This regulatory network was inferred from the input dataset. The miRNAs and mRNAs are
presented as round and rectangle nodes respectively. The numerical value popped up upon mouse over the gene node is the log2 transformed fold-change of the gene expression between the two groups. All of the nodes are clickable, and the detailed information of the miRNAs/mRNAs and related cancer pathway will be displayed in another window. The edges between nodes are supported by both interactions (predicted or experimentally verified) and correlations learnt from cancer dataset. The numerical value popped up upon mouse over the edge is the correlation beat value (effect size) between the two nodes. The experimental evidences of the edges reported in previous cancer studies are highlighted by red/orange color. All of these information can be accessed by the "mouse-over" action. This network shows a full map of the miRNA-mRNA regulation of the input gene list(s), and the hub miRNAs (with the high network degree/betweenness centrality) would be the potential cancer drivers or tumor suppressors. The full result table can be accessed in the "Regulations" tab.
"miRNACancerMAP" is also a network visualization tool for users to draw their regulatory network by personal customization. Users can set the complexity of the network by limiting the number of nodes or edges. And the color of the nodes can be defined by different categories of the mRNAs and miRNAs, such as Gene-Ontology, pathway, and expression status. Users can also select to use network degree or network betweenness centrality to define the node size. And edges can be black or colored by the correlation. Purple edge means negative correlation (mostly found between miRNA and mRNA), and blue edge means positive correlation (found in PPI or miRNA-miRNA sponge effect). We can also add the protein-protein interactions (PPI) into the network. This result will show the cluster of genes regulated by some specific miRNAs. Additionally, miRNA-miRNA edges can be added by the "miRNA sponge" button, presenting some clusters of miRNAs that have the interactions via sponge effect.
miRNA-gene regulations
| Num | microRNA | Gene | miRNA log2FC | miRNA pvalue | Gene log2FC | Gene pvalue | Interaction | Correlation beta | Correlation P-value | PMID | Reported in cancer studies |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | hsa-miR-194-5p | ADAMTS5 | 2.24 | 0 | 0.19 | 0.27716 | mirMAP | -0.1 | 0.00032 | NA | |
| 2 | hsa-miR-194-5p | ALDH1L2 | 2.24 | 0 | 0.23 | 0.32096 | mirMAP | -0.14 | 0.00018 | NA | |
| 3 | hsa-miR-194-5p | ANGPTL1 | 2.24 | 0 | -2.71 | 0 | MirTarget | -0.77 | 0 | NA | |
| 4 | hsa-miR-194-5p | ARHGAP24 | 2.24 | 0 | -0.93 | 0 | miRNATAP | -0.16 | 0 | NA | |
| 5 | hsa-miR-194-5p | ASAP1 | 2.24 | 0 | 0.66 | 0 | MirTarget; miRNATAP | -0.1 | 0 | NA | |
| 6 | hsa-miR-194-5p | BICD2 | 2.24 | 0 | -0.02 | 0.88611 | miRNATAP | -0.22 | 0 | NA | |
| 7 | hsa-miR-194-5p | BNC1 | 2.24 | 0 | -1.62 | 0.00071 | MirTarget | -0.53 | 0 | NA | |
| 8 | hsa-miR-194-5p | BNIP2 | 2.24 | 0 | -0.05 | 0.63907 | MirTarget | -0.15 | 0 | NA | |
| 9 | hsa-miR-194-5p | BRMS1L | 2.24 | 0 | -0.02 | 0.82317 | MirTarget | -0.11 | 0 | NA | |
| 10 | hsa-miR-194-5p | C1orf21 | 2.24 | 0 | -0.73 | 9.0E-5 | mirMAP; miRNATAP | -0.11 | 0.00076 | NA | |
| 11 | hsa-miR-194-5p | CADM1 | 2.24 | 0 | -0.9 | 0.00084 | miRNATAP | -0.28 | 0 | NA | |
| 12 | hsa-miR-194-5p | CAMK2G | 2.24 | 0 | -0.53 | 0.00041 | miRNATAP | -0.15 | 0 | NA | |
| 13 | hsa-miR-194-5p | CBX5 | 2.24 | 0 | 0.16 | 0.24446 | mirMAP | -0.13 | 0 | NA | |
| 14 | hsa-miR-194-5p | CDH2 | 2.24 | 0 | -0.68 | 0.04042 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.38 | 0 | 21845495; 20979124 | The results of real-time PCR and western blot highlighted that miR-194 interacted with N-cadherin and negatively regulated its expression at the translational level;The overexpression of miR-194 in liver mesenchymal-like cancer cells reduced the expression of the mesenchymal cell marker N-cadherin and suppressed invasion and migration of the mesenchymal-like cancer cells both in vitro and in vivo |
| 15 | hsa-miR-194-5p | CDK14 | 2.24 | 0 | -0.3 | 0.18672 | MirTarget | -0.26 | 0 | NA | |
| 16 | hsa-miR-194-5p | CDK5R2 | 2.24 | 0 | -1.28 | 0.01166 | mirMAP | -0.19 | 0.02733 | NA | |
| 17 | hsa-miR-194-5p | CEP170 | 2.24 | 0 | 0.94 | 0 | miRNATAP | -0.11 | 4.0E-5 | NA | |
| 18 | hsa-miR-194-5p | CHD5 | 2.24 | 0 | -0.92 | 0.01521 | mirMAP | -0.4 | 0 | NA | |
| 19 | hsa-miR-194-5p | CHRNB2 | 2.24 | 0 | -0.9 | 0.01463 | mirMAP | -0.18 | 0.00381 | NA | |
| 20 | hsa-miR-194-5p | CLASP1 | 2.24 | 0 | 0.09 | 0.35045 | miRNATAP | -0.14 | 0 | NA | |
| 21 | hsa-miR-194-5p | CLIP4 | 2.24 | 0 | -1.15 | 1.0E-5 | MirTarget | -0.44 | 0 | NA | |
| 22 | hsa-miR-194-5p | CLLU1 | 2.24 | 0 | -1.13 | 0.00877 | miRNATAP | -0.18 | 0.01381 | NA | |
| 23 | hsa-miR-194-5p | DAAM1 | 2.24 | 0 | -0.34 | 0.0105 | miRNATAP | -0.22 | 0 | NA | |
| 24 | hsa-miR-194-5p | DCBLD2 | 2.24 | 0 | -0.14 | 0.5524 | MirTarget | -0.29 | 0 | NA | |
| 25 | hsa-miR-194-5p | DCUN1D4 | 2.24 | 0 | -0.26 | 0.02159 | MirTarget | -0.11 | 0 | NA | |
| 26 | hsa-miR-194-5p | DIP2C | 2.24 | 0 | -0.44 | 0.01713 | miRNATAP | -0.22 | 0 | NA | |
| 27 | hsa-miR-194-5p | DMD | 2.24 | 0 | -1.51 | 0 | MirTarget; miRNATAP | -0.42 | 0 | NA | |
| 28 | hsa-miR-194-5p | DUSP10 | 2.24 | 0 | 0.51 | 0.00228 | miRNATAP | -0.11 | 8.0E-5 | NA | |
| 29 | hsa-miR-194-5p | EGR2 | 2.24 | 0 | 0.31 | 0.17079 | miRNATAP | -0.12 | 0.00109 | NA | |
| 30 | hsa-miR-194-5p | EHBP1 | 2.24 | 0 | -0.08 | 0.56084 | MirTarget; miRNATAP | -0.2 | 0 | NA | |
| 31 | hsa-miR-194-5p | EIF4E3 | 2.24 | 0 | -1.24 | 0 | miRNATAP | -0.19 | 0 | NA | |
| 32 | hsa-miR-194-5p | EMCN | 2.24 | 0 | -0.47 | 0.01631 | MirTarget | -0.18 | 0 | NA | |
| 33 | hsa-miR-194-5p | ENAH | 2.24 | 0 | -0.3 | 0.08986 | mirMAP; miRNATAP | -0.15 | 0 | NA | |
| 34 | hsa-miR-194-5p | EPHA3 | 2.24 | 0 | 0.18 | 0.53588 | miRNATAP | -0.24 | 0 | NA | |
| 35 | hsa-miR-194-5p | EPHA5 | 2.24 | 0 | -0.96 | 0.01733 | MirTarget | -0.43 | 0 | NA | |
| 36 | hsa-miR-194-5p | ERBB4 | 2.24 | 0 | -1.97 | 2.0E-5 | miRNATAP | -0.23 | 0.00317 | NA | |
| 37 | hsa-miR-194-5p | ERG | 2.24 | 0 | 0 | 0.99011 | MirTarget | -0.14 | 0 | NA | |
| 38 | hsa-miR-194-5p | EYA4 | 2.24 | 0 | 0.3 | 0.36876 | miRNATAP | -0.22 | 0.00012 | NA | |
| 39 | hsa-miR-194-5p | FAM63B | 2.24 | 0 | 0.07 | 0.61635 | miRNATAP | -0.14 | 0 | NA | |
| 40 | hsa-miR-194-5p | FOXO1 | 2.24 | 0 | -0.14 | 0.33874 | miRNATAP | -0.11 | 0 | 24631529 | Gα12gep oncogene inhibits FOXO1 in hepatocellular carcinoma as a consequence of miR 135b and miR 194 dysregulation; In addition Gα12QL repressed miR-194 cluster gene products 194/192/215 which contributed to MDM2-mediated FOXO1 repression; Moreover decrease of FOXO1 or miR-194 was statistically significant between stages T1 and T2 whereas increase of miR-135b discriminated tumor stage T3a versus T1/T2 |
| 41 | hsa-miR-194-5p | FOXP2 | 2.24 | 0 | -1.9 | 1.0E-5 | miRNATAP | -0.63 | 0 | NA | |
| 42 | hsa-miR-194-5p | FZD6 | 2.24 | 0 | 0.37 | 0.02474 | MirTarget | -0.11 | 4.0E-5 | NA | |
| 43 | hsa-miR-194-5p | GAS7 | 2.24 | 0 | -0.56 | 0.01602 | mirMAP | -0.35 | 0 | NA | |
| 44 | hsa-miR-194-5p | GLCCI1 | 2.24 | 0 | -1 | 0 | MirTarget | -0.12 | 1.0E-5 | NA | |
| 45 | hsa-miR-194-5p | GPM6B | 2.24 | 0 | -2.16 | 0 | miRNATAP | -0.56 | 0 | NA | |
| 46 | hsa-miR-194-5p | GRIK3 | 2.24 | 0 | -3.21 | 0 | mirMAP | -0.59 | 0 | NA | |
| 47 | hsa-miR-194-5p | GYG1 | 2.24 | 0 | -0.45 | 0.00051 | MirTarget; miRNATAP | -0.22 | 0 | NA | |
| 48 | hsa-miR-194-5p | HOOK3 | 2.24 | 0 | 0.27 | 0.09957 | MirTarget | -0.21 | 0 | NA | |
| 49 | hsa-miR-194-5p | IGF1R | 2.24 | 0 | 0.12 | 0.57112 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.14 | 6.0E-5 | NA | |
| 50 | hsa-miR-194-5p | ITGA9 | 2.24 | 0 | -0.83 | 0.00174 | miRNAWalker2 validate; miRTarBase | -0.3 | 0 | NA | |
| 51 | hsa-miR-194-5p | ITPKB | 2.24 | 0 | -0.7 | 0.00106 | MirTarget; miRNATAP | -0.36 | 0 | NA | |
| 52 | hsa-miR-194-5p | JMY | 2.24 | 0 | -0.27 | 0.06205 | mirMAP | -0.19 | 0 | NA | |
| 53 | hsa-miR-194-5p | KAT6B | 2.24 | 0 | -0.48 | 0.00018 | MirTarget; miRNATAP | -0.12 | 0 | NA | |
| 54 | hsa-miR-194-5p | KCNA1 | 2.24 | 0 | -2.36 | 0.00043 | mirMAP | -0.76 | 0 | NA | |
| 55 | hsa-miR-194-5p | KCTD20 | 2.24 | 0 | 0 | 0.97607 | MirTarget | -0.11 | 0 | NA | |
| 56 | hsa-miR-194-5p | KIAA1644 | 2.24 | 0 | -1.37 | 0.00031 | mirMAP | -0.49 | 0 | NA | |
| 57 | hsa-miR-194-5p | KIF1B | 2.24 | 0 | -0.12 | 0.32258 | mirMAP | -0.1 | 0 | NA | |
| 58 | hsa-miR-194-5p | KLF12 | 2.24 | 0 | -0.39 | 0.08931 | MirTarget | -0.26 | 0 | NA | |
| 59 | hsa-miR-194-5p | KLF15 | 2.24 | 0 | -2.91 | 0 | miRNATAP | -0.47 | 0 | NA | |
| 60 | hsa-miR-194-5p | KLF7 | 2.24 | 0 | -0.38 | 0.04874 | miRNATAP | -0.16 | 0 | NA | |
| 61 | hsa-miR-194-5p | LGI1 | 2.24 | 0 | -4.11 | 0 | MirTarget; miRNATAP | -0.62 | 0 | NA | |
| 62 | hsa-miR-194-5p | LHX6 | 2.24 | 0 | -0.09 | 0.69193 | miRNATAP | -0.14 | 0.00022 | NA | |
| 63 | hsa-miR-194-5p | LIMCH1 | 2.24 | 0 | -0.92 | 1.0E-5 | MirTarget | -0.17 | 0 | NA | |
| 64 | hsa-miR-194-5p | LNPEP | 2.24 | 0 | -0.18 | 0.24961 | mirMAP | -0.14 | 0 | NA | |
| 65 | hsa-miR-194-5p | LPP | 2.24 | 0 | -0.46 | 0.04475 | mirMAP | -0.28 | 0 | NA | |
| 66 | hsa-miR-194-5p | LRCH2 | 2.24 | 0 | -0.76 | 0.00191 | MirTarget | -0.36 | 0 | NA | |
| 67 | hsa-miR-194-5p | MAF | 2.24 | 0 | -1.26 | 0 | miRNATAP | -0.18 | 0 | NA | |
| 68 | hsa-miR-194-5p | MAN1C1 | 2.24 | 0 | -0.36 | 0.10568 | miRNATAP | -0.18 | 0 | NA | |
| 69 | hsa-miR-194-5p | MAP1B | 2.24 | 0 | -0.56 | 0.05021 | MirTarget | -0.43 | 0 | NA | |
| 70 | hsa-miR-194-5p | MAP2 | 2.24 | 0 | -0.5 | 0.06746 | MirTarget; miRNATAP | -0.36 | 0 | NA | |
| 71 | hsa-miR-194-5p | MAP3K3 | 2.24 | 0 | -0.29 | 0.01565 | miRNATAP | -0.21 | 0 | NA | |
| 72 | hsa-miR-194-5p | MBNL2 | 2.24 | 0 | -0.2 | 0.12802 | miRNATAP | -0.1 | 0 | NA | |
| 73 | hsa-miR-194-5p | MECP2 | 2.24 | 0 | -0.15 | 0.15366 | mirMAP; miRNATAP | -0.17 | 0 | NA | |
| 74 | hsa-miR-194-5p | MEF2C | 2.24 | 0 | -0.5 | 0.01448 | miRNATAP | -0.23 | 0 | NA | |
| 75 | hsa-miR-194-5p | MEIS2 | 2.24 | 0 | -0.88 | 4.0E-5 | MirTarget; miRNATAP | -0.17 | 0 | NA | |
| 76 | hsa-miR-194-5p | MPP2 | 2.24 | 0 | -0.98 | 0.00061 | mirMAP | -0.41 | 0 | NA | |
| 77 | hsa-miR-194-5p | MTSS1 | 2.24 | 0 | -0.81 | 0.00035 | MirTarget; miRNATAP | -0.22 | 0 | NA | |
| 78 | hsa-miR-194-5p | MYH10 | 2.24 | 0 | 0.37 | 0.10713 | miRNATAP | -0.26 | 0 | NA | |
| 79 | hsa-miR-194-5p | MYO5A | 2.24 | 0 | -0.09 | 0.61662 | mirMAP | -0.23 | 0 | NA | |
| 80 | hsa-miR-194-5p | NBEA | 2.24 | 0 | -1.41 | 0.00027 | miRNATAP | -0.5 | 0 | NA | |
| 81 | hsa-miR-194-5p | NFAT5 | 2.24 | 0 | 0.46 | 0.00066 | miRNATAP | -0.1 | 1.0E-5 | NA | |
| 82 | hsa-miR-194-5p | NLGN4Y | 2.24 | 0 | -0.85 | 0.31856 | MirTarget | -0.46 | 0.00146 | NA | |
| 83 | hsa-miR-194-5p | NR2F2 | 2.24 | 0 | -0.14 | 0.3979 | miRNATAP | -0.12 | 1.0E-5 | NA | |
| 84 | hsa-miR-194-5p | NRP1 | 2.24 | 0 | 0.58 | 0.00021 | miRNATAP | -0.12 | 0 | NA | |
| 85 | hsa-miR-194-5p | NTRK3 | 2.24 | 0 | -1.77 | 3.0E-5 | mirMAP | -0.46 | 0 | NA | |
| 86 | hsa-miR-194-5p | OPCML | 2.24 | 0 | -2.5 | 0 | miRNATAP | -0.32 | 0 | NA | |
| 87 | hsa-miR-194-5p | OSTM1 | 2.24 | 0 | -0.25 | 0.0298 | MirTarget | -0.16 | 0 | NA | |
| 88 | hsa-miR-194-5p | PACSIN1 | 2.24 | 0 | -0.62 | 0.06496 | mirMAP | -0.16 | 0.0047 | NA | |
| 89 | hsa-miR-194-5p | PAIP2 | 2.24 | 0 | -0.51 | 0 | MirTarget | -0.16 | 0 | NA | |
| 90 | hsa-miR-194-5p | PARVA | 2.24 | 0 | -0.48 | 0.00429 | MirTarget | -0.24 | 0 | NA | |
| 91 | hsa-miR-194-5p | PER3 | 2.24 | 0 | -1.21 | 0 | miRNATAP | -0.22 | 0 | NA | |
| 92 | hsa-miR-194-5p | PFN2 | 2.24 | 0 | -0.31 | 0.26876 | miRNATAP | -0.33 | 0 | NA | |
| 93 | hsa-miR-194-5p | PHF1 | 2.24 | 0 | -0.68 | 0 | miRNATAP | -0.17 | 0 | NA | |
| 94 | hsa-miR-194-5p | PHLDA1 | 2.24 | 0 | 0.06 | 0.78159 | MirTarget | -0.12 | 0.0005 | NA | |
| 95 | hsa-miR-194-5p | PIP4K2A | 2.24 | 0 | 0.05 | 0.65696 | MirTarget | -0.11 | 0 | NA | |
| 96 | hsa-miR-194-5p | PITPNM2 | 2.24 | 0 | -0.47 | 0.00216 | MirTarget; miRNATAP | -0.21 | 0 | NA | |
| 97 | hsa-miR-194-5p | PJA2 | 2.24 | 0 | -0.72 | 0 | MirTarget | -0.21 | 0 | NA | |
| 98 | hsa-miR-194-5p | PKD2 | 2.24 | 0 | -0.24 | 0.21741 | miRNATAP | -0.3 | 0 | NA | |
| 99 | hsa-miR-194-5p | PPFIBP1 | 2.24 | 0 | 0.08 | 0.54797 | MirTarget | -0.12 | 0 | NA | |
| 100 | hsa-miR-194-5p | PPP1R12B | 2.24 | 0 | -1.14 | 0.00076 | mirMAP | -0.44 | 0 | NA | |
| 101 | hsa-miR-194-5p | PPP2R2C | 2.24 | 0 | -0.96 | 0.07428 | miRNATAP | -0.5 | 0 | NA | |
| 102 | hsa-miR-194-5p | PRICKLE2 | 2.24 | 0 | -1.11 | 0.00011 | miRNATAP | -0.48 | 0 | NA | |
| 103 | hsa-miR-194-5p | PRKCB | 2.24 | 0 | -1.31 | 2.0E-5 | miRNATAP | -0.32 | 0 | NA | |
| 104 | hsa-miR-194-5p | PTPN13 | 2.24 | 0 | -1.21 | 0 | miRNAWalker2 validate; miRTarBase | -0.45 | 0 | NA | |
| 105 | hsa-miR-194-5p | PTPRD | 2.24 | 0 | -1.01 | 0.00064 | miRNATAP | -0.22 | 1.0E-5 | NA | |
| 106 | hsa-miR-194-5p | RAB11FIP2 | 2.24 | 0 | -0.26 | 0.0617 | MirTarget; miRNATAP | -0.12 | 0 | NA | |
| 107 | hsa-miR-194-5p | RAB6B | 2.24 | 0 | -1.2 | 0 | miRNATAP | -0.29 | 0 | NA | |
| 108 | hsa-miR-194-5p | RERE | 2.24 | 0 | -0.18 | 0.12411 | miRNATAP | -0.11 | 0 | NA | |
| 109 | hsa-miR-194-5p | REV3L | 2.24 | 0 | 0.06 | 0.62263 | miRNATAP | -0.12 | 0 | NA | |
| 110 | hsa-miR-194-5p | SAMD4A | 2.24 | 0 | -0.13 | 0.55532 | miRNATAP | -0.27 | 0 | NA | |
| 111 | hsa-miR-194-5p | SAMD8 | 2.24 | 0 | -0.11 | 0.40369 | MirTarget | -0.11 | 0 | NA | |
| 112 | hsa-miR-194-5p | SATB1 | 2.24 | 0 | -0.9 | 2.0E-5 | miRNATAP | -0.26 | 0 | NA | |
| 113 | hsa-miR-194-5p | SH3PXD2A | 2.24 | 0 | 0.1 | 0.49534 | mirMAP | -0.18 | 0 | NA | |
| 114 | hsa-miR-194-5p | SLC16A9 | 2.24 | 0 | -2.32 | 0 | miRNAWalker2 validate | -0.19 | 0.01671 | NA | |
| 115 | hsa-miR-194-5p | SLC25A27 | 2.24 | 0 | -0.59 | 0.08043 | MirTarget | -0.32 | 0 | NA | |
| 116 | hsa-miR-194-5p | SLC30A8 | 2.24 | 0 | -2.66 | 0 | MirTarget; miRNATAP | -0.5 | 0 | NA | |
| 117 | hsa-miR-194-5p | SLC35F3 | 2.24 | 0 | -1.63 | 5.0E-5 | miRNATAP | -0.28 | 4.0E-5 | NA | |
| 118 | hsa-miR-194-5p | SLC7A5 | 2.24 | 0 | 0.29 | 0.24318 | MirTarget | -0.13 | 0.00299 | NA | |
| 119 | hsa-miR-194-5p | SNX1 | 2.24 | 0 | -0.51 | 0 | MirTarget | -0.13 | 0 | NA | |
| 120 | hsa-miR-194-5p | SNX29 | 2.24 | 0 | -0.34 | 0.01371 | miRNATAP | -0.23 | 0 | NA | |
| 121 | hsa-miR-194-5p | SOCS2 | 2.24 | 0 | -1.1 | 0 | miRNAWalker2 validate; miRTarBase; miRNATAP | -0.15 | 0 | 22102710 | Functional studies indicated that CDKN1A/p21 and SOCS2 expression is directly regulated by miR-20a/-20b and miR-194 respectively; miR-20a/b and miR-194 target CDKN1A and SOCS2 in follicular lymphoma potentially contributing to tumor cell proliferation and survival |
| 122 | hsa-miR-194-5p | SOX5 | 2.24 | 0 | -1.09 | 4.0E-5 | miRNAWalker2 validate; miRTarBase | -0.31 | 0 | NA | |
| 123 | hsa-miR-194-5p | SPOCK3 | 2.24 | 0 | -2.32 | 0 | miRNATAP | -0.5 | 0 | NA | |
| 124 | hsa-miR-194-5p | STAT5B | 2.24 | 0 | -0.18 | 0.1082 | miRNATAP | -0.13 | 0 | NA | |
| 125 | hsa-miR-194-5p | TCEAL4 | 2.24 | 0 | -0.41 | 0.01183 | MirTarget | -0.22 | 0 | NA | |
| 126 | hsa-miR-194-5p | TDRD9 | 2.24 | 0 | -0.58 | 0.09038 | MirTarget | -0.13 | 0.02967 | NA | |
| 127 | hsa-miR-194-5p | TEAD1 | 2.24 | 0 | -0.18 | 0.334 | MirTarget | -0.27 | 0 | NA | |
| 128 | hsa-miR-194-5p | TJP1 | 2.24 | 0 | -0.13 | 0.31513 | miRNATAP | -0.13 | 0 | NA | |
| 129 | hsa-miR-194-5p | TMEM108 | 2.24 | 0 | -1.31 | 0 | MirTarget | -0.36 | 0 | NA | |
| 130 | hsa-miR-194-5p | TMEM43 | 2.24 | 0 | 0.01 | 0.8988 | MirTarget; miRNATAP | -0.18 | 0 | NA | |
| 131 | hsa-miR-194-5p | TRIM23 | 2.24 | 0 | -0.34 | 0.00212 | MirTarget | -0.17 | 0 | 26221053 | Transcripts encoding tripartite motif containing 23 TRIM23 a ubiquitin ligase involved in NF-κB activation and chromosome 21 open reading frame 91 C21ORF91 a protein of unknown function were identified as direct targets of miR-194 in HCC cells; knocking down either protein decreased the activity of a luciferase NF-κB reporter |
| 132 | hsa-miR-194-5p | TRPS1 | 2.24 | 0 | -0.19 | 0.44109 | MirTarget; miRNATAP | -0.41 | 0 | NA | |
| 133 | hsa-miR-194-5p | TSHZ1 | 2.24 | 0 | -0.61 | 3.0E-5 | miRNATAP | -0.17 | 0 | NA | |
| 134 | hsa-miR-194-5p | TUSC3 | 2.24 | 0 | -0.77 | 0.00052 | MirTarget | -0.32 | 0 | NA | |
| 135 | hsa-miR-194-5p | UNC80 | 2.24 | 0 | -1.93 | 0 | miRNATAP | -0.32 | 1.0E-5 | NA | |
| 136 | hsa-miR-194-5p | WTIP | 2.24 | 0 | -0.2 | 0.33494 | mirMAP | -0.18 | 0 | NA | |
| 137 | hsa-miR-194-5p | ZC3H6 | 2.24 | 0 | -0.42 | 0.00729 | mirMAP | -0.16 | 0 | NA | |
| 138 | hsa-miR-194-5p | ZCCHC5 | 2.24 | 0 | -0.95 | 0.05183 | miRNATAP | -0.58 | 0 | NA | |
| 139 | hsa-miR-194-5p | ZEB1 | 2.24 | 0 | -0.47 | 0.04764 | miRNATAP | -0.36 | 0 | NA | |
| 140 | hsa-miR-194-5p | ZFHX3 | 2.24 | 0 | 0.39 | 0.01363 | MirTarget; miRNATAP | -0.16 | 0 | NA | |
| 141 | hsa-miR-194-5p | ZFHX4 | 2.24 | 0 | 0.02 | 0.95574 | MirTarget; miRNATAP | -0.36 | 0 | NA | |
| 142 | hsa-miR-194-5p | ZNF25 | 2.24 | 0 | -0.52 | 0.00115 | miRNATAP | -0.26 | 0 | NA | |
| 143 | hsa-miR-194-5p | ZNF516 | 2.24 | 0 | -0.53 | 0.0017 | MirTarget; miRNATAP | -0.2 | 0 | NA | |
| 144 | hsa-miR-194-5p | ZNF532 | 2.24 | 0 | 0.41 | 0.01627 | miRNATAP | -0.24 | 0 | NA | |
| 145 | hsa-miR-194-5p | ZNF548 | 2.24 | 0 | -0.08 | 0.53946 | MirTarget | -0.12 | 0 | NA | |
| 146 | hsa-miR-194-5p | ZNF615 | 2.24 | 0 | -0.53 | 0.00435 | MirTarget | -0.2 | 0 | NA |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | NEUROGENESIS | 34 | 1402 | 3.543e-10 | 1.648e-06 |
| 2 | CENTRAL NERVOUS SYSTEM DEVELOPMENT | 23 | 872 | 8.784e-08 | 0.0002044 |
| 3 | MOVEMENT OF CELL OR SUBCELLULAR COMPONENT | 28 | 1275 | 1.384e-07 | 0.0002146 |
| 4 | FOREBRAIN DEVELOPMENT | 14 | 357 | 3.717e-07 | 0.0003578 |
| 5 | NEURON DIFFERENTIATION | 22 | 874 | 3.845e-07 | 0.0003578 |
| 6 | HEAD DEVELOPMENT | 19 | 709 | 1.03e-06 | 0.0007988 |
| 7 | REGULATION OF MULTICELLULAR ORGANISMAL DEVELOPMENT | 30 | 1672 | 3.287e-06 | 0.002185 |
| 8 | LOCOMOTION | 23 | 1114 | 5.952e-06 | 0.003462 |
| 9 | NEURON DEVELOPMENT | 17 | 687 | 1.134e-05 | 0.005646 |
| 10 | COGNITION | 10 | 251 | 1.61e-05 | 0.005646 |
| 11 | CELL PART MORPHOGENESIS | 16 | 633 | 1.612e-05 | 0.005646 |
| 12 | NEURON MIGRATION | 7 | 110 | 1.625e-05 | 0.005646 |
| 13 | CELL MORPHOGENESIS INVOLVED IN NEURON DIFFERENTIATION | 12 | 368 | 1.699e-05 | 0.005646 |
| 14 | CELL DEVELOPMENT | 26 | 1426 | 1.265e-05 | 0.005646 |
| 15 | DENDRITE DEVELOPMENT | 6 | 79 | 2.448e-05 | 0.007119 |
| 16 | CELL MORPHOGENESIS INVOLVED IN DIFFERENTIATION | 14 | 513 | 2.441e-05 | 0.007119 |
| 17 | REGULATION OF CELL DIFFERENTIATION | 26 | 1492 | 2.766e-05 | 0.007571 |
| 18 | CELL PROJECTION ORGANIZATION | 19 | 902 | 3.187e-05 | 0.008239 |
| 19 | REGULATION OF NERVOUS SYSTEM DEVELOPMENT | 17 | 750 | 3.466e-05 | 0.008489 |
| 20 | NEURON PROJECTION MORPHOGENESIS | 12 | 402 | 4.034e-05 | 0.008937 |
| 21 | REGULATION OF CELL DEVELOPMENT | 18 | 836 | 3.94e-05 | 0.008937 |
| 22 | NEURON PROJECTION DEVELOPMENT | 14 | 545 | 4.716e-05 | 0.009541 |
| 23 | TELENCEPHALON DEVELOPMENT | 9 | 228 | 4.642e-05 | 0.009541 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | NUCLEIC ACID BINDING TRANSCRIPTION FACTOR ACTIVITY | 29 | 1199 | 9.642e-09 | 8.958e-06 |
| 2 | RNA POLYMERASE II TRANSCRIPTION FACTOR ACTIVITY SEQUENCE SPECIFIC DNA BINDING | 17 | 629 | 3.563e-06 | 0.001655 |
| 3 | TRANSMEMBRANE RECEPTOR PROTEIN TYROSINE KINASE ACTIVITY | 6 | 64 | 7.223e-06 | 0.002237 |
| 4 | ZINC ION BINDING | 23 | 1155 | 1.071e-05 | 0.002487 |
| 5 | SEQUENCE SPECIFIC DNA BINDING | 21 | 1037 | 2.114e-05 | 0.003928 |
| 6 | TRANSMEMBRANE RECEPTOR PROTEIN KINASE ACTIVITY | 6 | 81 | 2.824e-05 | 0.003999 |
| 7 | CYTOSKELETAL PROTEIN BINDING | 18 | 819 | 3.013e-05 | 0.003999 |
| 8 | MACROMOLECULAR COMPLEX BINDING | 24 | 1399 | 7.534e-05 | 0.008748 |
| 9 | CHROMATIN BINDING | 12 | 435 | 8.577e-05 | 0.008854 |
| 10 | REGULATORY REGION NUCLEIC ACID BINDING | 17 | 818 | 0.0001008 | 0.009367 |
| Num | GO | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|
| 1 | SYNAPSE | 19 | 754 | 2.548e-06 | 0.001488 |
| 2 | CELL JUNCTION | 23 | 1151 | 1.013e-05 | 0.002957 |
Over-represented Pathway
| Num | Pathway | Pathview | Overlap | Size | P Value | Adj. P Value |
|---|---|---|---|---|---|---|
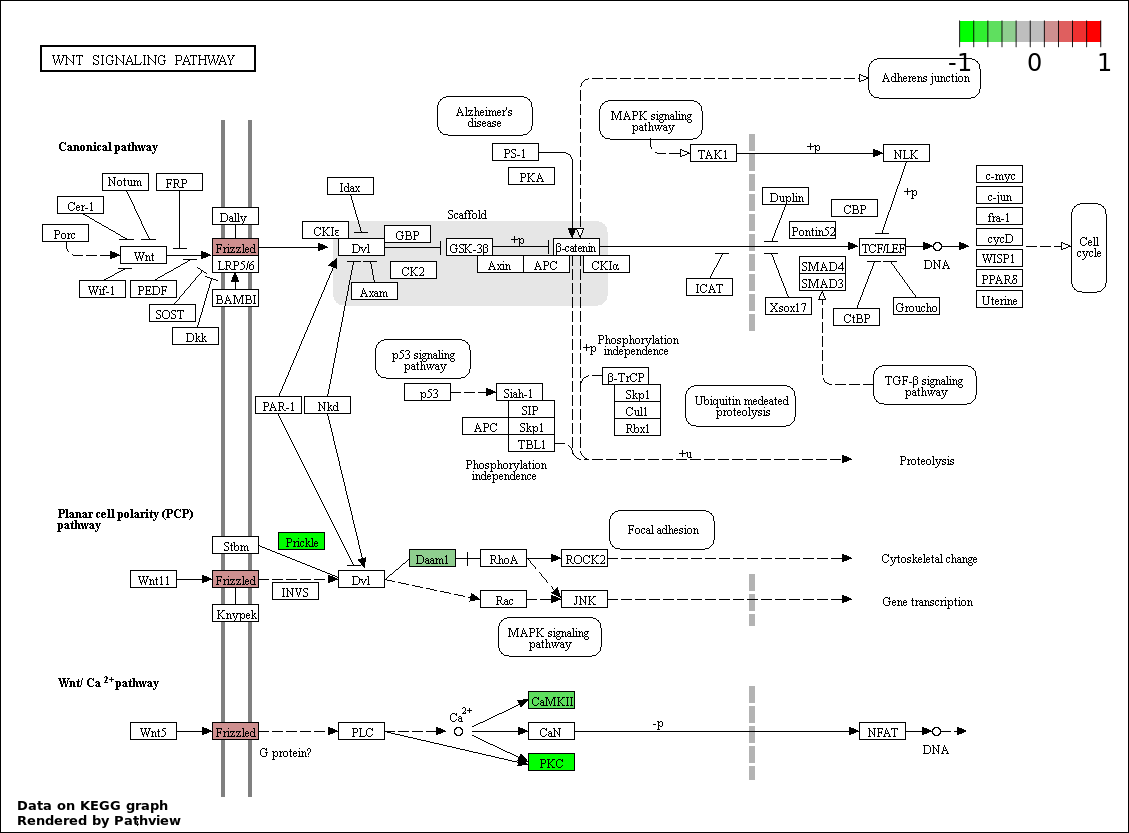
| 1 | hsa04310_Wnt_signaling_pathway | 6 | 151 | 0.0008537 | 0.1537 | |
| 2 | hsa04012_ErbB_signaling_pathway | 4 | 87 | 0.003796 | 0.3416 | |
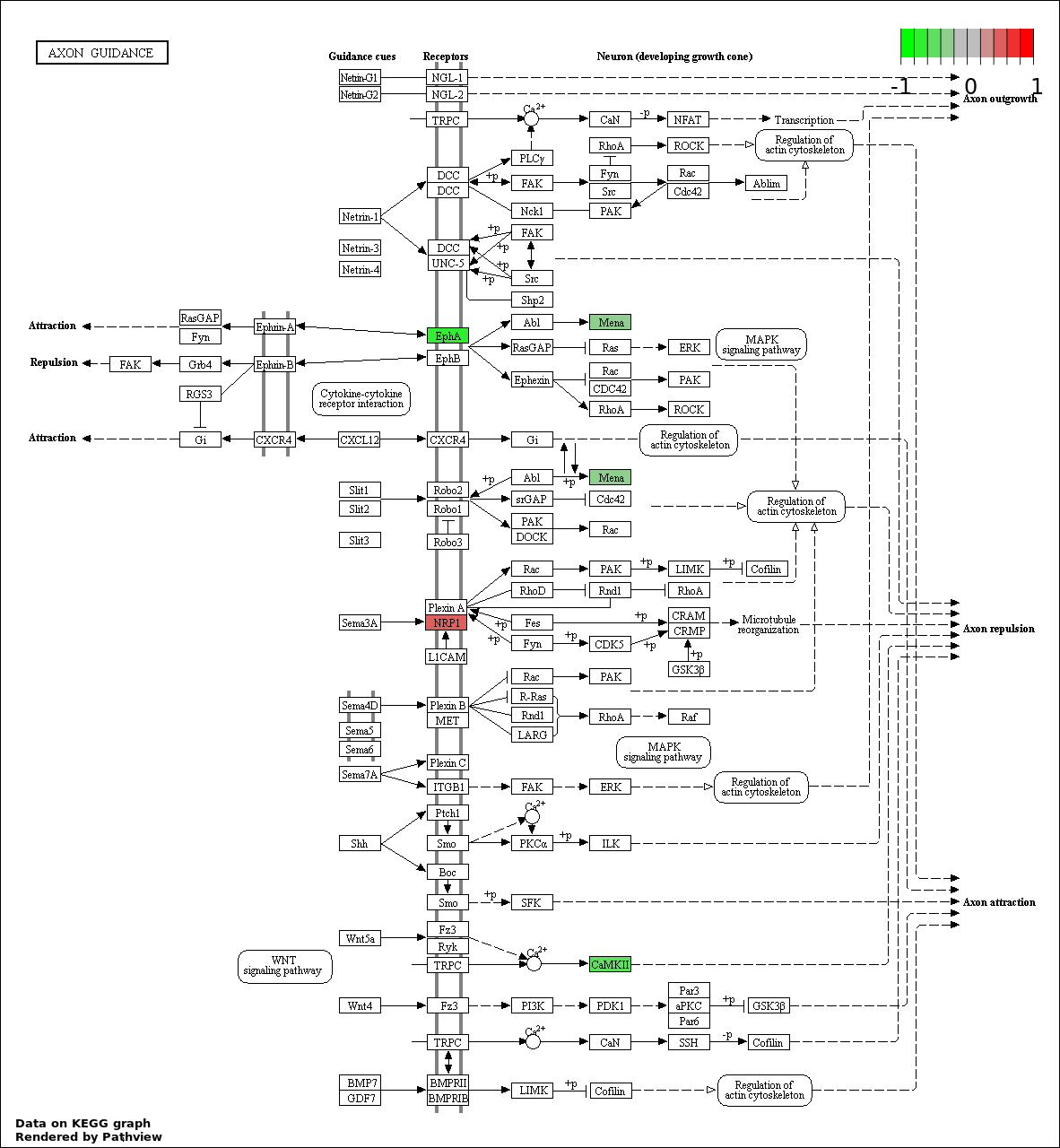
| 3 | hsa04360_Axon_guidance | 4 | 130 | 0.01524 | 0.6157 | |
| 4 | hsa04530_Tight_junction | 4 | 133 | 0.01644 | 0.6157 | |
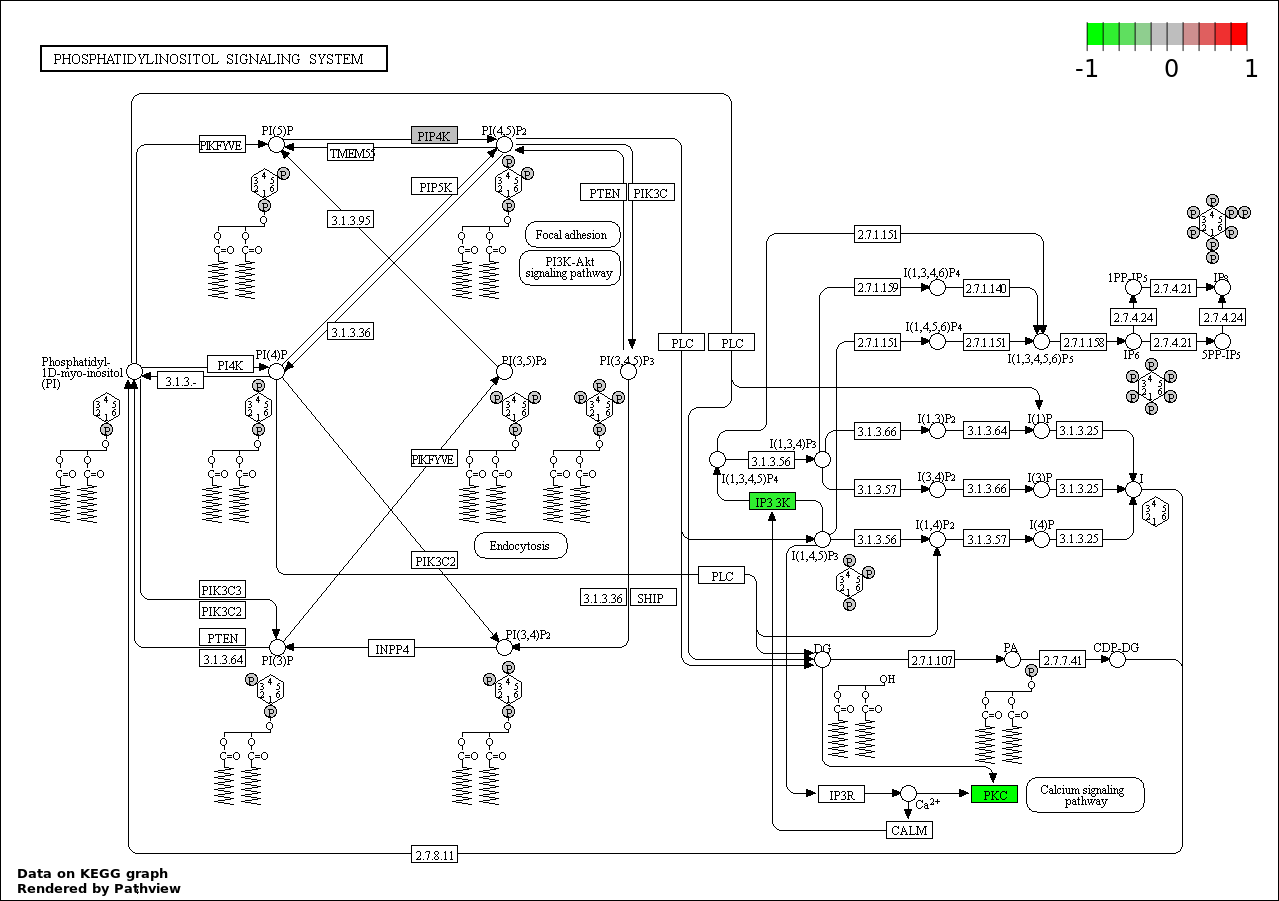
| 5 | hsa04070_Phosphatidylinositol_signaling_system | 3 | 78 | 0.01947 | 0.6157 | |
| 6 | hsa04810_Regulation_of_actin_cytoskeleton | 5 | 214 | 0.02052 | 0.6157 | |
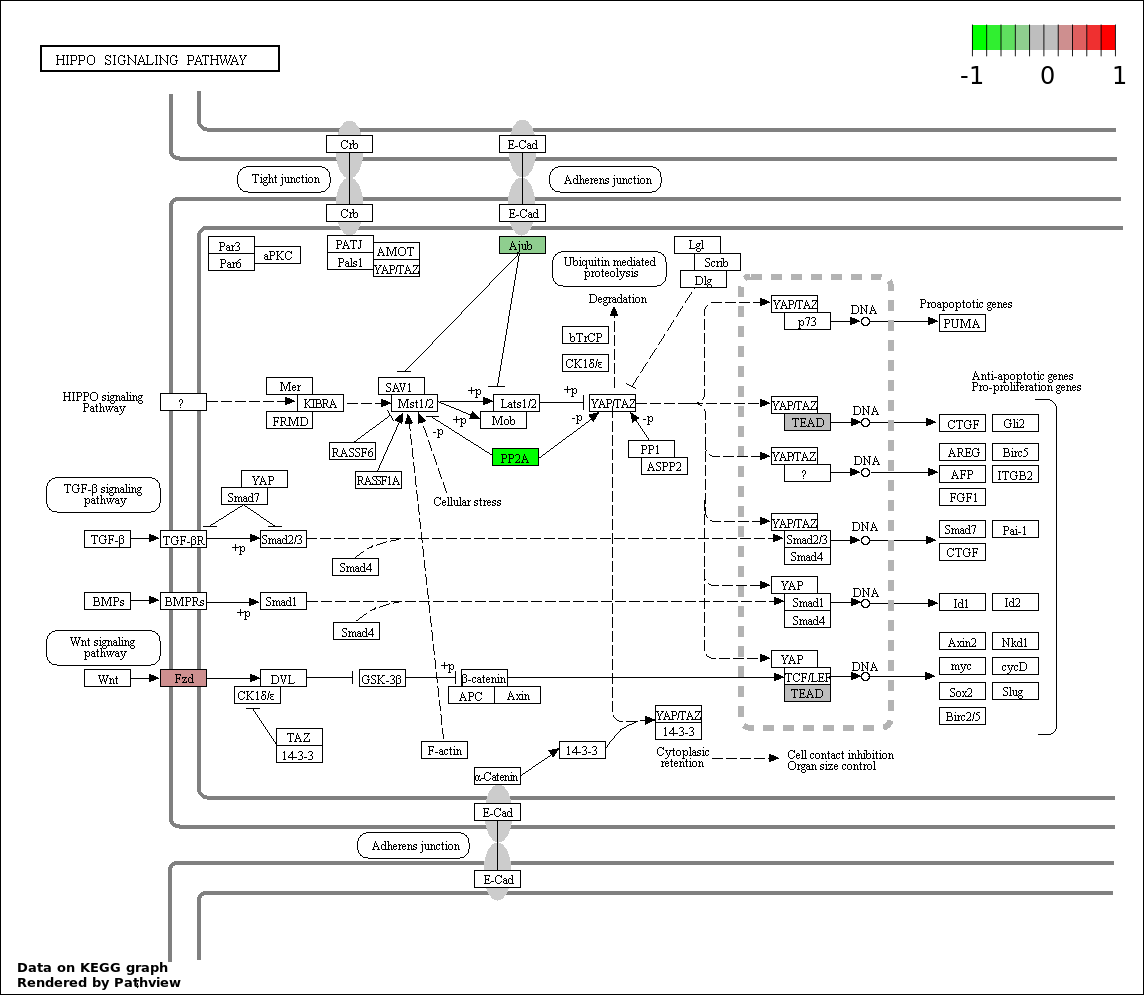
| 7 | hsa04390_Hippo_signaling_pathway | 4 | 154 | 0.02649 | 0.6811 | |
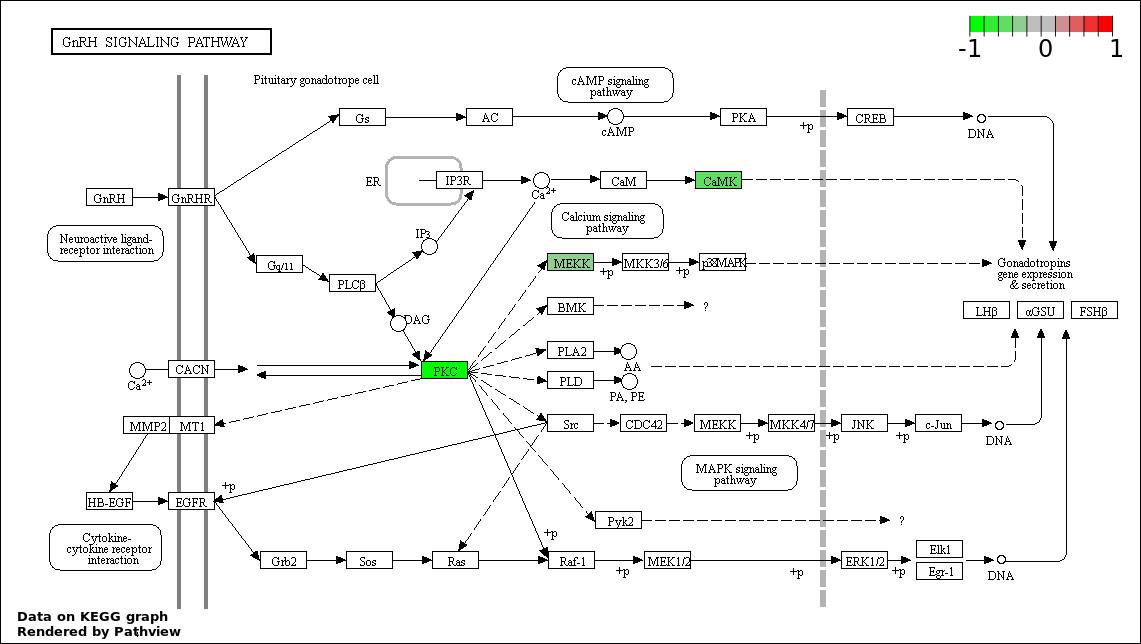
| 8 | hsa04912_GnRH_signaling_pathway | 3 | 101 | 0.03782 | 0.7371 | |
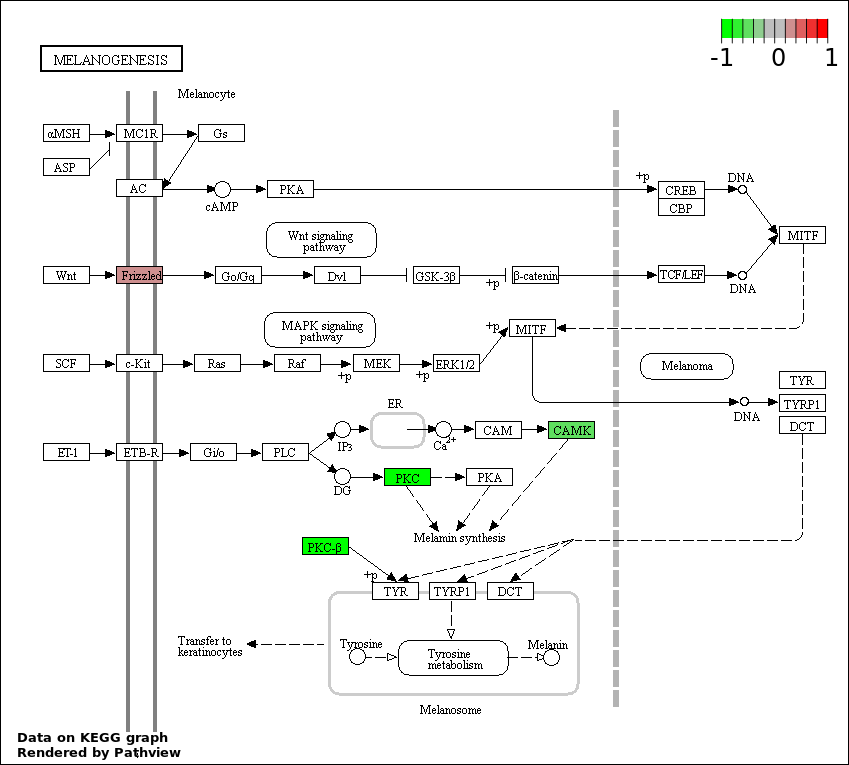
| 9 | hsa04916_Melanogenesis | 3 | 101 | 0.03782 | 0.7371 | |
| 10 | hsa04020_Calcium_signaling_pathway | 4 | 177 | 0.04095 | 0.7371 | |
| 11 | hsa00510_N.Glycan_biosynthesis | 2 | 49 | 0.04979 | 0.7937 | |
| 12 | hsa04510_Focal_adhesion | 4 | 200 | 0.05911 | 0.7937 | |
| 13 | hsa04144_Endocytosis | 4 | 203 | 0.06175 | 0.7937 | |
| 14 | hsa00562_Inositol_phosphate_metabolism | 2 | 57 | 0.0651 | 0.7937 | |
| 15 | hsa04722_Neurotrophin_signaling_pathway | 3 | 127 | 0.06614 | 0.7937 | |
| 16 | hsa04514_Cell_adhesion_molecules_.CAMs. | 3 | 136 | 0.07767 | 0.871 | |
| 17 | hsa04720_Long.term_potentiation | 2 | 70 | 0.09274 | 0.871 | |
| 18 | hsa04730_Long.term_depression | 2 | 70 | 0.09274 | 0.871 | |
| 19 | hsa04520_Adherens_junction | 2 | 73 | 0.09953 | 0.871 | |
| 20 | hsa04971_Gastric_acid_secretion | 2 | 74 | 0.1018 | 0.871 | |
| 21 | hsa04662_B_cell_receptor_signaling_pathway | 2 | 75 | 0.1041 | 0.871 | |
| 22 | hsa04370_VEGF_signaling_pathway | 2 | 76 | 0.1065 | 0.871 | |
| 23 | hsa04010_MAPK_signaling_pathway | 4 | 268 | 0.1332 | 0.9588 | |
| 24 | hsa04540_Gap_junction | 2 | 90 | 0.1403 | 0.9713 | |
| 25 | hsa04666_Fc_gamma_R.mediated_phagocytosis | 2 | 95 | 0.1529 | 0.9975 | |
| 26 | hsa04114_Oocyte_meiosis | 2 | 114 | 0.2024 | 1 | |
| 27 | hsa04270_Vascular_smooth_muscle_contraction | 2 | 116 | 0.2077 | 1 | |
| 28 | hsa04650_Natural_killer_cell_mediated_cytotoxicity | 2 | 136 | 0.2616 | 1 | |
| 29 | hsa04910_Insulin_signaling_pathway | 2 | 138 | 0.267 | 1 | |
| 30 | hsa04630_Jak.STAT_signaling_pathway | 2 | 155 | 0.3128 | 1 | |
| 31 | hsa04141_Protein_processing_in_endoplasmic_reticulum | 2 | 168 | 0.3475 | 1 | |
| 32 | hsa04062_Chemokine_signaling_pathway | 2 | 189 | 0.4022 | 1 | |
| 33 | hsa04151_PI3K_AKT_signaling_pathway | 3 | 351 | 0.4738 | 1 | |
| 34 | hsa04014_Ras_signaling_pathway | 2 | 236 | 0.5159 | 1 |