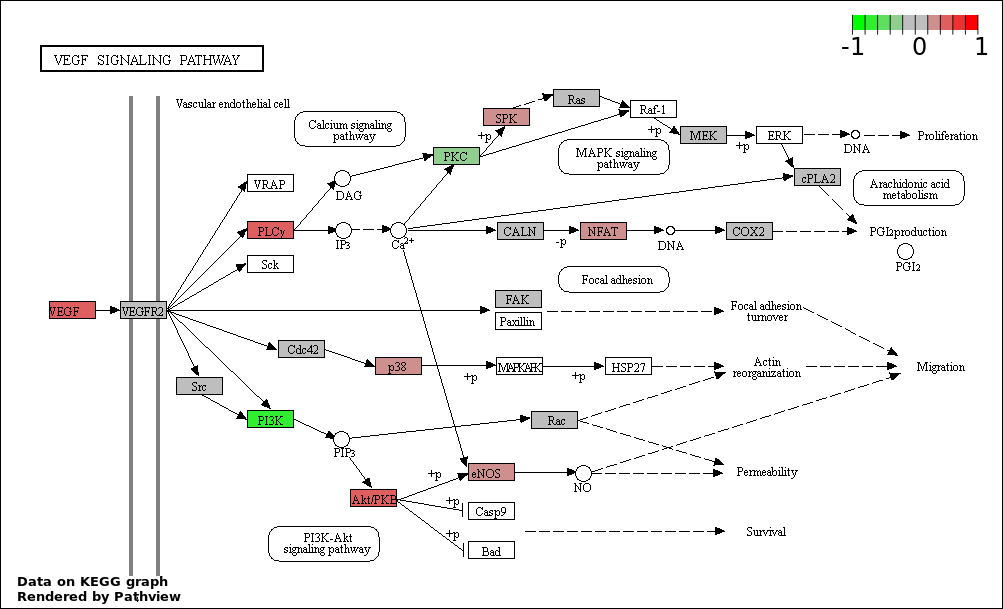
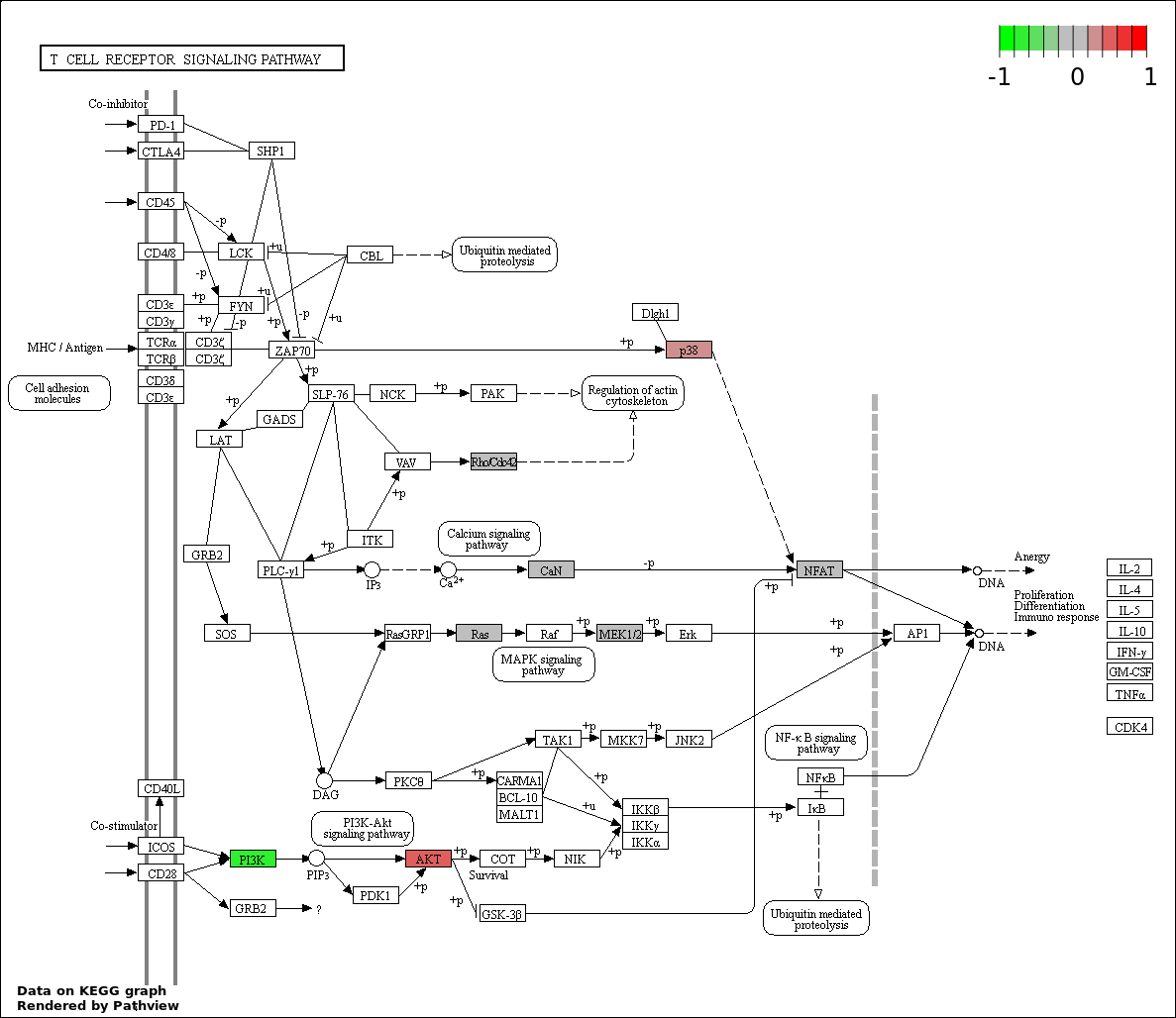
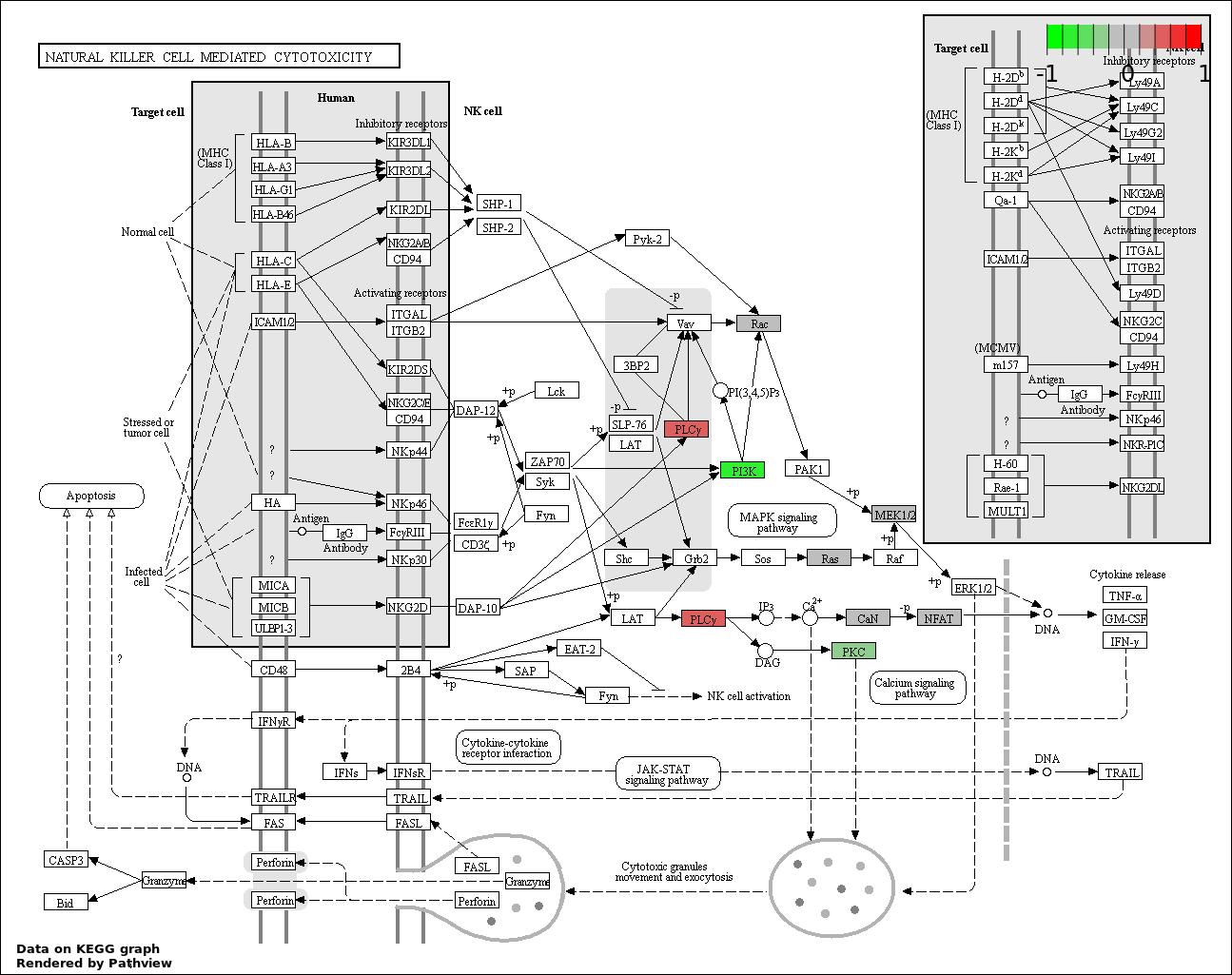
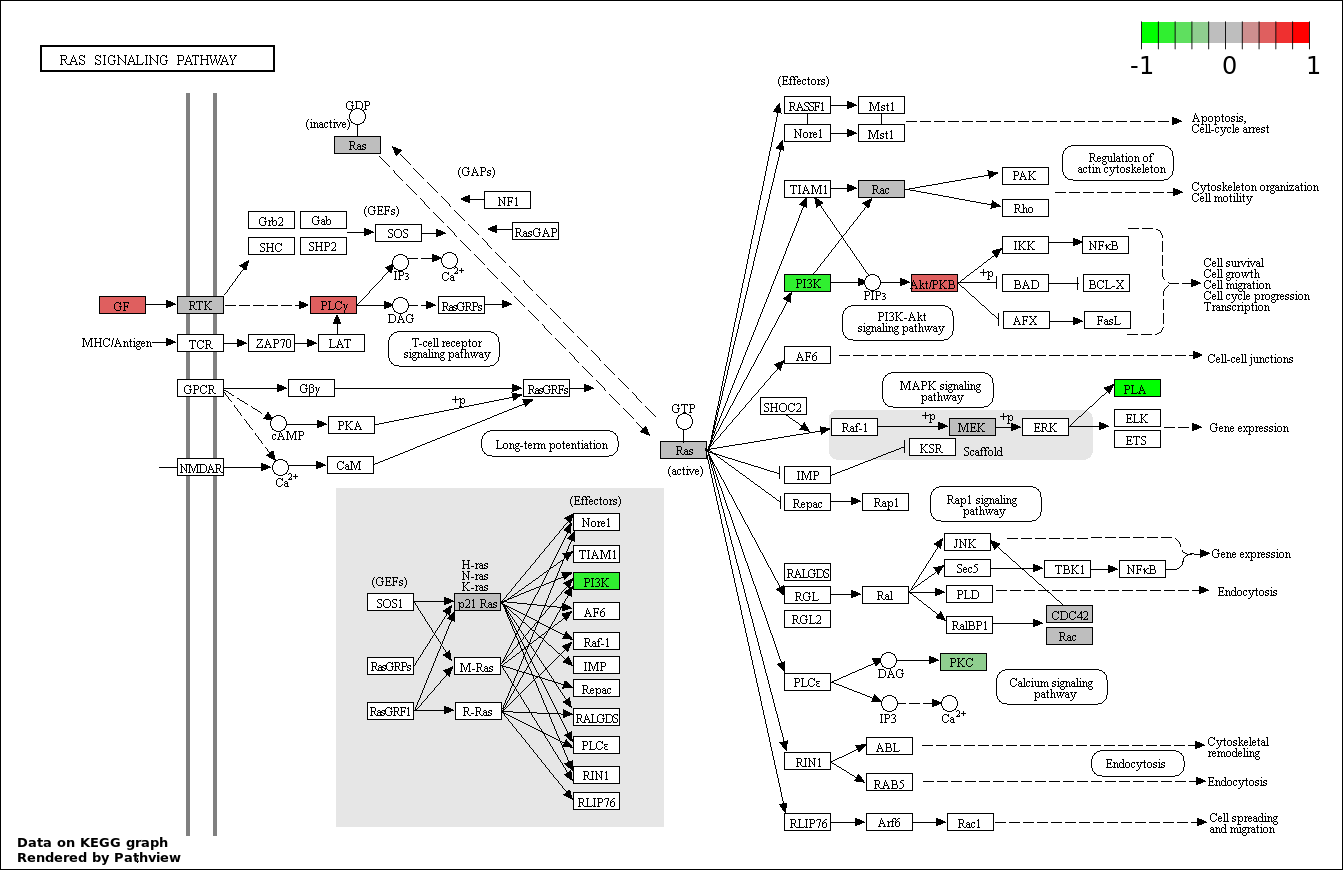
This regulatory network was inferred from the input dataset. The miRNAs and mRNAs are
presented as round and rectangle nodes respectively. The numerical value popped up upon mouse over the gene node is the log2 transformed fold-change of the gene expression between the two groups. All of the nodes are clickable, and the detailed information of the miRNAs/mRNAs and related cancer pathway will be displayed in another window. The edges between nodes are supported by both interactions (predicted or experimentally verified) and correlations learnt from cancer dataset. The numerical value popped up upon mouse over the edge is the correlation beat value (effect size) between the two nodes. The experimental evidences of the edges reported in previous cancer studies are highlighted by red/orange color. All of these information can be accessed by the "mouse-over" action. This network shows a full map of the miRNA-mRNA regulation of the input gene list(s), and the hub miRNAs (with the high network degree/betweenness centrality) would be the potential cancer drivers or tumor suppressors. The full result table can be accessed in the "Regulations" tab.
"miRNACancerMAP" is also a network visualization tool for users to draw their regulatory network by personal customization. Users can set the complexity of the network by limiting the number of nodes or edges. And the color of the nodes can be defined by different categories of the mRNAs and miRNAs, such as Gene-Ontology, pathway, and expression status. Users can also select to use network degree or network betweenness centrality to define the node size. And edges can be black or colored by the correlation. Purple edge means negative correlation (mostly found between miRNA and mRNA), and blue edge means positive correlation (found in PPI or miRNA-miRNA sponge effect). We can also add the protein-protein interactions (PPI) into the network. This result will show the cluster of genes regulated by some specific miRNAs. Additionally, miRNA-miRNA edges can be added by the "miRNA sponge" button, presenting some clusters of miRNAs that have the interactions via sponge effect.
miRNA-gene regulations
| Num | microRNA | Gene | miRNA log2FC | miRNA pvalue | Gene log2FC | Gene pvalue | Interaction | Correlation beta | Correlation P-value | PMID | Reported in cancer studies |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | hsa-miR-106a-5p | AKT3 | -0.2 | 0.80221 | 0.46 | 0.53933 | miRNATAP | -0.55 | 0 | NA | |
| 2 | hsa-miR-106b-5p | AKT3 | -0.3 | 0.86929 | 0.46 | 0.53933 | miRNATAP | -0.91 | 0 | NA | |
| 3 | hsa-miR-107 | AKT3 | -0.04 | 0.98836 | 0.46 | 0.53933 | PITA; miRanda | -0.55 | 0.00017 | NA | |
| 4 | hsa-miR-1275 | AKT3 | -0.83 | 0.08038 | 0.46 | 0.53933 | PITA | -0.27 | 0 | NA | |
| 5 | hsa-miR-142-3p | AKT3 | -0.15 | 0.9461 | 0.46 | 0.53933 | miRanda | -0.2 | 0.00627 | NA | |
| 6 | hsa-miR-15a-5p | AKT3 | -0.07 | 0.96484 | 0.46 | 0.53933 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.72 | 0 | NA | |
| 7 | hsa-miR-15b-5p | AKT3 | -0.27 | 0.87097 | 0.46 | 0.53933 | miRNATAP | -0.67 | 0 | NA | |
| 8 | hsa-miR-16-5p | AKT3 | 0.01 | 0.99448 | 0.46 | 0.53933 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.75 | 0 | NA | |
| 9 | hsa-miR-17-3p | AKT3 | 0.1 | 0.96011 | 0.46 | 0.53933 | miRNATAP | -0.52 | 0 | NA | |
| 10 | hsa-miR-17-5p | AKT3 | -0.18 | 0.93454 | 0.46 | 0.53933 | TargetScan; miRNATAP | -0.63 | 0 | NA | |
| 11 | hsa-miR-181b-5p | AKT3 | 0.16 | 0.93018 | 0.46 | 0.53933 | miRNATAP | -0.27 | 0.00963 | NA | |
| 12 | hsa-miR-20a-5p | AKT3 | -0.18 | 0.92812 | 0.46 | 0.53933 | miRNATAP | -0.53 | 0 | NA | |
| 13 | hsa-miR-28-3p | AKT3 | -0.23 | 0.93535 | 0.46 | 0.53933 | miRNATAP | -0.52 | 0.00719 | NA | |
| 14 | hsa-miR-29a-3p | AKT3 | 0.01 | 0.99698 | 0.46 | 0.53933 | miRNATAP | -0.81 | 0 | NA | |
| 15 | hsa-miR-29b-3p | AKT3 | -0.1 | 0.95899 | 0.46 | 0.53933 | miRNATAP | -0.68 | 0 | 26512921 | MicroRNA 29B mir 29b regulates the Warburg effect in ovarian cancer by targeting AKT2 and AKT3 |
| 16 | hsa-miR-32-3p | AKT3 | -0.57 | 0.13133 | 0.46 | 0.53933 | mirMAP | -0.56 | 0 | NA | |
| 17 | hsa-miR-320a | AKT3 | -0.42 | 0.8402 | 0.46 | 0.53933 | PITA; miRanda; miRNATAP | -0.33 | 0.00138 | NA | |
| 18 | hsa-miR-320b | AKT3 | -0.24 | 0.85922 | 0.46 | 0.53933 | PITA; miRanda; miRNATAP | -0.25 | 0.00855 | NA | |
| 19 | hsa-miR-335-3p | AKT3 | -0.24 | 0.8845 | 0.46 | 0.53933 | mirMAP | -0.58 | 0 | NA | |
| 20 | hsa-miR-33a-3p | AKT3 | -0.79 | 0.01052 | 0.46 | 0.53933 | mirMAP | -0.49 | 0 | NA | |
| 21 | hsa-miR-340-5p | AKT3 | -0 | 0.99757 | 0.46 | 0.53933 | mirMAP | -0.28 | 0.006 | NA | |
| 22 | hsa-miR-362-3p | AKT3 | -0.72 | 0.03459 | 0.46 | 0.53933 | miRanda | -0.42 | 0 | NA | |
| 23 | hsa-miR-362-5p | AKT3 | -0.27 | 0.75202 | 0.46 | 0.53933 | PITA; TargetScan; miRNATAP | -0.52 | 0 | NA | |
| 24 | hsa-miR-369-3p | AKT3 | 0.12 | 0.8323 | 0.46 | 0.53933 | mirMAP | -0.23 | 0.00901 | NA | |
| 25 | hsa-miR-374a-5p | AKT3 | -0.34 | 0.76692 | 0.46 | 0.53933 | mirMAP | -0.58 | 0 | NA | |
| 26 | hsa-miR-374b-5p | AKT3 | -0.29 | 0.8357 | 0.46 | 0.53933 | mirMAP | -0.68 | 0 | NA | |
| 27 | hsa-miR-421 | AKT3 | -0.18 | 0.7347 | 0.46 | 0.53933 | miRanda; mirMAP | -0.46 | 0 | NA | |
| 28 | hsa-miR-424-5p | AKT3 | 0.25 | 0.87015 | 0.46 | 0.53933 | miRNATAP | -0.24 | 0.00104 | 26315541 | Silencing Akt3 and E2F3 by siRNA pheno-copied the effect of ectopic miR-424 on HCC growth; Whereas overexpression of Akt3 and E2F3 attenuated the effect of miR-424 on HCC growth |
| 29 | hsa-miR-501-3p | AKT3 | -0.75 | 0.55276 | 0.46 | 0.53933 | miRNATAP | -0.5 | 0 | NA | |
| 30 | hsa-miR-502-3p | AKT3 | -0.48 | 0.5143 | 0.46 | 0.53933 | miRNATAP | -0.64 | 0 | NA | |
| 31 | hsa-miR-502-5p | AKT3 | -0.71 | 0.02613 | 0.46 | 0.53933 | PITA; miRNATAP | -0.41 | 0 | NA | |
| 32 | hsa-miR-505-3p | AKT3 | -0.47 | 0.69038 | 0.46 | 0.53933 | mirMAP | -0.77 | 0 | 22051041 | We also find that Akt3 correlate inversely with miR-505 modulates drug sensitivity in MCF7-ADR |
| 33 | hsa-miR-548o-3p | AKT3 | -0.19 | 0.51773 | 0.46 | 0.53933 | mirMAP | -0.31 | 8.0E-5 | NA | |
| 34 | hsa-miR-577 | AKT3 | -1.06 | 0.32606 | 0.46 | 0.53933 | mirMAP | -0.38 | 0 | NA | |
| 35 | hsa-miR-663b | AKT3 | 0.1 | 0.81499 | 0.46 | 0.53933 | PITA | -0.25 | 1.0E-5 | NA | |
| 36 | hsa-miR-769-5p | AKT3 | -0.31 | 0.7357 | 0.46 | 0.53933 | PITA; miRNATAP | -0.84 | 0 | NA | |
| 37 | hsa-miR-93-5p | AKT3 | -0.61 | 0.8253 | 0.46 | 0.53933 | miRNATAP | -0.76 | 0 | NA | |
| 38 | hsa-miR-143-5p | CDC42 | 0.11 | 0.93336 | -0.15 | 0.93702 | miRNATAP | -0.12 | 2.0E-5 | NA | |
| 39 | hsa-miR-125a-3p | KDR | 0.15 | 0.72476 | -0.04 | 0.969 | miRanda | -0.14 | 0.00574 | NA | |
| 40 | hsa-miR-149-5p | KDR | -0.03 | 0.96106 | -0.04 | 0.969 | miRNATAP | -0.18 | 0.00029 | NA | |
| 41 | hsa-miR-15a-5p | KDR | -0.07 | 0.96484 | -0.04 | 0.969 | miRNATAP | -0.25 | 0.00079 | NA | |
| 42 | hsa-miR-15b-5p | KDR | -0.27 | 0.87097 | -0.04 | 0.969 | miRNATAP | -0.33 | 0 | NA | |
| 43 | hsa-miR-16-5p | KDR | 0.01 | 0.99448 | -0.04 | 0.969 | miRTarBase; miRNATAP | -0.37 | 0 | 26934556 | The expression levels of two target genes Myb and VEGFR2 were affected significantly by miR-16 while glucose administration inhibited miR-16 expression and enhanced tumor cell proliferation and migration; Hyperglycemia can impact the clinical outcomes of CRC patients likely by inhibiting miR-16 expression and the expression of its downstream genes Myb and VEGFR2 |
| 44 | hsa-miR-200b-3p | KDR | -0.43 | 0.86396 | -0.04 | 0.969 | miRNAWalker2 validate; miRTarBase; TargetScan | -0.24 | 7.0E-5 | NA | |
| 45 | hsa-miR-200c-3p | KDR | -0.44 | 0.88712 | -0.04 | 0.969 | miRNATAP | -0.24 | 0.00011 | 24205206 | MiR 200c increases the radiosensitivity of non small cell lung cancer cell line A549 by targeting VEGF VEGFR2 pathway; MiR-200c at the nexus of epithelial-mesenchymal transition EMT is predicted to target VEGFR2; The purpose of this study is to test the hypothesis that regulation of VEGFR2 pathway by miR-200c could modulate the radiosensitivity of cancer cells; Bioinformatic analysis luciferase reporter assays and biochemical assays were carried out to validate VEGFR2 as a direct target of miR-200c; We identified VEGFR2 as a novel target of miR-200c |
| 46 | hsa-miR-335-5p | KDR | -0.03 | 0.97338 | -0.04 | 0.969 | miRNAWalker2 validate | -0.3 | 0 | NA | |
| 47 | hsa-miR-429 | KDR | -0.46 | 0.80624 | -0.04 | 0.969 | PITA; miRanda; miRNATAP | -0.23 | 0 | NA | |
| 48 | hsa-miR-590-3p | KDR | -0.28 | 0.59127 | -0.04 | 0.969 | miRanda | -0.32 | 0 | NA | |
| 49 | hsa-miR-590-5p | KDR | -0.55 | 0.47274 | -0.04 | 0.969 | miRanda | -0.33 | 0 | NA | |
| 50 | hsa-miR-126-5p | KRAS | 0.08 | 0.95664 | 0.08 | 0.95389 | MirTarget | -0.12 | 0.00133 | 22845403 | miR-126 is also known to target other crucial oncogenes in PDAC such as KRAS and CRK |
| 51 | hsa-miR-1271-5p | KRAS | -0.46 | 0.09563 | 0.08 | 0.95389 | MirTarget | -0.11 | 8.0E-5 | NA | |
| 52 | hsa-miR-181c-5p | KRAS | 0.04 | 0.96916 | 0.08 | 0.95389 | miRNAWalker2 validate; miRTarBase | -0.1 | 0.0009 | NA | |
| 53 | hsa-miR-199a-5p | KRAS | 0.16 | 0.9358 | 0.08 | 0.95389 | miRanda; miRNATAP | -0.11 | 0.0009 | NA | |
| 54 | hsa-miR-199b-5p | KRAS | -0.04 | 0.97717 | 0.08 | 0.95389 | miRanda; miRNATAP | -0.11 | 0.0001 | 27517624 | The miR-199b prognostic impact was particularly evident in both younger and KRAS wild-type subgroups |
| 55 | hsa-miR-30e-5p | KRAS | -0.07 | 0.97968 | 0.08 | 0.95389 | mirMAP; miRNATAP | -0.12 | 0.00857 | NA | |
| 56 | hsa-miR-101-3p | MAP2K1 | 0.01 | 0.99704 | -0.08 | 0.95198 | miRNAWalker2 validate | -0.18 | 2.0E-5 | NA | |
| 57 | hsa-miR-34c-5p | MAP2K1 | -0.02 | 0.95279 | -0.08 | 0.95198 | PITA; miRanda | -0.11 | 0.0001 | NA | |
| 58 | hsa-miR-497-5p | MAP2K1 | -0.01 | 0.98915 | -0.08 | 0.95198 | miRNAWalker2 validate | -0.15 | 1.0E-5 | NA | |
| 59 | hsa-miR-34c-5p | MAPK13 | -0.02 | 0.95279 | 0.09 | 0.95638 | miRanda; miRNATAP | -0.12 | 0.0038 | NA | |
| 60 | hsa-miR-125a-3p | MAPK14 | 0.15 | 0.72476 | 0.13 | 0.93083 | miRanda | -0.1 | 0 | NA | |
| 61 | hsa-miR-125a-5p | MAPK14 | -0 | 0.99916 | 0.13 | 0.93083 | PITA; miRanda; miRNATAP | -0.13 | 0.00034 | NA | |
| 62 | hsa-miR-142-5p | MAPK14 | -0.12 | 0.92967 | 0.13 | 0.93083 | PITA | -0.13 | 0 | NA | |
| 63 | hsa-miR-145-3p | MAPK14 | 0.13 | 0.91316 | 0.13 | 0.93083 | MirTarget; miRNATAP | -0.1 | 0.00177 | NA | |
| 64 | hsa-miR-22-3p | MAPK14 | 0.06 | 0.98656 | 0.13 | 0.93083 | MirTarget; miRNATAP | -0.17 | 0.00042 | NA | |
| 65 | hsa-miR-27a-3p | MAPK14 | -0.12 | 0.9588 | 0.13 | 0.93083 | MirTarget; miRNATAP | -0.12 | 0.00681 | NA | |
| 66 | hsa-miR-27b-3p | MAPK14 | -0.14 | 0.95194 | 0.13 | 0.93083 | MirTarget; miRNATAP | -0.14 | 0.00597 | NA | |
| 67 | hsa-miR-320b | MAPK14 | -0.24 | 0.85922 | 0.13 | 0.93083 | PITA; miRanda | -0.11 | 0.00016 | NA | |
| 68 | hsa-miR-342-3p | MAPK14 | -0.37 | 0.77314 | 0.13 | 0.93083 | miRanda | -0.14 | 0 | NA | |
| 69 | hsa-miR-132-3p | NFAT5 | -0.24 | 0.87175 | 0.24 | 0.86876 | miRNATAP | -0.3 | 0 | NA | |
| 70 | hsa-miR-140-3p | NFAT5 | -0.37 | 0.86476 | 0.24 | 0.86876 | miRNATAP | -0.23 | 0.00235 | NA | |
| 71 | hsa-miR-146b-5p | NFAT5 | -0.4 | 0.83751 | 0.24 | 0.86876 | miRanda | -0.2 | 3.0E-5 | NA | |
| 72 | hsa-miR-155-5p | NFAT5 | -0.41 | 0.82867 | 0.24 | 0.86876 | mirMAP; miRNATAP | -0.15 | 0.00026 | NA | |
| 73 | hsa-miR-21-3p | NFAT5 | -0.4 | 0.8782 | 0.24 | 0.86876 | MirTarget | -0.18 | 0.00029 | NA | |
| 74 | hsa-miR-21-5p | NFAT5 | -0.15 | 0.97024 | 0.24 | 0.86876 | miRNAWalker2 validate; mirMAP | -0.29 | 5.0E-5 | NA | |
| 75 | hsa-miR-511-5p | NFAT5 | -0.31 | 0.6072 | 0.24 | 0.86876 | MirTarget | -0.11 | 0.00293 | NA | |
| 76 | hsa-miR-582-3p | NFAT5 | -0.23 | 0.89768 | 0.24 | 0.86876 | MirTarget | -0.17 | 0.0001 | NA | |
| 77 | hsa-miR-582-5p | NFAT5 | -0.4 | 0.58123 | 0.24 | 0.86876 | MirTarget; PITA; miRNATAP | -0.15 | 0.0014 | NA | |
| 78 | hsa-miR-889-3p | NFAT5 | 0.17 | 0.84879 | 0.24 | 0.86876 | MirTarget | -0.17 | 0.00648 | NA | |
| 79 | hsa-miR-92b-3p | NFAT5 | -0.57 | 0.68932 | 0.24 | 0.86876 | miRNATAP | -0.18 | 0 | NA | |
| 80 | hsa-miR-421 | NFATC1 | -0.18 | 0.7347 | -0.12 | 0.85235 | miRanda | -0.58 | 0 | NA | |
| 81 | hsa-miR-940 | NFATC1 | -0.23 | 0.68006 | -0.12 | 0.85235 | PITA | -0.3 | 1.0E-5 | NA | |
| 82 | hsa-miR-130b-5p | NFATC2 | -0.65 | 0.3791 | 0.21 | 0.70733 | MirTarget | -0.37 | 0.00095 | NA | |
| 83 | hsa-miR-16-2-3p | NFATC2 | -0.37 | 0.54685 | 0.21 | 0.70733 | mirMAP | -0.32 | 0.0026 | NA | |
| 84 | hsa-miR-182-5p | NFATC2 | -0.1 | 0.97338 | 0.21 | 0.70733 | mirMAP | -0.27 | 0.00971 | NA | |
| 85 | hsa-miR-200b-5p | NFATC2 | -0.5 | 0.72226 | 0.21 | 0.70733 | mirMAP | -0.31 | 0.00378 | NA | |
| 86 | hsa-miR-26b-3p | NFATC2 | -0.36 | 0.68112 | 0.21 | 0.70733 | MirTarget | -0.32 | 0.00865 | NA | |
| 87 | hsa-miR-31-3p | NFATC2 | 0.2 | 0.76203 | 0.21 | 0.70733 | MirTarget | -0.24 | 1.0E-5 | NA | |
| 88 | hsa-miR-340-5p | NFATC2 | -0 | 0.99757 | 0.21 | 0.70733 | miRNATAP | -0.35 | 0.00984 | NA | |
| 89 | hsa-miR-484 | NFATC2 | -0.18 | 0.88633 | 0.21 | 0.70733 | MirTarget | -0.42 | 0.00431 | NA | |
| 90 | hsa-miR-590-3p | NFATC2 | -0.28 | 0.59127 | 0.21 | 0.70733 | mirMAP | -0.34 | 0.00047 | NA | |
| 91 | hsa-miR-7-1-3p | NFATC2 | -0.46 | 0.6659 | 0.21 | 0.70733 | mirMAP | -0.33 | 0.00324 | NA | |
| 92 | hsa-miR-7-5p | NFATC2 | 0.21 | 0.77371 | 0.21 | 0.70733 | MirTarget; miRNATAP | -0.22 | 0.00223 | NA | |
| 93 | hsa-miR-1254 | NFATC4 | -0.49 | 0.19552 | 0.23 | 0.8009 | MirTarget | -0.17 | 0.00079 | NA | |
| 94 | hsa-miR-130b-5p | NFATC4 | -0.65 | 0.3791 | 0.23 | 0.8009 | mirMAP | -0.23 | 0.00013 | NA | |
| 95 | hsa-miR-15b-3p | NFATC4 | -0.56 | 0.60918 | 0.23 | 0.8009 | mirMAP | -0.27 | 3.0E-5 | NA | |
| 96 | hsa-miR-29a-3p | NFATC4 | 0.01 | 0.99698 | 0.23 | 0.8009 | miRNATAP | -0.26 | 0.00188 | NA | |
| 97 | hsa-miR-29b-3p | NFATC4 | -0.1 | 0.95899 | 0.23 | 0.8009 | miRNATAP | -0.23 | 0.00033 | NA | |
| 98 | hsa-miR-320c | NFATC4 | -0.58 | 0.1136 | 0.23 | 0.8009 | miRanda; mirMAP | -0.14 | 0.00652 | NA | |
| 99 | hsa-miR-361-3p | NFATC4 | -0.29 | 0.84773 | 0.23 | 0.8009 | mirMAP | -0.24 | 0.00961 | NA | |
| 100 | hsa-miR-429 | NFATC4 | -0.46 | 0.80624 | 0.23 | 0.8009 | miRNATAP | -0.21 | 2.0E-5 | NA | |
| 101 | hsa-miR-532-5p | NFATC4 | -0.35 | 0.87895 | 0.23 | 0.8009 | mirMAP | -0.26 | 3.0E-5 | NA | |
| 102 | hsa-miR-625-5p | NFATC4 | -0.69 | 0.21031 | 0.23 | 0.8009 | mirMAP | -0.14 | 0.00216 | NA | |
| 103 | hsa-miR-629-3p | NFATC4 | -0.5 | 0.33397 | 0.23 | 0.8009 | mirMAP | -0.2 | 9.0E-5 | NA | |
| 104 | hsa-miR-429 | NOS3 | -0.46 | 0.80624 | 0.4 | 0.68809 | miRanda | -0.15 | 0.0031 | NA | |
| 105 | hsa-let-7e-5p | NRAS | 0.21 | 0.92234 | -0.12 | 0.93258 | MirTarget; miRNATAP | -0.13 | 0.00397 | NA | |
| 106 | hsa-let-7f-1-3p | PIK3CA | -0.31 | 0.69341 | 0.02 | 0.97942 | mirMAP | -0.12 | 0.00648 | NA | |
| 107 | hsa-miR-148a-5p | PIK3CA | -0.45 | 0.74842 | 0.02 | 0.97942 | mirMAP | -0.11 | 7.0E-5 | NA | |
| 108 | hsa-miR-148b-3p | PIK3CA | -0.2 | 0.91188 | 0.02 | 0.97942 | miRNAWalker2 validate | -0.12 | 0.00491 | NA | |
| 109 | hsa-miR-16-2-3p | PIK3CA | -0.37 | 0.54685 | 0.02 | 0.97942 | mirMAP | -0.1 | 9.0E-5 | NA | |
| 110 | hsa-miR-17-5p | PIK3CA | -0.18 | 0.93454 | 0.02 | 0.97942 | miRNAWalker2 validate | -0.12 | 0 | NA | |
| 111 | hsa-miR-186-5p | PIK3CA | -0.32 | 0.85413 | 0.02 | 0.97942 | mirMAP | -0.15 | 0.00027 | NA | |
| 112 | hsa-miR-32-3p | PIK3CA | -0.57 | 0.13133 | 0.02 | 0.97942 | mirMAP | -0.11 | 0 | NA | |
| 113 | hsa-miR-335-5p | PIK3CA | -0.03 | 0.97338 | 0.02 | 0.97942 | miRNAWalker2 validate | -0.1 | 0.0001 | NA | |
| 114 | hsa-miR-339-5p | PIK3CA | -0.3 | 0.71291 | 0.02 | 0.97942 | miRanda | -0.1 | 1.0E-5 | NA | |
| 115 | hsa-miR-374b-5p | PIK3CA | -0.29 | 0.8357 | 0.02 | 0.97942 | mirMAP | -0.14 | 0.00032 | NA | |
| 116 | hsa-miR-501-5p | PIK3CA | -0.83 | 0.05827 | 0.02 | 0.97942 | mirMAP | -0.11 | 0 | NA | |
| 117 | hsa-miR-576-5p | PIK3CA | -0.51 | 0.41719 | 0.02 | 0.97942 | PITA | -0.11 | 7.0E-5 | NA | |
| 118 | hsa-miR-582-3p | PIK3CA | -0.23 | 0.89768 | 0.02 | 0.97942 | miRNATAP | -0.14 | 0 | NA | |
| 119 | hsa-miR-590-3p | PIK3CA | -0.28 | 0.59127 | 0.02 | 0.97942 | miRanda; mirMAP | -0.12 | 0 | NA | |
| 120 | hsa-miR-590-5p | PIK3CA | -0.55 | 0.47274 | 0.02 | 0.97942 | miRanda | -0.14 | 0 | NA | |
| 121 | hsa-miR-96-5p | PIK3CA | 0.09 | 0.92309 | 0.02 | 0.97942 | TargetScan | -0.13 | 0 | NA | |
| 122 | hsa-miR-1468-5p | PIK3CD | -0.2 | 0.68841 | -0.05 | 0.94703 | MirTarget | -0.29 | 0.00025 | NA | |
| 123 | hsa-miR-29a-3p | PIK3CD | 0.01 | 0.99698 | -0.05 | 0.94703 | mirMAP | -0.35 | 0.00244 | NA | |
| 124 | hsa-miR-29b-3p | PIK3CD | -0.1 | 0.95899 | -0.05 | 0.94703 | mirMAP | -0.26 | 0.00266 | NA | |
| 125 | hsa-miR-30b-5p | PIK3CD | -0.01 | 0.99462 | -0.05 | 0.94703 | MirTarget | -0.4 | 0 | NA | |
| 126 | hsa-miR-30c-5p | PIK3CD | -0.3 | 0.86581 | -0.05 | 0.94703 | MirTarget; miRNATAP | -0.42 | 0.00071 | NA | |
| 127 | hsa-miR-30d-5p | PIK3CD | -0.17 | 0.95173 | -0.05 | 0.94703 | MirTarget; miRNATAP | -0.36 | 0.00123 | NA | |
| 128 | hsa-miR-421 | PIK3CD | -0.18 | 0.7347 | -0.05 | 0.94703 | miRanda | -0.49 | 0 | NA | |
| 129 | hsa-miR-484 | PIK3CD | -0.18 | 0.88633 | -0.05 | 0.94703 | MirTarget; miRNATAP | -0.35 | 0.00143 | NA | |
| 130 | hsa-miR-7-5p | PIK3CD | 0.21 | 0.77371 | -0.05 | 0.94703 | miRTarBase; MirTarget; miRNATAP | -0.2 | 0.00035 | 22234835 | We first screened and identified a novel miR-7 target phosphoinositide 3-kinase catalytic subunit delta PIK3CD; A correlation between miR-7 and PIK3CD expression was also confirmed in clinical samples of HCC; By targeting PIK3CD mTOR and p70S6K miR-7 efficiently regulates the PI3K/Akt pathway |
| 131 | hsa-miR-942-5p | PIK3CD | -0.51 | 0.50778 | -0.05 | 0.94703 | MirTarget | -0.19 | 0.00863 | NA | |
| 132 | hsa-miR-126-3p | PIK3CG | -0.11 | 0.96367 | -0.36 | 0.46972 | miRTarBase | -0.53 | 0.00048 | NA | |
| 133 | hsa-miR-26b-5p | PIK3CG | -0.02 | 0.99038 | -0.36 | 0.46972 | miRNAWalker2 validate | -0.8 | 0 | NA | |
| 134 | hsa-miR-29b-3p | PIK3CG | -0.1 | 0.95899 | -0.36 | 0.46972 | miRTarBase | -0.67 | 0 | NA | |
| 135 | hsa-miR-335-3p | PIK3CG | -0.24 | 0.8845 | -0.36 | 0.46972 | mirMAP | -0.56 | 0 | NA | |
| 136 | hsa-miR-421 | PIK3CG | -0.18 | 0.7347 | -0.36 | 0.46972 | miRanda | -0.44 | 0.00013 | NA | |
| 137 | hsa-miR-502-5p | PIK3CG | -0.71 | 0.02613 | -0.36 | 0.46972 | miRNATAP | -0.34 | 0.00065 | 26163264 | Phosphoinositide 3-kinase catalytic subunit gamma PIK3CG was identified as a direct downstream target of miR-502 in HCC cells; Notably overexpression of PIK3CG reversed the inhibitory effects of miR-502 in HCC cells |
| 138 | hsa-miR-590-3p | PIK3CG | -0.28 | 0.59127 | -0.36 | 0.46972 | miRanda | -0.44 | 1.0E-5 | NA | |
| 139 | hsa-miR-132-3p | PIK3R1 | -0.24 | 0.87175 | -0.11 | 0.93752 | MirTarget | -0.19 | 0.0073 | NA | |
| 140 | hsa-miR-15a-5p | PIK3R1 | -0.07 | 0.96484 | -0.11 | 0.93752 | MirTarget | -0.22 | 9.0E-5 | NA | |
| 141 | hsa-miR-15b-5p | PIK3R1 | -0.27 | 0.87097 | -0.11 | 0.93752 | MirTarget | -0.14 | 0.00644 | NA | |
| 142 | hsa-miR-16-5p | PIK3R1 | 0.01 | 0.99448 | -0.11 | 0.93752 | MirTarget | -0.19 | 0.00026 | NA | |
| 143 | hsa-miR-185-5p | PIK3R1 | -0.29 | 0.82059 | -0.11 | 0.93752 | miRNATAP | -0.24 | 0.00027 | NA | |
| 144 | hsa-miR-21-5p | PIK3R1 | -0.15 | 0.97024 | -0.11 | 0.93752 | miRNAWalker2 validate; MirTarget; miRNATAP | -0.32 | 4.0E-5 | 26676464 | PIK3R1 targeting by miR 21 suppresses tumor cell migration and invasion by reducing PI3K/AKT signaling and reversing EMT and predicts clinical outcome of breast cancer; Next we identified the PIK3R1 as a direct target of miR-21 and showed that it was negatively regulated by miR-21; Taken together we provide novel evidence that miR-21 knockdown suppresses cell growth migration and invasion partly by inhibiting PI3K/AKT activation via direct targeting PIK3R1 and reversing EMT in breast cancer |
| 145 | hsa-miR-212-3p | PIK3R1 | -0.02 | 0.97323 | -0.11 | 0.93752 | MirTarget | -0.23 | 5.0E-5 | NA | |
| 146 | hsa-miR-22-5p | PIK3R1 | -0.08 | 0.9317 | -0.11 | 0.93752 | mirMAP | -0.19 | 3.0E-5 | NA | |
| 147 | hsa-miR-221-3p | PIK3R1 | 0.09 | 0.95912 | -0.11 | 0.93752 | MirTarget | -0.2 | 0.00053 | NA | |
| 148 | hsa-miR-222-3p | PIK3R1 | -0.03 | 0.98401 | -0.11 | 0.93752 | MirTarget | -0.2 | 6.0E-5 | NA | |
| 149 | hsa-miR-369-3p | PIK3R1 | 0.12 | 0.8323 | -0.11 | 0.93752 | PITA | -0.17 | 0.00038 | NA | |
| 150 | hsa-miR-424-5p | PIK3R1 | 0.25 | 0.87015 | -0.11 | 0.93752 | MirTarget | -0.12 | 0.00204 | NA |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | POSITIVE REGULATION OF RESPONSE TO STIMULUS | 32 | 1929 | 8.839e-26 | 4.113e-22 |
| 2 | PHOSPHATE CONTAINING COMPOUND METABOLIC PROCESS | 31 | 1977 | 6.325e-24 | 1.471e-20 |
| 3 | INTRACELLULAR SIGNAL TRANSDUCTION | 28 | 1572 | 2.08e-22 | 3.226e-19 |
| 4 | IMMUNE RESPONSE REGULATING CELL SURFACE RECEPTOR SIGNALING PATHWAY | 17 | 323 | 1.44e-20 | 1.675e-17 |
| 5 | FC RECEPTOR SIGNALING PATHWAY | 15 | 206 | 2.984e-20 | 2.777e-17 |
| 6 | GLYCEROLIPID BIOSYNTHETIC PROCESS | 14 | 211 | 2.496e-18 | 1.936e-15 |
| 7 | GLYCEROPHOSPHOLIPID METABOLIC PROCESS | 15 | 297 | 7.621e-18 | 5.066e-15 |
| 8 | PHOSPHOLIPID BIOSYNTHETIC PROCESS | 14 | 235 | 1.147e-17 | 6.67e-15 |
| 9 | PHOSPHATIDYLINOSITOL METABOLIC PROCESS | 13 | 193 | 4.009e-17 | 2.073e-14 |
| 10 | GLYCEROLIPID METABOLIC PROCESS | 15 | 356 | 1.143e-16 | 5.32e-14 |
| 11 | PHOSPHOLIPID METABOLIC PROCESS | 15 | 364 | 1.591e-16 | 6.73e-14 |
| 12 | VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 10 | 74 | 1.982e-16 | 7.684e-14 |
| 13 | REGULATION OF IMMUNE SYSTEM PROCESS | 22 | 1403 | 1.164e-15 | 4.166e-13 |
| 14 | LIPID BIOSYNTHETIC PROCESS | 16 | 539 | 2.15e-15 | 7.145e-13 |
| 15 | REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 23 | 1656 | 2.511e-15 | 7.789e-13 |
| 16 | FC EPSILON RECEPTOR SIGNALING PATHWAY | 11 | 142 | 3.024e-15 | 8.794e-13 |
| 17 | POSITIVE REGULATION OF CELL COMMUNICATION | 22 | 1532 | 7.256e-15 | 1.986e-12 |
| 18 | PHOSPHATIDYLETHANOLAMINE ACYL CHAIN REMODELING | 7 | 23 | 1.76e-14 | 4.551e-12 |
| 19 | CELLULAR LIPID METABOLIC PROCESS | 18 | 913 | 2.758e-14 | 6.755e-12 |
| 20 | PHOSPHATIDYLCHOLINE ACYL CHAIN REMODELING | 7 | 26 | 4.703e-14 | 1.094e-11 |
| 21 | ORGANOPHOSPHATE BIOSYNTHETIC PROCESS | 14 | 450 | 9.491e-14 | 2.103e-11 |
| 22 | CALCIUM MEDIATED SIGNALING | 9 | 90 | 1.224e-13 | 2.589e-11 |
| 23 | REGULATION OF IMMUNE RESPONSE | 17 | 858 | 1.693e-13 | 3.283e-11 |
| 24 | VASCULATURE DEVELOPMENT | 14 | 469 | 1.668e-13 | 3.283e-11 |
| 25 | PHOSPHATIDYLGLYCEROL METABOLIC PROCESS | 7 | 31 | 1.866e-13 | 3.474e-11 |
| 26 | POSITIVE REGULATION OF BIOSYNTHETIC PROCESS | 22 | 1805 | 2.123e-13 | 3.799e-11 |
| 27 | POSITIVE REGULATION OF INTRACELLULAR SIGNAL TRANSDUCTION | 17 | 876 | 2.37e-13 | 4.083e-11 |
| 28 | PHOSPHORYLATION | 19 | 1228 | 3.141e-13 | 5.219e-11 |
| 29 | PHOSPHATIDYLGLYCEROL ACYL CHAIN REMODELING | 6 | 17 | 5.263e-13 | 8.444e-11 |
| 30 | SECOND MESSENGER MEDIATED SIGNALING | 10 | 160 | 5.576e-13 | 8.648e-11 |
| 31 | INOSITOL PHOSPHATE MEDIATED SIGNALING | 6 | 18 | 7.883e-13 | 1.183e-10 |
| 32 | LOCOMOTION | 18 | 1114 | 8.205e-13 | 1.193e-10 |
| 33 | ACTIVATION OF IMMUNE RESPONSE | 13 | 427 | 1.139e-12 | 1.606e-10 |
| 34 | REGULATION OF PHOSPHATIDYLINOSITOL 3 KINASE ACTIVITY | 7 | 40 | 1.306e-12 | 1.788e-10 |
| 35 | IMMUNE SYSTEM PROCESS | 22 | 1984 | 1.452e-12 | 1.93e-10 |
| 36 | LIPID METABOLIC PROCESS | 18 | 1158 | 1.578e-12 | 2.04e-10 |
| 37 | REGULATION OF TRANSPORT | 21 | 1804 | 2.499e-12 | 3.142e-10 |
| 38 | POSITIVE REGULATION OF IMMUNE SYSTEM PROCESS | 16 | 867 | 3.21e-12 | 3.93e-10 |
| 39 | POSITIVE REGULATION OF PHOSPHATE METABOLIC PROCESS | 17 | 1036 | 3.507e-12 | 4.079e-10 |
| 40 | POSITIVE REGULATION OF PHOSPHORUS METABOLIC PROCESS | 17 | 1036 | 3.507e-12 | 4.079e-10 |
| 41 | REGULATION OF PHOSPHORUS METABOLIC PROCESS | 20 | 1618 | 3.665e-12 | 4.16e-10 |
| 42 | REGULATION OF LIPID KINASE ACTIVITY | 7 | 48 | 5.099e-12 | 5.649e-10 |
| 43 | REGULATION OF LIPID METABOLIC PROCESS | 11 | 282 | 5.799e-12 | 6.275e-10 |
| 44 | POSITIVE REGULATION OF TRANSCRIPTION FACTOR IMPORT INTO NUCLEUS | 7 | 51 | 7.983e-12 | 8.075e-10 |
| 45 | REGULATION OF CELL DEATH | 19 | 1472 | 7.763e-12 | 8.075e-10 |
| 46 | TRANSMEMBRANE RECEPTOR PROTEIN TYROSINE KINASE SIGNALING PATHWAY | 13 | 498 | 7.89e-12 | 8.075e-10 |
| 47 | ANGIOGENESIS | 11 | 293 | 8.77e-12 | 8.683e-10 |
| 48 | POSITIVE REGULATION OF TRANSPORT | 16 | 936 | 1.018e-11 | 9.87e-10 |
| 49 | PROTEIN PHOSPHORYLATION | 16 | 944 | 1.157e-11 | 1.099e-09 |
| 50 | FC GAMMA RECEPTOR SIGNALING PATHWAY | 8 | 95 | 1.302e-11 | 1.212e-09 |
| 51 | CELL MOTILITY | 15 | 835 | 2.732e-11 | 2.398e-09 |
| 52 | LOCALIZATION OF CELL | 15 | 835 | 2.732e-11 | 2.398e-09 |
| 53 | POSITIVE REGULATION OF PROTEIN IMPORT | 8 | 104 | 2.725e-11 | 2.398e-09 |
| 54 | REGULATION OF PHOSPHOLIPID METABOLIC PROCESS | 7 | 61 | 2.965e-11 | 2.555e-09 |
| 55 | POSITIVE REGULATION OF MULTICELLULAR ORGANISMAL PROCESS | 18 | 1395 | 3.547e-11 | 3.001e-09 |
| 56 | POSITIVE REGULATION OF IMMUNE RESPONSE | 13 | 563 | 3.644e-11 | 3.027e-09 |
| 57 | REGULATION OF ANATOMICAL STRUCTURE MORPHOGENESIS | 16 | 1021 | 3.741e-11 | 3.053e-09 |
| 58 | CELL ACTIVATION | 13 | 568 | 4.066e-11 | 3.262e-09 |
| 59 | PHOSPHATIDYLCHOLINE METABOLIC PROCESS | 7 | 64 | 4.204e-11 | 3.315e-09 |
| 60 | ORGANOPHOSPHATE METABOLIC PROCESS | 15 | 885 | 6.194e-11 | 4.803e-09 |
| 61 | LEUKOCYTE MIGRATION | 10 | 259 | 6.718e-11 | 5.124e-09 |
| 62 | POSITIVE REGULATION OF MAPK CASCADE | 12 | 470 | 7.59e-11 | 5.696e-09 |
| 63 | POSITIVE REGULATION OF CELLULAR PROTEIN LOCALIZATION | 11 | 360 | 8.001e-11 | 5.909e-09 |
| 64 | REGULATION OF RESPONSE TO STRESS | 18 | 1468 | 8.235e-11 | 5.987e-09 |
| 65 | PHOSPHATIDYLINOSITOL BIOSYNTHETIC PROCESS | 8 | 120 | 8.688e-11 | 6.219e-09 |
| 66 | BLOOD VESSEL MORPHOGENESIS | 11 | 364 | 9.002e-11 | 6.346e-09 |
| 67 | POSITIVE REGULATION OF NUCLEOCYTOPLASMIC TRANSPORT | 8 | 121 | 9.289e-11 | 6.378e-09 |
| 68 | MOVEMENT OF CELL OR SUBCELLULAR COMPONENT | 17 | 1275 | 9.321e-11 | 6.378e-09 |
| 69 | PHOSPHATIDYLINOSITOL ACYL CHAIN REMODELING | 5 | 16 | 1.061e-10 | 7.156e-09 |
| 70 | PHOSPHATIDYLSERINE ACYL CHAIN REMODELING | 5 | 17 | 1.501e-10 | 9.978e-09 |
| 71 | POSITIVE REGULATION OF PROTEIN LOCALIZATION TO NUCLEUS | 8 | 129 | 1.556e-10 | 1.02e-08 |
| 72 | CARDIOVASCULAR SYSTEM DEVELOPMENT | 14 | 788 | 1.734e-10 | 1.105e-08 |
| 73 | CIRCULATORY SYSTEM DEVELOPMENT | 14 | 788 | 1.734e-10 | 1.105e-08 |
| 74 | POSITIVE REGULATION OF PROTEIN MODIFICATION PROCESS | 16 | 1135 | 1.799e-10 | 1.131e-08 |
| 75 | POSITIVE REGULATION OF ESTABLISHMENT OF PROTEIN LOCALIZATION | 12 | 514 | 2.122e-10 | 1.317e-08 |
| 76 | ETHANOLAMINE CONTAINING COMPOUND METABOLIC PROCESS | 7 | 85 | 3.24e-10 | 1.984e-08 |
| 77 | PLATELET ACTIVATION | 8 | 142 | 3.361e-10 | 2.031e-08 |
| 78 | ENZYME LINKED RECEPTOR PROTEIN SIGNALING PATHWAY | 13 | 689 | 4.391e-10 | 2.619e-08 |
| 79 | PHOSPHATIDYLINOSITOL 3 PHOSPHATE BIOSYNTHETIC PROCESS | 6 | 49 | 5.676e-10 | 3.343e-08 |
| 80 | REGULATION OF TRANSCRIPTION FACTOR IMPORT INTO NUCLEUS | 7 | 95 | 7.15e-10 | 4.158e-08 |
| 81 | LIPID PHOSPHORYLATION | 7 | 99 | 9.577e-10 | 5.501e-08 |
| 82 | LIPID CATABOLIC PROCESS | 9 | 247 | 1.132e-09 | 6.422e-08 |
| 83 | EPIDERMAL GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 6 | 55 | 1.166e-09 | 6.538e-08 |
| 84 | POSITIVE REGULATION OF GENE EXPRESSION | 18 | 1733 | 1.233e-09 | 6.828e-08 |
| 85 | PHOSPHATIDYLINOSITOL 3 KINASE SIGNALING | 5 | 25 | 1.274e-09 | 6.974e-08 |
| 86 | AMMONIUM ION METABOLIC PROCESS | 8 | 169 | 1.345e-09 | 7.275e-08 |
| 87 | POSITIVE REGULATION OF INTRACELLULAR PROTEIN TRANSPORT | 9 | 258 | 1.66e-09 | 8.877e-08 |
| 88 | REGULATION OF INTRACELLULAR TRANSPORT | 12 | 621 | 1.822e-09 | 9.635e-08 |
| 89 | REGULATION OF KINASE ACTIVITY | 13 | 776 | 1.865e-09 | 9.749e-08 |
| 90 | IMMUNE EFFECTOR PROCESS | 11 | 486 | 1.907e-09 | 9.86e-08 |
| 91 | REGULATION OF PROTEIN LOCALIZATION | 14 | 950 | 1.977e-09 | 1.011e-07 |
| 92 | SINGLE ORGANISM BIOSYNTHETIC PROCESS | 16 | 1340 | 2.04e-09 | 1.032e-07 |
| 93 | POSITIVE REGULATION OF MOLECULAR FUNCTION | 18 | 1791 | 2.094e-09 | 1.042e-07 |
| 94 | REGULATION OF CELL ADHESION | 12 | 629 | 2.105e-09 | 1.042e-07 |
| 95 | POSITIVE REGULATION OF DEVELOPMENTAL PROCESS | 15 | 1142 | 2.127e-09 | 1.042e-07 |
| 96 | ANATOMICAL STRUCTURE FORMATION INVOLVED IN MORPHOGENESIS | 14 | 957 | 2.173e-09 | 1.042e-07 |
| 97 | POSITIVE REGULATION OF INTRACELLULAR TRANSPORT | 10 | 370 | 2.152e-09 | 1.042e-07 |
| 98 | PHOSPHATIDYLSERINE METABOLIC PROCESS | 5 | 28 | 2.346e-09 | 1.114e-07 |
| 99 | POSITIVE REGULATION OF CELLULAR COMPONENT ORGANIZATION | 15 | 1152 | 2.397e-09 | 1.126e-07 |
| 100 | REGULATION OF PROTEIN IMPORT | 8 | 183 | 2.524e-09 | 1.174e-07 |
| 101 | CELLULAR RESPONSE TO OXYGEN CONTAINING COMPOUND | 13 | 799 | 2.654e-09 | 1.223e-07 |
| 102 | REGULATION OF BODY FLUID LEVELS | 11 | 506 | 2.906e-09 | 1.325e-07 |
| 103 | RESPONSE TO OXYGEN CONTAINING COMPOUND | 16 | 1381 | 3.153e-09 | 1.425e-07 |
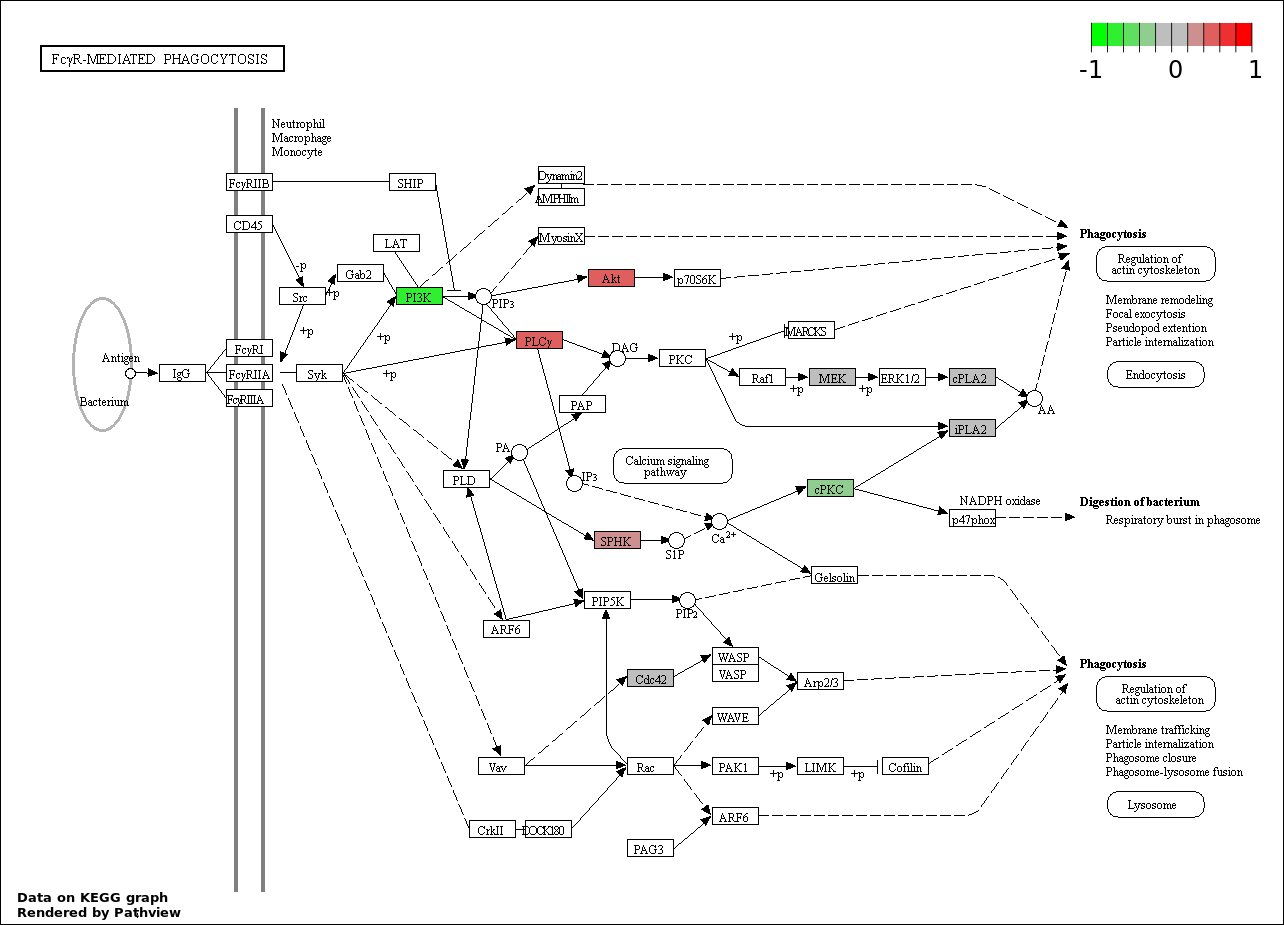
| 104 | PHAGOCYTOSIS | 8 | 190 | 3.394e-09 | 1.518e-07 |
| 105 | REGULATION OF MAPK CASCADE | 12 | 660 | 3.617e-09 | 1.603e-07 |
| 106 | LEUKOCYTE DIFFERENTIATION | 9 | 292 | 4.902e-09 | 2.152e-07 |
| 107 | PHOSPHATIDIC ACID METABOLIC PROCESS | 5 | 33 | 5.625e-09 | 2.446e-07 |
| 108 | RESPONSE TO NITROGEN COMPOUND | 13 | 859 | 6.337e-09 | 2.73e-07 |
| 109 | AMINE METABOLIC PROCESS | 7 | 131 | 6.854e-09 | 2.926e-07 |
| 110 | POSITIVE REGULATION OF LOCOMOTION | 10 | 420 | 7.239e-09 | 3.035e-07 |
| 111 | REGULATION OF CELLULAR PROTEIN LOCALIZATION | 11 | 552 | 7.175e-09 | 3.035e-07 |
| 112 | LIPID MODIFICATION | 8 | 210 | 7.451e-09 | 3.096e-07 |
| 113 | ALDITOL PHOSPHATE METABOLIC PROCESS | 5 | 35 | 7.671e-09 | 3.131e-07 |
| 114 | POSITIVE REGULATION OF VASCULATURE DEVELOPMENT | 7 | 133 | 7.618e-09 | 3.131e-07 |
| 115 | HEMOSTASIS | 9 | 311 | 8.481e-09 | 3.431e-07 |
| 116 | REGULATION OF CELL DIFFERENTIATION | 16 | 1492 | 9.567e-09 | 3.805e-07 |
| 117 | POSITIVE REGULATION OF PROTEIN METABOLIC PROCESS | 16 | 1492 | 9.567e-09 | 3.805e-07 |
| 118 | REGULATION OF CELLULAR LOCALIZATION | 15 | 1277 | 9.717e-09 | 3.832e-07 |
| 119 | REGULATION OF PROTEIN LOCALIZATION TO NUCLEUS | 8 | 218 | 9.984e-09 | 3.871e-07 |
| 120 | REGULATION OF CELL PROLIFERATION | 16 | 1496 | 9.94e-09 | 3.871e-07 |
| 121 | REGULATION OF NUCLEOCYTOPLASMIC TRANSPORT | 8 | 220 | 1.072e-08 | 4.118e-07 |
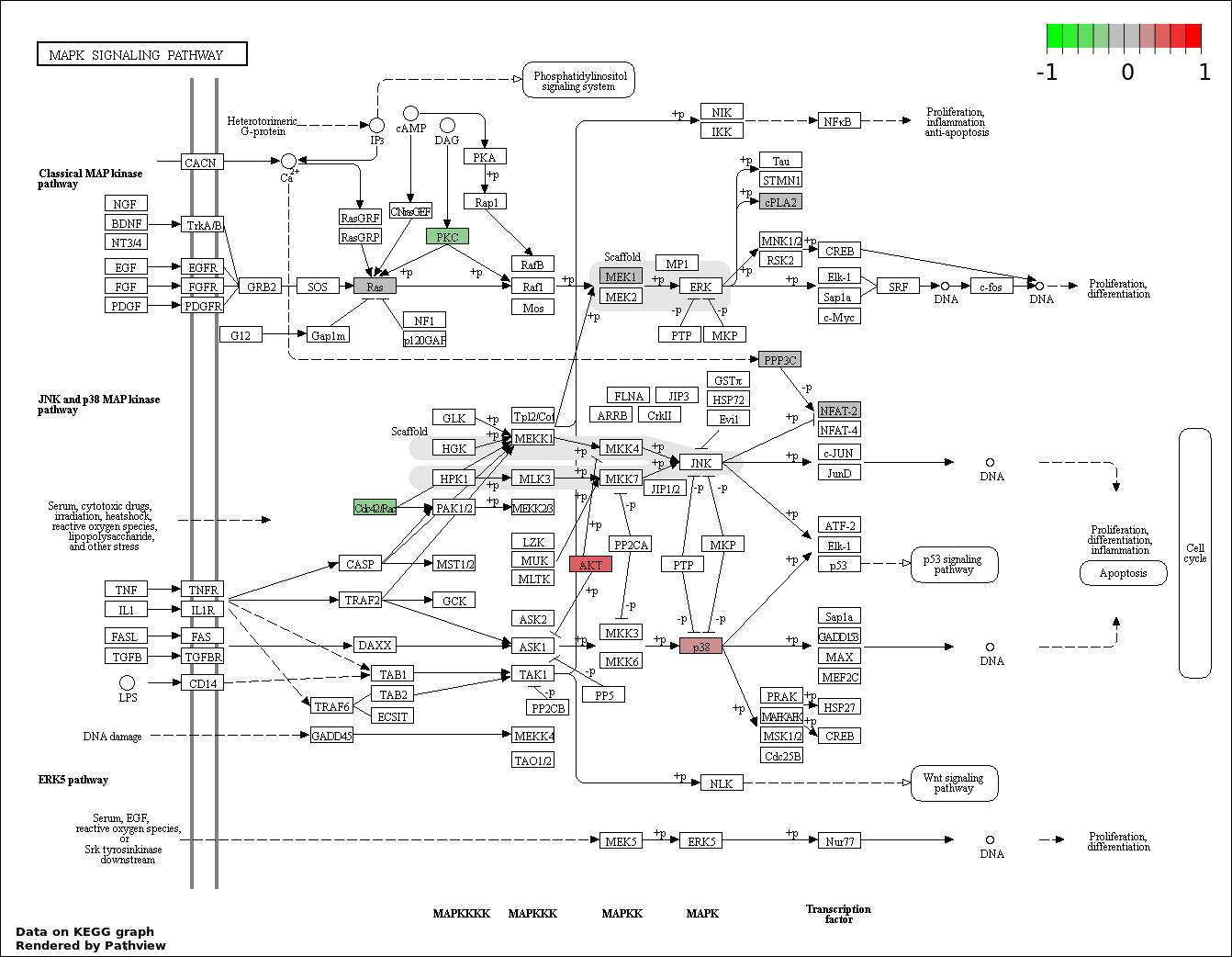
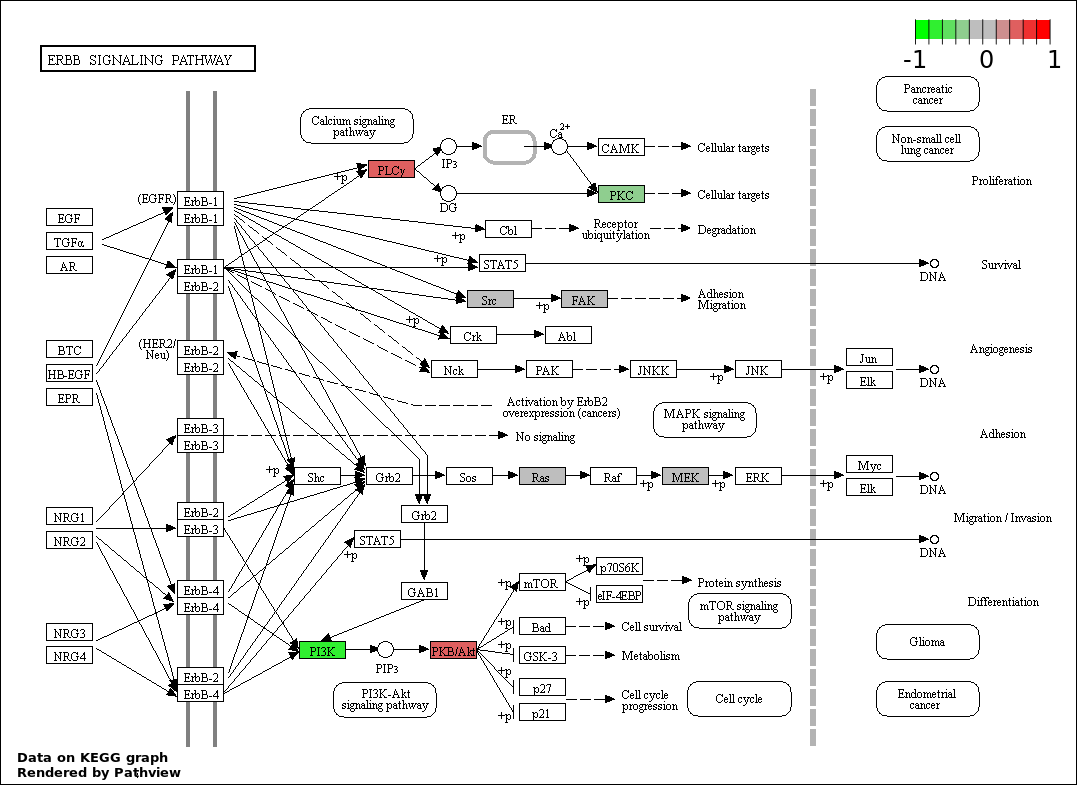
| 122 | ERBB SIGNALING PATHWAY | 6 | 79 | 1.08e-08 | 4.118e-07 |
| 123 | IMMUNE SYSTEM DEVELOPMENT | 11 | 582 | 1.239e-08 | 4.688e-07 |
| 124 | ERBB2 SIGNALING PATHWAY | 5 | 39 | 1.353e-08 | 5.075e-07 |
| 125 | HOMEOSTATIC PROCESS | 15 | 1337 | 1.803e-08 | 6.71e-07 |
| 126 | REGULATION OF TRANSFERASE ACTIVITY | 13 | 946 | 2e-08 | 7.386e-07 |
| 127 | WOUND HEALING | 10 | 470 | 2.104e-08 | 7.71e-07 |
| 128 | ALCOHOL METABOLIC PROCESS | 9 | 348 | 2.242e-08 | 8.149e-07 |
| 129 | REGULATION OF CYTOPLASMIC TRANSPORT | 10 | 481 | 2.618e-08 | 9.442e-07 |
| 130 | POSITIVE REGULATION OF KINASE ACTIVITY | 10 | 482 | 2.67e-08 | 9.555e-07 |
| 131 | POSITIVE REGULATION OF DEFENSE RESPONSE | 9 | 364 | 3.3e-08 | 1.172e-06 |
| 132 | REGULATION OF EPITHELIAL CELL MIGRATION | 7 | 166 | 3.539e-08 | 1.248e-06 |
| 133 | REGULATION OF CELL SUBSTRATE ADHESION | 7 | 173 | 4.703e-08 | 1.645e-06 |
| 134 | REGULATION OF MULTICELLULAR ORGANISMAL DEVELOPMENT | 16 | 1672 | 4.801e-08 | 1.667e-06 |
| 135 | REGULATION OF INTRACELLULAR PROTEIN TRANSPORT | 9 | 381 | 4.88e-08 | 1.682e-06 |
| 136 | POSITIVE REGULATION OF EPITHELIAL CELL MIGRATION | 6 | 103 | 5.363e-08 | 1.835e-06 |
| 137 | REGULATION OF PROTEIN MODIFICATION PROCESS | 16 | 1710 | 6.577e-08 | 2.234e-06 |
| 138 | POSITIVE REGULATION OF CYTOPLASMIC TRANSPORT | 8 | 282 | 7.358e-08 | 2.481e-06 |
| 139 | NEGATIVE REGULATION OF CELL DEATH | 12 | 872 | 7.908e-08 | 2.647e-06 |
| 140 | ENDOTHELIAL CELL MIGRATION | 5 | 57 | 9.581e-08 | 3.184e-06 |
| 141 | REGULATION OF RESPONSE TO WOUNDING | 9 | 413 | 9.71e-08 | 3.204e-06 |
| 142 | LEUKOCYTE ACTIVATION | 9 | 414 | 9.912e-08 | 3.225e-06 |
| 143 | REGULATION OF ENDOTHELIAL CELL MIGRATION | 6 | 114 | 9.844e-08 | 3.225e-06 |
| 144 | ANTIGEN RECEPTOR MEDIATED SIGNALING PATHWAY | 7 | 195 | 1.068e-07 | 3.426e-06 |
| 145 | POSITIVE REGULATION OF RESPONSE TO EXTERNAL STIMULUS | 8 | 296 | 1.068e-07 | 3.426e-06 |
| 146 | RESPONSE TO WOUNDING | 10 | 563 | 1.143e-07 | 3.618e-06 |
| 147 | REGULATION OF CELLULAR RESPONSE TO INSULIN STIMULUS | 5 | 59 | 1.142e-07 | 3.618e-06 |
| 148 | POSITIVE REGULATION OF CELLULAR RESPONSE TO INSULIN STIMULUS | 4 | 23 | 1.181e-07 | 3.714e-06 |
| 149 | REGULATION OF PROTEIN TARGETING | 8 | 307 | 1.412e-07 | 4.41e-06 |
| 150 | RESPONSE TO EXTERNAL STIMULUS | 16 | 1821 | 1.578e-07 | 4.894e-06 |
| 151 | POSITIVE REGULATION OF MAP KINASE ACTIVITY | 7 | 207 | 1.604e-07 | 4.943e-06 |
| 152 | INOSITOL LIPID MEDIATED SIGNALING | 6 | 124 | 1.624e-07 | 4.973e-06 |
| 153 | POSITIVE REGULATION OF BLOOD VESSEL ENDOTHELIAL CELL MIGRATION | 4 | 25 | 1.683e-07 | 5.118e-06 |
| 154 | CELLULAR RESPONSE TO ORGANIC SUBSTANCE | 16 | 1848 | 1.934e-07 | 5.842e-06 |
| 155 | POSITIVE REGULATION OF LIPID METABOLIC PROCESS | 6 | 128 | 1.961e-07 | 5.888e-06 |
| 156 | POSITIVE REGULATION OF ENDOTHELIAL CELL MIGRATION | 5 | 67 | 2.178e-07 | 6.455e-06 |
| 157 | SINGLE ORGANISM CATABOLIC PROCESS | 12 | 957 | 2.169e-07 | 6.455e-06 |
| 158 | TAXIS | 9 | 464 | 2.601e-07 | 7.661e-06 |
| 159 | POSITIVE REGULATION OF TRANSFERASE ACTIVITY | 10 | 616 | 2.629e-07 | 7.693e-06 |
| 160 | REGULATION OF PHOSPHATIDYLINOSITOL 3 KINASE SIGNALING | 6 | 138 | 3.062e-07 | 8.905e-06 |
| 161 | LYMPHOCYTE ACTIVATION | 8 | 342 | 3.216e-07 | 9.293e-06 |
| 162 | ORGANIC HYDROXY COMPOUND METABOLIC PROCESS | 9 | 482 | 3.582e-07 | 1.023e-05 |
| 163 | REGULATION OF VASCULATURE DEVELOPMENT | 7 | 233 | 3.577e-07 | 1.023e-05 |
| 164 | REGULATION OF CELL ACTIVATION | 9 | 484 | 3.709e-07 | 1.048e-05 |
| 165 | POSITIVE REGULATION OF CELL PROLIFERATION | 11 | 814 | 3.717e-07 | 1.048e-05 |
| 166 | RESPONSE TO ENDOGENOUS STIMULUS | 14 | 1450 | 4.02e-07 | 1.127e-05 |
| 167 | POSITIVE REGULATION OF CELL DIFFERENTIATION | 11 | 823 | 4.147e-07 | 1.155e-05 |
| 168 | RESPONSE TO ABIOTIC STIMULUS | 12 | 1024 | 4.486e-07 | 1.242e-05 |
| 169 | PEPTIDYL SERINE MODIFICATION | 6 | 148 | 4.627e-07 | 1.269e-05 |
| 170 | VESICLE MEDIATED TRANSPORT | 13 | 1239 | 4.636e-07 | 1.269e-05 |
| 171 | ENDOCYTOSIS | 9 | 509 | 5.654e-07 | 1.538e-05 |
| 172 | POSITIVE REGULATION OF CELL ADHESION | 8 | 376 | 6.586e-07 | 1.782e-05 |
| 173 | TISSUE MIGRATION | 5 | 84 | 6.791e-07 | 1.827e-05 |
| 174 | POSITIVE REGULATION OF CATALYTIC ACTIVITY | 14 | 1518 | 7.013e-07 | 1.875e-05 |
| 175 | REGULATION OF CELL CELL ADHESION | 8 | 380 | 7.132e-07 | 1.896e-05 |
| 176 | REGULATION OF CELL MATRIX ADHESION | 5 | 90 | 9.584e-07 | 2.534e-05 |
| 177 | WNT SIGNALING PATHWAY CALCIUM MODULATING PATHWAY | 4 | 39 | 1.072e-06 | 2.819e-05 |
| 178 | REGULATION OF CELL MORPHOGENESIS | 9 | 552 | 1.11e-06 | 2.901e-05 |
| 179 | POSITIVE REGULATION OF ERK1 AND ERK2 CASCADE | 6 | 172 | 1.117e-06 | 2.903e-05 |
| 180 | POSITIVE REGULATION OF NFAT PROTEIN IMPORT INTO NUCLEUS | 3 | 11 | 1.209e-06 | 3.126e-05 |
| 181 | REPRODUCTIVE SYSTEM DEVELOPMENT | 8 | 408 | 1.216e-06 | 3.126e-05 |
| 182 | MYELOID LEUKOCYTE DIFFERENTIATION | 5 | 96 | 1.321e-06 | 3.338e-05 |
| 183 | REGULATION OF RESPONSE TO EXTERNAL STIMULUS | 11 | 926 | 1.327e-06 | 3.338e-05 |
| 184 | ICOSANOID METABOLIC PROCESS | 5 | 96 | 1.321e-06 | 3.338e-05 |
| 185 | FATTY ACID DERIVATIVE METABOLIC PROCESS | 5 | 96 | 1.321e-06 | 3.338e-05 |
| 186 | POSITIVE REGULATION OF GLUCOSE TRANSPORT | 4 | 42 | 1.453e-06 | 3.635e-05 |
| 187 | POSITIVE REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 7 | 289 | 1.516e-06 | 3.773e-05 |
| 188 | REGULATION OF GLUCOSE TRANSPORT | 5 | 100 | 1.618e-06 | 3.983e-05 |
| 189 | POSITIVE REGULATION OF GLUCOSE IMPORT IN RESPONSE TO INSULIN STIMULUS | 3 | 12 | 1.61e-06 | 3.983e-05 |
| 190 | REGULATION OF DEFENSE RESPONSE | 10 | 759 | 1.761e-06 | 4.312e-05 |
| 191 | MODULATION OF SYNAPTIC TRANSMISSION | 7 | 301 | 1.987e-06 | 4.84e-05 |
| 192 | REGULATION OF CELLULAR COMPONENT MOVEMENT | 10 | 771 | 2.027e-06 | 4.913e-05 |
| 193 | ICOSANOID BIOSYNTHETIC PROCESS | 4 | 46 | 2.106e-06 | 5.052e-05 |
| 194 | FATTY ACID DERIVATIVE BIOSYNTHETIC PROCESS | 4 | 46 | 2.106e-06 | 5.052e-05 |
| 195 | REGULATION OF HOMOTYPIC CELL CELL ADHESION | 7 | 307 | 2.264e-06 | 5.385e-05 |
| 196 | DEVELOPMENTAL PROCESS INVOLVED IN REPRODUCTION | 9 | 602 | 2.268e-06 | 5.385e-05 |
| 197 | REGULATION OF MUSCLE SYSTEM PROCESS | 6 | 195 | 2.317e-06 | 5.472e-05 |
| 198 | REGULATION OF HOMEOSTATIC PROCESS | 8 | 447 | 2.401e-06 | 5.642e-05 |
| 199 | REGULATION OF MAP KINASE ACTIVITY | 7 | 319 | 2.918e-06 | 6.788e-05 |
| 200 | POSITIVE REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 11 | 1004 | 2.915e-06 | 6.788e-05 |
| 201 | REGULATION OF ADHERENS JUNCTION ORGANIZATION | 4 | 50 | 2.956e-06 | 6.818e-05 |
| 202 | POSITIVE REGULATION OF INFLAMMATORY RESPONSE | 5 | 113 | 2.96e-06 | 6.818e-05 |
| 203 | REGULATION OF BLOOD VESSEL ENDOTHELIAL CELL MIGRATION | 4 | 51 | 3.202e-06 | 7.34e-05 |
| 204 | CYTOKINE PRODUCTION | 5 | 120 | 3.978e-06 | 9.073e-05 |
| 205 | B CELL RECEPTOR SIGNALING PATHWAY | 4 | 54 | 4.035e-06 | 9.159e-05 |
| 206 | REGULATION OF CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 7 | 337 | 4.189e-06 | 9.462e-05 |
| 207 | REGULATION OF NFAT PROTEIN IMPORT INTO NUCLEUS | 3 | 17 | 4.943e-06 | 0.00011 |
| 208 | REGULATION OF GLUCOSE IMPORT IN RESPONSE TO INSULIN STIMULUS | 3 | 17 | 4.943e-06 | 0.00011 |
| 209 | NEGATIVE REGULATION OF ANOIKIS | 3 | 17 | 4.943e-06 | 0.00011 |
| 210 | CELLULAR HOMEOSTASIS | 9 | 676 | 5.834e-06 | 0.0001293 |
| 211 | OVULATION | 3 | 18 | 5.923e-06 | 0.0001294 |
| 212 | CELLULAR RESPONSE TO NITROGEN COMPOUND | 8 | 505 | 5.904e-06 | 0.0001294 |
| 213 | MAST CELL MEDIATED IMMUNITY | 3 | 18 | 5.923e-06 | 0.0001294 |
| 214 | REGULATION OF GLUCOSE IMPORT | 4 | 60 | 6.169e-06 | 0.000134 |
| 215 | CHEMICAL HOMEOSTASIS | 10 | 874 | 6.191e-06 | 0.000134 |
| 216 | REGULATION OF SYNAPSE STRUCTURE OR ACTIVITY | 6 | 232 | 6.303e-06 | 0.0001358 |
| 217 | CELLULAR RESPONSE TO STRESS | 13 | 1565 | 6.349e-06 | 0.0001361 |
| 218 | MUSCLE CELL DIFFERENTIATION | 6 | 237 | 7.121e-06 | 0.000152 |
| 219 | CIRCULATORY SYSTEM PROCESS | 7 | 366 | 7.193e-06 | 0.0001528 |
| 220 | REGULATION OF ERK1 AND ERK2 CASCADE | 6 | 238 | 7.294e-06 | 0.0001543 |
| 221 | RESPONSE TO HORMONE | 10 | 893 | 7.482e-06 | 0.0001575 |
| 222 | POSITIVE REGULATION OF CYTOKINE PRODUCTION | 7 | 370 | 7.721e-06 | 0.0001618 |
| 223 | PLACENTA DEVELOPMENT | 5 | 138 | 7.881e-06 | 0.0001644 |
| 224 | RESPONSE TO BACTERIUM | 8 | 528 | 8.175e-06 | 0.0001698 |
| 225 | NON CANONICAL WNT SIGNALING PATHWAY | 5 | 140 | 8.453e-06 | 0.0001748 |
| 226 | MAST CELL ACTIVATION | 3 | 21 | 9.614e-06 | 0.0001979 |
| 227 | REGULATION OF CELL JUNCTION ASSEMBLY | 4 | 68 | 1.018e-05 | 0.0002087 |
| 228 | CELLULAR RESPONSE TO INSULIN STIMULUS | 5 | 146 | 1.037e-05 | 0.0002106 |
| 229 | T CELL RECEPTOR SIGNALING PATHWAY | 5 | 146 | 1.037e-05 | 0.0002106 |
| 230 | REGULATION OF PEPTIDE TRANSPORT | 6 | 256 | 1.105e-05 | 0.0002236 |
| 231 | REGULATION OF CELL PROJECTION ORGANIZATION | 8 | 558 | 1.222e-05 | 0.0002461 |
| 232 | LYSOSOME LOCALIZATION | 3 | 23 | 1.277e-05 | 0.000256 |
| 233 | REGULATION OF CYTOKINE PRODUCTION | 8 | 563 | 1.303e-05 | 0.0002602 |
| 234 | AMEBOIDAL TYPE CELL MIGRATION | 5 | 154 | 1.343e-05 | 0.0002658 |
| 235 | REGULATION OF ORGANELLE ORGANIZATION | 11 | 1178 | 1.34e-05 | 0.0002658 |
| 236 | RESPONSE TO PEPTIDE | 7 | 404 | 1.366e-05 | 0.0002682 |
| 237 | SIGNAL TRANSDUCTION BY PROTEIN PHOSPHORYLATION | 7 | 404 | 1.366e-05 | 0.0002682 |
| 238 | CELL DEVELOPMENT | 12 | 1426 | 1.409e-05 | 0.0002755 |
| 239 | CELLULAR CHEMICAL HOMEOSTASIS | 8 | 570 | 1.425e-05 | 0.0002774 |
| 240 | REGULATION OF ANOIKIS | 3 | 24 | 1.457e-05 | 0.0002825 |
| 241 | CELLULAR GLUCOSE HOMEOSTASIS | 4 | 75 | 1.505e-05 | 0.0002905 |
| 242 | MULTICELLULAR ORGANISMAL HOMEOSTASIS | 6 | 272 | 1.559e-05 | 0.0002997 |
| 243 | CELLULAR RESPONSE TO PEPTIDE | 6 | 274 | 1.625e-05 | 0.0003111 |
| 244 | POSITIVE REGULATION OF RESPONSE TO WOUNDING | 5 | 162 | 1.715e-05 | 0.0003271 |
| 245 | LYMPHOCYTE COSTIMULATION | 4 | 78 | 1.758e-05 | 0.0003339 |
| 246 | VASCULAR PROCESS IN CIRCULATORY SYSTEM | 5 | 163 | 1.767e-05 | 0.0003342 |
| 247 | INSULIN RECEPTOR SIGNALING PATHWAY | 4 | 80 | 1.944e-05 | 0.0003662 |
| 248 | RESPONSE TO CARBOHYDRATE | 5 | 168 | 2.044e-05 | 0.0003819 |
| 249 | POSITIVE REGULATION OF PROTEIN KINASE B SIGNALING | 4 | 81 | 2.042e-05 | 0.0003819 |
| 250 | RESPONSE TO LITHIUM ION | 3 | 27 | 2.097e-05 | 0.0003902 |
| 251 | POSITIVE REGULATION OF CELL DEATH | 8 | 605 | 2.188e-05 | 0.0004057 |
| 252 | TISSUE HOMEOSTASIS | 5 | 171 | 2.225e-05 | 0.0004109 |
| 253 | STRIATED MUSCLE CELL DIFFERENTIATION | 5 | 173 | 2.353e-05 | 0.0004306 |
| 254 | SMALL MOLECULE METABOLIC PROCESS | 13 | 1767 | 2.36e-05 | 0.0004306 |
| 255 | POSITIVE REGULATION OF ACUTE INFLAMMATORY RESPONSE | 3 | 28 | 2.345e-05 | 0.0004306 |
| 256 | REGULATION OF INFLAMMATORY RESPONSE | 6 | 294 | 2.417e-05 | 0.0004394 |
| 257 | FATTY ACID METABOLIC PROCESS | 6 | 296 | 2.511e-05 | 0.0004546 |
| 258 | TISSUE DEVELOPMENT | 12 | 1518 | 2.638e-05 | 0.0004758 |
| 259 | LEUKOCYTE DEGRANULATION | 3 | 30 | 2.898e-05 | 0.0005187 |
| 260 | CELLULAR RESPONSE TO VASCULAR ENDOTHELIAL GROWTH FACTOR STIMULUS | 3 | 30 | 2.898e-05 | 0.0005187 |
| 261 | SINGLE ORGANISM CELL ADHESION | 7 | 459 | 3.101e-05 | 0.0005528 |
| 262 | REPRODUCTION | 11 | 1297 | 3.287e-05 | 0.0005838 |
| 263 | PEPTIDYL AMINO ACID MODIFICATION | 9 | 841 | 3.325e-05 | 0.0005883 |
| 264 | POSITIVE REGULATION OF LIPID KINASE ACTIVITY | 3 | 32 | 3.531e-05 | 0.0006223 |
| 265 | MYELOID CELL DIFFERENTIATION | 5 | 189 | 3.596e-05 | 0.0006307 |
| 266 | REGULATION OF PROTEIN SERINE THREONINE KINASE ACTIVITY | 7 | 470 | 3.606e-05 | 0.0006307 |
| 267 | RESPONSE TO MOLECULE OF BACTERIAL ORIGIN | 6 | 321 | 3.954e-05 | 0.000689 |
| 268 | FEMALE GAMETE GENERATION | 4 | 96 | 3.99e-05 | 0.0006927 |
| 269 | RESPONSE TO INORGANIC SUBSTANCE | 7 | 479 | 4.067e-05 | 0.0007035 |
| 270 | RESPONSE TO FLUID SHEAR STRESS | 3 | 34 | 4.248e-05 | 0.0007321 |
| 271 | POSITIVE REGULATION OF CELL SUBSTRATE ADHESION | 4 | 99 | 4.502e-05 | 0.000773 |
| 272 | RESPONSE TO INSULIN | 5 | 205 | 5.296e-05 | 0.0009059 |
| 273 | REGULATION OF CALCIUM ION TRANSPORT | 5 | 209 | 5.805e-05 | 0.0009857 |
| 274 | REGULATION OF SYSTEM PROCESS | 7 | 507 | 5.826e-05 | 0.0009857 |
| 275 | LYMPHOCYTE DIFFERENTIATION | 5 | 209 | 5.805e-05 | 0.0009857 |
| 276 | INNATE IMMUNE RESPONSE ACTIVATING CELL SURFACE RECEPTOR SIGNALING PATHWAY | 4 | 106 | 5.882e-05 | 0.0009915 |
| 277 | REGULATION OF CELL ADHESION MEDIATED BY INTEGRIN | 3 | 39 | 6.443e-05 | 0.001078 |
| 278 | PEPTIDYL TYROSINE AUTOPHOSPHORYLATION | 3 | 39 | 6.443e-05 | 0.001078 |
| 279 | WNT SIGNALING PATHWAY | 6 | 351 | 6.493e-05 | 0.001083 |
| 280 | UNSATURATED FATTY ACID METABOLIC PROCESS | 4 | 109 | 6.558e-05 | 0.00109 |
| 281 | NEUROGENESIS | 11 | 1402 | 6.706e-05 | 0.00111 |
| 282 | POSITIVE REGULATION OF HOMEOSTATIC PROCESS | 5 | 216 | 6.785e-05 | 0.00112 |
| 283 | ALCOHOL BIOSYNTHETIC PROCESS | 4 | 111 | 7.04e-05 | 0.001157 |
| 284 | CELLULAR MODIFIED AMINO ACID METABOLIC PROCESS | 5 | 220 | 7.4e-05 | 0.001212 |
| 285 | MYELOID CELL ACTIVATION INVOLVED IN IMMUNE RESPONSE | 3 | 41 | 7.495e-05 | 0.001224 |
| 286 | REGULATION OF APOPTOTIC SIGNALING PATHWAY | 6 | 363 | 7.817e-05 | 0.001272 |
| 287 | NEGATIVE REGULATION OF CELL ADHESION | 5 | 223 | 7.889e-05 | 0.001279 |
| 288 | POSITIVE REGULATION OF PHOSPHOLIPID METABOLIC PROCESS | 3 | 42 | 8.06e-05 | 0.001293 |
| 289 | REGULATION OF SEQUENCE SPECIFIC DNA BINDING TRANSCRIPTION FACTOR ACTIVITY | 6 | 365 | 8.058e-05 | 0.001293 |
| 290 | REGULATED EXOCYTOSIS | 5 | 224 | 8.057e-05 | 0.001293 |
| 291 | REGULATION OF SUBSTRATE ADHESION DEPENDENT CELL SPREADING | 3 | 43 | 8.652e-05 | 0.001379 |
| 292 | MYELOID LEUKOCYTE MEDIATED IMMUNITY | 3 | 43 | 8.652e-05 | 0.001379 |
| 293 | POSITIVE REGULATION OF SEQUENCE SPECIFIC DNA BINDING TRANSCRIPTION FACTOR ACTIVITY | 5 | 228 | 8.758e-05 | 0.001391 |
| 294 | NEGATIVE REGULATION OF CELL COMMUNICATION | 10 | 1192 | 8.919e-05 | 0.001412 |
| 295 | REGULATION OF PROTEIN KINASE B SIGNALING | 4 | 121 | 9.844e-05 | 0.001553 |
| 296 | SPROUTING ANGIOGENESIS | 3 | 45 | 9.921e-05 | 0.00156 |
| 297 | REGULATION OF NEURON DIFFERENTIATION | 7 | 554 | 0.0001015 | 0.001591 |
| 298 | REGULATION OF PROTEIN SECRETION | 6 | 389 | 0.0001143 | 0.001784 |
| 299 | REGULATION OF CELLULAR COMPONENT BIOGENESIS | 8 | 767 | 0.0001166 | 0.001809 |
| 300 | DEFENSE RESPONSE | 10 | 1231 | 0.0001166 | 0.001809 |
| 301 | CATABOLIC PROCESS | 12 | 1773 | 0.0001205 | 0.001862 |
| 302 | MUSCLE CELL DEVELOPMENT | 4 | 128 | 0.0001224 | 0.001885 |
| 303 | POSITIVE REGULATION OF ORGANELLE ORGANIZATION | 7 | 573 | 0.0001252 | 0.001923 |
| 304 | CELL DEATH | 9 | 1001 | 0.0001276 | 0.00195 |
| 305 | REGULATION OF TRANSCRIPTION FROM RNA POLYMERASE II PROMOTER | 12 | 1784 | 0.0001278 | 0.00195 |
| 306 | SYSTEM PROCESS | 12 | 1785 | 0.0001285 | 0.001954 |
| 307 | BRANCHING MORPHOGENESIS OF AN EPITHELIAL TUBE | 4 | 131 | 0.0001338 | 0.002028 |
| 308 | ARACHIDONIC ACID METABOLIC PROCESS | 3 | 50 | 0.0001361 | 0.002056 |
| 309 | B CELL ACTIVATION | 4 | 132 | 0.0001378 | 0.002075 |
| 310 | SECRETION | 7 | 588 | 0.0001469 | 0.002205 |
| 311 | REGULATION OF NEURON PROJECTION DEVELOPMENT | 6 | 408 | 0.0001482 | 0.002211 |
| 312 | LEUKOCYTE CELL CELL ADHESION | 5 | 255 | 0.000148 | 0.002211 |
| 313 | CELLULAR RESPONSE TO RADIATION | 4 | 137 | 0.000159 | 0.002364 |
| 314 | RESPONSE TO ORGANOPHOSPHORUS | 4 | 139 | 0.0001681 | 0.002484 |
| 315 | CELL ACTIVATION INVOLVED IN IMMUNE RESPONSE | 4 | 139 | 0.0001681 | 0.002484 |
| 316 | REGULATION OF SYNAPTIC PLASTICITY | 4 | 140 | 0.0001728 | 0.002545 |
| 317 | CELL CELL ADHESION | 7 | 608 | 0.0001805 | 0.00265 |
| 318 | MUSCLE STRUCTURE DEVELOPMENT | 6 | 432 | 0.0002021 | 0.002957 |
| 319 | POSITIVE REGULATION OF ENDOTHELIAL CELL CHEMOTAXIS | 2 | 11 | 0.0002121 | 0.003046 |
| 320 | CHEMICAL HOMEOSTASIS WITHIN A TISSUE | 2 | 11 | 0.0002121 | 0.003046 |
| 321 | REGULATION OF FEVER GENERATION | 2 | 11 | 0.0002121 | 0.003046 |
| 322 | CYCLOOXYGENASE PATHWAY | 2 | 11 | 0.0002121 | 0.003046 |
| 323 | REGULATION OF CELL DEVELOPMENT | 8 | 836 | 0.0002108 | 0.003046 |
| 324 | UNSATURATED FATTY ACID BIOSYNTHETIC PROCESS | 3 | 58 | 0.0002119 | 0.003046 |
| 325 | VASCULOGENESIS | 3 | 59 | 0.0002229 | 0.003192 |
| 326 | ACTIVATION OF PROTEIN KINASE ACTIVITY | 5 | 279 | 0.0002249 | 0.00321 |
| 327 | MATERNAL PROCESS INVOLVED IN FEMALE PREGNANCY | 3 | 60 | 0.0002343 | 0.003334 |
| 328 | POSITIVE REGULATION OF CYCLASE ACTIVITY | 3 | 61 | 0.0002461 | 0.003491 |
| 329 | ANATOMICAL STRUCTURE HOMEOSTASIS | 5 | 285 | 0.0002481 | 0.003509 |
| 330 | POSITIVE REGULATION OF EPITHELIAL CELL PROLIFERATION | 4 | 154 | 0.000249 | 0.003511 |
| 331 | REGULATION OF BROWN FAT CELL DIFFERENTIATION | 2 | 12 | 0.0002542 | 0.003573 |
| 332 | INFLAMMATORY RESPONSE | 6 | 454 | 0.0002641 | 0.003701 |
| 333 | REGULATION OF MUSCLE ADAPTATION | 3 | 63 | 0.0002708 | 0.003784 |
| 334 | RESPONSE TO PURINE CONTAINING COMPOUND | 4 | 158 | 0.0002746 | 0.003825 |
| 335 | CENTRAL NERVOUS SYSTEM DEVELOPMENT | 8 | 872 | 0.0002807 | 0.003899 |
| 336 | NEURON DIFFERENTIATION | 8 | 874 | 0.0002851 | 0.003948 |
| 337 | REGULATION OF VESICLE MEDIATED TRANSPORT | 6 | 462 | 0.00029 | 0.004004 |
| 338 | LIPOXYGENASE PATHWAY | 2 | 13 | 3e-04 | 0.004118 |
| 339 | LYMPH VESSEL MORPHOGENESIS | 2 | 13 | 3e-04 | 0.004118 |
| 340 | POSITIVE REGULATION OF CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 4 | 162 | 0.000302 | 0.004133 |
| 341 | HEART DEVELOPMENT | 6 | 466 | 0.0003037 | 0.004144 |
| 342 | CELLULAR RESPONSE TO UV | 3 | 66 | 0.0003108 | 0.004216 |
| 343 | POSITIVE REGULATION OF LIPID BIOSYNTHETIC PROCESS | 3 | 66 | 0.0003108 | 0.004216 |
| 344 | RESPONSE TO BIOTIC STIMULUS | 8 | 886 | 0.0003126 | 0.004228 |
| 345 | RESPONSE TO LIPID | 8 | 888 | 0.0003174 | 0.004281 |
| 346 | ESTABLISHMENT OF LOCALIZATION IN CELL | 11 | 1676 | 0.0003272 | 0.004401 |
| 347 | POSITIVE REGULATION OF CELL PROJECTION ORGANIZATION | 5 | 303 | 0.000329 | 0.004412 |
| 348 | RESPONSE TO GROWTH FACTOR | 6 | 475 | 0.0003363 | 0.004497 |
| 349 | POSITIVE REGULATION OF MITOCHONDRION ORGANIZATION | 4 | 167 | 0.000339 | 0.004499 |
| 350 | MORPHOGENESIS OF A BRANCHING STRUCTURE | 4 | 167 | 0.000339 | 0.004499 |
| 351 | POSITIVE REGULATION OF ENDOTHELIAL CELL PROLIFERATION | 3 | 68 | 0.0003394 | 0.004499 |
| 352 | VASCULAR ENDOTHELIAL GROWTH FACTOR SIGNALING PATHWAY | 2 | 14 | 0.0003496 | 0.004506 |
| 353 | ARACHIDONIC ACID SECRETION | 2 | 14 | 0.0003496 | 0.004506 |
| 354 | REGULATION OF FIBROBLAST APOPTOTIC PROCESS | 2 | 14 | 0.0003496 | 0.004506 |
| 355 | ESTABLISHMENT OF NUCLEUS LOCALIZATION | 2 | 14 | 0.0003496 | 0.004506 |
| 356 | T CELL MIGRATION | 2 | 14 | 0.0003496 | 0.004506 |
| 357 | CELLULAR COMPONENT MORPHOGENESIS | 8 | 900 | 0.0003474 | 0.004506 |
| 358 | CARDIOLIPIN METABOLIC PROCESS | 2 | 14 | 0.0003496 | 0.004506 |
| 359 | REGULATION OF PROTEIN KINASE C SIGNALING | 2 | 14 | 0.0003496 | 0.004506 |
| 360 | POSITIVE REGULATION OF SPROUTING ANGIOGENESIS | 2 | 14 | 0.0003496 | 0.004506 |
| 361 | ARACHIDONATE TRANSPORT | 2 | 14 | 0.0003496 | 0.004506 |
| 362 | CELL PROJECTION ORGANIZATION | 8 | 902 | 0.0003526 | 0.004532 |
| 363 | GLUCOSE HOMEOSTASIS | 4 | 170 | 0.0003627 | 0.004636 |
| 364 | CARBOHYDRATE HOMEOSTASIS | 4 | 170 | 0.0003627 | 0.004636 |
| 365 | EXOCYTOSIS | 5 | 310 | 0.0003653 | 0.004657 |
| 366 | POSITIVE REGULATION OF CELL ACTIVATION | 5 | 311 | 0.0003708 | 0.004702 |
| 367 | POSITIVE REGULATION OF APOPTOTIC SIGNALING PATHWAY | 4 | 171 | 0.0003708 | 0.004702 |
| 368 | CELL MIGRATION INVOLVED IN SPROUTING ANGIOGENESIS | 2 | 15 | 0.0004028 | 0.005049 |
| 369 | REGULATION OF HEAT GENERATION | 2 | 15 | 0.0004028 | 0.005049 |
| 370 | NEGATIVE REGULATION OF DENDRITE MORPHOGENESIS | 2 | 15 | 0.0004028 | 0.005049 |
| 371 | HOMEOSTASIS OF NUMBER OF CELLS | 4 | 175 | 0.0004048 | 0.005049 |
| 372 | RESPIRATORY BURST | 2 | 15 | 0.0004028 | 0.005049 |
| 373 | ORGANIC HYDROXY COMPOUND BIOSYNTHETIC PROCESS | 4 | 175 | 0.0004048 | 0.005049 |
| 374 | RESPONSE TO CORTICOSTEROID | 4 | 176 | 0.0004136 | 0.005146 |
| 375 | REGULATION OF SECRETION | 7 | 699 | 0.0004214 | 0.005229 |
| 376 | CELLULAR RESPONSE TO CARBOHYDRATE STIMULUS | 3 | 74 | 0.0004354 | 0.005374 |
| 377 | REGULATION OF ACUTE INFLAMMATORY RESPONSE | 3 | 74 | 0.0004354 | 0.005374 |
| 378 | REGULATION OF METAL ION TRANSPORT | 5 | 325 | 0.0004534 | 0.005566 |
| 379 | GRANULOCYTE MIGRATION | 3 | 75 | 0.0004529 | 0.005566 |
| 380 | MONOCARBOXYLIC ACID METABOLIC PROCESS | 6 | 503 | 0.000456 | 0.005584 |
| 381 | REGULATION OF EARLY ENDOSOME TO LATE ENDOSOME TRANSPORT | 2 | 16 | 0.0004598 | 0.005586 |
| 382 | HEAD DEVELOPMENT | 7 | 709 | 0.0004589 | 0.005586 |
| 383 | POSITIVE REGULATION OF VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR SIGNALING PATHWAY | 2 | 16 | 0.0004598 | 0.005586 |
| 384 | RESPONSE TO CYTOKINE | 7 | 714 | 0.0004786 | 0.0058 |
| 385 | RESPONSE TO METAL ION | 5 | 333 | 0.0005064 | 0.006105 |
| 386 | CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 6 | 513 | 0.000506 | 0.006105 |
| 387 | REGULATION OF RECEPTOR MEDIATED ENDOCYTOSIS | 3 | 78 | 0.0005081 | 0.00611 |
| 388 | PEPTIDYL TYROSINE MODIFICATION | 4 | 186 | 0.0005095 | 0.00611 |
| 389 | REGULATION OF CHROMATIN BINDING | 2 | 17 | 0.0005204 | 0.006193 |
| 390 | POSITIVE REGULATION OF FATTY ACID BIOSYNTHETIC PROCESS | 2 | 17 | 0.0005204 | 0.006193 |
| 391 | REGULATION OF ENDOTHELIAL CELL CHEMOTAXIS | 2 | 17 | 0.0005204 | 0.006193 |
| 392 | PROTEIN DEPHOSPHORYLATION | 4 | 190 | 0.000552 | 0.006552 |
| 393 | RESPONSE TO REACTIVE OXYGEN SPECIES | 4 | 191 | 0.000563 | 0.006665 |
| 394 | PROTEIN AUTOPHOSPHORYLATION | 4 | 192 | 0.0005741 | 0.00678 |
| 395 | REGULATION OF MITOCHONDRIAL DEPOLARIZATION | 2 | 18 | 0.0005848 | 0.006888 |
| 396 | POSITIVE REGULATION OF MUSCLE CELL DIFFERENTIATION | 3 | 84 | 0.0006312 | 0.00741 |
| 397 | RESPIRATORY SYSTEM DEVELOPMENT | 4 | 197 | 0.0006323 | 0.00741 |
| 398 | PROSTAGLANDIN BIOSYNTHETIC PROCESS | 2 | 19 | 0.0006527 | 0.007451 |
| 399 | CELLULAR RESPONSE TO FLUID SHEAR STRESS | 2 | 19 | 0.0006527 | 0.007451 |
| 400 | REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 7 | 750 | 0.0006416 | 0.007451 |
| 401 | REGULATION OF CELL MIGRATION INVOLVED IN SPROUTING ANGIOGENESIS | 2 | 19 | 0.0006527 | 0.007451 |
| 402 | SMALL GTPASE MEDIATED SIGNAL TRANSDUCTION | 5 | 352 | 0.0006511 | 0.007451 |
| 403 | EPHRIN RECEPTOR SIGNALING PATHWAY | 3 | 85 | 0.0006534 | 0.007451 |
| 404 | REGULATION OF MYOBLAST FUSION | 2 | 19 | 0.0006527 | 0.007451 |
| 405 | MACROPHAGE DIFFERENTIATION | 2 | 19 | 0.0006527 | 0.007451 |
| 406 | HEART PROCESS | 3 | 85 | 0.0006534 | 0.007451 |
| 407 | RESPONSE TO OXIDATIVE STRESS | 5 | 352 | 0.0006511 | 0.007451 |
| 408 | PROSTANOID BIOSYNTHETIC PROCESS | 2 | 19 | 0.0006527 | 0.007451 |
| 409 | NEURON PROJECTION DEVELOPMENT | 6 | 545 | 0.0006954 | 0.007911 |
| 410 | EMBRYONIC HEMOPOIESIS | 2 | 20 | 0.0007243 | 0.008044 |
| 411 | ICOSANOID TRANSPORT | 2 | 20 | 0.0007243 | 0.008044 |
| 412 | LONG CHAIN FATTY ACID METABOLIC PROCESS | 3 | 88 | 0.0007229 | 0.008044 |
| 413 | CELLULAR RESPONSE TO GROWTH HORMONE STIMULUS | 2 | 20 | 0.0007243 | 0.008044 |
| 414 | OVULATION CYCLE PROCESS | 3 | 88 | 0.0007229 | 0.008044 |
| 415 | NUCLEUS LOCALIZATION | 2 | 20 | 0.0007243 | 0.008044 |
| 416 | FATTY ACID DERIVATIVE TRANSPORT | 2 | 20 | 0.0007243 | 0.008044 |
| 417 | ACTIVATION OF INNATE IMMUNE RESPONSE | 4 | 204 | 0.0007205 | 0.008044 |
| 418 | LYMPH VESSEL DEVELOPMENT | 2 | 20 | 0.0007243 | 0.008044 |
| 419 | POSITIVE REGULATION OF PEPTIDYL SERINE PHOSPHORYLATION | 3 | 88 | 0.0007229 | 0.008044 |
| 420 | NEURON PROJECTION GUIDANCE | 4 | 205 | 0.0007338 | 0.008129 |
| 421 | CELLULAR RESPONSE TO ENDOGENOUS STIMULUS | 8 | 1008 | 0.0007374 | 0.00814 |
| 422 | MULTICELLULAR ORGANISM REPRODUCTION | 7 | 768 | 0.0007382 | 0.00814 |
| 423 | CELLULAR RESPONSE TO HORMONE STIMULUS | 6 | 552 | 0.0007433 | 0.008157 |
| 424 | TUBE DEVELOPMENT | 6 | 552 | 0.0007433 | 0.008157 |
| 425 | REGULATION OF PEPTIDE SECRETION | 4 | 209 | 0.0007887 | 0.008635 |
| 426 | DENDRITIC CELL MIGRATION | 2 | 21 | 0.0007996 | 0.008652 |
| 427 | CELLULAR RESPONSE TO LIGHT STIMULUS | 3 | 91 | 0.000797 | 0.008652 |
| 428 | DECIDUALIZATION | 2 | 21 | 0.0007996 | 0.008652 |
| 429 | POSITIVE REGULATION OF ADHERENS JUNCTION ORGANIZATION | 2 | 21 | 0.0007996 | 0.008652 |
| 430 | CELL MORPHOGENESIS INVOLVED IN NEURON DIFFERENTIATION | 5 | 368 | 0.0007954 | 0.008652 |
| 431 | POSITIVE REGULATION OF SECRETION | 5 | 370 | 0.000815 | 0.008799 |
| 432 | POSITIVE REGULATION OF PROTEIN SECRETION | 4 | 211 | 0.0008172 | 0.008802 |
| 433 | MULTI MULTICELLULAR ORGANISM PROCESS | 4 | 213 | 0.0008464 | 0.009095 |
| 434 | POSITIVE REGULATION OF BLOOD CIRCULATION | 3 | 93 | 0.000849 | 0.009102 |
| 435 | BIOLOGICAL ADHESION | 8 | 1032 | 0.00086 | 0.0092 |
| 436 | NEGATIVE REGULATION OF MUSCLE CONTRACTION | 2 | 22 | 0.0008784 | 0.00931 |
| 437 | POSITIVE REGULATION OF MUSCLE HYPERTROPHY | 2 | 22 | 0.0008784 | 0.00931 |
| 438 | POSITIVE REGULATION OF PROTEIN AUTOPHOSPHORYLATION | 2 | 22 | 0.0008784 | 0.00931 |
| 439 | POSITIVE REGULATION OF CARDIAC MUSCLE HYPERTROPHY | 2 | 22 | 0.0008784 | 0.00931 |
| 440 | REGULATION OF MITOCHONDRION ORGANIZATION | 4 | 218 | 0.0009227 | 0.009757 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | KINASE ACTIVITY | 17 | 842 | 1.248e-13 | 8.67e-11 |
| 2 | PHOSPHOLIPASE A2 ACTIVITY | 7 | 31 | 1.866e-13 | 8.67e-11 |
| 3 | TRANSFERASE ACTIVITY TRANSFERRING PHOSPHORUS CONTAINING GROUPS | 17 | 992 | 1.753e-12 | 5.427e-10 |
| 4 | PHOSPHOLIPASE ACTIVITY | 8 | 94 | 1.194e-11 | 2.774e-09 |
| 5 | PHOSPHATIDYLINOSITOL 3 KINASE ACTIVITY | 7 | 70 | 8.042e-11 | 1.245e-08 |
| 6 | LIPASE ACTIVITY | 8 | 117 | 7.081e-11 | 1.245e-08 |
| 7 | X1 PHOSPHATIDYLINOSITOL 3 KINASE ACTIVITY | 6 | 43 | 2.496e-10 | 3.313e-08 |
| 8 | PHOSPHATIDYLINOSITOL KINASE ACTIVITY | 6 | 51 | 7.288e-10 | 8.463e-08 |
| 9 | HYDROLASE ACTIVITY ACTING ON ESTER BONDS | 13 | 739 | 1.031e-09 | 1.065e-07 |
| 10 | RIBONUCLEOTIDE BINDING | 18 | 1860 | 3.836e-09 | 3.564e-07 |
| 11 | CARBOXYLIC ESTER HYDROLASE ACTIVITY | 7 | 135 | 8.453e-09 | 7.139e-07 |
| 12 | PROTEIN KINASE ACTIVITY | 11 | 640 | 3.285e-08 | 2.543e-06 |
| 13 | CALMODULIN BINDING | 7 | 179 | 5.943e-08 | 4.247e-06 |
| 14 | ENZYME BINDING | 16 | 1737 | 8.186e-08 | 5.432e-06 |
| 15 | KINASE REGULATOR ACTIVITY | 6 | 186 | 1.761e-06 | 0.0001091 |
| 16 | KINASE BINDING | 9 | 606 | 2.395e-06 | 0.0001309 |
| 17 | PROTEIN SERINE THREONINE KINASE ACTIVITY | 8 | 445 | 2.322e-06 | 0.0001309 |
| 18 | PROTEIN PHOSPHATASE BINDING | 5 | 120 | 3.978e-06 | 0.0001993 |
| 19 | PHOSPHATIDYLINOSITOL PHOSPHATE KINASE ACTIVITY | 3 | 16 | 4.076e-06 | 0.0001993 |
| 20 | ADENYL NUCLEOTIDE BINDING | 13 | 1514 | 4.41e-06 | 0.0002049 |
| 21 | PHOSPHATASE BINDING | 5 | 162 | 1.715e-05 | 0.0007587 |
| 22 | PHOSPHOPROTEIN PHOSPHATASE ACTIVITY | 5 | 178 | 2.698e-05 | 0.001139 |
| 23 | CALCIUM ION BINDING | 8 | 697 | 5.981e-05 | 0.002416 |
| 24 | PHOSPHORIC ESTER HYDROLASE ACTIVITY | 6 | 368 | 8.429e-05 | 0.003263 |
| 25 | PHOSPHATASE ACTIVITY | 5 | 275 | 0.0002103 | 0.007578 |
| 26 | INSULIN RECEPTOR SUBSTRATE BINDING | 2 | 11 | 0.0002121 | 0.007578 |
| 27 | CALCIUM DEPENDENT PROTEIN KINASE ACTIVITY | 2 | 12 | 0.0002542 | 0.008433 |
| 28 | MAP KINASE KINASE ACTIVITY | 2 | 12 | 0.0002542 | 0.008433 |
| 29 | PROTEIN SERINE THREONINE PHOSPHATASE ACTIVITY | 3 | 64 | 0.0002837 | 0.009089 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | PHOSPHATIDYLINOSITOL 3 KINASE COMPLEX | 7 | 20 | 5.591e-15 | 3.265e-12 |
| 2 | EXTRINSIC COMPONENT OF MEMBRANE | 10 | 252 | 5.132e-11 | 1.499e-08 |
| 3 | TRANSFERASE COMPLEX TRANSFERRING PHOSPHORUS CONTAINING GROUPS | 7 | 237 | 4.013e-07 | 7.812e-05 |
| 4 | MAST CELL GRANULE | 3 | 21 | 9.614e-06 | 0.001404 |
| 5 | CATALYTIC COMPLEX | 10 | 1038 | 2.766e-05 | 0.00323 |
Over-represented Pathway
| Num | Pathway | Pathview | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|---|
| 1 | hsa04370_VEGF_signaling_pathway | 40 | 76 | 4.793e-103 | 8.628e-101 | |
| 2 | hsa04664_Fc_epsilon_RI_signaling_pathway | 24 | 79 | 2.565e-49 | 2.309e-47 | |
| 3 | hsa04662_B_cell_receptor_signaling_pathway | 22 | 75 | 1.516e-44 | 9.096e-43 | |
| 4 | hsa04660_T_cell_receptor_signaling_pathway | 22 | 108 | 1.388e-40 | 6.247e-39 | |
| 5 | hsa04650_Natural_killer_cell_mediated_cytotoxicity | 22 | 136 | 3.565e-38 | 1.283e-36 | |
| 6 | hsa04014_Ras_signaling_pathway | 23 | 236 | 1.122e-34 | 3.365e-33 | |
| 7 | hsa04010_MAPK_signaling_pathway | 23 | 268 | 2.332e-33 | 5.996e-32 | |
| 8 | hsa04012_ErbB_signaling_pathway | 16 | 87 | 2.206e-28 | 4.964e-27 | |
| 9 | hsa04666_Fc_gamma_R.mediated_phagocytosis | 16 | 95 | 1.019e-27 | 2.037e-26 | |
| 10 | hsa04380_Osteoclast_differentiation | 17 | 128 | 1.321e-27 | 2.378e-26 | |
| 11 | hsa04912_GnRH_signaling_pathway | 16 | 101 | 2.932e-27 | 4.798e-26 | |
| 12 | hsa04510_Focal_adhesion | 17 | 200 | 3.635e-24 | 5.452e-23 | |
| 13 | hsa04722_Neurotrophin_signaling_pathway | 15 | 127 | 1.65e-23 | 2.285e-22 | |
| 14 | hsa04670_Leukocyte_transendothelial_migration | 14 | 117 | 5.032e-22 | 6.47e-21 | |
| 15 | hsa04730_Long.term_depression | 12 | 70 | 6.469e-21 | 7.763e-20 | |
| 16 | hsa04914_Progesterone.mediated_oocyte_maturation | 12 | 87 | 1.055e-19 | 1.187e-18 | |
| 17 | hsa04210_Apoptosis | 12 | 89 | 1.408e-19 | 1.491e-18 | |
| 18 | hsa04360_Axon_guidance | 13 | 130 | 2.073e-19 | 2.073e-18 | |
| 19 | hsa04960_Aldosterone.regulated_sodium_reabsorption | 10 | 42 | 4.241e-19 | 4.017e-18 | |
| 20 | hsa04062_Chemokine_signaling_pathway | 14 | 189 | 5.208e-19 | 4.688e-18 | |
| 21 | hsa04151_PI3K_AKT_signaling_pathway | 16 | 351 | 2.471e-18 | 2.118e-17 | |
| 22 | hsa04620_Toll.like_receptor_signaling_pathway | 11 | 102 | 7.13e-17 | 5.834e-16 | |
| 23 | hsa04973_Carbohydrate_digestion_and_absorption | 9 | 44 | 1.311e-16 | 1.026e-15 | |
| 24 | hsa04810_Regulation_of_actin_cytoskeleton | 13 | 214 | 1.558e-16 | 1.169e-15 | |
| 25 | hsa04070_Phosphatidylinositol_signaling_system | 10 | 78 | 3.452e-16 | 2.486e-15 | |
| 26 | hsa04150_mTOR_signaling_pathway | 9 | 52 | 6.726e-16 | 4.657e-15 | |
| 27 | hsa04910_Insulin_signaling_pathway | 11 | 138 | 2.194e-15 | 1.463e-14 | |
| 28 | hsa00592_alpha.Linolenic_acid_metabolism | 7 | 20 | 5.591e-15 | 3.594e-14 | |
| 29 | hsa04310_Wnt_signaling_pathway | 11 | 151 | 6.018e-15 | 3.735e-14 | |
| 30 | hsa04720_Long.term_potentiation | 9 | 70 | 1.159e-14 | 6.956e-14 | |
| 31 | hsa04270_Vascular_smooth_muscle_contraction | 10 | 116 | 2.124e-14 | 1.234e-13 | |
| 32 | hsa00591_Linoleic_acid_metabolism | 7 | 30 | 1.447e-13 | 8.14e-13 | |
| 33 | hsa00590_Arachidonic_acid_metabolism | 8 | 59 | 2.501e-13 | 1.364e-12 | |
| 34 | hsa04972_Pancreatic_secretion | 9 | 101 | 3.565e-13 | 1.887e-12 | |
| 35 | hsa00565_Ether_lipid_metabolism | 7 | 36 | 5.883e-13 | 3.025e-12 | |
| 36 | hsa04975_Fat_digestion_and_absorption | 7 | 46 | 3.718e-12 | 1.859e-11 | |
| 37 | hsa04020_Calcium_signaling_pathway | 9 | 177 | 5.864e-11 | 2.853e-10 | |
| 38 | hsa00564_Glycerophospholipid_metabolism | 7 | 80 | 2.101e-10 | 9.95e-10 | |
| 39 | hsa04630_Jak.STAT_signaling_pathway | 8 | 155 | 6.763e-10 | 3.121e-09 | |
| 40 | hsa04530_Tight_junction | 7 | 133 | 7.618e-09 | 3.428e-08 | |
| 41 | hsa04540_Gap_junction | 6 | 90 | 2.38e-08 | 1.045e-07 | |
| 42 | hsa04916_Melanogenesis | 5 | 101 | 1.7e-06 | 7.285e-06 | |
| 43 | hsa04114_Oocyte_meiosis | 5 | 114 | 3.091e-06 | 1.294e-05 | |
| 44 | hsa00562_Inositol_phosphate_metabolism | 4 | 57 | 5.018e-06 | 2.053e-05 | |
| 45 | hsa04520_Adherens_junction | 3 | 73 | 0.0004183 | 0.001673 | |
| 46 | hsa04320_Dorso.ventral_axis_formation | 2 | 25 | 0.001136 | 0.004447 | |
| 47 | hsa04621_NOD.like_receptor_signaling_pathway | 2 | 59 | 0.00621 | 0.02378 | |
| 48 | hsa04144_Endocytosis | 3 | 203 | 0.007724 | 0.02896 | |
| 49 | hsa04622_RIG.I.like_receptor_signaling_pathway | 2 | 71 | 0.008884 | 0.03263 | |
| 50 | hsa04971_Gastric_acid_secretion | 2 | 74 | 0.009619 | 0.03463 | |
| 51 | hsa04970_Salivary_secretion | 2 | 89 | 0.01369 | 0.0483 |
lncRNA-mediated sponge
| Num | lncRNA | miRNAs | miRNAs count | Gene | Sponge regulatory network | lncRNA log2FC | lncRNA pvalue | Gene log2FC | Gene pvalue | lncRNA-gene Pearson correlation |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | CECR7 |
hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-24-3p;hsa-miR-26b-5p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30b-5p;hsa-miR-30c-5p;hsa-miR-30d-5p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-361-5p;hsa-miR-429;hsa-miR-491-5p;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-590-5p;hsa-miR-7-1-3p | 24 | PRKCB | Sponge network | 0.551 | 0.56177 | -0.289 | 0.6299 | 0.583 |
| 2 | EMX2OS |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-32-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-374b-5p;hsa-miR-421;hsa-miR-424-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-502-5p;hsa-miR-505-3p;hsa-miR-577;hsa-miR-663b;hsa-miR-93-5p | 25 | AKT3 | Sponge network | 1.057 | 0.31716 | 0.457 | 0.53933 | 0.542 |
| 3 | CECR7 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-1275;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-32-3p;hsa-miR-335-3p;hsa-miR-33a-3p;hsa-miR-362-3p;hsa-miR-362-5p;hsa-miR-501-3p;hsa-miR-502-3p;hsa-miR-502-5p;hsa-miR-93-5p | 20 | AKT3 | Sponge network | 0.551 | 0.56177 | 0.457 | 0.53933 | 0.508 |
| 4 | EMX2OS |
hsa-miR-141-5p;hsa-miR-148a-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-27a-3p;hsa-miR-29a-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-452-3p;hsa-miR-484;hsa-miR-502-5p;hsa-miR-589-3p;hsa-miR-590-3p;hsa-miR-7-1-3p;hsa-miR-7-5p | 21 | PRKCB | Sponge network | 1.057 | 0.31716 | -0.289 | 0.6299 | 0.407 |
| 5 | MEG3 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-15a-5p;hsa-miR-15b-5p;hsa-miR-16-5p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-577;hsa-miR-93-5p | 12 | AKT3 | Sponge network | 0.433 | 0.33816 | 0.457 | 0.53933 | 0.405 |
| 6 | CECR7 |
hsa-let-7f-1-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-32-3p;hsa-miR-335-5p;hsa-miR-582-3p;hsa-miR-590-3p;hsa-miR-590-5p | 11 | PIK3CA | Sponge network | 0.551 | 0.56177 | 0.023 | 0.97942 | 0.368 |
| 7 | ZNF883 |
hsa-miR-106a-5p;hsa-miR-106b-5p;hsa-miR-107;hsa-miR-17-3p;hsa-miR-17-5p;hsa-miR-20a-5p;hsa-miR-29a-3p;hsa-miR-29b-3p;hsa-miR-335-3p;hsa-miR-33a-3p | 10 | AKT3 | Sponge network | 0.913 | 0.16772 | 0.457 | 0.53933 | 0.3 |
| 8 | MEG3 |
hsa-miR-141-5p;hsa-miR-16-2-3p;hsa-miR-186-5p;hsa-miR-194-5p;hsa-miR-200b-3p;hsa-miR-200c-3p;hsa-miR-26b-5p;hsa-miR-29b-1-5p;hsa-miR-30d-3p;hsa-miR-335-3p;hsa-miR-335-5p;hsa-miR-429;hsa-miR-590-5p;hsa-miR-7-1-3p | 14 | PRKCB | Sponge network | 0.433 | 0.33816 | -0.289 | 0.6299 | 0.266 |
| 9 | EMX2OS |
hsa-let-7f-1-3p;hsa-miR-148a-5p;hsa-miR-148b-3p;hsa-miR-16-2-3p;hsa-miR-17-5p;hsa-miR-186-5p;hsa-miR-32-3p;hsa-miR-335-5p;hsa-miR-339-5p;hsa-miR-374b-5p;hsa-miR-576-5p;hsa-miR-582-3p;hsa-miR-590-3p | 13 | PIK3CA | Sponge network | 1.057 | 0.31716 | 0.023 | 0.97942 | 0.254 |