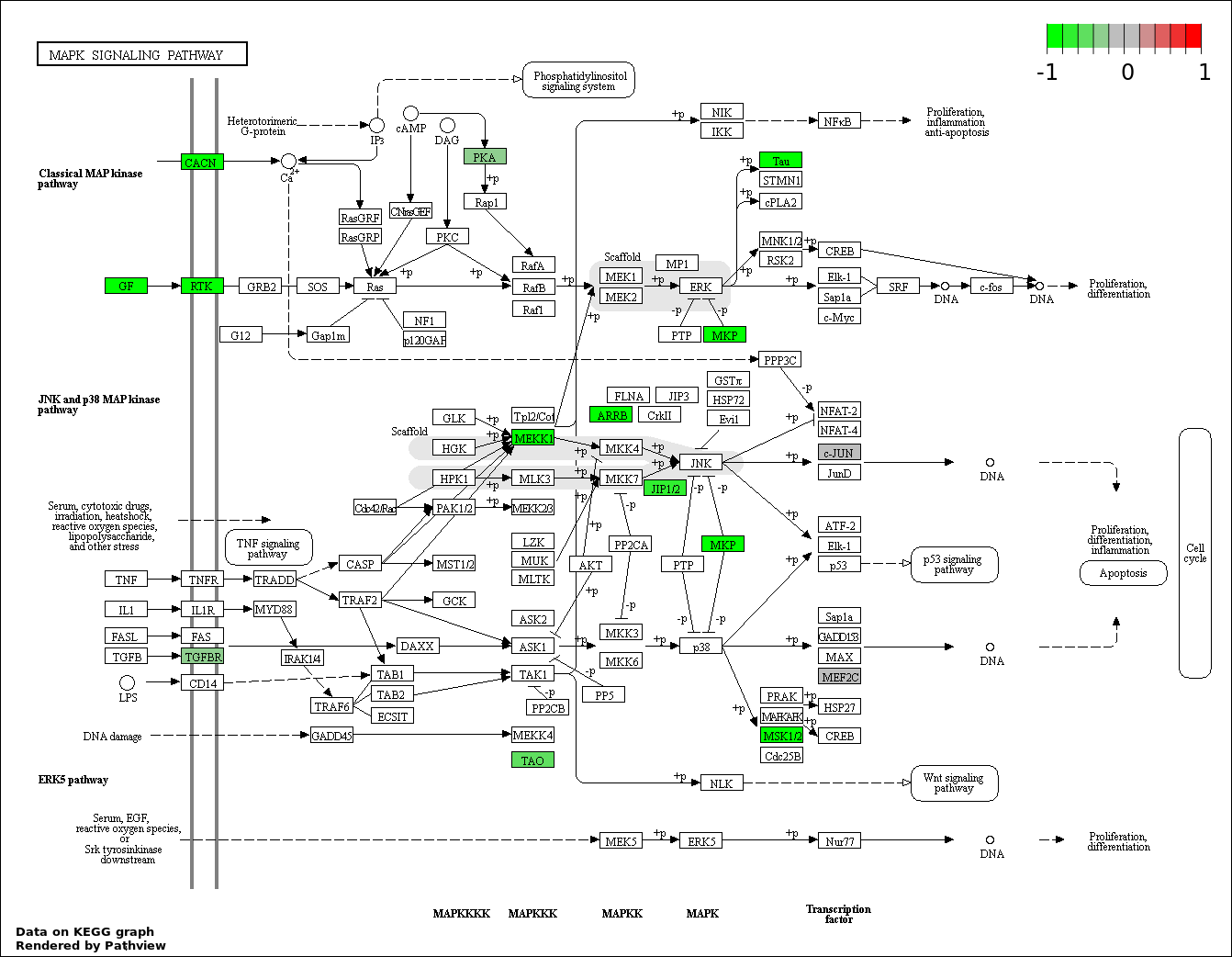
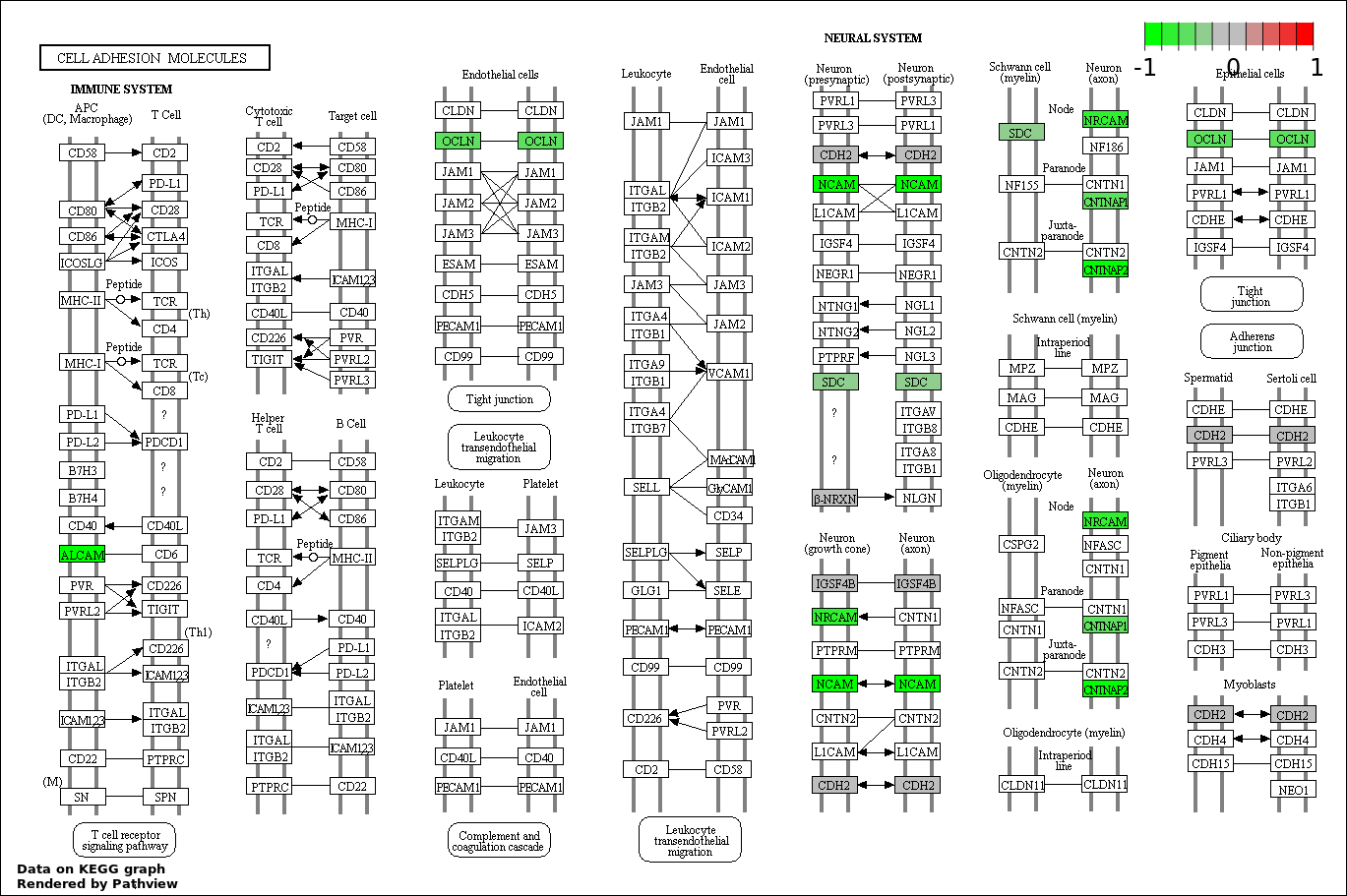
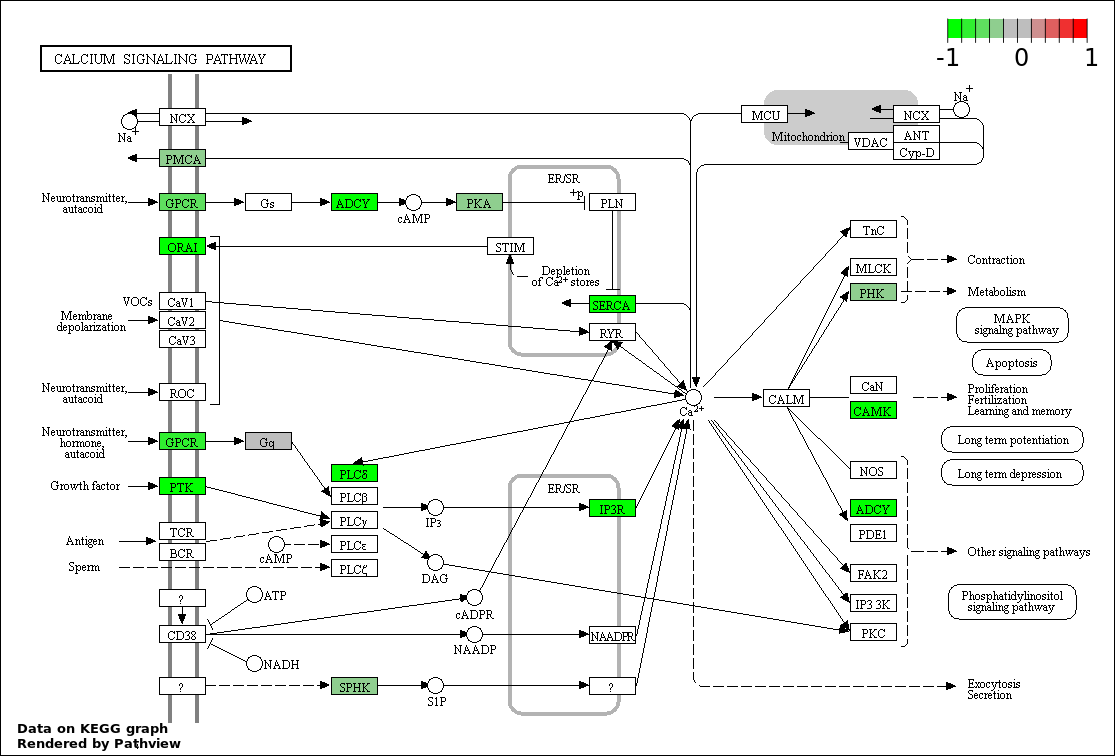
This regulatory network was inferred from the input dataset. The miRNAs and mRNAs are
presented as round and rectangle nodes respectively. The numerical value popped up upon mouse over the gene node is the log2 transformed fold-change of the gene expression between the two groups. All of the nodes are clickable, and the detailed information of the miRNAs/mRNAs and related cancer pathway will be displayed in another window. The edges between nodes are supported by both interactions (predicted or experimentally verified) and correlations learnt from cancer dataset. The numerical value popped up upon mouse over the edge is the correlation beat value (effect size) between the two nodes. The experimental evidences of the edges reported in previous cancer studies are highlighted by red/orange color. All of these information can be accessed by the "mouse-over" action. This network shows a full map of the miRNA-mRNA regulation of the input gene list(s), and the hub miRNAs (with the high network degree/betweenness centrality) would be the potential cancer drivers or tumor suppressors. The full result table can be accessed in the "Regulations" tab.
"miRNACancerMAP" is also a network visualization tool for users to draw their regulatory network by personal customization. Users can set the complexity of the network by limiting the number of nodes or edges. And the color of the nodes can be defined by different categories of the mRNAs and miRNAs, such as Gene-Ontology, pathway, and expression status. Users can also select to use network degree or network betweenness centrality to define the node size. And edges can be black or colored by the correlation. Purple edge means negative correlation (mostly found between miRNA and mRNA), and blue edge means positive correlation (found in PPI or miRNA-miRNA sponge effect). We can also add the protein-protein interactions (PPI) into the network. This result will show the cluster of genes regulated by some specific miRNAs. Additionally, miRNA-miRNA edges can be added by the "miRNA sponge" button, presenting some clusters of miRNAs that have the interactions via sponge effect.
miRNA-gene regulations
| Num | microRNA | Gene | miRNA log2FC | miRNA pvalue | Gene log2FC | Gene pvalue | Interaction | Correlation beta | Correlation P-value | PMID | Reported in cancer studies |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | hsa-miR-455-3p | ABCB1 | 1.82 | 0.0036 | -0.64 | 0.02964 | MirTarget | -0.32 | 0 | NA | |
| 2 | hsa-miR-505-5p | ABCB8 | 1.75 | 0 | -0.47 | 0.33037 | mirMAP | -0.16 | 0 | NA | |
| 3 | hsa-miR-224-5p | ABCC5 | 2.66 | 0 | -0.67 | 0.21962 | mirMAP | -0.12 | 0 | NA | |
| 4 | hsa-miR-224-5p | ABHD2 | 2.66 | 0 | -1.06 | 0.12031 | mirMAP | -0.14 | 0 | NA | |
| 5 | hsa-miR-452-5p | ABHD2 | 2.02 | 0.00026 | -1.06 | 0.12031 | mirMAP | -0.22 | 0 | NA | |
| 6 | hsa-miR-584-5p | ABHD2 | 1.8 | 0.00055 | -1.06 | 0.12031 | mirMAP | -0.23 | 0 | NA | |
| 7 | hsa-miR-584-5p | ABLIM3 | 1.8 | 0.00055 | -2.38 | 0 | mirMAP | -0.12 | 0.00693 | NA | |
| 8 | hsa-miR-18a-5p | ABR | 2.3 | 0 | -0.31 | 0.60278 | miRNAWalker2 validate; mirMAP | -0.11 | 0 | NA | |
| 9 | hsa-miR-224-5p | ACSS1 | 2.66 | 0 | -0.51 | 0.33926 | MirTarget | -0.16 | 0 | NA | |
| 10 | hsa-miR-452-5p | ACSS1 | 2.02 | 0.00026 | -0.51 | 0.33926 | MirTarget | -0.16 | 0 | NA | |
| 11 | hsa-miR-505-3p | ADAM22 | 1.83 | 3.0E-5 | -1.03 | 2.0E-5 | mirMAP | -0.15 | 0.00322 | NA | |
| 12 | hsa-miR-505-5p | ADAMTS14 | 1.75 | 0 | -0.3 | 0.28567 | mirMAP | -0.35 | 0 | NA | |
| 13 | hsa-miR-455-3p | ADAMTS18 | 1.82 | 0.0036 | -0.6 | 0.05192 | PITA | -0.26 | 0 | NA | |
| 14 | hsa-miR-18a-5p | ADAMTS2 | 2.3 | 0 | -0.29 | 0.51167 | mirMAP | -0.15 | 0 | NA | |
| 15 | hsa-miR-20a-3p | ADAMTS6 | 1.6 | 0 | -0.69 | 0.00023 | miRNATAP | -0.13 | 0 | NA | |
| 16 | hsa-miR-455-3p | ADAMTS8 | 1.82 | 0.0036 | -0.33 | 0.21979 | MirTarget; miRNATAP | -0.26 | 0 | NA | |
| 17 | hsa-miR-20a-3p | ADCY1 | 1.6 | 0 | -2.23 | 0 | mirMAP | -0.27 | 0 | NA | |
| 18 | hsa-miR-455-3p | ADCY1 | 1.82 | 0.0036 | -2.23 | 0 | PITA; miRNATAP | -0.28 | 0 | NA | |
| 19 | hsa-miR-455-3p | ADORA2A | 1.82 | 0.0036 | -0.46 | 0.20325 | MirTarget | -0.12 | 9.0E-5 | NA | |
| 20 | hsa-miR-505-3p | AFF1 | 1.83 | 3.0E-5 | -0.76 | 0.20112 | mirMAP | -0.13 | 0 | NA | |
| 21 | hsa-miR-224-5p | AFF3 | 2.66 | 0 | -4 | 0 | MirTarget | -0.49 | 0 | NA | |
| 22 | hsa-miR-18a-5p | AFF4 | 2.3 | 0 | -0.7 | 0.2756 | miRNAWalker2 validate | -0.22 | 0 | NA | |
| 23 | hsa-miR-584-5p | AFF4 | 1.8 | 0.00055 | -0.7 | 0.2756 | MirTarget | -0.1 | 0 | NA | |
| 24 | hsa-miR-224-5p | AGBL4 | 2.66 | 0 | -1.65 | 0 | mirMAP | -0.2 | 0 | NA | |
| 25 | hsa-miR-20a-3p | AGFG2 | 1.6 | 0 | -0.34 | 0.40473 | mirMAP | -0.12 | 0 | NA | |
| 26 | hsa-miR-455-3p | AGFG2 | 1.82 | 0.0036 | -0.34 | 0.40473 | mirMAP | -0.11 | 0 | NA | |
| 27 | hsa-miR-18a-5p | ALCAM | 2.3 | 0 | -1.44 | 0.0323 | MirTarget; miRNATAP | -0.34 | 0 | NA | |
| 28 | hsa-miR-455-3p | ALDH1A2 | 1.82 | 0.0036 | -0.35 | 0.24407 | PITA | -0.28 | 0 | NA | |
| 29 | hsa-miR-455-3p | ALX4 | 1.82 | 0.0036 | 0.15 | 0.6928 | mirMAP | -0.2 | 6.0E-5 | NA | |
| 30 | hsa-miR-455-3p | AMDHD2 | 1.82 | 0.0036 | -0.47 | 0.22828 | mirMAP | -0.16 | 0 | NA | |
| 31 | hsa-miR-224-5p | AMN1 | 2.66 | 0 | -0.48 | 0.12341 | MirTarget | -0.11 | 0 | NA | |
| 32 | hsa-miR-224-5p | AMZ1 | 2.66 | 0 | -1.47 | 0 | mirMAP | -0.23 | 0 | NA | |
| 33 | hsa-miR-505-3p | AMZ1 | 1.83 | 3.0E-5 | -1.47 | 0 | mirMAP | -0.38 | 0 | NA | |
| 34 | hsa-miR-20a-3p | ANGPTL1 | 1.6 | 0 | -0.93 | 0.00174 | MirTarget | -0.26 | 0 | NA | |
| 35 | hsa-miR-505-3p | ANKMY2 | 1.83 | 3.0E-5 | -0.58 | 0.15659 | MirTarget | -0.12 | 0 | NA | |
| 36 | hsa-miR-505-3p | ANKRD13D | 1.83 | 3.0E-5 | -0.13 | 0.79536 | miRNAWalker2 validate | -0.14 | 0 | NA | |
| 37 | hsa-miR-18a-5p | ANKRD50 | 2.3 | 0 | -0.73 | 0.17675 | MirTarget; miRNATAP | -0.18 | 0 | NA | |
| 38 | hsa-miR-452-5p | ANKS1B | 2.02 | 0.00026 | -1.36 | 0 | miRNATAP | -0.29 | 0 | NA | |
| 39 | hsa-miR-505-3p | APBB2 | 1.83 | 3.0E-5 | -1.32 | 0.0295 | mirMAP | -0.26 | 0 | NA | |
| 40 | hsa-miR-942-5p | APBB2 | 1.66 | 0 | -1.32 | 0.0295 | MirTarget | -0.27 | 0 | NA | |
| 41 | hsa-miR-20a-3p | APC2 | 1.6 | 0 | -0.54 | 0.01393 | mirMAP | -0.14 | 1.0E-5 | NA | |
| 42 | hsa-miR-505-5p | APC2 | 1.75 | 0 | -0.54 | 0.01393 | mirMAP | -0.17 | 0 | NA | |
| 43 | hsa-miR-18a-5p | APOLD1 | 2.3 | 0 | -0.38 | 0.4521 | mirMAP | -0.16 | 0 | NA | |
| 44 | hsa-miR-505-3p | AR | 1.83 | 3.0E-5 | -4.4 | 0 | mirMAP | -0.88 | 0 | NA | |
| 45 | hsa-miR-505-5p | AR | 1.75 | 0 | -4.4 | 0 | MirTarget | -0.68 | 0 | NA | |
| 46 | hsa-miR-18a-5p | ARFGEF2 | 2.3 | 0 | -0.4 | 0.50575 | MirTarget | -0.12 | 0 | NA | |
| 47 | hsa-miR-18a-5p | ARHGAP5 | 2.3 | 0 | -0.05 | 0.93513 | MirTarget | -0.12 | 0 | NA | |
| 48 | hsa-miR-452-5p | ARID2 | 2.02 | 0.00026 | -0.69 | 0.16232 | MirTarget | -0.14 | 0 | NA | |
| 49 | hsa-miR-18a-5p | ARL15 | 2.3 | 0 | -0.82 | 0.02217 | MirTarget; miRNATAP | -0.26 | 0 | NA | |
| 50 | hsa-miR-224-5p | ARMCX2 | 2.66 | 0 | -0.5 | 0.33207 | MirTarget | -0.11 | 0 | NA | |
| 51 | hsa-miR-455-3p | ARRB1 | 1.82 | 0.0036 | -0.81 | 0.09105 | MirTarget | -0.21 | 0 | NA | |
| 52 | hsa-miR-455-3p | ARX | 1.82 | 0.0036 | -0.74 | 0.09906 | miRNATAP | -0.38 | 0 | NA | |
| 53 | hsa-miR-452-5p | ASAH1 | 2.02 | 0.00026 | -1 | 0.17531 | MirTarget | -0.1 | 0 | NA | |
| 54 | hsa-miR-20a-3p | ASPA | 1.6 | 0 | -0.95 | 0.00059 | mirMAP | -0.24 | 0 | NA | |
| 55 | hsa-miR-18a-5p | ASTN1 | 2.3 | 0 | 0.11 | 0.73119 | MirTarget | -0.25 | 0 | NA | |
| 56 | hsa-miR-18a-5p | ASXL2 | 2.3 | 0 | -0.04 | 0.92196 | miRNATAP | -0.17 | 0 | NA | |
| 57 | hsa-miR-18a-5p | ATF7 | 2.3 | 0 | -0.31 | 0.57742 | mirMAP | -0.14 | 0 | NA | |
| 58 | hsa-miR-18a-5p | ATM | 2.3 | 0 | -0.05 | 0.92324 | miRNAWalker2 validate; miRTarBase; MirTarget | -0.12 | 0 | 23857602; 23437304; 25963391; 23229340 | Furthermore we used antisense oligonucleotides against micro RNAs miRNA or miRNA overexpression plasmids to study the role of miR-18a and -106a on ATM expression; Furthermore we identified that ERα activates miR-18a and -106a to downregulate ATM expression; We reveal a novel mechanism involving ERα and miR-18a and -106a regulation of ATM in breast cancer;MicroRNA 18a attenuates DNA damage repair through suppressing the expression of ataxia telangiectasia mutated in colorectal cancer; Through in silico search the 3'UTR of Ataxia telangiectasia mutated ATM contains a conserved miR-18a binding site; Expression of ATM was down-regulated in CRC tumors p<0.0001 and inversely correlated with miR-18a expression r = -0.4562 p<0.01; This was further confirmed by the down-regulation of ATM protein by miR-18a; As ATM is a key enzyme in DNA damage repair we evaluated the effect of miR-18a on DNA double-strand breaks; miR-18a attenuates cellular repair of DNA double-strand breaks by directly suppressing ATM a key enzyme in DNA damage repair;However the upregulation of miR-18a suppressed the level of ataxia-telangiectasia mutated and attenuated DNA double-strand break repair after irradiation which re-sensitized the cervical cancer cells to radiotherapy by promoting apoptosis;MicroRNA 18a upregulates autophagy and ataxia telangiectasia mutated gene expression in HCT116 colon cancer cells; Previous studies showed that certain microRNAs including miR-18a potentially regulate ATM in cancer cells; However the mechanisms behind the modulation of ATM by miR-18a remain to be elucidated in colon cancer cells; In the present study we explored the impact of miR-18a on the autophagy process and ATM expression in HCT116 colon cancer cells; Western blotting and luciferase assays were implemented to explore the impact of miR-18a on ATM gene expression in HCT116 cells; Moreover miR-18a overexpression led to the upregulation of ATM expression and suppression of mTORC1 activity; Results of the present study pertaining to the role of miR-18a in regulating autophagy and ATM gene expression in colon cancer cells revealed a novel function for miR-18a in a critical cellular event and on a crucial gene with significant impacts in cancer development progression treatment and in other diseases |
| 59 | hsa-miR-18a-5p | ATP1B2 | 2.3 | 0 | -0.53 | 0.01045 | mirMAP | -0.23 | 0 | NA | |
| 60 | hsa-miR-455-3p | ATP2A3 | 1.82 | 0.0036 | -0.97 | 0.08995 | mirMAP | -0.16 | 0 | NA | |
| 61 | hsa-miR-18a-5p | ATP2B1 | 2.3 | 0 | -0.32 | 0.5434 | miRNATAP | -0.12 | 0 | NA | |
| 62 | hsa-miR-18a-5p | ATP2C2 | 2.3 | 0 | -0.99 | 0.0117 | MirTarget | -0.2 | 0 | NA | |
| 63 | hsa-miR-505-3p | ATP6V1G1 | 1.83 | 3.0E-5 | -0.39 | 0.54218 | MirTarget | -0.12 | 0 | NA | |
| 64 | hsa-miR-455-3p | ATP8A2 | 1.82 | 0.0036 | -1.24 | 1.0E-5 | mirMAP | -0.15 | 0.00038 | NA | |
| 65 | hsa-miR-20a-3p | ATRNL1 | 1.6 | 0 | -1.76 | 0 | MirTarget; mirMAP | -0.21 | 0.00197 | NA | |
| 66 | hsa-miR-18a-5p | ATXN1 | 2.3 | 0 | -0.48 | 0.35126 | MirTarget; miRNATAP | -0.16 | 0 | NA | |
| 67 | hsa-miR-455-3p | BAG3 | 1.82 | 0.0036 | -0.37 | 0.51919 | miRNATAP | -0.1 | 0 | NA | |
| 68 | hsa-miR-18a-5p | BBX | 2.3 | 0 | -0.06 | 0.91654 | MirTarget; miRNATAP | -0.11 | 0 | NA | |
| 69 | hsa-miR-452-5p | BCAS1 | 2.02 | 0.00026 | -4.85 | 0 | mirMAP | -0.65 | 0 | NA | |
| 70 | hsa-miR-20a-3p | BCL2 | 1.6 | 0 | -1.88 | 0.00098 | mirMAP | -0.14 | 0.0001 | NA | |
| 71 | hsa-miR-224-5p | BCL2 | 2.66 | 0 | -1.88 | 0.00098 | mirMAP | -0.24 | 0 | 24796455 | In addition the expressions of Bcl2 mRNA and protein were 1.05 ± 0.04 and 0.21 ± 0.03 in the miR-224 ASO group significantly lower than that in the control group 4.87 ± 0.96 and 0.88 ± 0.09 P < 0.01 |
| 72 | hsa-miR-452-5p | BCL2 | 2.02 | 0.00026 | -1.88 | 0.00098 | mirMAP | -0.28 | 0 | NA | |
| 73 | hsa-miR-505-5p | BCL2L1 | 1.75 | 0 | -0.51 | 0.42908 | miRNATAP | -0.11 | 0 | NA | |
| 74 | hsa-miR-584-5p | BCOR | 1.8 | 0.00055 | -0.5 | 0.33033 | MirTarget | -0.15 | 0 | NA | |
| 75 | hsa-miR-18a-5p | BHLHE22 | 2.3 | 0 | 0.39 | 0.0481 | MirTarget; miRNATAP | -0.1 | 0.00101 | NA | |
| 76 | hsa-miR-942-5p | BHLHE40 | 1.66 | 0 | -1.28 | 0.10105 | MirTarget | -0.32 | 0 | NA | |
| 77 | hsa-miR-505-3p | BTBD9 | 1.83 | 3.0E-5 | -0.32 | 0.4633 | mirMAP | -0.11 | 0 | NA | |
| 78 | hsa-miR-455-3p | BTG2 | 1.82 | 0.0036 | -1.49 | 0.04452 | MirTarget; PITA; miRNATAP | -0.31 | 0 | NA | |
| 79 | hsa-miR-224-5p | BTRC | 2.66 | 0 | -0.93 | 0.05784 | MirTarget | -0.13 | 0 | NA | |
| 80 | hsa-miR-505-3p | BTRC | 1.83 | 3.0E-5 | -0.93 | 0.05784 | MirTarget | -0.2 | 0 | NA | |
| 81 | hsa-miR-452-5p | BZW1 | 2.02 | 0.00026 | -0.47 | 0.51928 | MirTarget; miRNATAP | -0.11 | 0 | NA | |
| 82 | hsa-miR-20a-3p | C14orf132 | 1.6 | 0 | -1.68 | 0.00016 | mirMAP | -0.26 | 0 | NA | |
| 83 | hsa-miR-18a-5p | C1orf21 | 2.3 | 0 | -1.31 | 0.03763 | mirMAP | -0.35 | 0 | NA | |
| 84 | hsa-miR-20a-3p | C1orf21 | 1.6 | 0 | -1.31 | 0.03763 | mirMAP | -0.22 | 0 | NA | |
| 85 | hsa-miR-224-5p | C1orf21 | 2.66 | 0 | -1.31 | 0.03763 | mirMAP | -0.13 | 0 | NA | |
| 86 | hsa-miR-452-5p | C1orf21 | 2.02 | 0.00026 | -1.31 | 0.03763 | mirMAP | -0.13 | 0 | NA | |
| 87 | hsa-miR-505-3p | C1orf21 | 1.83 | 3.0E-5 | -1.31 | 0.03763 | mirMAP | -0.4 | 0 | NA | |
| 88 | hsa-miR-505-5p | C1orf21 | 1.75 | 0 | -1.31 | 0.03763 | mirMAP | -0.28 | 0 | NA | |
| 89 | hsa-miR-18a-5p | C1orf226 | 2.3 | 0 | -0.84 | 0.08628 | mirMAP | -0.29 | 0 | NA | |
| 90 | hsa-miR-505-3p | C2orf72 | 1.83 | 3.0E-5 | -0 | 0.99424 | miRNATAP | -0.15 | 0.04229 | NA | |
| 91 | hsa-miR-18a-5p | C5orf30 | 2.3 | 0 | -1.64 | 0.00035 | miRNATAP | -0.22 | 0 | NA | |
| 92 | hsa-miR-18a-5p | CA12 | 2.3 | 0 | -4.61 | 0 | miRNAWalker2 validate; MirTarget | -0.85 | 0 | NA | |
| 93 | hsa-miR-455-3p | CACNA2D2 | 1.82 | 0.0036 | -3.3 | 0 | PITA; miRNATAP | -0.43 | 0 | NA | |
| 94 | hsa-miR-505-3p | CACNB4 | 1.83 | 3.0E-5 | -0.64 | 0.00531 | miRNATAP | -0.17 | 9.0E-5 | NA | |
| 95 | hsa-miR-18a-5p | CACNG4 | 2.3 | 0 | -3.36 | 0 | mirMAP | -0.48 | 0 | NA | |
| 96 | hsa-miR-20a-3p | CADM2 | 1.6 | 0 | -2.36 | 0 | MirTarget; miRNATAP | -0.15 | 0.01139 | NA | |
| 97 | hsa-miR-224-5p | CADM2 | 2.66 | 0 | -2.36 | 0 | mirMAP | -0.16 | 1.0E-5 | NA | |
| 98 | hsa-miR-452-5p | CADM2 | 2.02 | 0.00026 | -2.36 | 0 | MirTarget | -0.13 | 0.00422 | NA | |
| 99 | hsa-miR-505-5p | CADM2 | 1.75 | 0 | -2.36 | 0 | MirTarget; miRNATAP | -0.29 | 1.0E-5 | NA | |
| 100 | hsa-miR-584-5p | CADM2 | 1.8 | 0.00055 | -2.36 | 0 | mirMAP | -0.19 | 0.00291 | NA | |
| 101 | hsa-miR-455-3p | CADM3 | 1.82 | 0.0036 | 0.16 | 0.66511 | mirMAP | -0.4 | 0 | NA | |
| 102 | hsa-miR-20a-3p | CADPS2 | 1.6 | 0 | -1.09 | 0.0296 | MirTarget | -0.14 | 0 | NA | |
| 103 | hsa-miR-18a-5p | CALN1 | 2.3 | 0 | -1.14 | 0.00178 | mirMAP | -0.42 | 0 | NA | |
| 104 | hsa-miR-20a-3p | CALN1 | 1.6 | 0 | -1.14 | 0.00178 | mirMAP | -0.26 | 0 | NA | |
| 105 | hsa-miR-18a-5p | CAMK2B | 2.3 | 0 | -1.56 | 0 | mirMAP | -0.27 | 0 | NA | |
| 106 | hsa-miR-18a-5p | CAMK2N1 | 2.3 | 0 | -1.69 | 0.00251 | miRNATAP | -0.3 | 0 | NA | |
| 107 | hsa-miR-455-3p | CAMK2N1 | 1.82 | 0.0036 | -1.69 | 0.00251 | PITA | -0.3 | 0 | NA | |
| 108 | hsa-miR-505-5p | CAPSL | 1.75 | 0 | -2.76 | 0 | MirTarget | -0.54 | 0 | NA | |
| 109 | hsa-miR-452-5p | CASC1 | 2.02 | 0.00026 | -2.56 | 0 | MirTarget | -0.45 | 0 | NA | |
| 110 | hsa-miR-452-5p | CASD1 | 2.02 | 0.00026 | -0.96 | 0.05329 | MirTarget; miRNATAP | -0.16 | 0 | NA | |
| 111 | hsa-miR-18a-5p | CASZ1 | 2.3 | 0 | -0.88 | 0.0914 | mirMAP | -0.22 | 0 | NA | |
| 112 | hsa-miR-455-3p | CBLN2 | 1.82 | 0.0036 | -1.61 | 0.00089 | MirTarget; miRNATAP | -0.27 | 0.00024 | NA | |
| 113 | hsa-miR-505-3p | CBX7 | 1.83 | 3.0E-5 | -0.47 | 0.36872 | mirMAP | -0.27 | 0 | NA | |
| 114 | hsa-miR-505-3p | CCDC125 | 1.83 | 3.0E-5 | -1.49 | 0.00066 | MirTarget | -0.37 | 0 | NA | |
| 115 | hsa-miR-942-5p | CCND1 | 1.66 | 0 | -1.64 | 0.03536 | MirTarget | -0.26 | 0 | NA | |
| 116 | hsa-miR-224-5p | CCNG2 | 2.66 | 0 | -1.09 | 0.07868 | mirMAP | -0.12 | 0 | NA | |
| 117 | hsa-miR-452-5p | CCNO | 2.02 | 0.00026 | -1.97 | 0 | MirTarget | -0.27 | 0 | NA | |
| 118 | hsa-miR-455-3p | CDA | 1.82 | 0.0036 | -0.04 | 0.79435 | miRNATAP | -0.13 | 0 | NA | |
| 119 | hsa-miR-452-5p | CDH2 | 2.02 | 0.00026 | -0.03 | 0.91735 | miRNATAP | -0.14 | 4.0E-5 | NA | |
| 120 | hsa-miR-20a-3p | CDH6 | 1.6 | 0 | -0.69 | 0.00035 | mirMAP | -0.12 | 1.0E-5 | NA | |
| 121 | hsa-miR-455-3p | CDKL2 | 1.82 | 0.0036 | -0.87 | 0.00567 | MirTarget | -0.27 | 0 | NA | |
| 122 | hsa-miR-455-3p | CDKN1A | 1.82 | 0.0036 | -0.33 | 0.59485 | MirTarget | -0.12 | 0 | NA | |
| 123 | hsa-miR-505-5p | CDKN1A | 1.75 | 0 | -0.33 | 0.59485 | miRNAWalker2 validate; MirTarget | -0.11 | 6.0E-5 | NA | |
| 124 | hsa-miR-942-5p | CDKN1A | 1.66 | 0 | -0.33 | 0.59485 | miRNAWalker2 validate | -0.11 | 5.0E-5 | NA | |
| 125 | hsa-miR-505-3p | CDKN1B | 1.83 | 3.0E-5 | -0.32 | 0.60215 | MirTarget | -0.11 | 0 | NA | |
| 126 | hsa-miR-18a-5p | CDON | 2.3 | 0 | -0.93 | 0.03701 | mirMAP | -0.27 | 0 | NA | |
| 127 | hsa-miR-452-5p | CDON | 2.02 | 0.00026 | -0.93 | 0.03701 | mirMAP | -0.11 | 0 | NA | |
| 128 | hsa-miR-505-3p | CDON | 1.83 | 3.0E-5 | -0.93 | 0.03701 | mirMAP | -0.17 | 0 | NA | |
| 129 | hsa-miR-505-5p | CENPP | 1.75 | 0 | -0.69 | 0.00969 | mirMAP | -0.19 | 0 | NA | |
| 130 | hsa-miR-584-5p | CENPP | 1.8 | 0.00055 | -0.69 | 0.00969 | mirMAP | -0.3 | 0 | NA | |
| 131 | hsa-miR-20a-3p | CHN2 | 1.6 | 0 | -1.74 | 0 | MirTarget; miRNATAP | -0.27 | 0 | NA | |
| 132 | hsa-miR-455-3p | CHRD | 1.82 | 0.0036 | -2.64 | 0 | miRNATAP | -0.39 | 0 | NA | |
| 133 | hsa-miR-455-3p | CHRDL1 | 1.82 | 0.0036 | 0.7 | 0.18687 | PITA; miRNATAP | -0.28 | 1.0E-5 | NA | |
| 134 | hsa-miR-18a-5p | CLASP2 | 2.3 | 0 | -0.44 | 0.39581 | miRNATAP | -0.14 | 0 | NA | |
| 135 | hsa-miR-505-5p | CLCN7 | 1.75 | 0 | -0.29 | 0.6398 | mirMAP | -0.11 | 0 | NA | |
| 136 | hsa-miR-942-5p | CLDN12 | 1.66 | 0 | -0.42 | 0.41121 | MirTarget | -0.14 | 0 | NA | |
| 137 | hsa-miR-942-5p | CNTD1 | 1.66 | 0 | -1.28 | 0 | MirTarget | -0.34 | 0 | NA | |
| 138 | hsa-miR-20a-3p | CNTNAP1 | 1.6 | 0 | -0.52 | 0.07934 | MirTarget | -0.1 | 0 | NA | |
| 139 | hsa-miR-584-5p | CNTNAP2 | 1.8 | 0.00055 | -2.6 | 0 | mirMAP | -0.79 | 0 | NA | |
| 140 | hsa-miR-20a-3p | COL14A1 | 1.6 | 0 | -1.29 | 0.04415 | miRNATAP | -0.36 | 0 | NA | |
| 141 | hsa-miR-224-5p | COQ7 | 2.66 | 0 | -0.87 | 0.0466 | mirMAP | -0.11 | 0 | NA | |
| 142 | hsa-miR-20a-3p | CPE | 1.6 | 0 | -1.43 | 0.02439 | MirTarget | -0.21 | 0 | NA | |
| 143 | hsa-miR-20a-3p | CPEB2 | 1.6 | 0 | -1.44 | 0.00246 | miRNATAP | -0.18 | 0 | NA | |
| 144 | hsa-miR-505-3p | CPEB2 | 1.83 | 3.0E-5 | -1.44 | 0.00246 | miRNATAP | -0.38 | 0 | NA | |
| 145 | hsa-miR-18a-5p | CREBL2 | 2.3 | 0 | -0.55 | 0.34569 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.19 | 0 | NA | |
| 146 | hsa-miR-18a-5p | CRIM1 | 2.3 | 0 | -0.31 | 0.6343 | MirTarget; miRNATAP | -0.27 | 0 | NA | |
| 147 | hsa-miR-20a-3p | CRTC1 | 1.6 | 0 | -0.37 | 0.38434 | mirMAP | -0.11 | 0 | NA | |
| 148 | hsa-miR-942-5p | CRY2 | 1.66 | 0 | -0.72 | 0.18723 | MirTarget | -0.23 | 0 | NA | |
| 149 | hsa-miR-455-3p | CSDC2 | 1.82 | 0.0036 | -1.43 | 0 | mirMAP | -0.33 | 0 | NA | |
| 150 | hsa-miR-452-5p | CSMD3 | 2.02 | 0.00026 | -1.2 | 0.03461 | MirTarget | -0.22 | 0 | NA |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | HOMOPHILIC CELL ADHESION VIA PLASMA MEMBRANE ADHESION MOLECULES | 28 | 153 | 3.021e-13 | 1.406e-09 |
| 2 | BIOLOGICAL ADHESION | 82 | 1032 | 6.741e-13 | 1.568e-09 |
| 3 | CELL CELL ADHESION VIA PLASMA MEMBRANE ADHESION MOLECULES | 31 | 204 | 2.827e-12 | 4.385e-09 |
| 4 | NEUROGENESIS | 94 | 1402 | 1.801e-10 | 2.095e-07 |
| 5 | SYNAPSE ORGANIZATION | 22 | 145 | 4.31e-09 | 4.011e-06 |
| 6 | NEURON DIFFERENTIATION | 64 | 874 | 7.22e-09 | 5.599e-06 |
| 7 | CELL DEVELOPMENT | 90 | 1426 | 9.151e-09 | 6.083e-06 |
| 8 | CELL CELL ADHESION | 49 | 608 | 2.447e-08 | 1.423e-05 |
| 9 | ORGAN MORPHOGENESIS | 60 | 841 | 5.554e-08 | 2.872e-05 |
| 10 | BEHAVIOR | 43 | 516 | 7.055e-08 | 3.283e-05 |
| 11 | CENTRAL NERVOUS SYSTEM DEVELOPMENT | 61 | 872 | 8.607e-08 | 3.641e-05 |
| 12 | SINGLE ORGANISM BEHAVIOR | 35 | 384 | 1.414e-07 | 4.687e-05 |
| 13 | REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 96 | 1656 | 1.712e-07 | 4.687e-05 |
| 14 | RESPONSE TO ENDOGENOUS STIMULUS | 87 | 1450 | 1.616e-07 | 4.687e-05 |
| 15 | CELLULAR RESPONSE TO ENDOGENOUS STIMULUS | 67 | 1008 | 1.326e-07 | 4.687e-05 |
| 16 | TISSUE DEVELOPMENT | 90 | 1518 | 1.639e-07 | 4.687e-05 |
| 17 | REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 102 | 1784 | 1.299e-07 | 4.687e-05 |
| 18 | HEAD DEVELOPMENT | 52 | 709 | 1.895e-07 | 4.898e-05 |
| 19 | GROWTH | 36 | 410 | 2.359e-07 | 5.505e-05 |
| 20 | RESPONSE TO ABIOTIC STIMULUS | 67 | 1024 | 2.366e-07 | 5.505e-05 |
| 21 | UROGENITAL SYSTEM DEVELOPMENT | 29 | 299 | 4.646e-07 | 9.827e-05 |
| 22 | DEVELOPMENTAL GROWTH | 31 | 333 | 4.627e-07 | 9.827e-05 |
| 23 | REGULATION OF GROWTH | 47 | 633 | 5.276e-07 | 0.0001067 |
| 24 | NEGATIVE REGULATION OF CELL COMMUNICATION | 73 | 1192 | 8.343e-07 | 0.0001618 |
| 25 | REGULATION OF MEMBRANE POTENTIAL | 31 | 343 | 8.769e-07 | 0.0001632 |
| 26 | REGULATION OF MULTICELLULAR ORGANISMAL DEVELOPMENT | 94 | 1672 | 9.169e-07 | 0.0001641 |
| 27 | PATTERN SPECIFICATION PROCESS | 35 | 418 | 1.055e-06 | 0.0001819 |
| 28 | REGULATION OF SYNAPSE ORGANIZATION | 16 | 113 | 1.436e-06 | 0.0002386 |
| 29 | NEURON DEVELOPMENT | 48 | 687 | 2.232e-06 | 0.0003387 |
| 30 | REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 51 | 750 | 2.347e-06 | 0.0003387 |
| 31 | MAMMARY GLAND DEVELOPMENT | 16 | 117 | 2.298e-06 | 0.0003387 |
| 32 | REGULATION OF PROTEIN MODIFICATION PROCESS | 94 | 1710 | 2.402e-06 | 0.0003387 |
| 33 | REGULATION OF CELLULAR COMPONENT MOVEMENT | 52 | 771 | 2.366e-06 | 0.0003387 |
| 34 | SKELETAL SYSTEM DEVELOPMENT | 36 | 455 | 2.769e-06 | 0.000379 |
| 35 | RESPONSE TO STEROID HORMONE | 38 | 497 | 3.312e-06 | 0.0004403 |
| 36 | REGULATION OF CELL DIFFERENTIATION | 84 | 1492 | 3.546e-06 | 0.0004583 |
| 37 | REGULATION OF CELLULAR RESPONSE TO GROWTH FACTOR STIMULUS | 23 | 229 | 3.941e-06 | 0.0004914 |
| 38 | EMBRYONIC SKELETAL SYSTEM DEVELOPMENT | 16 | 122 | 4.013e-06 | 0.0004914 |
| 39 | NEGATIVE REGULATION OF RESPONSE TO STIMULUS | 78 | 1360 | 4.177e-06 | 0.0004983 |
| 40 | REGULATION OF PHOSPHORUS METABOLIC PROCESS | 89 | 1618 | 4.499e-06 | 0.0005234 |
| 41 | REGULATION OF SYNAPSE STRUCTURE OR ACTIVITY | 23 | 232 | 4.906e-06 | 0.0005567 |
| 42 | REGULATION OF SYSTEM PROCESS | 38 | 507 | 5.272e-06 | 0.000584 |
| 43 | GLAND DEVELOPMENT | 32 | 395 | 6.074e-06 | 0.0006573 |
| 44 | RESPONSE TO HORMONE | 56 | 893 | 8.602e-06 | 0.0009096 |
| 45 | RESPONSE TO OXYGEN LEVELS | 27 | 311 | 9.285e-06 | 0.0009392 |
| 46 | REGIONALIZATION | 27 | 311 | 9.285e-06 | 0.0009392 |
| 47 | POSITIVE REGULATION OF DEVELOPMENTAL PROCESS | 67 | 1142 | 1.013e-05 | 0.001003 |
| 48 | REGULATION OF SMALL GTPASE MEDIATED SIGNAL TRANSDUCTION | 25 | 278 | 1.081e-05 | 0.001048 |
| 49 | REGULATION OF CELL DEVELOPMENT | 53 | 836 | 1.115e-05 | 0.001059 |
| 50 | ENZYME LINKED RECEPTOR PROTEIN SIGNALING PATHWAY | 46 | 689 | 1.185e-05 | 0.001103 |
| 51 | NEGATIVE REGULATION OF LOCOMOTION | 24 | 263 | 1.265e-05 | 0.001155 |
| 52 | REGULATION OF CELL GROWTH | 31 | 391 | 1.299e-05 | 0.001163 |
| 53 | REGULATION OF CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 28 | 337 | 1.445e-05 | 0.001259 |
| 54 | POSTSYNAPTIC MEMBRANE ORGANIZATION | 7 | 25 | 1.461e-05 | 0.001259 |
| 55 | POSITIVE REGULATION OF MOLECULAR FUNCTION | 94 | 1791 | 1.584e-05 | 0.00134 |
| 56 | REGULATION OF EPITHELIAL CELL PROLIFERATION | 25 | 285 | 1.656e-05 | 0.001376 |
| 57 | ORGAN GROWTH | 11 | 68 | 1.748e-05 | 0.001427 |
| 58 | REGULATION OF PHOSPHATIDYLINOSITOL 3 KINASE SIGNALING | 16 | 138 | 1.97e-05 | 0.001503 |
| 59 | CELLULAR RESPONSE TO STEROID HORMONE STIMULUS | 21 | 218 | 1.943e-05 | 0.001503 |
| 60 | CARDIOVASCULAR SYSTEM DEVELOPMENT | 50 | 788 | 1.941e-05 | 0.001503 |
| 61 | CIRCULATORY SYSTEM DEVELOPMENT | 50 | 788 | 1.941e-05 | 0.001503 |
| 62 | REGULATION OF DEVELOPMENTAL GROWTH | 25 | 289 | 2.097e-05 | 0.001574 |
| 63 | POSITIVE REGULATION OF PHOSPHATE METABOLIC PROCESS | 61 | 1036 | 2.315e-05 | 0.001683 |
| 64 | POSITIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS | 61 | 1036 | 2.315e-05 | 0.001683 |
| 65 | REGULATION OF HEART CONTRACTION | 21 | 221 | 2.386e-05 | 0.001708 |
| 66 | NEURON PROJECTION DEVELOPMENT | 38 | 545 | 2.68e-05 | 0.00189 |
| 67 | REGULATION OF ANATOMICAL STRUCTURE MORPHOGENESIS | 60 | 1021 | 2.864e-05 | 0.00196 |
| 68 | STEM CELL DIFFERENTIATION | 19 | 190 | 2.85e-05 | 0.00196 |
| 69 | CELLULAR RESPONSE TO ORGANIC SUBSTANCE | 95 | 1848 | 3.194e-05 | 0.002154 |
| 70 | NEGATIVE REGULATION OF NITROGEN COMPOUND METABOLIC PROCESS | 81 | 1517 | 3.623e-05 | 0.002321 |
| 71 | POSITIVE REGULATION OF GENE EXPRESSION | 90 | 1733 | 3.641e-05 | 0.002321 |
| 72 | REGULATION OF SMOOTH MUSCLE CELL MIGRATION | 9 | 49 | 3.58e-05 | 0.002321 |
| 73 | RHYTHMIC PROCESS | 25 | 298 | 3.501e-05 | 0.002321 |
| 74 | CONNECTIVE TISSUE DEVELOPMENT | 19 | 194 | 3.805e-05 | 0.002393 |
| 75 | MODULATION OF SYNAPTIC TRANSMISSION | 25 | 301 | 4.13e-05 | 0.002562 |
| 76 | CARTILAGE DEVELOPMENT | 16 | 147 | 4.32e-05 | 0.002645 |
| 77 | CELLULAR RESPONSE TO LIPID | 33 | 457 | 4.568e-05 | 0.002761 |
| 78 | NEGATIVE REGULATION OF CELL PROLIFERATION | 42 | 643 | 4.693e-05 | 0.0028 |
| 79 | LOCOMOTORY BEHAVIOR | 18 | 181 | 4.943e-05 | 0.002911 |
| 80 | NEGATIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS | 37 | 541 | 5.128e-05 | 0.002946 |
| 81 | NEGATIVE REGULATION OF PHOSPHATE METABOLIC PROCESS | 37 | 541 | 5.128e-05 | 0.002946 |
| 82 | POSITIVE REGULATION OF SMOOTH MUSCLE CELL MIGRATION | 7 | 30 | 5.334e-05 | 0.002955 |
| 83 | POSITIVE REGULATION OF PROTEIN MODIFICATION PROCESS | 64 | 1135 | 5.335e-05 | 0.002955 |
| 84 | DEVELOPMENTAL GROWTH INVOLVED IN MORPHOGENESIS | 13 | 104 | 5.311e-05 | 0.002955 |
| 85 | ADULT BEHAVIOR | 15 | 135 | 5.855e-05 | 0.003203 |
| 86 | NEGATIVE REGULATION OF DEVELOPMENTAL PROCESS | 49 | 801 | 5.921e-05 | 0.003203 |
| 87 | NEGATIVE REGULATION OF GENE EXPRESSION | 79 | 1493 | 6.171e-05 | 0.003235 |
| 88 | SKELETAL SYSTEM MORPHOGENESIS | 19 | 201 | 6.178e-05 | 0.003235 |
| 89 | RECEPTOR CLUSTERING | 8 | 41 | 6.187e-05 | 0.003235 |
| 90 | NEGATIVE REGULATION OF GROWTH | 21 | 236 | 6.273e-05 | 0.003243 |
| 91 | REGULATION OF AXONOGENESIS | 17 | 168 | 6.388e-05 | 0.003266 |
| 92 | NEGATIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 46 | 740 | 6.832e-05 | 0.003455 |
| 93 | VOCALIZATION BEHAVIOR | 5 | 14 | 7.04e-05 | 0.003522 |
| 94 | REGULATION OF NEURON PROJECTION DEVELOPMENT | 30 | 408 | 7.237e-05 | 0.003583 |
| 95 | CELLULAR RESPONSE TO HORMONE STIMULUS | 37 | 552 | 7.782e-05 | 0.003811 |
| 96 | REGULATION OF BLOOD CIRCULATION | 24 | 295 | 8.092e-05 | 0.003922 |
| 97 | RESPONSE TO DRUG | 31 | 431 | 8.287e-05 | 0.003975 |
| 98 | POSITIVE REGULATION OF CELL COMMUNICATION | 80 | 1532 | 8.576e-05 | 0.004072 |
| 99 | MESENCHYME DEVELOPMENT | 18 | 190 | 9.298e-05 | 0.00437 |
| 100 | RESPONSE TO AUDITORY STIMULUS | 6 | 23 | 9.447e-05 | 0.004396 |
| 101 | NEURON MIGRATION | 13 | 110 | 9.551e-05 | 0.0044 |
| 102 | FOREBRAIN DEVELOPMENT | 27 | 357 | 0.000103 | 0.0047 |
| 103 | REGULATION OF DENDRITIC SPINE DEVELOPMENT | 9 | 56 | 0.0001067 | 0.004822 |
| 104 | RESPONSE TO LIPID | 52 | 888 | 0.0001078 | 0.004822 |
| 105 | NEUROMUSCULAR PROCESS | 12 | 97 | 0.0001131 | 0.00501 |
| 106 | INTRACELLULAR SIGNAL TRANSDUCTION | 81 | 1572 | 0.00012 | 0.005266 |
| 107 | REGULATION OF POSITIVE CHEMOTAXIS | 6 | 24 | 0.0001223 | 0.00532 |
| 108 | ENDOCHONDRAL BONE MORPHOGENESIS | 8 | 45 | 0.0001238 | 0.005333 |
| 109 | REGULATION OF GTPASE ACTIVITY | 42 | 673 | 0.0001292 | 0.005516 |
| 110 | RESPONSE TO ESTRADIOL | 15 | 146 | 0.0001431 | 0.005975 |
| 111 | NEURON CELL CELL ADHESION | 5 | 16 | 0.0001451 | 0.005975 |
| 112 | POSITIVE REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 51 | 876 | 0.0001446 | 0.005975 |
| 113 | PROTEIN LOCALIZATION TO SYNAPSE | 5 | 16 | 0.0001451 | 0.005975 |
| 114 | OSSIFICATION | 21 | 251 | 0.0001504 | 0.006137 |
| 115 | MEMBRANE ASSEMBLY | 6 | 25 | 0.0001564 | 0.006219 |
| 116 | STARTLE RESPONSE | 6 | 25 | 0.0001564 | 0.006219 |
| 117 | PROTEIN LOCALIZATION | 90 | 1805 | 0.0001555 | 0.006219 |
| 118 | CELL PROJECTION ORGANIZATION | 52 | 902 | 0.0001587 | 0.006257 |
| 119 | REGULATION OF CELL MORPHOGENESIS | 36 | 552 | 0.0001664 | 0.006508 |
| 120 | VASCULATURE DEVELOPMENT | 32 | 469 | 0.0001696 | 0.006577 |
| 121 | POSITIVE REGULATION OF CATALYTIC ACTIVITY | 78 | 1518 | 0.0001764 | 0.006783 |
| 122 | CHONDROCYTE DIFFERENTIATION | 9 | 60 | 0.0001845 | 0.006872 |
| 123 | RESPONSE TO ESTROGEN | 19 | 218 | 0.0001811 | 0.006872 |
| 124 | REGULATION OF CELL PROLIFERATION | 77 | 1496 | 0.0001846 | 0.006872 |
| 125 | RENAL SYSTEM PROCESS | 12 | 102 | 0.0001836 | 0.006872 |
| 126 | REPLACEMENT OSSIFICATION | 6 | 26 | 0.0001975 | 0.007152 |
| 127 | MESENCHYMAL CELL DIFFERENTIATION | 14 | 134 | 0.0001979 | 0.007152 |
| 128 | MAMMARY GLAND ALVEOLUS DEVELOPMENT | 5 | 17 | 0.0001998 | 0.007152 |
| 129 | MAMMARY GLAND LOBULE DEVELOPMENT | 5 | 17 | 0.0001998 | 0.007152 |
| 130 | ENDOCHONDRAL OSSIFICATION | 6 | 26 | 0.0001975 | 0.007152 |
| 131 | REGULATION OF MUSCLE TISSUE DEVELOPMENT | 12 | 103 | 0.0002014 | 0.007155 |
| 132 | REGULATION OF KINASE ACTIVITY | 46 | 776 | 0.0002041 | 0.007194 |
| 133 | CELL GROWTH | 14 | 135 | 0.000214 | 0.007486 |
| 134 | MULTI ORGANISM BEHAVIOR | 10 | 75 | 0.0002234 | 0.007757 |
| 135 | REGULATION OF DENDRITE DEVELOPMENT | 13 | 120 | 0.0002313 | 0.00797 |
| 136 | POSITIVE REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 30 | 437 | 0.0002417 | 0.008091 |
| 137 | RESPONSE TO ORGANIC CYCLIC COMPOUND | 52 | 917 | 0.0002367 | 0.008091 |
| 138 | REGULATION OF CELL MATRIX ADHESION | 11 | 90 | 0.0002409 | 0.008091 |
| 139 | POSITIVE REGULATION OF PHOSPHATIDYLINOSITOL 3 KINASE SIGNALING | 9 | 62 | 0.0002383 | 0.008091 |
| 140 | CIRCADIAN RHYTHM | 14 | 137 | 0.0002496 | 0.008289 |
| 141 | NEGATIVE REGULATION OF CELLULAR RESPONSE TO GROWTH FACTOR STIMULUS | 13 | 121 | 0.0002512 | 0.008289 |
| 142 | NEGATIVE REGULATION OF NEURON DEATH | 16 | 171 | 0.0002583 | 0.008465 |
| 143 | REGULATION OF ADHERENS JUNCTION ORGANIZATION | 8 | 50 | 0.0002654 | 0.008636 |
| 144 | ORGAN MATURATION | 5 | 18 | 0.000269 | 0.008691 |
| 145 | POSITIVE REGULATION OF DEVELOPMENTAL GROWTH | 15 | 156 | 0.0002969 | 0.009528 |
| 146 | NEGATIVE REGULATION OF PHOSPHORYLATION | 29 | 422 | 0.0002999 | 0.009558 |
| 147 | NEURON PROJECTION MORPHOGENESIS | 28 | 402 | 0.0003051 | 0.009658 |
| 148 | POSITIVE REGULATION OF SMALL GTPASE MEDIATED SIGNAL TRANSDUCTION | 7 | 39 | 0.0003088 | 0.009707 |
| 149 | CELLULAR RESPONSE TO ORGANIC CYCLIC COMPOUND | 31 | 465 | 0.0003189 | 0.009959 |
| 150 | EMBRYONIC SKELETAL SYSTEM MORPHOGENESIS | 11 | 93 | 0.0003219 | 0.009984 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | CALCIUM ION BINDING | 56 | 697 | 2.624e-09 | 2.437e-06 |
| 2 | NUCLEIC ACID BINDING TRANSCRIPTION FACTOR ACTIVITY | 75 | 1199 | 2.568e-07 | 0.0001193 |
| 3 | RNA POLYMERASE II TRANSCRIPTION FACTOR ACTIVITY SEQUENCE SPECIFIC DNA BINDING | 47 | 629 | 4.404e-07 | 0.0001364 |
| 4 | TRANSCRIPTION FACTOR BINDING | 40 | 524 | 1.903e-06 | 0.000442 |
| 5 | TRANSCRIPTIONAL ACTIVATOR ACTIVITY RNA POLYMERASE II TRANSCRIPTION REGULATORY REGION SEQUENCE SPECIFIC BINDING | 28 | 315 | 4.064e-06 | 0.0007551 |
| 6 | REGULATORY REGION NUCLEIC ACID BINDING | 53 | 818 | 6.045e-06 | 0.0009359 |
| 7 | ENZYME BINDING | 92 | 1737 | 1.387e-05 | 0.001841 |
| 8 | TRANSCRIPTION FACTOR ACTIVITY RNA POLYMERASE II CORE PROMOTER PROXIMAL REGION SEQUENCE SPECIFIC BINDING | 27 | 328 | 2.412e-05 | 0.002791 |
| 9 | ZINC ION BINDING | 66 | 1155 | 2.704e-05 | 0.002791 |
| 10 | TRANSCRIPTIONAL ACTIVATOR ACTIVITY RNA POLYMERASE II CORE PROMOTER PROXIMAL REGION SEQUENCE SPECIFIC BINDING | 21 | 226 | 3.33e-05 | 0.003093 |
| 11 | MACROMOLECULAR COMPLEX BINDING | 75 | 1399 | 6.329e-05 | 0.005345 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | NEURON PART | 100 | 1265 | 1.884e-15 | 1.1e-12 |
| 2 | SYNAPSE | 68 | 754 | 2.353e-13 | 6.229e-11 |
| 3 | NEURON PROJECTION | 78 | 942 | 3.2e-13 | 6.229e-11 |
| 4 | CELL PROJECTION | 111 | 1786 | 3.033e-10 | 4.428e-08 |
| 5 | SYNAPTIC MEMBRANE | 32 | 261 | 3.934e-10 | 4.595e-08 |
| 6 | SYNAPSE PART | 52 | 610 | 1.323e-09 | 1.288e-07 |
| 7 | PLASMA MEMBRANE REGION | 64 | 929 | 7.129e-08 | 5.947e-06 |
| 8 | DENDRITE | 39 | 451 | 1.107e-07 | 8.08e-06 |
| 9 | AXON | 37 | 418 | 1.308e-07 | 8.49e-06 |
| 10 | CELL JUNCTION | 73 | 1151 | 2.216e-07 | 1.294e-05 |
| 11 | MAIN AXON | 12 | 58 | 4.751e-07 | 2.221e-05 |
| 12 | SOMATODENDRITIC COMPARTMENT | 48 | 650 | 4.664e-07 | 2.221e-05 |
| 13 | MEMBRANE REGION | 71 | 1134 | 5.323e-07 | 2.221e-05 |
| 14 | AXON PART | 24 | 219 | 5.091e-07 | 2.221e-05 |
| 15 | NEURON PROJECTION MEMBRANE | 9 | 36 | 2.437e-06 | 9.489e-05 |
| 16 | AXOLEMMA | 6 | 14 | 3.655e-06 | 0.0001334 |
| 17 | CELL BODY | 37 | 494 | 7.116e-06 | 0.0002445 |
| 18 | POTASSIUM CHANNEL COMPLEX | 13 | 90 | 1.104e-05 | 0.0003556 |
| 19 | CELL PROJECTION PART | 58 | 946 | 1.157e-05 | 0.0003556 |
| 20 | POSTSYNAPSE | 29 | 378 | 4.472e-05 | 0.001306 |
| 21 | CATION CHANNEL COMPLEX | 17 | 167 | 5.925e-05 | 0.001648 |
| 22 | POSTSYNAPTIC MEMBRANE | 19 | 205 | 8.056e-05 | 0.002138 |
| 23 | PRESYNAPSE | 23 | 283 | 0.0001149 | 0.002917 |
| 24 | EXCITATORY SYNAPSE | 18 | 197 | 0.0001474 | 0.003587 |
| 25 | PLASMA MEMBRANE PROTEIN COMPLEX | 34 | 510 | 0.0001665 | 0.003891 |
| 26 | LEADING EDGE MEMBRANE | 14 | 134 | 0.0001979 | 0.004444 |
| 27 | EXTRACELLULAR MATRIX | 29 | 426 | 0.0003503 | 0.007576 |
| 28 | PLATELET DENSE TUBULAR NETWORK | 4 | 11 | 0.0003652 | 0.007617 |
| 29 | RECEPTOR COMPLEX | 24 | 327 | 0.000381 | 0.007672 |
| 30 | CELL LEADING EDGE | 25 | 350 | 0.0004354 | 0.008475 |
Over-represented Pathway
| Num | Pathway | Pathview | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|---|
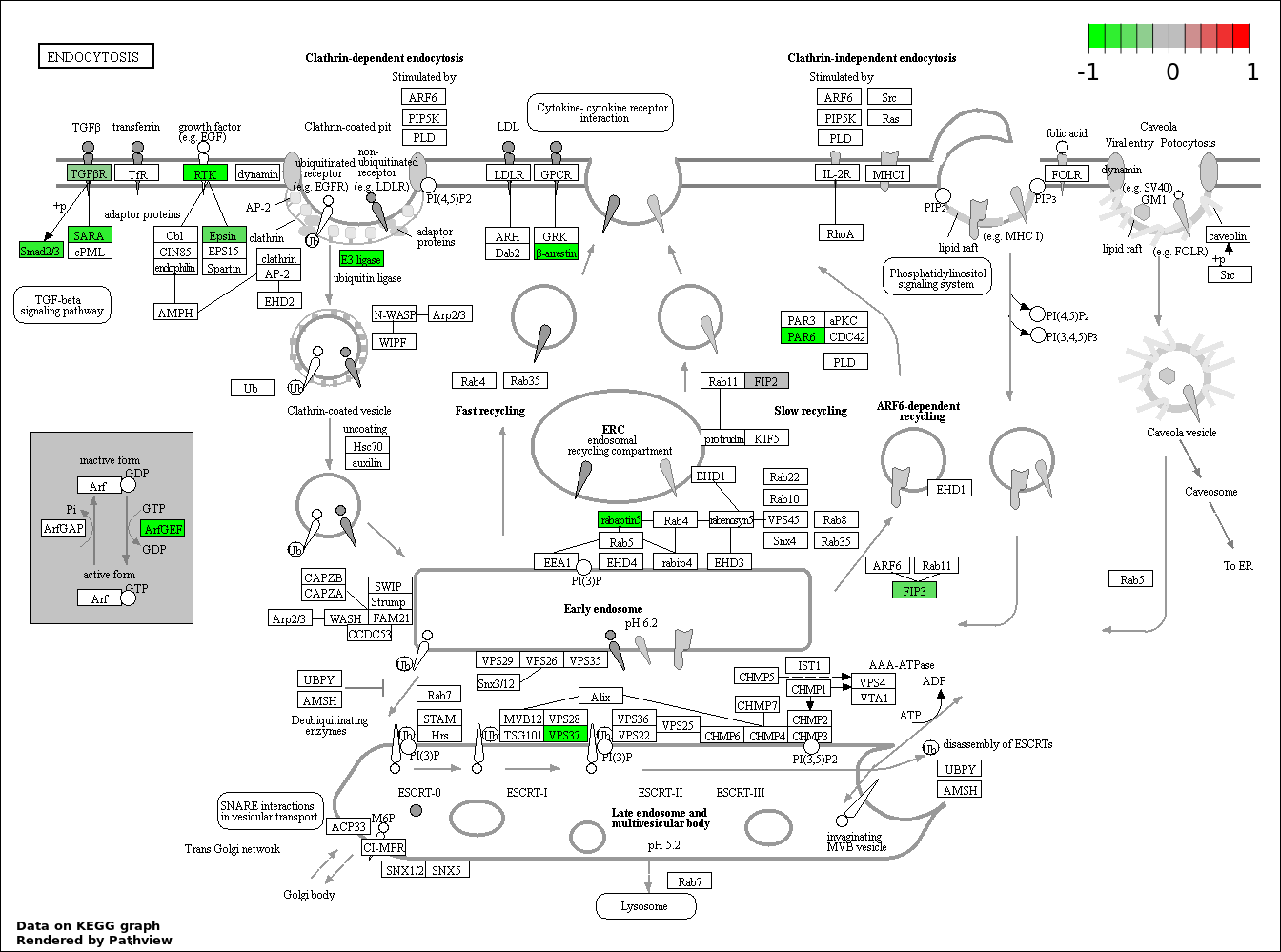
| 1 | hsa04144_Endocytosis | 19 | 203 | 7.062e-05 | 0.006834 | |
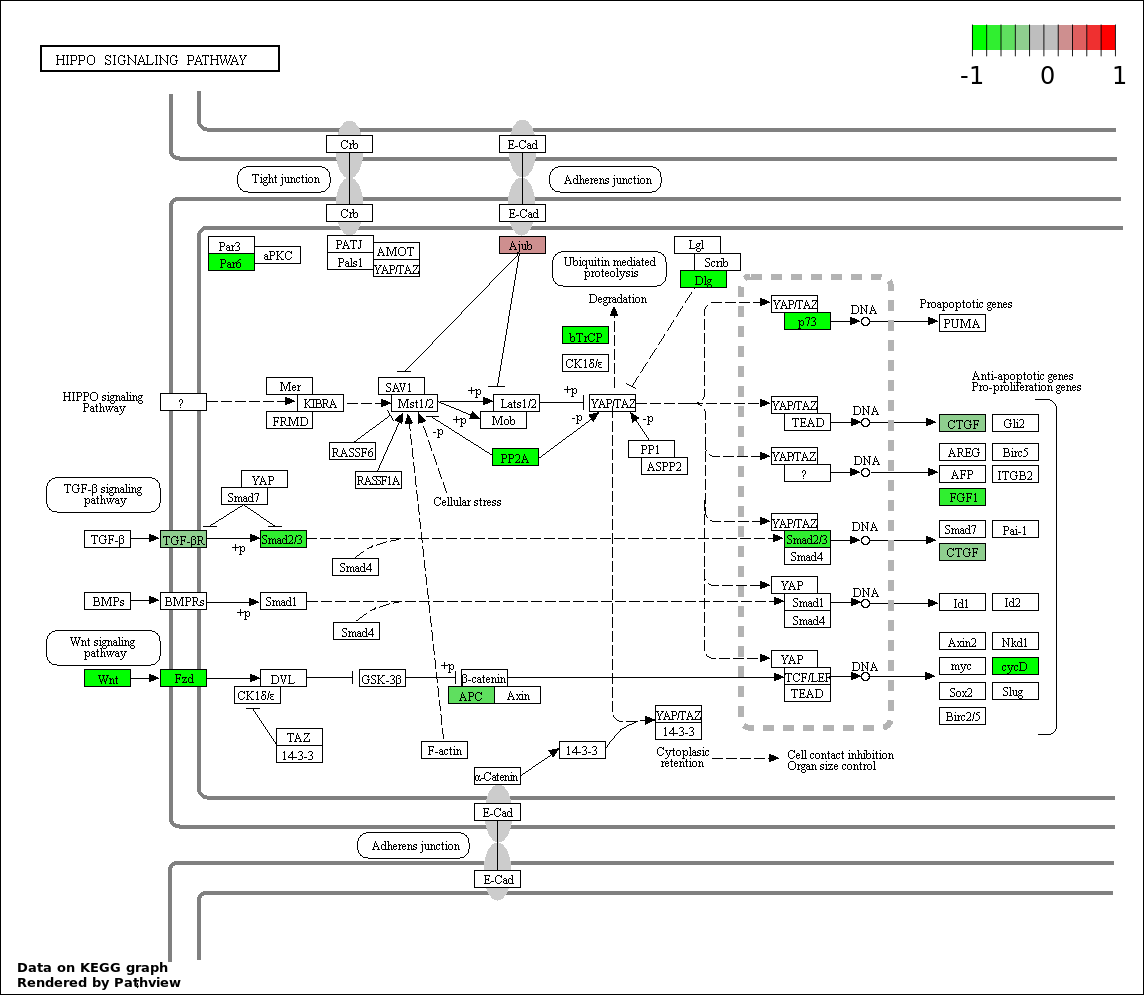
| 2 | hsa04390_Hippo_signaling_pathway | 16 | 154 | 7.593e-05 | 0.006834 | |
| 3 | hsa04115_p53_signaling_pathway | 9 | 69 | 0.0005388 | 0.03233 | |
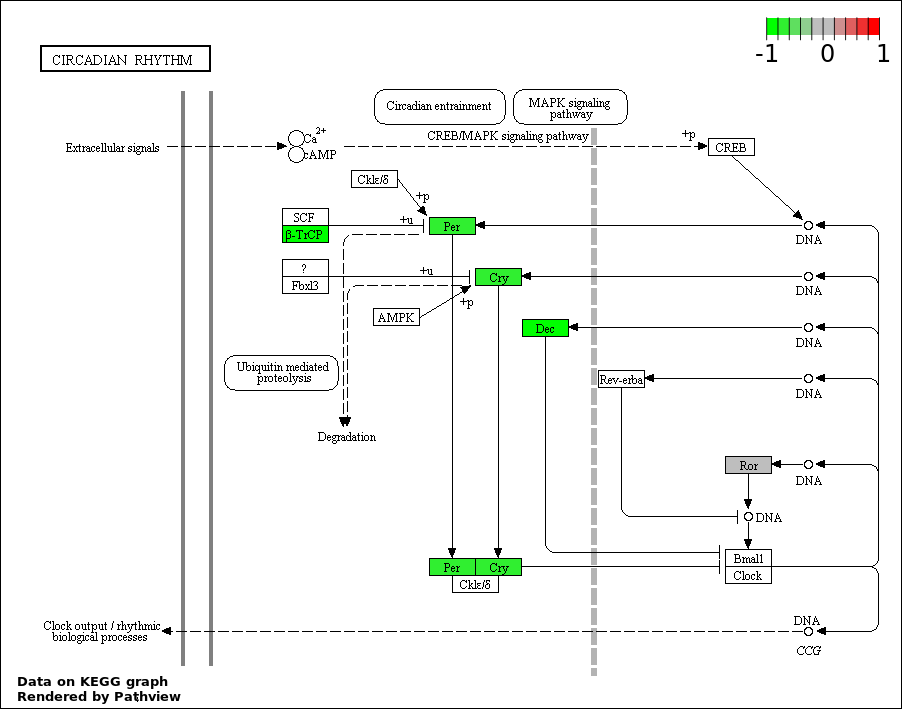
| 4 | hsa04710_Circadian_rhythm_._mammal | 5 | 23 | 0.0009171 | 0.04127 | |
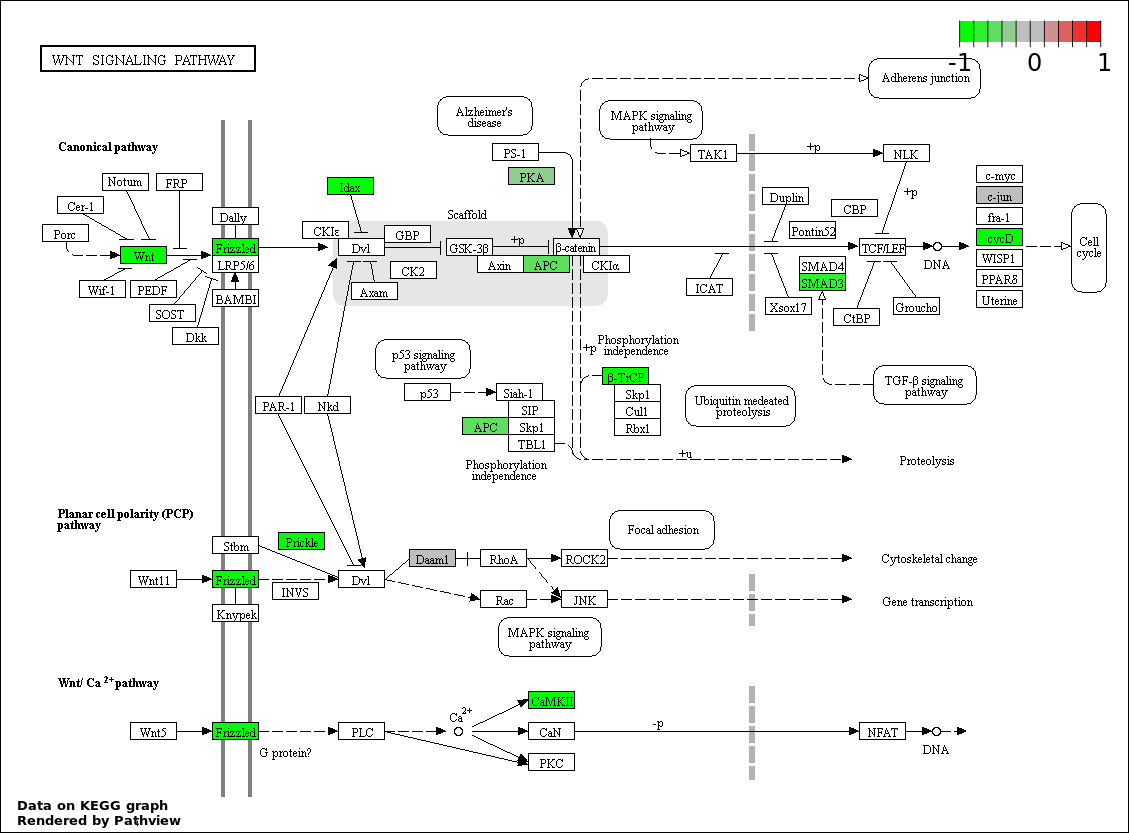
| 5 | hsa04310_Wnt_signaling_pathway | 13 | 151 | 0.002027 | 0.06634 | |
| 6 | hsa04010_MAPK_signaling_pathway | 19 | 268 | 0.002211 | 0.06634 | |
| 7 | hsa04514_Cell_adhesion_molecules_.CAMs. | 11 | 136 | 0.006931 | 0.1754 | |
| 8 | hsa04020_Calcium_signaling_pathway | 13 | 177 | 0.007798 | 0.1754 | |
| 9 | hsa04151_PI3K_AKT_signaling_pathway | 21 | 351 | 0.00942 | 0.1884 | |
| 10 | hsa04970_Salivary_secretion | 8 | 89 | 0.01098 | 0.1915 | |
| 11 | hsa04722_Neurotrophin_signaling_pathway | 10 | 127 | 0.0117 | 0.1915 | |
| 12 | hsa04114_Oocyte_meiosis | 9 | 114 | 0.01606 | 0.2409 | |
| 13 | hsa04120_Ubiquitin_mediated_proteolysis | 10 | 139 | 0.02083 | 0.2885 | |
| 14 | hsa04012_ErbB_signaling_pathway | 7 | 87 | 0.02902 | 0.3731 | |
| 15 | hsa04912_GnRH_signaling_pathway | 7 | 101 | 0.05732 | 0.6456 | |
| 16 | hsa00534_Glycosaminoglycan_biosynthesis_._heparan_sulfate | 3 | 26 | 0.05739 | 0.6456 | |
| 17 | hsa04350_TGF.beta_signaling_pathway | 6 | 85 | 0.07018 | 0.7431 | |
| 18 | hsa00561_Glycerolipid_metabolism | 4 | 50 | 0.09028 | 0.9028 | |
| 19 | hsa04270_Vascular_smooth_muscle_contraction | 7 | 116 | 0.1019 | 0.965 | |
| 20 | hsa04971_Gastric_acid_secretion | 5 | 74 | 0.1081 | 0.9728 | |
| 21 | hsa04340_Hedgehog_signaling_pathway | 4 | 56 | 0.1234 | 0.9829 | |
| 22 | hsa04070_Phosphatidylinositol_signaling_system | 5 | 78 | 0.1273 | 0.9829 | |
| 23 | hsa04916_Melanogenesis | 6 | 101 | 0.1311 | 0.9829 | |
| 24 | hsa04972_Pancreatic_secretion | 6 | 101 | 0.1311 | 0.9829 | |
| 25 | hsa00564_Glycerophospholipid_metabolism | 5 | 80 | 0.1375 | 0.9902 | |
| 26 | hsa04510_Focal_adhesion | 10 | 200 | 0.147 | 1 | |
| 27 | hsa00600_Sphingolipid_metabolism | 3 | 40 | 0.155 | 1 | |
| 28 | hsa04360_Axon_guidance | 7 | 130 | 0.1564 | 1 | |
| 29 | hsa04914_Progesterone.mediated_oocyte_maturation | 5 | 87 | 0.1758 | 1 | |
| 30 | hsa04014_Ras_signaling_pathway | 11 | 236 | 0.1842 | 1 | |
| 31 | hsa02010_ABC_transporters | 3 | 44 | 0.1889 | 1 | |
| 32 | hsa04540_Gap_junction | 5 | 90 | 0.1933 | 1 | |
| 33 | hsa04720_Long.term_potentiation | 4 | 70 | 0.2161 | 1 | |
| 34 | hsa04730_Long.term_depression | 4 | 70 | 0.2161 | 1 | |
| 35 | hsa04976_Bile_secretion | 4 | 71 | 0.2233 | 1 | |
| 36 | hsa00030_Pentose_phosphate_pathway | 2 | 27 | 0.2345 | 1 | |
| 37 | hsa00340_Histidine_metabolism | 2 | 29 | 0.26 | 1 | |
| 38 | hsa04260_Cardiac_muscle_contraction | 4 | 77 | 0.268 | 1 | |
| 39 | hsa04110_Cell_cycle | 6 | 128 | 0.2719 | 1 | |
| 40 | hsa00512_Mucin_type_O.Glycan_biosynthesis | 2 | 30 | 0.2728 | 1 | |
| 41 | hsa04630_Jak.STAT_signaling_pathway | 7 | 155 | 0.2783 | 1 | |
| 42 | hsa00562_Inositol_phosphate_metabolism | 3 | 57 | 0.3078 | 1 | |
| 43 | hsa04512_ECM.receptor_interaction | 4 | 85 | 0.3296 | 1 | |
| 44 | hsa00051_Fructose_and_mannose_metabolism | 2 | 36 | 0.3488 | 1 | |
| 45 | hsa04210_Apoptosis | 4 | 89 | 0.3607 | 1 | |
| 46 | hsa04960_Aldosterone.regulated_sodium_reabsorption | 2 | 42 | 0.4219 | 1 | |
| 47 | hsa04962_Vasopressin.regulated_water_reabsorption | 2 | 44 | 0.4454 | 1 | |
| 48 | hsa04520_Adherens_junction | 3 | 73 | 0.4558 | 1 | |
| 49 | hsa04530_Tight_junction | 5 | 133 | 0.4767 | 1 | |
| 50 | hsa00520_Amino_sugar_and_nucleotide_sugar_metabolism | 2 | 48 | 0.4906 | 1 | |
| 51 | hsa00510_N.Glycan_biosynthesis | 2 | 49 | 0.5016 | 1 | |
| 52 | hsa04910_Insulin_signaling_pathway | 5 | 138 | 0.5092 | 1 | |
| 53 | hsa04150_mTOR_signaling_pathway | 2 | 52 | 0.5335 | 1 | |
| 54 | hsa00230_Purine_metabolism | 5 | 162 | 0.6513 | 1 | |
| 55 | hsa00010_Glycolysis_._Gluconeogenesis | 2 | 65 | 0.655 | 1 | |
| 56 | hsa04610_Complement_and_coagulation_cascades | 2 | 69 | 0.6869 | 1 | |
| 57 | hsa03320_PPAR_signaling_pathway | 2 | 70 | 0.6945 | 1 | |
| 58 | hsa03018_RNA_degradation | 2 | 71 | 0.702 | 1 | |
| 59 | hsa04810_Regulation_of_actin_cytoskeleton | 6 | 214 | 0.7417 | 1 | |
| 60 | hsa04974_Protein_digestion_and_absorption | 2 | 81 | 0.7683 | 1 | |
| 61 | hsa03015_mRNA_surveillance_pathway | 2 | 83 | 0.7799 | 1 | |
| 62 | hsa04380_Osteoclast_differentiation | 3 | 128 | 0.8165 | 1 | |
| 63 | hsa04141_Protein_processing_in_endoplasmic_reticulum | 4 | 168 | 0.8289 | 1 | |
| 64 | hsa00240_Pyrimidine_metabolism | 2 | 99 | 0.8558 | 1 | |
| 65 | hsa04062_Chemokine_signaling_pathway | 4 | 189 | 0.8896 | 1 | |
| 66 | hsa04145_Phagosome | 3 | 156 | 0.9046 | 1 | |
| 67 | hsa04670_Leukocyte_transendothelial_migration | 2 | 117 | 0.9121 | 1 | |
| 68 | hsa00190_Oxidative_phosphorylation | 2 | 132 | 0.9425 | 1 | |
| 69 | hsa04740_Olfactory_transduction | 2 | 388 | 1 | 1 |