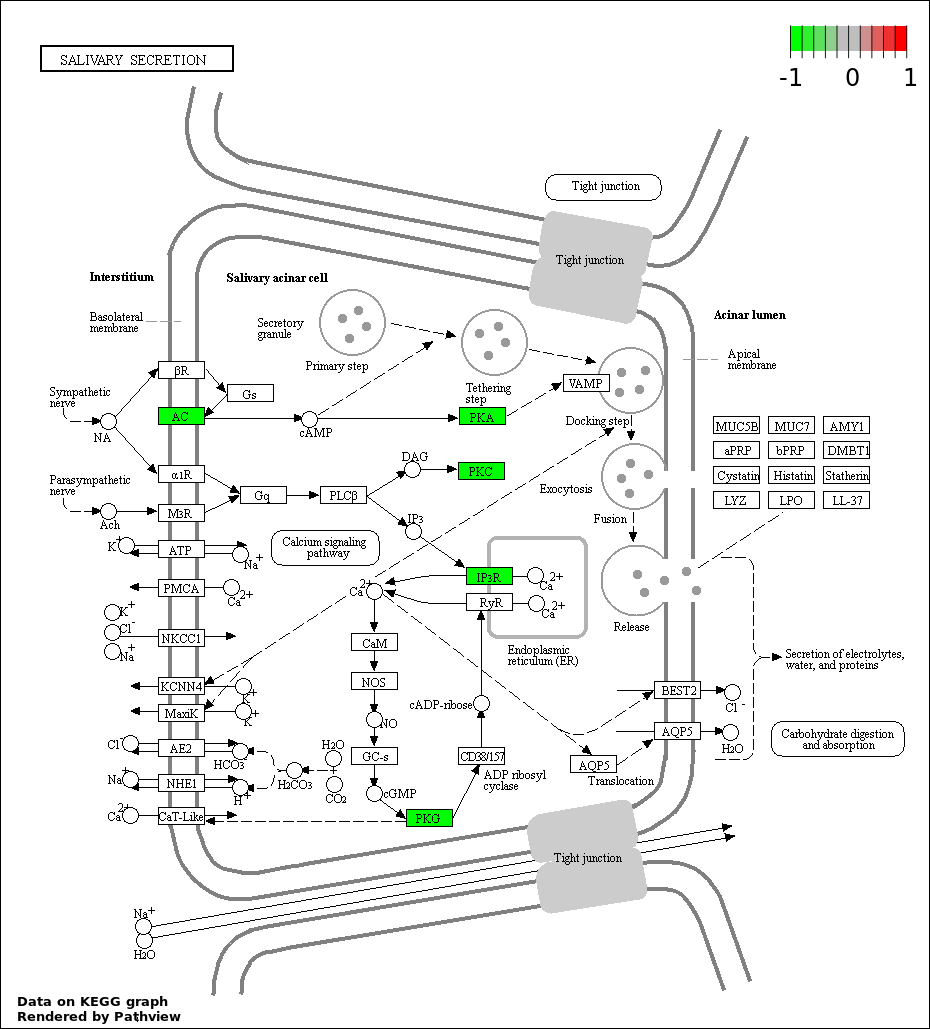
| 8 | hsa04970_Salivary_secretion | 7 | 89 | 0.0003472 | 0.007811 | |
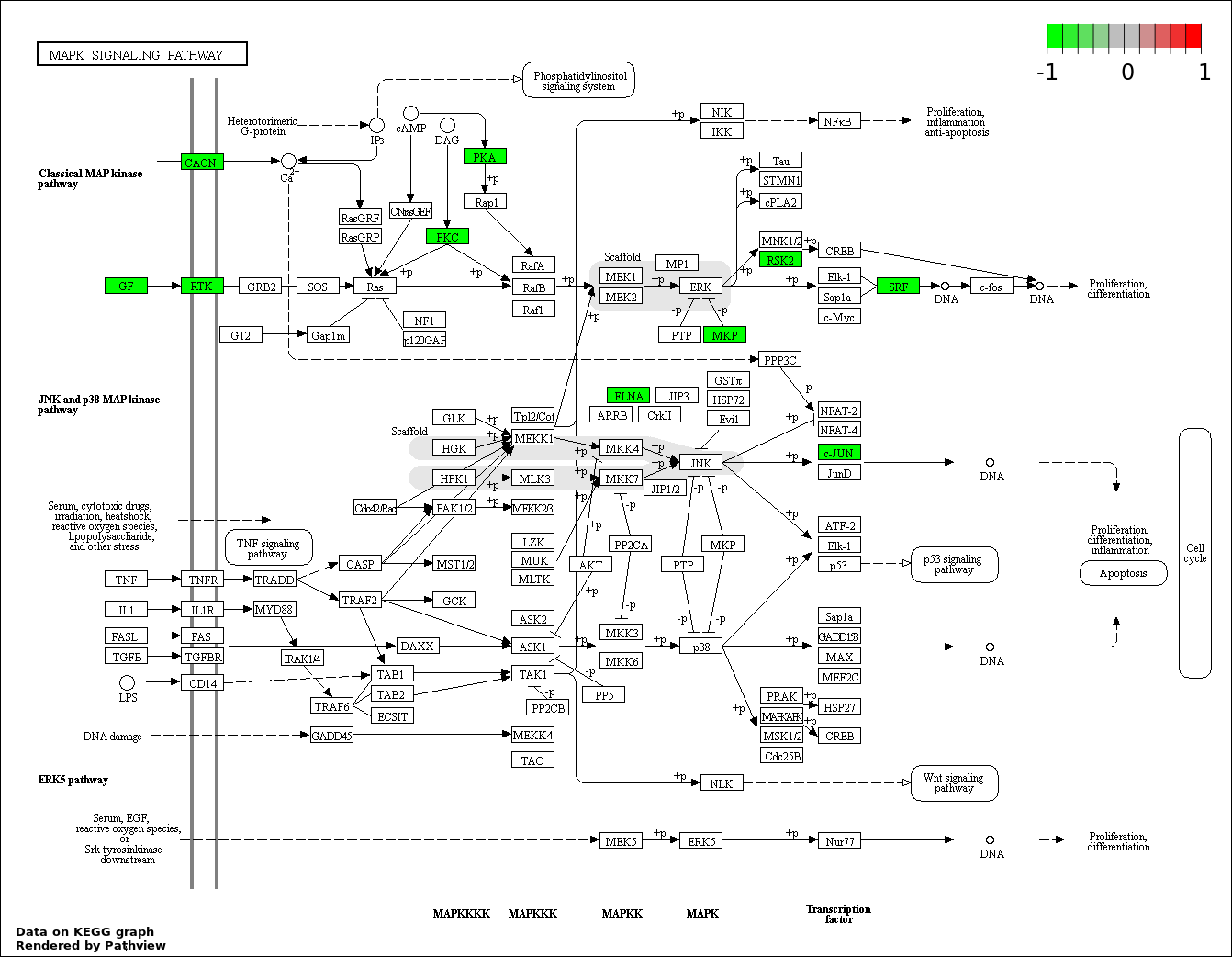
| 9 | hsa04010_MAPK_signaling_pathway | 12 | 268 | 0.0006658 | 0.01306 | |
| 10 | hsa04916_Melanogenesis | 7 | 101 | 0.000746 | 0.01306 | |
| 11 | hsa04014_Ras_signaling_pathway | 11 | 236 | 0.0007979 | 0.01306 | |
| 12 | hsa04512_ECM.receptor_interaction | 6 | 85 | 0.001614 | 0.02421 | |
| 13 | hsa04012_ErbB_signaling_pathway | 6 | 87 | 0.001819 | 0.02518 | |
| 14 | hsa04730_Long.term_depression | 5 | 70 | 0.003738 | 0.04806 | |
| 15 | hsa04020_Calcium_signaling_pathway | 8 | 177 | 0.004858 | 0.05829 | |
| 16 | hsa00534_Glycosaminoglycan_biosynthesis_._heparan_sulfate | 3 | 26 | 0.006425 | 0.07228 | |
| 17 | hsa04390_Hippo_signaling_pathway | 7 | 154 | 0.007983 | 0.08452 | |
| 18 | hsa04666_Fc_gamma_R.mediated_phagocytosis | 5 | 95 | 0.01331 | 0.1331 | |
| 19 | hsa04810_Regulation_of_actin_cytoskeleton | 8 | 214 | 0.01438 | 0.1363 | |
| 20 | hsa04972_Pancreatic_secretion | 5 | 101 | 0.01696 | 0.1526 | |
| 21 | hsa04520_Adherens_junction | 4 | 73 | 0.02281 | 0.1955 | |
| 22 | hsa04310_Wnt_signaling_pathway | 6 | 151 | 0.02472 | 0.2022 | |
| 23 | hsa04962_Vasopressin.regulated_water_reabsorption | 3 | 44 | 0.02697 | 0.2024 | |
| 24 | hsa04114_Oocyte_meiosis | 5 | 114 | 0.02699 | 0.2024 | |
| 25 | hsa04070_Phosphatidylinositol_signaling_system | 4 | 78 | 0.02825 | 0.2034 | |
| 26 | hsa04380_Osteoclast_differentiation | 5 | 128 | 0.04132 | 0.2861 | |
| 27 | hsa04514_Cell_adhesion_molecules_.CAMs. | 5 | 136 | 0.05124 | 0.3416 | |
| 28 | hsa04910_Insulin_signaling_pathway | 5 | 138 | 0.05392 | 0.3466 | |
| 29 | hsa04630_Jak.STAT_signaling_pathway | 5 | 155 | 0.0799 | 0.4959 | |
| 30 | hsa04370_VEGF_signaling_pathway | 3 | 76 | 0.1022 | 0.6129 | |
| 31 | hsa04664_Fc_epsilon_RI_signaling_pathway | 3 | 79 | 0.1114 | 0.6468 | |
| 32 | hsa04722_Neurotrophin_signaling_pathway | 4 | 127 | 0.1187 | 0.6679 | |
| 33 | hsa04360_Axon_guidance | 4 | 130 | 0.1263 | 0.6738 | |
| 34 | hsa04960_Aldosterone.regulated_sodium_reabsorption | 2 | 42 | 0.1273 | 0.6738 | |
| 35 | hsa04530_Tight_junction | 4 | 133 | 0.134 | 0.6873 | |
| 36 | hsa04914_Progesterone.mediated_oocyte_maturation | 3 | 87 | 0.1375 | 0.6873 | |
| 37 | hsa04210_Apoptosis | 3 | 89 | 0.1443 | 0.7019 | |
| 38 | hsa04150_mTOR_signaling_pathway | 2 | 52 | 0.1787 | 0.8246 | |
| 39 | hsa04340_Hedgehog_signaling_pathway | 2 | 56 | 0.2 | 0.8372 | |
| 40 | hsa04660_T_cell_receptor_signaling_pathway | 3 | 108 | 0.2137 | 0.8704 | |
| 41 | hsa00230_Purine_metabolism | 4 | 162 | 0.2176 | 0.8704 | |
| 42 | hsa04670_Leukocyte_transendothelial_migration | 3 | 117 | 0.2488 | 0.9736 | |
| 43 | hsa04920_Adipocytokine_signaling_pathway | 2 | 68 | 0.2653 | 1 | |
| 44 | hsa04976_Bile_secretion | 2 | 71 | 0.2818 | 1 | |
| 45 | hsa04662_B_cell_receptor_signaling_pathway | 2 | 75 | 0.3036 | 1 | |
| 46 | hsa04062_Chemokine_signaling_pathway | 4 | 189 | 0.3047 | 1 | |
| 47 | hsa04260_Cardiac_muscle_contraction | 2 | 77 | 0.3145 | 1 | |
| 48 | hsa04650_Natural_killer_cell_mediated_cytotoxicity | 3 | 136 | 0.3249 | 1 | |
| 49 | hsa04145_Phagosome | 3 | 156 | 0.4051 | 1 | |
| 50 | hsa04142_Lysosome | 2 | 121 | 0.5351 | 1 | |
| 51 | hsa04144_Endocytosis | 3 | 203 | 0.5783 | 1 | |
| 52 | hsa04141_Protein_processing_in_endoplasmic_reticulum | 2 | 168 | 0.7117 | 1 | |
| 53 | hsa04740_Olfactory_transduction | 2 | 388 | 0.9795 | 1 |