This regulatory network was inferred from the input dataset. The miRNAs and mRNAs are
presented as round and rectangle nodes respectively. The numerical value popped up upon mouse over the gene node is the log2 transformed fold-change of the gene expression between the two groups. All of the nodes are clickable, and the detailed information of the miRNAs/mRNAs and related cancer pathway will be displayed in another window. The edges between nodes are supported by both interactions (predicted or experimentally verified) and correlations learnt from cancer dataset. The numerical value popped up upon mouse over the edge is the correlation beat value (effect size) between the two nodes. The experimental evidences of the edges reported in previous cancer studies are highlighted by red/orange color. All of these information can be accessed by the "mouse-over" action. This network shows a full map of the miRNA-mRNA regulation of the input gene list(s), and the hub miRNAs (with the high network degree/betweenness centrality) would be the potential cancer drivers or tumor suppressors. The full result table can be accessed in the "Regulations" tab.
"miRNACancerMAP" is also a network visualization tool for users to draw their regulatory network by personal customization. Users can set the complexity of the network by limiting the number of nodes or edges. And the color of the nodes can be defined by different categories of the mRNAs and miRNAs, such as Gene-Ontology, pathway, and expression status. Users can also select to use network degree or network betweenness centrality to define the node size. And edges can be black or colored by the correlation. Purple edge means negative correlation (mostly found between miRNA and mRNA), and blue edge means positive correlation (found in PPI or miRNA-miRNA sponge effect). We can also add the protein-protein interactions (PPI) into the network. This result will show the cluster of genes regulated by some specific miRNAs. Additionally, miRNA-miRNA edges can be added by the "miRNA sponge" button, presenting some clusters of miRNAs that have the interactions via sponge effect.
miRNA-gene regulations
| Num | microRNA | Gene | miRNA log2FC | miRNA pvalue | Gene log2FC | Gene pvalue | Interaction | Correlation beta | Correlation P-value | PMID | Reported in cancer studies |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | hsa-miR-106a-5p | TGFBR2 | 0.57 | 0.00015 | -2.4 | 0 | miRTarBase; miRNATAP | -0.17 | 0 | 22912877 | MiR-106a inhibits the expression of transforming growth factor-β receptor 2 TGFBR2 leading to increased CRC cell migration and invasion |
| 2 | hsa-miR-106b-5p | TGFBR2 | 1.22 | 0 | -2.4 | 0 | miRNATAP | -0.58 | 0 | NA | |
| 3 | hsa-miR-107 | TGFBR2 | 0.68 | 0 | -2.4 | 0 | miRanda; miRNATAP | -0.53 | 0 | NA | |
| 4 | hsa-miR-130b-3p | TGFBR2 | 1.38 | 0 | -2.4 | 0 | miRNAWalker2 validate; miRNATAP | -0.36 | 0 | 25024357 | Follow-up experiments showed two miRNAs miR-9-5p and miR-130b-3p in this module had increased expression while their target gene TGFBR2 had decreased expression in a cohort of human NSCLC |
| 5 | hsa-miR-17-5p | TGFBR2 | 0.6 | 1.0E-5 | -2.4 | 0 | miRNAWalker2 validate; miRTarBase; TargetScan; miRNATAP | -0.34 | 0 | 25011053; 27120811 | We demonstrate that miR-17 overexpression interferes with the TGFβ-EMT axis and hinders RCC sphere formation; and validated TGFBR2 as a direct and biologically relevant target during this process;MiR-17-5p was found to bind to the 3'UTR of TGFBR2 mRNA and further validation of this specific binding was performed through a reporter assay; An inverse correlation between miR-17-5p and TGFBR2 protein was observed in gastric cancer tissues; Cell studies revealed that miR-17-5p negatively regulated TGFBR2 expression by directly binding to the 3'UTR of TGFBR2 mRNA thereby promoting cell growth and migration; The results of our study suggest a novel regulatory network in gastric cancer mediated by miR-17-5p and TGFBR2 and may indicate that TGFBR2 could serve as a new therapeutic target in gastric cancer |
| 6 | hsa-miR-18a-5p | TGFBR2 | 0.97 | 0 | -2.4 | 0 | miRNAWalker2 validate | -0.23 | 0 | NA | |
| 7 | hsa-miR-19a-3p | TGFBR2 | 0.82 | 0 | -2.4 | 0 | miRNAWalker2 validate; miRNATAP | -0.3 | 0 | NA | |
| 8 | hsa-miR-19b-3p | TGFBR2 | 0.14 | 0.25566 | -2.4 | 0 | miRNAWalker2 validate; miRNATAP | -0.24 | 0 | NA | |
| 9 | hsa-miR-20a-5p | TGFBR2 | 0.45 | 0.0006 | -2.4 | 0 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.35 | 0 | NA | |
| 10 | hsa-miR-20b-5p | TGFBR2 | 0.78 | 9.0E-5 | -2.4 | 0 | miRNATAP | -0.1 | 1.0E-5 | NA | |
| 11 | hsa-miR-21-5p | TGFBR2 | 2.32 | 0 | -2.4 | 0 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.54 | 0 | 24037531 | Androgen receptor and microRNA 21 axis downregulates transforming growth factor beta receptor II TGFBR2 expression in prostate cancer; Our results revealed that miR-21 suppresses TGFBR2 levels by binding to its 3'-UTR and AR signaling further potentiates this effect in both untransformed and transformed human prostate epithelial cells as well as in human prostate cancers; Manipulation of androgen signaling or the expression levels of AR or miR-21 negatively altered TGFBR2 expression in untransformed and transformed human prostate epithelial cells human prostate cancer xenografts and mouse prostate glands; Together these results suggest that the AR and miR-21 axis exerts its oncogenic effects in prostate tumors by downregulating TGFBR2 hence inhibiting the tumor-suppressive activity of TGFβ pathway |
| 12 | hsa-miR-301a-3p | TGFBR2 | 1.32 | 0 | -2.4 | 0 | miRNATAP | -0.32 | 0 | 25551793 | MicroRNA 301a promotes migration and invasion by targeting TGFBR2 in human colorectal cancer; TGFBR2 was identified to be the downstream target of miR-301a; Knockdown of TGFBR2 in cells treated by miR-301a inhibitor elevated the previously abrogated migration and invasion; Our data indicated that miR-301a correlated with the metastatic and invasive ability in human colorectal cancers and miR-301a exerted its role as oncogene by targeting TGFBR2 |
| 13 | hsa-miR-320b | TGFBR2 | 0.41 | 0.00113 | -2.4 | 0 | miRanda; miRNATAP | -0.21 | 0 | NA | |
| 14 | hsa-miR-340-5p | TGFBR2 | 1.09 | 0 | -2.4 | 0 | mirMAP | -0.31 | 0 | NA | |
| 15 | hsa-miR-429 | TGFBR2 | 2.84 | 0 | -2.4 | 0 | miRNATAP | -0.4 | 0 | NA | |
| 16 | hsa-miR-454-3p | TGFBR2 | 1.28 | 0 | -2.4 | 0 | miRNATAP | -0.38 | 0 | NA | |
| 17 | hsa-miR-590-3p | TGFBR2 | 1.27 | 0 | -2.4 | 0 | miRanda | -0.34 | 0 | NA | |
| 18 | hsa-miR-590-5p | TGFBR2 | 0.75 | 0 | -2.4 | 0 | miRNAWalker2 validate; miRTarBase; PITA; miRanda; miRNATAP | -0.29 | 0 | NA | |
| 19 | hsa-miR-9-5p | TGFBR2 | 0.73 | 0.00262 | -2.4 | 0 | miRNATAP | -0.12 | 0 | 25024357 | Follow-up experiments showed two miRNAs miR-9-5p and miR-130b-3p in this module had increased expression while their target gene TGFBR2 had decreased expression in a cohort of human NSCLC |
| 20 | hsa-miR-92a-3p | TGFBR2 | -0.34 | 0.00073 | -2.4 | 0 | miRNAWalker2 validate | -0.22 | 0 | NA | |
| 21 | hsa-miR-93-5p | TGFBR2 | 1.02 | 0 | -2.4 | 0 | miRNAWalker2 validate; miRNATAP | -0.61 | 0 | NA |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | REGULATION OF CELL DEATH | 71 | 1472 | 3.664e-30 | 1.705e-26 |
| 2 | REGULATION OF PROTEIN MODIFICATION PROCESS | 70 | 1710 | 2.154e-25 | 5.012e-22 |
| 3 | CELLULAR RESPONSE TO STRESS | 65 | 1565 | 9.314e-24 | 1.183e-20 |
| 4 | REGULATION OF PHOSPHORUS METABOLIC PROCESS | 66 | 1618 | 1.017e-23 | 1.183e-20 |
| 5 | REGULATION OF TRANSFERASE ACTIVITY | 51 | 946 | 2.38e-23 | 2.129e-20 |
| 6 | REGULATION OF CELL CYCLE | 51 | 949 | 2.745e-23 | 2.129e-20 |
| 7 | RESPONSE TO ENDOGENOUS STIMULUS | 61 | 1450 | 1.794e-22 | 1.192e-19 |
| 8 | REGULATION OF KINASE ACTIVITY | 45 | 776 | 8.147e-22 | 4.739e-19 |
| 9 | RESPONSE TO ORGANIC CYCLIC COMPOUND | 48 | 917 | 2.1e-21 | 1.085e-18 |
| 10 | CELLULAR RESPONSE TO ORGANIC SUBSTANCE | 67 | 1848 | 2.864e-21 | 1.333e-18 |
| 11 | CELL CYCLE | 54 | 1316 | 2.802e-19 | 1.185e-16 |
| 12 | POSITIVE REGULATION OF RESPONSE TO STIMULUS | 65 | 1929 | 6.438e-19 | 2.304e-16 |
| 13 | POSITIVE REGULATION OF PROTEIN METABOLIC PROCESS | 57 | 1492 | 6.124e-19 | 2.304e-16 |
| 14 | RESPONSE TO ABIOTIC STIMULUS | 47 | 1024 | 1.217e-18 | 4.045e-16 |
| 15 | POSITIVE REGULATION OF MOLECULAR FUNCTION | 62 | 1791 | 1.481e-18 | 4.595e-16 |
| 16 | REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 59 | 1656 | 3.431e-18 | 9.978e-16 |
| 17 | NEGATIVE REGULATION OF PROTEIN METABOLIC PROCESS | 47 | 1087 | 1.291e-17 | 3.535e-15 |
| 18 | CELL DEATH | 45 | 1001 | 1.683e-17 | 4.351e-15 |
| 19 | CELLULAR RESPONSE TO DNA DAMAGE STIMULUS | 38 | 720 | 4.492e-17 | 1.1e-14 |
| 20 | POSITIVE REGULATION OF CELL DEATH | 35 | 605 | 5.508e-17 | 1.281e-14 |
| 21 | PHOSPHORYLATION | 49 | 1228 | 5.843e-17 | 1.295e-14 |
| 22 | NEGATIVE REGULATION OF CELL CYCLE | 30 | 433 | 9.191e-17 | 1.944e-14 |
| 23 | NEGATIVE REGULATION OF CELL DEATH | 41 | 872 | 1.248e-16 | 2.524e-14 |
| 24 | DNA METABOLIC PROCESS | 38 | 758 | 2.43e-16 | 4.712e-14 |
| 25 | RESPONSE TO OXYGEN CONTAINING COMPOUND | 51 | 1381 | 2.838e-16 | 5.282e-14 |
| 26 | POSITIVE REGULATION OF PROTEIN MODIFICATION PROCESS | 46 | 1135 | 3.612e-16 | 6.464e-14 |
| 27 | NEGATIVE REGULATION OF CELL COMMUNICATION | 47 | 1192 | 4.612e-16 | 7.947e-14 |
| 28 | RESPONSE TO ALCOHOL | 27 | 362 | 5.801e-16 | 9.64e-14 |
| 29 | REGULATION OF RESPONSE TO STRESS | 52 | 1468 | 7.547e-16 | 1.211e-13 |
| 30 | REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 30 | 470 | 8.513e-16 | 1.32e-13 |
| 31 | CELL CYCLE PROCESS | 44 | 1081 | 1.552e-15 | 2.33e-13 |
| 32 | TRANSFORMING GROWTH FACTOR BETA RECEPTOR SIGNALING PATHWAY | 16 | 95 | 1.909e-15 | 2.775e-13 |
| 33 | ENZYME LINKED RECEPTOR PROTEIN SIGNALING PATHWAY | 35 | 689 | 2.909e-15 | 4.082e-13 |
| 34 | PHOSPHATE CONTAINING COMPOUND METABOLIC PROCESS | 60 | 1977 | 2.983e-15 | 4.082e-13 |
| 35 | POSITIVE REGULATION OF BIOSYNTHETIC PROCESS | 57 | 1805 | 3.341e-15 | 4.442e-13 |
| 36 | RESPONSE TO STEROID HORMONE | 30 | 497 | 3.803e-15 | 4.915e-13 |
| 37 | POSITIVE REGULATION OF TRANSFERASE ACTIVITY | 33 | 616 | 4.384e-15 | 5.513e-13 |
| 38 | TRANSMEMBRANE RECEPTOR PROTEIN SERINE THREONINE KINASE SIGNALING PATHWAY | 20 | 190 | 6.367e-15 | 7.797e-13 |
| 39 | POSITIVE REGULATION OF PHOSPHATE METABOLIC PROCESS | 42 | 1036 | 8.868e-15 | 1.032e-12 |
| 40 | POSITIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS | 42 | 1036 | 8.868e-15 | 1.032e-12 |
| 41 | PROTEIN PHOSPHORYLATION | 40 | 944 | 1.014e-14 | 1.151e-12 |
| 42 | INTRACELLULAR SIGNAL TRANSDUCTION | 52 | 1572 | 1.228e-14 | 1.329e-12 |
| 43 | REGULATION OF CELL CYCLE PROCESS | 31 | 558 | 1.218e-14 | 1.329e-12 |
| 44 | POSITIVE REGULATION OF CATALYTIC ACTIVITY | 51 | 1518 | 1.283e-14 | 1.357e-12 |
| 45 | REGULATION OF CELL CYCLE ARREST | 16 | 108 | 1.569e-14 | 1.622e-12 |
| 46 | CELL CYCLE PHASE TRANSITION | 22 | 255 | 1.733e-14 | 1.702e-12 |
| 47 | REGULATION OF CATABOLIC PROCESS | 35 | 731 | 1.704e-14 | 1.702e-12 |
| 48 | CELLULAR RESPONSE TO ENDOGENOUS STIMULUS | 41 | 1008 | 1.756e-14 | 1.702e-12 |
| 49 | REGULATION OF CELLULAR RESPONSE TO STRESS | 34 | 691 | 1.928e-14 | 1.831e-12 |
| 50 | NEGATIVE REGULATION OF RESPONSE TO STIMULUS | 47 | 1360 | 6.482e-14 | 6.032e-12 |
| 51 | POSITIVE REGULATION OF CELL COMMUNICATION | 50 | 1532 | 7.667e-14 | 6.995e-12 |
| 52 | DNA REPAIR | 28 | 480 | 7.966e-14 | 7.128e-12 |
| 53 | CELL CYCLE CHECKPOINT | 19 | 194 | 1.168e-13 | 1.025e-11 |
| 54 | POSITIVE REGULATION OF GENE EXPRESSION | 53 | 1733 | 1.52e-13 | 1.31e-11 |
| 55 | RESPONSE TO LIPID | 37 | 888 | 1.982e-13 | 1.639e-11 |
| 56 | INTERSPECIES INTERACTION BETWEEN ORGANISMS | 32 | 662 | 2.008e-13 | 1.639e-11 |
| 57 | SYMBIOSIS ENCOMPASSING MUTUALISM THROUGH PARASITISM | 32 | 662 | 2.008e-13 | 1.639e-11 |
| 58 | SIGNAL TRANSDUCTION BY P53 CLASS MEDIATOR | 16 | 127 | 2.127e-13 | 1.707e-11 |
| 59 | RESPONSE TO DRUG | 26 | 431 | 3.07e-13 | 2.421e-11 |
| 60 | POSITIVE REGULATION OF KINASE ACTIVITY | 27 | 482 | 6.012e-13 | 4.663e-11 |
| 61 | REGULATION OF DNA METABOLIC PROCESS | 23 | 340 | 7.461e-13 | 5.691e-11 |
| 62 | CELL CYCLE G2 M PHASE TRANSITION | 16 | 138 | 7.911e-13 | 5.937e-11 |
| 63 | MITOTIC CELL CYCLE CHECKPOINT | 16 | 139 | 8.861e-13 | 6.544e-11 |
| 64 | SIGNAL TRANSDUCTION IN RESPONSE TO DNA DAMAGE | 14 | 96 | 9.047e-13 | 6.577e-11 |
| 65 | RESPONSE TO HORMONE | 36 | 893 | 1.16e-12 | 8.304e-11 |
| 66 | REGULATION OF PROTEOLYSIS | 32 | 711 | 1.368e-12 | 9.647e-11 |
| 67 | RESPONSE TO TRANSFORMING GROWTH FACTOR BETA | 16 | 144 | 1.541e-12 | 1.07e-10 |
| 68 | MITOTIC CELL CYCLE | 33 | 766 | 1.929e-12 | 1.32e-10 |
| 69 | REGULATION OF CELL PROLIFERATION | 47 | 1496 | 2.021e-12 | 1.363e-10 |
| 70 | RESPONSE TO GROWTH FACTOR | 26 | 475 | 2.819e-12 | 1.874e-10 |
| 71 | TISSUE DEVELOPMENT | 47 | 1518 | 3.385e-12 | 2.219e-10 |
| 72 | POSITIVE REGULATION OF CELL CYCLE | 22 | 332 | 3.654e-12 | 2.361e-10 |
| 73 | REGULATION OF MAPK CASCADE | 30 | 660 | 5.812e-12 | 3.705e-10 |
| 74 | RESPONSE TO RADIATION | 24 | 413 | 5.912e-12 | 3.717e-10 |
| 75 | NEGATIVE REGULATION OF CELL CYCLE PROCESS | 18 | 214 | 7.126e-12 | 4.421e-10 |
| 76 | RESPONSE TO ESTROGEN | 18 | 218 | 9.734e-12 | 5.959e-10 |
| 77 | REGULATION OF MITOTIC CELL CYCLE | 25 | 468 | 1.297e-11 | 7.835e-10 |
| 78 | NEGATIVE REGULATION OF MOLECULAR FUNCTION | 38 | 1079 | 1.464e-11 | 8.734e-10 |
| 79 | NEGATIVE REGULATION OF MITOTIC CELL CYCLE | 17 | 199 | 2.181e-11 | 1.285e-09 |
| 80 | POSITIVE REGULATION OF CELLULAR COMPONENT ORGANIZATION | 39 | 1152 | 2.473e-11 | 1.438e-09 |
| 81 | NEGATIVE REGULATION OF PROTEIN MODIFICATION PROCESS | 28 | 616 | 3.173e-11 | 1.823e-09 |
| 82 | NEGATIVE REGULATION OF NITROGEN COMPOUND METABOLIC PROCESS | 45 | 1517 | 4.563e-11 | 2.589e-09 |
| 83 | RESPONSE TO TOXIC SUBSTANCE | 18 | 241 | 5.184e-11 | 2.906e-09 |
| 84 | REPRODUCTION | 41 | 1297 | 5.825e-11 | 3.227e-09 |
| 85 | POSITIVE REGULATION OF CELL CYCLE PROCESS | 18 | 247 | 7.778e-11 | 4.258e-09 |
| 86 | CELL DEVELOPMENT | 43 | 1426 | 8.105e-11 | 4.385e-09 |
| 87 | POSITIVE REGULATION OF IMMUNE RESPONSE | 26 | 563 | 1.214e-10 | 6.493e-09 |
| 88 | RESPONSE TO INORGANIC SUBSTANCE | 24 | 479 | 1.281e-10 | 6.772e-09 |
| 89 | POSITIVE REGULATION OF ORGANELLE ORGANIZATION | 26 | 573 | 1.777e-10 | 9.289e-09 |
| 90 | INTRACELLULAR RECEPTOR SIGNALING PATHWAY | 15 | 168 | 1.835e-10 | 9.485e-09 |
| 91 | REGULATION OF ORGANELLE ORGANIZATION | 38 | 1178 | 1.881e-10 | 9.616e-09 |
| 92 | RESPONSE TO IONIZING RADIATION | 14 | 145 | 2.668e-10 | 1.349e-08 |
| 93 | NEGATIVE REGULATION OF CELL CYCLE PHASE TRANSITION | 14 | 146 | 2.924e-10 | 1.448e-08 |
| 94 | DNA INTEGRITY CHECKPOINT | 14 | 146 | 2.924e-10 | 1.448e-08 |
| 95 | DNA BIOSYNTHETIC PROCESS | 13 | 121 | 3.1e-10 | 1.518e-08 |
| 96 | CELL DIVISION | 23 | 460 | 3.332e-10 | 1.608e-08 |
| 97 | NEGATIVE REGULATION OF CATALYTIC ACTIVITY | 31 | 829 | 3.352e-10 | 1.608e-08 |
| 98 | MITOTIC DNA INTEGRITY CHECKPOINT | 12 | 100 | 4.149e-10 | 1.97e-08 |
| 99 | RESPONSE TO OXYGEN LEVELS | 19 | 311 | 4.62e-10 | 2.171e-08 |
| 100 | REGULATION OF PROTEIN MODIFICATION BY SMALL PROTEIN CONJUGATION OR REMOVAL | 18 | 280 | 5.991e-10 | 2.788e-08 |
| 101 | POSITIVE REGULATION OF DNA METABOLIC PROCESS | 15 | 185 | 7.119e-10 | 3.28e-08 |
| 102 | REGULATION OF CELL CYCLE PHASE TRANSITION | 19 | 321 | 7.854e-10 | 3.583e-08 |
| 103 | RESPONSE TO NITROGEN COMPOUND | 31 | 859 | 7.944e-10 | 3.588e-08 |
| 104 | RESPONSE TO CYTOKINE | 28 | 714 | 9.212e-10 | 4.121e-08 |
| 105 | REGULATION OF APOPTOTIC SIGNALING PATHWAY | 20 | 363 | 9.614e-10 | 4.26e-08 |
| 106 | POSITIVE REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 18 | 289 | 9.952e-10 | 4.328e-08 |
| 107 | POSITIVE REGULATION OF CELL CYCLE ARREST | 11 | 85 | 9.946e-10 | 4.328e-08 |
| 108 | REGULATION OF MULTICELLULAR ORGANISMAL DEVELOPMENT | 45 | 1672 | 1.061e-09 | 4.572e-08 |
| 109 | CELL PROLIFERATION | 27 | 672 | 1.122e-09 | 4.791e-08 |
| 110 | NEGATIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS | 24 | 541 | 1.477e-09 | 6.193e-08 |
| 111 | NEGATIVE REGULATION OF PHOSPHATE METABOLIC PROCESS | 24 | 541 | 1.477e-09 | 6.193e-08 |
| 112 | REGULATION OF HYDROLASE ACTIVITY | 39 | 1327 | 1.522e-09 | 6.323e-08 |
| 113 | AGING | 17 | 264 | 1.787e-09 | 7.358e-08 |
| 114 | EMBRYO DEVELOPMENT | 31 | 894 | 2.068e-09 | 8.441e-08 |
| 115 | CELLULAR RESPONSE TO ORGANIC CYCLIC COMPOUND | 22 | 465 | 2.308e-09 | 9.337e-08 |
| 116 | HEART DEVELOPMENT | 22 | 466 | 2.401e-09 | 9.63e-08 |
| 117 | ACTIVATION OF IMMUNE RESPONSE | 21 | 427 | 2.755e-09 | 1.096e-07 |
| 118 | RESPONSE TO ESTRADIOL | 13 | 146 | 3.211e-09 | 1.266e-07 |
| 119 | TRANSCRIPTION COUPLED NUCLEOTIDE EXCISION REPAIR | 10 | 73 | 3.313e-09 | 1.284e-07 |
| 120 | G1 DNA DAMAGE CHECKPOINT | 10 | 73 | 3.313e-09 | 1.284e-07 |
| 121 | NEGATIVE REGULATION OF TRANSFERASE ACTIVITY | 19 | 351 | 3.448e-09 | 1.326e-07 |
| 122 | REGULATION OF PROTEIN CATABOLIC PROCESS | 20 | 393 | 3.762e-09 | 1.435e-07 |
| 123 | NEGATIVE REGULATION OF GENE EXPRESSION | 41 | 1493 | 3.885e-09 | 1.47e-07 |
| 124 | POSITIVE REGULATION OF CELL PROLIFERATION | 29 | 814 | 3.98e-09 | 1.494e-07 |
| 125 | POSITIVE REGULATION OF IMMUNE SYSTEM PROCESS | 30 | 867 | 4.08e-09 | 1.519e-07 |
| 126 | ERBB2 SIGNALING PATHWAY | 8 | 39 | 4.609e-09 | 1.689e-07 |
| 127 | NUCLEOTIDE EXCISION REPAIR DNA INCISION | 8 | 39 | 4.609e-09 | 1.689e-07 |
| 128 | NEGATIVE REGULATION OF CELL CYCLE G1 S PHASE TRANSITION | 11 | 98 | 4.657e-09 | 1.693e-07 |
| 129 | REGULATION OF MAP KINASE ACTIVITY | 18 | 319 | 4.754e-09 | 1.715e-07 |
| 130 | REGULATION OF NEURON DEATH | 16 | 252 | 6.684e-09 | 2.392e-07 |
| 131 | APOPTOTIC SIGNALING PATHWAY | 17 | 289 | 6.983e-09 | 2.48e-07 |
| 132 | REPRODUCTIVE SYSTEM DEVELOPMENT | 20 | 408 | 7.099e-09 | 2.502e-07 |
| 133 | GROWTH | 20 | 410 | 7.709e-09 | 2.697e-07 |
| 134 | REGENERATION | 13 | 161 | 1.057e-08 | 3.669e-07 |
| 135 | REGULATION OF SIGNAL TRANSDUCTION BY P53 CLASS MEDIATOR | 13 | 162 | 1.139e-08 | 3.925e-07 |
| 136 | NEGATIVE REGULATION OF PHOSPHORYLATION | 20 | 422 | 1.251e-08 | 4.281e-07 |
| 137 | NUCLEOTIDE EXCISION REPAIR PREINCISION COMPLEX ASSEMBLY | 7 | 29 | 1.292e-08 | 4.39e-07 |
| 138 | RESPONSE TO EXTERNAL STIMULUS | 45 | 1821 | 1.486e-08 | 5.011e-07 |
| 139 | CELL CYCLE G1 S PHASE TRANSITION | 11 | 111 | 1.754e-08 | 5.828e-07 |
| 140 | G1 S TRANSITION OF MITOTIC CELL CYCLE | 11 | 111 | 1.754e-08 | 5.828e-07 |
| 141 | NEGATIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 31 | 983 | 1.9e-08 | 6.271e-07 |
| 142 | POSITIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 38 | 1395 | 2.005e-08 | 6.571e-07 |
| 143 | MACROMOLECULAR COMPLEX ASSEMBLY | 38 | 1398 | 2.123e-08 | 6.908e-07 |
| 144 | REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 44 | 1784 | 2.372e-08 | 7.666e-07 |
| 145 | POSITIVE REGULATION OF NEURON DEATH | 9 | 67 | 2.462e-08 | 7.813e-07 |
| 146 | ERROR FREE TRANSLESION SYNTHESIS | 6 | 19 | 2.468e-08 | 7.813e-07 |
| 147 | ERROR PRONE TRANSLESION SYNTHESIS | 6 | 19 | 2.468e-08 | 7.813e-07 |
| 148 | REGULATION OF TRANSMEMBRANE RECEPTOR PROTEIN SERINE THREONINE KINASE SIGNALING PATHWAY | 14 | 207 | 2.741e-08 | 8.618e-07 |
| 149 | EPITHELIUM DEVELOPMENT | 30 | 945 | 2.862e-08 | 8.938e-07 |
| 150 | REGULATION OF BINDING | 16 | 283 | 3.43e-08 | 1.064e-06 |
| 151 | NEGATIVE REGULATION OF DEVELOPMENTAL PROCESS | 27 | 801 | 4.536e-08 | 1.398e-06 |
| 152 | CELLULAR CATABOLIC PROCESS | 36 | 1322 | 5.104e-08 | 1.562e-06 |
| 153 | NEURON APOPTOTIC PROCESS | 7 | 35 | 5.289e-08 | 1.608e-06 |
| 154 | EMBRYO DEVELOPMENT ENDING IN BIRTH OR EGG HATCHING | 22 | 554 | 5.384e-08 | 1.627e-06 |
| 155 | REGULATION OF CYCLIN DEPENDENT PROTEIN KINASE ACTIVITY | 10 | 97 | 5.429e-08 | 1.63e-06 |
| 156 | RESPONSE TO METAL ION | 17 | 333 | 5.608e-08 | 1.673e-06 |
| 157 | CATABOLIC PROCESS | 43 | 1773 | 5.76e-08 | 1.707e-06 |
| 158 | DNA SYNTHESIS INVOLVED IN DNA REPAIR | 9 | 74 | 5.985e-08 | 1.762e-06 |
| 159 | RESPONSE TO UV | 11 | 126 | 6.595e-08 | 1.93e-06 |
| 160 | POSTREPLICATION REPAIR | 8 | 54 | 6.842e-08 | 1.99e-06 |
| 161 | POSITIVE REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 28 | 876 | 7.588e-08 | 2.18e-06 |
| 162 | EXTRACELLULAR MATRIX DISASSEMBLY | 9 | 76 | 7.581e-08 | 2.18e-06 |
| 163 | POSITIVE REGULATION OF PROTEIN LOCALIZATION TO NUCLEUS | 11 | 129 | 8.41e-08 | 2.401e-06 |
| 164 | REGULATION OF NEURON APOPTOTIC PROCESS | 13 | 192 | 8.6e-08 | 2.44e-06 |
| 165 | PROTEIN LOCALIZATION | 43 | 1805 | 9.558e-08 | 2.695e-06 |
| 166 | POSITIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 30 | 1004 | 1.082e-07 | 3.034e-06 |
| 167 | REGULATION OF CELL DIFFERENTIATION | 38 | 1492 | 1.16e-07 | 3.233e-06 |
| 168 | NUCLEOTIDE EXCISION REPAIR DNA GAP FILLING | 6 | 24 | 1.174e-07 | 3.251e-06 |
| 169 | CHROMOSOME ORGANIZATION | 30 | 1009 | 1.205e-07 | 3.28e-06 |
| 170 | REGULATION OF WNT SIGNALING PATHWAY | 16 | 310 | 1.205e-07 | 3.28e-06 |
| 171 | RESPONSE TO TUMOR NECROSIS FACTOR | 14 | 233 | 1.201e-07 | 3.28e-06 |
| 172 | CARDIOVASCULAR SYSTEM DEVELOPMENT | 26 | 788 | 1.254e-07 | 3.373e-06 |
| 173 | CIRCULATORY SYSTEM DEVELOPMENT | 26 | 788 | 1.254e-07 | 3.373e-06 |
| 174 | NEGATIVE REGULATION OF CELLULAR COMPONENT ORGANIZATION | 24 | 684 | 1.304e-07 | 3.487e-06 |
| 175 | REGULATION OF CELLULAR PROTEIN CATABOLIC PROCESS | 15 | 274 | 1.419e-07 | 3.772e-06 |
| 176 | EPITHELIAL CELL APOPTOTIC PROCESS | 6 | 25 | 1.531e-07 | 4.048e-06 |
| 177 | ORGAN REGENERATION | 9 | 83 | 1.645e-07 | 4.301e-06 |
| 178 | CELLULAR RESPONSE TO OXYGEN CONTAINING COMPOUND | 26 | 799 | 1.642e-07 | 4.301e-06 |
| 179 | POSITIVE REGULATION OF CELLULAR PROTEIN LOCALIZATION | 17 | 360 | 1.714e-07 | 4.456e-06 |
| 180 | ACTIVATION OF INNATE IMMUNE RESPONSE | 13 | 204 | 1.744e-07 | 4.507e-06 |
| 181 | REGULATION OF IMMUNE RESPONSE | 27 | 858 | 1.814e-07 | 4.662e-06 |
| 182 | POSITIVE REGULATION OF APOPTOTIC SIGNALING PATHWAY | 12 | 171 | 1.853e-07 | 4.738e-06 |
| 183 | PROTEOLYSIS | 33 | 1208 | 1.87e-07 | 4.754e-06 |
| 184 | TRANSMEMBRANE RECEPTOR PROTEIN TYROSINE KINASE SIGNALING PATHWAY | 20 | 498 | 1.88e-07 | 4.755e-06 |
| 185 | POSITIVE REGULATION OF PROTEOLYSIS | 17 | 363 | 1.928e-07 | 4.85e-06 |
| 186 | FC RECEPTOR SIGNALING PATHWAY | 13 | 206 | 1.952e-07 | 4.883e-06 |
| 187 | POSITIVE REGULATION OF MAP KINASE ACTIVITY | 13 | 207 | 2.065e-07 | 5.137e-06 |
| 188 | REGULATION OF CELLULAR LOCALIZATION | 34 | 1277 | 2.167e-07 | 5.345e-06 |
| 189 | NEUROGENESIS | 36 | 1402 | 2.181e-07 | 5.345e-06 |
| 190 | DNA REPLICATION | 13 | 208 | 2.183e-07 | 5.345e-06 |
| 191 | REGULATION OF IMMUNE SYSTEM PROCESS | 36 | 1403 | 2.219e-07 | 5.407e-06 |
| 192 | DEVELOPMENTAL PROCESS INVOLVED IN REPRODUCTION | 22 | 602 | 2.268e-07 | 5.496e-06 |
| 193 | NUCLEOTIDE EXCISION REPAIR | 10 | 113 | 2.329e-07 | 5.614e-06 |
| 194 | CELLULAR RESPONSE TO OXYGEN LEVELS | 11 | 143 | 2.413e-07 | 5.787e-06 |
| 195 | REGULATION OF RELEASE OF CYTOCHROME C FROM MITOCHONDRIA | 7 | 44 | 2.79e-07 | 6.622e-06 |
| 196 | REGULATION OF INTRINSIC APOPTOTIC SIGNALING PATHWAY | 11 | 145 | 2.777e-07 | 6.622e-06 |
| 197 | CELLULAR MACROMOLECULE LOCALIZATION | 33 | 1234 | 3.04e-07 | 7.18e-06 |
| 198 | NUCLEOTIDE BINDING DOMAIN LEUCINE RICH REPEAT CONTAINING RECEPTOR SIGNALING PATHWAY | 6 | 28 | 3.176e-07 | 7.421e-06 |
| 199 | POSITIVE REGULATION OF RELEASE OF CYTOCHROME C FROM MITOCHONDRIA | 6 | 28 | 3.176e-07 | 7.421e-06 |
| 200 | REGULATION OF CELL CYCLE G1 S PHASE TRANSITION | 11 | 147 | 3.19e-07 | 7.421e-06 |
| 201 | CELLULAR COMPONENT DISASSEMBLY | 20 | 515 | 3.206e-07 | 7.423e-06 |
| 202 | RESPONSE TO KETONE | 12 | 182 | 3.64e-07 | 8.386e-06 |
| 203 | RHYTHMIC PROCESS | 15 | 298 | 4.171e-07 | 9.561e-06 |
| 204 | NEURON DEATH | 7 | 47 | 4.462e-07 | 1.018e-05 |
| 205 | REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY | 11 | 153 | 4.771e-07 | 1.083e-05 |
| 206 | IMMUNE SYSTEM PROCESS | 44 | 1984 | 4.917e-07 | 1.111e-05 |
| 207 | NEGATIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 24 | 740 | 5.427e-07 | 1.22e-05 |
| 208 | NEGATIVE REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 18 | 437 | 5.51e-07 | 1.233e-05 |
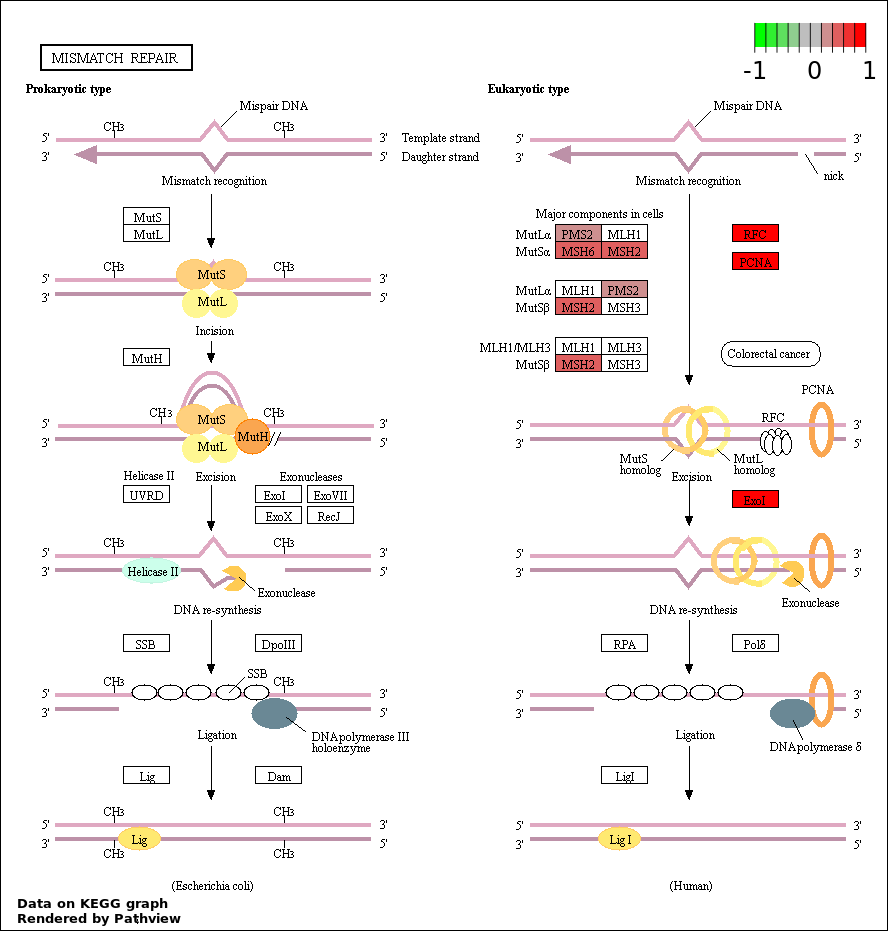
| 209 | MISMATCH REPAIR | 6 | 31 | 6.051e-07 | 1.347e-05 |
| 210 | RESPONSE TO REACTIVE OXYGEN SPECIES | 12 | 191 | 6.108e-07 | 1.353e-05 |
| 211 | RESPONSE TO EXTRACELLULAR STIMULUS | 18 | 441 | 6.286e-07 | 1.386e-05 |
| 212 | RESPONSE TO OXIDATIVE STRESS | 16 | 352 | 6.649e-07 | 1.459e-05 |
| 213 | POSITIVE REGULATION OF BINDING | 10 | 127 | 6.936e-07 | 1.515e-05 |
| 214 | EXTRINSIC APOPTOTIC SIGNALING PATHWAY | 9 | 99 | 7.534e-07 | 1.623e-05 |
| 215 | REGULATION OF TRANSFORMING GROWTH FACTOR BETA RECEPTOR SIGNALING PATHWAY | 9 | 99 | 7.534e-07 | 1.623e-05 |
| 216 | REGULATION OF CELLULAR RESPONSE TO TRANSFORMING GROWTH FACTOR BETA STIMULUS | 9 | 99 | 7.534e-07 | 1.623e-05 |
| 217 | NEURON PROJECTION DEVELOPMENT | 20 | 545 | 7.787e-07 | 1.67e-05 |
| 218 | REGULATION OF DNA REPLICATION | 11 | 161 | 7.935e-07 | 1.694e-05 |
| 219 | MACROMOLECULE CATABOLIC PROCESS | 27 | 926 | 8.067e-07 | 1.706e-05 |
| 220 | POSITIVE REGULATION OF PROTEIN MODIFICATION BY SMALL PROTEIN CONJUGATION OR REMOVAL | 12 | 196 | 8.042e-07 | 1.706e-05 |
| 221 | REGULATION OF LIGASE ACTIVITY | 10 | 130 | 8.606e-07 | 1.812e-05 |
| 222 | SIGNAL TRANSDUCTION IN ABSENCE OF LIGAND | 6 | 33 | 8.95e-07 | 1.859e-05 |
| 223 | EXTRINSIC APOPTOTIC SIGNALING PATHWAY IN ABSENCE OF LIGAND | 6 | 33 | 8.95e-07 | 1.859e-05 |
| 224 | CYTOPLASMIC PATTERN RECOGNITION RECEPTOR SIGNALING PATHWAY | 6 | 33 | 8.95e-07 | 1.859e-05 |
| 225 | REGULATION OF CELLULAR PROTEIN LOCALIZATION | 20 | 552 | 9.49e-07 | 1.962e-05 |
| 226 | POSITIVE REGULATION OF NF KAPPAB TRANSCRIPTION FACTOR ACTIVITY | 10 | 132 | 9.906e-07 | 2.032e-05 |
| 227 | POSITIVE REGULATION OF TRANSPORT | 27 | 936 | 9.914e-07 | 2.032e-05 |
| 228 | NEGATIVE REGULATION OF APOPTOTIC SIGNALING PATHWAY | 12 | 200 | 9.962e-07 | 2.033e-05 |
| 229 | CELLULAR RESPONSE TO CYTOKINE STIMULUS | 21 | 606 | 1.024e-06 | 2.081e-05 |
| 230 | CARTILAGE DEVELOPMENT INVOLVED IN ENDOCHONDRAL BONE MORPHOGENESIS | 5 | 19 | 1.062e-06 | 2.149e-05 |
| 231 | NEGATIVE REGULATION OF CELL DIFFERENTIATION | 21 | 609 | 1.108e-06 | 2.225e-05 |
| 232 | RESPONSE TO LIGHT STIMULUS | 14 | 280 | 1.11e-06 | 2.225e-05 |
| 233 | REGULATION OF CELLULAR COMPONENT MOVEMENT | 24 | 771 | 1.122e-06 | 2.241e-05 |
| 234 | IMMUNE RESPONSE REGULATING CELL SURFACE RECEPTOR SIGNALING PATHWAY | 15 | 323 | 1.151e-06 | 2.288e-05 |
| 235 | REGULATION OF PROTEIN BINDING | 11 | 168 | 1.209e-06 | 2.394e-05 |
| 236 | RESPONSE TO ETHANOL | 10 | 136 | 1.303e-06 | 2.568e-05 |
| 237 | REGULATION OF PROTEIN LOCALIZATION | 27 | 950 | 1.316e-06 | 2.584e-05 |
| 238 | INNATE IMMUNE RESPONSE ACTIVATING CELL SURFACE RECEPTOR SIGNALING PATHWAY | 9 | 106 | 1.344e-06 | 2.629e-05 |
| 239 | ERBB SIGNALING PATHWAY | 8 | 79 | 1.381e-06 | 2.689e-05 |
| 240 | CYTOSKELETON ORGANIZATION | 25 | 838 | 1.423e-06 | 2.759e-05 |
| 241 | POSITIVE REGULATION OF LOCOMOTION | 17 | 420 | 1.462e-06 | 2.82e-05 |
| 242 | POSITIVE REGULATION OF INNATE IMMUNE RESPONSE | 13 | 246 | 1.467e-06 | 2.82e-05 |
| 243 | ORGAN MORPHOGENESIS | 25 | 841 | 1.517e-06 | 2.905e-05 |
| 244 | MEMBRANE ORGANIZATION | 26 | 899 | 1.53e-06 | 2.915e-05 |
| 245 | DNA DAMAGE RESPONSE DETECTION OF DNA DAMAGE | 6 | 36 | 1.535e-06 | 2.915e-05 |
| 246 | APOPTOTIC MITOCHONDRIAL CHANGES | 7 | 57 | 1.722e-06 | 3.256e-05 |
| 247 | NEGATIVE REGULATION OF CYTOKINE PRODUCTION | 12 | 211 | 1.749e-06 | 3.296e-05 |
| 248 | POSITIVE REGULATION OF LIGASE ACTIVITY | 9 | 110 | 1.836e-06 | 3.444e-05 |
| 249 | CELLULAR GLUCAN METABOLIC PROCESS | 7 | 58 | 1.941e-06 | 3.584e-05 |
| 250 | GLUCAN METABOLIC PROCESS | 7 | 58 | 1.941e-06 | 3.584e-05 |
| 251 | FC EPSILON RECEPTOR SIGNALING PATHWAY | 10 | 142 | 1.931e-06 | 3.584e-05 |
| 252 | REGULATION OF CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY | 12 | 213 | 1.931e-06 | 3.584e-05 |
| 253 | REGULATION OF CYTOPLASMIC TRANSPORT | 18 | 481 | 2.166e-06 | 3.984e-05 |
| 254 | REGULATION OF CELL CYCLE G2 M PHASE TRANSITION | 7 | 59 | 2.184e-06 | 4e-05 |
| 255 | DEVELOPMENT OF PRIMARY SEXUAL CHARACTERISTICS | 12 | 216 | 2.234e-06 | 4.076e-05 |
| 256 | NUCLEOTIDE EXCISION REPAIR DNA DUPLEX UNWINDING | 5 | 22 | 2.347e-06 | 4.265e-05 |
| 257 | POSITIVE REGULATION OF CHROMATIN MODIFICATION | 8 | 85 | 2.419e-06 | 4.379e-05 |
| 258 | REGULATION OF PROTEIN LOCALIZATION TO NUCLEUS | 12 | 218 | 2.458e-06 | 4.434e-05 |
| 259 | NOTCH SIGNALING PATHWAY | 9 | 114 | 2.475e-06 | 4.446e-05 |
| 260 | REGULATION OF NUCLEOCYTOPLASMIC TRANSPORT | 12 | 220 | 2.703e-06 | 4.821e-05 |
| 261 | REGULATION OF PEPTIDASE ACTIVITY | 16 | 392 | 2.704e-06 | 4.821e-05 |
| 262 | OVARIAN FOLLICLE DEVELOPMENT | 7 | 61 | 2.745e-06 | 4.875e-05 |
| 263 | PEPTIDYL SERINE MODIFICATION | 10 | 148 | 2.809e-06 | 4.97e-05 |
| 264 | REGULATION OF PROTEIN STABILITY | 12 | 221 | 2.833e-06 | 4.993e-05 |
| 265 | FEMALE SEX DIFFERENTIATION | 9 | 116 | 2.86e-06 | 5.022e-05 |
| 266 | EXTRACELLULAR STRUCTURE ORGANIZATION | 14 | 304 | 2.913e-06 | 5.095e-05 |
| 267 | REGULATION OF NUCLEASE ACTIVITY | 5 | 23 | 2.974e-06 | 5.177e-05 |
| 268 | GLAND DEVELOPMENT | 16 | 395 | 2.982e-06 | 5.177e-05 |
| 269 | NEURON DIFFERENTIATION | 25 | 874 | 3.008e-06 | 5.204e-05 |
| 270 | POSITIVE REGULATION OF PROTEIN CATABOLIC PROCESS | 13 | 263 | 3.072e-06 | 5.294e-05 |
| 271 | MYELOID CELL HOMEOSTASIS | 8 | 88 | 3.148e-06 | 5.385e-05 |
| 272 | OVULATION CYCLE PROCESS | 8 | 88 | 3.148e-06 | 5.385e-05 |
| 273 | POSITIVE REGULATION OF CHROMOSOME ORGANIZATION | 10 | 150 | 3.17e-06 | 5.403e-05 |
| 274 | WNT SIGNALING PATHWAY | 15 | 351 | 3.208e-06 | 5.447e-05 |
| 275 | TUMOR NECROSIS FACTOR MEDIATED SIGNALING PATHWAY | 9 | 118 | 3.297e-06 | 5.578e-05 |
| 276 | TRANSLESION SYNTHESIS | 6 | 41 | 3.396e-06 | 5.725e-05 |
| 277 | SEX DIFFERENTIATION | 13 | 266 | 3.479e-06 | 5.844e-05 |
| 278 | MORPHOGENESIS OF AN EPITHELIUM | 16 | 400 | 3.502e-06 | 5.861e-05 |
| 279 | INTRINSIC APOPTOTIC SIGNALING PATHWAY | 10 | 152 | 3.571e-06 | 5.955e-05 |
| 280 | NEURON PROJECTION MORPHOGENESIS | 16 | 402 | 3.732e-06 | 6.201e-05 |
| 281 | TUBE DEVELOPMENT | 19 | 552 | 3.794e-06 | 6.282e-05 |
| 282 | POSITIVE REGULATION OF SEQUENCE SPECIFIC DNA BINDING TRANSCRIPTION FACTOR ACTIVITY | 12 | 228 | 3.909e-06 | 6.449e-05 |
| 283 | REGULATION OF INNATE IMMUNE RESPONSE | 15 | 357 | 3.942e-06 | 6.482e-05 |
| 284 | NEGATIVE REGULATION OF CELLULAR RESPONSE TO GROWTH FACTOR STIMULUS | 9 | 121 | 4.058e-06 | 6.648e-05 |
| 285 | REGULATION OF CELLULAR RESPONSE TO GROWTH FACTOR STIMULUS | 12 | 229 | 4.088e-06 | 6.675e-05 |
| 286 | CELLULAR RESPONSE TO NITROGEN COMPOUND | 18 | 505 | 4.271e-06 | 6.949e-05 |
| 287 | NEGATIVE REGULATION OF CELLULAR CATABOLIC PROCESS | 10 | 156 | 4.506e-06 | 7.28e-05 |
| 288 | CELLULAR RESPONSE TO INORGANIC SUBSTANCE | 10 | 156 | 4.506e-06 | 7.28e-05 |
| 289 | REGULATION OF CELL DEVELOPMENT | 24 | 836 | 4.534e-06 | 7.3e-05 |
| 290 | NEGATIVE REGULATION OF CELLULAR RESPONSE TO TRANSFORMING GROWTH FACTOR BETA STIMULUS | 7 | 66 | 4.695e-06 | 7.507e-05 |
| 291 | NEGATIVE REGULATION OF TRANSFORMING GROWTH FACTOR BETA RECEPTOR SIGNALING PATHWAY | 7 | 66 | 4.695e-06 | 7.507e-05 |
| 292 | ENDOCYTOSIS | 18 | 509 | 4.763e-06 | 7.59e-05 |
| 293 | GROWTH PLATE CARTILAGE DEVELOPMENT | 4 | 12 | 4.786e-06 | 7.6e-05 |
| 294 | PEPTIDYL AMINO ACID MODIFICATION | 24 | 841 | 5.016e-06 | 7.938e-05 |
| 295 | REGULATION OF CYTOKINE PRODUCTION | 19 | 563 | 5.037e-06 | 7.945e-05 |
| 296 | REGULATION OF SEQUENCE SPECIFIC DNA BINDING TRANSCRIPTION FACTOR ACTIVITY | 15 | 365 | 5.154e-06 | 8.102e-05 |
| 297 | STEROID HORMONE MEDIATED SIGNALING PATHWAY | 9 | 125 | 5.304e-06 | 8.309e-05 |
| 298 | POSITIVE REGULATION OF ESTABLISHMENT OF PROTEIN LOCALIZATION | 18 | 514 | 5.449e-06 | 8.508e-05 |
| 299 | REGULATION OF STRESS ACTIVATED PROTEIN KINASE SIGNALING CASCADE | 11 | 197 | 5.677e-06 | 8.805e-05 |
| 300 | REGULATION OF NECROTIC CELL DEATH | 5 | 26 | 5.671e-06 | 8.805e-05 |
| 301 | ENDOCHONDRAL BONE MORPHOGENESIS | 6 | 45 | 5.948e-06 | 9.194e-05 |
| 302 | POSITIVE REGULATION OF INTRACELLULAR TRANSPORT | 15 | 370 | 6.071e-06 | 9.354e-05 |
| 303 | POSITIVE REGULATION OF MAPK CASCADE | 17 | 470 | 6.576e-06 | 0.0001007 |
| 304 | POSITIVE REGULATION OF CYTOPLASMIC TRANSPORT | 13 | 282 | 6.564e-06 | 0.0001007 |
| 305 | RESPONSE TO TOPOLOGICALLY INCORRECT PROTEIN | 10 | 163 | 6.658e-06 | 0.0001016 |
| 306 | REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY IN ABSENCE OF LIGAND | 6 | 46 | 6.783e-06 | 0.0001031 |
| 307 | POSITIVE REGULATION OF PROTEIN IMPORT INTO NUCLEUS TRANSLOCATION | 4 | 13 | 6.858e-06 | 0.0001033 |
| 308 | HEPATOCYTE APOPTOTIC PROCESS | 4 | 13 | 6.858e-06 | 0.0001033 |
| 309 | REGULATION OF CELL ADHESION | 20 | 629 | 6.816e-06 | 0.0001033 |
| 310 | RESPONSE TO VITAMIN | 8 | 98 | 7.068e-06 | 0.0001061 |
| 311 | NEGATIVE REGULATION OF PROTEOLYSIS | 14 | 329 | 7.223e-06 | 0.0001081 |
| 312 | NEGATIVE REGULATION OF CATABOLIC PROCESS | 11 | 203 | 7.555e-06 | 0.0001127 |
| 313 | INTRACELLULAR STEROID HORMONE RECEPTOR SIGNALING PATHWAY | 7 | 71 | 7.681e-06 | 0.0001138 |
| 314 | INTRINSIC APOPTOTIC SIGNALING PATHWAY IN RESPONSE TO DNA DAMAGE | 7 | 71 | 7.681e-06 | 0.0001138 |
| 315 | POSITIVE REGULATION OF NEURON APOPTOTIC PROCESS | 6 | 47 | 7.71e-06 | 0.0001139 |
| 316 | ENERGY RESERVE METABOLIC PROCESS | 7 | 72 | 8.435e-06 | 0.0001242 |
| 317 | APPENDAGE DEVELOPMENT | 10 | 169 | 9.161e-06 | 0.000134 |
| 318 | LIMB DEVELOPMENT | 10 | 169 | 9.161e-06 | 0.000134 |
| 319 | CELLULAR RESPONSE TO ARSENIC CONTAINING SUBSTANCE | 4 | 14 | 9.525e-06 | 0.0001389 |
| 320 | REGULATION OF PROTEIN UBIQUITINATION INVOLVED IN UBIQUITIN DEPENDENT PROTEIN CATABOLIC PROCESS | 8 | 103 | 1.023e-05 | 0.0001482 |
| 321 | STRESS ACTIVATED PROTEIN KINASE SIGNALING CASCADE | 8 | 103 | 1.023e-05 | 0.0001482 |
| 322 | NUCLEUS ORGANIZATION | 9 | 136 | 1.055e-05 | 0.0001524 |
| 323 | REGULATION OF CELL SIZE | 10 | 172 | 1.069e-05 | 0.000154 |
| 324 | POSITIVE REGULATION OF PROTEIN IMPORT | 8 | 104 | 1.098e-05 | 0.0001573 |
| 325 | CELLULAR RESPONSE TO REACTIVE OXYGEN SPECIES | 8 | 104 | 1.098e-05 | 0.0001573 |
| 326 | REGULATION OF TUMOR NECROSIS FACTOR MEDIATED SIGNALING PATHWAY | 6 | 50 | 1.112e-05 | 0.0001588 |
| 327 | NEGATIVE REGULATION OF PROTEIN MODIFICATION BY SMALL PROTEIN CONJUGATION OR REMOVAL | 9 | 139 | 1.258e-05 | 0.000179 |
| 328 | SOMATIC DIVERSIFICATION OF IMMUNE RECEPTORS VIA SOMATIC MUTATION | 4 | 15 | 1.289e-05 | 0.0001822 |
| 329 | POSITIVE REGULATION OF NUCLEASE ACTIVITY | 4 | 15 | 1.289e-05 | 0.0001822 |
| 330 | DNA RECOMBINATION | 11 | 215 | 1.299e-05 | 0.0001831 |
| 331 | MULTI ORGANISM REPRODUCTIVE PROCESS | 24 | 891 | 1.313e-05 | 0.0001846 |
| 332 | ANAPHASE PROMOTING COMPLEX DEPENDENT CATABOLIC PROCESS | 7 | 77 | 1.319e-05 | 0.0001846 |
| 333 | POSITIVE REGULATION OF CATABOLIC PROCESS | 15 | 395 | 1.321e-05 | 0.0001846 |
| 334 | ESTABLISHMENT OF LOCALIZATION IN CELL | 36 | 1676 | 1.345e-05 | 0.0001873 |
| 335 | POSITIVE REGULATION OF DEVELOPMENTAL PROCESS | 28 | 1142 | 1.361e-05 | 0.0001891 |
| 336 | POSITIVE REGULATION OF INTRACELLULAR PROTEIN TRANSPORT | 12 | 258 | 1.369e-05 | 0.0001895 |
| 337 | CELLULAR RESPONSE TO HORMONE STIMULUS | 18 | 552 | 1.434e-05 | 0.0001975 |
| 338 | MOVEMENT OF CELL OR SUBCELLULAR COMPONENT | 30 | 1275 | 1.435e-05 | 0.0001975 |
| 339 | POSTTRANSCRIPTIONAL REGULATION OF GENE EXPRESSION | 16 | 448 | 1.449e-05 | 0.0001989 |
| 340 | REGULATION OF MITOCHONDRION ORGANIZATION | 11 | 218 | 1.479e-05 | 0.0002018 |
| 341 | CELLULAR RESPONSE TO STEROID HORMONE STIMULUS | 11 | 218 | 1.479e-05 | 0.0002018 |
| 342 | PATTERN RECOGNITION RECEPTOR SIGNALING PATHWAY | 8 | 109 | 1.551e-05 | 0.0002099 |
| 343 | CELLULAR COMPONENT MORPHOGENESIS | 24 | 900 | 1.548e-05 | 0.0002099 |
| 344 | RESPONSE TO HYDROGEN PEROXIDE | 8 | 109 | 1.551e-05 | 0.0002099 |
| 345 | PROTEIN LOCALIZATION TO ORGANELLE | 18 | 556 | 1.579e-05 | 0.0002129 |
| 346 | REGULATION OF PROTEIN TARGETING | 13 | 307 | 1.625e-05 | 0.0002185 |
| 347 | GLOBAL GENOME NUCLEOTIDE EXCISION REPAIR | 5 | 32 | 1.652e-05 | 0.0002216 |
| 348 | POLYSACCHARIDE METABOLIC PROCESS | 7 | 80 | 1.699e-05 | 0.0002271 |
| 349 | REGULATION OF MITOCHONDRIAL MEMBRANE POTENTIAL | 6 | 54 | 1.748e-05 | 0.0002331 |
| 350 | ESTABLISHMENT OF PROTEIN LOCALIZATION | 32 | 1423 | 1.781e-05 | 0.0002368 |
| 351 | PROTEIN MATURATION | 12 | 265 | 1.787e-05 | 0.0002368 |
| 352 | EMBRYONIC ORGAN DEVELOPMENT | 15 | 406 | 1.822e-05 | 0.0002409 |
| 353 | CELLULAR RESPONSE TO LIPID | 16 | 457 | 1.849e-05 | 0.000243 |
| 354 | NUCLEAR ENVELOPE ORGANIZATION | 7 | 81 | 1.844e-05 | 0.000243 |
| 355 | RESPONSE TO WOUNDING | 18 | 563 | 1.865e-05 | 0.0002432 |
| 356 | IN UTERO EMBRYONIC DEVELOPMENT | 13 | 311 | 1.862e-05 | 0.0002432 |
| 357 | T CELL RECEPTOR SIGNALING PATHWAY | 9 | 146 | 1.866e-05 | 0.0002432 |
| 358 | CARTILAGE DEVELOPMENT | 9 | 147 | 1.97e-05 | 0.0002561 |
| 359 | JNK CASCADE | 7 | 82 | 1.999e-05 | 0.0002583 |
| 360 | REGULATION OF INTRACELLULAR TRANSPORT | 19 | 621 | 1.996e-05 | 0.0002583 |
| 361 | OVULATION CYCLE | 8 | 113 | 2.019e-05 | 0.0002602 |
| 362 | RESPONSE TO TRANSITION METAL NANOPARTICLE | 9 | 148 | 2.08e-05 | 0.0002673 |
| 363 | MEMBRANE PROTEIN INTRACELLULAR DOMAIN PROTEOLYSIS | 4 | 17 | 2.211e-05 | 0.0002834 |
| 364 | PROTEASOMAL PROTEIN CATABOLIC PROCESS | 12 | 271 | 2.229e-05 | 0.000285 |
| 365 | RESPONSE TO ACID CHEMICAL | 13 | 319 | 2.429e-05 | 0.0003096 |
| 366 | NEURON DEVELOPMENT | 20 | 687 | 2.44e-05 | 0.0003102 |
| 367 | ORGANONITROGEN COMPOUND METABOLIC PROCESS | 37 | 1796 | 2.505e-05 | 0.0003176 |
| 368 | TOLL LIKE RECEPTOR SIGNALING PATHWAY | 7 | 85 | 2.529e-05 | 0.0003189 |
| 369 | EPHRIN RECEPTOR SIGNALING PATHWAY | 7 | 85 | 2.529e-05 | 0.0003189 |
| 370 | MICROTUBULE BASED PROCESS | 17 | 522 | 2.541e-05 | 0.0003195 |
| 371 | POSITIVE REGULATION OF WNT SIGNALING PATHWAY | 9 | 152 | 2.57e-05 | 0.0003224 |
| 372 | CELL PART MORPHOGENESIS | 19 | 633 | 2.595e-05 | 0.0003242 |
| 373 | RESPONSE TO MINERALOCORTICOID | 5 | 35 | 2.599e-05 | 0.0003242 |
| 374 | RESPONSE TO NUTRIENT | 10 | 191 | 2.652e-05 | 0.0003299 |
| 375 | TRANSCRIPTION INITIATION FROM RNA POLYMERASE II PROMOTER | 9 | 153 | 2.707e-05 | 0.0003359 |
| 376 | RESPONSE TO ENDOPLASMIC RETICULUM STRESS | 11 | 233 | 2.74e-05 | 0.0003391 |
| 377 | REGULATION OF TRANSPORT | 37 | 1804 | 2.758e-05 | 0.0003404 |
| 378 | TUBE MORPHOGENESIS | 13 | 323 | 2.765e-05 | 0.0003404 |
| 379 | POSITIVE REGULATION OF CELLULAR PROTEIN CATABOLIC PROCESS | 10 | 192 | 2.773e-05 | 0.0003404 |
| 380 | CENTRAL NERVOUS SYSTEM DEVELOPMENT | 23 | 872 | 2.797e-05 | 0.0003425 |
| 381 | REGULATION OF CHROMOSOME ORGANIZATION | 12 | 278 | 2.864e-05 | 0.0003498 |
| 382 | REGULATION OF CELLULAR RESPONSE TO INSULIN STIMULUS | 6 | 59 | 2.924e-05 | 0.0003562 |
| 383 | ACTIVATION OF PROTEIN KINASE ACTIVITY | 12 | 279 | 2.967e-05 | 0.0003604 |
| 384 | POSITIVE REGULATION OF ERBB SIGNALING PATHWAY | 5 | 36 | 2.993e-05 | 0.0003627 |
| 385 | CONNECTIVE TISSUE DEVELOPMENT | 10 | 194 | 3.03e-05 | 0.0003662 |
| 386 | EYE DEVELOPMENT | 13 | 326 | 3.044e-05 | 0.0003669 |
| 387 | ANTIGEN RECEPTOR MEDIATED SIGNALING PATHWAY | 10 | 195 | 3.166e-05 | 0.0003806 |
| 388 | PROTEIN LOCALIZATION TO MEMBRANE | 14 | 376 | 3.2e-05 | 0.0003838 |
| 389 | REGULATION OF PATHWAY RESTRICTED SMAD PROTEIN PHOSPHORYLATION | 6 | 60 | 3.222e-05 | 0.0003854 |
| 390 | TISSUE MORPHOGENESIS | 17 | 533 | 3.303e-05 | 0.0003932 |
| 391 | CARBOHYDRATE BIOSYNTHETIC PROCESS | 8 | 121 | 3.313e-05 | 0.0003932 |
| 392 | POSITIVE REGULATION OF NUCLEOCYTOPLASMIC TRANSPORT | 8 | 121 | 3.313e-05 | 0.0003932 |
| 393 | POSITIVE REGULATION OF PROTEIN COMPLEX ASSEMBLY | 10 | 197 | 3.454e-05 | 0.0004079 |
| 394 | NEGATIVE REGULATION OF WNT SIGNALING PATHWAY | 10 | 197 | 3.454e-05 | 0.0004079 |
| 395 | HORMONE MEDIATED SIGNALING PATHWAY | 9 | 158 | 3.489e-05 | 0.000411 |
| 396 | CELLULAR RESPONSE TO TOPOLOGICALLY INCORRECT PROTEIN | 8 | 122 | 3.515e-05 | 0.000413 |
| 397 | REGULATION OF CELL CELL ADHESION | 14 | 380 | 3.592e-05 | 0.0004209 |
| 398 | NEGATIVE REGULATION OF INTRINSIC APOPTOTIC SIGNALING PATHWAY | 7 | 90 | 3.668e-05 | 0.0004278 |
| 399 | REGULATION OF JNK CASCADE | 9 | 159 | 3.666e-05 | 0.0004278 |
| 400 | REGULATION OF INTRACELLULAR PROTEIN TRANSPORT | 14 | 381 | 3.696e-05 | 0.0004299 |
| 401 | POSITIVE REGULATION OF MITOTIC CELL CYCLE | 8 | 123 | 3.727e-05 | 0.0004324 |
| 402 | EMBRYONIC MORPHOGENESIS | 17 | 539 | 3.799e-05 | 0.0004369 |
| 403 | HEAD DEVELOPMENT | 20 | 709 | 3.802e-05 | 0.0004369 |
| 404 | MULTICELLULAR ORGANISM REPRODUCTION | 21 | 768 | 3.796e-05 | 0.0004369 |
| 405 | DEVELOPMENTAL GROWTH | 13 | 333 | 3.791e-05 | 0.0004369 |
| 406 | ANATOMICAL STRUCTURE FORMATION INVOLVED IN MORPHOGENESIS | 24 | 957 | 4.154e-05 | 0.0004761 |
| 407 | REGULATION OF ANATOMICAL STRUCTURE MORPHOGENESIS | 25 | 1021 | 4.2e-05 | 0.0004802 |
| 408 | MITOCHONDRIAL MEMBRANE ORGANIZATION | 7 | 92 | 4.228e-05 | 0.0004822 |
| 409 | REGULATION OF CELLULAR COMPONENT SIZE | 13 | 337 | 4.286e-05 | 0.0004876 |
| 410 | BONE GROWTH | 4 | 20 | 4.395e-05 | 0.0004988 |
| 411 | MULTICELLULAR ORGANISM METABOLIC PROCESS | 7 | 93 | 4.533e-05 | 0.0005132 |
| 412 | SENSORY ORGAN DEVELOPMENT | 16 | 493 | 4.608e-05 | 0.0005204 |
| 413 | GENERATION OF PRECURSOR METABOLITES AND ENERGY | 12 | 292 | 4.619e-05 | 0.0005204 |
| 414 | REGULATION OF AUTOPHAGY | 11 | 249 | 5.02e-05 | 0.0005642 |
| 415 | CANONICAL WNT SIGNALING PATHWAY | 7 | 95 | 5.198e-05 | 0.0005828 |
| 416 | REGULATION OF PROTEIN IMPORT INTO NUCLEUS TRANSLOCATION | 4 | 21 | 5.386e-05 | 0.0005996 |
| 417 | POSITIVE REGULATION OF MITOCHONDRION ORGANIZATION | 9 | 167 | 5.38e-05 | 0.0005996 |
| 418 | CELLULAR MACROMOLECULAR COMPLEX ASSEMBLY | 20 | 727 | 5.387e-05 | 0.0005996 |
| 419 | ANDROGEN RECEPTOR SIGNALING PATHWAY | 5 | 41 | 5.71e-05 | 0.0006341 |
| 420 | REGULATION OF INTERFERON GAMMA PRODUCTION | 7 | 97 | 5.941e-05 | 0.0006574 |
| 421 | MICROTUBULE CYTOSKELETON ORGANIZATION | 13 | 348 | 5.948e-05 | 0.0006574 |
| 422 | NUCLEIC ACID PHOSPHODIESTER BOND HYDROLYSIS | 11 | 254 | 6.006e-05 | 0.0006622 |
| 423 | NEGATIVE REGULATION OF CELL GROWTH | 9 | 170 | 6.177e-05 | 0.0006794 |
| 424 | HEART MORPHOGENESIS | 10 | 212 | 6.415e-05 | 0.000704 |
| 425 | LOCOMOTION | 26 | 1114 | 6.481e-05 | 0.0007095 |
| 426 | REGULATION OF PROTEIN HOMODIMERIZATION ACTIVITY | 4 | 22 | 6.531e-05 | 0.0007117 |
| 427 | RELEASE OF CYTOCHROME C FROM MITOCHONDRIA | 4 | 22 | 6.531e-05 | 0.0007117 |
| 428 | NEGATIVE REGULATION OF CELL DEVELOPMENT | 12 | 303 | 6.589e-05 | 0.000713 |
| 429 | MULTI ORGANISM TRANSPORT | 6 | 68 | 6.585e-05 | 0.000713 |
| 430 | MULTI ORGANISM LOCALIZATION | 6 | 68 | 6.585e-05 | 0.000713 |
| 431 | SKELETAL SYSTEM DEVELOPMENT | 15 | 455 | 6.708e-05 | 0.0007242 |
| 432 | SIGNAL TRANSDUCTION BY PROTEIN PHOSPHORYLATION | 14 | 404 | 6.945e-05 | 0.0007481 |
| 433 | POSITIVE REGULATION OF TRANSMEMBRANE RECEPTOR PROTEIN SERINE THREONINE KINASE SIGNALING PATHWAY | 7 | 100 | 7.216e-05 | 0.0007736 |
| 434 | REGULATION OF INSULIN RECEPTOR SIGNALING PATHWAY | 5 | 43 | 7.214e-05 | 0.0007736 |
| 435 | I KAPPAB KINASE NF KAPPAB SIGNALING | 6 | 70 | 7.757e-05 | 0.000826 |
| 436 | HOMEOSTASIS OF NUMBER OF CELLS | 9 | 175 | 7.725e-05 | 0.000826 |
| 437 | REGULATION OF MEMBRANE PERMEABILITY | 6 | 70 | 7.757e-05 | 0.000826 |
| 438 | ENERGY DERIVATION BY OXIDATION OF ORGANIC COMPOUNDS | 10 | 217 | 7.792e-05 | 0.0008278 |
| 439 | RESPONSE TO CORTICOSTEROID | 9 | 176 | 8.071e-05 | 0.0008554 |
| 440 | NEGATIVE REGULATION OF TRANSMEMBRANE RECEPTOR PROTEIN SERINE THREONINE KINASE SIGNALING PATHWAY | 7 | 102 | 8.184e-05 | 0.0008654 |
| 441 | REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 20 | 750 | 8.254e-05 | 0.0008708 |
| 442 | PROTEIN COMPLEX BIOGENESIS | 26 | 1132 | 8.421e-05 | 0.0008845 |
| 443 | PROTEIN COMPLEX ASSEMBLY | 26 | 1132 | 8.421e-05 | 0.0008845 |
| 444 | HOMEOSTATIC PROCESS | 29 | 1337 | 8.765e-05 | 0.0009185 |
| 445 | PROTEIN CATABOLIC PROCESS | 17 | 579 | 9.155e-05 | 0.0009572 |
| 446 | POSITIVE REGULATION OF DEFENSE RESPONSE | 13 | 364 | 9.358e-05 | 0.0009763 |
| 447 | REGULATION OF ALPHA BETA T CELL DIFFERENTIATION | 5 | 46 | 0.0001002 | 0.001043 |
| 448 | PROTEIN FOLDING | 10 | 224 | 0.0001014 | 0.001051 |
| 449 | POSITIVE REGULATION OF CELL DIFFERENTIATION | 21 | 823 | 0.0001014 | 0.001051 |
| 450 | NEGATIVE REGULATION OF CELL PROLIFERATION | 18 | 643 | 0.0001037 | 0.001072 |
| 451 | REGULATION OF PROTEIN IMPORT | 9 | 183 | 0.0001088 | 0.001122 |
| 452 | GLYCOGEN BIOSYNTHETIC PROCESS | 4 | 25 | 0.0001103 | 0.00113 |
| 453 | GLUCAN BIOSYNTHETIC PROCESS | 4 | 25 | 0.0001103 | 0.00113 |
| 454 | PHOSPHATIDYLINOSITOL 3 KINASE SIGNALING | 4 | 25 | 0.0001103 | 0.00113 |
| 455 | MEMBRANE DISASSEMBLY | 5 | 47 | 0.0001112 | 0.001135 |
| 456 | NUCLEAR ENVELOPE DISASSEMBLY | 5 | 47 | 0.0001112 | 0.001135 |
| 457 | REGULATION OF CELLULAR COMPONENT BIOGENESIS | 20 | 767 | 0.0001117 | 0.001138 |
| 458 | CELLULAR CARBOHYDRATE METABOLIC PROCESS | 8 | 144 | 0.0001136 | 0.001151 |
| 459 | CELLULAR RESPONSE TO OXIDATIVE STRESS | 9 | 184 | 0.0001134 | 0.001151 |
| 460 | PROTEIN DNA COMPLEX SUBUNIT ORGANIZATION | 10 | 229 | 0.0001215 | 0.001229 |
| 461 | POSITIVE REGULATION OF PATHWAY RESTRICTED SMAD PROTEIN PHOSPHORYLATION | 5 | 48 | 0.0001232 | 0.001243 |
| 462 | NEGATIVE REGULATION OF PROTEIN CATABOLIC PROCESS | 7 | 109 | 0.0001244 | 0.001246 |
| 463 | POSITIVE REGULATION OF LEUKOCYTE MIGRATION | 7 | 109 | 0.0001244 | 0.001246 |
| 464 | ORGANIC CYCLIC COMPOUND CATABOLIC PROCESS | 14 | 427 | 0.0001245 | 0.001246 |
| 465 | SINGLE ORGANISM CELLULAR LOCALIZATION | 22 | 898 | 0.0001239 | 0.001246 |
| 466 | REGULATION OF PROTEIN COMPLEX ASSEMBLY | 13 | 375 | 0.0001259 | 0.001257 |
| 467 | NEGATIVE REGULATION OF STRIATED MUSCLE CELL DIFFERENTIATION | 4 | 26 | 0.0001293 | 0.001288 |
| 468 | CELL PROJECTION ORGANIZATION | 22 | 902 | 0.000132 | 0.001312 |
| 469 | REGULATION OF CELL ACTIVATION | 15 | 484 | 0.0001329 | 0.001319 |
| 470 | MALE SEX DIFFERENTIATION | 8 | 148 | 0.0001374 | 0.00136 |
| 471 | POSITIVE REGULATION OF HYDROLASE ACTIVITY | 22 | 905 | 0.0001384 | 0.001367 |
| 472 | IMMUNE EFFECTOR PROCESS | 15 | 486 | 0.0001391 | 0.001371 |
| 473 | INTRACELLULAR PROTEIN TRANSPORT | 20 | 781 | 0.0001423 | 0.0014 |
| 474 | ZYMOGEN ACTIVATION | 7 | 112 | 0.0001474 | 0.001447 |
| 475 | RESPONSE TO PROGESTERONE | 5 | 50 | 0.0001499 | 0.001469 |
| 476 | REGULATION OF HISTONE H3 K4 METHYLATION | 4 | 27 | 0.0001506 | 0.001469 |
| 477 | REGULATION OF TRANSCRIPTION INVOLVED IN G1 S TRANSITION OF MITOTIC CELL CYCLE | 4 | 27 | 0.0001506 | 0.001469 |
| 478 | MULTICELLULAR ORGANISMAL MACROMOLECULE METABOLIC PROCESS | 6 | 79 | 0.0001525 | 0.001481 |
| 479 | BONE MORPHOGENESIS | 6 | 79 | 0.0001525 | 0.001481 |
| 480 | REGULATION OF CANONICAL WNT SIGNALING PATHWAY | 10 | 236 | 0.0001554 | 0.001504 |
| 481 | NEGATIVE REGULATION OF GROWTH | 10 | 236 | 0.0001554 | 0.001504 |
| 482 | PROTEIN FOLDING IN ENDOPLASMIC RETICULUM | 3 | 11 | 0.0001601 | 0.001546 |
| 483 | REGULATION OF REACTIVE OXYGEN SPECIES METABOLIC PROCESS | 8 | 152 | 0.0001651 | 0.001584 |
| 484 | REGULATION OF CHROMATIN ORGANIZATION | 8 | 152 | 0.0001651 | 0.001584 |
| 485 | CELLULAR CARBOHYDRATE BIOSYNTHETIC PROCESS | 5 | 51 | 0.0001649 | 0.001584 |
| 486 | NEGATIVE REGULATION OF ORGANELLE ORGANIZATION | 13 | 387 | 0.0001716 | 0.001643 |
| 487 | SENSORY ORGAN MORPHOGENESIS | 10 | 239 | 0.0001722 | 0.001645 |
| 488 | REGULATION OF CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 12 | 337 | 0.000179 | 0.001706 |
| 489 | CELL CYCLE ARREST | 8 | 154 | 0.0001807 | 0.001719 |
| 490 | REGULATION OF RESPONSE TO EXTERNAL STIMULUS | 22 | 926 | 0.0001913 | 0.001817 |
| 491 | MAMMARY GLAND DEVELOPMENT | 7 | 117 | 0.0001934 | 0.001829 |
| 492 | VIRAL LIFE CYCLE | 11 | 290 | 0.0001934 | 0.001829 |
| 493 | REGULATION OF CYTOSKELETON ORGANIZATION | 15 | 502 | 0.0001975 | 0.001864 |
| 494 | POSITIVE REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY | 5 | 53 | 0.0001981 | 0.001866 |
| 495 | REGULATION OF ERBB SIGNALING PATHWAY | 6 | 83 | 0.0002002 | 0.001874 |
| 496 | SOMATIC DIVERSIFICATION OF IMMUNOGLOBULINS | 4 | 29 | 0.0002006 | 0.001874 |
| 497 | NIK NF KAPPAB SIGNALING | 6 | 83 | 0.0002002 | 0.001874 |
| 498 | RESPONSE TO ARSENIC CONTAINING SUBSTANCE | 4 | 29 | 0.0002006 | 0.001874 |
| 499 | AMYLOID PRECURSOR PROTEIN METABOLIC PROCESS | 3 | 12 | 0.0002119 | 0.001976 |
| 500 | NEGATIVE REGULATION OF DEVELOPMENTAL GROWTH | 6 | 84 | 0.0002138 | 0.00199 |
| 501 | POSITIVE REGULATION OF CANONICAL WNT SIGNALING PATHWAY | 7 | 119 | 0.0002148 | 0.001995 |
| 502 | SINGLE ORGANISM BIOSYNTHETIC PROCESS | 28 | 1340 | 0.0002185 | 0.002026 |
| 503 | NEGATIVE REGULATION OF HYDROLASE ACTIVITY | 13 | 397 | 0.00022 | 0.002036 |
| 504 | DNA TEMPLATED TRANSCRIPTION INITIATION | 9 | 202 | 0.0002287 | 0.002111 |
| 505 | MYD88 INDEPENDENT TOLL LIKE RECEPTOR SIGNALING PATHWAY | 4 | 30 | 0.0002296 | 0.002111 |
| 506 | RESPONSE TO X RAY | 4 | 30 | 0.0002296 | 0.002111 |
| 507 | POSITIVE REGULATION OF DNA REPLICATION | 6 | 86 | 0.0002432 | 0.002232 |
| 508 | NEGATIVE REGULATION OF KINASE ACTIVITY | 10 | 250 | 0.0002475 | 0.002267 |
| 509 | NEGATIVE REGULATION OF CANONICAL WNT SIGNALING PATHWAY | 8 | 162 | 0.0002552 | 0.002329 |
| 510 | NEURON PROJECTION GUIDANCE | 9 | 205 | 0.0002551 | 0.002329 |
| 511 | SMAD PROTEIN SIGNAL TRANSDUCTION | 5 | 56 | 0.0002573 | 0.002343 |
| 512 | NEGATIVE REGULATION OF CELLULAR RESPONSE TO INSULIN STIMULUS | 4 | 31 | 0.0002615 | 0.002377 |
| 513 | POSITIVE REGULATION OF CELLULAR AMINE METABOLIC PROCESS | 3 | 13 | 0.0002734 | 0.00246 |
| 514 | REGULATION OF IRE1 MEDIATED UNFOLDED PROTEIN RESPONSE | 3 | 13 | 0.0002734 | 0.00246 |
| 515 | POSITIVE REGULATION OF CELLULAR COMPONENT BIOGENESIS | 13 | 406 | 0.0002732 | 0.00246 |
| 516 | REGULATION OF HISTONE PHOSPHORYLATION | 3 | 13 | 0.0002734 | 0.00246 |
| 517 | POSITIVE REGULATION OF INTERLEUKIN 13 PRODUCTION | 3 | 13 | 0.0002734 | 0.00246 |
| 518 | REGULATION OF DEFENSE RESPONSE | 19 | 759 | 0.000279 | 0.002506 |
| 519 | REGULATION OF CELLULAR AMIDE METABOLIC PROCESS | 12 | 354 | 0.0002808 | 0.002518 |
| 520 | REGULATION OF GENERATION OF PRECURSOR METABOLITES AND ENERGY | 6 | 89 | 0.0002931 | 0.002623 |
| 521 | GERM CELL DEVELOPMENT | 9 | 209 | 0.0002942 | 0.002628 |
| 522 | MYD88 DEPENDENT TOLL LIKE RECEPTOR SIGNALING PATHWAY | 4 | 32 | 0.0002965 | 0.002633 |
| 523 | PATTERNING OF BLOOD VESSELS | 4 | 32 | 0.0002965 | 0.002633 |
| 524 | REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER IN RESPONSE TO HYPOXIA | 4 | 32 | 0.0002965 | 0.002633 |
| 525 | FOREBRAIN DEVELOPMENT | 12 | 357 | 0.0003031 | 0.002687 |
| 526 | NEGATIVE REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 7 | 126 | 0.000305 | 0.002698 |
| 527 | REGULATION OF HOMOTYPIC CELL CELL ADHESION | 11 | 307 | 0.0003152 | 0.002783 |
| 528 | WOUND HEALING | 14 | 470 | 0.0003328 | 0.002933 |
| 529 | MITOTIC NUCLEAR DIVISION | 12 | 361 | 0.0003352 | 0.002943 |
| 530 | G2 DNA DAMAGE CHECKPOINT | 4 | 33 | 0.0003348 | 0.002943 |
| 531 | POSITIVE REGULATION OF TYPE 2 IMMUNE RESPONSE | 3 | 14 | 0.0003453 | 0.003009 |
| 532 | POSITIVE REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY IN ABSENCE OF LIGAND | 3 | 14 | 0.0003453 | 0.003009 |
| 533 | POSITIVE REGULATION OF CELL SIZE | 3 | 14 | 0.0003453 | 0.003009 |
| 534 | POST EMBRYONIC ORGAN DEVELOPMENT | 3 | 14 | 0.0003453 | 0.003009 |
| 535 | VESICLE MEDIATED TRANSPORT | 26 | 1239 | 0.0003499 | 0.003043 |
| 536 | POSITIVE REGULATION OF CELL ACTIVATION | 11 | 311 | 0.0003518 | 0.00305 |
| 537 | TUBE FORMATION | 7 | 129 | 0.0003519 | 0.00305 |
| 538 | NEGATIVE REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 10 | 262 | 0.0003592 | 0.003107 |
| 539 | NEGATIVE REGULATION OF NEURON DEATH | 8 | 171 | 0.0003675 | 0.003172 |
| 540 | ESTABLISHMENT OF PROTEIN LOCALIZATION TO MEMBRANE | 10 | 264 | 0.0003814 | 0.003286 |
| 541 | CELLULAR RESPONSE TO HYDROGEN PEROXIDE | 5 | 61 | 0.0003846 | 0.003308 |
| 542 | ACTIVATION OF CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY | 6 | 95 | 0.0004169 | 0.003579 |
| 543 | NEGATIVE REGULATION OF INTERFERON GAMMA PRODUCTION | 4 | 35 | 0.0004216 | 0.0036 |
| 544 | TRANSCRIPTION FROM RNA POLYMERASE I PROMOTER | 4 | 35 | 0.0004216 | 0.0036 |
| 545 | MEMBRANE PROTEIN PROTEOLYSIS | 4 | 35 | 0.0004216 | 0.0036 |
| 546 | CELLULAR RESPONSE TO NITRIC OXIDE | 3 | 15 | 0.0004284 | 0.003644 |
| 547 | DNA DAMAGE RESPONSE SIGNAL TRANSDUCTION RESULTING IN TRANSCRIPTION | 3 | 15 | 0.0004284 | 0.003644 |
| 548 | NEGATIVE REGULATION OF NEURON APOPTOTIC PROCESS | 7 | 135 | 0.0004632 | 0.003912 |
| 549 | MITOCHONDRIAL TRANSPORT | 8 | 177 | 0.0004626 | 0.003912 |
| 550 | POSITIVE REGULATION OF STRESS ACTIVATED PROTEIN KINASE SIGNALING CASCADE | 7 | 135 | 0.0004632 | 0.003912 |
| 551 | CELL GROWTH | 7 | 135 | 0.0004632 | 0.003912 |
| 552 | DEOXYRIBONUCLEOTIDE METABOLIC PROCESS | 4 | 36 | 0.0004706 | 0.00396 |
| 553 | POSITIVE REGULATION OF PROTEIN ACETYLATION | 4 | 36 | 0.0004706 | 0.00396 |
| 554 | NEGATIVE REGULATION OF CELLULAR PROTEIN CATABOLIC PROCESS | 5 | 64 | 0.000481 | 0.004038 |
| 555 | POSITIVE REGULATION OF CELL ADHESION | 12 | 376 | 0.0004825 | 0.004038 |
| 556 | REGULATION OF CELL DIVISION | 10 | 272 | 0.0004819 | 0.004038 |
| 557 | SEXUAL REPRODUCTION | 18 | 730 | 0.0004836 | 0.004038 |
| 558 | EYE MORPHOGENESIS | 7 | 136 | 0.0004842 | 0.004038 |
| 559 | NEGATIVE REGULATION OF EXTRINSIC APOPTOTIC SIGNALING PATHWAY | 6 | 98 | 0.0004925 | 0.004099 |
| 560 | DNA CONFORMATION CHANGE | 10 | 273 | 0.0004959 | 0.00412 |
| 561 | CELLULAR RESPONSE TO RADIATION | 7 | 137 | 0.000506 | 0.004189 |
| 562 | ACTIVATION OF MAPK ACTIVITY | 7 | 137 | 0.000506 | 0.004189 |
| 563 | REGULATION OF MAMMARY GLAND EPITHELIAL CELL PROLIFERATION | 3 | 16 | 0.0005234 | 0.004244 |
| 564 | DNA DEPENDENT DNA REPLICATION | 6 | 99 | 0.0005199 | 0.004244 |
| 565 | NOTCH RECEPTOR PROCESSING | 3 | 16 | 0.0005234 | 0.004244 |
| 566 | SOMATIC RECOMBINATION OF IMMUNOGLOBULIN GENES INVOLVED IN IMMUNE RESPONSE | 3 | 16 | 0.0005234 | 0.004244 |
| 567 | OOGENESIS | 5 | 65 | 0.0005168 | 0.004244 |
| 568 | POSITIVE REGULATION OF HISTONE H3 K4 METHYLATION | 3 | 16 | 0.0005234 | 0.004244 |
| 569 | RNA CAPPING | 4 | 37 | 0.0005235 | 0.004244 |
| 570 | ISOTYPE SWITCHING | 3 | 16 | 0.0005234 | 0.004244 |
| 571 | SOMATIC DIVERSIFICATION OF IMMUNOGLOBULINS INVOLVED IN IMMUNE RESPONSE | 3 | 16 | 0.0005234 | 0.004244 |
| 572 | VIRION ASSEMBLY | 4 | 37 | 0.0005235 | 0.004244 |
| 573 | X7 METHYLGUANOSINE RNA CAPPING | 4 | 37 | 0.0005235 | 0.004244 |
| 574 | POSITIVE REGULATION OF ALPHA BETA T CELL DIFFERENTIATION | 4 | 37 | 0.0005235 | 0.004244 |
| 575 | MAINTENANCE OF LOCATION | 7 | 138 | 0.0005285 | 0.004277 |
| 576 | REGULATION OF PROTEASOMAL PROTEIN CATABOLIC PROCESS | 8 | 181 | 0.0005364 | 0.004333 |
| 577 | REGULATION OF CELL MORPHOGENESIS | 15 | 552 | 0.0005373 | 0.004333 |
| 578 | LIMBIC SYSTEM DEVELOPMENT | 6 | 100 | 0.0005484 | 0.004415 |
| 579 | TELENCEPHALON DEVELOPMENT | 9 | 228 | 0.0005542 | 0.004454 |
| 580 | ORGANELLE FISSION | 14 | 496 | 0.0005678 | 0.004555 |
| 581 | REGULATION OF EXTENT OF CELL GROWTH | 6 | 101 | 0.0005782 | 0.004631 |
| 582 | POSITIVE REGULATION OF DNA REPAIR | 4 | 38 | 0.0005805 | 0.004633 |
| 583 | REGULATION OF CD4 POSITIVE ALPHA BETA T CELL ACTIVATION | 4 | 38 | 0.0005805 | 0.004633 |
| 584 | CELL AGING | 5 | 67 | 0.0005945 | 0.004728 |
| 585 | REGULATION OF DNA TEMPLATED TRANSCRIPTION IN RESPONSE TO STRESS | 5 | 67 | 0.0005945 | 0.004728 |
| 586 | REGULATION OF CELL PROJECTION ORGANIZATION | 15 | 558 | 0.0006005 | 0.004768 |
| 587 | CELLULAR RESPONSE TO LITHIUM ION | 3 | 17 | 0.0006308 | 0.005 |
| 588 | REGULATION OF ALPHA BETA T CELL ACTIVATION | 5 | 68 | 0.0006364 | 0.005028 |
| 589 | POSITIVE REGULATION OF CELL CYCLE PHASE TRANSITION | 5 | 68 | 0.0006364 | 0.005028 |
| 590 | REGULATION OF MUSCLE TISSUE DEVELOPMENT | 6 | 103 | 0.0006415 | 0.005044 |
| 591 | MEIOTIC CELL CYCLE | 8 | 186 | 0.0006417 | 0.005044 |
| 592 | REGULATION OF MUSCLE ORGAN DEVELOPMENT | 6 | 103 | 0.0006415 | 0.005044 |
| 593 | REGULATION OF CELL GROWTH | 12 | 391 | 0.0006809 | 0.005335 |
| 594 | REGULATION OF RESPONSE TO CYTOKINE STIMULUS | 7 | 144 | 0.000681 | 0.005335 |
| 595 | ANATOMICAL STRUCTURE HOMEOSTASIS | 10 | 285 | 0.0006917 | 0.005409 |
| 596 | APICAL JUNCTION ASSEMBLY | 4 | 40 | 0.0007074 | 0.005513 |
| 597 | NEGATIVE REGULATION OF TYPE I INTERFERON PRODUCTION | 4 | 40 | 0.0007074 | 0.005513 |
| 598 | MYELOID CELL DIFFERENTIATION | 8 | 189 | 0.0007126 | 0.005544 |
| 599 | CELLULAR RESPONSE TO INSULIN STIMULUS | 7 | 146 | 0.000739 | 0.00574 |
| 600 | POSITIVE REGULATION OF CELL CYCLE G2 M PHASE TRANSITION | 3 | 18 | 0.0007513 | 0.005807 |
| 601 | REGULATION OF INTERLEUKIN 13 PRODUCTION | 3 | 18 | 0.0007513 | 0.005807 |
| 602 | POSITIVE REGULATION OF T HELPER CELL DIFFERENTIATION | 3 | 18 | 0.0007513 | 0.005807 |
| 603 | SINGLE ORGANISM CATABOLIC PROCESS | 21 | 957 | 0.0007589 | 0.005856 |
| 604 | REGULATION OF GROWTH | 16 | 633 | 0.0007672 | 0.005911 |
| 605 | REGULATION OF DEVELOPMENTAL GROWTH | 10 | 289 | 0.0007696 | 0.005919 |
| 606 | NUCLEOSIDE MONOPHOSPHATE METABOLIC PROCESS | 9 | 239 | 0.0007758 | 0.005957 |
| 607 | CELLULAR RESPONSE TO ESTROGEN STIMULUS | 4 | 41 | 0.0007777 | 0.005962 |
| 608 | ORGANONITROGEN COMPOUND CATABOLIC PROCESS | 11 | 343 | 0.0007939 | 0.006076 |
| 609 | PROTEIN COMPLEX SUBUNIT ORGANIZATION | 29 | 1527 | 0.0007978 | 0.006095 |
| 610 | REGULATION OF LEUKOCYTE MIGRATION | 7 | 149 | 0.0008331 | 0.006355 |
| 611 | POLYSACCHARIDE BIOSYNTHETIC PROCESS | 4 | 42 | 0.0008528 | 0.006431 |
| 612 | POSITIVE REGULATION OF DNA BINDING | 4 | 42 | 0.0008528 | 0.006431 |
| 613 | SOMATIC DIVERSIFICATION OF IMMUNE RECEPTORS | 4 | 42 | 0.0008528 | 0.006431 |
| 614 | REGULATION OF CELLULAR CARBOHYDRATE CATABOLIC PROCESS | 4 | 42 | 0.0008528 | 0.006431 |
| 615 | REGULATION OF HEART GROWTH | 4 | 42 | 0.0008528 | 0.006431 |
| 616 | REGULATION OF ORGAN MORPHOGENESIS | 9 | 242 | 0.0008474 | 0.006431 |
| 617 | REGULATION OF CARBOHYDRATE CATABOLIC PROCESS | 4 | 42 | 0.0008528 | 0.006431 |
| 618 | REGULATION OF MICROTUBULE BASED PROCESS | 9 | 243 | 0.0008724 | 0.006558 |
| 619 | POSITIVE REGULATION OF CELL CELL ADHESION | 9 | 243 | 0.0008724 | 0.006558 |
| 620 | POSITIVE REGULATION OF PROTEIN BINDING | 5 | 73 | 0.0008803 | 0.006561 |
| 621 | REGULATION OF INTERLEUKIN 5 PRODUCTION | 3 | 19 | 0.0008855 | 0.006561 |
| 622 | POSITIVE REGULATION OF ADAPTIVE IMMUNE RESPONSE | 5 | 73 | 0.0008803 | 0.006561 |
| 623 | CELLULAR RESPONSE TO FLUID SHEAR STRESS | 3 | 19 | 0.0008855 | 0.006561 |
| 624 | ERYTHROCYTE HOMEOSTASIS | 5 | 73 | 0.0008803 | 0.006561 |
| 625 | VENTRICULAR CARDIAC MUSCLE CELL DIFFERENTIATION | 3 | 19 | 0.0008855 | 0.006561 |
| 626 | HIPPOCAMPUS DEVELOPMENT | 5 | 73 | 0.0008803 | 0.006561 |
| 627 | RESPONSE TO SALT STRESS | 3 | 19 | 0.0008855 | 0.006561 |
| 628 | CELLULAR RESPONSE TO REACTIVE NITROGEN SPECIES | 3 | 19 | 0.0008855 | 0.006561 |
| 629 | CELL MOTILITY | 19 | 835 | 0.0008888 | 0.006564 |
| 630 | LOCALIZATION OF CELL | 19 | 835 | 0.0008888 | 0.006564 |
| 631 | REGULATION OF ESTABLISHMENT OF PLANAR POLARITY | 6 | 110 | 0.0009064 | 0.006684 |
| 632 | IMMUNE SYSTEM DEVELOPMENT | 15 | 582 | 0.0009205 | 0.006777 |
| 633 | NEGATIVE REGULATION OF PEPTIDASE ACTIVITY | 9 | 245 | 0.0009242 | 0.006794 |
| 634 | CARDIAC MUSCLE CELL DIFFERENTIATION | 5 | 74 | 0.0009365 | 0.006873 |
| 635 | REGULATION OF TYPE I INTERFERON PRODUCTION | 6 | 111 | 0.0009503 | 0.006963 |
| 636 | PALLIUM DEVELOPMENT | 7 | 153 | 0.0009731 | 0.007119 |
| 637 | REGULATION OF DNA REPAIR | 5 | 75 | 0.0009952 | 0.007262 |
| 638 | RESPONSE TO AMINO ACID | 6 | 112 | 0.0009957 | 0.007262 |
| 639 | NEGATIVE REGULATION OF ERBB SIGNALING PATHWAY | 4 | 44 | 0.001018 | 0.007413 |
| 640 | EMBRYONIC HEMOPOIESIS | 3 | 20 | 0.001034 | 0.007494 |
| 641 | NEGATIVE REGULATION OF CELL CYCLE ARREST | 3 | 20 | 0.001034 | 0.007494 |
| 642 | TRACHEA DEVELOPMENT | 3 | 20 | 0.001034 | 0.007494 |
| 643 | SKELETAL SYSTEM MORPHOGENESIS | 8 | 201 | 0.001062 | 0.007682 |
| 644 | BONE DEVELOPMENT | 7 | 156 | 0.00109 | 0.007874 |
| 645 | CENTROSOME CYCLE | 4 | 45 | 0.001109 | 0.007985 |
| 646 | PROTEIN LOCALIZATION TO CHROMOSOME | 4 | 45 | 0.001109 | 0.007985 |
| 647 | REGULATION OF ANATOMICAL STRUCTURE SIZE | 13 | 472 | 0.001112 | 0.007986 |
| 648 | POSITIVE REGULATION OF CELL DEVELOPMENT | 13 | 472 | 0.001112 | 0.007986 |
| 649 | CELLULAR PROCESS INVOLVED IN REPRODUCTION IN MULTICELLULAR ORGANISM | 9 | 252 | 0.001126 | 0.008071 |
| 650 | B CELL HOMEOSTASIS | 3 | 21 | 0.001197 | 0.008519 |
| 651 | CHONDROCYTE DEVELOPMENT | 3 | 21 | 0.001197 | 0.008519 |
| 652 | REGULATION OF MEMBRANE PROTEIN ECTODOMAIN PROTEOLYSIS | 3 | 21 | 0.001197 | 0.008519 |
| 653 | SOMATIC RECOMBINATION OF IMMUNOGLOBULIN GENE SEGMENTS | 3 | 21 | 0.001197 | 0.008519 |
| 654 | RESPONSE TO NITRIC OXIDE | 3 | 21 | 0.001197 | 0.008519 |
| 655 | RESPONSE TO INSULIN | 8 | 205 | 0.001204 | 0.008545 |
| 656 | PEPTIDYL THREONINE MODIFICATION | 4 | 46 | 0.001205 | 0.008545 |
| 657 | PATTERN SPECIFICATION PROCESS | 12 | 418 | 0.00121 | 0.008573 |
| 658 | NEGATIVE REGULATION OF CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 6 | 117 | 0.001248 | 0.008829 |
| 659 | MESODERM DEVELOPMENT | 6 | 118 | 0.001304 | 0.00921 |
| 660 | POSITIVE REGULATION OF LYMPHOCYTE DIFFERENTIATION | 5 | 80 | 0.001331 | 0.009382 |
| 661 | LEUKOCYTE MIGRATION | 9 | 259 | 0.001361 | 0.009583 |
| 662 | BETA CATENIN DESTRUCTION COMPLEX DISASSEMBLY | 3 | 22 | 0.001376 | 0.009629 |
| 663 | REGULATION OF CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY INVOLVED IN APOPTOTIC SIGNALING PATHWAY | 3 | 22 | 0.001376 | 0.009629 |
| 664 | POSITIVE REGULATION OF INTERLEUKIN 4 PRODUCTION | 3 | 22 | 0.001376 | 0.009629 |
| 665 | MEMBRANE PROTEIN ECTODOMAIN PROTEOLYSIS | 3 | 22 | 0.001376 | 0.009629 |
| 666 | CELL MORPHOGENESIS INVOLVED IN NEURON DIFFERENTIATION | 11 | 368 | 0.001399 | 0.009773 |
| 667 | REGULATION OF NUCLEAR DIVISION | 7 | 163 | 0.001405 | 0.009784 |
| 668 | DNA GEOMETRIC CHANGE | 5 | 81 | 0.001407 | 0.009784 |
| 669 | REGULATION OF JUN KINASE ACTIVITY | 5 | 81 | 0.001407 | 0.009784 |
| 670 | REGULATION OF CARDIAC MUSCLE TISSUE DEVELOPMENT | 4 | 48 | 0.001414 | 0.009805 |
| 671 | IMMUNOGLOBULIN PRODUCTION | 4 | 48 | 0.001414 | 0.009805 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | ENZYME BINDING | 61 | 1737 | 1.536e-18 | 1.427e-15 |
| 2 | KINASE BINDING | 36 | 606 | 7.99e-18 | 3.711e-15 |
| 3 | RECEPTOR BINDING | 49 | 1476 | 7.651e-14 | 2.369e-11 |
| 4 | KINASE ACTIVITY | 34 | 842 | 5.137e-12 | 1.193e-09 |
| 5 | CYCLIN DEPENDENT PROTEIN KINASE ACTIVITY | 9 | 34 | 4.031e-11 | 7.49e-09 |
| 6 | IDENTICAL PROTEIN BINDING | 39 | 1209 | 1.036e-10 | 1.375e-08 |
| 7 | TRANSFERASE ACTIVITY TRANSFERRING PHOSPHORUS CONTAINING GROUPS | 35 | 992 | 1.01e-10 | 1.375e-08 |
| 8 | MACROMOLECULAR COMPLEX BINDING | 41 | 1399 | 5.761e-10 | 6.69e-08 |
| 9 | PROTEIN KINASE ACTIVITY | 26 | 640 | 1.864e-09 | 1.924e-07 |
| 10 | PROTEIN SERINE THREONINE KINASE ACTIVITY | 21 | 445 | 5.709e-09 | 5.304e-07 |
| 11 | KINASE REGULATOR ACTIVITY | 14 | 186 | 7.012e-09 | 5.785e-07 |
| 12 | RNA POLYMERASE II CARBOXY TERMINAL DOMAIN KINASE ACTIVITY | 6 | 16 | 7.472e-09 | 5.785e-07 |
| 13 | CYTOKINE RECEPTOR BINDING | 16 | 271 | 1.871e-08 | 1.337e-06 |
| 14 | ADENYL NUCLEOTIDE BINDING | 39 | 1514 | 5.661e-08 | 3.756e-06 |
| 15 | ANDROGEN RECEPTOR BINDING | 7 | 39 | 1.169e-07 | 7.239e-06 |
| 16 | STEROID HORMONE RECEPTOR BINDING | 9 | 81 | 1.329e-07 | 7.716e-06 |
| 17 | PROTEIN DIMERIZATION ACTIVITY | 32 | 1149 | 1.915e-07 | 1.046e-05 |
| 18 | PROTEIN COMPLEX BINDING | 28 | 935 | 2.886e-07 | 1.49e-05 |
| 19 | PROTEIN HOMODIMERIZATION ACTIVITY | 24 | 722 | 3.489e-07 | 1.706e-05 |
| 20 | PROTEIN DOMAIN SPECIFIC BINDING | 22 | 624 | 4.169e-07 | 1.937e-05 |
| 21 | ENZYME REGULATOR ACTIVITY | 28 | 959 | 4.804e-07 | 2.125e-05 |
| 22 | TRANSFORMING GROWTH FACTOR BETA RECEPTOR BINDING | 7 | 50 | 6.906e-07 | 2.916e-05 |
| 23 | UNFOLDED PROTEIN BINDING | 9 | 100 | 8.207e-07 | 3.177e-05 |
| 24 | MOLECULAR FUNCTION REGULATOR | 34 | 1353 | 8.196e-07 | 3.177e-05 |
| 25 | HORMONE RECEPTOR BINDING | 11 | 168 | 1.209e-06 | 4.494e-05 |
| 26 | RIBONUCLEOTIDE BINDING | 41 | 1860 | 1.547e-06 | 5.529e-05 |
| 27 | MISMATCHED DNA BINDING | 4 | 12 | 4.786e-06 | 0.0001647 |
| 28 | TRANSCRIPTION FACTOR BINDING | 18 | 524 | 7.094e-06 | 0.0002354 |
| 29 | CYCLIN DEPENDENT PROTEIN SERINE THREONINE KINASE REGULATOR ACTIVITY | 5 | 28 | 8.334e-06 | 0.000267 |
| 30 | RNA BINDING | 35 | 1598 | 1.195e-05 | 0.0003701 |
| 31 | PROTEIN C TERMINUS BINDING | 10 | 186 | 2.111e-05 | 0.0006132 |
| 32 | POLY A RNA BINDING | 28 | 1170 | 2.112e-05 | 0.0006132 |
| 33 | CYCLIN BINDING | 4 | 19 | 3.544e-05 | 0.0009977 |
| 34 | GROWTH FACTOR ACTIVITY | 9 | 160 | 3.851e-05 | 0.001052 |
| 35 | DAMAGED DNA BINDING | 6 | 63 | 4.264e-05 | 0.001132 |
| 36 | EPHRIN RECEPTOR BINDING | 4 | 24 | 9.337e-05 | 0.00241 |
| 37 | DNA DEPENDENT ATPASE ACTIVITY | 6 | 79 | 0.0001525 | 0.003828 |
| 38 | TAU PROTEIN KINASE ACTIVITY | 3 | 12 | 0.0002119 | 0.00518 |
| 39 | PROTEIN HETERODIMERIZATION ACTIVITY | 14 | 468 | 0.0003188 | 0.007594 |
| 40 | DNA N GLYCOSYLASE ACTIVITY | 3 | 15 | 0.0004284 | 0.009256 |
| 41 | RIBONUCLEOPROTEIN COMPLEX BINDING | 6 | 95 | 0.0004169 | 0.009256 |
| 42 | CYTOKINE ACTIVITY | 9 | 219 | 0.0004142 | 0.009256 |
| 43 | CYSTEINE TYPE ENDOPEPTIDASE ACTIVITY INVOLVED IN APOPTOTIC PROCESS | 3 | 15 | 0.0004284 | 0.009256 |
| 44 | ATPASE ACTIVITY | 13 | 427 | 0.0004415 | 0.009323 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | PROTEIN KINASE COMPLEX | 15 | 90 | 1.743e-14 | 1.018e-11 |
| 2 | TRANSFERASE COMPLEX TRANSFERRING PHOSPHORUS CONTAINING GROUPS | 17 | 237 | 3.411e-10 | 9.96e-08 |
| 3 | CHROMOSOME | 31 | 880 | 1.419e-09 | 2.763e-07 |
| 4 | PERINUCLEAR REGION OF CYTOPLASM | 26 | 642 | 1.99e-09 | 2.905e-07 |
| 5 | CATALYTIC COMPLEX | 32 | 1038 | 1.856e-08 | 2.167e-06 |
| 6 | MICROTUBULE CYTOSKELETON | 32 | 1068 | 3.609e-08 | 3.513e-06 |
| 7 | CYTOSKELETON | 45 | 1967 | 1.45e-07 | 1.209e-05 |
| 8 | CYCLIN DEPENDENT PROTEIN KINASE HOLOENZYME COMPLEX | 6 | 31 | 6.051e-07 | 4.417e-05 |
| 9 | TRANSFERASE COMPLEX | 23 | 703 | 8.2e-07 | 4.789e-05 |
| 10 | APICAL JUNCTION COMPLEX | 10 | 128 | 7.458e-07 | 4.789e-05 |
| 11 | PIGMENT GRANULE | 9 | 103 | 1.055e-06 | 5.6e-05 |
| 12 | CHROMOSOMAL REGION | 15 | 330 | 1.502e-06 | 7.144e-05 |
| 13 | MICROTUBULE ORGANIZING CENTER | 21 | 623 | 1.59e-06 | 7.144e-05 |
| 14 | EXTRACELLULAR MATRIX | 17 | 426 | 1.773e-06 | 7.395e-05 |
| 15 | CARBOXY TERMINAL DOMAIN PROTEIN KINASE COMPLEX | 5 | 22 | 2.347e-06 | 9.136e-05 |
| 16 | CENTROSOME | 18 | 487 | 2.578e-06 | 9.409e-05 |
| 17 | CYTOSKELETAL PART | 34 | 1436 | 3.086e-06 | 0.0001021 |
| 18 | CELL BODY | 18 | 494 | 3.146e-06 | 0.0001021 |
| 19 | HOLO TFIIH COMPLEX | 4 | 12 | 4.786e-06 | 0.0001471 |
| 20 | NUCLEAR CHROMOSOME | 18 | 523 | 6.911e-06 | 0.0002018 |
| 21 | TRANSCRIPTION FACTOR COMPLEX | 13 | 298 | 1.186e-05 | 0.0003297 |
| 22 | CELL JUNCTION | 28 | 1151 | 1.571e-05 | 0.000417 |
| 23 | ENDOCYTIC VESICLE LUMEN | 4 | 17 | 2.211e-05 | 0.0005614 |
| 24 | SPINDLE | 12 | 289 | 4.18e-05 | 0.001017 |
| 25 | ENDOCYTIC VESICLE | 11 | 256 | 6.444e-05 | 0.001505 |
| 26 | CHROMOSOME CENTROMERIC REGION | 9 | 174 | 7.392e-05 | 0.001619 |
| 27 | PROTEINACEOUS EXTRACELLULAR MATRIX | 13 | 356 | 7.486e-05 | 0.001619 |
| 28 | CYTOSOLIC PART | 10 | 223 | 9.768e-05 | 0.002037 |
| 29 | SOMATODENDRITIC COMPARTMENT | 18 | 650 | 0.0001187 | 0.00239 |
| 30 | EXTRACELLULAR SPACE | 29 | 1376 | 0.0001442 | 0.002806 |
| 31 | ENDOPLASMIC RETICULUM CHAPERONE COMPLEX | 3 | 11 | 0.0001601 | 0.002833 |
| 32 | CELL CELL JUNCTION | 13 | 383 | 0.000155 | 0.002833 |
| 33 | IKAPPAB KINASE COMPLEX | 3 | 11 | 0.0001601 | 0.002833 |
| 34 | CONDENSED CHROMOSOME | 9 | 195 | 0.0001757 | 0.003019 |
| 35 | CONDENSED CHROMOSOME OUTER KINETOCHORE | 3 | 12 | 0.0002119 | 0.003535 |
| 36 | SPINDLE POLE | 7 | 126 | 0.000305 | 0.004948 |
| 37 | MYELIN SHEATH | 8 | 168 | 0.0003263 | 0.005151 |
| 38 | APICAL PART OF CELL | 12 | 361 | 0.0003352 | 0.005152 |
| 39 | MISMATCH REPAIR COMPLEX | 3 | 14 | 0.0003453 | 0.005171 |
| 40 | PRONUCLEUS | 3 | 15 | 0.0004284 | 0.006255 |
| 41 | INTRACELLULAR VESICLE | 26 | 1259 | 0.0004461 | 0.006354 |
| 42 | MITOCHONDRION | 31 | 1633 | 0.0005218 | 0.007256 |
| 43 | CYTOPLASMIC REGION | 10 | 287 | 0.0007298 | 0.009162 |
| 44 | VESICLE LUMEN | 6 | 106 | 0.0007465 | 0.009162 |
| 45 | CELL SURFACE | 18 | 757 | 0.0007371 | 0.009162 |
| 46 | MEMBRANE MICRODOMAIN | 10 | 288 | 0.0007495 | 0.009162 |
| 47 | CELL CORTEX | 9 | 238 | 0.0007531 | 0.009162 |
| 48 | ENVELOPE | 23 | 1090 | 0.0007247 | 0.009162 |
Over-represented Pathway
| Num | Pathway | Pathview | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|---|
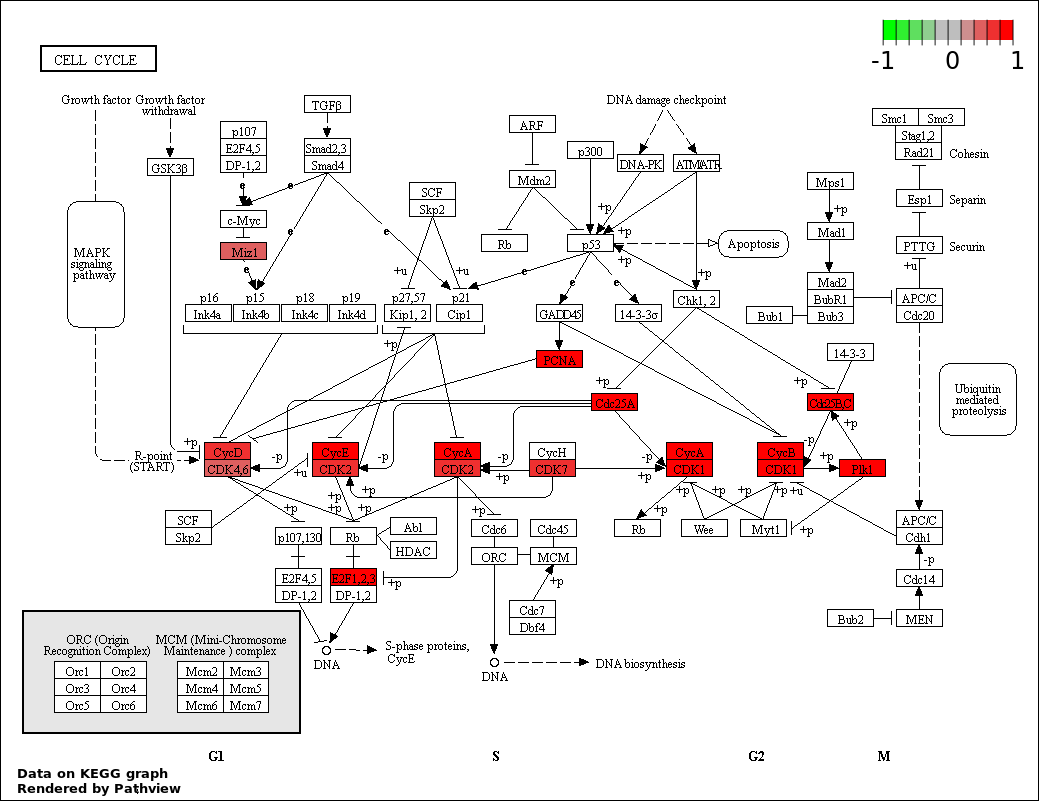
| 1 | hsa04110_Cell_cycle | 16 | 128 | 2.41e-13 | 4.265e-11 | |
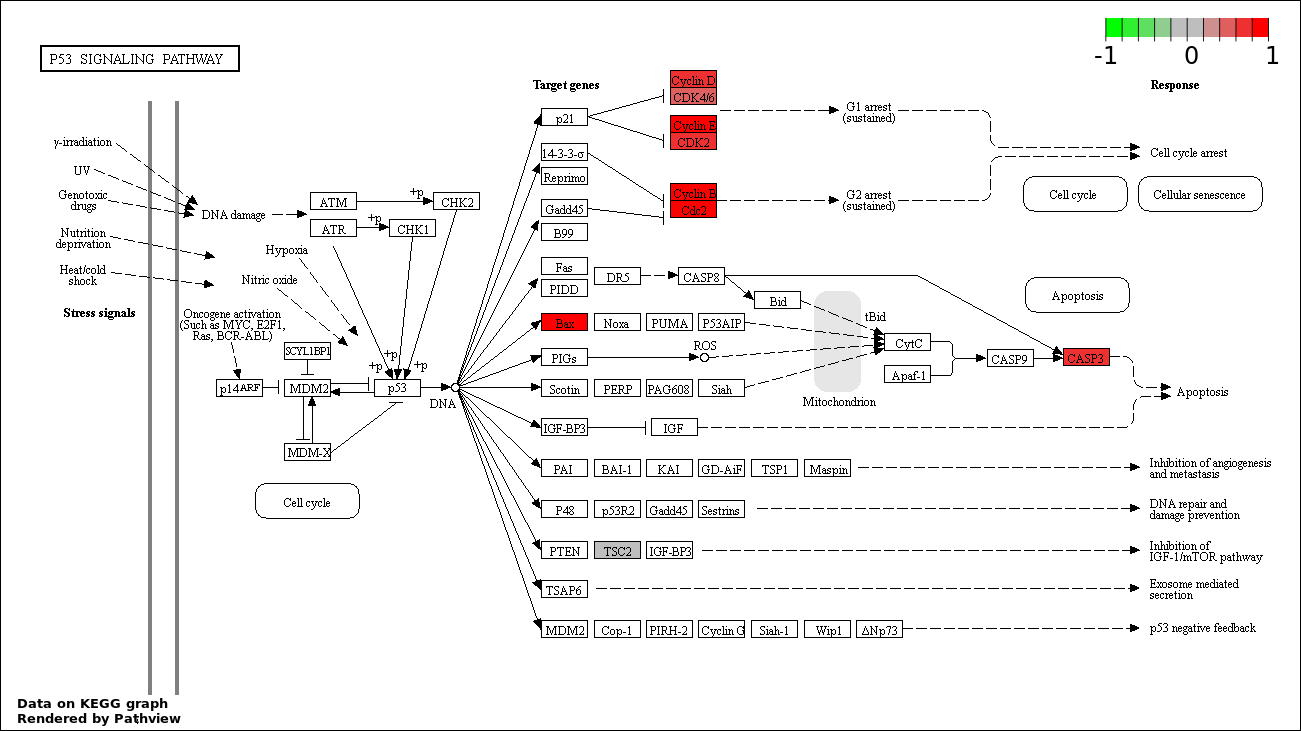
| 2 | hsa04115_p53_signaling_pathway | 10 | 69 | 1.878e-09 | 1.261e-07 | |
| 3 | hsa03430_Mismatch_repair | 7 | 23 | 2.138e-09 | 1.261e-07 | |
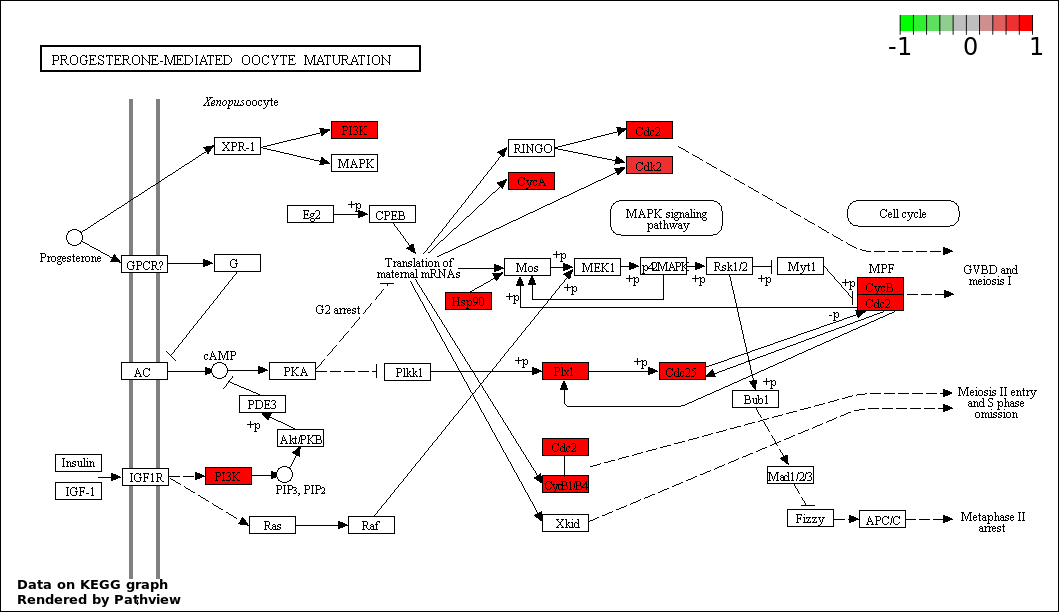
| 4 | hsa04914_Progesterone.mediated_oocyte_maturation | 10 | 87 | 1.886e-08 | 8.345e-07 | |
| 5 | hsa04621_NOD.like_receptor_signaling_pathway | 8 | 59 | 1.396e-07 | 4.943e-06 | |
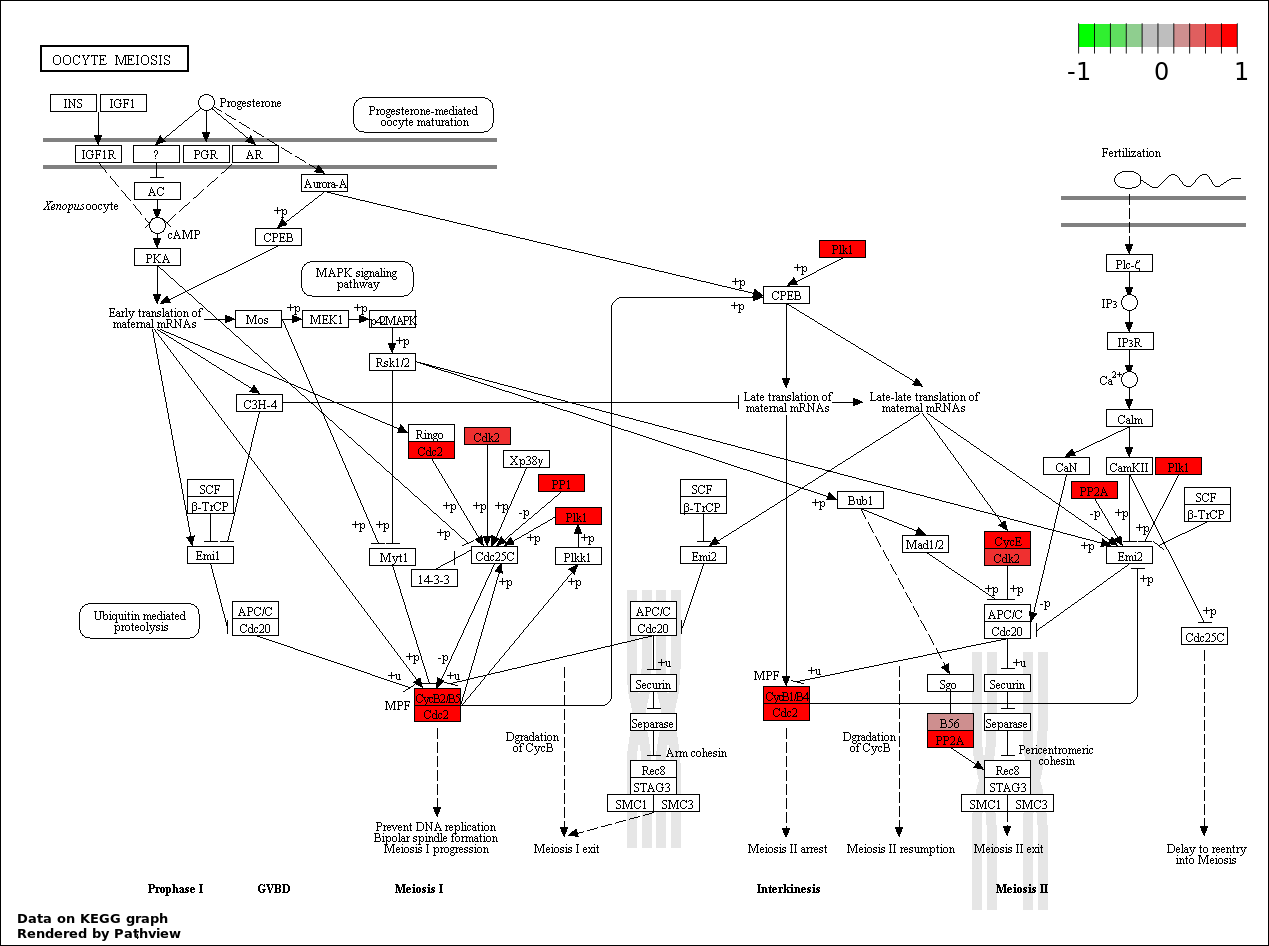
| 6 | hsa04114_Oocyte_meiosis | 10 | 114 | 2.53e-07 | 7.465e-06 | |
| 7 | hsa03420_Nucleotide_excision_repair | 7 | 45 | 3.275e-07 | 8.282e-06 | |
| 8 | hsa04530_Tight_junction | 10 | 133 | 1.062e-06 | 2.349e-05 | |
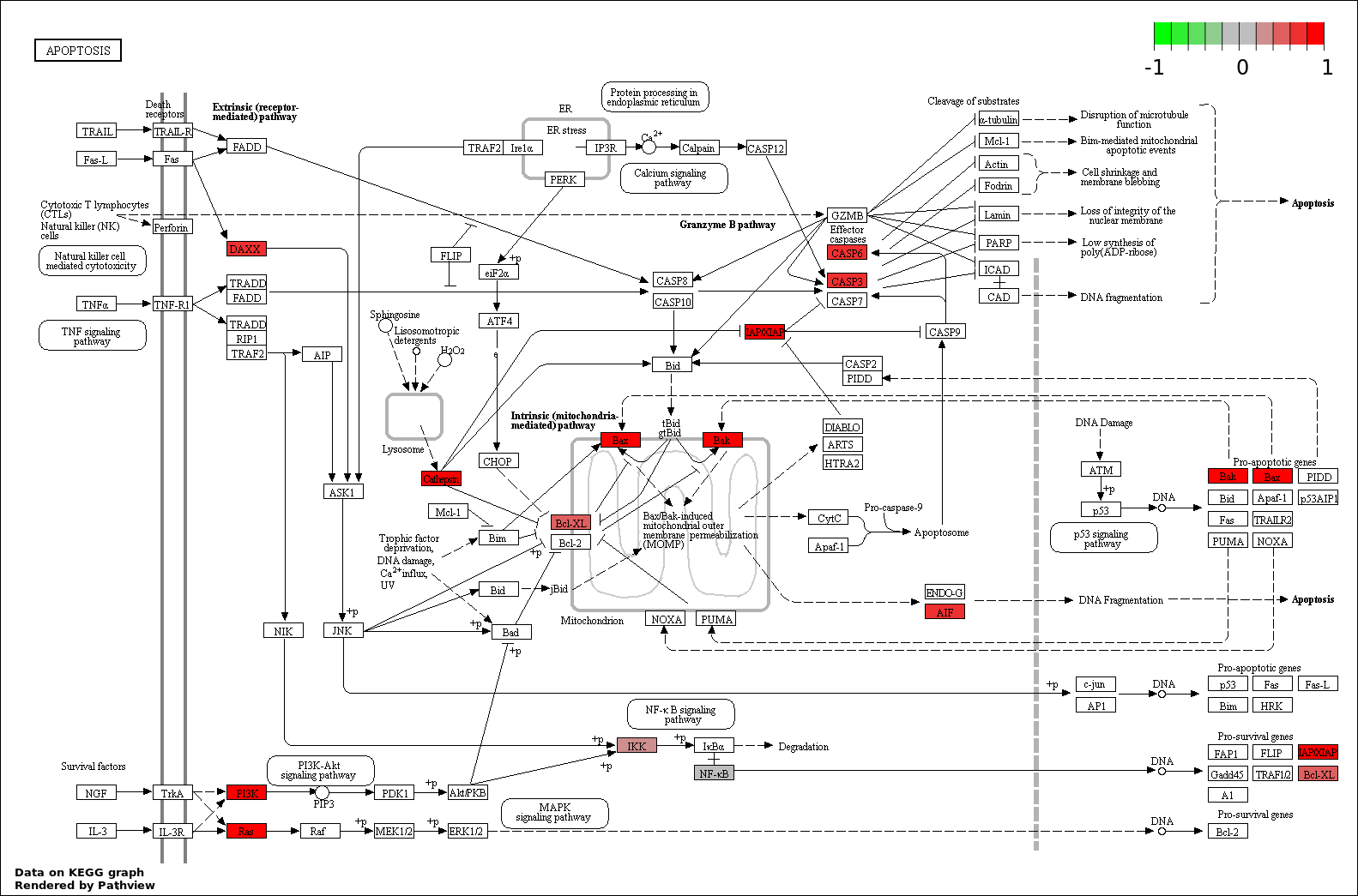
| 9 | hsa04210_Apoptosis | 8 | 89 | 3.429e-06 | 6.744e-05 | |
| 10 | hsa04910_Insulin_signaling_pathway | 9 | 138 | 1.187e-05 | 0.0002101 | |
| 11 | hsa04350_TGF.beta_signaling_pathway | 7 | 85 | 2.529e-05 | 0.0004069 | |
| 12 | hsa04510_Focal_adhesion | 10 | 200 | 3.927e-05 | 0.0005792 | |
| 13 | hsa04722_Neurotrophin_signaling_pathway | 8 | 127 | 4.686e-05 | 0.000638 | |
| 14 | hsa04141_Protein_processing_in_endoplasmic_reticulum | 9 | 168 | 5.635e-05 | 0.0007125 | |
| 15 | hsa04062_Chemokine_signaling_pathway | 8 | 189 | 0.0007126 | 0.008408 | |
| 16 | hsa04660_T_cell_receptor_signaling_pathway | 6 | 108 | 0.0008234 | 0.009109 | |
| 17 | hsa04310_Wnt_signaling_pathway | 7 | 151 | 0.000901 | 0.00936 | |
| 18 | hsa04662_B_cell_receptor_signaling_pathway | 5 | 75 | 0.0009952 | 0.00936 | |
| 19 | hsa00400_Phenylalanine._tyrosine_and_tryptophan_biosynthesis | 2 | 5 | 0.001005 | 0.00936 | |
| 20 | hsa04144_Endocytosis | 8 | 203 | 0.001131 | 0.01001 | |
| 21 | hsa04330_Notch_signaling_pathway | 4 | 47 | 0.001306 | 0.01101 | |
| 22 | hsa00480_Glutathione_metabolism | 4 | 50 | 0.001647 | 0.01325 | |
| 23 | hsa04010_MAPK_signaling_pathway | 9 | 268 | 0.001721 | 0.01325 | |
| 24 | hsa04012_ErbB_signaling_pathway | 5 | 87 | 0.001932 | 0.01425 | |
| 25 | hsa03410_Base_excision_repair | 3 | 34 | 0.004891 | 0.03463 | |
| 26 | hsa03030_DNA_replication | 3 | 36 | 0.005749 | 0.03914 | |
| 27 | hsa04370_VEGF_signaling_pathway | 4 | 76 | 0.007483 | 0.04906 | |
| 28 | hsa04612_Antigen_processing_and_presentation | 4 | 78 | 0.008191 | 0.05049 | |
| 29 | hsa00350_Tyrosine_metabolism | 3 | 41 | 0.008272 | 0.05049 | |
| 30 | hsa04380_Osteoclast_differentiation | 5 | 128 | 0.009924 | 0.05855 | |
| 31 | hsa00360_Phenylalanine_metabolism | 2 | 17 | 0.01261 | 0.07201 | |
| 32 | hsa04150_mTOR_signaling_pathway | 3 | 52 | 0.01581 | 0.08747 | |
| 33 | hsa04623_Cytosolic_DNA.sensing_pathway | 3 | 56 | 0.01926 | 0.1033 | |
| 34 | hsa04620_Toll.like_receptor_signaling_pathway | 4 | 102 | 0.02025 | 0.1054 | |
| 35 | hsa04920_Adipocytokine_signaling_pathway | 3 | 68 | 0.0319 | 0.1613 | |
| 36 | hsa04730_Long.term_depression | 3 | 70 | 0.03434 | 0.1673 | |
| 37 | hsa04142_Lysosome | 4 | 121 | 0.03496 | 0.1673 | |
| 38 | hsa04520_Adherens_junction | 3 | 73 | 0.03817 | 0.1778 | |
| 39 | hsa00250_Alanine._aspartate_and_glutamate_metabolism | 2 | 32 | 0.04166 | 0.1891 | |
| 40 | hsa04664_Fc_epsilon_RI_signaling_pathway | 3 | 79 | 0.04646 | 0.2056 | |
| 41 | hsa04650_Natural_killer_cell_mediated_cytotoxicity | 4 | 136 | 0.05002 | 0.2159 | |
| 42 | hsa00270_Cysteine_and_methionine_metabolism | 2 | 36 | 0.05155 | 0.2159 | |
| 43 | hsa03015_mRNA_surveillance_pathway | 3 | 83 | 0.05245 | 0.2159 | |
| 44 | hsa04512_ECM.receptor_interaction | 3 | 85 | 0.05557 | 0.2235 | |
| 45 | hsa04540_Gap_junction | 3 | 90 | 0.06376 | 0.2508 | |
| 46 | hsa03010_Ribosome | 3 | 92 | 0.06718 | 0.256 | |
| 47 | hsa03013_RNA_transport | 4 | 152 | 0.06943 | 0.256 | |
| 48 | hsa04630_Jak.STAT_signaling_pathway | 4 | 155 | 0.07344 | 0.2653 | |
| 49 | hsa04912_GnRH_signaling_pathway | 3 | 101 | 0.08357 | 0.2901 | |
| 50 | hsa04916_Melanogenesis | 3 | 101 | 0.08357 | 0.2901 | |
| 51 | hsa00983_Drug_metabolism_._other_enzymes | 2 | 52 | 0.0978 | 0.3266 | |
| 52 | hsa00330_Arginine_and_proline_metabolism | 2 | 54 | 0.1042 | 0.3415 | |
| 53 | hsa04340_Hedgehog_signaling_pathway | 2 | 56 | 0.1107 | 0.3562 | |
| 54 | hsa04670_Leukocyte_transendothelial_migration | 3 | 117 | 0.1164 | 0.3679 | |
| 55 | hsa04610_Complement_and_coagulation_cascades | 2 | 69 | 0.1552 | 0.4634 | |
| 56 | hsa03320_PPAR_signaling_pathway | 2 | 70 | 0.1588 | 0.4634 | |
| 57 | hsa04720_Long.term_potentiation | 2 | 70 | 0.1588 | 0.4634 | |
| 58 | hsa00980_Metabolism_of_xenobiotics_by_cytochrome_P450 | 2 | 71 | 0.1623 | 0.4634 | |
| 59 | hsa04622_RIG.I.like_receptor_signaling_pathway | 2 | 71 | 0.1623 | 0.4634 | |
| 60 | hsa00982_Drug_metabolism_._cytochrome_P450 | 2 | 73 | 0.1695 | 0.4688 | |
| 61 | hsa04810_Regulation_of_actin_cytoskeleton | 4 | 214 | 0.1735 | 0.4724 | |
| 62 | hsa04974_Protein_digestion_and_absorption | 2 | 81 | 0.1987 | 0.5329 | |
| 63 | hsa00230_Purine_metabolism | 3 | 162 | 0.2273 | 0.583 | |
| 64 | hsa00240_Pyrimidine_metabolism | 2 | 99 | 0.266 | 0.654 | |
| 65 | hsa03040_Spliceosome | 2 | 128 | 0.3738 | 0.818 | |
| 66 | hsa04360_Axon_guidance | 2 | 130 | 0.381 | 0.818 | |
| 67 | hsa00190_Oxidative_phosphorylation | 2 | 132 | 0.3882 | 0.818 | |
| 68 | hsa04514_Cell_adhesion_molecules_.CAMs. | 2 | 136 | 0.4025 | 0.8381 | |
| 69 | hsa04120_Ubiquitin_mediated_proteolysis | 2 | 139 | 0.4131 | 0.8502 | |
| 70 | hsa04020_Calcium_signaling_pathway | 2 | 177 | 0.5384 | 1 |
lncRNA-mediated sponge
| Num | lncRNA | miRNAs | miRNAs count | Gene | Sponge regulatory network | lncRNA log2FC | lncRNA pvalue | Gene log2FC | Gene pvalue | lncRNA-gene Pearson correlation |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | RP11-1024P17.1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-9-5p | 17 | TGFBR2 | Sponge network | -2.541 | 0 | -2.395 | 0 | 0.887 |
| 2 | MAGI2-AS3 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 19 | TGFBR2 | Sponge network | -2.333 | 0 | -2.395 | 0 | 0.865 |
| 3 | RP11-175K6.1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-590-5p;hsa-miR-9-5p;hsa-miR-93-5p | 17 | TGFBR2 | Sponge network | -3.312 | 0 | -2.395 | 0 | 0.838 |
| 4 | RP11-389C8.2 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-5p | 14 | TGFBR2 | Sponge network | -1.579 | 0 | -2.395 | 0 | 0.827 |
| 5 | ADAMTS9-AS2 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p | 16 | TGFBR2 | Sponge network | -4.039 | 0 | -2.395 | 0 | 0.77 |
| 6 | RP11-166D19.1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p | 19 | TGFBR2 | Sponge network | -1.912 | 0 | -2.395 | 0 | 0.738 |
| 7 | MIR143HG | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-9-5p;hsa-miR-93-5p | 19 | TGFBR2 | Sponge network | -4.451 | 0 | -2.395 | 0 | 0.733 |
| 8 | MIR22HG | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-454-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 16 | TGFBR2 | Sponge network | -1.551 | 0 | -2.395 | 0 | 0.724 |
| 9 | LINC00968 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-340-5p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-9-5p;hsa-miR-93-5p | 16 | TGFBR2 | Sponge network | -4.245 | 0 | -2.395 | 0 | 0.706 |
| 10 | RP11-1101K5.1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-9-5p;hsa-miR-93-5p | 15 | TGFBR2 | Sponge network | -8.619 | 0 | -2.395 | 0 | 0.67 |
| 11 | RP11-276H19.1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-5p | 16 | TGFBR2 | Sponge network | -2.749 | 0 | -2.395 | 0 | 0.667 |
| 12 | TRHDE-AS1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-9-5p;hsa-miR-93-5p | 18 | TGFBR2 | Sponge network | -7.569 | 0 | -2.395 | 0 | 0.661 |
| 13 | MEG3 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-5p;hsa-miR-9-5p;hsa-miR-93-5p | 18 | TGFBR2 | Sponge network | -2.353 | 0 | -2.395 | 0 | 0.657 |
| 14 | EMX2OS | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-9-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 17 | TGFBR2 | Sponge network | -2.548 | 0 | -2.395 | 0 | 0.656 |
| 15 | CTD-2013N24.2 | hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-93-5p | 10 | TGFBR2 | Sponge network | -1.842 | 0 | -2.395 | 0 | 0.638 |
| 16 | ADAMTS9-AS1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-9-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 21 | TGFBR2 | Sponge network | -4.696 | 0 | -2.395 | 0 | 0.638 |
| 17 | LINC00924 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-93-5p | 16 | TGFBR2 | Sponge network | -3.023 | 0 | -2.395 | 0 | 0.635 |
| 18 | RP11-384P7.7 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-9-5p;hsa-miR-93-5p | 16 | TGFBR2 | Sponge network | -4.533 | 0 | -2.395 | 0 | 0.62 |
| 19 | ALDH1L1-AS2 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-9-5p;hsa-miR-93-5p | 13 | TGFBR2 | Sponge network | -7.428 | 0 | -2.395 | 0 | 0.62 |
| 20 | WDR86-AS1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-20b-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-93-5p | 11 | TGFBR2 | Sponge network | -3.263 | 0 | -2.395 | 0 | 0.615 |
| 21 | LINC00702 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 17 | TGFBR2 | Sponge network | -1.308 | 0 | -2.395 | 0 | 0.614 |
| 22 | AL035610.1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-5p | 16 | TGFBR2 | Sponge network | -6.728 | 0 | -2.395 | 0 | 0.608 |
| 23 | HOXA-AS2 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-93-5p | 13 | TGFBR2 | Sponge network | -2.242 | 0 | -2.395 | 0 | 0.598 |
| 24 | ADIPOQ-AS1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-340-5p;hsa-miR-590-3p;hsa-miR-9-5p;hsa-miR-93-5p | 13 | TGFBR2 | Sponge network | -8 | 0 | -2.395 | 0 | 0.588 |
| 25 | RP11-92A5.2 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-340-5p;hsa-miR-590-3p | 13 | TGFBR2 | Sponge network | -9.601 | 0 | -2.395 | 0 | 0.576 |
| 26 | RP4-575N6.4 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p | 11 | TGFBR2 | Sponge network | -2.602 | 0 | -2.395 | 0 | 0.57 |
| 27 | NR2F1-AS1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 16 | TGFBR2 | Sponge network | -0.993 | 0 | -2.395 | 0 | 0.565 |
| 28 | TBX5-AS1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-590-5p;hsa-miR-92a-3p | 12 | TGFBR2 | Sponge network | -0.804 | 0 | -2.395 | 0 | 0.56 |
| 29 | RP11-180N14.1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-9-5p | 12 | TGFBR2 | Sponge network | -2.382 | 0 | -2.395 | 0 | 0.557 |
| 30 | ACTA2-AS1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-590-3p | 14 | TGFBR2 | Sponge network | -3.065 | 0 | -2.395 | 0 | 0.547 |
| 31 | RP11-54A9.1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-9-5p;hsa-miR-93-5p | 13 | TGFBR2 | Sponge network | -2.207 | 0 | -2.395 | 0 | 0.547 |
| 32 | DIO3OS | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p | 12 | TGFBR2 | Sponge network | -3.064 | 0 | -2.395 | 0 | 0.543 |
| 33 | MIR497HG | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-590-5p;hsa-miR-9-5p;hsa-miR-93-5p | 15 | TGFBR2 | Sponge network | -2.527 | 0 | -2.395 | 0 | 0.541 |
| 34 | RP11-38H17.1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-301a-3p;hsa-miR-93-5p | 10 | TGFBR2 | Sponge network | -3.941 | 0 | -2.395 | 0 | 0.539 |
| 35 | RP11-822E23.8 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-93-5p | 14 | TGFBR2 | Sponge network | -2.406 | 0 | -2.395 | 0 | 0.534 |
| 36 | LINC00641 | hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 11 | TGFBR2 | Sponge network | -1.721 | 0 | -2.395 | 0 | 0.534 |
| 37 | AP000473.5 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-301a-3p;hsa-miR-454-3p | 10 | TGFBR2 | Sponge network | -2.465 | 0 | -2.395 | 0 | 0.533 |
| 38 | WDFY3-AS2 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p | 16 | TGFBR2 | Sponge network | -1.618 | 0 | -2.395 | 0 | 0.531 |
| 39 | AC108142.1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-93-5p | 15 | TGFBR2 | Sponge network | -2.247 | 0 | -2.395 | 0 | 0.516 |
| 40 | AC003991.3 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-590-3p;hsa-miR-9-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 18 | TGFBR2 | Sponge network | -2.021 | 0 | -2.395 | 0 | 0.514 |
| 41 | CTD-2078B5.2 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-301a-3p;hsa-miR-93-5p | 10 | TGFBR2 | Sponge network | -2.992 | 0 | -2.395 | 0 | 0.51 |
| 42 | AF131215.2 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-454-3p | 13 | TGFBR2 | Sponge network | -2.008 | 0 | -2.395 | 0 | 0.509 |
| 43 | RBPMS-AS1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-93-5p | 12 | TGFBR2 | Sponge network | -2.131 | 0 | -2.395 | 0 | 0.506 |
| 44 | PDZRN3-AS1 | hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-590-5p | 10 | TGFBR2 | Sponge network | -2.387 | 0 | -2.395 | 0 | 0.505 |
| 45 | FZD10-AS1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 16 | TGFBR2 | Sponge network | -1.884 | 0 | -2.395 | 0 | 0.501 |
| 46 | LINC01088 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-93-5p | 11 | TGFBR2 | Sponge network | -1.675 | 0 | -2.395 | 0 | 0.481 |
| 47 | RP11-276H19.2 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-93-5p | 13 | TGFBR2 | Sponge network | -3.719 | 0 | -2.395 | 0 | 0.48 |
| 48 | AC144831.1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-9-5p;hsa-miR-93-5p | 14 | TGFBR2 | Sponge network | -1.421 | 0 | -2.395 | 0 | 0.48 |
| 49 | HOXB-AS1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-429 | 11 | TGFBR2 | Sponge network | -2.063 | 0 | -2.395 | 0 | 0.475 |
| 50 | RP11-251M1.1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-301a-3p;hsa-miR-320b | 10 | TGFBR2 | Sponge network | -2.455 | 0 | -2.395 | 0 | 0.468 |
| 51 | AC004947.2 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 15 | TGFBR2 | Sponge network | -3.692 | 0 | -2.395 | 0 | 0.465 |
| 52 | DNM3OS | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-9-5p;hsa-miR-93-5p | 17 | TGFBR2 | Sponge network | -0.276 | 0.07533 | -2.395 | 0 | 0.458 |
| 53 | LL22NC03-86G7.1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-9-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 20 | TGFBR2 | Sponge network | -1.159 | 0 | -2.395 | 0 | 0.438 |
| 54 | TPTEP1 | hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-9-5p;hsa-miR-93-5p | 13 | TGFBR2 | Sponge network | -2.33 | 0 | -2.395 | 0 | 0.435 |
| 55 | AF131215.9 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-590-3p | 11 | TGFBR2 | Sponge network | -1.776 | 0 | -2.395 | 0 | 0.428 |
| 56 | AGAP11 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-590-5p;hsa-miR-93-5p | 15 | TGFBR2 | Sponge network | -2.72 | 0 | -2.395 | 0 | 0.428 |
| 57 | AC007743.1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-590-5p;hsa-miR-93-5p | 14 | TGFBR2 | Sponge network | -2.327 | 0 | -2.395 | 0 | 0.42 |
| 58 | RP1-34H18.1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p | 10 | TGFBR2 | Sponge network | -2.546 | 0 | -2.395 | 0 | 0.417 |
| 59 | RP4-798P15.3 | hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-340-5p;hsa-miR-590-3p;hsa-miR-590-5p | 12 | TGFBR2 | Sponge network | -1.167 | 0 | -2.395 | 0 | 0.416 |
| 60 | HOXD-AS2 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-454-3p;hsa-miR-93-5p | 15 | TGFBR2 | Sponge network | -1.529 | 0 | -2.395 | 0 | 0.415 |
| 61 | CTD-2154I11.2 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-340-5p | 10 | TGFBR2 | Sponge network | -1.917 | 0 | -2.395 | 0 | 0.415 |
| 62 | AC034187.2 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-93-5p | 10 | TGFBR2 | Sponge network | -1.611 | 0 | -2.395 | 0 | 0.415 |
| 63 | CTD-2647L4.4 | hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-93-5p | 10 | TGFBR2 | Sponge network | -1.068 | 0 | -2.395 | 0 | 0.412 |
| 64 | RP11-116O18.1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 15 | TGFBR2 | Sponge network | -3.1 | 0 | -2.395 | 0 | 0.397 |
| 65 | SOCS2-AS1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-9-5p;hsa-miR-93-5p | 16 | TGFBR2 | Sponge network | -1.697 | 0 | -2.395 | 0 | 0.395 |
| 66 | RP11-890B15.3 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-340-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-93-5p | 14 | TGFBR2 | Sponge network | -0.935 | 0 | -2.395 | 0 | 0.384 |
| 67 | RP11-545I5.3 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-340-5p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 16 | TGFBR2 | Sponge network | -1.007 | 0 | -2.395 | 0 | 0.384 |
| 68 | TPRG1-AS1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 12 | TGFBR2 | Sponge network | -1.972 | 0 | -2.395 | 0 | 0.382 |
| 69 | MIRLET7DHG | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-93-5p | 11 | TGFBR2 | Sponge network | -1.664 | 0 | -2.395 | 0 | 0.377 |
| 70 | RP11-228B15.4 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-93-5p | 12 | TGFBR2 | Sponge network | -1.572 | 0 | -2.395 | 0 | 0.376 |
| 71 | ZNF582-AS1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-590-5p;hsa-miR-93-5p | 11 | TGFBR2 | Sponge network | -1.307 | 0 | -2.395 | 0 | 0.374 |
| 72 | LINC00648 | hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p | 10 | TGFBR2 | Sponge network | -5.143 | 0 | -2.395 | 0 | 0.373 |
| 73 | RP11-401P9.4 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-590-5p;hsa-miR-93-5p | 14 | TGFBR2 | Sponge network | -0.963 | 0 | -2.395 | 0 | 0.371 |
| 74 | RP11-244H3.1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-590-3p | 13 | TGFBR2 | Sponge network | -0.877 | 0 | -2.395 | 0 | 0.369 |
| 75 | RP11-760H22.2 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-454-3p | 11 | TGFBR2 | Sponge network | -1.096 | 0 | -2.395 | 0 | 0.365 |
| 76 | RP11-67L2.2 | hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-93-5p | 11 | TGFBR2 | Sponge network | -0.677 | 0 | -2.395 | 0 | 0.36 |
| 77 | RP11-400K9.4 | hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-93-5p | 10 | TGFBR2 | Sponge network | -1.058 | 0 | -2.395 | 0 | 0.355 |
| 78 | RP11-5C23.1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-340-5p;hsa-miR-590-3p | 11 | TGFBR2 | Sponge network | -0.816 | 0 | -2.395 | 0 | 0.353 |
| 79 | CTC-366B18.4 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-93-5p | 11 | TGFBR2 | Sponge network | -0.778 | 0 | -2.395 | 0 | 0.35 |
| 80 | RP11-130L8.1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 14 | TGFBR2 | Sponge network | -1.674 | 0 | -2.395 | 0 | 0.347 |
| 81 | LINC00654 | hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-5p | 10 | TGFBR2 | Sponge network | -0.902 | 0 | -2.395 | 0 | 0.346 |
| 82 | CTD-2619J13.17 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-93-5p | 12 | TGFBR2 | Sponge network | -0.985 | 0 | -2.395 | 0 | 0.344 |
| 83 | RP11-486M23.1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-93-5p | 10 | TGFBR2 | Sponge network | -2.239 | 0 | -2.395 | 0 | 0.343 |
| 84 | SNHG18 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-93-5p | 13 | TGFBR2 | Sponge network | -0.741 | 0 | -2.395 | 0 | 0.342 |
| 85 | PWAR6 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-9-5p;hsa-miR-93-5p | 20 | TGFBR2 | Sponge network | -1.696 | 0 | -2.395 | 0 | 0.34 |
| 86 | RP11-378A13.1 | hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-93-5p | 11 | TGFBR2 | Sponge network | -0.486 | 0 | -2.395 | 0 | 0.337 |
| 87 | LINC01085 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-429;hsa-miR-590-3p;hsa-miR-93-5p | 16 | TGFBR2 | Sponge network | -3.703 | 0 | -2.395 | 0 | 0.336 |
| 88 | CTD-3247F14.2 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-9-5p;hsa-miR-93-5p | 12 | TGFBR2 | Sponge network | -2.833 | 0 | -2.395 | 0 | 0.335 |
| 89 | RP11-978I15.10 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-9-5p | 10 | TGFBR2 | Sponge network | -3.591 | 0 | -2.395 | 0 | 0.335 |
| 90 | CTC-523E23.4 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-9-5p | 16 | TGFBR2 | Sponge network | -1.207 | 0 | -2.395 | 0 | 0.333 |
| 91 | RP11-158M2.3 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p | 10 | TGFBR2 | Sponge network | -5.833 | 0 | -2.395 | 0 | 0.331 |
| 92 | RP11-834C11.4 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 13 | TGFBR2 | Sponge network | -0.637 | 0 | -2.395 | 0 | 0.328 |
| 93 | RP11-573G6.6 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-93-5p | 11 | TGFBR2 | Sponge network | -1.242 | 0 | -2.395 | 0 | 0.326 |
| 94 | RP11-2B6.2 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-590-3p | 11 | TGFBR2 | Sponge network | -1.878 | 0 | -2.395 | 0 | 0.326 |
| 95 | AC141928.1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-454-3p | 12 | TGFBR2 | Sponge network | -1.583 | 0 | -2.395 | 0 | 0.324 |
| 96 | CTC-559E9.4 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-340-5p;hsa-miR-590-5p;hsa-miR-93-5p | 11 | TGFBR2 | Sponge network | -1.065 | 0 | -2.395 | 0 | 0.317 |
| 97 | CKMT2-AS1 | hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 11 | TGFBR2 | Sponge network | -0.724 | 0 | -2.395 | 0 | 0.316 |
| 98 | H19 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-454-3p;hsa-miR-93-5p | 10 | TGFBR2 | Sponge network | -0.301 | 0.07845 | -2.395 | 0 | 0.312 |
| 99 | RP11-359E10.1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-340-5p;hsa-miR-93-5p | 13 | TGFBR2 | Sponge network | -1.868 | 0 | -2.395 | 0 | 0.311 |
| 100 | RP11-403B2.7 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-590-3p | 12 | TGFBR2 | Sponge network | -1.946 | 0 | -2.395 | 0 | 0.31 |
| 101 | GRIK1-AS1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-590-3p | 12 | TGFBR2 | Sponge network | -3.557 | 0 | -2.395 | 0 | 0.308 |
| 102 | AC009950.2 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-93-5p | 13 | TGFBR2 | Sponge network | -0.716 | 0.0005 | -2.395 | 0 | 0.308 |
| 103 | SNHG14 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 19 | TGFBR2 | Sponge network | -1.168 | 0 | -2.395 | 0 | 0.306 |
| 104 | RP11-403I13.7 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-590-5p | 13 | TGFBR2 | Sponge network | -0.691 | 0.00056 | -2.395 | 0 | 0.305 |
| 105 | RP11-78O7.2 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-590-3p;hsa-miR-93-5p | 10 | TGFBR2 | Sponge network | -0.963 | 0 | -2.395 | 0 | 0.3 |
| 106 | RP11-456K23.1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-454-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-92a-3p;hsa-miR-93-5p | 16 | TGFBR2 | Sponge network | -0.777 | 0 | -2.395 | 0 | 0.3 |
| 107 | CTB-92J24.3 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20b-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-340-5p;hsa-miR-429;hsa-miR-93-5p | 14 | TGFBR2 | Sponge network | -3.42 | 0 | -2.395 | 0 | 0.298 |
| 108 | RP11-403I13.8 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b;hsa-miR-590-3p;hsa-miR-590-5p | 13 | TGFBR2 | Sponge network | -0.541 | 0.0007 | -2.395 | 0 | 0.294 |
| 109 | GAS6-AS2 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-21-5p;hsa-miR-429;hsa-miR-590-5p | 12 | TGFBR2 | Sponge network | -0.768 | 0 | -2.395 | 0 | 0.285 |
| 110 | RP11-507B12.2 | hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p;hsa-miR-320b | 10 | TGFBR2 | Sponge network | -2.391 | 9.0E-5 | -2.395 | 0 | 0.284 |
| 111 | RP11-293M10.6 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-93-5p | 14 | TGFBR2 | Sponge network | -0.601 | 0.00056 | -2.395 | 0 | 0.261 |
| 112 | AC012360.6 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-20b-5p;hsa-miR-301a-3p;hsa-miR-429;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 15 | TGFBR2 | Sponge network | -0.873 | 9.0E-5 | -2.395 | 0 | 0.259 |
| 113 | RP1-78O14.1 | hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-301a-3p | 10 | TGFBR2 | Sponge network | 0.343 | 0.08978 | -2.395 | 0 | 0.255 |
| 114 | AC007246.3 | hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-19b-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-590-3p;hsa-miR-92a-3p;hsa-miR-93-5p | 12 | TGFBR2 | Sponge network | -0.496 | 0 | -2.395 | 0 | 0.254 |
| 115 | CTB-107G13.1 | hsa-miR-106b-5p;hsa-miR-107;hsa-miR-130b-3p;hsa-miR-17-5p;hsa-miR-18a-5p;hsa-miR-19a-3p;hsa-miR-20a-5p;hsa-miR-21-5p;hsa-miR-301a-3p;hsa-miR-590-3p | 10 | TGFBR2 | Sponge network | -3.353 | 0 | -2.395 | 0 | 0.251 |